Abstract
Borrelia burgdorferi, the pathogen of Lyme borreliosis, persists in nature through a tick-rodent transmission cycle. A selective assessment of the microbial transcriptome, limited to gene encoding putative membrane proteins, reveals that bba52 transcription in vivo is strictly confined to the vector-specific portion of microbial life cycle with highest expression levels in feeding ticks and swift downregulation in mice. bba52 deletion did not affect murine disease as assessed by the genesis of arthritis and carditis or long-term pathogen persistence in mice or ticks. However, bba52 deficiency did impair microbial transitions between hosts and vector, defects that could be fully rescued when bba52 expression was genetically restored to the original genomic locus. These studies establish that BBA52 facilitates vector-host transitions by the pathogen and as such, is a potential antigenic target for interference with B. burgdorferi transmission from ticks to mammalian hosts.
Keywords: Borrelia burgdorferi, Lyme disease, pathogen transmission
INTRODUCTION
Lyme borreliosis, caused by Borrelia burgdorferi, is a vector-borne zoonosis prevalent in North America and Europe [1]. When feeding on an infected host, usually wild rodents, immature Ixodes ticks acquire the pathogen, transstadially maintain the infection and, during a subsequent blood meal, transmit the pathogen to mammals. Once infected with B. burgdorferi, humans develop a wide array of clinical complications including the characteristic skin rash erythema migrans, arthritis, carditis and a variety of neurological disorders [2]. As wild rodents are the natural reservoir hosts of B. burgdorferi, certain inbred mice, such as C3H mice are considered excellent models of pathogenesis and are use to study transmission cycle of spirochetes [3]. Genome sequencing of B. burgdorferi [4, 5], studies on the expression and regulation of borrelial genes [6-17] and advances in genetic manipulation techniques [18] have all greatly contributed to our understanding of the unique biology and enzootic infection cycle of this spirochete. However, a human vaccine against B. burgdorferi is currently unavailable, and thus, the development of effective preventive measures remains one of the major focuses of Lyme disease research.
B. burgdorferi may persist in a host or vector for months to years, shuffling between locations during short episodes of tick feeding [19]. During migration from an infected tick to a host, B. burgdorferi invades salivary glands and transmits along with tick saliva [19]. Many salivary gland proteins (Salps) are feeding-induced, soluble and can influence the spirochete transition between vector and host [20, 21]. A few I. scapularis Salps have been identified [22-24] that play important roles in spirochete infection cycle. Further characterization of the interactions of Salps with borrelial antigens and their contributions to infectivity will aid in our understanding of poorly-understood aspects of borrelial transmission.
It is clear that the identification of borrelial antigens that play important roles in spirochete survival in ticks or enable vector-host transitions is the key to blocking pathogen transmission. As extracellularly exposed membrane proteins may directly interact with different environments during transmission or dissemination thus contributing to pathogen adaptation, we sought to assess the expression of selected putative membrane proteins in feeding ticks and mice. We further studied one of these genes, bba52, which displayed vector-specific expression. BBA52, annotated as an outer membrane protein of no assigned function [4, 5], is encoded by the linear plasmid (lp) lp54, which is a stable extra-chromosomal element and is considered to be a necessary part of the spirochete genome [25]. We show that BBA52 facilitates vector-host transitions of B. burgdorferi and is a potential antigenic target to interfere with transmission of Lyme borreliosis.
METERIALS AND METHODS
Mice, Borrelia and ticks
An infectious isolate of Borrelia burgdorferi B31, clone A3, was used throughout the study [26]. For in vitro studies, spirochetes were harvested from the log phase of growth (107 cells/ml). Six- to eight-week-old female C3H/HeN mice were purchased from the National Institutes of Health. The ticks used in this study were reared in the laboratory as described [27]. All animal experiments were performed according to the guidelines of the Institutional Biosafety Committee and the Institutional Animal Care and Use Committee.
PCR
Following B. burgdorferi target genes were selected based on their predicted localization to the spirochete membrane according to database annotation (www.tigr.org) and PSORT in silico analysis as recently described [28]: bb0019, bb0027, bb0028, bb0108, bb0144, bb0155, bb0213, bb0258, bb0262, bb0298, bb0319, bb0323, bb0328, bb0353, bb0379, bb0381, bb0469, bb0586, bb0656, bb0678, bb0679, bb0735, bb0744, bb0769, bb0843, bba03, bba33, bba52, bba62, bba64, bba73, bba74, bbb27, bbe31, bbg01, bbi16, bbj23, bbj27, bbk33, bbk45, bbl23, bbm38, bbn26, bbn38, bbn39. Nucleotide sequences of each of the gene-specific primers will be available on request. For gene expression analysis, groups of C3H mice (3 animals/group) were infected with B. burgdorferi (105 cells/mouse), and following two weeks of infection, dermal samples were collected. For analysis of gene expression in feeding ticks during transmission, naturally infected nymphs (25 ticks/mouse) were allowed to engorge on naïve mice, and were collected at 24, 48 and 96 hours after attachment and pooled together. Two independent experiments used the same parameters of gene expression analysis to ensure the reproducibility of the assay. For analysis of bba52 expression, RNA was isolated from mice (10 animals/group) at 1, 2 and 3 weeks after infection, from ticks that parasitized on infected mice (25 ticks/mice, 3 animals/group) and from infected nymphs (10 nymphs/mouse, 3 animals/group) that engorged on naïve mice. Total RNA was isolated from murine and tick samples, and RT-PCR or quantitative RT-PCR (qRT-PCR) analysis was performed as described previously [28].
Generation of recombinant proteins and antisera
The primers used for amplification of bba52 without the respective signal peptides, are shown in Table 1. Purification of recombinant BBA52 and generation of murine BBA52 antisera were performed as described [28]. In addition, using a commercial source (GenScript Corporation), affinity-purified polyclonal antibodies against a BBA52 peptide sequence (EFLDDPSQESDELEC) of predicted immunogenicity was generated in rabbits. Generated BBA52 antibodies specifically detected 33-kDa native B. burgdorferi BBA52 and did not cross-react with other spirochete proteins.
Table 1.
