Abstract
Fli1 is a member of the Ets family of transcription factors and is preferentially expressed in hematopoietic cell lineages. Its expression level is linked to the pathogenesis of lupus. In this study, we identified mechanisms involved in the transcriptional regulation of the mouse and human Fli1 promoters. We demonstrate that the Fli1 promoter is upregulated by Ets factors Ets1, Ets2, Fli1 and Elf1 either alone or in combination with GATA factors, but is inhibited by Tel. In vitro binding studies demonstrate Elf1, Tel and Fli1 in T cells bind the three Ets binding sites in the murine Fli1 proximal promoter. We identified transcription factor binding sites in the human Fli1 promoter region that function in T cells in a similar manner to those in the mouse promoter. Furthermore, we demonstrate similar binding of Ets factors to the endogenous mouse and human Fli1 promoters in T cells and knocking down Ets1 results in an upregulation of Fli1 expression. Together, these results suggest the human and mouse genes are regulated similarly and that Ets1 may be important in preventing over-expression of Fli1 in T cells. This report lays the groundwork for identifying targets for manipulating Fli1 expression as a possible therapeutic approach.
Keywords: Fli1, Ets, gene expression, transcription factors, lymphocytes, lupus
Introduction
Fli1 is a member of the Ets family of transcription factors that share a highly conserved Ets DNA binding domain and bind to Ets binding sites (EBS) within target genes. First identified as a proto-oncogene, Fli1 is normally expressed in hematopoietic tissues, as well as lung, heart and ovary 1–4. During lymphoid development, Fli1 is expressed highly in mature B cells, pre-T cells, and resting, mature T cells 5.
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease characterized by the production of autoantibodies, formation of immune complexes and subsequent deposition in target tissues resulting in local inflammation and organ damage 6. A key feature of lupus pathogenesis is aberrant activation and proliferation of lymphocytes and over-expression of transcription factors 7. Fli1 is over-expressed in splenic T cells of lupus prone NZB/NZW f1 mice, the spleen of lupus prone MRL/lpr mice and in PBMCs of SLE patients 8; 9. When Fli1 is over-expressed in healthy, non-autoimmune prone mice, they develop autoreactive lymphocytes, autoantibodies and an immune complex mediated kidney disease similar to that observed in lupus prone mice 10. Interestingly, genetically lowering the levels of Fli1 by 50% in MRL/lpr lupus prone mice resulted in decreased autoantibody production, improved kidney disease, and significantly prolonged survival demonstrating an unequivocal association between Fli1 expression and lupus pathogenesis 9. Therefore, understanding how the Fli1 gene is regulated will be important for determining how it becomes over-expressed in lupus.
Recently, we demonstrated that activity of the mouse Fli1 (mFli1) promoter is regulated by four EBSs, a GATA binding site and two STAT binding sites within the proximal promoter in B and T cells 11. We observed in splenic extracts that Elf1, Fli1 and Tel bind three of the EBSs in vitro. Together, with the fact that many Ets factors are co-expressed with Fli1 in lymphocytes, these studies indicated several Ets factors are likely important for regulating Fli1 transcription in lymphocytes. Although the mouse and human 5′ upstream regions are highly homologous, especially within the distal and proximal promoters, the transcriptional regulation of the human Fli1 gene (hFli1) in lymphocytes has not been investigated.
To further our understanding of the mechanisms involved in regulating the expression of mouse and human Fli1 in lymphocytes. We identified cis-regulatory elements in the human Fli1 promoter and examined binding of Ets and GATA factors to the endogenous mouse and human Fli1 promoters in T cells. We demonstrate that the same cis-regulatory elements within the proximal mouse promoter are present in the human promoter and function in a similar manner in T cells. We demonstrate that five different Ets factors can activate the Fli1 proximal promoter individually or in conjunction with GATA factors in an additive or synergistic manner. Importantly, we demonstrate similar binding of Ets factors Ets1, Ets2, Fli1 and Elf1 to the endogenous promoter of both human and mouse Fli1 genes and demonstrate that lowering Ets1 levels results in an upregulation of Fli1 expression in T cells. This report furthers our knowledge of the mechanisms involved in the regulation of Fli1 expression and suggests murine lymphocytes will be an important model for studying hFli1 dysregulation as it relates to patients with SLE.
Results
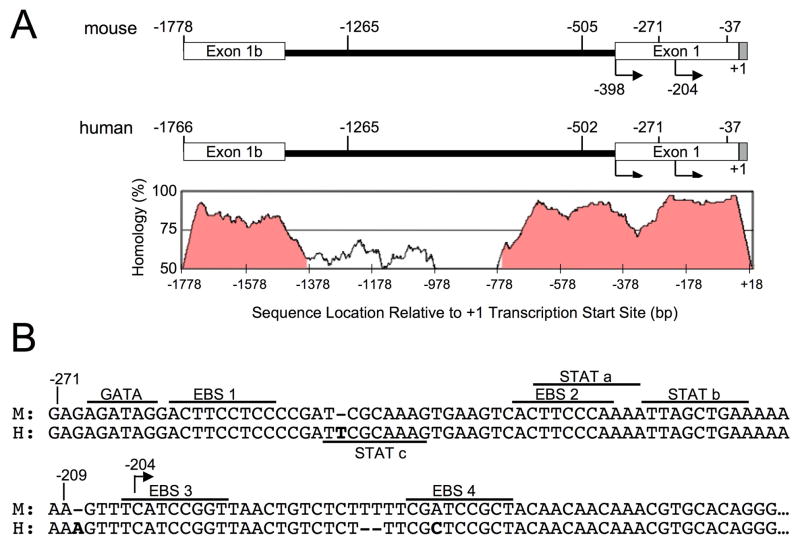
The mouse and human Fli1 upstream regulatory regions are highly homologous as illustrated in Figure 1A. Sequence alignment was performed and percent homology calculated using the mVISTA program of the VISTA family of tools developed and hosted by the Genomics Division of Lawrence Berkeley National Laboratory 12. The VISTA map below the schematic of the regions illustrates the high percentage of homology between the mouse and human sequences in the exon 1b and exon 1 regions (Fig. 1A). Specifically, the distal and proximal promoters are 83% and 88% homologous, respectfully, and 95% homologous in the region from −271 to −37. This homology suggests they may be regulated in a similar manner. Cis-regulatory elements within the human −271 to −37 sequence were identified by scanning for known transcription factor binding sites and comparing to the same region in the mouse Fli1 gene (Fig. 1B). Putative binding sites analyzed include four Ets binding sites (EBS1-4), a GATA binding site, and two STAT binding sites (STATa and STATb), all of which are nearly perfectly conserved in the mouse sequence (Fig. 1B). An additional putative STAT (STATc) site was identified, which is not present in the mouse promoter.
Figure 1. A schematic of the human and mouse Fli1−1766 to +18 regions.
(A) Comparison of the upstream regulatory region encompassing the human Fli1 proximal and distal promoter regions from −1766 to +18 with the equivalent region in the mouse (−1778 to +18). Below and aligned with the schematic is a VISTA map illustrating the percent of homology across the region. (B) Sequence alignment of human and mouse Fli1 promoter regions showing common putative transcription factor binding sites are underlined and labeled above the sequence. Unique to the human Fli1 promoter, a STATc binding site is underlined and labeled below the sequence. Numbering is relative to the +1 start site of the human sequence.
