Abstract
Autophagy is a highly conserved cellular degradation process in which portions of cytosol and organelles are sequestered into a double-membrane vesicle, an autophagosome, and delivered into a degradative organelle, the vacuole/lysosome, for breakdown and eventual recycling of the resulting macromolecules. This process relieves the cell from various stress conditions. Autophagy plays a critical role during cellular development and differentiation, functions in tumor suppression, and may be linked to life span extension. Autophagy also has diverse roles in innate and adaptive immunity, such as resistance to pathogen invasion. Substantial progress has been made in the identification of many autophagy-related (ATG) genes that are essential to drive this cellular process, including both selective and nonselective types of autophagy. Identification of the ATG genes in yeast, and the finding of orthologs in other organisms, reveals the conservation of the autophagic machinery in all eukaryotes. Here, we summarize our current knowledge about the machinery and molecular mechanism of autophagy.
1 Introduction
Autophagy, “self-eating” at the subcellular level, has gained tremendous attention in the past few years, and our knowledge concerning the mechanism of autophagy has expanded dramatically (Yorimitsu and Klionsky 2005b). There are three major types of autophagy in eukaryotic cells—macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA)—and they are mechanistically different from each other (Klionsky 2005; Massey et al. 2004). Both macro- and microautophagy involve dynamic membrane rearrangement to engulf portions of the cytoplasm, and they have the capacity for the sequestration of large structures, such as entire organelles. Microautophagy involves the direct engulfment of cytoplasm at the lysosome surface by invagination, protrusion, and septation of the lysosome membrane. In contrast, during macroautophagy, portions of cytoplasm are sequestered into a de novo-formed double-membrane vesicle termed an autophagosome. Subsequently, the completed autophagosome fuses with the lysosome/vacuole and the inner single-membrane vesicle is released into the lumen. In either case, the membrane of the resulting autophagic body is lysed to allow the contents to be broken down, and the resulting macromolecules are transported back into the cytosol through membrane permeases for reuse. In contrast, CMA does not involve a similar type of membrane rearrangement; instead, it translocates unfolded, soluble proteins directly across the limiting membrane of the lysosome.
In this chapter, we will focus on macroautophagy, hereafter referred to as autophagy. Autophagy is an evolutionarily conserved process that occurs ubiquitously in all eukaryotic cells (Reggiori and Klionsky 2002) and has many physiological roles. Autophagy is active at a basal level for the turnover of long-lived proteins and also for the removal of superfluous or damaged organelles. This latter function might provide a connection to autophagy's proposed role in life span extension (Levine and Klionsky 2004). On the other hand, autophagy is induced as a cellular response to various stress conditions, such as nutrient limitation, heat, and oxidative stress. Autophagy also plays a role in cellular development and differentiation (Levine and Klionsky 2004). Moreover, autophagy is implicated in a wide range of diseases (Huang and Klionsky 2007; Mizushima et al. 2008; Shintani and Klionsky 2004a), including cancer and neurodegenerative disorders such as Alzheimer', Parkinson' and Huntington' diseases. In addition, autophagy has diverse roles in innate and adaptive immunity (Levine and Deretic 2007). For example, autophagy can eliminate invasive pathogens, including viruses, parasites and bacteria; autophagy also promotes MHC class II presentation of microbial (and self) antigens. Finally, in the absence of apoptosis, autophagy may participate in a type of programmed cell death (type II) that is distinct from apoptosis, although the physiological relevance of the former is not clear (Levine and Yuan 2005).
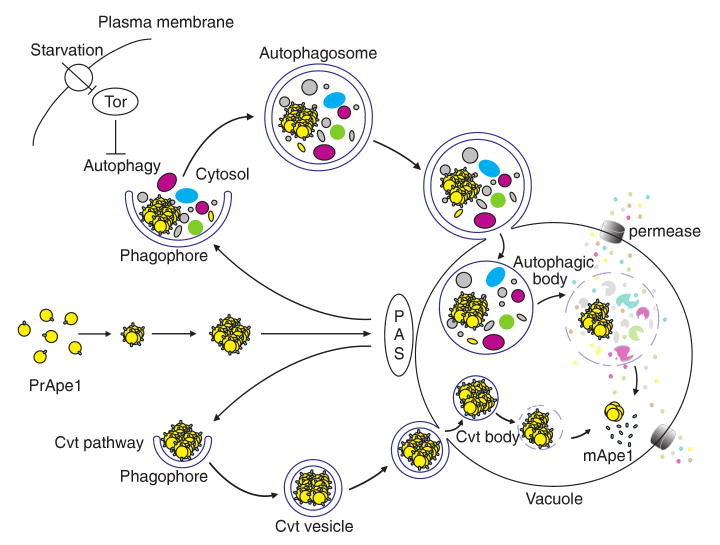
The morphology of autophagy was first identified in mammalian cells in the 1950s, and extensive morphological and pharmacological studies defined the basic steps of this process. Subsequent work in various fungi starting in the 1990s allowed the identification of individual molecular components that participate in autophagy. To date, there are 31 autophagy-related (ATG) genes (Huang and Klionsky 2007; Klionsky et al. 2003). The ATG genes were discovered from genetic screens for mutants that affected protein turnover (nonspecific autophagy), peroxisome degradation (pexophagy) and delivery of a resident vacuolar hydrolase (the cytoplasm to vacuole targeting (Cvt) pathway). Although the Cvt, pexophagy, and autophagy pathways are morphologically and mechanistically similar and share most of the Atg components, they differ in several aspects (Fig. 1). Autophagy and pexophagy are degradative, whereas the Cvt pathway is biosynthetic. Autophagy is generally considered nonselective, whereas pexophagy and the Cvt pathway are highly selective. The Cvt pathway is used to deliver two resident vacuolar hydrolases, aminopeptidase I (Ape1) and α-mannosidase (Ams1) (Hutchins and Klionsky 2001; Scott et al. 1997). A double-membrane vesicle that sequesters these two proteins is termed a Cvt vesicle; this is relatively consistent in size but significantly smaller than the autophagosome, being 140–160 nm in diameter compared to 300–900 nm for the autophagosome (Baba et al. 1997). Similarly, the vesicle formed during pexophagy, the pexophagosome, is also larger than the Cvt vesicle in order to accommodate its specific cargos, peroxisomes (Hutchins et al. 1999). In contrast to the autophagosome, both the Cvt vesicle and pexophagosome appear to closely enwrap the cargo and exclude bulk cytoplasm.
