Abstract
Mounting evidence suggests that the proinflammatory cytokine macrophage migration inhibitory factor (MIF) may serve as an important link between chronic inflammation and carcinogenesis as evidenced by the increase in serum MIF found in patients with various cancers. The present study identifies anti-thrombin III (ATIII) as an endogenous MIF binding protein, which reduces MIF biological activity. Serum MIF in bladder cancer patients (TCC stage II, n=50) was increased when compared to normal patients (n=50), while ATIII-MIF complexes were decreased in bladder cancer patient serum. These data suggest that increased circulating levels of bioactive MIF are present in bladder cancer patient serum.
Keywords: Cytokines, protein-complexes, erk1/2
1. Introduction
The inability to down-regulate inflammatory responses is a hallmark of numerous chronic inflammatory pathologies, including cancer [1]. Many cancers arise from sites of infection, chronic irritation, and inflammation, with the resulting tumor cells exploiting inflammatory signal transduction to aid in tumor cell invasion, migration and metastasis [2]. Integration and titration of cytokine signals are important for the control of these inflammatory pathologies. In general, cytokine activity is regulated by the bioavailability of ligands and/or receptors. While receptor bioavailability is primarily achieved by an increase or a decrease in the cell-surface expression of the functional receptor, control of the ligand often occurs by interaction of the cytokine with endogenous molecules capable of sequestering the ligand from its cognate receptor [3].
Macrophage migration inhibitory factor (MIF) is a ubiquitous proinflammatory cytokine that is produced in various inflammatory disorders and functions as a central regulator of the innate immune system [4]. Current consensus based on extensive experimental evidence is that MIF directly participates in the activation of intracellular cell signaling pathways, and that it is a principal initiator of the immune and inflammatory responses [5,6]. To date, the mechanism by which MIF acts as an immune mediator has not been completely defined. Experimental studies using MIF knockout mice, neutralizing anti-MIF antibodies or specific small molecule inhibitors has shown an attenuation of numerous disease pathologies including bladder inflammation, sepsis, autoimmune diabetes, arthritis, bladder and prostate cancer [7–9].
Since baseline levels of MIF are found in the serum and the urine of normal patients without underlying inflammatory conditions [10–12] a number of regulatory mechanisms may explain why MIF is not pro-inflammatory under these circumstances. For instance, 1) MIF levels may need to reach a certain threshold to initiate proinflammatory signal transduction. Evidence for this mechanism include the documented increase in MIF expression in a number of inflammatory and neoplastic conditions [5,6,11,13]. 2) The MIF receptor, recently identified as CD74, is present in low levels on cells under non-inflammatory conditions and thus it is unable to initiate pro-inflammatory signal transduction. We recently reported increased amounts of urothelial cell surface CD74 in a rat model of bladder inflammation, which lends support to this hypothesis [14]. 3) MIF exists in different forms; for instance, active versus inactive, which modulates the bioactivity of the molecule. We previously described that MIF can associate with ceruloplasmin, albumin, alpha-2-macroglobulin and uromodulin to form high molecular weight complexes in the urine of patients with urinary tract infections [12]. However it remains unclear whether these complexes in fact modulate MIF activity. Therefore, the types and changes in MIF complexes that are associated with inflammatory conditions may be critical for determining the biologically relevant form(s) of MIF in vivo and may provide closer insight into the MIF mechanism of action.
MIF is constitutively expressed and secreted by numerous cell types, and significant levels can be found in the blood, urine and other body fluids [15–17]. Therefore, circulating or extracellular MIF is readily present under basal (“normal”) conditions. The ubiquitous nature of this cytokine suggests that regulating the bioavailability of MIF may be important in controlling the cellular response. Therefore, the binding of MIF to endogenous molecules may be an important feature in the control of MIF activity in vivo. The current study was undertaken to identify the major MIF binding proteins in human serum obtained from normal subjects and those with bladder cancer and to assess their role in regulating MIF activity. We report herein that serum anti-thrombin III (ATIII) binds MIF and that each protein influences the other’s biologic activity. Serum MIF levels are increased, while ATIII-MIF complexes were decreased in bladder cancer patient serum.
2. Materials and Methods
2.1. Human subjects approvals
Approval for all facets of this study was obtained from the Bay Pines VAMC Institutional Review Board. Serum samples were obtained with waiver of informed consent under section 46.116(d) of the Department of Health and Human Services human subject regulations at 45 CFR 46.
2.2. Purification of MIF complexes from human serum
Serum (5 ml, pooled normal samples) was filtered through a 0.45 µm filter and then pre-purified by removing albumin (HiTrap Blue HP, GE Healthcare, Piscataway, NJ) and immunoglobulin (HiTrap Protein A HP, GE Healthcare) using sequential chromatography columns with 20 mM sodium phosphate buffer, pH 7.4 and the ÄKTA Purifier fast pressure liquid chromatography (FPLC) system (GE Healthcare). The non-binding proteins were concentrated in volume using a 3-kDa molecular weight cut off membrane (Amicon, Millipore, Billerica, MA), desalted (Zeba spin columns, ThermoScientific, Rockford, IL) and size fractionated on a HiPrep Sephacryl 16/60 S-300 (GE Healthcare) using 0.05M Sodium phosphate, 0.15M NaCl, pH 7.2. Fractions corresponding to the 5 isolated peaks were pooled, desalted and concentrated to 1.5 ml total volume. MIF Western blotting on all of the fractions was performed as described previously using an affinity purified biotinylated MIF antibody (AF-289-PB, R&D Systems, Minneapolis, MN) and a near-infrared 800 continuous wave (CW) labeled strepavidin [12]. The resulting signal was detected using Odyssey infrared imaging system (LI-COR, Lincoln, NB).
Proteins in fraction 5 were separated by 2-dimensional gel electrophoresis (2D gel) by first removing impurities using the 2-D Cleanup Kit (GEHealtcare). The protein pellets were resuspended in rehydration buffer (8 M urea, 20 mM dithiothreitol (DTT), 2% CHAPS, 0.5% ampholytes, 0.02% bromophenol blue), and linear immobilized pH gradient strips (pH 3–10, ZOOM strips, Invitrogen, Carlsbad, CA) were rehydrated overnight according to the manufacturer’s guidelines. Isoelectric focusing (IEF) was performed for a total of 1.37 kV/h at room temperature. The strips then were equilibrated for 15 mins in 1× sample buffer containing 50 mM DTT (NuPAGE, Invitrogen) followed by incubation in 1× sample buffer containing 125 mM iodoacetamide, and the proteins were further separated using a 4–12% Bis-Tris PAGE gel (Invitrogen). Duplicate gels were run; one gel was Coomassie stained and the other used for Western blotting. The Western blot was used to identify MIF containing spots, which were excised from Coomassie-stained, two-dimensional gels and subjected to mass spectroscopy analysis (University of Florida, Interdisciplinary Center for Biotechnology Research, Gainesville, Florida). Two samples from two separate 2D gels were independently analyzed.
