Abstract
The breast and ovarian cancer susceptibility gene BRCA1 encodes a tumor suppressor. BRCA1 protein, which is involved in DNA damage response, has been thought to be found primarily in cell nuclei. In the present investigation, immunohistological studies of BRCA1 protein in frozen breast cancer tissue and MCF7 and HeLa cell lines revealed BRCA1 expression in both nucleoli and nucleoplasmic foci. Immunoelectron microscopic studies of estrogen-stimulated MCF7 cells demonstrated BRCA1 protein localization in the granular components of the nucleolus. Moreover, immunofluorescence of BRCA1 and nucleolin double-labeling showed colocalization in both nucleoli and nucleoplasmic foci in breast tumor cells and asynchronously growing MCF7 and HeLa cells. Multiparameter analysis of BRCA1 and nucleolin in relation to cell cycle position (DNA content) showed expression during G1−S and persistence of BRCA1 during G2/M. After γ-irradiation of MCF7 cells, BRCA1 protein dispersed from nucleoli and nucleoplasmic foci to other nucleoplasmic sites, which did not colocalize with nucleolin. Small interfering RNA-mediated knockdown of BRCA1 protein resulted in decreased immunofluorescence staining, which was confirmed by Western blotting. The observed colocalization of BRCA1 and nucleolin raises new possibilities for the nucleoplasm-nucleolus pathways of these proteins and their functional significance.
Breast cancer rates have been increasing in the United States; by age 70, an American woman's life-time risk for developing breast cancer is about 10%.1 Mutations in the breast cancer tumor suppressor genes BRCA12 and BRCA23 have been linked to both familial breast and ovarian cancer. Development of tumors in carriers of one mutant allele of either BRCA1 or BRCA2 requires the somatic loss of the wild-type allele, which is a widespread occurrence in breast tumorigenesis.4
The majority of known cancer-causing BRCA1 mutations induce protein truncation, highlighting a requirement for the BRCA1 C-terminal domain repeats in mediating BRCA1 tumor suppressor function. However, somatic mutations in BRCA1 have not been found in sporadic breast cancer tumor tissue.5 Instead it is thought that BRCA1 participates in the tumorigenesis of sporadic breast cancer through reduction in BRCA1 mRNA and protein levels, as compared with normal tissue.6–10 Functionally, BRCA1 participates in many signaling pathways involved in transcription and checkpoint control, and is recruited for the formation of DNA repair complexes, in association with proteins such as Mre11-Nbs1-Rad50, and BRCA2.11 Cell cycle studies have shown that BRCA1 protein is found in nuclear foci (dots) during S-phase, and after γ-irradiation BRCA1 colocalizes with BRCA1-associated ring domain and Rad51-containing foci.12
Our immunohistological studies of frozen tissue sections from breast carcinomas and transmission electron microscopic studies of estrogen-stimulated MCF7 cells have shown nuclear, nucleolar, and cytoplasmic BRCA1 protein staining.13,14 With transmission electron microscopy, we found the BRCA1 nuclear staining on the periphery of dots, around nucleoli, and also in the cytoplasm in multivesicular bodies near the Golgi apparatus.14 Since the BRCA1 protein localization was largely studied by photonic or confocal microscopy, only few studies on its subcellular localization observed by transmission electron microscopy were published. However, confocal microscopy and immunogold electron microscopy have demonstrated the colocalization of BRCA1 protein and β-tubulin in microtubules of the mitotic spindle and in centrosomes.15 Coene et al,16 using both confocal microscopy and transmission electron microscopy with small interfering (si)RNA-mediated knockdown of BRCA1, have found that it is localized in mitochondria, as well as the nucleus. Ganesan et al,17 and Silver et al,18 have found that BRCA1 protein shows overlapping staining for XIST gene on the inactive X chromosome.
In the present study, we further demonstrate the localization of BRCA1 in the granular components (GCs) of the nucleolus by transmission electron microscopy, and colocalization of BRCA1 protein and nucleolin in nucleoli and nuclear speckles by confocal microscopy. In addition, we show BRCA1 and nucleolin co-expression during G1−S phases of the cell cycle by laser scanning cytometry (LSC), relocalization of BRCA1 from nucleoli, and nuclear speckles to irradiation-induced nuclear foci after γ-irradiation. These results were validated using siRNA-mediated knockdown of nuclear and nucleolar BRCA1.
Materials and Methods
Patients and Tumor Tissue
This study was approved by the Institutional Review Board of the Mount Sinai School of Medicine. We randomly selected 18 breast tumors from patients submitted to the surgical pathology division of the Department of Pathology between 1996 and 2000 and snap froze them in liquid nitrogen. The tumors were classified and graded according to modified Bloom-Scarff-Richardson criteria.19 Family history, histopathological diagnosis, age of onset, lymph node status, and estrogen and progesterone receptor status were recorded for each patient and entered into a database. Once the clinical data were collected, each patient and corresponding specimen was assigned a number, to preserve confidentiality.
Immunohistology
The methodology for preparing the frozen sections has been described previously.13 Briefly, tissue previously snap frozen in liquid N2 was mounted at about −8°C in oil (an approximately eutectic mixture of aliphatic esters with a freezing point of about −9°C), frozen at about −25°C, and sectioned using a special adhesive tape to capture the section. The sections were transferred to a −13°C microscope slide coated with a UV-polymerizable adhesive and treated with a flash of UV to polymerize the adhesive and adhere the section (Instrumedics, St. Louis, MO). The slide-mounted, 6-μm frozen sections were melted at room temperature and air dried for 1 hour before being dipped for 30 to 60 seconds in −20°C methanol, and then dried for 5 minutes before being used.
Cell Culture
For the immunofluorescence studies, MCF7 cells were cultured in modified Eagle's medium plus 8% calf serum and 5 μg/ml bovine insulin. HeLa cells were grown in modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum, 50U/ml penicillin, 50 μg/ml streptomycin, and 2 mmol/L l-glutamine. For the irradiation experiments, MCF7 cells were γ-irradiated (4Gy) and fixed for analysis 1 to 2 hours later.
For immunoperoxidase studies (electron microscopy), the MCF7 human breast carcinoma cell line from the Michigan Cancer Foundation (Detroit, MI) was maintained in Dulbecco's modified Eagle's medium (Gibco BRL, Paisley, Scotland, UK), supplemented with 10% fetal calf serum and 0.5% gentamicin. MCF7 cells, grown on glass coverslips for photonic microscopy or plated in 16-mm wells (24-well dishes; Linbro, UK) for transmission electron microscopy, were maintained for 2 days in phenol red-free Dulbecco's modified Eagle's medium with 5% charcoal-stripped fetal calf serum and 0.5% gentamicin before treatment with 10 nmol/L E2 or the control ethanol vehicle. In all experiments (before and during treatment), medium was changed three times a week. E2 (17β-estradiol) had been provided by Roussel-Uclaf (Paris, France). Stock solutions of steroids were prepared as 1000-fold concentrates in absolute ethanol.
Antibodies
Mouse monoclonal BRCA1 antibodies20 MS110 r 1-304, MS 13 r 1-304, AP 16 r 1313-1863, and SG 11 r-1846-1863 (Oncogene Research Products, La Jolla, CA), and K-18 antibody21 r-70-89 (Santa Cruz Technology, CA), were prepared in a 1:20 dilution. Normal goat serum was used to block nonspecific immunoglobulin binding and the sections were incubated at 4°C overnight. As controls, nonrelevant mouse monoclonal antibodies, such as mouse plasmocytoma cell (MOPC) antibody21, purchased from Letton Bionetics Inc., (Kensington, MD) or rabbit polyclonal antibody were used. The specificity of the K-18 antibody binding was checked by pre-absorption with the peptide13 (Santa Cruz Technology, CA) and stained in parallel with a positive control. Nucleolin mouse monoclonal antibody 7G222 (kind gift from S. Pinol-Roma) was used in a 1:2 dilution.
Immunofluorescence
MCF7 or HeLa cells grown on chamber slides, were washed three times in PBS, excess fluid was removed, and the slides were immersed in liquid N2 for 13 seconds, followed by 20 minutes fixation in −20°C methanol. HeLa cells were also fixed in a freshly prepared mixture of 4% paraformaldehyde in PBS for 10 minutes, and permeabilized with 0.02% Triton X100. The frozen section or cell culture slides were immersed in PBS, and then incubated in primary rabbit or mouse antibodies, or simultaneously with both antibodies for colocalization, in a humidified atmosphere at 4°C for 16 hours. Following three washes in PBS, the slides were incubated with species-specific, fluorescein isothiocyanate (FITC) or thromboxane A2 receptor (TXR)-conjugated secondary antibodies for 30 minutes, at a 1:100 dilution, washed extensively in PBS, and mounted with Vectashield with or without 4,6-diamidino-2-phenylindole (DAPI) (Vector Labs, Burlingame, CA). Fluorescently stained cells or tissues were observed using a Zeiss axiophot microscope equipped with a ×40 Plan-Neofluar objective, and a Leica TCS-SP (UV) confocal laser scanning microscope.