Fig. 1.
Schematic overview of autophagy and the Cvt pathway in yeast. Both pathways involve the engulfment of cargos within distinct double-membrane vesicles, which are thought to originate from the phagophore assembly site (PAS). The Cvt pathway is biosynthetic and is used for the delivery of two resident vacuolar hydrolases, aminopeptidase I (Ape1), and α-mannosidase (Ams1), and it occurs under vegetative conditions. The Cvt vesicle is approximately 140–160 nm in diameter and appears to closely enwrap the specific cargo, the Cvt complex (consisting of prApe1 and the Atg19 receptor), and exclude bulk cytoplasm. In contrast, autophagy is degradative and is induced by inactivation of Tor kinase upon nutrient starvation. The autophagosome, which is 300–900 nm in diameter, sequesters cytoplasm, including organelles, and can also specifically sequester the Cvt complex. Once completed, the double-membrane vesicles dock and fuse with the vacuole, and release the inner single-membrane vesicles (autophagic or Cvt body) into the lumen. Subsequently, these vesicles are broken down, allowing the maturation of prApe1 and the degradation of cytoplasm, with recycling of the resulting macromolecules through vacuolar permeases. This figure is modified from Fig. 1 of Yorimitsu and Klionsky (2005b)
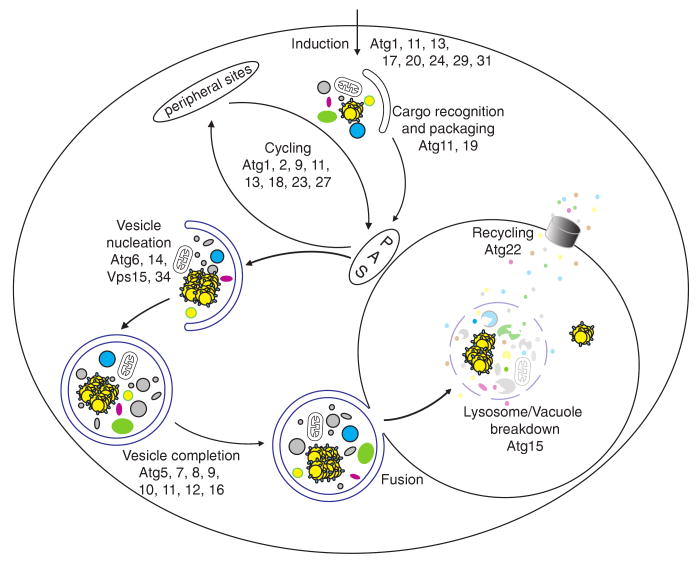
These dynamic pathways can be broken down into a series of steps (Fig. 2), including induction, cargo recognition and packaging, vesicle nucleation, vesicle expansion and completion, Atg protein cycling, vesicle fusion with the vacuole/lysosome, vesicle breakdown, and recycling of the resulting macromolecules (Huang and Klionsky 2007). Thus, the Atg proteins can be classified into several different groups according to their functions at the different steps of the pathway. Many orthologs of the ATG gene products have also been identified and studied in higher eukaryotes, such as worms, insects, plants and mammals, and they have essentially similar roles as those in yeast (Xie and Klionsky 2007; Yorimitsu and Klionsky 2005b). Continued investigation of functions of the ATG gene products in yeast will greatly expand our understanding of autophagy. In this chapter, we will mainly discuss the molecular machinery of autophagy, with an emphasis on yeast.
Fig. 2.
Schematic representation of autophagy and autophagy-related pathways. These dynamic pathways can be broken down into a series of steps including induction, cargo recognition and packaging, vesicle nucleation, vesicle expansion and completion, Atg protein cycling, vesicle fusion with the vacuole/lysosome, vesicle breakdown and recycling of the resulting macromolecules. The Atg proteins can be classified into several different groups according to their functions at the different steps of the pathway. The Atg1 complex may act at multiple steps of the pathway, including induction and Atg protein cycling. During the vesicle formation process, several Atg proteins are involved in cycling between the peripheral sites and the PAS. PAS, phagophore assembly site; thought to be the organizing site for phagophore formation. This figure is modified from Fig. 2 of Huang and Klionsky (2007)
2 Molecular Mechanism of Autophagy
2.1 Induction and Regulation of Autophagy
Insufficient autophagy can be deleterious (Komatsu et al. 2007a; Kuma et al. 2004), but excessive levels may also be harmful. Accordingly, autophagy is a tightly regulated process in all eukaryotes. The induction and regulation of autophagy have been studied extensively in yeast, mammalian cells and Drosophila. Several signaling pathways, as summarized in the following, are involved in the control of autophagy.
TORC1
The protein target of rapamycin, Tor, plays a major regulatory role in autophagy induction (Carrera 2004). Tor forms two functionally distinct protein complexes, Tor complex 1 and 2 (TORC1 and TORC2) (Loewith et al. 2002), and TORC1 has the primary role in regulating autophagy. Under nutrient-rich conditions, TORC1 is active and inhibits autophagy, whereas TORC1 is inhibited upon nutrient deprivation, allowing an increase in autophagic activity (Noda and Ohsumi 1998).
