Abstract
Currently, there is no reliable system for regulated gene expression and regulated gene knockdown in cells with finite lifespan. In this manuscript, we describe a vector system, consisting of a retrovirus for the delivery of rtTA, and a lentivirus for the delivery of either a transgene or a miR-shRNA for the modification of primary cells. Primary rat pulmonary microvascular endothelial cells (PMVEC) modified by these vectors for the inducible expression of Gaussia luciferase or DsRed Express demonstrated greater than 100-fold induction of the transgene expression with doxycycline. The system works reliably in both sequential and simultaneous infection modes, with about 95% of the sells selected with two antibiotics being inducible in each mode. The lentiviral vector for gene knockdown allows for the direct cloning of shRNA oligos using alpha-complementation, and for the monitoring of induction of RNA interference with fluorescent reporter, mCherry. The gene knockdown vector was validated by knocking down β-actin expression in PMVECs, with two of the four constructs showing 59 and 75% knockdown, respectively, compared to uninduced controls. The vectors described here were successfully used for the modification of various primary and established cell lines for regulated gene expression and regulated knockdown.
Keywords: Retroviral vector, lentiviral vector, Tet-On Advanced system, gene knockdown, regulated expression, endothelial cells, primary cells
Tetracycline-regulated systems are an important tool for gene expression in mammalian cells. Apart from temporal control over transgene expression, these systems frequently allow for the expression of genes whose products are cytotoxic. Therefore, it is often possible to achieve inducible expression of a transgene when attempts at generating transfectants with constitutive expression have failed [1, 2]. Currently, the time consuming generation of a cell line derivatives expressing a regulatory variant of Tet-repressor protein is a major limitation of available tetracycline-regulated systems. This generation typically involves screening of a number of clones obtained by stable transfection with a tet-regulator plasmid to obtain clones with optimal induction properties. As a result, the utility of these systems for regulated gene expression in cells with finite replicative lifespan, is limited. Here, we describe a retro-lentiviral Tet-On Advanced system designed to overcome this limitation by transcriptionally linking expression of rtTA Advanced to blasticidin resistance gene in a retroviral vector, and by using lentiviral vectors to deliver transgene into target cells. We also describe a modification of this system, which allows for the doxycycline- inducible gene knockdown.
Materials and Methods
All molecular biology procedures were performed according to established protocols [3].
Cell lines and culture conditions
The cultures of the primary rat pulmonary microvascular endothelial cells (PMVECs) were generated as described previously [4] and provided by a Cell Culture Core. PMVECs were maintained in DMEM medium supplemented with 10% Fetal Calf Serum, 1 mM pyruvate, and 50 μg/mL of gentamicin at 37°C in the atmosphere of 5% CO2.
Plasmids
pTet-On Advanced, pTRE-tight and pDsRed Express were from Clontech, Mountain View, CA; pSF91-GCSh-gp91 (phox) was described earlier [5]; pNEBR-X1GLuc Control plasmid was from New England Biolabs, Ipswich, MA; pPRIME-CMV-GFP-FF3 [6] was kindly provided by S.J. Elledge, the pcDNA6.2-GW/EmGFP-miR was from Invitrogen (Invitrogen, Carlsbad, CA), psPAX2 and pMD2.G were from Addgene, Cambridge, MA.
Production of retro- and lentiviral supernatants and infection of target cells
Retro- and lentivirus containing supernatants were produced by CaPO4-mediated transfection of the Phoenix ampho and HEK293FT cell lines, respectively, using established protocols [7, 8]. Gag, Pol and Env functions for lentiviral constructs were provided in trans by cotransfection of the vector plasmid with two helper plasmids, psPAX2 and pMD2.G. Target cells were infected with retroviruses expressing rtTA in 35-mm dishes at 20% confluence by incubating them overnight with corresponding supernatant in the presence of 8 μg/mL polybrene (Sigma-Aldrich Corp., St. Louis, MO). The next day, supernatant was removed and cells were allowed to recover for 24h in DMEM, after which cells were trypsinized, transferred into 140-mm dishes and blasticidin selection (30 μg/mL) was applied for 5 days. Lentivirus infection of rtTA expressing cells was conducted similarly, except puromycin selection (4 μg/mL) was applied for 3 days when necessary. For luciferase assays, cells were induced with doxycycline (3 μg/mL) immediately after infection without puromycin selection.