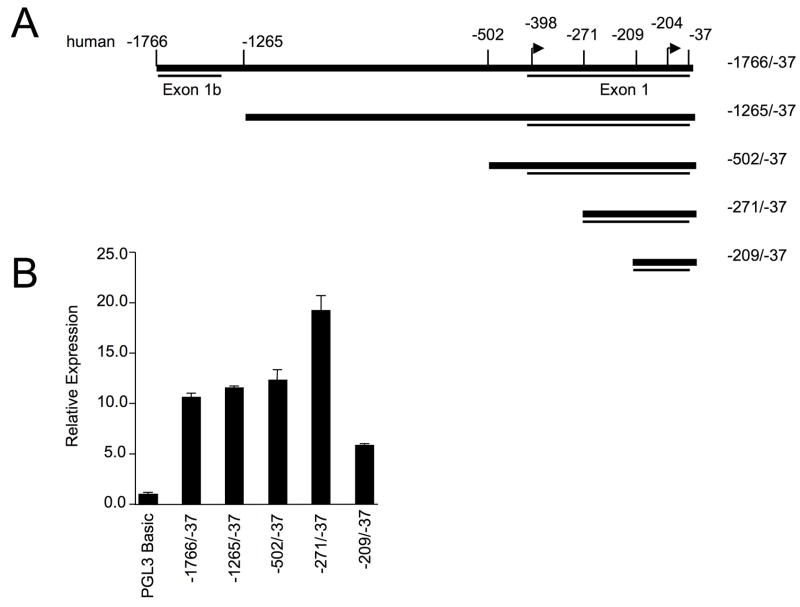
A human Fli1 P/R construct containing the region from −1766 to −37 (equivalent to the mouse region of −1778 to −37) was amplified from genomic DNA and ligated into the pGL3Basic reporter vector. To define the sequences necessary to drive expression in T lymphocytes, successive deletions from the 5′ end of the hFli1 sequence were generated using the −1766/−37 construct (Fig. 2A) and transiently transfected into the Jurkat T cell line (Fig. 2B). Similar to the mouse promoter, sequences from −1766 to −502 were not required for promoter activity, deletion of sequences from −502 to −271 resulted in increased promoter activity, while deletion from −271 to −209 significantly decreased promoter activity (Fig. 2B). These results suggest the region between −271 to −209 contains regulatory elements necessary for promoter activity in T cells and the −502 to −271 region may contain a negative regulatory element, similar to results observed with the mouse promoter 11.
Figure 2. 5′ deletion analysis of hFli1 promoter activity in human T cells.
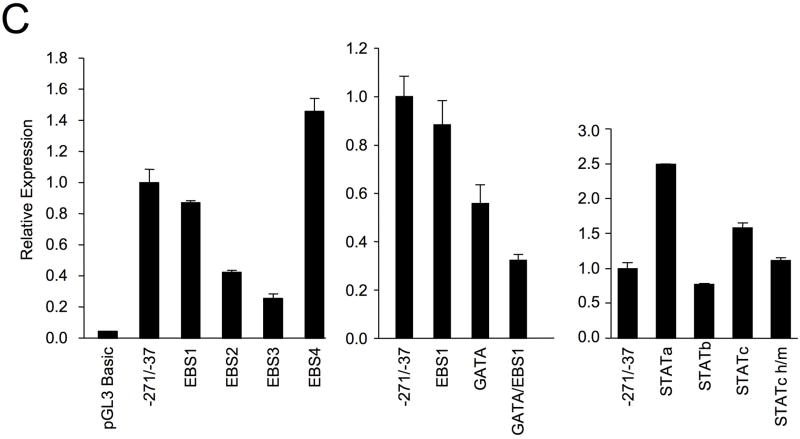
(A) Schematic of the human Fli1 −1766 to −37 region. Deletions were made from the 5′ end of the pGL3 human Fli1 −271/−37 as indicated to generate a series of promoter/reporter constructs. (B) Transient transfection of constructs into the Jurkat human T cell line. Expression is presented relative to the pGL3 Basic empty vector, which was set to 1. (C) Transient transfection of pGL3 human Fli1 −271/−37 containing mutations in each of the putative transcription factor binding sites indicated in Fig. 1B. Results are representative of at least three independent transfections performed with two independently derived clones.
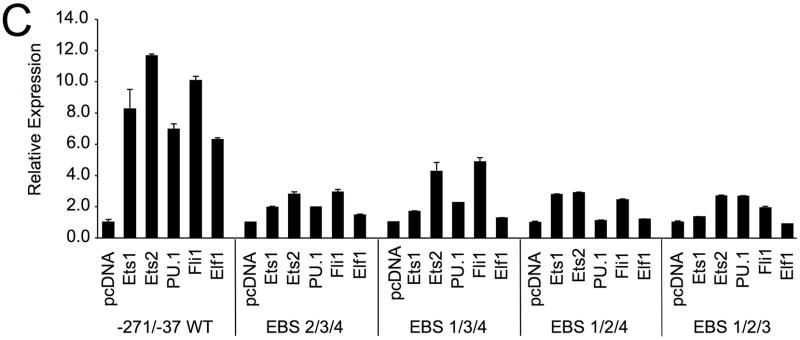
To determine whether the regulatory elements, identified in Figure 1B, are important for human promoter activity in T cells, each binding site was mutated by site-directed mutagenesis within the −271/−37 P/R construct and transfected into the Jurkat T cell line. The mutations were identical to those generated in the mouse construct 11. The EBS1 site is located adjacent to a GATA site resulting in a possible GATA/EBS dual element 13. Mutating the EBS1 site had little effect while the GATA mutation resulted in a 40% decease in promoter activity (Fig. 2C). Interestingly, mutating both the GATA and EBS1 sites resulted in a 70% decease in promoter activity, suggesting a cooperative dual element in the human Fli1 promoter. Mutations in either EBS2 or EBS3 resulted in a 60–70% decrease in promoter activity, while mutating EBS4 resulted in a 40% increase in promoter activity (Fig. 2C).
The STATa and EBS2 sites overlap and the EBS2 mutation may disrupt binding to the STATa binding site as well. Therefore, a mutation in the STATa site was engineered to disrupt the binding of the STATa site and leave the EBS2 site intact. The STATa mutation resulted in a 2.5-fold increase in promoter activity (Fig. 2C), indicating that the STATa site is a negative regulatory element and the EBS2 is a positive regulatory element. A STATb mutation resulted in a slight decrease in activity. These results are similar to those observed in the mouse promoter in a murine T cell line, suggesting that all of the sites identified in the mouse are functional sites in the human promoter as well 11. In the human promoter region, an additional putative STAT site (STATc) was identified, which is not present in the mouse promoter (Fig. 1B). Mutation of the STATc site resulted in a 1.5-fold increase in promoter activity, suggesting it is a functional element (Fig. 2C). A second mutant of the STATc site, STATc h/m, was constructed. The STATc h/m mutation is a one base pair deletion converting the human sequence to match the mouse sequence. This mutation had no effect on promoter activity compared to the wild-type construct, demonstrating that the STATc site does not require the deleted base pair and suggests this site in the mouse promoter may be a functional STAT site. Together, these results suggest that the STATa and STATc sites are negative regulatory elements while the STATb site acts as a weak positive regulatory element in T cells.