In yeast, TORC1 acts on autophagy in two ways (Klionsky 2005). First, TORC1 regulates the Atg1–Atg13–Atg17 kinase complex (Fig. 3a). The formation of this ternary complex correlates with an increase in autophagic activity. Atg1, a serine/threonine kinase, is one of the key Atg proteins required for both autophagy and the Cvt pathway (Matsuura et al. 1997). Based on yeast two-hybrid data and affinity isolation, Atg1 is found to be in a complex with Atg13 and Atg17 (Kamada et al. 2000; Kabeya et al. 2005). The observation that Atg17 interacts with Atg13 in the absence of Atg1 but not vice versa suggests that Atg13 mediates the interaction between Atg1 and Atg17. TORC1 regulates (directly or indirectly) the Atg13 phosphorylation state. Under nutrient-rich conditions, Atg13 is highly phosphorylated, and has a lower affinity for Atg1 and Atg17. Upon the inactivation of TORC1 by rapamycin or nutrient deprivation, Atg13 is rapidly and partially dephosphorylated, leading to a higher affinity for Atg1 and Atg17. The identities of the phosphatase(s) that control Atg13 phosphorylation are currently unknown. The interaction of Atg1 with hypophosphorylated Atg13 and Atg17 allows the activation of Atg1 kinase activity. Loss of interaction between Atg1 and Atg13 or between Atg13 and Atg17 leads to a decrease in Atg1 kinase activity and decreased autophagy. The kinase activity of Atg1 is essential for both autophagy and the Cvt pathway, although a higher level of kinase activity appears to be needed for the Cvt pathway (Kamada et al. 2000; Kabeya et al. 2005; Cheong et al. 2008; Abeliovich et al. 2003). It is possible that the kinase activity of Atg1 is critical to the magnitude of autophagy but not its initiation (Nair and Klionsky 2005). The downstream substrate of Atg1 kinase is unclear, and it is still a matter of debate as to whether Atg1 primarily acts on autophagy through its kinase activity or through a structural role during autophagic complex formation. However, one role of the Atg1–Atg13–Atg17 ternary complex is thought to be that of regulating the switch between autophagy and the Cvt pathway in response to environmental changes.
Fig. 3a–c.

Regulation of autophagy induction in yeast and mammalian cells. a Regulatory complex for autophagy induction in yeast. In yeast, autophagy is mainly a starvation response, and Tor kinase complex 1 (TORC1) regulates the induction of autophagy upon sensing the nutrient conditions. Atg1 kinase, which is essential for both autophagy and the Cvt pathway, forms a putative complex with several Atg proteins that are primarily required for autophagy (in green) or the Cvt pathway (in purple). Under nutrient-rich conditions, TORC1 is active and Atg13 is highly phosphorylated, and this hyperphosphorylated Atg13 has a lower affinity for Atg1 and Atg17. Upon inactivation of TORC1 by nutrient starvation, Atg13 is rapidly and partially dephosphorylated, leading to a higher affinity for Atg1 and Atg17. The formation of the Atg1–Atg13–Atg17 ternary complex allows the activation of Atg1 kinase activity, which may regulate the switch between autophagy and the Cvt pathway in response to environmental changes. The function of additional components of the putative complex depicted here, including Atg20, Atg24, Atg29, Atg31 and Vac8, are not known. Atg11 may function in part as a scaffold protein. This figure is modified from Fig. 2 of Yorimitsu and Klionsky (2005b) b Multiple nutrient-sensing kinase signaling pathways converge on autophagy in yeast. TORC1 plays a major role in the regulation of autophagy. Ras is active under nutrient-rich conditions and allows the activation of PKA, which inhibits autophagy. The PKA and Sch9 signaling pathways cooperatively regulate the induction of autophagy in parallel with Tor, although Sch9 is also a direct substrate of TORC1. The eIF2α kinase signaling pathway positively regulates autophagy, and Gcn2 might be another target of TORC1. Snf1 and Pho85 are additional positive and negative regulatory components, respectively, of autophagy in yeast. c Regulation of autophagy in mammalian cells. mTor activation depends on several inputs, including nutrients (amino acids), energy (ATP) and growth factor (insulin/IGF). In response to insulin receptor stimulation, a class I phosphoinositide 3-kinase (PtdIns3K) is activated and generates PtdIns(3,4)P2 and PtdIns(3,4,5)P3 at the plasma membrane, and the latter two activate 3-phosphoinositide-dependent protein kinase 1 (PDK1) and protein kinase B (PKB)/Akt. PKB phosphorylates and inhibits the GTPase-activating protein complex TSC1–TSC2, leading to the stabilization of Rheb-GTP, which stimulates mTor, causing inhibition of autophagy. PTEN, a 3′-phosphoinositide phosphatase, antagonizes PKB and has a stimulatory effect on autophagy. Both mTor and PDK1 stimulate p70S6 kinase (p70S6K). In one model, under nutrient-rich conditions, activation of S6K directly stimulates autophagy, or it is stimulated indirectly through inhibition of PtdIns3K, allowing a basal level of autophagy for homeostatic purposes. Under starvation conditions, inhibition of mTor prevents further activation of S6K, which limits and prevents excessive autophagy. Both ATP and amino acid deprivation result in mTor inactivation independent of the insulin signaling pathway. Amino acids activate mTor via inhibition of the TSC1–TSC2 complex or are sensed by mTor directly. Energy stress causes activation of the LKB1–AMPK pathway, which inhibits mTor by activating TSC1–TSC2. AMPK phosphorylates and stabilizes p27, a cyclin-kinase inhibitor, leading to activation of autophagy. An antiapoptotic protein, Bcl-2, associates with Beclin 1, the mammalian homolog of Atg6, and inhibits a class III PtdIns3K complex, whereas the latter serves a stimulatory role in autophagy. Also shown is the notion that Atg1 overexpression negatively feeds back on Tor activity in Drosophila
Homologs of Atg1 are involved in autophagy in various multicellular organisms such as Dictyostelium discoideum (Otto et al. 2004), Drosophila melanogaster (Scott et al. 2004), C. elegans (Melendez et al. 2003), Arabidopsis thaliana (Hanaoka et al. 2002), and mammals (Yan et al. 1998, 1999). In Drosophila, Atg1 activity is modulated by TORC1 as in yeast, because the induction of autophagy that results from the overexpression of Atg1 is suppressed when the TORC1 signaling pathway is impaired (Scott et al. 2007). Normally, a feedback mechanism may occur in which Atg1 downregulates Tor activity, resulting in a further activation of Atg1 and a further increase in autophagy (Fig. 3c). Because these studies are based on over-expressed Atg1, however, the physiological significance is not clear at present. In mammals, the uncoordinated 51-like kinases 1 and 2 (Ulk1 and Ulk2) appear to be the functionally equivalent mammalian homologs of yeast Atg1. Knockdown of ULK1 inhibits the induction of autophagy by rapamycin treatment, indicating that Ulk1 functions downstream of mTOR in autophagy regulation (Chan et al. 2007). In contrast to the result in Drosophila, overexpression of ULK1 or ULK2 suppresses autophagy. Furthermore, moderate expression of kinase-dead ULK mutants also efficiently suppresses autophagy, indicating that kinase activity of the Ulk proteins is critical during this process (Hara et al. 2008). FIP200 is a recently identified Ulk-interacting protein that is required for autophagy (Hara et al. 2008). Ulk and FIP200 function together and form a complex that is essential during an early step in autophagosome formation; FIP200 is thus thought to be a counterpart of yeast Atg17. Further identification and analysis of a functional homolog of mammalian Atg13 might help to clarify the functional relationship between the yeast and mammalian Atg1 complex.