Luciferase assays
Gaussia luciferase activity was assayed using Gaussia Luciferase assay kit (New England Biolabs) according to the manufacturer's recommendations.
FACS Analysis
Fluorescence Activated Cell Sorting analysis was Performed on Becton Dickinson FACSVantage with DIVA software.
Western Blotting
For Western Blotting, cells were grown in the presence or absence of Doxycycline (3 μg/mL) for 48h, lysed in the buffer containing 10 mM Tris-HCl, pH=8.0, 1 mM EDTA, 0.5% SDS and 1x EDTA-free protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). The lysates were sonicated for 15 sec in microfuge tubes using Branson Digital Sonifier model S-450D (Branson, Danbury, CT) at 15% output. Protein concentration was determined using BCA Protein Assay Kit (Thermo Fisher Scientific, Rockford, IL), and samples were adjusted to the same concentration with lysis buffer. Proteins were separated in 10% acrylamide gel, transferred to PVDF membrane, blocked with 5% skim milk in TBS-Tween (0.1%), and probed with anti-β-actin mouse monoclonal antibodies (Sigma-Aldrich Corp, St. Louis, MO) and with anti-HSP60 (loading control, BD Biosciences, San Jose, CA). After washing, membranes were incubated with secondary HRP-conjugated goat anti-mouse antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), and visualized with SuperSignal West Pico kit (Thermo Fisher Scientific, Rockford, IL). The images of the membrane were analyzed using Fujifilm LAS-1000 Luminescent Image Analysis System (Fujufilm Life Science, Woodbridge, CT).
Results and Discussion
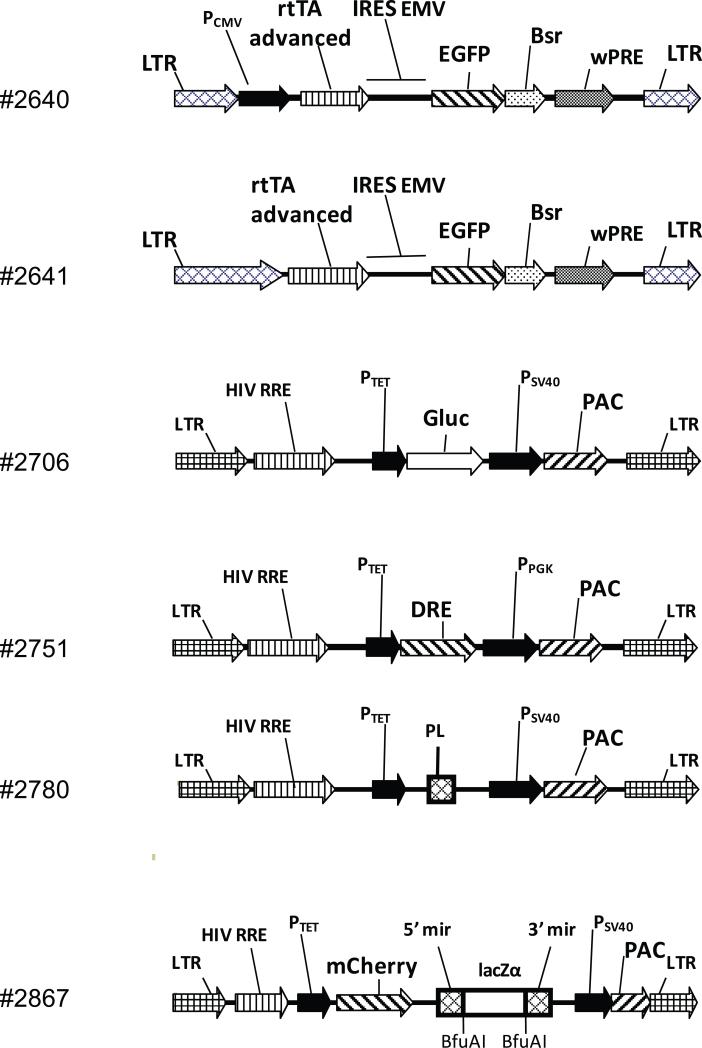
To generate a delivery vehicle for the Tet-On reverse transactivator protein (rtTA), the retroviral vector pSF91-GCSh-gp91 (phox) [5], was modified as follows: a) the sequences between 5’ and 3’ Long Terminal Repeats (LTRs) were removed and replaced with a cassette encompassing an Internal Ribosome Entry Site (IRES) from encephalomyelocarditis virus (EMV) followed by a fusion between Enhanced Green Fluorescent Protein (EGFP) and Blasticidin resistance gene (Bsr) and by a woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) and b) an rtTA Advanced gene was inserted upstream of IRES in thus generated retroviral vector either under control of the strong cytomegalovirus (CMV) promoter, or alone. The resulting constructs were designated #2640 and #2641, respectively (Fig.1). In these constructs, expression of rtTA is driven by the CMV or retroviral LTR promoters, respectively and is transcriptionally linked to the expression of the EGFP-Bsr fusion.
Fig. 1.