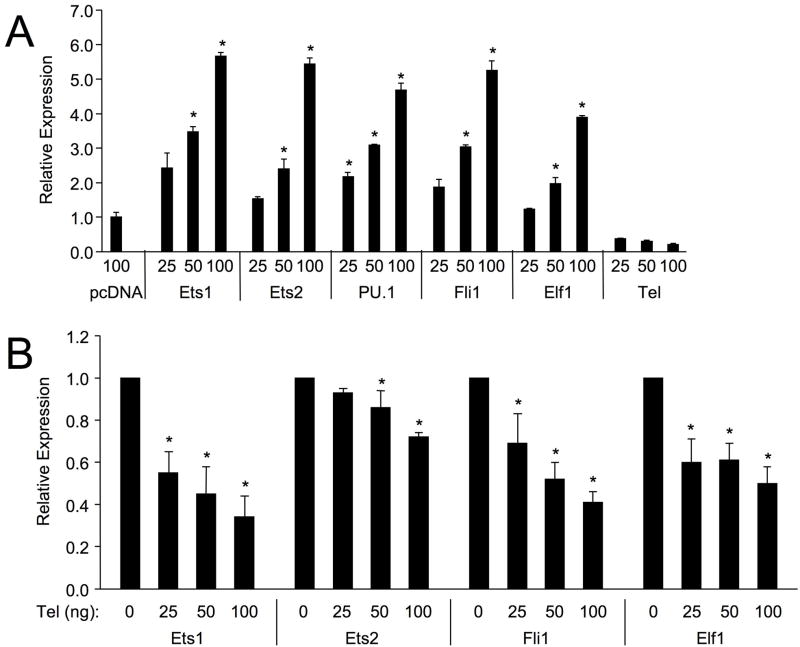
Fli1 is expressed highly in mature B cells and resting mature T cells along with at least 7 other Ets factors 5. To determine which co-expressed Ets factors transactivate from the four sites identified in the proximal promoter, co-transfection experiments with the mFli1 −271/−37 P/R vector, and Ets expression vectors were performed in HeLa cells. HeLa cells express very low levels of most Ets factors 14, reducing effects on the promoter/reporter (P/R) construct by endogenous Ets expression. Cells were co-transfected with a constant amount of the mFli1 −271/−37 P/R vector and either an empty expression vector or increasing amounts of Ets1, Ets2, PU.1, Fli1, Elf1, or Tel expression vectors. Ets1, Ets2, PU.1, Fli1 and Elf1 all activated the promoter in a dose-dependent manner; however, Tel not only failed to stimulate, but inhibited the basal expression of the mFli1 −271/−37 P/R construct (Fig. 3A). Moreover, Tel inhibited Ets1, Ets2, Fli1 and Elf1 activation of the mouse promoter in a dose dependent manner (Fig. 3B). Similarly, Tel inhibited the activation of the human Fli1 −502/−37 and −271/−37 P/R constructs by Ets1, Ets2, Fli1 and Elf1 (data not shown). Tel may be inhibiting transactivation of the Fli1 promoter by the other Ets factors through competition for binding to available EBS or by direct physical interaction as Tel was shown to directly interact with Fli1 in 293 cells 15.
Figure 3. Transactivation of mFli1 Promoter by Ets Factors.
HeLa cells were co-transfected with 1 ug of mFli1 −271/−37 P/R vector and either 100 ng of an empty expression vector (pcDNA3) or increasing amounts of Ets1, Ets2, PU.1, Fli1, Elf1 or Tel (A) expression vectors. (B) HeLa cells were co-transfected with 1.0 ug of Fli1 −271/−37 P/R vector and either 100 ng of Ets1, Ets2, Fli1 or Elf1 along with an increasing amount of Tel expression vector. (C) HeLa cells were co-transfected with various Ets factors and either the Fli1 −271/−37 P/R alone or the P/R containing mutations in 3 of the 4 Ets binding sites indicated. (A and C) Expression is presented relative to the P/R vector co-transfected with the empty expression vector pcDNA3, which was set to 1. (B) Expression is presented relative to the P/R vector co-transfected with Ets1, Ets2, Fli1 or Elf1, which was set to 1. Results are representative of at least three separate transfections. Students T test was used to compare relative expression to mFli1 −271/−37 P/R plus pcDNA3 (A) or mFli1 −271/−37 P/R plus Ets1, Ets2, Fli1 or Elf1 (B). P-values < 0.05 considered significant as indicated by (*).
Through site directed mutagenesis of each individual EBS, we demonstrated that EBS1, 2 and 3 are positive regulatory elements and EBS4 is a negative regulatory element in the mouse11 and human Fli1 promoters (Fig. 2). Co-transfection of Ets factors with the mFli1 −271/−37 construct containing a mutation in EBS1, 2 or 3 in HeLa cells resulted in slight decreases in promoter activity compared to the wildtype P/R (data not shown). However, the differences were not statistically significant, indicating that each of these factors is able to transactivate from any one of the three positive EBS. To determine precisely through which EBS each Ets factor may preferentially transactivate the Fli1 promoter, HeLa cells were co-transfected with the mFli1 −271/−37 P/R containing mutations within three of the four Ets binding sites, resulting in the availability of only one of the EBS (Fig. 3C). When only one positive EBS was available, transactivation of the mutated P/R construct by each Ets factor was markedly reduced compared to the wild-type P/R, indicating that the stimulation of the promoter by Ets1, Ets2, PU.1, Fli1 and Elf1 is dependent on the three positive Ets binding sites, EBS1, 2, and 3. Furthermore, these results suggest that the Ets factors are highly promiscuous and that binding and transactivation through the three positive EBS is additive and that all three sites are necessary for optimal promoter activityin this cellular context.
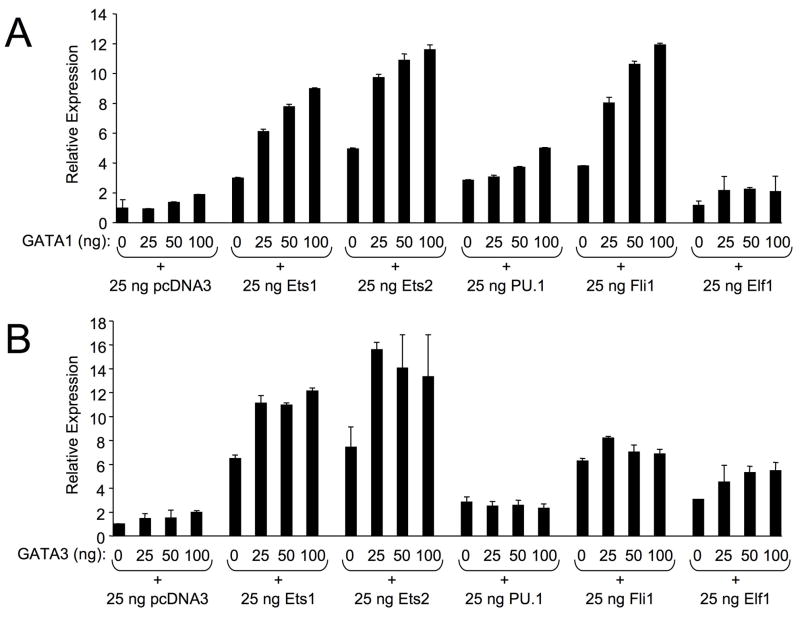
GATA/EBS dual elements function cooperatively in the activation of other genes 13; 16. To determine whether Ets and GATA proteins can synergistically activate the Fli1 promoter through the GATA/EBS1 dual element, the −271/−37 mFli1 P/R vector and expression vectors for Ets1, Ets2, PU.1, Fli1, and Elf1 were co-transfected into HeLa cells with increasing amounts of a GATA1 or GATA3 expression vector. A low level of 25ng of each Ets factor, which has minimal effect on promoter activity alone, was used in order to observe any additive or synergistic effects in conjunction with the GATA factors. Results from these experiments demonstrated that GATA1 or GATA3 alone activates the mFli1 promoter to a maximum of two- and three-fold, respectively (Fig. 4A and 4B). However, co-transfection of Ets1, Ets2 or Fli1 with increasing amounts of GATA1 resulted in a more than additive activation of the mFli1 promoter (Fig. 4A), suggesting synergistic activity. Synergistic activity was also observed with GATA3 and Ets1 or Ets2, but GATA3 had only an additive effect in conjunction with Fli1 (Fig. 4B). On the other hand, Elf1 synergized with GATA3, but had only a slight additive effect in conjunction with GATA1 (Fig. 4A and 4B). Neither GATA1 nor GATA3 could synergize in conjunction with PU.1. In fact, GATA3 inhibited PU.1 stimulation of the promoter (Fig. 4B). Interestingly, promoter activation by all of the Ets factors tested, except Elf1, in the presence of GATA3 decreased as GATA3 levels increased (Fig. 4B), indicating Fli1 promoter activity may be sensitive to the precise levels of GATA3, the GATA factor which is preferentially expressed in T cells 17; 18.