Second, TORC1 acts through its downstream effectors to control autophagy. Several, but not all, TORC1 readouts, including autophagy, are regulated through protein phosphatase type 2A (PP2A) and/or 2A-related protein phosphatase (Sit4) (De Virgilio and Loewith 2006). PP2A and Sit4 are in distinct complexes containing Tap42. Under nutrient-rich conditions, Tap42 is phosphorylated and tightly associates with PP2A and Sit4. Starvation or rapamycin treatment causes dephosphorylation and dissociation of Tap42 or a change in conformation, resulting in the activation of Sit4. TORC1 may directly phosphorylate Tap42, or it may indirectly regulate Tap42 via Tip41. Upon the inactivation of TORC1, Tip41 is dephosphorylated and has a high affinity for Tap42, resulting in the inhibition of the latter. One report suggests that Tap42 does not transmit the signal from TORC1 to regulate autophagy (Kamada et al. 2000). However, more recent data indicate a role for Tap42 in the negative regulation of this process (Yorimitsu et al. 2009).
The conserved Tor protein in mammalian cells (mTor) also senses nutrient status and modulates autophagy, but the mechanism of regulation is more complex than in fungi, which are not responsive to hormones. As shown in Fig. 3c, the regulatory cascade upstream of mTor includes an insulin receptor, insulin-receptor substrates 1 and 2, class I phosphoinositide 3-kinase (PtdIns3K), 3-phosphoinositide-dependent protein kinase 1 (PDK1), and protein kinase B (PKB)/Akt (Meijer and Codogno 2006). mTor activity is controlled by the heterodimer TSC1–TSC2, which acts as a GTPase-activating protein (GAP) for the GTPase Rheb. The GDP-bound form of Rheb inhibits mTor, whereas the GTP-bound form stimulates the enzyme. PKB phosphorylates and inhibits the TSC1–TSC2 complex, leading to the activation of mTor signaling. PTEN, a 3′-phosphoinositide phosphatase, antagonizes PKB, and has a stimulatory effect on autophagy (Arico et al. 2001). The best characterized signaling pathway, located downstream of mTor, includes components such as ribosomal subunit S6 kinase (p70S6K). In one model, S6K exerts a negative feedback on mTor signaling by phosphorylating IRS1 to downregulate insulin signaling, leading to a decline in PtdIns(3,4,5)P3, an inhibitor of autophagy; this feedback regulation may ensure a basal level of autophagy even under nutrient-rich conditions (Klionsky et al. 2005).
Ras/cAMP-dependent protein kinase A (PKA)
In addition to TORC1, the Ras/PKA signaling pathway also regulates autophagy from yeast to mammals (Budovskaya et al. 2004; Furuta et al. 2004; Mavrakis et al. 2006; Schmelzle et al. 2004; Yorimitsu et al. 2007). Under nutrient-rich conditions in yeast, two redundant small GTPases (Ras1 and Ras2) are activated, and stimulate adenylyl cyclase to produce cAMP. cAMP binds to the PKA regulatory subunit (Bcy1) and allows its dissociation from the three PKA catalytic subunits (Tpk1, Tpk2, and Tpk3), resulting in the activation of PKA (Thevelein and de Winde 1999). Constitutive activation of PKA through a dominant hyperactive allele of RAS2, RAS2G19V, or deletion of BCY1 prevents the induction of autophagy by nutrient starvation or rapamycin, whereas inactivation of PKA by a dominant negative allele of RAS2, RAS2G22A, induces autophagy under rich conditions without rapamycin (Budovskaya et al. 2004; Schmelzle et al. 2004). Thus, in addition to TORC1, Ras/PKA is another negative regulator of autophagy (Fig. 3b). Among the Atg proteins, Atg1, Atg13, Atg18 and Atg21 contain PKA phosphorylation sites. However, it is still unclear whether the phosphorylation of these Atg proteins by PKA has any functional link to autophagy (Budovskaya et al. 2005).
Sch9 is a homolog of mammalian PKB or p70S6 kinase (Urban et al. 2007). A recent report shows that PKA and Sch9 signaling pathways cooperatively regulate the induction of autophagy (Yorimitsu et al. 2007). Simultaneous inactivation of PKA and Sch9 triggers the induction of autophagy under rich conditions independent of effects on TORC1, whereas further inactivation of TORC1 causes an additive effect. These observations suggest a model wherein PKA, Sch9, and TORC1, at least in part, regulate autophagy in parallel (Fig. 3b). This model is supported by the finding that TORC1 and Ras/PKA function as two parallel pathways that independently act in regulating cell growth (Zurita-Martinez and Cardenas 2005). However, Sch9 is a direct substrate of TORC1 (Urban et al. 2007); furthermore, it is also suggested that TORC1 transmits signals through the Ras/PKA pathway to its downstream targets (Schmelzle et al. 2004). Therefore, the connection between PKA, Sch9, and TORC1 with regard to their effects in autophagy regulation is still not clear.