2.3. LC- MS/MS
The enzymatically-digested samples were injected onto a capillary trap (LC Packings, PepMap, Dionex, Sunnyvale, CA) and desalted for 5 mins with a flow rate 10 ml/min of 0.1% v/v acetic acid. The samples were loaded onto a C18 Pep Map HPLC column (Dionex). The elution gradient of the HPLC column started at 3% solvent A, 97% solvent B and finished at 60% solvent A, 40% solvent B for 60 min for protein identification. Solvent A consisted of 0.1% v/v acetic acid, 3% v/v acetonitrile, and 96.9% v/v H2O. Solvent B consisted of 0.1% v/v acetic acid, 96.9% v/v acetonitrile, and 3% v/v H2O. Peptide peaks were monitored by UV at 280 nm. Peptide fractions were collected and pooled into fractions of equal peptide content for mass spectrometry analysis. LC-MS/MS analysis was carried out on a hybrid quadrupole-time of flight, mass spectrometer (ABI QSTAR XL, Applied Biosystems, Framingham, MA). The focusing potential and ion spray voltage was set to 275 V and 2600 V, respectively. The information-dependent acquisition (IDA) mode of operation was employed in which a survey scan from m/z 400–1200 was acquired followed by collision-induced dissociation (CID) of the three most intense ions. Survey and MS/MS spectra for each IDA cycle were accumulated for 1 and 3 s, respectively.
2.4. Protein Search Algorithm
Tandem mass spectra were extracted by ABI Analyst version 1.1. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.0.01). Mascot was set up to search the National Center for Biotechnology Information (NCBI, 03/02/2008) database assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0.30 Da and a parent ion tolerance of 0.30 Da. Iodoacetamide derivatives of cysteine deamidation of asparagine and glutamine and oxidation of methionine are specified in Mascot as variable modifications. Scaffold (version Scaffold-01-06-03, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm [18]. Protein identifications were accepted if they can be established at greater than 99.0% probability and contain at least 2 identified unique peptides. Protein probabilities were assigned by the Protein Prophet algorithm [19].
2.5. Preparation of MIF-AT III complexes in vitro
ATIII (human plasma, Calbiochem, EMD Biosciences, La Jolla, CA) and MIF (human recombinant, bioactive) were mixed in a 1:2 molar ratio (MIF, 50.4 µg/ml; ATIII 110.9 µg/ml) and incubated for 28 hrs at 37 °C in Tris Heparin buffer (50 mM Tris-HCl pH 8.0, 175 mM NaCl, 7.5 mM sodium EDTA) with 4 IE units of heparin/ml. The formation of ATIII-MIF complexes was determined by increases in the ATIII band molecular weight along with the absence of MIF bands in reducing 4–12% Bis-Tris PAGE gels (Invitrogen).
2.6. Kinetic analysis of MIF tautomerase activity inhibition and AT-III inhibition by complex formation
In vitro ATIII-MIF complexes were formed as described above and used in the following enzymatic assays. To establish whether ATIII-MIF complex formation is involved in the regulation of functional specificities of both proteins, we first analyzed the effect of ATIII on MIF activity using a D-dopachrome tautomerase assay. MIF activity in solutions containing MIF only (4M solution), ATIII only (2M solution) and ATIII-MIF complexes (1:2 molar ratio) was determined using a modified tautomerase assay as described previously [20]. Briefly, D-dopachrome methylester substrate was prepared in situ by oxidation of L-dopamethyl ester with sodium periodate. Catalytic activities were analyzed by pipeting samples into individual wells in a standard 96 well ELISA plate. The substrate (100 nM final concentration) diluted in 10 mM sodium phosphate/1 mM EDTA (pH 6.2) was added to each well at room temperature and activity measured by monitoring change in absorbance at 475 nm over a period of 2 mins using a plate reader (BioTek, Winooski, VT).
ATIII inactivation of thrombin activity in solutions containing MIF only (4M solution), ATIII only (2M solutions) and ATIII-MIF complexes (1:2 molar ratio) was determined using a thrombin assay [21]. Thrombin hydrolysis of a colorimetric substrate (#02-32-0078, EMD Biosciences) was monitored for 10 mins in a microtiter plates by measuring change in absorbance at 405 nm.
2.7. Biological activity of ATIII-MIF complexes
In vitro ATIII-MIF complexes were formed as described above and used in the following assays to determine the biological activity of the MIF in the ATIII-MIF complexes. RAW 264.7 macrophage cultures (American Type Culture Collection, Manassas, VA) were routinely cultured in DMEM with Glutamax supplemented with 10% FBS (standard growth medium) without antibiotics in a 5% CO2, 37°C humidified incubator.
Activation of extracellular signal regulated kinases (ERK)1/2 in RAW 264.7 macrophages was determined by separately adding either MIF only (4M solution), ATIII only (2M solution) or ATIII-MIF complexes (final concentration 1:2 molar ratio) to confluent cultures in DMEM with 1% FBS for 1 hr at 37 °C. The ratio of phosphorylated ERK1/2 to total ERK1/2 in the resulting cell lysates was analyzed by using the Bio-Plex 200 system (BioRad, Hercules, CA). Individual bead kits for phosphorylated ERK1/2 and total ERK1/2 and appropriate cell lysate solutions were purchased from the manufacturer. Overnight incubations were done on a microtiter plate shaker (IKA, Staufen, Germany) at 550 rpm. Filtration after incubations was performed with a vacuum filtration manifold (BioRad). Total bead counts for p-ERK1/2 and total pERK1/2 were determined separately.
RAW macrophages (5×104 per well of a 96 well plate) were plated in standard growth medium for 18 h and then starved overnight in DMEM with 0.05% BSA. MIF stimulation of RAW 264.7 macrophage cell proliferation was determined by separately adding either MIF only (4M), ATIII only (2M) or ATIII-MIF complexes (1:2 molar ratio) to the sub confluent cell cultures and incubating for an additional 24 h. Cell proliferation was evaluated using a WST-1 (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzenedisulfonate) to formazan assay (Millipore, Billerica, MA). WST-1 reagent was added to the culture medium (1/10 dilution), and absorbance was measured at 450 nm following incubation at 37°C for 3 h.
2.8. Quantification of ATIII-MIF complexes in human serum
Serum samples remaining after routine clinical evaluation and prior to being discarded were collected until 50 control and 50 transitional cell carcinoma (TCC, AJCC stage II, documented by computerized record search) samples were obtained. The sera were stored at −80 °C prior to analysis. Control patients were defined as those without either documented cancer diagnosis or chronic inflammatory diseases. Sera were assayed for total MIF by ELISA using the protocol described by the manufacturer (DuoSet ELISA, R&D Systems). As a control, addition of human plasma ATIII was tested and determined not to inhibit anti-MIF capture of recombinant MIF in the total MIF ELISA (data not shown).
ATIII-MIF complexes were quantified using a MIF capture-ATIII detection ELISA. The ability of the MIF and ATIII antibodies to recognize ATIII-MIF complexes was verified by preliminary western-blots (data not shown). The MIF capture antibody (mouse monoclonal clone 4E4, Genway, San Diego, CA) was plated overnight at a concentration of 1 µg/ml in high binding ELISA plates (Microlite 2, ThermoScientific, Waltham, MA, USA) at room temperature. Plates were blocked (1% BSA in PBS pH 7.4) for 1 hr at room temperature. The samples were applied in duplicate and incubated 2 hrs at room temperature. Wells were then washed three times with wash buffer (PBS containing 0.05% Tween-20, pH 7.4) using an automated plate washer (ThermoScientific). Detection antibody (biotinylated-goat anti-AT-III, BAF1267, R&D Systems) was added at a final concentration of 200 ng/ml and incubated 2 hrs at room temperature. The wells were washed as described above, 100 µl streptavidin-horseradish peroxidase (1:200 dilution in reagent diluent, DY998, R&D Systems) added to each well and the plate incubated for 20 mins at room temperature. The wells were washed as described above, 100 µl peroxidase substrate (R&D Systems; DY999) was added, and each well read using a plate reader (Biotek, Winooski, VT, USA). Data are expressed as OD450 nm.