Vector maps. Abbreviations: 5’ and 3’ mir, 5’ and 3’ flanking sequences derived from the murine mir-155 micro-RNA gene; Bsr, blasticidin resistance gene; DRE, DsRed express; EGFP, enhanced green fluorescent protein; Gluc, (secreted) Gaussia luciferase; HIV RRE, human immunodeficiency virus rev response element; IRES EMV, encephalomyelocarditis virus internal ribosome entry site; lacZα, gene encoding the alpha-fragment of E. coli β-galactosidase gene; LTR, retro/lentiviral long terminal repeat; PAC, puromycin resistance gene; PL, polylinker. Unique sites 5’-EcoRI, XhoI, ApaI, MluI, NotI, XbaI, SpeI, BamHI, SmaI, PstI-3’; PPGK, phosphoglycerate kinase promoter; PCMV, CMV promoter; PSV40, SV40 promoter; PTet, doxycycline-regulated promoter; rtTA, reverse tetracycline-controlled transactivator protein; wPRE, woodchuck hepatitis virus posttranscriptional regulatory element.
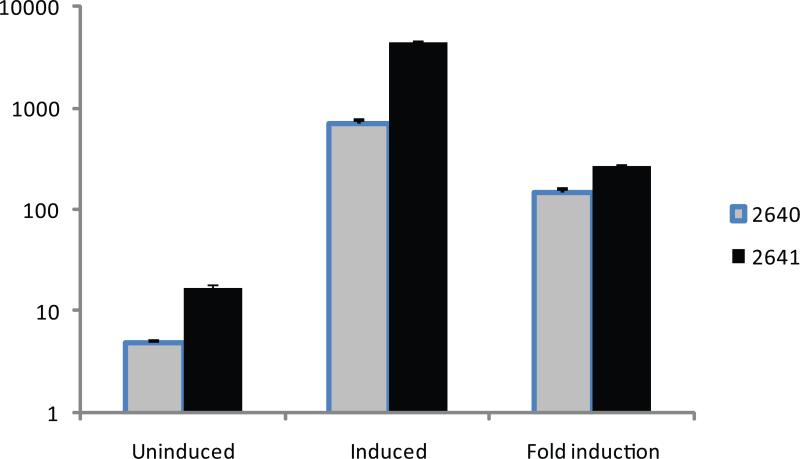
In order to validate constructs #2640 and #2641-modified PMVECs for doxycycline-regulated gene expression, two lentiviral reporter vectors containing Tet-responsive promoter sequence (TRE)-driven Gaussia princeps luciferase (Gluc) [9] or DsRed Express (DRE) genes were generated. The distinctive feature of Gaussia luciferase is that this is a secreted protein which is fairly stable in the tissue culture media. Therefore, enzyme activity assays do not involve cell lysis, and are performed by sampling growth media. Initially, pBABE-puro-derived retroviral vectors were engineered for the delivery of the reporter construct. However, doubly infected PMVECs had high background expression of luciferase (and therefore, low inducibility) irrespective of the orientation of TRE-Gluc construct relative to the LTRs. We rationalized that this phenomenon may be due to the enhancer activity of the LTR(s), and thus lentiviral vectors with their highly attenuated LTRs may improve inducibility. The lentiviral reporter construct was derived from the pPRIME-CMV-GFP-FF3 [6] by removing sequences between the HIV RRE and the 3'LTR and replacing them with TRE-Gluc reporter followed by SV40-promoter driven puromycin resistance gene and WPRE. The resulting construct was designated #2706 (Fig. 1). We first evaluated if strong CMV promoter offers any advantages vs. weak LTR promoter for the expression of rtTA. While robust expression mediated by CMV promoter could be advantageous, this promoter is susceptible to silencing by CpG methylation in both mouse [10] and rat [11] cells. As a consequence, a stable relatively low-level expression driven by LTR promoter may prove to be superior to high level, but unstable expression provided by CMV. Indeed, as it can be seen in Fig. 2, strong CMV promoter present in construct #2640 offers no advantage over the LTR promoter present in construct #2641.