eIF2α kinase signaling and GCN4 general control
In response to amino acid starvation, budding yeast initiates a general amino acid control to induce the transcription of numerous genes. Central to this response is Gcn4, a master transcriptional activator of gene expression (Hinnebusch 2005). Gcn4 synthesis is mainly regulated at the translational level. Derepression of GCN4 mRNA translation requires a protein kinase, Gcn2, whose only known substrate is the α subunit of translation initiation factor 2 (eIF2). The eIF2α kinase signaling pathway is also involved in the regulation of autophagy from yeast to mammals (Fig. 3b) (Talloczy et al. 2002). Upon the loss of Gcn2 or Gcn4, or in the presence of the eIF2α nonphosphorylatable mutant SUI2-S51A, autophagic activity is impaired. Intriguingly, TORC1 is implicated in the eIF2α kinase signaling pathway because rapamycin activates Gcn2, at least in part, through dephosphorylation of Ser577 (Kubota et al. 2003). Thus, Gcn2 might be another target of TORC1.
Other signaling pathways controlling autophagy
Snf1, the closest yeast homolog of the mammalian AMP-activated protein kinase, and Pho85, a cyclin-dependent kinase (CDK), antagonistically control autophagy in yeast (Fig. 3b) (Wang et al. 2001b). Snf1, which is activated upon glucose depletion to allow transcription of glucose-repressed genes, is required for starvation-induced autophagy. Pho85, which has multiple functions through associations with its ten different cyclins (Pcls), is a negative regulator of autophagy, although the functions of the various Pcl proteins and the pathways that they regulate are currently unknown (Carroll and O'Shea 2002).
In mammalian cells, AMPK is also required for autophagy (Meley et al. 2006). During energy stress, AMP accumulation causes activation of the LKB1-AMPK pathway, which inhibits mTor by activating TSC1/TSC2 (Hoyer-Hansen and Jaattela 2007). Furthermore, AMPK phosphorylates p27, a cyclin-kinase inhibitor, thereby stabilizing p27, whereas ectopic expression of wild-type or a stabilized p27 mutant induces autophagy (Liang et al. 2007).
2.2 The Cvt Pathway and Other Selective Types of Autophagy
Although autophagy is generally considered to be a nonselective pathway for the degradation of bulk cytoplasmic components, recent findings indicate that there are many types of selective autophagy in both yeast and higher eukaryotes. In yeast, even bulk autophagy can be selective; cytosolic acetaldehyde dehydrogenase, Ald6, is preferentially sequestered into autophagosomes relative to other cytosolic proteins (Onodera and Ohsumi 2004). Several organelles are selectively degraded through autophagy. For example, the selective degradation of mitochondria is termed mitophagy (Kim et al. 2007). This type of selective process is thought to play a crucial role in mitochondrial homeostasis; however, the mechanism underlying mitophagy remains unclear. The use of electron microscopy to observe mitochondrial degradation indicates that mitophagy occurs both selectively and nonselectively. A recent report demonstrates that mature ribosomes are rapidly degraded by autophagy in yeast through a process termed ribophagy. This degradation involves a type of selective autophagy in that it specifically requires catalytic activity of the Ubp3/Bre5 ubiquitin protease (Kraft et al. 2008).
In fungi such as Saccharomyces cerevisiae, Pichia pastoris, Hansenula polymorpha, and Yarrowia lipolytica, peroxisomes are selectively engulfed and degraded through two morphologically distinct autophagic degradation processes, micro- and macropexophagy (Gunkel et al. 1999; Hutchins and Klionsky 2001; Sakai et al. 2006; Tuttle et al. 1993; Veenhuis et al. 1983). When fungi grow on specific carbon sources, such as oleic acid or methanol, peroxisome proliferation is induced to adapt to the new physiological conditions that require peroxisome metabolism. When peroxisome proliferation becomes unnecessary and peroxisomes become superfluous (as occurs after shifting to a preferred carbon source such as glucose), peroxisomes are rapidly and specifically degraded. The two main modes of pexophagy, micro- and macropexophagy share most of the molecular components with nonspecific autophagy. However, the presence of Pex14 at the peroxisomal membrane is necessary for the specific recognition of the organelle by the macropexophagy machinery (Bellu et al. 2001). A specificity factor, Atg11, which is required for the Cvt pathway, is also essential for the selective transport of peroxisomes to the vacuole (Kim et al. 2001). A recently identified pexophagy-specific protein, PpAtg30, functions as a peroxisome receptor through interactions with PpPex3, PpPex14, PpAtg11, and PpAtg17 to deliver peroxisomes to the site for pexophagosome formation (Farre et al. 2008). Furthermore, a fully functional actin cytoskeleton is required for selective autophagy, including the Cvt pathway and pexophagy, but not for nonselective autophagy (Reggiori et al. 2005a).
The Cvt pathway is a unique type of specific autophagy. The mechanism of the selective recognition and packaging of prApe1 has been relatively well clarified (Fig. 4). The Ape1 protein is synthesized in the cytoplasm as a precursor form (prApe1) (Klionsky et al. 1992). After synthesis, prApe1 assembles into a dodecamer (Kim et al. 1997), which is further packaged into a larger oligomeric structure called the Ape1 complex (Shintani et al. 2002). The prApe1 propeptide contains vacuolar targeting information (Martinez et al. 1997; Oda et al. 1996). In addition, the propeptide also mediates the interaction between prApe1 and its receptor protein, Atg19, to form the Cvt complex in the cytosol (Scott et al. 2001). Another Cvt cargo, Ams1, also binds Atg19 via a site that is distinct from the prApe1 binding site and is concentrated at the Cvt complex (Shintani et al. 2002). The Cvt complex is subsequently enwrapped by a double membrane that forms a Cvt vesicle. The Cvt complex can be also sequestered within autophagosomes, depending on the nutrient conditions (Baba et al. 1997), but this still occurs through a selective process that involves Atg19.
Fig. 4.