2.9. Statistical analyses
Statistical analysis was performed using Prism statistical analysis software version 5.0 (GraphPad Software, Inc., San Diego, CA).
3. Results
3.1 Isolation of AT-III MIF complexes in human serum
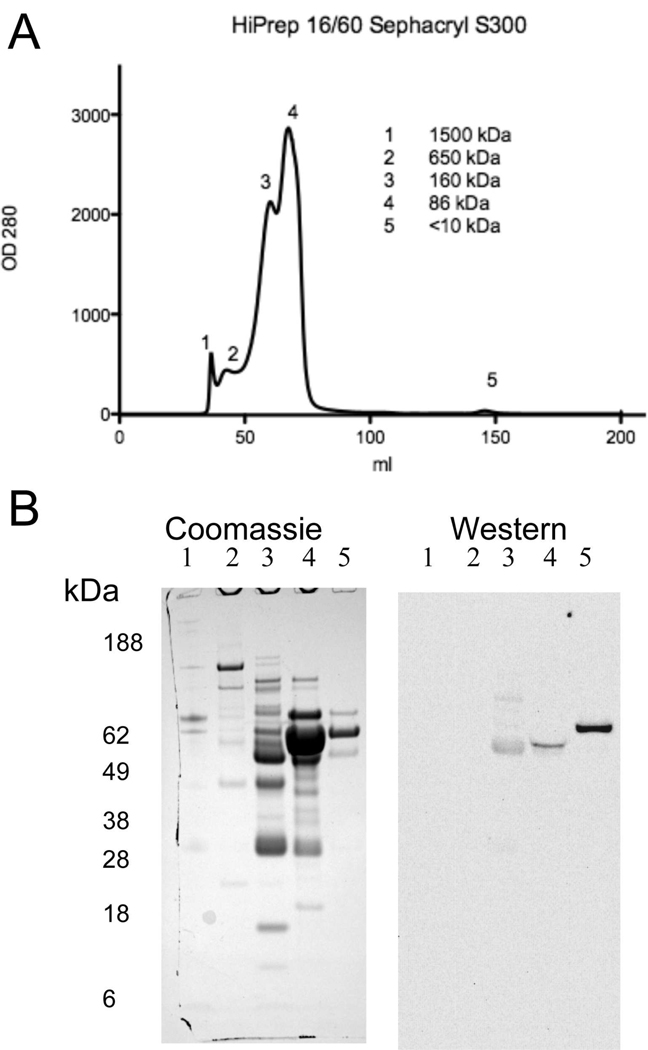
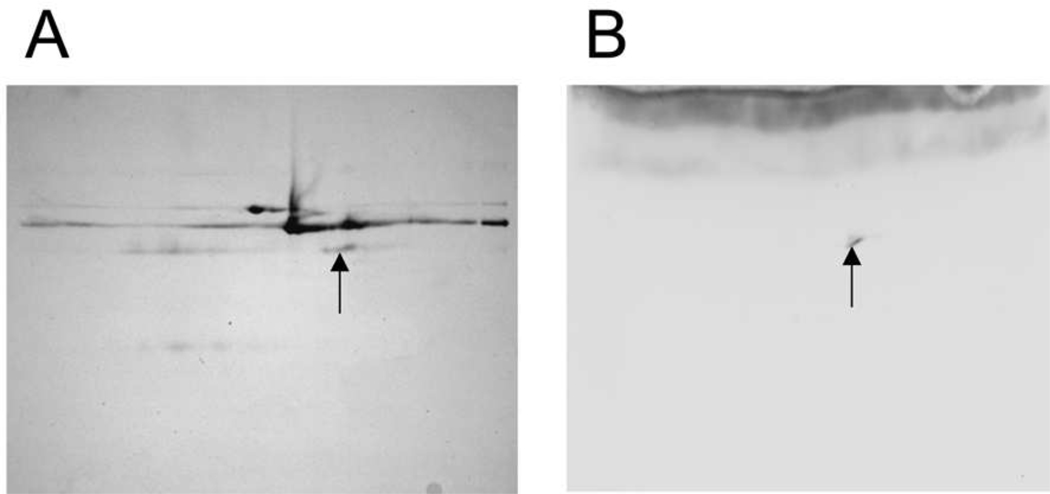
Pooled serum was pre-purified by collecting binding buffer until the OD280 returned to baseline following application of serum sample to the HiTrap Blue and HiTrap Protein A columns. Neither MIF nor MIF containing complexes were found in the elution fractions of these affinity resins as determined by Western blotting (data not shown). The serum binding buffer fractions were pooled and subsequently fractionated by size exclusion chromatography resulting in five protein peaks that ranged in size from approximately 1500 kDa to less than 10 kDa (Fig. 1A). Each of these fractions was separated by SDS-PAGE with fractions 1–5 exhibiting multiple protein bands (Fig. 1B). MIF containing protein complexes, as identified by Western blotting with anti-MIF antibody, were found in fractions 3, 4 and 5. Fraction 5 was selected for further analysis since a single intense MIF reactive band of approximately 62 kDa was identified in the Western blot (Fig. 1B). The proteins in this fraction interact with the Sephacryl resin since this fraction eluted in a volume greater than the total column volume. The presence of 0.15 M NaCl in the elution buffer precludes an ionic interaction. Therefore these proteins exhibit a hydrophobic interaction with the Sephacryl resin. Fractionation using lower ionic strength conditions (0.05M Sodium phosphate, 0.05M NaCl pH 8.0) resulted in loss of fraction 5 (data not shown). The proteins in fraction 5 were separated in duplicate 2 D gels (pH 3–10, 4–12% Bis-Tris PAGE) with one gel stained for total protein with Coomassie and the other gel used for MIF Western blot. The Western blot identified a single protein spot, which was identified as antithrombin III (ATIII) by mass spectroscopy (Fig. 2, Table 1).
Fig. 1.
Gel exclusion size chromatography of human serum following the removal of albumin and immunoglobulin. A) Serum following the removal of IgG and albumin was size fractionated on a HiPrep Sephacryl 16/60 S-300 column under native conditions resulting in five peaks. OD280 determined using Monitore UPC-900 flow through cell and the ÄKTA fast pressure liquid chromatography system (GE Healthcare, Piscataway, NJ). B) Sephacryl fractions corresponding to each peak were pooled, concentrated to a final volume of 1.5 ml and 15 µl aliquots analyzed by SDS-PAGE and Coomassie stained for total protein (left panel). Coomassie stained gels were imaged using the 700 nm channel of the Odyssey infrared imager (LI-COR). Duplicate gels were prepared and analyzed by Western blotting for MIF (right panel). MIF biotinylated detection antibody and strepavidin 800 CW complexes binding to immobilized MIF complexes on the blots were detected using the 800 nm channel of the Odyssey infrared imager. Fraction 5 was chosen for further analysis since the fraction had a single intense MIF containing band on the Western blot (right panel) and only three main protein bands (approximately 62 kDa) detected by Coomassie staining (left panel). Lane 1–5, fractions 1–5 respectively.