Oligonucleotide primers used in the current study
| Sequence (5′ – 3′) | Purpose |
|---|---|
| GGGAGCTCAAAAGACAAAATCGCTTTGC | Primer P1, 5′ PCR of the left arm for constructing bba52 mutant. A SacI site (italicized) is attached for the purpose of cloning. |
| AAGGATCCAATATTCTCCTAATATTTAGATGT | Primer P2, 3′ PCR of the left arm for constructing bba52 mutant. A BamHI (italicized) is attached for the purpose of cloning. |
| GGGTCGACTGATTTGCTTTGGAAGTTTT | Primer P3, 5′ PCR of the right arm for constructing bba52 mutant. A Sal1 site (italicized) is attached for the purpose of cloning. |
| GCGGTACCTTAATCCTTTTGGCGAGTT | Primer P4, 3′PCR of the right arm for constructing bba52 mutant. A KpnI site (italicized) is attached for the purpose of cloning. |
| GGTTGCATTCGATTCCTGTT | Primer P5, upstream 5′ PCR primer for the detection of intended integration of pflaB-Kan cassette in bba52 locus |
| AAGTAAAATCACCTCATCTTCTGCTGTT | Primer P6, downstream 3′ PCR primer for the detection of intended integration of pflaB-kan cassette in bba52 genomic locus |
| AGTGTTGCAAGACCATTTGATTTTA | Primer P7, bba52 forward primer |
| TTAAATAAACTGATCTTCAAGAGAA | Primer P8, bba52 reverse primer |
| ATGAATAAGCAAGAGATTGCGAC | Primer P9, kanamycin internal forward primer |
| ATTCCGACTCGTCCAACATC | Primer P10, kanamycin internal reverse primer |
| GTAAGCTCAGCCCGTGCA | Forward primer for RT-PCR of bba51 |
| GCTGTAATAAACCCCCAGATTAA | Reverse primer for RT-PCR of bba51 |
| TGACGAAGAGATTGCAGTCAA | Forward primer for RT-PCR of bba53 |
| CTACCTTTGCTTTTTGGCTTT | Reverse primer for RT-PCR of bba53 |
| GCTCAAATAAGAGGTTTGTC | Forward primer for RT-PCR of flaB |
| ATTCCAAGCTCTTCAGCTG | Reverse primer for RT-PCR of flaB |
| TTGCTGATCAAGCTCAATATAACCA | Forward primer for Quantitative RT-PCR of flaB |
| TTGAGACCCTGAAAGTGATGC | Reverse primer for Quantitative RT-PCR of flaB |
| AGAGGGAAATCGTGCGTGAC | Forward primer for Quantitative RT-PCR of mouse β-actin |
| CAATAGTGATGACCTGGCCGT | Reverse primer for Quantitative RT-PCR of mouse β -actin: |
| GGTATCGTGCTCGACTC | Forward primer for Quantitative RT-PCR of tick β –actin |
| ATCAGGTAGTCGGTCAGG | Reverse primer for Quantitative RT-PCR of tick β -actin: |
| CCAAAAGCCCACAAGGTGTA | Forward primer for Quantitative RT-PCR of bba52 |
| TCTCTTTCCCCATCATCTGG | Reverse primer for Quantitative RT-PCR of bba52 |
| CGGAATTCTTAAATAAACTGATCTTCAAGAG | Primer P11, 3′ PCR of the left arm for bba52 complementation. A EcoRI site (italicized) is attached for cloning. |
| CGGAATTCCGAGCTTCAAGGAAGA | Primer P12, Forward primer for amplification of flgB-aadA cassette in pKFSS1 vector. A EcoRI site (italicized) is attached for cloning. |
| CGCGGATCCATTATTTGCCGACTACC | Primer P13, primer for amplification of flgB-aadA cassette in pKFSS1 vector. A BamHI site (italicized) is attached for cloning. |
| GAGGATCCAGTGTTGCAAGACCATTTGATTTTA | Forward primer for recombinant BBA52 production in E. coli. A BamHI site (italicized) is attached for cloning. |
| GGCTCGAGTTAAATAAACTGATCTTCAAGAGAA | Reverse primer for recombinant BBA52 production in E. coli. A XhoI site (italicized) is attached for cloning. |
Confocal immunofluorescence microscopy
Confocal immunofluorescence of tick salivary glands was performed using LSM-510 laser scanning microscope (Zeiss) as detailed earlier [29, 30]. Samples for each time point of analysis were dissected from a minimum of 5 ticks, and whole organs were scanned at 0.6 μm intervals through the full tissue thickness. Spirochetes were detected using FITC-labeled anti-B. burgdorferi goat IgG (KPL), whereas tick salivary glands were labeled with Texas Red-phalloidin (Invitrogen), respectively.
Genetic manipulation of B. burgdorferi
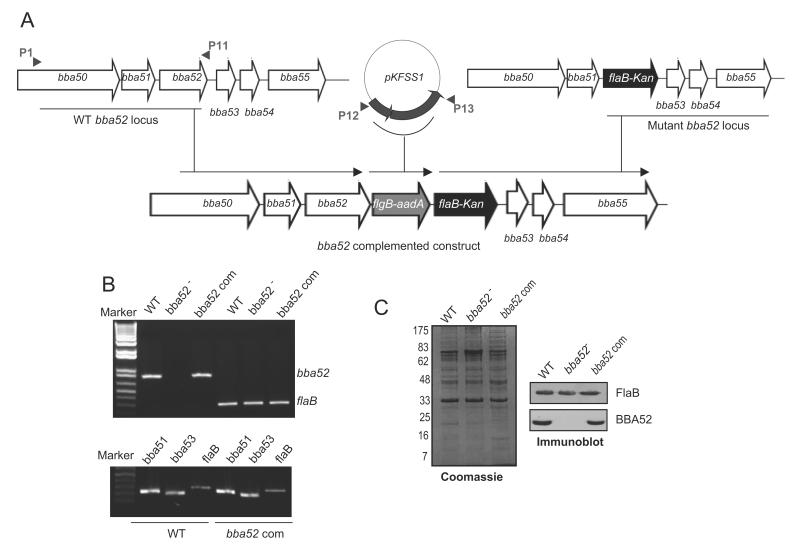
BBA52-deficient B. burgdorferi was generated via homologous recombination by replacing the entire bba52 open reading frame (ORF) with a flaBp-kan cassette as described [28], using the oligonucleotide primers as detailed in Table 1. For genetic complementation, initial efforts to restore bba52 expression used our published strategies [28]; however, transformation strategies using either a DNA insert representing the upstream of the bba52 ORF with the intergenic region (40 nucleotides) or flaB promoter, failed to yield any transformants. We therefore, devised a new strategy to accomplish bba52 complementation in cis by re-inserting a wild-type copy of bba52 ORF into the original gene locus in plasmid lp54. To achieve this, we amplified and assembled two DNA inserts, one using P1-P11 encompassing bba52 ORF and the other using the primers P12 and P13 that included the aadA cassette [31], which confers resistance to streptomycin used for the selection of transformants, and the flgB promoter (figure 5A). The insert carrying P1-P13 amplicon was replaced with the P1-P2 flanking region of the original mutagenic construct pXLF-P1P4 to obtain bba52 complemented construct. The construct was checked for identity and 25μg DNA was transformed into bba52 mutant. Twelve clones grew in the presence of both kanamycin (350μg/ml) and streptomycin (100μg/ml). One of the clones that restored bba52 mRNA and protein, and also retained a comparable plasmid profile to the mutant, except for loss of the non-essential plasmid lp5 [32], was used for further studies.
Figure 5. Genetic complementation of bba52 mutant B. burgdorferi.