LSC
For LSC, MCF7 cells were indirectly stained with polyclonal K-18 BRCA1 antibody and FITC-labeled goat anti-rabbit (green), and the slides were re-suspended in 5 μg/ml propidium iodide (PI) (Molecular Probes, Carlsbad, CA) to counterstain cellular DNA. For nucleolin staining, MCF7 cells were indirectly stained with the mouse monoclonal nucleolin antibody and FITC-labeled goat anti-mouse antibody (green) and their DNA was counterstained with PI (red). LSC methodology (Compucyte, Cambridge, MA) has been previously reported23; briefly, cellular fluorescence of PI and FITC was excited at 488 nm and emission of these fluorochromes was measured using the standard long-pass (570 nm) or bandpass (530 nm) filters, respectively. The triggering threshold is depicted in red, whereas the total nuclear fluorescence contour is set on green. At least 1500 cells were measured per slide. The relocation feature of LSC was used to identify individual cells and to reveal their morphology.
Electron Microscopy
MCF7 cells were fixed in a freshly prepared mixture of 4% paraformaldehyde in PBS for 1 hour. They were then permeabilized for 25 minutes with 0.02% Triton X100 in 0.05% Tween-PBS, blocked with 2.5% normal horse serum from the Vectastain ABC kit (Vector Laboratories) in 0.02% Triton X100 in 0.05% Tween-PBS. They were incubated for 60 minutes with BRCA1 monoclonal antibody MS110 (Ab-1) (1/40) or with the control monoclonal antibody (MOPC21) at the same concentration. Cells were then washed and incubated for 30 minutes with biotin-conjugated horse antibody to mouse IgG (Vectastain, ABC kit). At the end of the secondary antibody incubation, and after quenching endogenous peroxidase activity in 0.1% hydrogen peroxide in PBS, cells were incubated in streptavidin-biotin peroxidase complex (Vectastain, ABC kit). All incubations of cells with primary antibodies, secondary antibodies, and streptavidin-biotin peroxidase complex were performed with 0.02% Triton in 1% bovine gamma globulin-PBS. All washes were done in 0.02% Triton X100 in 0.05% Tween-PBS. For photonic microscopy, after washing in PBS, and incubation in 3,3′ diaminobenzidine (Sigma-Aldrich, St. Louis, MO) solution, the cells were counterstained in hematoxylin, dehydrated with alcohol and xylene, and then mounted on slides using a permanent mounting medium. Slides were observed and photographed using a Leitz DMRB light microscope and a 3-CCD DXC-950P color video camera connected to a microcomputer. For transmission electron microscopy, the immunocytochemical analysis was performed as described above with two additional post-fixation steps, one with 1% glutaraldehyde in PBS (30 minutes) before incubation in 3,3′ diaminobenzidine solution, and one with 1% OsO4 in PBS (30 minutes) after 3,3′ diaminobenzidine incubation. Cells were then stained with 3% aqueous uranyl acetate, dehydrated, and embedded in EMbed 812. Ultrathin sections of cells sectioned parallel to the growth substrate were observed under a Hitachi 7110 electron microscope.
Immunostaining Quantification
BRCA1 staining intensity in 21 fields observed at ×20 magnifications was quantified with an image analyzer (SAMBA TITN, Unilog, Grenoble, France) adapted to a Leitz DMRB light microscope and a 3-CCD DXC-950P color video camera connected to a microcomputer. The percentage of BRCA1-stained nuclear and cellular (nucleus plus cytoplasm) surface was determined. Staining intensity value was the averaged of the integrated photonic densities of stained surfaces measured in control or E2-treated cells. The results were expressed by a quantitative immunohistochemical score given in arbitrary units, taking into account the number of stained cells and staining intensity (quantitative immunohistochemical score = percentage of stained surface × mean staining intensity × 10).
Statistical Analysis
Mann Whitney nonparametric test was used to compare paired group of data.
Western Blotting
Gel electrophoresis of 180 μg of whole cell lysates obtained from control (C) and E2 treated cells (E2) for 7 days was performed using 8% SDS-polyacrylamide gel electrophoresis. Subcellular extraction of MCF7 cells was performed by using the ProteoExtract Subcellular Proteome Extraction Kit (Merck, Chimie SAS, Fontenay-Sous-Bois, France) according to manufacturer's instructions. Gel electrophoresis of 100 μg of whole cell lysates or 24 μg of cytoplasmic extracts and 55 μg per lane of nuclear fractions from control cells (treated for 7 days with ethanol vehicle) was performed using 12% SDS-polyacrylamide gel electrophoresis. In both experiments, the proteins were blotted onto a polyvinylidene difluoride membrane, blocked with 5% nonfat milk powder for 1 hour in 0.1% Tween 20 PBS, washed and probed overnight at 4°C or 3 hours at room temperature with the primary antibody MS110 (Ab-1) 1:50 or K-18 1:50 in 5% not-fat dry milk. After washing, membranes were incubated with sheep anti-mouse or goat anti-rabbit horseradish peroxidase-labeled secondary antibody and protein bands were detected using the bioluminescence technique and Hyperfilm enhanced chemiluminescence development. α-tubulin (kind gift from Ned Lamb) and c-jun (N) (Santa Cruz Technology) polyclonal antibodies were used as loading or fraction purity controls for cytoplasmic and nuclear fractions, respectively.
Gel electrophoresis of 40 μg per lane of HeLa whole cell lysate from 1) empty vector transfected cells, 2) si-green fluorescent protein (GFP) transfected cells, and 3) siHBRCA1 transfected cells was performed using 6% SDS-polyacrylamide gel electrophoresis. The proteins were blotted onto a nitrocellulose membrane, blocked for 1 hour in 5% milk powder/0.1% Triton X100/PBS at room temperature, and incubated with the primary antibody MS110 (Ab-1) 1:100 in 5% milk powder overnight at 4°C on a rocking platform; washed, blocked, and then finally detected with secondary antibody goat anti-mouse using the bioluminescence technique and Hyperfilm ECL development. We used α-tubulin (Cell Signaling Technology, Beverly, MA) as a loading control.
Small Interfering RNA
For the siRNA experiments, two constructs were made into pSUPER vectors.24 For silencing BRCA1, the pSUPER plasmid contained nucleotides 215 to 233 (genbank #U14680) 5′-GGAACCTGTCTCCACAAAG-3′. The control pSUPER plasmid contains a nucleotide sequence generated against GFP. As further controls, we used the empty vector, or plasmid pcDNA3.1-GFP. For the staining experiments, HeLa cells were plated onto chamber slides (3 × 105 cells per well). Twenty-four hours later, the medium was exchanged to remove the antibiotics and 1 μg of the selected construct was transfected per well of the chamber slide, and left overnight. The cells in the chamber slides were then fixed for immunostaining. Similarly, for biochemical analysis, HeLa cells were plated on 100 mm culture dishes (106 cells) per dish, and transfected with 5 μg of construct. After 24 hours, the cells were rinsed in ice cold PBS, lyzed in radioimmunoprecipitation assay buffer supplemented with protease and phosphatase inhibitors.25
Results
We have previously shown nuclear and nucleolar localization of BRCA1 in frozen tissue sections of human infiltrating mammary adenocarcinomas and MCF7 cells.13,14 In the present study, we compared the immunoperoxidase staining of four mouse monoclonal BRCA1 antibodies, MS110, MS13, AP16, and SG11, and the rabbit polyclonal K-18 BRCA1 antibody in methanol-fixed frozen tissue sections of a poorly differentiated mammary adenocarcinoma from patient 1 (Figure 1, A−F). Although all of the BRCA1 antibodies stained the nuclei of epithelial cells of the tumor, the AP16 antibody stained many more tumor cell nuclei intensely (Figure 1D), and the other antibodies stained different numbers of tumor nuclei with varying intensities.
Figure 1.