Figure 4. Activation of the mFli1 Promoter by GATA and Ets factors.
HeLa cells were co-transfected with 1.0 ug of Fli1 −271/−37 P/R vector and an empty expression vector (pcDNA3) or Ets1, Ets2, Pu.1, Fli1, or Elf1 expression vectors as indicated, along with increasing amounts of an empty expression vector (pSG5) or a GATA1 (A) or GATA3 (B) expression vector. Expression is presented relative to the P/R vector co-transfected with the empty expression vector pcDNA3, which was set to 1. Results are representative of at least three separate transfections.
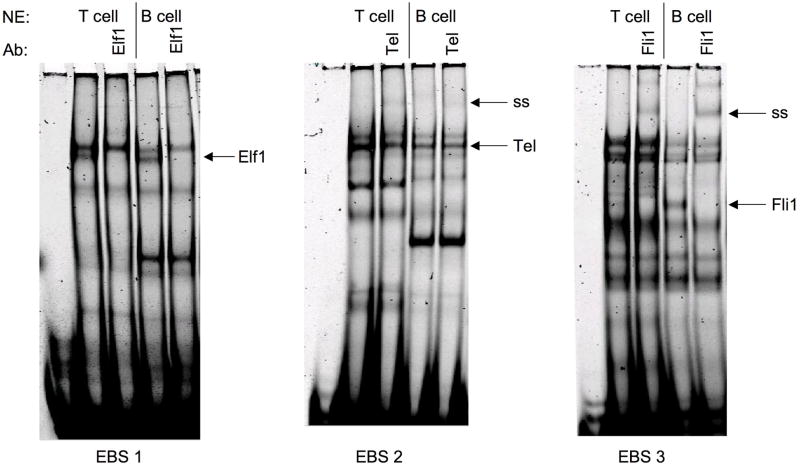
Although most of the Ets factors examined can stimulate the mFli1 promoter activity in HeLa cells, the dynamics in primary T cells, such as relative expression levels, availability of co-factors, and/or availability of binding partners likely determine which Ets factor will bind and transactivate from a specific EBS. Previously we demonstrated that Elf1, Tel and Fli1 bind in vitro to EBS1, 2, and 3, respectfully in extracts from whole mouse spleen 11. We further demonstrate here that this binding is mirrored specifically in primary murine T and B cells (Fig. 5). Binding of Elf1 to GATA/EBS1, Tel to EBS2, and Fli1 to EBS3 probes was identified by the disappearance or supershift of specific DNA-protein complexes following addition of Ets-specific antibodies in both B and T cell nuclear extracts from spleens of non-autoimmune prone BALB/c mice. Similar results were observed in purified B and T cells from another non-autoimmune prone strain, C57BL/6 (data not shown).
Figure 5. Binding of Ets Factors to EBS1, 2, and 3 of the mFli1 Promoter.
Labeled oligos containing the GATA/EBS1, EBS2, or EBS3 cis-regulatory elements were incubated with nuclear extract prepared from naive T and B lymphocytes isolated from BALB/c spleens in the absence or presence of the antibody indicated. Elf1, Elf1 binding; Tel, Tel binding; Fli1, Fli1 binding; and ss, supershift are indicated to the right of each gel. Addition of Elf1 antibody resulted in loss of DNA-Elf1 complex. Each experiment was performed at least three times using two independently derived extracts with similar results.
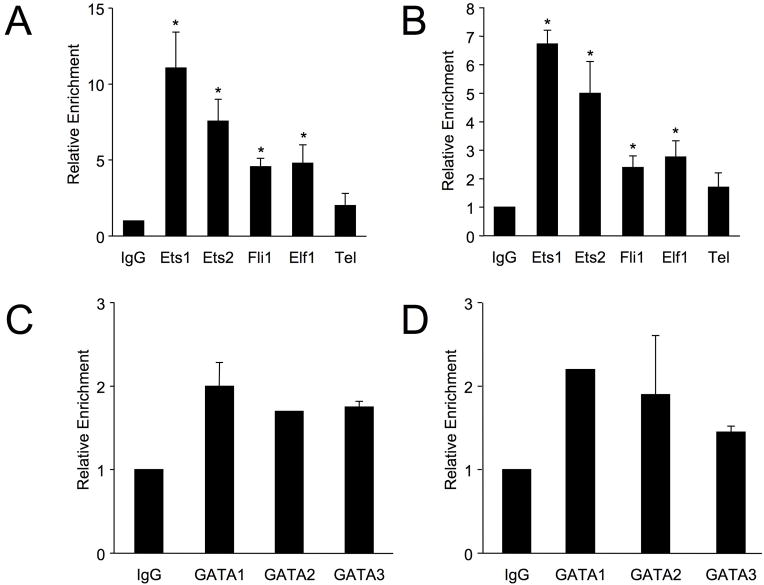
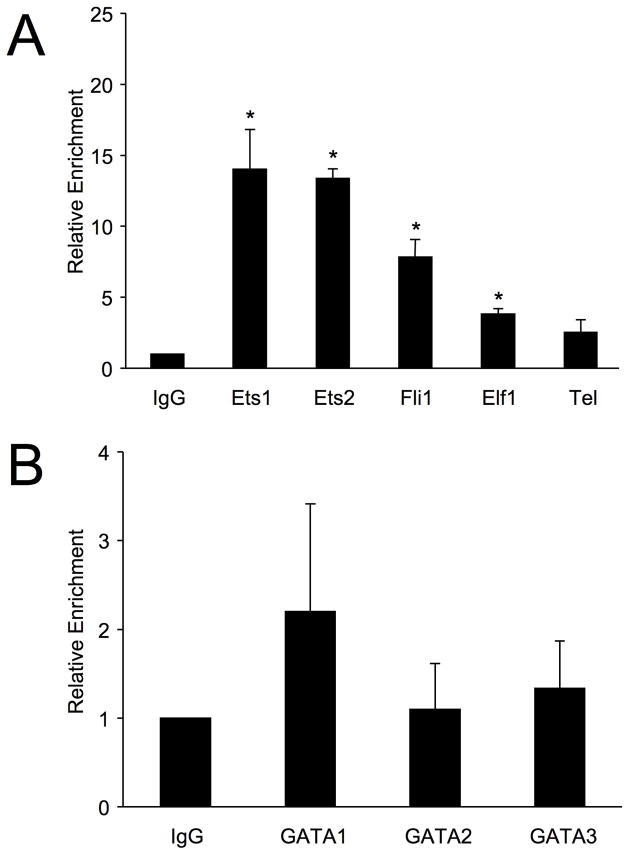
Ets1 and Ets2 do not readily bind to DNA in vitro due to the presence of auto-inhibitory domains 19, and proteins that bind a particular site in vitro may not preferentially bind the same site in vivo. Therefore, to identify the Ets factors that bind the Fli1 promoter in live T cells, in vivo binding assays using chromatin-immunoprecipitation (ChIP) were performed in T cells with antibodies specific for various Ets factors. In primary, naive T cells from C57BL/6 mice, antibodies specific for Ets1, Ets2, Fli1, or Elf1 resulted in significant enrichment of the mFli1 proximal promoter compared to IgG, with Ets1 and Ets2 resulting in the greatest relative enrichment (Fig. 6A). ChIP assays also were performed in primary, naive T cells from C57BL/6 mice using antibodies against GATA1, 2 and 3 (Fig. 6C). Although GATA1 and GATA3 can cooperatively activate the Fli1 promoter in conjunction with Ets1, Ets2, Elf1 and Fli1, no significant enrichment of the mFli1 promoter was observed with GATA1, 2 and 3 antibodies in naive T cells. These results were replicated in T cells from BALB/c mice (Fig. 6B and 6D), demonstrating that the relative binding of Ets factors to the mFli1 promoter is not strain specific. As a negative control, all ChIP templates were used in PCR with primers specific for the Exon3 region of the murine Fli1 gene, which is located 75 Kb downstream of the promoter, and not expected to bind Ets factors. No enrichment of Exon3 was observed with any of the antibodies used (data not shown).