Fig. 2.
Isolation of MIF containing protein complexes in Sephacryl fraction 5. Fraction 5 was chosen for further analysis since this fraction had a single intense MIF containing band as determined by Western blot. A) Proteins were separated by 2D-gel electrophoresis (IEF linear gradient pH 3 – 10) followed by SDS-PAGE (4–12% Bis-Tris gel) and Coomassie staining. Coomassie staining bands were imaged using the 700 nm channel of the Odyssey infrared imager. B) Duplicate gels were prepared for MIF Western blotting. Images were obtained using strepavidin conjugated 800 CW infrared dye and the Odyssey imager. The matched region on the Coomassie stained gel (arrows) was excised and subjected to mass spec analysis (Hybrid quadrupole-time of flight, ABI QSTAR XL), which identified the MIF protein as Serpinc1 also known as anti-thrombin III (ATIII, See Table 1).
Table I.
Mass spectroscopy identification of a MIF complex in the 2D gel band.
| Fraction 5 | Serpinc1 Serine peptidase inhibitor, clade C (Antithrombin), member 1 | ||||
|---|---|---|---|---|---|
| Peptide | Accession # | Protein Identification Probability |
Peptide sequence | Mascot Identity Score |
Calculated Peptide Mass (daltons) |
| 1 | IPI00372372 | 100% | EQLQDMGLVDLFSPEK | 31.5 | 1864.9006 |
| 2 | IPI00372372 | 100% | LQPLDFK | 34.6 | 860.4883 |
| 3 | IPI00372372 | 100% | NDNDNIFLSPLSISTAFAMTK | 30.5 | 2315.1228 |
Fragment ion data generated by Information Dependent Acquisition via QSTAR ® were searched against National Center for Biotechnology Information nr sequence database using Mascot (Matrix Science, Boston, Massachusetts) database search engine. Probability based MOWSE (Molecular Weight Search) scores above default significant value were considered for protein identification, in addition to validation by manual interpretation of tandem MS data. Column 1, peptide number. Column 2, National Center for Biotechnology Information reference number for identified protein. Column 3, probability of accurate protein identification. Column 4, peptide sequence. Column 5, Mascot identity score. Probability is defined as the observed database match is random event. An individual score >30 indicates identity or extensive homology, Column 6, calculated peptide mass.
3.2. In vitro complex formation
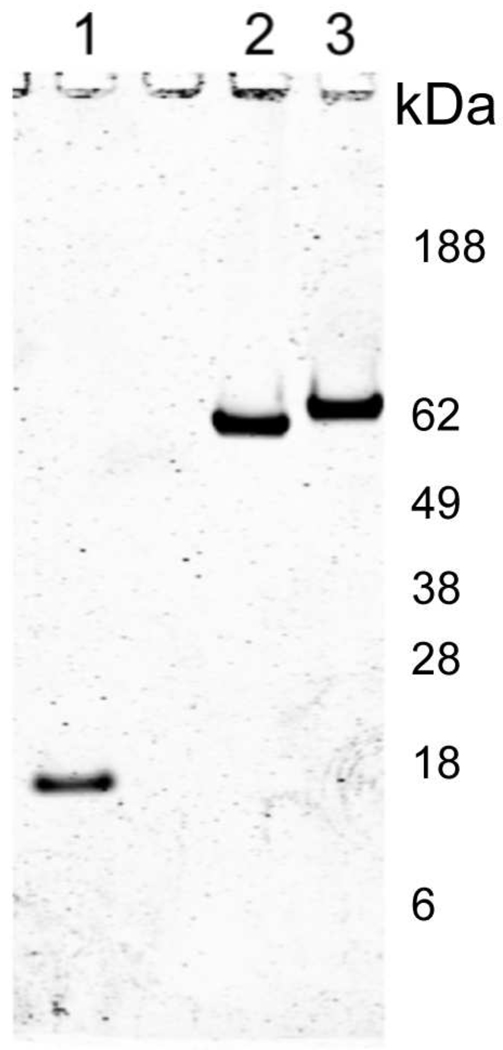
AT-III MIF incubation mixtures were analyzed by 4–12% Bis-Tris PAGE gels under reducing conditions and subsequently stained with Coomassie Brilliant Blue R-250 (Fig. 3). AT-III MIF complex occurred only after the addition of heparin and incubation for 28 hrs at 37 °C (data not shown), indicating that complex formation required activated AT-III. AT-III MIF complexes were approximately 62 kDa, which corresponds to the molecular weight of the MIF containing complex found in fraction 5 (Fig 3) and suggests that the complex may be formed from a monomer of ATIII (52,000) and a monomer of MIF (12,000).
Fig. 3.
ATIII-MIF complexes. In vitro ATIII-MIF complexes were formed by mixing recombinant ATIII and MIF in a 1:2 molar ratio and incubating for 28 hrs at 37°C in Tris Heparin buffer (50 mM Tris-HCl pH 8.0, 175 mM NaCl, 7.5 mM sodium EDTA with 4 IE units of heparin/ml). The formation of ATIII-MIF complexes was determined by increases in ATIII band molecular weight along with the absence of MIF bands in Coomassie stained reducing SDS polyacrylamide gels. Coomassie stained bands were imaged using 700 nm channel of the Odyssey infrared imager. Lane 1, recombinant MIF; Lane 2, purified plasma ATIII; Lane 3, ATIII-MIF complex.