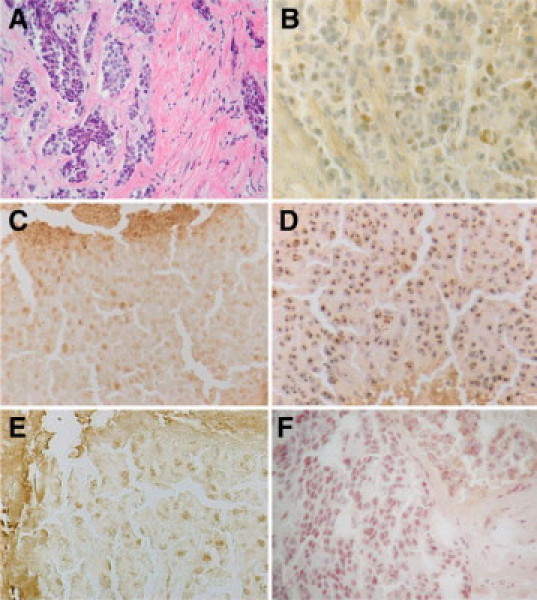
Comparison of mouse monoclonal BRCA1 antibodies: MS110, MS13, SG11, and AP16, and the rabbit polyclonal K-18 antibody in frozen tissue sections of a poorly differentiated adenocarcinoma from patient number 1. A: H&E-stained frozen section Original magnification ×270. B: Sequential section of the same tumor stained with MS110 mouse monoclonal BRCA1 antibody and anti-mouse IgG and avidin-biotin peroxidase detection system. Scattered tumor cell nuclei show BRCA1 staining Original magnification ×540. C: Sequential section stained with MS13 antibody shows scattered nuclear staining as well as some nonspecific staining of stromal material. Original magnification ×540. D: Sequential section stained with AP16 mouse monoclonal antibody shows focal nuclear staining. Original magnification ×540. E: Sequential section stained with SG11 antibody shows scattered tumor cell nuclear staining and nonspecific stromal staining. Original magnification ×540. F: Sequential section stained with the rabbit polyclonal K-18 antibody and anti-rabbit IgG and avidin-biotin peroxidase detection system shows nuclear staining of tumor cells. Original magnification ×540.
To obtain a more precise understanding of BRCA1 localization, we stained MCF7 cells treated with 10 nmol/L estradiol with the MS110 antibody (Figure 2A). BRCA1 labeled dots localized by photonic microscopy showed enhanced staining in nuclei, nucleoli, and the cytoplasm of treated cells compared with the untreated cells. BRCA1 staining was observed at the periphery of nucleoli as indicated by arrows, among other stained granules (Figure 2, A and B), while it is principally near but external to the nucleoli in control cells (Figure 2, C and D, arrows). Dark blue nucleoli (1 to 3) were clearly visible in nuclei of control cells stained with the nonrelevant monoclonal antibody MOPC 21 and counterstained with hematoxylin (Figure 2F). At a lower magnification, it appeared that there were a few groups of nontreated cells with labeled nuclei stained with MS110 antibody (Figure 2E).
Figure 2.
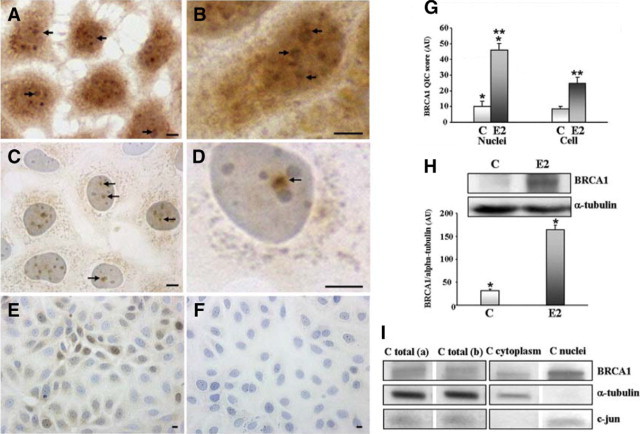
Nucleolar BRCA1 protein localization in MCF7 cells. MCF7 cells were treated (A, B) or not (C−F) with 10 nmol/L of 17 β-estradiol for 7 days. MCF7 cells were cultured and prepared as described in Materials and Methods, and immunostained with BRCA1 monoclonal antibody MS110 (Ab-1). A and B: Labeled nuclear dots for BRCA1 protein were observed in the nucleoplasm or in the periphery of the nucleoli (arrows), as it is better observed at higher magnification (B). C−F: In nontreated cells, labeled dots were mainly observed in the nucleoplasm, near nucleoli (C, arrows), as shown at higher magnification (D). Only some groups of few cells with labeled nuclei were observed at lower magnification (E) and no labeling was observed in cells incubated with the non relevant monoclonal antibody MOPC 21 (F). G: BRCA1 staining intensity quantified with an image analyzer in nuclei alone or nuclei plus cytoplasm of the same cells showed a significant increase in E2-treated cells’ nuclei (*P < 0.002) or with nuclei plus cytoplasm of E2 treated cells (**P < 0.0001). H: BRCA1 protein detected with MS110 antibody is significantly lower in whole cell lysates of control MCF7 cells compared with E2 treated cells. The histogram represents the BRCA1 concentration normalized with the reciprocal concentration of the α-tubulin (*P < 0.0001). I: In whole cell lysates of control MCF7 cells, the same results were observed with K-18 (C total a) and MS110 (C total b) primary antibodies. The presence of BRCA1 protein was observed also in cytoplasm and nuclear fractions of control cells at similar concentrations after protein normalization with MS110 antibody. α-tubulin and c-jun expression was used as a control for cytoplasm and nuclear fraction purity, respectively. Scale bars = 10 μm.
In control and E2 treated MCF7 cells, the percentage of cells containing labeled nuclei versus total cells was calculated in 21 fields observed at ×20 magnification to count about 2000 total cells. The mean of percentages was significantly different (P < 0.0001) and evaluated as 16% in control cells and 74.7% (4.6-fold increase) in E2 treated cells. To measure BRCA1 concentration, a semiautomatic quantification of nuclear and total (nuclear plus cytoplasm) cell BRCA1 staining was realized in 21 fields at ×20 magnification with a Samba analyzer (Figure 2G). The results were expressed by a quantitative immunohistochemical score given in arbitrary units. Histograms of mean values were represented and a similar 4.6-fold increase in BRCA1 protein staining (P < 0.002) was observed in E2-treated cell nuclei. No significant difference was observed between BRCA1 staining when nuclear alone or nuclear plus cytoplasmic surface of the same control cells were analyzed, indicating that BRCA1 protein staining in the cytoplasm represented quite the same quantity as in the nucleus. Otherwise, it was significantly decreased in E2 treated cells, confirming a higher level of BRCA1 in nuclei.
We used Western blotting to show lower levels of BRCA1 protein in control cells, compared with treated cells. There was a clearly visible 220-kDa BRCA1 protein in whole cell lysates from E2 treated MCF7 cells (5.2-fold increase) compared with the much less staining (just visible) 220-kDa BRCA1 protein in whole cell lysates from control MCF7 cells (Figure 2H). After normalization with the reciprocal concentration of the control α-tubulin, BRCA1 concentrations expressed in arbitrary units were represented in the histogram. Western blotting of whole cell lysates, and of cytoplasm, and nuclear fractions of control cells was performed to determine the specific localization of BRCA1 with MS110 antibody (Figure 2, I, C total b, C cytoplasm, C nuclei). We used α-tubulin and c-jun (N) polyclonal antibodies as purity controls of cytoplasm and nuclear fractions, respectively (Figure 2I). The quantification of BRCA1 protein in the two fractions, normalized by the protein quantity, gave similar results (189 and 211) for the two compartments confirming the SAMBA quantification data in control cells. To validate both the BRCA1 antibodies, after dehybridization of the same membrane, BRCA1 was also detected with K-18 antibody (Figure 2I, C total a) and gave the same results.
We resolved the localization of the nuclear granules visualized by light microscopy, by preparing ultrathin sections in epoxy-included MCF7 cells for analysis using transmission electron microscopy (Figure 3). BRCA1 appears localized on the periphery of spherical nuclear dots and labeling was observed around nucleoli, sometimes in close contact with nucleolus fibers (Figure 3A) or GCs of the periphery of nucleoli (Figure 3, C and D). There was also labeling in the multivesicular bodies in the cytoplasm, but not the Golgi (Figure 3A).14 This confirmed the presence of BRCA1 protein in the nucleus and nucleolus since we observed here ultrathin sections of the cells. No labeled dots were observed in nuclei or nucleoli of cells incubated with the nonrelevant monoclonal antibody MOPC21 (Figure 3B).