Figure 6. Association of Ets and GATA Factors with the Endogenous Fli1 Promoter in Primary Mouse T Cells.
T cells isolated from three 8-week old C57BL6 (A, C) and BALB/c (B, D) spleens were subjected to Chromatin Immunoprecipitation assays with antibodies against Ets factors (A, B) or GATA factors (C, D), as indicated. Eluted chromatin from IPs was used in real-time PCR using primers specific for the Fli1 promoter region in Exon1. Immunoprecipitations were performed on three sets of chromatin, represented as an average in graph, and PCR was run twice in triplicate for each set. Error bars represent standard error. Students T test was used to compare IPs to non-specific IgG within each mouse strain. P-values < 0.05 was considered significant as indicated by (*).
To further determine whether the human and mouse genes are similarly regulated in T cells, in vivo binding of Ets factors to the human promoter was examined. ChIP assays were performed in the Jurkat T cell line using antibodies specific for various Ets factors. Antibodies specific for Ets1, Ets2, Fli1, and Elf1 resulted in significant enrichment of the human Fli1 proximal promoter compared to IgG (Fig. 7A). Overall, the relative amount of bound Ets factors to the human promoter region in human T cells in vivo was similar to that observed in the mouse promoter in mouse T cells in vivo with the exception of Elf1 (Table 1). ChIP assays also were performed with the human promoter in Jurkat T cells using antibodies against GATA1, 2 and 3. As observed with the mouse promoter, no significant enrichment of the human promoter with GATA1, 2 or 3 antibodies was observed (Fig. 7B).
Figure 7. Association of Ets and GATA Factors with the Endogenous Fli1 Promoter in Human T Cells.
The Jurkat human T cells were subjected to Chromatin Immunoprecipitation assays with antibodies against Ets factors (A) or GATA factors (B), as indicated above. Eluted chromatin from IPs was used in real-time PCR using primers specific for the human Fli1 promoter region in Exon1. Immunoprecipitations were performed on three sets of chromatin, represented as an average in graph, and PCR was run twice in triplicate for each set. Error bars represent standard error. Students T test was used to compare IPs to non-specific IgG. P-values < 0.05 was considered significant as indicated by (*).
Table 1. Ratio of bound Ets factors to endogenous Fli1 Promoter region in Human and Mouse T lymphocytes.
| T Cell | Ets1 | Ets2 | Fli1 | Elf1 | Tel |
|---|---|---|---|---|---|
| C57BL6 | 1 | 0.69 | 0.41 | 0.43 | 0.18 |
| BALB/c | 1 | 0.74 | 0.36 | 0.41 | 0.25 |
| Jurkat | 1 | 0.96 | 0.56 | 0.27 | 0.18 |
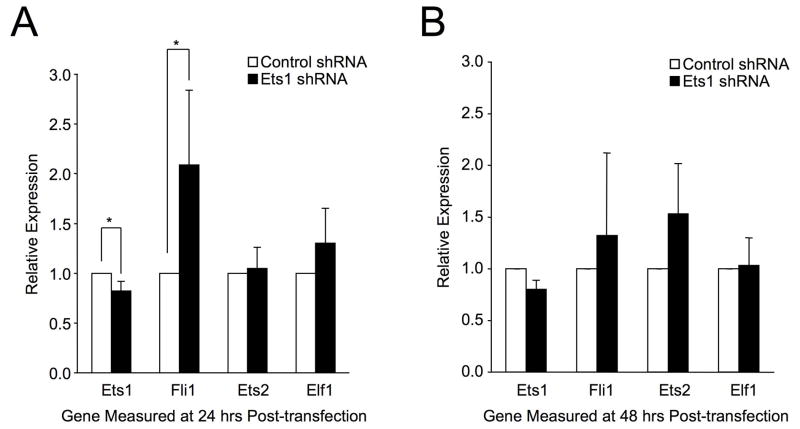
ChIP analysis demonstrated that Ets factors bind the in vivo Fli1 promoter region in T cells from mouse and human. In both species, Ets1 antibodies resulted in the greatest enrichment of this region. To determine whether reducing the levels of Ets1 has an effect on Fli1 expression, RNA interference for Ets1 was performed. S1A T cells were transfected with shRNA plasmids specific for mouse Ets1 or a scrambled sequence. Transfection efficiency was determined to be ~55% (data not shown). RNA was prepared 24 and 48 hrs after transfection and subjected to real-time PCR and Western Blotting for verification of Ets1 knockdown. Due to the low transfection efficiency of T cells we observed only a 20% decrease in Ets1 message levels (Fig. 8) and protein levels (data not shown) compared to cells transfected with shRNA scrambled negative control at both 24 hours (Fig. 8A) and 48 hours (Fig. 8B) post-transfection. Importantly, this reduction was reproducible and statistically significant at 24 hours and resulted in a reproducible and statistically significant 2.1-fold increase of Fli1 message levels at 24 hours (Fig. 8A). Fli1 expression also increased 1.3-fold at 48 hours but did not reach statistical significance (Fig. 8B). As an added control, message levels of Ets2 and Elf1, which are co-expressed with Fli1 in T cells, were measured in these samples. Reducing Ets1 had no significant effect on Ets2 and Elf1 message levels at 24 hours (Fig. 8A) or 48 hours (Fig. 8B) post-transfection, demonstrating that reducing Ets1 expression had a specific effect on Fli1 message levels. Together, these results further support a role for transcriptional regulation of endogenous Fli1 by Ets factors in T lymphocytes.
Figure 8. Targeted Reduction of Ets1 Increases Fli1 Transcription.
The mouse S1A T cell line was transfected with shRNA plasmids targeting mouse Ets1 or a scrambled sequence as a negative control. Total RNA was collected and subjected to real-time RTPCR using primers against mouse Ets1, Fli1, Ets2 and Elf1 relative to GAPDH at 24 hrs (A) and 48 hrs (B) post-transfection. Transfections were performed three times for the 24 hr time point and twice for the 48 hr time point. Real-time PCR was performed in triplicate on two independently derived sets of cDNA prepared from each set of transfected cells. Graphs represent an average of all real-time results obtained. Error bars represent standard deviation. Students T test was used to compare relative expression compared to controls using the most stringent settings. P-values < 0.05 was considered significant as indicated by (*).
Discussion
T lymphocytes are known to play a significant role in the pathogenesis of lupus 20 and it is clear that altering Fli1 expression has profound effects on the pathogenesis of lupus 8–10. Identifying the mechanisms involved in controlling Fli1 expression in T lymphocytes may result in new avenues for treating autoimmune diseases like lupus. This report provides further description of the mechanisms involved in the transcriptional regulation of the mouse Fli1 gene and the first description of mechanisms involved in the regulation of the human Fli1 gene in T lymphocytes.