(A) Construction of the bba52-complemented construct for re-insertion of bba52 in cis, in the original gene locus of the lp54 plasmid. A new 5′ arm was generated using primers P1-P11 and P12-P13, which were used to amplify and assemble two DNA inserts surrounding bba52 and aadA cassette with the flgB promoter, respectively. The insert representing P1-P13 amplicon (new 5′ arm) was fused with the flaB-Kan cassette carrying the old 3′ arm (as generated using P3-P4, figure 3A) to obtain bba52 complemented construct, and integrated in B. burgdorferi lp54 locus via homologous recombination. (B) RT-PCR analysis of the bba52 complemented isolate. Total RNA was isolated from either the wild type (WT), bba52 mutant (bba52-) or bba52-complemented B. burgdorferi (bba52 Com), converted to cDNA, then subjected to PCR analysis with flaB and bba52 primers, and analyzed on a 1.5% agarose gel (upper panel). bba52 complemented isolates did not display polar effects on the transcription of genes surrounding bba52 locus (bba51 and bba53) (lower panel). (C) Production of BBA52 protein in the complemented B. burgdorferi. Spirochete lysates were separated on a SDS-PAGE gel, stained with Coomassie blue (left panel), or transferred to nitrocellulose membrane and probed with BBA52 and FlaB antibodies (right panels).
Phenotypic analysis of genetically-manipulated B. burgdorferi
To ascertain the phenotype, the mutants and wild-type spirochetes (105 cells/mouse) were separately inoculated into groups of mice (15 animals/group). Skin, heart, joint and bladder, samples were isolated at 1, 2, 3, 4 and 12 weeks following infection. For each time point, samples from 3 mice were pooled by the tissue type and pathogen burdens were assessed by qRT-PCR analysis of flaB mRNA and normalized against murine or β-actin gene as described [28, 33]. Three micrograms of RNA per tissue sample was used for qRT-PCR analysis. At time of euthanasia, mice (3 animal/group) were assessed for swelling of the tibiotarsal joints. Histopathology of joints and hearts collected at weeks 0, 2, 3 and 4 following infection were performed as detailed [28, 34]. Portions of heart and spleen were also cultured in BSK-H medium. For acquisition studies, mice were infected (105 spirochetes/mouse, 3 animals/group). After two weeks, nymphs (25 ticks/mouse) were allowed to engorge on the mice and collected either during 24 and 48 hours of feeding or as repleted ticks. For transmission studies, infected nymphs were generated by allowing larva to feed on wild-type or genetically-manipulated B. burgdorferi-infected mice as described [29]. Infected nymphs (10 ticks/mouse, 3 animals/group) were allowed to fed on naïve mice and B. burgdorferi burdens in partially fed ticks (48 and 60h) or engorged ticks were determined by qRT-PCR. As one tick was sufficient in transmitting the infection to mice [35], single infected nymphs were allowed to feed on separate groups of naïve mouse (3 mice/group). At day 7 following tick feeding, all the mice were sacrificed, and the skin, joints, heart and bladder tissues were isolated and assessed for spirochete burden. Portions of heart and spleen tissues were cultured in BSK-H medium.
Statistical analysis
Results are expressed as the mean ± standard error (SEM). The significance of the difference between the mean values of the groups was evaluated by two-tailed Student’s t-test.
RESULTS
B. burgdorferi genes upregulated during transmission
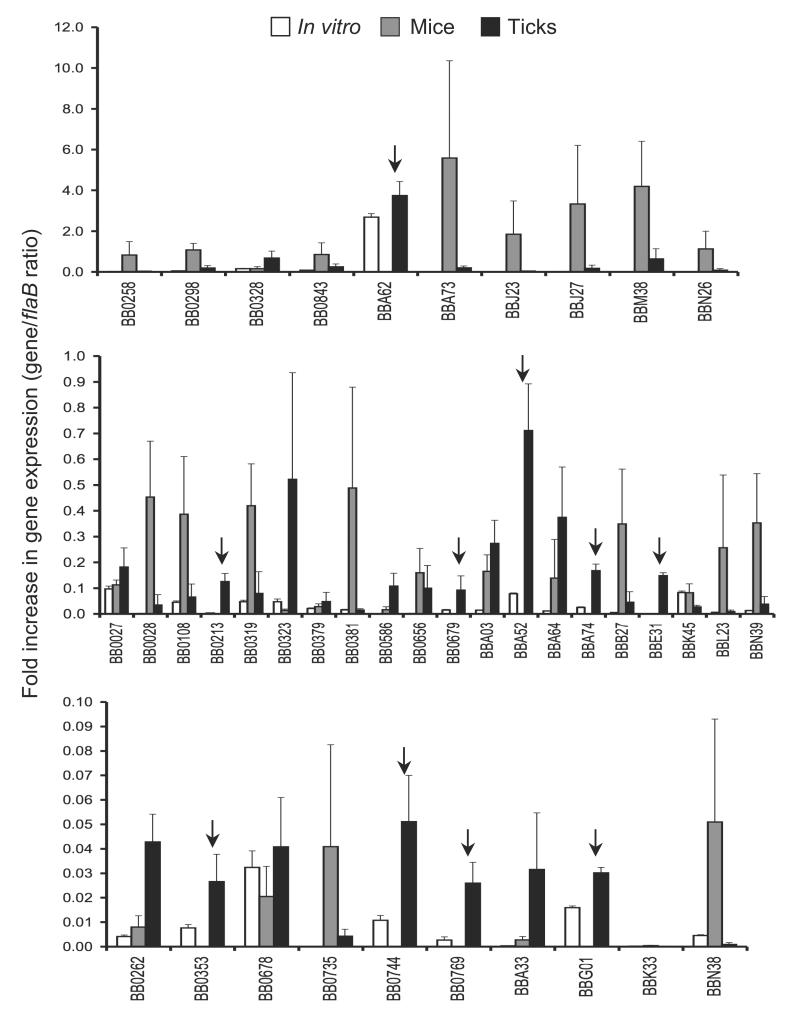
B. burgdorferi faces new environments during host-vector transitions and presumably alters antigenic expression to complete the transition and persist in the new environment. In order to identify microbial genes that are differentially expressed in feeding ticks during transmission, the transcript levels of selected spirochete genes in nymphs and murine dermis were compared using quantitative RT-PCR (qRT-PCR) analysis. The genes were selected due to their putative membrane localization as determined by database annotation and in silico analysis for extracellular exposure. Naïve mice (5 animals/group) were infested with B. burgdorferi-infected nymphs (25 ticks/group), and ticks were collected at 24-96 hours of feeding and pooled together. Dermis samples were collected from groups of mice (5 animals/group) two weeks after infection. Both murine and tick samples were subjected to qRT-PCR analysis as detailed in the Materials and Methods section. The results are represented as fold increase in individual gene transcript levels relative to flaB expression. bba52, along with a few other B. burgdorferi genes, is highly expressed in feeding ticks during transmission, in comparison to its transcript levels in mice (figure 1). We choose to focus on bba52 based on previous studies showing likely tick-specific expression [14, 36], its annotation as a non-paralogous and outer membrane protein of unknown function [4, 5] and unique genomic location as an insertion into the stable plasmid lp54, which otherwise contains many redundant sequences [4].
Figure 1. Relative expression levels of selected B. burgdorferi genes in feeding ticks during transmission.