Fig. 2.
Luciferase inducibility of PMVECs. PMVECs were infected with retrovirus 2640 or 2641, selected for 5 days with 30 μg/ml blasticidin, infected with lentivirus #2706, and induced with 3 μg/ml of doxycycline for 24h. Luciferase assays were performed as described in Materials and Methods. Luciferase activity is expressed in relative light units. The data are mean ± SEM (n=4).
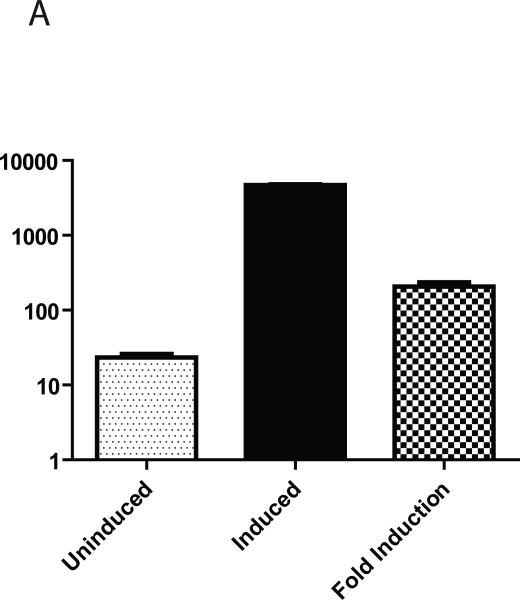
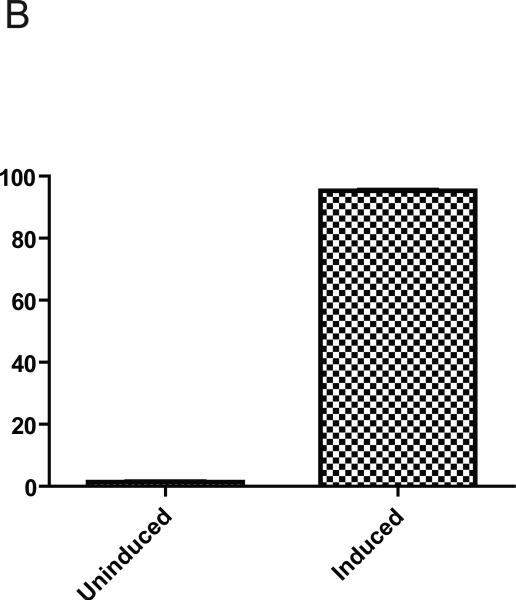
Two parameters are crucial for regulated gene expression in cells with limited proliferative capacity: inducibility and percent responsive cells. The first parameter characterizes fold increase in target gene expression upon the addition of the inducer (doxycycline), and the second characterizes the number of cells, that respond to addition of the doxycycline with increased transgene expression. The low percentage of responsive cells in the population is likely to complicate the interpretation of results even if the average level of transgene expression, as evaluated by, e.g. Western Blotting, is significant. Luciferase assays provide a reliable means to measure inducibility (Fig. 2). Simultaneous infection of PMVECs with retrovirus #2641 and lentivirus #2751 allowed us to independently obtain a measure of inducibility and to evaluate the percent responsive cells by means of FACS analysis. Inducibility, measured by DRE fluorescence was similar to that determined by luciferase assay (207.3 ± 27.2 and 272.2 ± 7.6 fold, respectively, Figs. 3A and 2). The great majority of selected cells (95.3 ± 0.2%, n=3, Fig. 3B) responded to the addition of doxycycline with increased fluorescence.
Fig. 3.
Inducibility of DRE and percent responsive PMVECs as reported by FACS. A, inducibility. B, percent responsive cells. PMVECs were simultaneously infected with retrovirus #2641 and lentivirus #2706 overnight, allowed to recover for 24h in DMEM, and selected with 30 μg/ml blasticidin for 2 days followed by selection with 30 μg/ml blasticidin plus 5 μg/ml puromycin for 1 day and finally with blasticidin and puromycin ± 3 μg/ml of doxycycline for 2 days. At the end of experiment, cells were collected by trypsinization and subjected to FACS analysis. The data are mean ± SEM (n=3).