The human and mouse Fli1 proximal promoters are 95% homologous within the −271/−37 region, the region we identified to contain optimal activity in T cells. We demonstrated that the previously identified cis-regulatory elements in the −271/−37 region of the mouse promoter are functionally conserved in the human promoter and identified an additional STAT site that functions as a negative regulatory element. Previously, we demonstrated Elf1, Tel and Fli1 binding to EBS1, 2, and 3, respectfully of the mFli1 promoter in nuclear extracts made from whole spleen 11. Here we confirmed this in vitro binding specifically in primary B and T cells. We also demonstrated that Ets1, Ets2, PU.1, Elf1, and Fli1 itself, which are co-expressed with Fli1 in lymphocytes, could transactivate the murine Fli1 proximal promoter.
To confirm our in vitro results, we performed in vivo binding assays with the mouse and human promoters. We demonstrated that Ets1, Ets2, Elf1, and Fli1 bind the endogenous Fli1 promoter in both human and mouse, suggesting the genes are regulated in a similar manner. For purposes of discussion, we compared the levels of Ets1 to the other Ets factors bound to the mouse and human promoters in the in vivo binding studies (summarized in Table 1). The relative binding to the mouse promoter in BALB/c and C57BL/6 was nearly identical. In comparing the binding to the mouse and human promoters, both species showed relatively little binding of Tel compared to Ets1. However, we observed lower levels of Elf1 and increased levels of Ets2 relative to Ets1 in human cells compared to mice. Despite these subtle differences, the overall trend of associated Ets factors with the Fli1 promoter is similar in both human and mouse T cells, indicating a key role of the Ets transcription factor family in regulating Fli1 in T cells.
Interestingly, Ets1 and Ets2 resulted in the greatest enrichment in both mouse and human indicating they may be key in vivo regulators of the Fli1 promoter in T cells. Ets1 may be a ubiquitous transactivator of Fli1 as it also has been implicated in regulating Fli1 expression in non-lymphoid tissues 21. In light of recent studies, binding of Ets1 to the endogenous Fli1 promoter poses an interesting scenario. Ets1 knockout mice exhibit a lupus-like autoimmune phenotype 22. These mice have an elevated proportion of T cells with an effector memory phenotype, similar to that observed in MRL/lpr lupus prone mice 23. Together with the known over-expression of Fli1 in lupus mice 8; 9, we hypothesize the Ets1 knockout mouse phenotype may be due in part to the dysregulation of Fli1 expression in these mice. Loss of Ets1 may allow other Ets factors that are stronger transactivators to bind, leading to over-expression of Fli1. This notion is further supported by our observation of increased transcription of Fli1 following RNA interference of Ets1 in S1A T cells.
The observed association of Elf1 with the Fli1 promoter in vivo is also of significant interest. Elf1 activation requires post-translational phosphorylation and glycosylation for nuclear transport and binding, which has been shown to be defective in T cells of lupus patients 24. Our results suggest Elf1 may be a key regulator of Fli1 expression in T cells and the Elf1 defect in human lupus may be one mechanism by which Fli1 becomes over-expressed in lupus T cells. Loss of Elf1 binding may allow other more potent Ets transactivators to bind, such as Fli1, leading to over-expression.
We demonstrated in this study the relative association of Fli1 with its own promoter in both human and mouse T cells in vivo. Over-expression or ectopic expression of Fli1 has been observed to upregulate endogenous Fli1 expression and perturb normal lymphoid cell function and programmed cell death 10. Previously, we demonstrated that Fli1 can upregulate its own expression in a T cell line 11. Together with the activation of the Fli1 promoter by Fli1 itself observed in this study, these results suggest a potential positive feedback loop for Fli1 expression in T cells.
In contrast to the other Ets factors tested, Tel, which is also co-expressed with Fli1 in T lymphocytes, not only failed to transactivate the proximal promoter it inhibited the basal expression of the promoter in HeLa cells. Tel also inhibited the stimulation of the promoter by Ets1, Ets2, Fli1 or Elf1, indicating Tel may act as a negative regulator of Fli1 under certain cellular conditions. It was also noted that we observed no significant in vivo binding of Tel to the Fli1 promoter in human or mouse T cells. This conflicts with our previous observation where over-expression of Tel resulted in an increase in endogenous Fli1 mRNA expression in a T cell line 11. Tel is normally expressed at very low levels in both naive and activated T cells. Therefore, it is likely that over-expression of Tel in this cellular context could result in aberrant expression of Fli1 or Tel may be acting through an as yet unidentified Ets binding site outside of the proximal promoter. Tel activity also may not be properly regulated in the T cell line used. In some cellular contexts, Tel must become sumoylated to act as a negative transcriptional repressor 15; 25. Regardless, our results indicate very little association of Tel with the endogenous proximal promoter in primary naive T cells, indicating that Tel may only play a small role, if any, in regulating Fli1 in naive T cells. However, based on the ability of Tel in primary T cell extracts to bind to the EBS2 site of the proximal promoter in vitro, we speculate that Tel may play a role in down-regulating Fli1 when T cells become activated.
Competition for available EBS may result in changes in relative association of Ets factors to the proximal promoter depending on cellular environment or state of disease. Overall levels of promoter bound Ets factors or availability of binding partners and cofactors may also change during pathogenesis. For example, steric constraints resulting in antagonism between Ets and GATA factors at the Fli1 promoter where both factors engage the DNA simultaneously may occur. This scenario may have precedent. Traditional thinking was that GATA3 specifically drove T helper type 2 cell differentiation, but has now emerged as having multiple roles in T lymphocyte development 26. How this single transcription factor that is continuously expressed during T cell development can perform different functions has yet to be understood. Competition and collaboration with other factors like Ets1 and Ets2 may partially explain this phenomenon. Ets1 and Ets2 cooperate with GATA3 to activate the interleukin-5 gene in both T and myeloid cells 27. In our current study, we observed Fli1 promoter/reporter transactivation by Ets1, Ets2, GATA3 or GATA1 individually, while synergistic transactivation resulted when these Ets and GATA factors were co-transfected. In naive T cells we were unable to pull down the in vivo Fli1 promoter region with antibodies against GATA factors, indicating GATA/Ets cooperation may not be important in this cellular context. GATA3 expression increases in activated T cells following immune stimuli. Therefore, GATA/Ets interactions may then become relevant in regulating the Fli1 promoter 28. Thus, balanced expression and activity of GATA and Ets factors could selectively promote Fli1 expression in certain physiologic states.
We hypothesize precise levels of Ets factors are important in regulating Fli1 expression. Loss of a single Ets factor could change the dynamics of Ets DNA binding, and/or interactions with binding partners or cofactors that may lead to an increase, as seen in lupus T cells, or a decrease, as seen in fibrosis, of Fli1 expression and contribute to pathogenesis. Future studies will be aimed at dissecting the dynamics of Ets factor binding and levels in regulating Fli1 and to identify potential targets and/or biomarkers for these diseases.
Materials and Methods
Mice
All mice were maintained at the Ralph H. Johnson VAMC Animal Care Facility (Charleston, SC) and experiments were approved by the Institutional Animal Care and Use Committee. BALB/c and C57BL/6 mice were purchased from Jackson Laboratory, Bar Harbor, ME. Spleens were harvested from euthanized mice. Splenic T and B lymphocytes were isolated by negative selection using the MACS T Cell and B Cell Isolation Kits (Miltenyi Biotec) according to the manufacturers recommendations. Cell populations were analyzed by flow cytometry and found to be 85–95% pure.
Cell Lines
HeLa cells were maintained in DMEM/F12 (50/50) supplemented with 5% FBS. S1A.TB.4.8.2 (S1A) and Jurkat T cells were maintained in RPMI 1640 supplemented with 10% FBS.