Total RNA was isolated from cultured spirochetes, from pooled B. burgdorferi infected nymphs collected at 24, 48 and 96 hours of feeding on naïve mice and from murine skin following 2 weeks of B. burgdorferi infection, and converted to cDNA for measuring gene-specific transcripts using qRT-PCR. Fold increase in the expression of individual genes in each of the tick or murine samples was calculated based on threshold cycle (Ct) values using the 2−ΔΔCt method normalized against flaB Ct values. Upper, middle and lower panels represent genes with the highest, moderate and lowest expression ratios relative to that of flaB. Bars indicate the mean ± SD from four qRT-PCR analyses of two independent infection experiments. Arrows indicates genes that are highly expressed in feeding ticks but remain undetectable in the murine dermis.
bba52 is upregulated during spirochete entry and exit through ticks
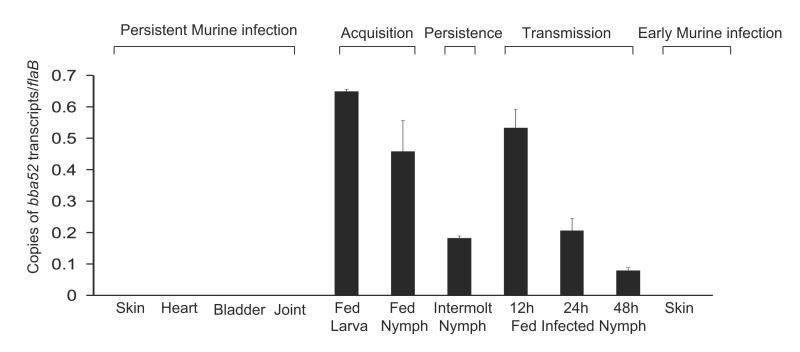
We assessed bba52 expression in detail during representative phases of the B. burgdorferi infection cycle. Mice (10 animals/group) were infected with B. burgdorferi (105 cells/mouse). Total RNA was isolated from skin, joint, heart and bladder samples at 1, 2 and 3 weeks post-infection and pooled by the tissue type. Ticks were parasitized on parallel groups of two-week-old-infected mice (25 nymphs or 30 larvae/mouse), and engorged ticks were isolated. Fed intermolt nymphs were analyzed at 25 days after feeding while larvae were allowed to molt to nymphs. Newly molted infected nymphs were allowed to feed on naïve mice (10 ticks/mice), and were collected following 12-48 hours of feeding. Skin samples were also collected from mice following 5 days of tick engorgement. Total RNA was prepared and subjected to qRT-PCR analysis to measure bba52 transcripts and normalized against flaB. The results showed that bba52 transcripts were undetectable in mice during persistent infection or early tick-borne infection, but were obvious at all tested stages of B. burgdorferi infection in ticks, with the highest levels during tick feeding (figure 2).
Figure 2. bba52 expression is vector-specific.
bba52 expression is analyzed at various stages of murine and tick infectivity. RNA was isolated from mice (10 animals/group) at 1, 2 and 3 weeks after B. burgdorferi infection and pooled by the tissue types (skin, heart, joint and bladder). Naïve larvae and nymphs were allowed to feed on B. burgdorferi-infected mice (25 ticks/mice) and collected at 96 hours (fed larva and fed nymph) or 25 days following feeding (intermolt nymph). B. burgdorferi-infected nymphs (10 nymphs/mouse) were allowed to feed on naïve mice and collected at 12, 24 and 48 hours of feeding (fed infected nymph). Murine skin samples were collected following 5 days of tick engorgement. RNA samples from murine and tick samples were analyzed by qRT-PCR and presented as copies of bba52 transcript per copy of flaB transcript. Error bars represent the mean ± SEM from four qRT-PCR analyses of two independent murine-tick infection experiments.
Construction of bba52 mutant B. burgdorferi
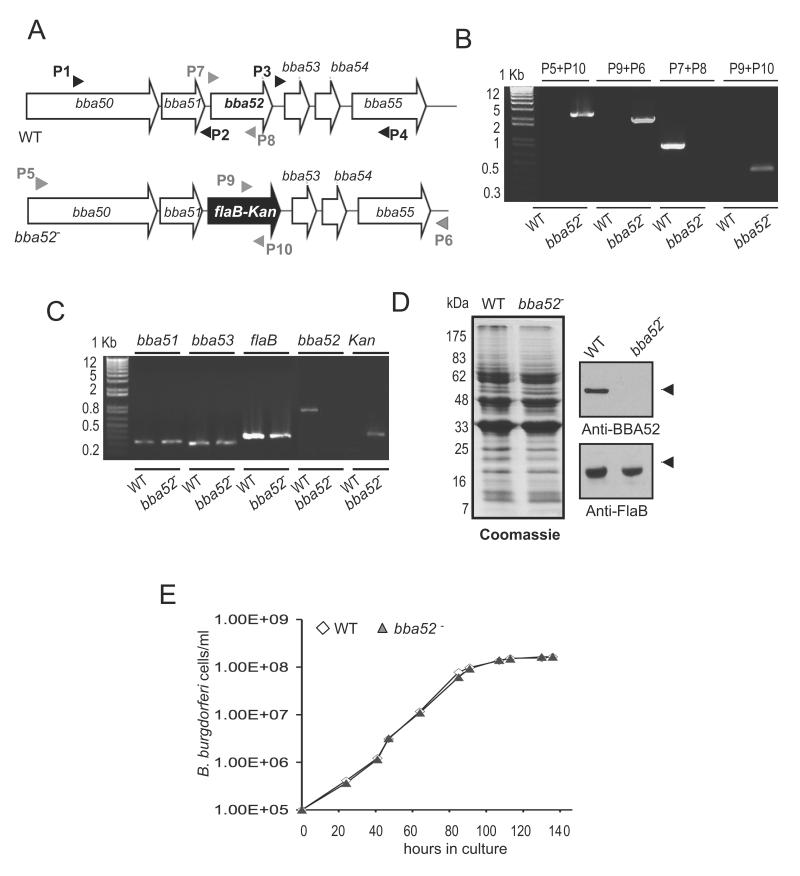
To understand the role of BBA52 in spirochete infectious cycle, we sought to create BBA52-deficient B. burgdorferi using an infectious clone. A suicide plasmid was created to replace the entire open reading frame of bba52 with an antibiotic resistance gene using homologous recombination (figure 3A). The construct was transformed into an infectious clone of B. burgdorferi, and a clone with intended recombination event was selected (figure 3B). The isolated mutant retained the same set of plasmids as in the wild-type spirochetes, but failed to produce bba52 mRNA (figure 3C) or protein (figure 3D). The genetic manipulation did not introduce unwanted polar effects as the mutant expressed the immediately neighboring genes, bba51 and bba53, at similar levels as the wild-type isolate (figure 3C). Deletion of bba52 did not alter B. burgdorferi growth kinetics in vitro (figure 3E).
Figure 3. Construction and analysis of bba52 mutant B. burgdorferi.