In recent years, gene knockdown has become a standard tool in experiments aimed at elucidation of gene function. The efficiency of siRNA-mediated knockdowns is typically limited by the efficiency of synthetic RNA delivery. Therefore, vector-mediated RNA interference offers distinct advantages as it allows for the generation of cell lines stably expressing shRNA. Historically, RNA-polymerase III (Pol III) promoter-driven short hairpin RNAs (shRNAs) were the first to be successfully used for this purpose [12]. However, knockdown of essential genes may have cytotoxic consequences, and Pol III promoters do not lend themselves easily to modification towards regulated expression. Hence, another approach was developed whereby oligonucleotides encoding a stem-loop structure are inserted into micro RNA context. The advantages of this approach are twofold: 1) it improves the efficiency of shRNA processing into siRNA and 2) it allows for the use of numerous regulated Pol II promoter systems. The examples of the latter approach are the use of human miR-30 [6] and mouse miR-155 (Invitrogen, Carlsbad, CA). Each of these two examples has its advantages and drawbacks. The first is easy to implement as two unique common restriction endonuclease sites have been engineered into miR-30, but the efficiency of knockdown correlates inversely with multiplicity of infection, thus diminishing the usefulness of this strategy in cells with limited proliferative capacity. Some other drawbacks of this system are that it makes use of relatively long oligonucleotides and the lack of a reliable algorithm for oligonucleotide design. In the Invitrogen's version of the system, a very efficient algorithm for shRNA design has been implemented. Also, in this version oligonucleotides are seamlessly integrated into miR-155 context by replacing 60 nucleotides of miR-155 rather than being inserted into it as in the first version. The drawback of the Invitrogen's version is that shRNA oligonucleotides cannot be cloned directly into the viral vector, which necessitates the use of intermediate plasmid for oligonucleotide cloning and subsequent transfer into the viral vector. Another side effect of seamless integration is that vector cannot be regenerated, which necessitates the purchase of precut vector thus driving up the cost of knockdown experiments. We combined the advantages of each miR-based shRNA approach in a single system by engineering an excisable stuffer fragment encoding lacZα into miR-155, followed by fusing the resulting construct to the 3’ terminus of the synthetic gene for the red fluorescent protein mCherry [13] and by moving this fusion into a lentiviral vector under control of the doxycycline-regulated promoter thus generating construct #2867 (Fig. 1). Digestion of this construct with BfuAI removes the lacZα and re-creates ends suitable for the cloning of oligonucleotides developed using Invitrogen's Block-iT RNAi Designer program directly into lentiviral vector.