Plasmids and Transfections
Cloning of the Fli1 promoter sequence and all mouse Fli1 promoter/reporter (P/R) constructs were described previously 11. pcDNAPU.1 was constructed by moving the PU.1 cDNA from pCMV5PU.1 into the pcDNA3 vector. pcDNAEts1, pcDNAEts2, and pCMV5PU.1 expression vectors were obtained from Dr. Angie Rizzino (University of Nebraska Medical Center). pcDNAFli1, pcDNAElf1, and pSG5GATA1 were obtained from Dr. Dennis Watson (Medical University of South Carolina) and pcDNATel and pCTAPGATA3 expression vectors were obtained from Dr. Maria Trojanowska (Medical University of South Carolina).
The sequence encompassing the human Fli1 proximal and distal promoter regions from −1766 to −37 (equivalent to the mouse region of −1778 to −37) was amplified from genomic DNA from an unaffected control in the Carolina Lupus Study cohort. The amplified product was ligated into the SmaI and BglII sites of the pGL3 Basic luciferase reporter vector (Promega) to generate the pGL3 hFli1−1766/−37 P/R construct. Deletions of the 5′ end of the −1766/−37 sequence were generated by restriction enzyme digestion and religation at −1265 (MluI) and −505 (NheI), to generate pGL3 hFli1−1265/−37 and pGL3 hFli1−502/−37 P/R constructs. The pGL3 hFli1 −271/−37 and pGL3 hFli1 −209/−37 P/R constructs were generated by PCR using primers to amplify the entire pGL3 hFli1 −1766/−37 plasmid excluding the regions from −1766 to −271 and −1766 to −209, respectively. Mutations within the four EBS, the GATA site and two of the three STAT sites (a and b) of the pGL3 hFli1 −271/−37 construct were generated by site-directed mutagenesis as described previously, incorporating the same base pair changes as described for the mouse Fli1 P/R mutant constructs11. The STATc and h/m STATc mutations were generated by site-directed mutagenesis as described previously11. The STATc mutant contains a two base pair change within the STATc site that destroys a TfiI site used for screening. The h/mSTATc mutant contains a deletion of the ‘T’ not present in the mouse sequence and also destroys a TfiI site used for screening. Two clones of each construct were generated and tested in transfections. Deletions and mutations were confirmed by direct sequencing of all clones. All transfection of human clones were performed in duplicate in Jurkat T cell line with Fugene reagent (Roche) according to manufacturers instructions. The mouse Fli1 −271/−37 P/R construct or human Fli1 −502/−37 and −271−37 P/R constructs were co-transfected into HeLa cells with either an empty expression vector (pcDNA3) or pcDNAEts1, pcDNAEts2, pcDNAFli1, pcDNAElf1, pcDNATel or pcDNAPU.1 expression vectors and/or pSG5GATA1 or pCTAPGATA3 expression vectors in the amounts indicated in the figures. All transfections were performed in duplicate with Fugene reagent (Roche) according to manufacturers instructions. Luciferase activity was measured and normalized to expression from a co-transfected renilla luciferase expression vector (Promega). Total amount of plasmid was kept constant. Expression is presented relative to the P/R vector co-transfected with the empty vector, which was set to 1.
SureSilencing shRNA plasmids targeting mouse Ets1 were purchased from SABiosciences (Frederick, MD). Clones were expanded and tested according to the manufacturer’s instructions. 4ug of the shRNA-Ets1 plasmids or scrambled control plasmids were transiently transfected into 1×106 cells of the S1A mouse T cell line using an Amaxa nucleofection device (Lonza Group Ltd.), program T-16, according to the manufacturer’s instructions. Cells were cultured as described above for 24 and 48 hours before extracting total RNA. Real-time PCR was performed as described below.
Nuclear Extracts, Electrophoretic Mobility Shift Assays (EMSA) and Protein Immunoblotting
Nuclear extracts were prepared from whole spleen, primary T cells and primary B cells from 12 week old BALB/c or C57BL/6 mice or from the S1A T cell line, as indicated in figure legends, using the NE-PER kit (Pierce). Extracts were incubated with fluorescently labeled probes consisting of synthesized, annealed oligos containing murine sequences surrounding the cis-regulatory elements indicated, and separated by electrophoresis on polyacrylamide gels, as described previously11. Binding was visualized by scanning the gels on the Odyssey Infrared Imaging System (Li-Cor). Specific factors bound to the cis-regulatory elements in the EMSAs were identified by addition of antibodies against the Ets transcription factors indicated in the figures. Antibodies were added to the binding reaction prior to addition of the probe.
Chromatin Immunoprecipitation (ChIP)
Nuclear proteins were crosslinked to chromatin by addition of 1% formaldehyde to the cells for 10 minutes. Chromatin from lysed cells was sheared to an average length of 500 bp using a Branson-450 Sonifier (Branson Ultrasonics Corp., Danbury, CT) and pre-cleared with blocked Protein A Dynabeads (Invitrogen). Immunoprecipitations were performed using antibodies against the factors indicated in figures. IgG was used as a negative control. All antibodies were purchased from Santa Cruz Biotechnology, Inc. Antibody/chromatin complexes were then bound to Protein A Dynabeads and washed on a magnetic separation stand. Eluted complexes were decrosslinked, proteins degraded with Proteinase K solution (Shelton Scientific), and immunoprecipitated DNA was purified with a PCR Purification Kit (Qiagen).
Real-time PCR
Purified DNA from ChIP assays was used as template in real-time PCR for either the mouse or human Fli1 promoter. Primers for the Exon1 promoter region of mouse Fli1 are as follows: Ex1ChIPUp 5′-TCC CCG ATG GCA AAG TG-3′ and Ex1ChIPDn 5′-TCC CCT GTG CAC GTT TGT-3′. Primers for the Exon 1 promoter region of human Fli1 are as follows: hEx1ChIPUp 5′-CCC CGA TTC GCA AAG TG-3′ and same Ex1ChIPDn sequence used for mouse region. Primers for mouse Fli1 Exon3 region are as follows: Ex3ChIPUp 5′-GCC AAC CCC ATG AAC TAT-3′ and Ex3ChIPDn 5′-GCA GGC ACA ATG ACT CTC-3′. Primers for human Exon 3 region are as follows: hEx3ChIPUp 5′-TGG ACT GCA GCG TTA GC-3′ and hEx3ChIPDn 5′-TGG TCA TGT TGG GAG GA-3′. PCR was quantified on a BioRad MyiQ real-time PCR SYBR Green detection system in triplicate in two independent runs. Fold-enrichment was compared to immunoprecipitations with IgG, which was set to 1.
Purified RNA from S1A T cells was extracted using Qiagen’s RNeasy kit. cDNA was prepared using iScript cDNA synthesis kit (BioRad). The cDNA served as a template for real-time PCR using iQ SYBR Green Supermix (BioRad) to measure Ets1, Fli1, Ets2, Elf1 and GAPDH expression. Primers used to measure mouse Ets1, Ets2 and Elf1 were described previously in Landry et al., 2005 29 and primers used to measure mouse Fli1 and GAPDH were described previously in Nowling et al., 2008 11. The threshold cycle number (Ct) for each gene in each sample was measured using the MyiQ Real-time PCR Detection system and software (BioRad). Relative expression levels were calculated using the Gene Expression Macro software (Biorad). Briefly, Ct values were normalized to GAPDH expression for each sample by subtracting the GAPDH Ct from the Fli1, Ets1, Ets2 or Elf1 Ct (gene of interest) to give the ΔCt. A relative fold change was calculated using the calculation 2(ΔΔCt) where ΔΔCt is the difference between the ΔCt of the sample and the ΔCt of the reference sample, which was set to 1. The cells transfected with the scrambled shRNA was used as the reference sample. Message levels for all genes were measured in triplicate within the same cDNA sample and two sets of cDNA were prepared from each RNA sample.