(A) Schematic representation of wild type (WT) and bba52 mutant (bba52-) B. burgdorferi at the bba52 locus. Genes bba50-bba55 (white box arrows) and the kanamycin-resistance cassette driven by the B. burgdorferi flaB promoter (flaB-Kan, black box arrow) are indicated. The regions up- and down-stream of the bba52 locus were amplified using primers P1-P4 (black arrow-heads) and ligated on either side of the flaB-Kan cassette to obtain the mutagenic construct, as detailed in the text. (B) Integration of the mutagenic construct, flaB-Kan, in the intended genomic locus. Primers 5-10 (gray arrowheads, positions indicated in figure 3A) were used for PCR analysis using isolated DNA from wild type (WT) or mutant B. burgdorferi (bba52-) and subjected to gel electrophoresis. The combination of primers used for PCR is indicated at the top, and migration of the DNA ladder is shown on the left. (C) RT-PCR assessment of bba52 transcripts and the polar effects of mutagenesis. Total RNA was isolated from wild-type B. burgdorferi (WT) and bba52 mutant (bba52-), converted to cDNA and used to amplify regions within bba52, flaB, kanamycin and genes surrounding the bba52 locus (bba51 and bba53) and visualized on a gel. (D) Protein analysis of wild-type B. burgdorferi (WT) and bba52 mutant (bba52-). Equal amounts of protein were separated on an SDS-PAGE gel, and either stained with Coomassie blue (left panel) or transferred onto a nitrocellulose membrane and probed with BBA52 and FlaB antibodies (right panels). Migration of protein standards is shown to the left in kDa. (E) Growth curves for the wild-type and bba52 mutant B. burgdorferi. Spirochetes were diluted to a density of 105 cells/ml and grown at 34°C in BSK-H medium. Triplicate samples were counted under a dark-field microscope using a Petroff-Hausser cell counter. Differences between wild type and bba52 mutant numbers were insignificant at all times of growth (P > 0.05).
BBA52-deficient B. burgdorferi remains infectious in mice
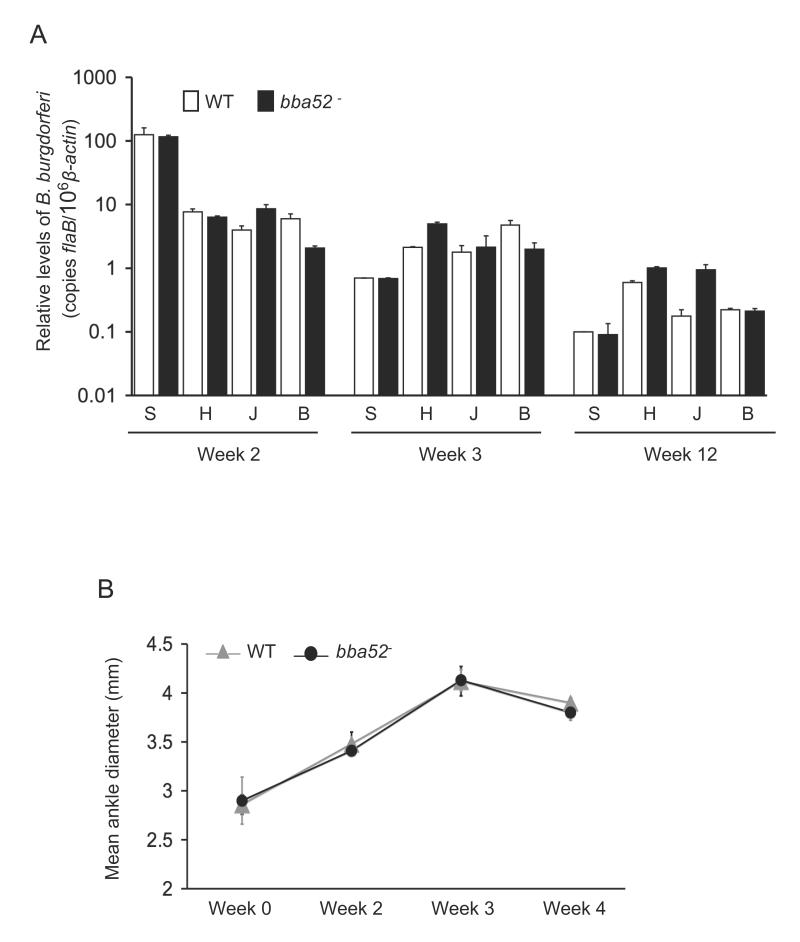
To determine whether bba52 deletion interferes with the ability of B. burgdorferi to persist in mice and induce inflammation, C3H mice (15 animals/group) were injected intradermally with the wild-type spirochetes or bba52 mutants (105 cells/mouse). Mice were sacrificed at weeks 1, 2, 3, 4 and 12 following infection. Pathogen burdens were assessed in isolated skin, heart, joint and bladder samples by qRT-PCR analysis using flaB as a surrogate marker, and normalized against murine β-actin levels. The results showed that both parental and bba52 mutants persisted at similar levels in all time points and tissues, and data from weeks 2, 3 and 12 are presented (figure 4A). Both bba52 mutant and wild-type spirochetes could be cultured from spleen tissues collected following one week of infection (data not shown). In agreement with the comparable pathogen loads, mice infected with either wild type or bba52 mutant B. burgdorferi developed similar disease, as evaluated by the development of ankle swelling (figure 4B) and histopathological observation of arthritis and carditis (data not shown). Taken together, these results suggest that BBA52 is not essential for persistence and virulence of B. burgdorferi in mice.
Figure 4. bba52 mutant B. burgdorferi retain full infectivity in mice.
(A) The pathogen burdens in multiple tissues of infected mice are shown. Mice (15 animals/group) were infected with wild type or the bba52 mutant isolates and spirochete burdens were analyzed in skin (S), heart (H), joint (J) and bladder (B) samples by measuring copies of B. burgdorferi flaB RNA at 2, 3 and 12 weeks of infection. Amounts of murine β-actin were determined in each sample and used to normalize the quantities of spirochete RNA. Bars represent the mean measurements ± SEM of qRT-PCR analyses from two independent infection experiments. The difference between wild type and bba52 mutant levels was statistically insignificant at all time points and tissues (P > 0.05). (B) Assessment of joint swelling in B. burgdorferi-infected mice. Groups of mice (3 animals/group) were infected with wild type or bba52 mutant and examined for joint swelling, using a digital caliper at 0, 2, 3 and 4 weeks after spirochete challenge. Data represent the mean ± SEM from two independent infection experiments. No difference in the ability of the wild type and bba52 mutant to induce joint swelling was recorded (P > 0.05).
bba52 mutant B. burgdorferi displays significant defects during transition between mice and ticks
Although both wild-type spirochetes and bba52 mutants persisted at similar levels in the murine dermis throughout the infection (figure 4A), the mutant was significantly impaired in its ability to transmit to naïve ticks. To ensure the specificity of the result, we sought to complement the mutants with a wild-type copy of the bba52 gene in cis, and use this isolate in mouse-tick transmission studies. The native promoter for bba52 is undefined and heterologous flaB promoter failed to drive bba52 expression in B. burgdorferi. We therefore, devised a new strategy (figure 5A) to re-insert bba52 in the original gene locus using homologous recombination, as detailed in the Material and Methods section. The complemented construct was transformed into mutants, and isolates were selected using antibiotics. PCR analysis confirmed that one of the bba52-complemented isolates retained all endogenous, except for the loss of the non-essential plasmid, lp5 (data not shown). The bba52-complemented isolate produced both bba52 mRNA (figure 5B, upper panel) and BBA52 protein (figure 5C). As expected, the genetic manipulation process did not introduce polar effects in complemented isolates as assessed by the transcription of surrounding genes, bba51 and bba53 (figure 5B, lower panel).