Temporal order of action of cargo recognition, packaging and sequestration in the Cvt pathway. A selective type of autophagy, the Cvt pathway, specifically transports the vacuolar hydrolases precursor Ape1 and Ams1 into the vacuole. Precursor Ape1 is synthesized in the cytosol, assembled into a dodecamer, and then further packaged into a larger oligomeric structure, called the Ape1 complex. Atg19 binds to the propeptide of prApe1 to form the Cvt complex in the cytosol; Ams1 is also incorporated into this complex via binding to Atg19. Atg11 subsequently associates with Atg19, acting as an adapter to bring the Cvt complex to the phagophore assembly site or PAS, a potential site for Cvt vesicle formation. The PAS may organize the formation of the sequestering vesicle, or it may literally become the sequestering vesicle as shown. Atg11 assembles with the Cvt complex before targeting to the PAS, and it forms a homodimer or homo-oligomer at the PAS, although it is not clear whether this self-interaction occurs before or after the arrival at this site. Several Atg components, including Atg8, are recruited to the PAS independent of Atg11. Atg8 is conjugated into Atg8—PE for subsequent vesicle formation. Atg8—PE interacts with Atg19, and allows the correct incorporation of the Cvt complex into the forming vesicle. Atg19 is delivered into the vacuole together with the cargo proteins and degraded there. The scaffold protein Atg11, however, dissociates from the Cvt complex before vesicle completion, although the exact timing and mechanism of its release remain to be resolved. This figure is modified from Fig. 3 of Yorimitsu and Klionsky (2005b)
Atg11 subsequently associates with Atg19, acting like an adapter or tethering protein to bring the Cvt complex to the phagophore assembly site (PAS), a potential site for the formation of the Cvt vesicle and autophagosome. Several lines of evidence support the idea that Atg11 assembles with the Cvt complex before targeting to the PAS (Yorimitsu and Klionsky 2005a). However, how Atg11 guides the Cvt complex to the PAS is still unclear. A C-terminal coiled-coil domain of Atg11 is critical for interaction with the C terminus of Atg19, whereas the N-terminal and/or central coiled-coil domains contain information necessary for the Cvt complex to be targeted to the PAS (Yorimitsu and Klionsky 2005a). Besides Atg19, Atg11 has several other interacting partners, including Atg1, Atg9, Atg17, Atg20, and itself (Chang and Huang 2007; He et al. 2006; Yorimitsu and Klionsky 2005a). Atg9, the only characterized transmembrane protein that is required for sequestering vesicle formation, interacts with Atg11 independent of Atg1 or Atg19, suggesting that there are distinct and multiple populations of Atg11 within the cell. Atg11 homo-oligomerization may allow various Atg11 populations, along with its various interacting partners, to be delivered to the PAS (Yorimitsu and Klionsky 2005a). A point mutation (H192L) in Atg9 disrupts the interaction with Atg11, preventing movement of Atg9 to the PAS and blocking the Cvt pathway, but not bulk autophagy (He et al. 2006), in agreement with the finding that Atg11 is not required for nonspecific autophagy (Kim et al. 2001).
After the arrival of the Atg11–Atg19-cargo complex at the PAS, Atg19 interacts with Atg8—PE to allow the transfer of the Atg19-cargo complex to the forming Cvt vesicle (or autophagosome); interaction between these two proteins may ensure the incorporation of the Cvt complex into the Cvt vesicle (Shintani et al. 2002). Unlike most receptors that recycle between donor and acceptor membranes, Atg19 is delivered into the vacuole together with the cargo proteins and degraded there. The scaffold protein Atg11, however, does not appear to remain associated with the Cvt complex; rather, it is thought to be released from Atg19 after delivery to the PAS and to dissociate from the complex before vesicle formation (Kim et al. 2001). It remains unknown whether there is a role for Atg11 during the process of Cvt vesicle completion, and the exact timing and mechanism of its release remain to be resolved. However, disassembly of the homo-oligomerized Atg11 requires the Atg1–Atg13–Atg17 kinase complex (Yorimitsu and Klionsky 2005a).
Increasing evidence indicates that selective autophagy also occurs in mammals. For example, the p62/SQSTM1/sequestosome protein preferentially recognizes polyubiquitinated protein aggregates and connects these with the autophagic machinery through interaction with the Atg8 mammalian homolog, LC3 (Bjørkøy et al. 2005; Komatsu et al. 2007b). Thus, p62 could function as a receptor protein similar to Atg19 to link polyubiquitinated proteins to autophagosomes. Another recent example of selective autophagy is seen with the clearance of mitochondria and ribosomes during reticulocyte maturation (Kundu et al. 2008). In this case, Ulk1 plays a critical role in selective autophagy, but is not essential for the induction of starvation-induced bulk autophagy. Selectivity is also seen with the degradation of peroxisomes in mammalian cells (Iwata et al. 2006). Finally, some pathogens are selectively targeted by autophagy, such as Mycobacterium tuberculosis and Streptococcus pyogenes (Gutierrez et al. 2004; Nakagawa et al. 2004). It is important to note, however, that other microbes (including bacteria and viruses) regulate autophagy for their own survival (Nakagawa et al. 2004; Ogawa et al. 2005; Orvedahl and Levine 2008). Shigella, an invasive bacteria, is able to escape autophagy by secreting IcsB on the bacterial surface. The IcsB protein interacts with VirG, which prevents the latter from binding Atg5 and triggering specific autophagic sequestration (Ogawa et al. 2005).