Acknowledgments
This work was supported by NIH grants DK072306 and AR053376. The authors would like to thank Dr. Angie Rizzino, Dr. Dennis Watson and Dr. Maria Trojanowska for their kind gifts of Ets and GATA expression vectors. A special thanks also goes to Dr. Gary Gilkeson for critical reading of the manuscript and providing valuable comments.
References
- 1.Ben-David Y, Giddens EB, Bernstein A. Identification and mapping of a common proviral integration site Fli-1 in erythroleukemia cells induced by Friend murine leukemia virus. Proc Natl Acad Sci U S A. 1990;87(4):1332–1336. doi: 10.1073/pnas.87.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-David Y, Giddens EB, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev. 1991;5(6):908–918. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- 3.Melet F, Motro B, Rossi DJ, Zhang L, Bernstein A. Generation of a novel Fli-1 protein by gene targeting leads to a defect in thymus development and a delay in Friend virus-induced erythroleukemia. Molecular and cellular biology. 1996;16(6):2708–2718. doi: 10.1128/mcb.16.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson DK, Smyth FE, Thompson DM, Cheng JQ, Testa JR, Papas TS, et al. The ERGB/Fli-1 gene: isolation and characterization of a new member of the family of human ETS transcription factors. Cell Growth Differ. 1992;3(10):705–713. [PubMed] [Google Scholar]
- 5.Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126(14):3131–3148. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 6.Gilkeson GS. Glomerular binding antibodies in systemic lupus erythematosus. Lupus: Molecular and Cellular Pathogenesis. 1999;7:448–470. [Google Scholar]
- 7.Oates JC, Gilkeson GS. Mediators of injury in lupus nephritis. Curr Opin Rheumatol. 2002;14(5):498–503. doi: 10.1097/00002281-200209000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Georgiou P, Markoulakou IG, Green JE, Dantis P, Romano-Spica V, Kottardid S, Lautenberger JA, Watson DK, Papas TS, Fischinger PJ, Bhat NK. Expression of ets family of genes in systemic lupus erythematosus and Sjogren’s syndrome. International Journal of Oncology. 1996;9:9–18. [PubMed] [Google Scholar]
- 9.Zhang XK, Gallant S, Molano I, Moussa OM, Ruiz P, Spyropoulos DD, et al. Decreased expression of the Ets family transcription factor Fli-1 markedly prolongs survival and significantly reduces renal disease in MRL/lpr mice. J Immunol. 2004;173(10):6481–6489. doi: 10.4049/jimmunol.173.10.6481. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Eddy A, Teng YT, Fritzler M, Kluppel M, Melet F, et al. An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Molecular and cellular biology. 1995;15(12):6961–6970. doi: 10.1128/mcb.15.12.6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowling TK, Fulton JD, Chike-Harris K, Gilkeson GS. Ets factors and a newly identified polymorphism regulate Fli1 promoter activity in lymphocytes. Molecular immunology. 2008;45(1):1–12. doi: 10.1016/j.molimm.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, et al. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13(4):721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henkel G, Brown MA. PU.1 and GATA: components of a mast cell-specific interleukin 4 intronic enhancer. Proc Natl Acad Sci U S A. 1994;91(16):7737–7741. doi: 10.1073/pnas.91.16.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32(18):5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwiatkowski BA, Bastian LS, Bauer TR, Jr, Tsai S, Zielinska-Kwiatkowska AG, Hickstein DD. The ets family member Tel binds to the Fli-1 oncoprotein and inhibits its transcriptional activity. The Journal of biological chemistry. 1998;273(28):17525–17530. doi: 10.1074/jbc.273.28.17525. [DOI] [PubMed] [Google Scholar]
- 16.Galson DL, Hensold JO, Bishop TR, Schalling M, D’Andrea AD, Jones C, et al. Mouse beta-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Molecular and cellular biology. 1993;13(5):2929–2941. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen ML, Kuo CL. A conserved sequence block in the murine and human T cell receptor Jalpha loci interacts with developmentally regulated nucleoprotein complexes in vitro and associates with GATA-3 and octamer-binding factors in vivo. Eur J Immunol. 2001;31(6):1696–1705. doi: 10.1002/1521-4141(200106)31:6<1696::aid-immu1696>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 18.Lavenu-Bombled C, Trainor CD, Makeh I, Romeo PH, Max-Audit I. Interleukin-13 gene expression is regulated by GATA-3 in T cells: role of a critical association of a GATA and two GATG motifs. The Journal of biological chemistry. 2002;277(21):18313–18321. doi: 10.1074/jbc.M110013200. [DOI] [PubMed] [Google Scholar]
- 19.Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, et al. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. The Journal of biological chemistry. 2005;280(8):7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 20.Tenbrock K, Juang YT, Kyttaris VC, Tsokos GC. Altered signal transduction in SLE T cells. Rheumatology (Oxford, England) 2007;46(10):1525–1530. doi: 10.1093/rheumatology/kem154. [DOI] [PubMed] [Google Scholar]
- 21.Lelievre E, Lionneton F, Mattot V, Spruyt N, Soncin F. Ets-1 regulates fli-1 expression in endothelial cells. Identification of ETS binding sites in the fli-1 gene promoter. The Journal of biological chemistry. 2002;277(28):25143–25151. doi: 10.1074/jbc.M201628200. [DOI] [PubMed] [Google Scholar]
- 22.Wang D, John SA, Clements JL, Percy DH, Barton KP, Garrett-Sinha LA. Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. International immunology. 2005;17(9):1179–1191. doi: 10.1093/intimm/dxh295. [DOI] [PubMed] [Google Scholar]
- 23.Clements JL, John SA, Garrett-Sinha LA. Impaired generation of CD8+ thymocytes in Ets-1-deficient mice. J Immunol. 2006;177(2):905–912. doi: 10.4049/jimmunol.177.2.905. [DOI] [PubMed] [Google Scholar]
- 24.Tsokos GC, Nambiar MP, Juang YT. Activation of the Ets transcription factor Elf-1 requires phosphorylation and glycosylation: defective expression of activated Elf-1 is involved in the decreased TCR zeta chain gene expression in patients with systemic lupus erythematosus. Ann N Y Acad Sci. 2003;987:240–245. doi: 10.1111/j.1749-6632.2003.tb06054.x. [DOI] [PubMed] [Google Scholar]
- 25.Lopez RG, Carron C, Oury C, Gardellin P, Bernard O, Ghysdael J. TEL is a sequence-specific transcriptional repressor. The Journal of biological chemistry. 1999;274(42):30132–30138. doi: 10.1074/jbc.274.42.30132. [DOI] [PubMed] [Google Scholar]
- 26.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 27.Blumenthal SG, Aichele G, Wirth T, Czernilofsky AP, Nordheim A, Dittmer J. Regulation of the human interleukin-5 promoter by Ets transcription factors. Ets1 and Ets2, but not Elf-1, cooperate with GATA3 and HTLV-I Tax1. The Journal of biological chemistry. 1999;274(18):12910–12916. doi: 10.1074/jbc.274.18.12910. [DOI] [PubMed] [Google Scholar]
- 28.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89(4):587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 29.Landry JR, Kinston S, Knezevic K, Donaldson IJ, Green AR, Gottgens B. Fli1, Elf1, and Ets1 regulate the proximal promoter of the LMO2 gene in endothelial cells. Blood. 2005;106(8):2680–2687. doi: 10.1182/blood-2004-12-4755. [DOI] [PubMed] [Google Scholar]