Figure 3.
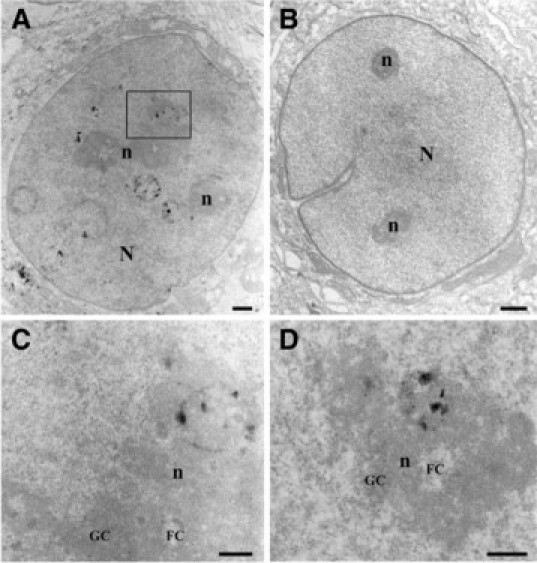
Nucleolar localization of BRCA1 protein using transmission electron microscopy. MCF7 cells were immunostained with BRCA1 monoclonal antibody MS110 (Ab-1) or non-relevant monoclonal antibody MOPC21. A: Labeled nuclear dots for BRCA1 protein were observed around nucleoli, sometimes in close contact with nucleolus fibers or within electron dense granular components (GC) of the nucleolar periphery. BRCA1-labeled protein was also observed in the cytoplasm, near the nuclear membrane in multivesicular bodies near the Golgi. The box plotted in A is represented at a higher magnification in C and the nucleolus of another cell at a high magnification in D. BRCA1 staining was detected in GCs of nucleoli surrounding the fibrillar center (FC). B: No labeled dots were observed in nuclei or nucleoli of cells incubated with the nonrelevant monoclonal antibody. N: nucleus, n: nucleolus. Scale bar = 0.25 μm.
We further analyzed the BRCA1 protein compartmentalization in frozen sections of a poorly differentiated mammary adenocarcinoma from patient number 16 by costaining with antibodies to BRCA1 and nucleolin (Figure 4, A−D). The rabbit polyclonal BRCA1 K-18 antibody stains many tumor cell nuclei (green) (Figure 4A). The mouse monoclonal nucleolin antibody stains nucleoli (red) (Figure 4C). BRCA1 protein and nucleolin frequently colocalize in tumor cell nuclei (yellow) (Figure 4D). Some of the tumor cell nuclei showed clumped fragmented BRCA1 staining, which might represent apoptotic cells (Figure 4, A and D).
Figure 4.
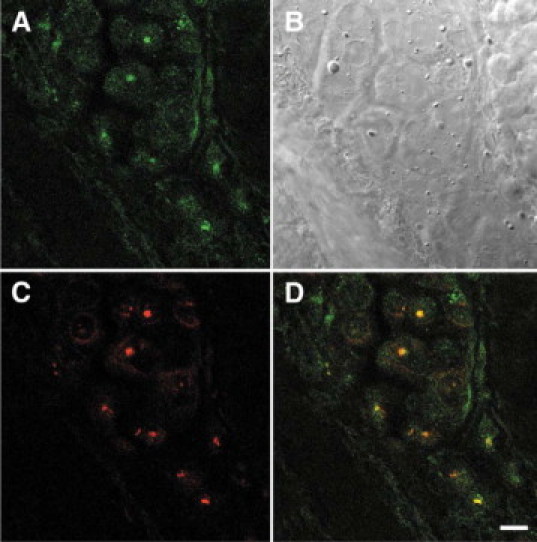
Colocalization of BRCA1 and nucleolin proteins in frozen tissue sections from poorly differentiated mammary adenocarcinoma from patient number 16. A: Rabbit polyclonal BRCA1 K-18 antibody stains many tumor cell nuclei (green). B: DIC Nomarski image of the same field as A, C, and D. C: Mouse monoclonal nucleolin antibody stains nucleoli (red). D: DIC image of C, nuclei stained for BRCA1 (green) merge with nucleolin (red) to show colocalization (yellow). There is also clumped BRCA1 staining (green). Scale bar = 10 μm.
We also studied BRCA1 protein and nucleolin staining in asynchronously growing MCF7 cells. Cells exhibited varying staining patterns with both antibodies (Figure 5). With the BRCA1 antibody, the nuclear patterns included nuclei with many small dot-like speckles, nuclei with punctate bodies and speckles, and nuclei with round centrally placed bodies (Figure 5A). On the same field, the mouse monoclonal antibody to nucleolin-stained nucleoli and many dot-like nuclear speckles (Figure 5B). The colocalization of BRCA1 protein and nucleolin occurred frequently in nucleoli (which are phase dense in Figure 5D) and in some nuclear speckles (Figure 5C). The fact that BRCA1 and nucleolin staining does not always coincide in nucleoli in asynchronous cells suggests that their colocalization may be dependent on the phase of the cycle of each cell. In addition, the absence of nucleolar BRCA1 staining in some cells (Figure 5D) shows that we have an internal control for BRCA1 specificity.
Figure 5.
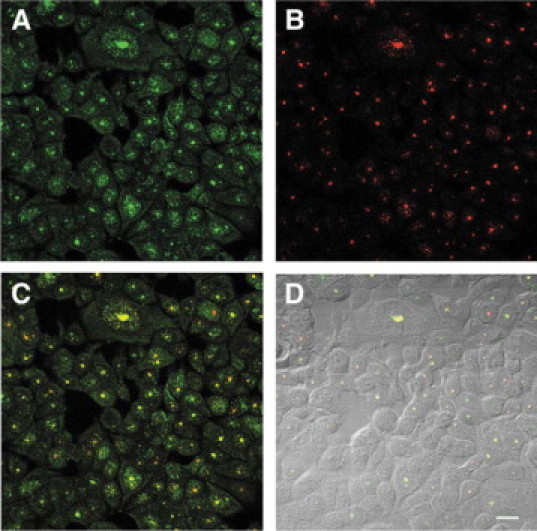
Colocalization of BRCA1 and nucleolin in an asynchronous culture of MCF7 cells. A: The rabbit polyclonal K-18 BRCA1 antibody stains many cell nuclei (green). B: Mouse monoclonal antibody to nucleolin stains nucleoli and many dot-like speckles (red). C: DIC image of B. Nuclei stain for BRCA1 (green) merge with nucleolin (red) to show colocalization (yellow). The merge occurs both in nucleoli (which are phase dense in D) and in some nuclear speckles. Not all of the nucleoli show BRCA1 colocalization, which provides an internal control for BRCA1 staining specificity. D: Nomarski image of the same field as A−C. Scale bar = 20 μm.
To test this possibility, we studied their pattern of expression versus DNA content in MCF7 cells with LSC (Figure 6). Fluorescence microscopic examination of the BRCA1 stained cells revealed many cells expressing nuclear BRCA1 and some negative cells (only a red nuclear signal corresponding to the PI counterstaining). Similarly, fluorescence microscopic detection of nucleolin staining in MCF7 cells showed that many cells expressed nucleolin staining, but that some cells were negative (only PI staining). Using a cytometric program for fluorescence in situ hybridization (Compucyte Corp.), we were able to count the number of BRCA1 and nucleolin “spots” present inside the nucleus of each individual cell and measured the DNA content by PI counterstaining. The cells were categorized based on the number of spots identified per cell and given a region number, ie, for BRCA1 staining, region 1 included 3.8% of cells displaying only one spot; region 2 included 0.5% cells displaying 2 spots (Figure 6A). In Figure 6B, the DNA content shows distribution of cells in the G1, S, and G2/M phases of the cell cycle. Maximum BRCA1 protein expression, as shown by peak BRCA1 immunofluorescence, was found during G1 and S phases, and was not completely degraded during G2/M phase (Figure 6, A and B). We also plotted nucleolin expression versus DNA content during cell cycle progression. There were 29.9% of MCF7 cells showing nucleolin staining of one spot in region 1; region 2 included 2.3% of cells with two spots (Figure 6C). Maximum nucleolin expression in the nucleolus was found in single and double nucleolar spots or bodies during the G1 and S phases of the cell cycle (Figure 6D).
Figure 6.