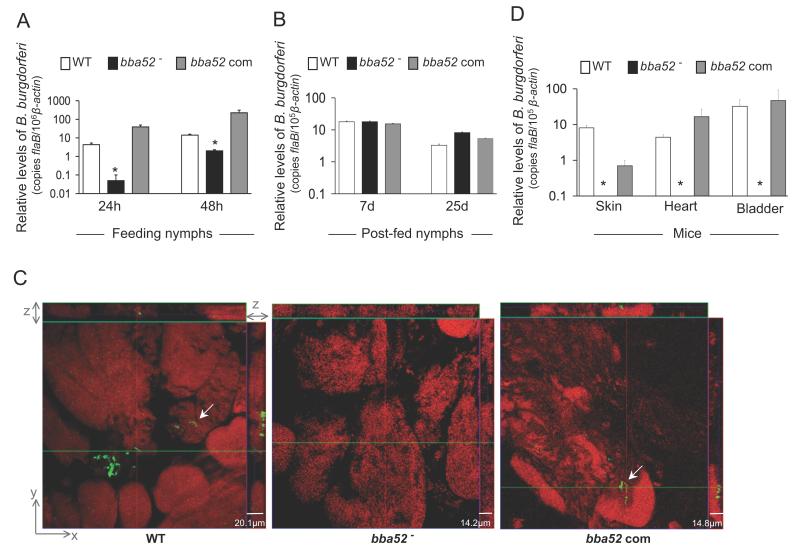
We then assessed whether BBA52 is required for B. burgdorferi entry, persistence and transmission through ticks. To examine the effect of the bba52 deletion on spirochete acquisition by ticks, larval and naïve nymphal ticks were allowed to parasitize mice that had been infected with wild type, bba52 mutant or bba52-complemented isolates. Partially-fed nymphs were forcibly removed 24 and 48 hours after the onset of feeding and parallel groups of larvae or nymphs were collected as fully-engorged ticks. The spirochete burden was assessed by qRT-PCR analysis of flaB normalized against tick β-actin levels. Compared to wild type or bba52-complemented isolates, the levels of bba52 mutants were significantly lower in feeding ticks analyzed at 24 hours (P < 0.002) and 48 hours (P < 0.02) of host attachment (figure 6A). However, analysis of fully engorged larva or nymphs at 7 and 25 days post-feeding (figure 6B) showed similar burdens of wild type and mutant spirochetes, suggesting bba52 deletion only transiently affected B. burgdorferi acquisition by ticks, without significant influence on microbial persistence in the ticks. We then compared the ability of the bba52 mutants to transmit back from infected ticks to naïve mice. Separate groups of nymphs naturally infected with wild type or mutant isolates were allowed to feed on naïve C3H mice (9 animals/group) and were collected as partially-fed (48 and 60 hours) or fully-engorged ticks. Spirochete burdens in ticks were assessed by qRT-PCR and confocal immunofluorescence analyses. Following 7 days of feeding, mouse infection was assessed by culture analysis of the heart and spleen samples and qRT-PCR analysis of skin, heart and bladder tissues. Results indicated that burdens of wild type and bba52 mutants were similar in fed tick gut (data not shown); however, the bba52 mutant was highly impaired in its ability to migrate to salivary glands (figure 6C) and transmit to mice (figure 6D). Both wild type and bba52 complemented isolates were recovered by culture analysis of murine spleen and heart samples. In contrast, the bba52 mutant remained undetectable in all of the 6 individual mouse spleens analyzed, but was recovered from the four of the 6 heart samples. This observation indicates a minor proportion of bba52 mutants that remained untraceable in immunofluorescence and qRT-PCR analyses are still capable of transmission. Collectively, these data establish that BBA52 function is nonessential for B. burgdorferi persistence in murine hosts or ticks but facilitates B. burgdorferi transitions between hosts and vector.
Figure 6. bba52 mutant B. burgdorferi is impaired in its ability to transit between murine hosts and ticks.
(A) B. burgdorferi burdens in ticks during acquisition from infected mice. Mice were infected with B. burgdorferi (3 mice/group) and, following 2 weeks of infection, naïve I. scapularis larvae or nymphs (25 ticks/mouse) were allowed to feed on mice. B. burgdorferi burdens in ticks were analyzed at the indicated time intervals following feeding by measuring copies of the B. burgdorferi flaB RNA. Amounts of tick β-actin were determined in each sample and used to normalize the quantities of spirochete RNA. Bars represent the mean ± SEM of eight qRT-PCR analyses derived from two independent infection experiments. Differences in the spirochete burdens in ticks infected with bba52 mutant and those with the bba52-complemented isolates or wild-type spirochetes were significant both at 24 and (*P < 0.002) 48 hours (*P < 0.02). (B) B. burgdorferi burdens in post-fed ticks. Nymphs were allowed to engorge on infected mice as described in figure 6A and B. burgdorferi burdens in post-fed ticks were analyzed at the indicated time intervals by measuring copies of the B. burgdorferi flaB RNA and normalize against tick β-actin RNA. Bars represent the mean ± SEM of eight qRT-PCR analyses derived from two independent infection experiments. Similar burdens of bba52 mutants, wild type and bba52-complemented isolates were evident at day 7 or day 25 (P > 0.05). (C) B. burgdorferi localization in infected salivary glands during transmission. A representative image showing confocal orthogonal display of infected salivary glands in the XZ and YZ axis revealing the distribution of spirochetes through the full thickness of the 60-hour fed salivary glands is shown. The spirochetes (arrow) were labeled with FITC-labeled goat anti-B. burgdorferi antibody (shown in green) and gland morphology were revealed by labeling of acinar actin filaments using Texas Red-phalloidin (shown in red). While wild type and complemented B. burgdorferi (bba52 Com) were occasionally observed within the gland, bba52 mutant (bba52-) was consistently undetected. (D) B. burgdorferi transmission from infected ticks to mice. B. burgdorferi-infected nymphs were generated by feeding larvae on mice infected with wild type and genetically-manipulated spirochetes, as described in the text. Newly-molted B. burgdorferi-infected nymphs were allowed to feed on naïve mice (1 tick/mouse, 3 animals/group). B. burgdorferi burdens were assessed in the indicated murine tissues after one week of tick feeding by measuring copies of the B. burgdorferi flaB RNA and normalized against mouse β-actin levels. Bars represent the mean ± SEM of four qRT-PCR analyses derived from two independent animal infection experiments. * bba52 mutants were undetectable.