2.3 Phosphatidylinositol 3-Kinase Complex
The class III phosphatidylinositol 3-kinase (PtdIns3K) is known to participate in various membrane trafficking events. Vps34 is the only PtdIns3K in yeast, and it forms at least two distinct complexes, complex I and II (Fig. 5). Each complex contains three common components, Vps34, Vps15, and Vps30/Atg6 (Kihara et al. 2001). The function of Vps34 is dependent on a serine/threonine kinase, Vps15, which is required for Vps34 membrane association and activity (Stack et al. 1995). The role of Vps30/Atg6 within these PtdIns3K complexes is not well understood. These three common proteins are involved in autophagy, the Cvt pathway and the sorting of carboxypeptidase Y (CPY), which is normally transported from the late Golgi to the vacuole through the CPY pathway. In addition, each complex contains another specific component, Atg14 (complex I) or Vps38 (complex II), which is thought to act as a connector between Vps30 and Vps15–Vps34. The region containing the coiled-coil domains I and II within the N-terminal half of Atg14 is responsible for the interaction between Vps34 and Vps30/Atg6. Loss of Atg14 disrupts complex I and causes a defect only in autophagy and the Cvt pathway, whereas Vps38 deletion disrupts complex II and blocks only the CPY pathway. The association of Atg14 or Vps38 confers functional specificity on the two PtdIns3K complexes by targeting Vps34 to distinct compartments, thus regulating different protein trafficking events. Vps15–Vps34 complexed with Vps30 and Atg14 localizes to the PAS, and functions in autophagy and the Cvt pathway; Vps15–Vps34 complexed with Vps30 and Vps38 localizes to endosomes, and functions in the CPY pathway (Obara et al. 2006).
Fig. 5.

Two phosphatidylinositol 3-kinase (PtdIns3K) complexes in yeast. Each complex contains three common components, Vps15, Vps34, and Vps30/Atg6. Vps34 is the PtdIns3K enzyme, and Vps15 is thought to be a regulatory component; the function of Vps30/Atg6 is not known. In addition, each complex contains another specific component, Atg14 (complex I) or Vps38 (complex II), which is thought to act as a connector between Vps30 and Vps15–Vps34. Complex I functions in autophagy and the Cvt pathway, whereas complex II acts in the Vps pathway, including the CPY and MVB pathways. This figure is modified from Fig. 5a of Yorimitsu and Klionsky (2005b)
PtdIns3K is a lipid kinase and the kinase activity of Vps34 is essential for autophagy and the Cvt pathway. One possible role of PtdIns3K is to produce PtdIns(3)P at the PAS to recruit PtdIns(3)P-binding proteins, which in turn recruit additional downstream effectors to the PAS. PtdIns(3)P is bound by proteins that have specific binding sites, such as the PX (phox homology) domain and the FYVE (for conserved in Fab1, YOTB, Vac1, and EEA1) zinc finger domain (Ellson et al. 2002; Stenmark et al. 2002). Two PX domain-containing proteins, Atg20 and Atg24, bind to PtdIns(3)P (Nice et al. 2002). These proteins are essential only for the Cvt pathway, not bulk autophagy. Their functional PX domains are necessary for membrane localization to the PAS and the endosome, which in turn depend on PtdIns3K complexes I and II, respectively. The role of endosomal localization is unknown since the CPY pathway is normal in the absence of Atg20 or Atg24; however, the endosomal localization is not necessary for Cvt transport. Atg20 and Atg24 interact with each other, and Atg24 and possibly Atg20 interact with Atg17 (Nice et al. 2002). In addition, Atg20 interacts with Atg11 (Yorimitsu and Klionsky 2005a). Thus, the Atg20–Atg24 complex might be part of the Atg1 kinase complex. Atg18 and Atg21 are also PtdIns(3)P-binding proteins, although neither of them contain known phosphoinositide-binding domains. Both proteins are recruited to the PAS in a manner dependent on PtdIns3K complex I (Guan et al. 2001; Stromhaug et al. 2004). Atg18 is needed for the correct movement of Atg9, but the function of Atg21 is not known.
In contrast to yeast, there are two types of PtdIns3K in mammalian cells: class I and class III PtdIns3K. Mammalian class III PtdIns3K, hVps34—similar to yeast Vps34—generates PtdIns(3)P, and plays a stimulatory role in autophagy (Fig. 3c). It forms a complex with its regulator, p150, the homolog of Vps15, and its accessory protein Beclin 1, the homolog of Vps30/Atg6 (Liang et al. 1999; Panaretou et al. 1997). Class I PtdIns3K uses PtdIns(4,5)P2 as substrate to yield PtdIns(3,4,5)P3. It functions at the plasma membrane and acts through an insulin signaling cascade to activate mTOR and PKB; hence it has an inhibitory effect on autophagy (Jacinto and Hall 2003). A major pathway by which amino acids control mTor is not mediated through class I PtdIns3K but instead through activation of the class III PtdIns3K, hVps34 (Nobukuni et al. 2005). Thus, hVps34 might also have an inhibitory effect on autophagy in mammalian cells. The specific function of PtdIns(3)P in mammalian cells has not yet been clarified, but it could function similar to that in yeast. Moreover, the effectors of PtdIns(3)P are also not clear. Atg20 and Atg24 do not have mammalian homologs. Atg18 has a human homolog and binds to PtdIns(3)P, but its role in autophagy has not yet been elucidated (Jeffries et al. 2004).
2.4 Two Ubiquitin-Like Protein Conjugation Systems
There are two protein conjugation systems that function in selective and nonselective autophagy, and they include the ubiquitin-like proteins Atg12 and Atg8 (Fig. 6) (Ohsumi 2001). Both conjugation systems are evolutionarily conserved from yeast to humans. Although Atg12 and Atg8 do not have apparent sequence homology with ubiquitin, each of them contains a ubiquitin fold at the C terminus, based on the crystal structures of Atg12 and Atg8 homologs from plants and mammals, respectively (Paz et al. 2000; Suzuki et al. 2005).
Fig. 6.