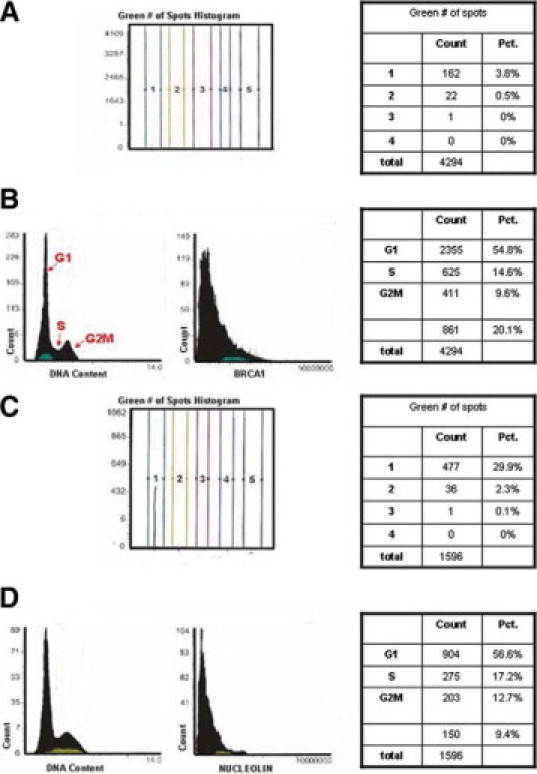
Detection of BRCA1 and nucleolin proteins during the cell cycle by LSC. For BRCA1 staining, MCF7 cells were indirectly stained with polyclonal rabbit K-18 BRCA1 antibody and FITC-labeled goat anti-rabbit (green). For nucleolin staining, the cells were indirectly stained with the mouse monoclonal nucleolin antibody and FITC-labeled goat anti-mouse antibody (green). Their DNA was counterstained with PI (red), as described in Materials and Methods. The triggering threshold is depicted in red, whereas the total nuclear fluorescence contour is set on green. The LSC software allows counting of the number of fluorescent bodies (“spots”) present inside the nucleus of each individual cell. The cells were categorized based on the number of spots identified per cell and given a region number. The green # of spots histogram summarizes the number of spots per cell in a given region number. The DNA content histogram shows distribution of cells in the G1, S, and G2/M phases of the cell cycle by region number. A: Fluorescence microscopic examination of MCF7 cells revealed many cells expressed nuclear BRCA1 protein and some cells were negative (only a red nuclear signal corresponding to the PI counterstaining). Region 1 included 3.8% of cells displaying only one spot; region 2 included 0.5% cells displaying 2 spots. B: Cytometric determination of BRCA1 protein during the cell cycle. The DNA content histogram shows cells in G1, S, and G2/M cell cycle phases. BRCA1 protein immunofluorescence peaked during G1−S phase and its expression was not completely degraded during G2/M. C: Fluorescence microscopic detection of nucleolin staining in MCF7 cells showed 29.9% of cells displaying only one spot in region 1; 2.3% of cells with two spots in region 2. D: Cytometric determination of nucleolin protein during the cell cycle. The DNA content histogram shows G1, S, and G2/M cell cycle phases. The nucleolin expression peak is found during the G1−S phase of the cell cycle.
Next, we investigated whether DNA damage influenced nucleolar localization. BRCA1 nucleolar localization was changed by γ-irradiation of MCF7 cells (Figure 7, A−D) compared with MCF7 cells in the absence of damage (Figure 5). One hour after γ-irradiation, BRCA1 protein was distributed in coarse granular nuclear foci in many nuclei, without apparent nucleolar staining, although some cells showed cytoplasmic staining (Figure 7A). In contrast, nucleolin staining remained in many nucleoli (Figure 7C) and did not colocalize with BRCA1 protein after irradiation (Figure 7B).
Figure 7.
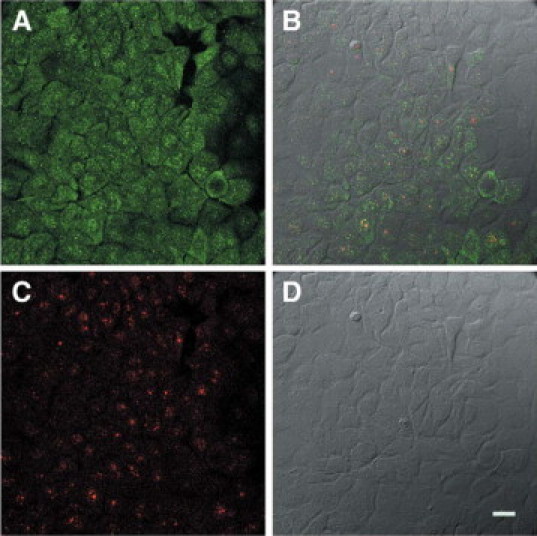
Confocal microscopy of γ-irradiated MCF7 cells immunostained with BRCA1 and nucleolin antibodies. A: Rabbit polyclonal BRCA1 antibody, K-18, stains multiple coarse granular foci (green) in many cell nuclei. There are some cells, which show predominantly cytoplasmic staining. B: DIC image of C. Nuclei that stain for BRCA1 (green) and nuclei that stain for nucleolin (red) are predominantly not colocalized. C: Mouse monoclonal antibody to nucleolin stains nucleoli (which are phase dense in D) and many dot-like speckles (red). D: Nomarski image of the same field as in A−C. Scale bar = 20 μm.
To determine whether the colocalization with nucleolin was also present in different cell types, we analyzed BRCA1 staining in asynchronously growing HeLa cells (Figure 8, A−H). In addition, to validate our results we compared BRCA1 staining generated by the MS110 and K-18 antibodies. With the BRCA1 monoclonal antibody MS110, HeLa cells showed nuclear speckles in the nucleoplasm on the periphery of nucleoli, or within the boundaries of nucleoli (Figure 8A). HeLa cells stained with the K-18 BRCA1 rabbit polyclonal antibody show punctate granular and nucleolar staining (Figure 8B). Simultaneous MS110 and K-18 staining showed colocalization in the nucleoplasm and the nucleoli (Figure 8D). When we simultaneously stained HeLa cells with BRCA1 and nucleolin antibodies, the localization of BRCA1 protein (green, Figure 8E) and nucleolin (red, Figure 8F) occurred frequently in nucleoli (yellow, Figure 8H).
Figure 8.
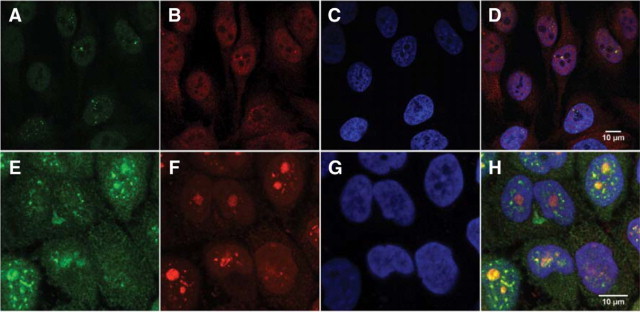
BRCA1 protein localization in HeLa cells. A: HeLa cells stained with the mouse monoclonal MS110 antibody (green) show nuclear speckles that are localized in the nucleoplasm on the periphery of nucleoli, although some are superimposed above nucleoli. B: HeLa cells stained with the rabbit polyclonal K-18 antibody (red) show nuclear speckles and nucleolar staining. C: DAPI staining of HeLa cells. D: Simultaneous MS110 and K-18 staining, counterstained with DAPI, shows colocalization (yellow) in the nucleoplasm and nucleoli. Scale bar = 10 μm. E: The rabbit polyclonal K-18 BRCA1 antibody stains many cell nuclei (green), as well as a BRCA1-staining centrosome. F: The mouse monoclonal antibody to nucleolin stains nucleoli (red). G: HeLa cells counterstained with DAPI. H: The overlay of BRCA1, nucleolin, and DAPI shows the colocalization of BRCA1 (green), merging with nucleolin (red) to show colocalization (yellow) in nucleoli and speckles in some HeLa cell nuclei In this asynchronous HeLa cell culture, however, not all of the nucleoli show BRCA1 colocalization with nucleolin; some nucleoli remain lightly stained with nucleolin, although the centrosome shows BRCA1 staining. Scale bar = 10 μm.
We obtained further validation of our results using siRNA-mediated knockdown of BRCA1 (Figure 9). Simultaneous BRCA1 and nucleolin staining were used in the siRNA experiments. Compared with the intense punctate nuclear staining of HeLa cells transfected with pcDNA3.1 GFP (Figure 9A), or the pSUPER GFP plasmid (Figure 9B), cells transfected with the pSUPER HBRCA1 plasmid show much diminished BRCA1 protein staining (Figure 9C). Colocalization of BRCA1 and nucleolin staining (yellow) was most pronounced in the control cells transfected with pcDNA3.1GFP (Figure 9A), compared the pSUPER GFP plasmid (Figure 9B), in which there was less overlap. We normalized the panel by selecting images with the same intensity settings.