DISCUSSION
B. burgdorferi undergoes remarkable changes in antigenic composition as it invades and colonizes diverse tissues in arthropods and mammals [10, 28, 37-41]. These changes, at least in part, are mediated by regulatory networks involving Rrp2-RpoN/RpoS or Rrp1–Hpk1 TCS and c-di-GMP [7, 13, 17, 42, 43], in addition to intergenic recombination-based mechanism involving the vlsE locus [44, 45]. Microarray analyses of transcriptional alterations in cultured spirochetes identified a large number of genes that are differentially expressed, including bba52, which responded to physiochemical alterations including variations in temperature, the addition of blood, or growth in a dialysis membrane chamber (DMC) implanted within the murine host [6, 7, 14, 36]. In agreement, our data show that selected B. burgdorferi genes are also variably expressed in vivo and highly transcribed in feeding ticks during transmission. The expression pattern of many of these genes (bb0323, bba52, bba62, bba74 and bbe31) agreed with previous studies involving cultured spirochetes that predicted preferential expression in ticks [6, 7, 11, 14]. Specifically, bba74 [46] and bba62 [33] were recently identified as being expressed in feeding ticks. The majority of these genes, however, encoded proteins of unknown functions that are possibly relevant for pathogen transmission from feeding ticks or the establishment of early mammalian infection.
The systematic identification of B. burgdorferi gene products important for infectivity is possible due to the seminal discovery of a borrelial genetic transformation process [47] and further progress in the gene manipulation process [18]. However, the unusual organization of the spirochete genome and the lack of promoter information in a large number of spirochete genes poses serious challenges for mutagenesis, especially for genetic complementation, as we encountered for bba52. The promoter of bba52 is undefined and ORF of 14 upstream genes are unidirectional possessing overlapping or short intergenic regions indicating potentially linked expression. The latter speculation is also suggested by their similar temperature regulation, such as enhanced transcript levels in spirochetes grown at 23°C relative to 37°C [36]. bba52 shares a short (40 base pair) intergenic region with bba51; however, use of this intergenic sequence as a native promoter to drive bba52 expression in B. burgdorferi was unsuccessful (data not shown). Although constitutively-active borrelial promoter flaB was able to produce BBA52 in E. coli (data not shown), the same promoter failed to restore bba52 expression in B. burgdorferi suggesting that the constitutive expression of bba52 may be detrimental to spirochetes. Thus, genetic complementation of regulated borrelial genes without a promoter identity remains technically challenging. In this case, we were able to complement bba52 by replacing the gene at its native cis location without polar effects, which fully restored the wild-type phenotype. This strategy could be helpful for the complementation of other regulated borrelial genes, particularly those lacking discernible promoters.
BBA52 is encoded by the linear plasmid (lp) lp54, a core part of the spirochete genome [25] and retains 65-68% amino acid identity to orthologs in B. afzelii and B. garinii. Our mutagenesis studies suggest that BBA52 is involved in spirochete transmission from ticks to mice. Following tick-borne transmission, bba52 mutants were undetectable in mice, but sometimes recoverable by culture analysis of murine tissues. Therefore, a basal level of spirochete transmission occurs independent of BBA52, possibly suggesting multiple pathways of transmission available to the spirochete. Alternatively, BBA52, along with other borrelial proteins, such as OspC or Lp6.6 [30, 33], could have complementary but non-essential roles in transmission process, as these antigens are all localized in the outer membrane [48, 49] and co-expressed in feeding ticks [33, 40]. Although the precise function of BBA52 in spirochete biology remains unknown, further characterization of antigenic determinants required for microbial transition between hosts and vectors may contribute to the development of novel transmission-blocking vaccines against vector-borne diseases.
ACKNOWLEDGEMENTS
This work was supported by funding from the National Institute Of Allergy And Infectious Diseases (Award Number R21AI076684 and R01AI080615 to U.P). We sincerely thank Kamoltip Promnares, Deborah Y. Shroder, Ireen Dryburgh-Barry, Xinyue Zhang and John F. Anderson for their excellent help with this study.
Footnotes
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Piesman J, Eisen L. Prevention of Tick-Borne Diseases. Annu Rev Entomol. 2008;53:323–343. doi: 10.1146/annurev.ento.53.103106.093429. [DOI] [PubMed] [Google Scholar]
- 2.Steere AC, Coburn J, Glickstein L. The emergence of Lyme disease. J Clin Invest. 2004;113:1093–101. doi: 10.1172/JCI21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthold SW, DeSouza M, Fikrig E, Persing DH. Lyme borreliosis in the laboratory mouse. In: Schuster SE, editor. Lyme disease. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1992. pp. 223–242. [Google Scholar]
- 4.Casjens S, Palmer N, van Vugt R, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 5.Fraser CM, Casjens S, Huang WM, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–6. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 6.Brooks CS, Hefty PS, Jolliff SE, Akins DR. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect Immun. 2003;71:3371–83. doi: 10.1128/IAI.71.6.3371-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caimano MJ, Iyer R, Eggers CH, et al. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol Microbiol. 2007;65:1193–217. doi: 10.1111/j.1365-2958.2007.05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher MA, Grimm D, Henion AK, et al. Borrelia burgdorferi sigma54 is required for mammalian infection and vector transmission but not for tick colonization. Proc Natl Acad Sci U S A. 2005;102:5162–7. doi: 10.1073/pnas.0408536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang FT, Nelson FK, Fikrig E. DNA Microarray Assessment of Putative Borrelia burgdorferi Lipoprotein Genes. Infect Immun. 2002;70:3300–3. doi: 10.1128/IAI.70.6.3300-3303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narasimhan S, Caimano MJ, Liang FT, et al. Borrelia burgdorferi transcriptome in the central nervous system of non-human primates. Proc Natl Acad Sci U S A. 2003;100:15953–8. doi: 10.1073/pnas.2432412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ojaimi C, Brooks C, Akins D, et al. Borrelia burgdorferi gene expression profiling with membrane-based arrays. Methods Enzymol. 2002;358:165–77. doi: 10.1016/s0076-6879(02)58088-5. [DOI] [PubMed] [Google Scholar]
- 12.Revel AT, Talaat AM, Norgard MV. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci U S A. 2002;99:1562–7. doi: 10.1073/pnas.032667699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rogers EA, Terekhova D, Zhang HM, Hovis KM, Schwartz I, Marconi RT. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol Microbiol. 2009;71:1551–73. doi: 10.1111/j.1365-2958.2009.06621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokarz R, Anderton JM, Katona LI, Benach JL. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect Immun. 2004;72:5419–32. doi: 10.1128/IAI.72.9.5419-5432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blevins JS, Xu H, He M, Norgard MV, Reitzer L, Yang XF. Rrp2, a sigma54-dependent transcriptional activator of Borrelia burgdorferi, activates rpoS in an enhancer-independent manner. J Bacteriol. 2009;191:2902–5. doi: 10.1128/JB.01721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He M, Oman T, Xu H, Blevins J, Norgard MV, Yang XF. Abrogation of ospAB constitutively activates the Rrp2-RpoN-RpoS pathway (sigmaN-sigmaS cascade) in Borrelia burgdorferi. Mol Microbiol. 2008;70:1453–64. doi: 10.1111/j.1365-2958.2008.06491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang XF, Alani SM, Norgard MV. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2003;100:11001–6. doi: 10.1073/pnas.1834315100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosa PA, Tilly K, Stewart PE. The burgeoning molecular genetics of the Lyme disease spirochaete. Nat Rev Microbiol. 2005;3:129–43. doi: 10.1038/nrmicro1086. [DOI] [PubMed] [Google Scholar]