3.3. AT-III MIF complex biological activity
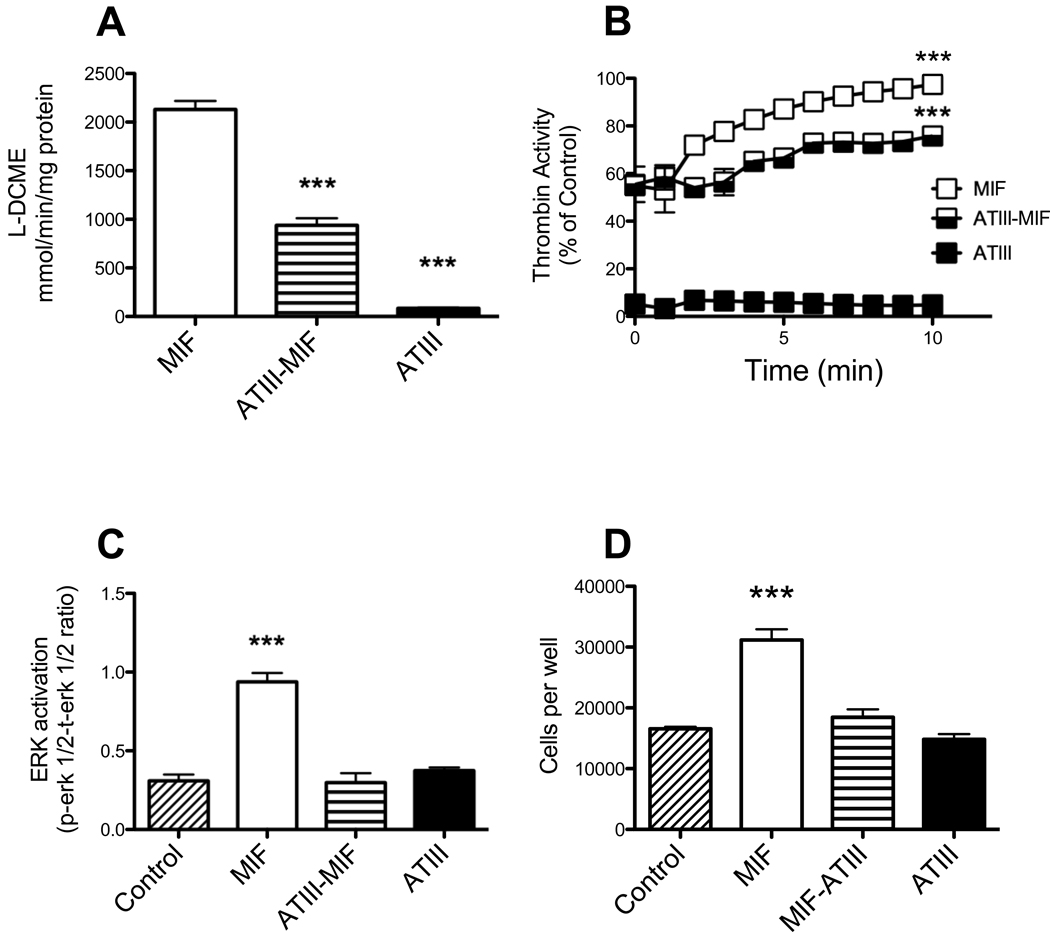
Bioactive MIF contains a catalytically active tautomerase site, which is a useful target of MIF function because this activity can be measured by in vitro assay. In vitro recombinant MIF alone demonstrated measurable tautomerase activity (2130±87.8 mmol L-DCME/min/mg MIF). The tautomerase activity in the ATIII-MIF complexes was reduced greater than two fold compared to the recombinant MIF alone (937.9±74.1 mmol L-DCME/min/mg MIF, p<0.001, Fig. 4A). ATIII alone exhibited negligible tautomerase activity (83.1±4.7 mmol L-DCME/min/mg ATIII) when compared to the natural decay of the substrate (natural decay 75.6±3.5 mmol L-DCME/min).
Fig. 4.
Activity of ATIII-MIF complexes. A) MIF activity – MIF enzymatic activity was determined using a modified tautomerase assay. 200 µl of substrate [(L-dopachrome methyl ester in 10 mM sodium phosphate/1 mM EDTA (pH 6.2)] was added to 60 µl of sample (either MIF alone, MIF-ATIII complex or ATIII alone). The samples were mixed and the change in A475 due to dopachrome tautomerization was monitored for 2 min. Kruskal-Wallis, Dunns multiple comparison test MIF activity compared with that of ATIII or ATIII-MIF complexes, ***, p=0.0005) B) ATIII activity - ATIII activity in either MIF alone, MIF-ATIII complex or ATIII alone was determined using a thrombin hydrolysis assay by mixing 60 µl of the sample with a colorimetric substrate (#02-32-0078, EMD Biosciences) and monitoring the change in absorbance at 405 nm for 10 min. Two way ANOVA, Bonferroni posttest comparison of MIF and ATIII-MIF complexes to ATIII, ***, p<0.0001. C) MIF bioactivity as measured by ERK 1/2 phosphorylation in RAW macrophages in response to either no treatment (control), MIF alone, MIF-ATIII complex or ATIII alone. Phospho- and total ERK1/2 amounts were determined by multiplex phosphoprotein assay (BioRad). One way ANOVA, Dunnett's Multiple Comparison Test phospho to total ERK 1/2 ratio of RAW macrophages incubated with either MIF, ATIII-MIF complexes or ATIII was compared to that in untreated controls, *** - p<0.0001. D) Proliferation of RAW macrophages in response to either no treatment (control), MIF alone, MIF-ATIII complex or ATIII alone. RAW macrophages (5×104 per well of a 96 well plate) were plated in DMEM with 10% FBS for 18 h and then starved overnight in DMEM with 0.05% BSA. Cell proliferation was stimulated by adding either MIF only (4M), ATIII only (2M) or ATIII-MIF complexes (1:2 molar ratio) and incubating for an additional 24 h. Cell numbers were estimated by the addition of WST-1 (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzenedisulfonate) to the culture medium (1/10 dilution) and incubating at 37°C for 3 h Absorbance was measured at 450 nm following incubation. One way ANOVA, Dunnett's Multiple Comparison Test of number of macrophages incubated with either MIF, ATIII-MIF complexes or ATIII was compared to that in untreated controls, *** - p<0.0001.
To assess the biological activity of ATIII in the ATIII-MIF complexes we performed thrombin assays. The inhibitory action of ATIII on thrombin activity in the ATIII-MIF complexes was reduced over 12-fold when compared with ATIII alone (p<0.001), while MIF alone did not reduce thrombin activity (Fig. 4B).
The response of RAW 264.7 macrophages to MIF alone and MIF in the ATIII-MIF complex was measured using phosphorylated ERK1/2 (phospho-ERK) as an indicator of biological MIF activity. A small amount of phospho-ERK can be detected in unstimulated RAW macrophages (0.310±0.04 ratio of phospho-ERK to total-ERK, Fig. 4C). The amount of phospho-ERK is increased threefold over the basal levels by recombinant MIF (0.939±0.06 ratio of phospho-ERK to total-ERK, p<0.001, Fig. 4C). The ATIII-MIF complexes and ATIII alone did not stimulate phospho-ERK production over the basal levels detected in the unstimulated RAW macrophage control cultures (Fig. 4C).
Recombinant MIF treatment results in the activation of macrophages resulting in an increased rate of cell proliferation. Change in the proliferation rate of RAW 264.7 macrophages to MIF alone and MIF in the ATIII-MIF complex was measured using WST-1 dye metabolism as an indicator of cell numbers. Recombinant MIF treatment results in a 1.9 fold increase in cell numbers compared with the unstimulated control cultures (p<0.001, Fig. 4D). The ATIII-MIF complexes and ATIII alone had no effect RAW macrophage cell proliferation (Fig. 4D).