Figure 9.
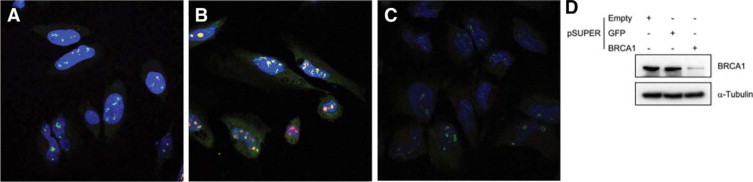
Confocal and biochemical analysis of siRNA-mediated knockdown of BRCA1 protein in HeLa cells. A: Cells transfected with plasmid pcDNA3.1 GFP show punctate nuclear staining for BRCA1 protein (green), and for nucleolin (red), which is predominantly colocalized (yellow). Original magnification ×630. B: Cells transfected with the pSUPER GFP plasmid show punctate nuclear staining, in which BRCA1 (green) staining is sometimes, but not always colocalized with nucleolin (yellow). Original magnification ×630. C: BRCA1 protein staining was diminished in cells transfected with the pSUPER HBRCA1 plasmid; counterstained with DAPI. Original magnification ×630. D: The 220-kDa BRCA1 protein is significantly knocked down in whole cell lysates from HeLa cells transfected with pSUPER HBRCA1 plasmid (lane 3); whereas the 220-kDa BRCA1 protein is clearly visible in control cells transfected with empty pSUPER plasmid (lane 1), or pSUPER GFP plasmid (lane 2). The loading control α-tubulin, shows equivalent concentrations in the lysates, and is not diminished in the siRNA-HBRCA1-treated cells.
We confirmed the knockdown of BRCA1 protein shown by immunofluorescence by Western blotting (Figure 9D). There is a clearly visible 220 kDa BRCA1 protein in cells transfected with empty vector, or pSUPER GFP plasmid, and much less staining of the 220-kDa BRCA1 protein in whole cell lysates from HeLa cells transfected with pSUPER HBRCA1 plasmid. The loading control, α-tubulin, shows equivalent concentrations in the lysates, and is not diminished in the siRNA-HBRCA1-treated cells.
Discussion
In the present study, we have extended our immunoperoxidase data that BRCA1 is localized in the nucleus and nucleolus in breast carcinoma tissues, and MCF7 cells,13,14 to show that BRCA1 protein and nucleolin colocalize in nucleoli and a subset of nuclear speckles. With photon microscopy of immunoperoxidase-stained, estrogen-stimulated MCF7 cells, we observed enhanced BRCA1 staining in the nucleus and the cytoplasm. We further resolved BRCA1 protein localization with transmission electron microscopy; and found labeled nuclear dots around nucleoli, sometimes in close contact with nucleolus fibers, or within electron dense regions corresponding to GCs of nucleoli, and in multivesicular bodies in the cytoplasm, close to the Golgi apparatus.
We studied and compared staining using the monoclonal BRCA1 antibodies MS110, MS13, AP16, and SG11, as well as the polyclonal rabbit K-18 antibody. In addition, we tested −20°C methanol and paraformaldehyde fixation procedures. The AP16 antibody provided the darkest staining. In a previous light and electron microscopic study of estrogen-stimulated MCF7 cells14 using an extensive array of BRCA1 antibodies, we have shown a nuclear, nucleolar, and cytoplasmic BRCA1 localization. However, Coene et al16 and Hadjisavvas et al26 found nuclear staining, but failed to observe nucleolar localization by light and electron microscopy. Since we found varying numbers of tumor cell nuclear staining with the different antibodies in sequential sections from the same tumor, it is likely that the differences can be ascribed to differences in accessibility of the BRCA1 epitopes rather than antibody specificity. The almost fivefold increase in BRCA1 levels due to estrogen treatment of MCF7 cells14 would explain the enhanced BRCA1 staining with the MS110 antibody on paraformaldehyde-fixed, triton-permeabilized, estradiol-treated MCF7 cells. We conclude that a combination of variables, including differences in the antibodies, fixation, permeabilization, cell origin, and hormonal treatment, may explain the differences in our results from previous data. Nonetheless, the verification of BRCA1 localization in nuclei and nucleoli by light and electron microscopy using different fixation protocols and antibodies argues strongly for the specificity of the antibody staining.
We addressed questions about the validation of the rabbit polyclonal K-18 antibody by pre-absorption with peptide, and showed colocalization of the MS110 and K-18 antibodies in nuclei and nucleoli. In addition, Aglipay et al27 using the rabbit polyclonal K-18 antibody have shown that a BRCA1-interacting protein, IF116, colocalizes with nucleolin; however, they did not show colocalization of BRCA1 and nucleolin. De Petrocellis et al21 also used the K-18 antibody to show that the endogenous cannabinoid, anandamide, inhibits human breast cancer cell proliferation, and a change in BRCA1 protein, mediated through the prolactin receptor. The overlap in staining between the MS110, AP16, and K-18 BRCA1 antibodies in nuclei and nucleoli, in a series of experiments, indicates specificity in BRCA1 protein localization.
In our present studies, we found that BRCA1 protein colocalized with nucleolin in nucleoli and a subset of speckles in asynchronously growing MCF7 cells. Using LSC, we also demonstrated that BRCA1 is found in single “spots” corresponding to nucleoli in G1 and S phases of the cell cycle and that nucleolin is maximally expressed in single “spots” in a greater percentage of MCF7 cells during G1 and S phases. This partial synchronicity of BRCA1 and nucleolin expression in nucleolar localization and cell cycle progression suggests cell cycle-dependent nucleolar BRCA1 protein shuttling.
To ascertain the changes in BRCA1 localization due to DNA damage, we subjected MCF7 cells to γ-irradiation. After 1 hour γ-irradiation of MCF7 cells, we found that BRCA1 is predominantly re-localized to coarse granular foci, rather than to nucleoli and speckles. BRCA1, which had colocalized with nucleolin in nucleoli and subsets of speckles in the absence of damage was no longer colocalized after irradiation, although nucleolin appeared to remain in nucleoli. Scully et al12,20 found that BRCA1 localizes to discrete nuclear foci (dots) during S phase in human breast and ovarian cancer cells, but that S phase nuclear foci underwent a major structural change in the face of genotoxic insult, such as hydroxyurea or UV-irradiation and appeared to disperse to foci or sites of “abnormal” (nonduplex) DNA structure in S phase cells associated with damaged replicating DNA.
It is important to clarify some confusion in terminology between “nuclear granules” and “nuclear speckles.” In our data on untreated MCF7 cells, we describe nuclei in which BRCA1 and nucleolin colocalize in nucleoli and in some but not all “nuclear speckles.” However, BRCA1 immunoperoxidase staining with the MS110 antibody on MCF7 cells using light microscopy show numerous “granules staining in the nucleus,” which may include tiny nucleoli that form during telophase, and rapidly coalescence to one large nucleolus during G1 and S phases,28 which would correspond to the electron microscopy results. Using immunofluorescence and immunoelectron microscopy, Shiohama et al29 have described the nucleolar protein, DGCR8, which colocalizes with nucleolin in the nucleolus, and in “nucleolin speckles” in the nucleoplasm. It is thus possible that the BRCA1-nucleolin complexes, which we describe as “nuclear speckles” might comprise a subset resembling the “nucleolin speckles,” which are small RNA containing foci in the nucleoplasm, adjacent to “splicing speckles” and would require further studies to analyze.
Monneron and Bernhard30 described these latter nucleoplasmic bodies as interchromatin granule clusters, which were associated with anti-Sm and anti-SC-35 antibodies.31
In our studies of BRCA1 protein localization and siRNA-mediated knockdown of BRCA1 protein in HeLa cells, we found diminished BRCA1 immunofluorescence staining in nuclei and nucleoli in cells treated with the siRNA against HBRCA1, but not in control transfections. Western blotting confirmed these results; there is a clearly visible 220-kDa BRCA1 protein in cells transfected with empty plasmid, or pSUPER GFP plasmid, and much less staining in whole cell lysates from HeLa cells transfected with pSUPER HBRCA1 plasmid. These data provide further evidence for the specificity of BRCA1 protein immunostaining.