- 19.de Silva AM, Tyson KR, Pal U. Molecular characterization of the tick-Borrelia interface. Front Biosci. 2009;14:3051–63. doi: 10.2741/3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das S, Banerjee G, DePonte K, Marcantonio N, Kantor FS, Fikrig E. Salp25D, an Ixodes scapularis antioxidant, is 1 of 14 immunodominant antigens in engorged tick salivary glands. J Infect Dis. 2001;184:1056–64. doi: 10.1086/323351. [DOI] [PubMed] [Google Scholar]
- 21.Rosa P. Lyme disease agent borrows a practical coat. Nat Med. 2005;11:831–2. doi: 10.1038/nm0805-831. [DOI] [PubMed] [Google Scholar]
- 22.Narasimhan S, Sukumaran B, Bozdogan U, et al. A tick antioxidant facilitates the Lyme disease agent’s successful migration from the mammalian host to the arthropod vector. Cell Host Microbe. 2007;2:7–18. doi: 10.1016/j.chom.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramamoorthy R, McClain NA, Gautam A, Scholl-Meeker D. Expression of the bmpB gene of Borrelia burgdorferi is modulated by two distinct transcription termination events. J Bacteriol. 2005;187:2592–600. doi: 10.1128/JB.187.8.2592-2600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyson K, Elkins C, Patterson H, Fikrig E, de Silva A. Biochemical and functional characterization of Salp20, an Ixodes scapularis tick salivary protein that inhibits the complement pathway. Insect Mol Biol. 2007;16:469–79. doi: 10.1111/j.1365-2583.2007.00742.x. [DOI] [PubMed] [Google Scholar]
- 25.Terekhova D, Iyer R, Wormser GP, Schwartz I. Comparative genome hybridization reveals substantial variation among clinical isolates of Borrelia burgdorferi sensu stricto with different pathogenic properties. J Bacteriol. 2006;188:6124–34. doi: 10.1128/JB.00459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elias AF, Stewart PE, Grimm D, et al. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun. 2002;70:2139–50. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman AS, Yang X, Kumar M, et al. Borrelia burgdorferi complement regulator-acquiring surface protein 2 does not contribute to complement resistance or host infectivity. PLoS ONE. 2008;3:3010e. doi: 10.1371/journal.pone.0003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Coleman AS, Anguita J, Pal U. A chromosomally encoded virulence factor protects the Lyme disease pathogen against host-adaptive immunity. PLoS Pathog. 2009;5:e1000326. doi: 10.1371/journal.ppat.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pal U, Li X, Wang T, et al. TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell. 2004;119:457–68. doi: 10.1016/j.cell.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 30.Pal U, Yang X, Chen M, et al. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J Clin Invest. 2004;113:220–30. doi: 10.1172/JCI19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frank KL, Bundle SF, Kresge ME, Eggers CE, Samuels DS. aadA Confers Streptomycin Resistance in Borrelia burgdorferi. J Bacteriol. 2003;185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purser JE, Norris SJ. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2000;97:13865–70. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Promnares K, Kumar M, Shroder DY, Zhang X, Anderson JF, Pal U. Borrelia burgdorferi small lipoprotein Lp6.6 is a member of multiple protein complexes in the outer membrane and facilitates pathogen transmission from ticks to mice. Mol Microbiol. 2009;74:112–25. doi: 10.1111/j.1365-2958.2009.06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X, Yang X, Kumar M, Pal U. BB0323 function is essential for Borrelia burgdorferi virulence and persistence through tick-rodent transmission cycle. J Infect Dis. 2009;200:1318–30. doi: 10.1086/605846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Donahue J, Piesman J, Spielman A. Reservoir competence of white-footed mice for Lyme disease spirochetes. Am J Trop Med Hyg. 1987;36:92–96. doi: 10.4269/ajtmh.1987.36.92. [DOI] [PubMed] [Google Scholar]
- 36.Ojaimi C, Brooks C, Casjens S, et al. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun. 2003;71:1689–705. doi: 10.1128/IAI.71.4.1689-1705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fingerle V, Rauser S, Hammer B, et al. Dynamics of dissemination and outer surface protein expression of different European Borrelia burgdorferi sensu lato strains in artificially infected Ixodes ricinus nymphs. J Clin Microbiol. 2002;40:1456–63. doi: 10.1128/JCM.40.4.1456-1463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang FT, Nelson FK, Fikrig E. Molecular adaptation of Borrelia burgdorferi in the murine host. J Exp Med. 2002;196:275–80. doi: 10.1084/jem.20020770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narasimhan S, Santiago F, Koski RA, et al. Examination of the Borrelia burgdorferi transcriptome in Ixodes scapularis during feeding. J Bacteriol. 2002;184:3122–5. doi: 10.1128/JB.184.11.3122-3125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci U S A. 1995;92:2909–13. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tilly K, Rosa PA, Stewart PE. Biology of infection with Borrelia burgdorferi. Infect Dis Clin North Am. 2008;22:217–34. doi: 10.1016/j.idc.2007.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boardman BK, He M, Ouyang Z, Xu H, Pang X, Yang XF. Essential role of the response regulator Rrp2 in the infectious cycle of Borrelia burgdorferi. Infect Immun. 2008;76:3844–53. doi: 10.1128/IAI.00467-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci U S A. 2001;98:12724–9. doi: 10.1073/pnas.231442498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coutte L, Botkin DJ, Gao L, Norris SJ. Detailed analysis of sequence changes occurring during vlsE antigenic variation in the mouse model of Borrelia burgdorferi infection. PLoS Pathog. 2009;5:e1000293. doi: 10.1371/journal.ppat.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang JR, Hardham JM, Barbour AG, Norris SJ. Antigenic variation in Lyme disease Borreliae by promiscuous recombination of Vmp-like sequence cassettes. Cell. 1997;89:275–85. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 46.Mulay VB, Caimano MJ, Iyer R, et al. Borrelia burgdorferi bba74 is expressed exclusively during tick feeding and is regulated by both arthropod- and mammalian host-specific signals. J Bacteriol. 2009;91:2783–94. doi: 10.1128/JB.01802-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samuels DS. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol Biol. 1995;47:253–9. doi: 10.1385/0-89603-310-4:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lahdenne P, Porcella SF, Hagman KE, et al. Molecular characterization of a 6.6-kilodalton Borrelia burgdorferi outer membrane-associated lipoprotein (lp6.6) which appears to be downregulated during mammalian infection. Infect. Immun. 1997;65:412–421. doi: 10.1128/iai.65.2.412-421.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilske B, Preac-Mursic V, Jauris S, et al. Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect Immun. 1993;61:2182–91. doi: 10.1128/iai.61.5.2182-2191.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]