3.4. Serum AT-III MIF complex in bladder cancer patients compared to control
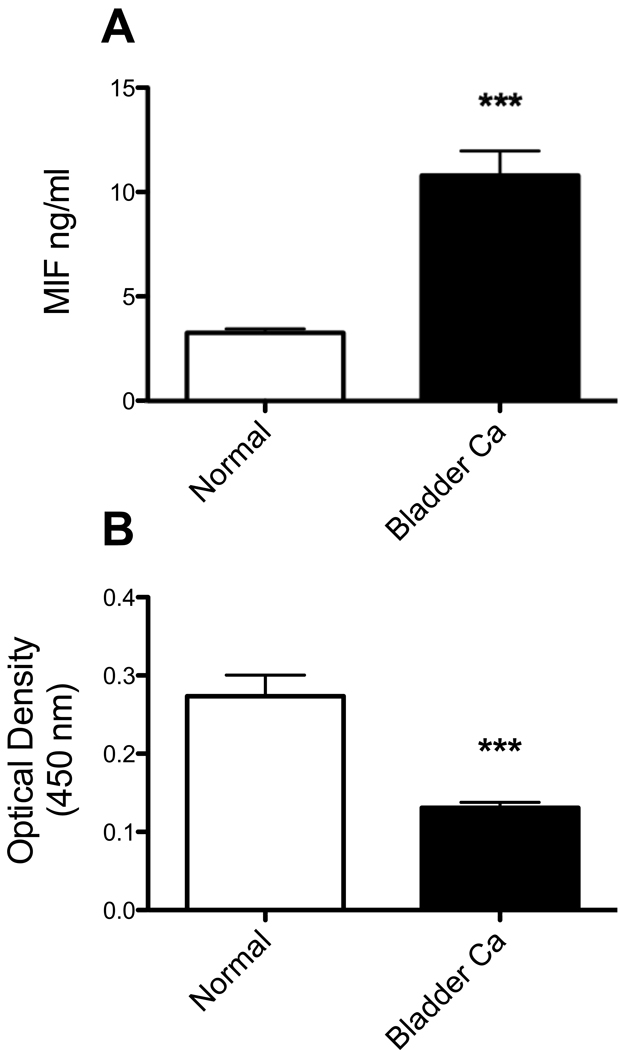
Total serum MIF and serum ATIII-MIF complexes were measured in serum samples from aged matched control (n=50, mean age 56.2 yrs) and TCC (n=50, mean age 60.8 yrs) patients by ELISA. ATIII did not affect anti-MIF capture and determination of total MIF in the serum (data not shown). Total serum MIF amounts were greater in TCC patients (10.8±1.2 ng/ml) compared to controls (3.3±1.5 ng/ml, two-tailed unpaired t-test, p<0.001, Fig. 5).
Fig. 5.
Serum MIF and serum ATIII-MIF complexes in bladder cancer patients. Serum from aged matched control (n=50, mean age 56.2 yrs) and TCC (n=50, mean age 60.8 yrs, stage II) patients were evaluated for total serum MIF (A) and serum MIF-ATIII complexes (B). Total serum MIF amounts were greater in TCC patients (10.8 ± 1.2 ng/ml) compared to controls (3.3 ± 1.5 ng/ml). Serum MIF-ATIII complex amounts (MIF capture ATIII detection) were lower in TCC patients (mean OD450 0.131 ± 0.001) compared to controls (mean OD450 0.273 ± 0.027). Two-tailed unpaired t-test, *** - p<0.0001.
Serum ATIII-MIF complex amounts (MIF capture ATIII detection, arbitrary OD450 units) were reduced greater than two-fold in TCC patients (0.131±0.01 OD450 units) compared to controls (0.273±0.028 OD450 units, two-tailed unpaired t-test, p<0.001, Fig. 5).
4. Discussion
MIF functions as a key regulator of the innate immune response, thus any shift towards increased MIF expression will result in an increased local or systemic inflammatory response [22]. Understanding the regulatory mechanisms of MIF activity within tissues requires identification of endogenous MIF binding proteins. In human serum, we isolated a novel MIF containing complex and identified antithrombin III (ATIII) as a new MIF binding protein. It was unexpected that the complex formed was not broken under reducing conditions. Similar complexes resistant to reducing conditions are formed between ATIII and prostate specific antigen and human glandular kallikrein-2 [23]. In vitro complex formation between biologically active recombinant MIF and ATIII purified from human plasma provided an additional, independent verification of this interaction. The data presented here demonstrate that biological activities of both MIF and ATIII are blocked by this interaction, suggesting that the binding of MIF to ATIII may attenuate MIF bioactivity and may also reduce MIF-dependent pro-inflammatory responses.
The serpin antithrombin (ATIII) controls a number of important coagulation enzymes including Factor Xa and thrombin with the aid of the co-factor, heparin (the physiologically active co-factors are heparan sulfate proteoglycans). Heparin activates antithrombin by inducing conformational changes in the protein that specifically enhances binding [24]. While the classical function of ATIII is as an anticoagulant regulator of blood clotting proteinases such as thrombin, recent studies demonstrate its ability to attenuate inflammatory responses by inhibiting cytokines and other inflammatory mediators found within serum and tissue [25, 26]. In particular, ATIII prevents LPS induced p42 ERK phosphorylation in murine macrophages (RAW 264.7) [25] and activation of NF-κβ in human peripheral blood mononuclear cells and human umbilical cord vein endothelial cells [27]. Our present results establish MIF as an additional cytokine whose activity is modulated by binding to ATIII. Previous studies of LPS activation of RAW 264.7 macrophages have documented MIF release and an autocrine effect, which includes an increase in ERK phosphorylation [28, 29]. Taken together and in light of the results presented here, these data identify MIF binding to ATIII as a biological mechanism that may serve to attenuate MIF activity. While the MIF binding site on ATIII remains to be identified the reduction in both ATIII and MIF activity in the formed complex suggests that MIF binding to ATIII may either block the ATIII heparin binding site, thereby reducing anti-thrombin activity by preventing the formation of the reactive site loop, or MIF binds to heparin activated ATIII at or near the exosites required for thrombin binding [30]. The reduction in MIF activity observed in the ATIII-MIF complexes also suggests that MIF binding to ATIII blocks the reactive N-terminal proline required for MIF activity [31].
Interestingly in vitro experiments identified thrombin and Factor Xa (FXa) as inducers of MIF expression in human endothelial cells and this induction was reduced by blocking with hirudin in the case of thrombin and ATIII in the case of FXa [32]. In addition, a more recent study by Wadgaonkar, et al. demonstrated that thrombin stimulates endothelial cell MIF secretion and a subsequent second phase of ERK phosphorylation that is dependent upon the secreted MIF [33]. Together this body of evidence suggests that MIF could play a critical functional role in linking the cytokine network with the coagulation cascade. Activated ATIII could act as a biological inhibitor of MIF activity by both inactivating thrombin-induced MIF upregulation, and by binding secreted MIF. This inhibition could occur locally at the site of inflammation and the resultant ATIII-MIF complex may removed from the circulation by binding to a specific receptor present on hepatocytes as is the case for ATIII clotting factor complexes [34]. Within tissues, ATIII is associated with heparin-like substances and exists in a high-affinity state; the inhibitor rapidly binds proteinases such as thrombin and as documented here, MIF. Once the complex forms, its affinity for heparin is decreased compared with ATIII alone, allowing the complex to dissociate from the cell surface for rapid clearance by the liver.
Decreased plasma ATIII levels have been described in a variety of different cancers, including lung, colon, ovary, and prostate [35–39]. Glycosaminoglycans on urothelial cell surfaces, as determined using a human bladder carcinoma (J82) cell line can activate ATIII and potentially play an important role in regulating protease activity by ATIII under normal conditions [40]. Recent studies by Kurata et al. suggest that ATIII prevented hepatic metastasis of colon cancer cells in a rat model by blocking TNF-alpha production [41]. MIF induces TNF-alpha secretion by a variety of different normal and neoplastic cell types (for a review see Bach et al. [6]) and has an established role in tumorigenesis. We recently established that treatments directed against MIF resulted in decreased prostate cancer cell proliferation and invasion [20]. Prior studies in this laboratory determined that MIF is synthesized and secreted by rat bladder, expressed in human urothelium, and secreted by HT-1376 human transitional cell carcinoma cells [7, 42]. Additionally, we established increased MIF serum levels in prostate cancer patients [10, 11]. Here we extend these findings and show that TCC patients have elevated serum MIF. Interestingly, along with the increased serum MIF we detected decreased amounts of ATIII-MIF complexes in TCC patients. If ATIII complexed MIF is biologically inactive then, by inference, TCC patient serum has increased amounts of bioactive MIF. Inactivation of MIF by ATIII may provide an additional therapy to cancer patients by reducing the availability of MIF to the tumor cells.