The functional significance of BRCA1 protein localization in the nucleolus and nuclear speckles remains to be fully elucidated; however, BRCA1 protein localization in the nucleolus may suggest other possible roles. The mammalian nucleolus can be morphologically divided into several discrete regions-the fibrillar center, which contains the ribosomal genes residues, the dense fibrillar center where ribosomal (r)RNA processing and ribosome assembly occurs, and the GCs, which contains preribosomes in the final stages of maturation before translocation to the cytoplasm.32,33 Although our electron microscopy data showed that BRCA1 protein was localized in the GCs, the precise role cannot be inferred because many nucleolar proteins are involved in coordinating and regulating cell-cycle control events and stress responses and do not necessarily have functions in ribosome biogenesis.34,35
The finding that BRCA1 protein localizes in the nucleolus might also suggest other possible functions in mRNA and rRNA biogenesis and repair of ribosomal genes. Literature curation reveals that BRCA1 interacts with over 65 proteins with 30 of them being classified by Gene Ontology Slim (http://www.informatics.jax.org/gotools/MGI_GO_Slim_Chart.html) as involved in RNA metabolism processes. This category is enriched 2.6-fold when compared with the full set of genes in the genome (Monteiro, unpublished). Interestingly, 15 BRCA1-interacting proteins were found to be components of the nucleolus by proteomic analysis.36 Many of these (such as SMARCA4, RBBB4, HDAC1, HDAC2, and DHX9) are involved in regulation of mRNA transcription and chromatin remodeling. Thus, it is likely that some functions performed by BRCA1 in the nucleolus reflect its prominent role in the regulation of transcription.37 Additionally, Guo et al38 have shown that Flap endonuclease 1 (FEN 1), a DNA repair protein involved in the resolution of stalled replication and homologous recombination, is enriched in nucleoli (among others such as WRN, BLM, XPG, and Rad 52) and may be involved in the repair of ribosomal genes.
Our immunohistochemical results show that BRCA1 colocalizes with nucleolin, and thus provide evidence for its possible involvement in processes of ribosome biogenesis, regulation of transcription by RNA polymerase I, modulation of chromatin conformation in the nucleolus, and binding to nascent rRNA.22 The notion that BRCA1 may play a role in ribosomal biogenesis is supported by evidence that BRCA1 has been shown to interact with RNA pol II39 via RPB2 and RPB10,40 which are also found in RNA pol I41 although the presence of BRCA1 in RNA pol I complexes has not been formally demonstrated.
Hölzel et al42 have demonstrated that a BRCA1 C-terminal domain within the mammalian Pes1 protein (which also exist in the yeast Pes1 homologue Nop7p)43 is quite unique for an rRNA processing factor. A significant number of documented BRCA1 C-terminal domain missense mutations tested in vitro lead to protein destabilization and may be associated with disease.44 A defect in the nucleolar localization and the loss of function of the BRCA1 protein in rRNA processing due to nonsense mutations, which remove residues of the second BRCA1 C-terminal domain repeat, could also contribute to the onset of breast cancer.
BRCA1 protein has also been shown to be involved in regulation of p53-dependent gene expression, and to interact with p53.45,46 Interestingly, p53 inhibits pol I-dependent gene expression and rRNA synthesis.47,48 Another human nucleolar protein, P14ARF activates p53-dependant checkpoints by binding to Mdm2, a p53 antagonist.49,50 These reported interactions between BRCA1, p53, Mdm2, and p14ARF proteins and our data on the interaction of BRCA1 and nucleolin give credence to a possible BRCA1 function that, by inhibiting rRNA synthesis, would reduce cell growth and proliferation, consistent with its tumor suppressor properties Our data on BRCA1 protein localization in the nucleolus would make it further possible to visualize and evaluate interconnections and regulations in these different pathways.
Acknowledgements
We dedicate this paper to the memory of Rafael Mira y Lopez, colleague and friend, whose humanity, kindness, and high standards, are sorely missed.
We thank Jean-Yves Cance for artwork and Michel Gleizes for cell culture and immunocytochemical preparation. The electron transmission microscopy work was performed at the CRIC (Centre Régional d’Imagerie Cellulaire–Montpellier, France), and we thank Florence Tribillac for ultrathin sections, Paul Paulet for electron microscopy photography, and Jérôme Taki for technical assistance on the electron transmission microscope. We thank Joseph Samet for help with photography and computer graphics, and Onur Kara for technical assistance.
Footnotes
Supported by Inserm and Université de Montpellier I. Confocal microscopy was performed at the MSSM-Microscopy Shared Research Facility supported, in part, with funding from NIH-NCI shared resources grant (1R24CAO95823-01). Work in the Monteiro Lab is funded by NIH grants CA92309 and CA116167 and by grants from the Florida Breast Cancer Coalition Foundation.
References
- 1.Newman B, Mu H, Butler LM, Millikan RC, Moorman PG, King MC. Frequency of breast cancer attributable to BRCA1 in a population based series of American women. JAMA. 1998;279:915–921. doi: 10.1001/jama.279.12.915. [DOI] [PubMed] [Google Scholar]
- 2.Miki Y, Swensen J, Shattuck-Eidens D, Futreal A, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal JA, Tran T, McClure M, Frye C, Hattier T, Phelps R, Haugen-Strano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Christian W, Bogden R, Dayananth P, Ward J, Tonin P, Narod S, Bristow PK, Norris FH, Helvering L, Morrison P, Rosteck P, Lei M, Barrett JC, Lewis C, Neuhausen S, Cannon-Albright L, Goldgar D, Wiseman R, Kamb A, Skolnick MH. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 3.Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppa-Lyonnet D, Belanger C, Bell R, Berry S, Bogden R, Chen Q, Davis T, Dumont M, Frye C, Hattier T, Jammulapati S, Janecki T, Jiang P, Kehrer R, Leblanc JF, Mitchell JT, McArthur-Morrison J, Nguyen K, Peng Y, Samson C, Schroeder M, Snyder SC, Steele L, Stringfellow M, Stroup C, Swedlund B, Swense J, Teng D, Thomas A, Tran T, Tranchant M, Weaver-Feldhaus J, Wong AK, Shizuya H, Eyfjord JE, Cannon-Albright L, Tranchant M, Labrie F, Skolnick MH, Weber B, Kamb A, Goldgar DE. The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet. 1996;12:333–337. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 4.Merajver SD, Frank TS, Xu J, Pham T, Calzone KA, Bennett-Baker P, Chamberlain J, Boyd J, Garber JE, Collins FS, Weber BL. Germline BRCA1 mutations and loss of the wild-type allele in tumors from families with early onset breast and ovarian cancer. Clin Cancer Res. 1995;1:539–544. [PubMed] [Google Scholar]
- 5.Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett LM, Haugen-Strano A, Swensen J, Miki Y, Eddington K, McClure M, Frye C, Weaver-Feldhaus J, Ding W, Gholami Z, Soderkvist P, Terry L, Jhanwar S, Berchuck A, Iglehart D, Marks J, Ballinger DG, Barrett JC, Skolnick MH, Kamb A, Wiseman R. bRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 6.Ozcelik H, To MD, Couture J, Bull SB, Andrulis IL. Preferential allelic expression can lead to reduced expression of BRCA1 in sporadic breast cancers. Int J Cancer. 1998;77:1–6. doi: 10.1002/(sici)1097-0215(19980703)77:1<1::aid-ijc1>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 7.Sourvinos G, Spandidos DA. Decreased BRCA1 expression levels may arrest the cell cycle through activation of p53 checkpoint in human sporadic breast tumors. Biochem Biophys Res Commun. 1998;245:75–80. doi: 10.1006/bbrc.1998.8379. [DOI] [PubMed] [Google Scholar]
- 8.Rio PG, Maurizis J-C, de Latour MP, Bignon YJ, Bernard-Gallon DJ. Quantification of BRCA1 protein in sporadic breast carcinoma with or without loss of heterozygosity of the BRCA1 gene. Int J Cancer. 1999;80:823–826. doi: 10.1002/(sici)1097-0215(19990315)80:6<823::aid-ijc5>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Rakha EA, El-Sheikh SE, Kandil MA, El-Sayed ME, Green AR, Ellis IO. Expression of BRCA1 protein in breast cancer and its prognostic significance. Hum Pathol. 2008;39:857–865. doi: 10.1016/j.humpath.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Wilson CA, Payton MN, Pekar SK, Zhang K, Pacific RE, Gudas JL, Thukral S, Calzone FJ, Reese DM, Slamon DI. BRCA1 protein products: antibody specificity. Nat Genet. 1996;13:264–265. doi: 10.1038/ng0796-264. [DOI] [PubMed] [Google Scholar]