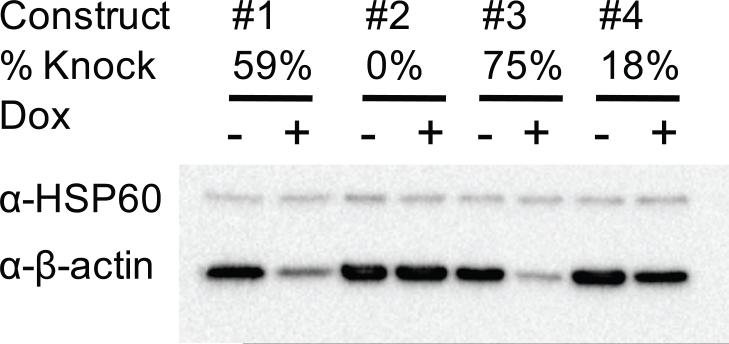
To validate vector #2687, a knockdown of the β-actin in PMVECs was attempted. Four constructs were generated targeting four different regions of the rat β-actin gene. After infection of the PMVECs with retrovirus #2641 and one of the knockdown constructs, double transfectants were selected with blasticidin (30 μg/mL) and puromycin (5 μg/mL). Two wells of 6-well plate were seeded with each transfectant, with one of the wells receiving doxycycline (3 μg/mL). After 48h induction period, cells were collected, lysed, and subjected to Western Blot Analysis. Two of the four constructs produced significant β-actin knockdowns upon induction (59% and 75%) compared to uninduced controls (Fig. 4) thus demonstrating utility of the vectors. At the time of submission, vectors described in Fig. 1 were used for almost a year in our Gene Delivery Core, and demonstrated their versatility and utility. During this period, a number of established and primary cell lines were successfully engineered for doxycycline-regulated expression (Table 1).
Fig. 4.
Knockdown of β-actin in PMVECs. For knockdown experiments, four lentiviral constructs were generated, which targeted four regions of the rat β-actin mRNA (target sequences: #1= gaagatttggcaccacacttt, #2=caggctgtgttgtccctgtat, #3= gaaatcgtgcgtgacattaaa, and 4= cctgacagactacctcatgaa). PMVECs were infected first with #2641 (selection for blasticidin resistance, 30 ug/ml, 5 days), and then with one of the knockdown constructs (selected for 3 days with with 5 ug/ml of puromycin). Cells were either induced (+) or not (−) with 3 μg/ml of doxycycline (Dox). Western Blotting was performed as described in Materials and Methods, chemiluminescense was documented with Fujifilm LAS-1000 cooled CCD camera, and analyzed with MultiGauge 3.0 program.
Table 1.
Inducibility of Gluc expression in various cell lines.