In summary this study identifies ATIII as a functional MIF binding protein in human serum. Shifts in the equilibrium between thrombin, ATIII and MIF at the site of inflammation may serve to regulate the local inflammatory response. In addition, ATIII-MIF complexes may circulate in the serum to the liver for degradation without eliciting an inflammatory response. ATIII-MIF complexes exhibit decreased MIF and ATIII bioactivity, suggesting that this is a potential mechanism for the previously documented anti-inflammatory action of ATIII. Conversely, high MIF levels may contribute to a pro-thrombotic state by inhibiting ATIII activity.
Acknowledgments
Gary A. Smith Jr., provided excellent technical assistance. The mass spectroscopy and identification of anti-thrombin III was performed at the Proteomics Division of University of Florida Interdisciplinary Center for Biotechnological Research in Gainesville, Florida, USA.
Role of the Funding Source
This material is based upon work supported (or supported in part) by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development (PLV; KLMS), and the NIH (LL, RB). This work was also supported by the National Institute of Diabetes and Digestive and Kidney Diseases DK075059 (PLV; KLMS), and the Bay Pines Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
We confirm that all authors fulfill all conditions required for authorship. We also confirm that there is no potential conflict of interest or financial dependence regarding this publication, as described in the Instruction for Authors. All authors have read and approved the manuscript.
References
- 1.Feldmann M. Many cytokines are very useful therapeutic targets in disease. J Clin Invest. 2008;118:3533–3536. doi: 10.1172/JCI37346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 3.Mantovani A, Garlanda C, Locati M, Rodriguez TV, Feo SG, Savino B, Vecchi A. Regulatory pathways in inflammation. Autoimmun Rev. 2007;7:8–11. doi: 10.1016/j.autrev.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Flaster H, Bernhagen J, Calandra T, Bucala R. The MIF-Glucocorticoid Dyad: Regulation of Inflammation and Immunity. Mol Endocrinol. 2007 doi: 10.1210/me.2007-0065. [DOI] [PubMed] [Google Scholar]
- 5.Flaster H, Bernhagen J, Calandra T, Bucala R. The macrophage migration inhibitory factor-glucocorticoid dyad: regulation of inflammation and immunity. Mol Endocrinol. 2007;21:1267–1280. doi: 10.1210/me.2007-0065. [DOI] [PubMed] [Google Scholar]
- 6.Bach JP, Rinn B, Meyer B, Dodel R, Bacher M. Role of MIF in inflammation and tumorigenesis. Oncology. 2008;75:127–133. doi: 10.1159/000155223. [DOI] [PubMed] [Google Scholar]
- 7.Meyer-Siegler KL, Vera PL. Intraluminal antibodies to macrophage migration inhibitory factor decrease substance P induced inflammatory changes in the rat bladder and prostate. J Urol. 2004;172:1504–1509. doi: 10.1097/01.ju.0000140213.54457.97. [DOI] [PubMed] [Google Scholar]
- 8.Al-Abed Y, Dabideen D, Aljabari B, Valster A, Messmer D, Ochani M, Tanovic M, Ochani K, Bacher M, Nicoletti F, Metz C, Pavlov VA, Miller EJ, Tracey KJ. ISO-1 binding to the tautomerase active site of MIF inhibits its pro-inflammatory activity and increases survival in severe sepsis. J Biol Chem. 2005;280:36541–36544. doi: 10.1074/jbc.C500243200. [DOI] [PubMed] [Google Scholar]
- 9.Stosic-Grujicic S, Stojanovic I, Maksimovic-Ivanic D, Momcilovic M, Popadic D, Harhaji L, Miljkovic D, Metz C, Mangano K, Papaccio G, Al-Abed Y, Nicoletti F. Macrophage migration inhibitory factor (MIF) is necessary for progression of autoimmune diabetes mellitus. J Cell Physiol. 2008;215:665–675. doi: 10.1002/jcp.21346. [DOI] [PubMed] [Google Scholar]
- 10.Meyer-Siegler KL, Bellino MA, Tannenbaum M. Macrophage migration inhibitory factor evaluation compared with prostate specific antigen as a biomarker in patients with prostate carcinoma. Cancer. 2002;94:1449–1456. doi: 10.1002/cncr.10354. [DOI] [PubMed] [Google Scholar]
- 11.Meyer-Siegler KL, Iczkowski KA, Vera PL. Further evidence for increased macrophage migration inhibitory factor expression in prostate cancer. BMC Cancer. 2005;5:73. doi: 10.1186/1471-2407-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer-Siegler KL, Iczkowski KA, Vera PL. Macrophage migration inhibitory factor is increased in the urine of patients with urinary tract infection: macrophage migration inhibitory factor-protein complexes in human urine. J Urol. 2006;175:1523–1528. doi: 10.1016/S0022-5347(05)00650-6. [DOI] [PubMed] [Google Scholar]
- 13.Javeed A, Zhao Y, Zhao Y. Macrophage-migration inhibitory factor: role in inflammatory diseases and graft rejection. Inflamm Res. 2008;57:45–50. doi: 10.1007/s00011-007-7110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer-Siegler KL, Xia S-L, Vera PL. Substance P increases cell-surface expression of CD74 (receptor for macrophage migration inhibitory factor): In vivo biotinylation of urothelial cell-surface proteins. Mediators of Inflammation 2009. 2009 doi: 10.1155/2009/535348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lolis E, Bucala R. Macrophage migration inhibitory factor. Expert Opin Ther Targets. 2003;7:153–164. doi: 10.1517/14728222.7.2.153. [DOI] [PubMed] [Google Scholar]
- 16.Petrovsky N, Socha L, Silva D, Grossman AB, Metz C, Bucala R. Macrophage migration inhibitory factor exhibits a pronounced circadian rhythm relevant to its role as a glucocorticoid counter-regulator. Immunol Cell Biol. 2003;81:137–143. doi: 10.1046/j.0818-9641.2002.01148.x. [DOI] [PubMed] [Google Scholar]
- 17.Brown FG, Nikolic-Paterson DJ, Chadban SJ, Dowling J, Jose M, Metz CN, Bucala R, Atkins RC. Urine macrophage migration inhibitory factor concentrations as a diagnostic tool in human renal allograft rejection. Transplantation. 2001;71:1777–1783. doi: 10.1097/00007890-200106270-00013. [DOI] [PubMed] [Google Scholar]
- 18.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 19.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 20.Meyer-Siegler KL, Iczkowski KA, Leng L, Bucala R, Vera PL. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J Immunol. 2006;177:8730–8739. doi: 10.4049/jimmunol.177.12.8730. [DOI] [PubMed] [Google Scholar]