- 11.Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- 12.Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 13.Tulchin N, Ornstein L, Bleiweiss IJ, Mendlowitz M, Weinstein E, Dikman SH. Nucleolar localization of BRCA1 protein in human breast cancer. Int J Oncol. 1998;13:513–518. [PubMed] [Google Scholar]
- 14.Chambon M, Nirde P, Gleizes M, Roger P, Vignon F. Localization of BRCA1 protein in human breast cancer cells. Breast Cancer Res Treat. 2003;79:107–119. doi: 10.1023/a:1023382223768. [DOI] [PubMed] [Google Scholar]
- 15.Lotti LV, Ottini L, D'Amico C, Gradini R, Belleudi CA, Frati L, Tomsi MR, Mariani-Costantini R. Subcellular localization of the BRCA1 gene product in mitotic cells. Genes Chromosomes Cancer. 2002;35:193–203. doi: 10.1002/gcc.10105. [DOI] [PubMed] [Google Scholar]
- 16.Coene ED, Hollinshead MS, Waeytens AT, Schelfhout VRJ, Eechaute WP, Shaw MK, Van Oostveldt PMV, Vaux DJ. Phosphorylated BRCA1 is predominantly located in the nucleus and mitochondria. Mol Biol Cell. 2005;16:997–1010. doi: 10.1091/mbc.E04-10-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganesan S, Silver DP, Greenberg RA, Avni D, Drapkin R, Miron A, Mok SC, Randrianarison V, Brodie S, Salstrom J, Rasmussen TP, Klimke A, Marrese C, Marahrens Y, Deng CX, Feunteun J, Livingston DM. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell. 2002;111:393–405. doi: 10.1016/s0092-8674(02)01052-8. [DOI] [PubMed] [Google Scholar]
- 18.Silver DP, Dimitrov SD, Feunteun J, Gelman R, Drapkin R, Lu SD, Shestakova E, Velmurugan S, DeNunzio N, Dragomir S, Mar J, Liu X, Rottenberg, Jonkers J, Ganesan S, Livingston DM. Further evidence for BRCA1 communication with the inactive X chromosome. Cell. 2007;128:991–1002. doi: 10.1016/j.cell.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 19.Association of Directors of Anatomic and Surgical Pathology Recommendations for the reporting of breast carcinoma. Mod Pathol. 1996;9:77–81. [PubMed] [Google Scholar]
- 20.Scully R, Ganesan S, Brown M, De Caprio JA, Cannistra SA, Feunteun J. Location of BRCA1 in human breast and ovarian cancer cells. Science. 1996;272:123–125. doi: 10.1126/science.272.5258.123. [DOI] [PubMed] [Google Scholar]
- 21.De Petrocellis L, Melck D, Palmisano A, Bisogno T, LaezzA C, Bifulco M, Di Marzo V. The endogenous cannabinoid anandamide inhibits human breast cancer cell proliferation. Proc Natl Acad Sci USA. 1998;95:8375–8380. doi: 10.1073/pnas.95.14.8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinol-Roma S. Association of non-ribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol Biol Cell. 1999;10:77–90. doi: 10.1091/mbc.10.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juan G, Cordon-Cardo C. Intranuclear compartmentalization of cyclin E during the cell cycle: disruption of the nucleoplasm-nucleolar shuttling of cyclin E in bladder cancer. Cancer Res. 2001;61:1220–1226. [PubMed] [Google Scholar]
- 24.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 25.Thomas JE, Smith M, Rubinfeld B, Gutowski M, Beckmann RP, Polakis P. Subcellular localization and analysis of apparent 180kD and 220kD protein of the breast cancer susceptibility gene. BRCA1 J Biol Chem. 1996;271:28630–28635. doi: 10.1074/jbc.271.45.28630. [DOI] [PubMed] [Google Scholar]
- 26.Hadjisavvas A, Mikellidou C, Zenios A, Zouvani I, Adamou A, Christodoulous C, Kyriacou K. Correlation between morphology, immunohistochemistry, and molecular pathology in hereditary and sporadic breast cancer cases. Ultrastruct Pathol. 2002;26:237–244. doi: 10.1080/01913120290104494. [DOI] [PubMed] [Google Scholar]
- 27.Aglipay JA, Lee SW, Okada S, Fujiuchi N, Ohtsuka T, Kwak JC, Wang Y, Johnstone RW, Deng C, Qin J, Ouchi T. A member of the pyrin family. IF116, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene. 2003;22:8931–8936. doi: 10.1038/sj.onc.1207057. [DOI] [PubMed] [Google Scholar]
- 28.Alberts B, Wilson J, Johnson H, Lewis J, Raff M, Roberts K. Molecular Biology of the Cell. Fifth Ed. Garland Science; 2007. p. 382. [Google Scholar]
- 29.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 30.Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastructural Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- 31.Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersen JS, Lyon CE, Fox AH, Leung AKL, Lam YW, Steen MM, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 33.Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez J-C, Greco A, Hochstrasser D, Diaz J-J. Functional proteomic analysis of human nucleolus. Mol Biol Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leung AK, Andersen JS, Mann M, Lamond AI. Bioinformatic analysis of the nucleolus. Biochem J. 2003;376:553–569. doi: 10.1042/BJ20031169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Politz JCR, Polena I, Trask I, Bazett-Jones DP, Pederson T. A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol Biol Cell. 2005;16:3401–3410. doi: 10.1091/mbc.E05-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 37.Monteiro AN. BRCA1: exploring the links to transcription. Trends Biochem Sci. 2000;25:469–474. doi: 10.1016/s0968-0004(00)01632-7. [DOI] [PubMed] [Google Scholar]
- 38.Guo Z, Qian L, Liu R, Dai H, Zhou M, Zheng L, Shen B. Nucleolar localization and dynamic roles of flap endonuclease I in ribosomal DNA replication and damage repair. Mol Biol Cell. 2008;28:4310–4319. doi: 10.1128/MCB.00200-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scully R, Anderson SF, Chao DM, Wei W, Ye L, Young RA, Livingston DM. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 1997;94:5605–5610. doi: 10.1073/pnas.94.11.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlegel BP, Green VJ, Ladias JA, Parvin JD. BRCA1 interaction with RNA polymerase II reveals a role for hRPB2 and hRB10α in activated transcription. Proc Natl Acad Sci USA. 2000;97:3148–3153. doi: 10.1073/pnas.070452397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shpakovski GV, Acker J, Wintzerith M, Lacroix JF, Thuriaux P, Vigneron M. Four subunits that are shared by three classes of RNA polymerase are functionally interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol Cell Biol. 1995;9:4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hölzel M, Grimm T, Rohrmoser M, Malamoussi A, Harasim T, Gruber-Eber A, Kremmer E, Eick D. The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Res. 2007;35:789–800. doi: 10.1093/nar/gkl1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams CC, Jakovljevic J, Roman J, Hampichamchai P, Woolford JL. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA. 2002;8:150–165. doi: 10.1017/s1355838202010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carvalho MA, Marsillac SM, Karchin R, Manoukian S, Grist S, Swaby RF, Urmenyi TP, Rondinelli E, Silva R, Gayol L, Baumbach L, Sutphen R, Pickard-Brzosowicz JL, Nathanson KL, Sali A, Goldgar D, Couch FJ, Radice P, Monteiro AN. Determination of cancer risk associated with germ line BRCA1 missense variants by functional analysis. Cancer Res. 2007;67:1494–1501. doi: 10.1158/0008-5472.CAN-06-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouchi T, Monteiro AN, August A, Aaronson SA, Hanafusa H. BRCA1 regulates p-53 dependent gene expression. Proc Natl Acad Sci USA. 1998;95:2302–2306. doi: 10.1073/pnas.95.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Somasundaram K, Peng Y, Tian H, Zhang H, Bi D, Weber BL, El-Deiry WS. BRCA1 physically associates with p53 and stimulates its transcriptional activity. Oncogene. 1998;16:1713–1721. doi: 10.1038/sj.onc.1201932. [DOI] [PubMed] [Google Scholar]
- 47.Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- 48.Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20:5930–5938. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somasundaram K, MacLachlan TK, Burns TF, Sgagias M, Cowan KH, Weber BL, el-Deiry WS. BRCA1 signals ARF-dependent stabilization and coactivation of p53. Oncogene. 1999;18:6605–6614. doi: 10.1038/sj.onc.1203284. [DOI] [PubMed] [Google Scholar]
- 50.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]