| cell line | Origin | fold inductionb | pooled or clonedc |
|---|---|---|---|
| PMVEC | primary rat endothelial | 272 ± 8 (n=4) | pooled |
| 143B | human osteosarcoma | 639 ± 62 (327-1029; n=14) | cloned |
| 143B ρo,a | human osteosarcoma | 689 ± 123 (329-1206; n=8) | cloned |
| MEF | mouse embryonic fibroblasts | 668 ± 37 (n=3) | pooled |
| MEF | mouse embryonic fibroblasts | 106± 12 (1-218; n=24) | cloned |
| C6 | rat glyoma | 955 ± 108 (n=3) | pooled |
| HeLa | human cervical carcinoma | 811 ± 156 (n=3) | pooled |
| NIH-3T3 | mouse fibroblasts | 447 ± 68 (171-794; n=8) | cloned |
| NIH-3T3 ρo,a | mouse fibroblasts | 524 ± 115 (110-943; n=6) | cloned |
Cells lacking mitochondrial DNA
Mean ± SEM (range; n). The range is only shown for cloned cell lines
Pooled, cells were infected en masse with both #2641 and #2706; cloned, cells were infected with #2641, clonally selected, and then random clones were assayed with #2706
Acknowledgements
This research was supported by grants PO1 HL06629907 and R21RR02396101
References
- 1.Pohjoismaki Jl, Wanrooij S, Hyvarinen Ak, et al. Alterations to the expression level of mitochondrial transcription factor A, TFAM, modify the mode of mitochondrial DNA replication in cultured human cells. Nucleic acids research. 2006;34(20):5815–5828. doi: 10.1093/nar/gkl703. doi:gkl703 [pii] 10.1093/nar/gkl703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pastukh V, Shokolenko I, Wang B, et al. Human mitochondrial transcription factor A possesses multiple subcellular targeting signals. FEBS J. 2007;274(24):6488–6499. doi: 10.1111/j.1742-4658.2007.06167.x. doi:EJB6167 [pii] 10.1111/j.1742-4658.2007.06167.x. [DOI] [PubMed] [Google Scholar]
- 3.Sambrook J, Russel Dw. A laboratory manual. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular Cloning. [Google Scholar]
- 4.King J, Hamil T, Creighton J, et al. Structural and functional characteristics of lung macro- and microvascular endothelial cell phenotypes. Microvasc Res. 2004;67(2):139–151. doi: 10.1016/j.mvr.2003.11.006. doi:10.1016/j.mvr.2003.11.006 S002628620300116X [pii] [DOI] [PubMed] [Google Scholar]
- 5.Rappa G, Anzanello F, Alexeyev M, et al. Gamma-glutamylcysteine synthetase-based selection strategy for gene therapy of chronic granulomatous disease and graft-vs.-host disease. Eur J Haematol. 2007;78(5):440–448. doi: 10.1111/j.1600-0609.2007.00833.x. doi:EJH833 [pii] 10.1111/j.1600-0609.2007.00833.x. [DOI] [PubMed] [Google Scholar]
- 6.Stegmeier F, Hu G, Rickles Rj, et al. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci U S A. 2005;102(37):13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zufferey R, Nagy D, Mandel Rj, et al. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15(9):871–875. doi: 10.1038/nbt0997-871. doi:10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 8.Pear Ws, Nolan Gp, Scott Ml, et al. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90(18):8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wurdinger T, Badr C, Pike L, et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5(2):171–173. doi: 10.1038/nmeth.1177. doi:nmeth.1177 [pii] 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loser P, Jennings Gs, Strauss M, et al. Reactivation of the previously silenced cytomegalovirus major immediate-early promoter in the mouse liver: involvement of NFkappaB. J Virol. 1998;72(1):180–190. doi: 10.1128/jvi.72.1.180-190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks Ar, Harkins Rn, Wang P, et al. Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. J Gene Med. 2004;6(4):395–404. doi: 10.1002/jgm.516. doi:10.1002/jgm.516. [DOI] [PubMed] [Google Scholar]
- 12.Paddison Pj, Caudy Aa, Bernstein E, et al. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16(8):948–958. doi: 10.1101/gad.981002. doi:10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastukh V, Shokolenko In, Wilson Gl, et al. Mutations in the passenger polypeptide can affect its partitioning between mitochondria and cytoplasm : Mutations can impair the mitochondrial import of DsRed. Mol Biol Rep. 2008;35(2):215–223. doi: 10.1007/s11033-007-9073-7. doi:10.1007/s11033-007-9073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]