- 21.Frigola A, Angeloni S, Cerqueti AR. Determination of antithrombin activity by an amidolytic and a clotting procedure. J Clin Pathol. 1979;32:21–25. doi: 10.1136/jcp.32.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calandra T, Echtenacher B, Roy DL, Pugin J, Metz CN, Hultner L, Heumann D, Mannel D, Bucala R, Glauser MP. Protection from septic shock by neutralization of macrophage migration inhibitory factor. Nat Med. 2000;6:164–170. doi: 10.1038/72262. [DOI] [PubMed] [Google Scholar]
- 23.Cao Y, Lundwall A, Gadaleanu V, Lilja H, Bjartell A. Anti-thrombin is expressed in the benign prostatic epithelium and in prostate cancer and is capable of forming complexes with prostate-specific antigen and human glandular kallikrein 2. Am J Pathol. 2002;161:2053–2063. doi: 10.1016/S0002-9440(10)64484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ersdal-Badju E, Lu A, Zuo Y, Picard V, Bock SC. Identification of the antithrombin III heparin binding site. J Biol Chem. 1997;272:19393–19400. doi: 10.1074/jbc.272.31.19393. [DOI] [PubMed] [Google Scholar]
- 25.Hagiwara S, Iwasaka H, Matsumoto S, Noguchi T. High dose antithrombin III inhibits HMGB1 and improves endotoxin-induced acute lung injury in rats. Intensive Care Med. 2008;34:361–367. doi: 10.1007/s00134-007-0887-5. [DOI] [PubMed] [Google Scholar]
- 26.Minnema MC, Chang AC, Jansen PM, Lubbers YT, Pratt BM, Whittaker BG, Taylor FB, Hack CE, Friedman B. Recombinant human antithrombin III improves survival and attenuates inflammatory responses in baboons lethally challenged with Escherichia coli. Blood. 2000;95:1117–1123. [PubMed] [Google Scholar]
- 27.Oelschlager C, Romisch J, Staubitz A, Stauss H, Leithauser B, Tillmanns H, Holschermann H. Antithrombin III inhibits nuclear factor kappaB activation in human monocytes and vascular endothelial cells. Blood. 2002;99:4015–4020. doi: 10.1182/blood.v99.11.4015. [DOI] [PubMed] [Google Scholar]
- 28.Roger T, Froidevaux C, Martin C, Calandra T. Macrophage migration inhibitory factor (MIF) regulates host responses to endotoxin through modulation of Toll-like receptor 4 (TLR4) J Endotoxin Res. 2003;9:119–123. doi: 10.1179/096805103125001513. [DOI] [PubMed] [Google Scholar]
- 29.Roger T, Chanson AL, Knaup-Reymond M, Calandra T. Macrophage migration inhibitory factor promotes innate immune responses by suppressing glucocorticoid-induced expression of mitogen-activated protein kinase phosphatase-1. Eur J Immunol. 2005;35:3405–3413. doi: 10.1002/eji.200535413. [DOI] [PubMed] [Google Scholar]
- 30.Olson ST, Chuang YJ. Heparin activates antithrombin anticoagulant function by generating new interaction sites (exosites) for blood clotting proteinases. Trends Cardiovasc Med. 2002;12:331–338. doi: 10.1016/s1050-1738(02)00183-4. [DOI] [PubMed] [Google Scholar]
- 31.Fingerle-Rowson G, Kaleswarapu DR, Schlander C, Kabgani N, Brocks T, Reinart N, Busch R, Schutz A, Lue H, Du X, Liu A, Xiong H, Chen Y, Nemajerova A, Hallek M, Bernhagen J, Leng L, Bucala R. A Tautomerase-null MIF Gene Knock-in Mouse Reveals that Protein Interactions and not Enzymatic Activity Mediate MIF-dependent Growth Regulation. Mol Cell Biol. 2009 doi: 10.1128/MCB.01907-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu T, Nishihira J, Watanabe H, Abe R, Honda A, Ishibashi T, Shimizu H. Macrophage migration inhibitory factor is induced by thrombin and factor Xa in endothelial cells. J Biol Chem. 2004;279:13729–13737. doi: 10.1074/jbc.M400150200. [DOI] [PubMed] [Google Scholar]
- 33.Wadgaonkar R, Somnay K, Garcia JG. Thrombin induced secretion of macrophage migration inhibitory factor (MIF) and its effect on nuclear signaling in endothelium. J Cell Biochem. 2008;105:1279–1288. doi: 10.1002/jcb.21928. [DOI] [PubMed] [Google Scholar]
- 34.Pizzo SV. Serpin receptor 1: a hepatic receptor that mediates the clearance of antithrombin III-proteinase complexes. Am J Med. 1989;87:10S–14S. doi: 10.1016/0002-9343(89)80524-8. [DOI] [PubMed] [Google Scholar]
- 35.Mulder AB, Zwaveling JH, Smid WM, Maring JK, van Ginkel RJ, Girbes AR, Schraffordt Koops H, van der Meer J. Augmented procoagulant activity in cancer patients, treated with recombinant interferon-gamma in addition to recombinant tumor necrosis factor-alpha and melphalan. Thromb Haemost. 1996;76:897–901. [PubMed] [Google Scholar]
- 36.Honegger H, Anderson N, Hewitt LA, Tullis JL. Antithrombin III profiles in malignancy, relationship primary tumors and metastatic sites. Thromb Haemost. 1981;46:500–503. [PubMed] [Google Scholar]
- 37.Bick RL. Antithrombin III and factor VIII in patients with neoplasms. Am J Clin Pathol. 1978;69:194–195. doi: 10.1093/ajcp/69.2.194a. [DOI] [PubMed] [Google Scholar]
- 38.Buller HR, Boon TA, Henny CP, Dabhoiwala NF, ten Cate JW. Estrogen-induced deficiency and decrease in antithrombin III activity in patients with prostatic cancer. J Urol. 1982;128:72–74. doi: 10.1016/s0022-5347(17)52762-7. [DOI] [PubMed] [Google Scholar]
- 39.Dowling P, O'Driscoll L, Meleady P, Henry M, Roy S, Ballot J, Moriarty M, Crown J, Clynes M. 2-D difference gel electrophoresis of the lung squamous cell carcinoma versus normal sera demonstrates consistent alterations in the levels of ten specific proteins. Electrophoresis. 2007;28:4302–4310. doi: 10.1002/elps.200700246. [DOI] [PubMed] [Google Scholar]
- 40.Hamamoto T, Kisiel W. The effect of cell surface glycosaminoglycans (GAGs) on the inactivation of factor VIIa--tissue factor activity by antithrombin III. Int J Hematol. 1998;68:67–78. doi: 10.1016/s0925-5710(98)00034-6. [DOI] [PubMed] [Google Scholar]
- 41.Kurata M, Okajima K, Kawamoto T, Uchiba M, Ohkohchi N. Antithrombin reduces reperfusion-induced hepatic metastasis of colon cancer cells. World J Gastroenterol. 2006;12:60–65. doi: 10.3748/wjg.v12.i1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer-Siegler KL, Leifheit EC, Vera PL. Inhibition of macrophage migration inhibitory factor decreases proliferation and cytokine expression in bladder cancer cells. BMC Cancer. 2004;4:34. doi: 10.1186/1471-2407-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]