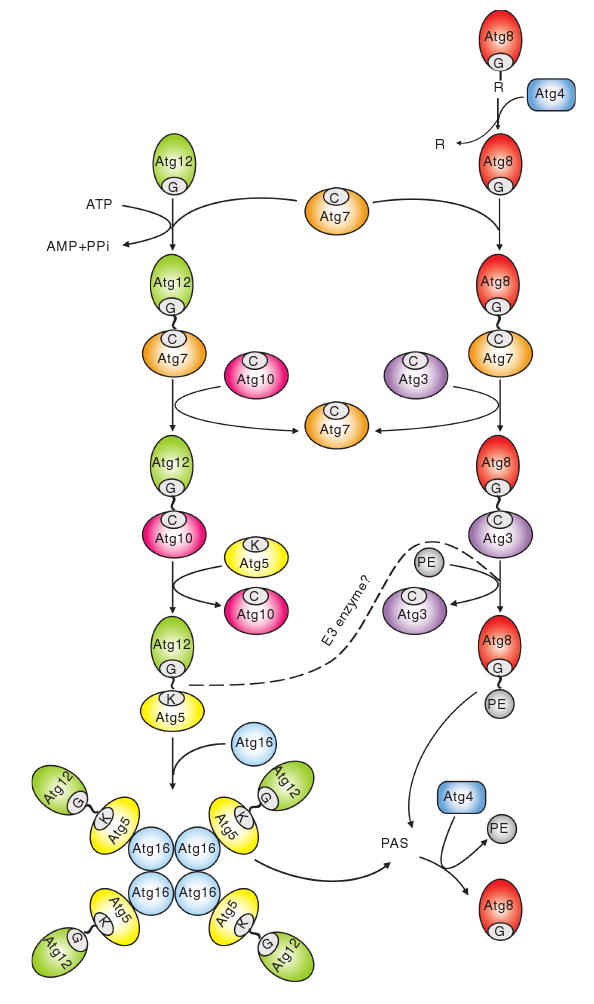
Two ubiquitin-like protein conjugation systems. The conjugation of Atg12 to Atg5 starts with activation by Atg7, which is homologous to the E1 ubiquitin-activating enzyme. Atg7 hydrolyzes ATP, resulting in the activation of Atg12 via the formation of a thioester bond between the C-terminal glycine of Atg12 and the active site cysteine of Atg7; subsequently, the activated Atg12 is transferred to the active site cysteine of Atg10, an E2-like enzyme, which catalyzes the conjugation of Atg12 to Atg5 through the formation of an isopeptide bond between the activated glycine of Atg12 and an internal lysine residue of Atg5. Atg12—Atg5 is finally assembled with Atg16. Atg16 forms a tetramer to allow the formation of an Atg12—Atg5—Atg16 multimeric structure. The conjugation of Atg8—PE starts with the cleavage of the C-terminal arginine of Atg8 by the protease Atg4. The exposed glycine of Atg8 is then bound to the active site cysteine of the same E1-like enzyme, Atg7. The activated Atg8 is then transferred to another E2-like enzyme, Atg3. Eventually, Atg3 catalyzes the conjugation of Atg8 to form Atg8—PE. The Atg12—Atg5 conjugate might function as an E3, ubiquitin ligase-like enzyme, to promote Atg8—PE conjugation. Both the Atg12—Atg5—Atg16 complex and Atg8—PE localize to the PAS to facilitate vesicle formation. The Atg8—PE that resides on the outer face of the sequestering vesicle is released from the membrane by a second Atg4-dependent cleavage. This figure is modified from Fig. 4 of Yorimitsu and Klionsky (2005b)
Atg12 is covalently attached to Atg5 through an isopeptide bond between a C-terminal glycine of Atg12 and an internal lysine residue of Atg5. The conjugation reaction is catalyzed by two additional proteins, Atg7 and Atg10 (Mizushima et al. 1998a). Atg7 is homologous to the E1 ubiquitin-activating enzyme, Uba1, in the ATP-binding region and the active cysteine residue, but not in terms of its overall structure (Tanida et al. 1999). Atg10 functions as an E2 ubiquitin-conjugating enzyme, although Atg10 shows no homology to the E2 enzymes that participate in the ubiquitin system (Shintani et al. 1999). As occurs during ubiquitination, Atg7 hydrolyzes ATP, resulting in the activation of Atg12 via the formation of a high-energy thioester bond between the C-terminal glycine of Atg12 and the active cysteine 507 of Atg7; subsequently, the activated Atg12 is directly transferred to the active cysteine 133 of Atg10 to form an Atg12—Atg10 thioester; finally, Atg12 is transferred to the target protein Atg5 to form the final conjugate. Atg5 is further bound noncovalently to another coiled-coil protein, Atg16, to form an Atg12—Atg5—Atg16 multimeric structure through homo-oligomerization of Atg16. This multimer has a molecular mass of approximately 350 kDa in yeast, and is predicted to represent a tetramer of the Atg12—Atg5—Atg16 complex. This is functionally essential for autophagy (Kuma et al. 2002). The Atg16 complex has recently been shown to specify the site of LC3 lipidation for membrane biogenesis in mammalian autophagy (Fujita et al. 2008).
A second ubiquitin-like protein, Atg8, is conjugated to a membrane lipid, phosphatidylethanolamine (PE) (Ichimura et al. 2000). The C-terminal arginine 117 residue of newly synthesized Atg8 is initially proteolytically cleaved by a cysteine protease, Atg4, exposing a glycine (Kirisako et al. 2000). The glycine is then bound to the active cysteine 507 of Atg7, the same E1-like enzyme used in the Atg12—Atg5 conjugation system. The activated Atg8 is then transferred to another E2-like enzyme (Atg3) at the active cysteine 234 residue via a thioester bond. The region around cysteine 234 of Atg3 shows partial homology to the corresponding region surrounding cysteine 133 of Atg10. Eventually, Atg8 is conjugated to PE through an amide bond between the C-terminal glycine and the amino group of PE. Atg8—PE is tightly associated with membranes, being an integral membrane protein. An in vitro reconstitution of the Atg8—PE conjugation process, using purified Atg7, Atg3, and Atg8ΔR (Atg8 lacking the last arginine residue), demonstrates that Atg7 and Atg3 are minimal catalysts (Ichimura et al. 2004). Unlike the Atg12—Atg5 conjugate, Atg8—PE conjugation is a reversible process in which Atg4 liberates Atg8 from its target lipid. The liberated Atg8 is recycled and used in another conjugation reaction to allow efficient progression of autophagy and the Cvt pathway (Kirisako et al. 2000).
Both the Atg12 and the Atg8 conjugation systems are evolutionarily conserved. The mammalian homologs for each component of the yeast Atg12—Atg5 conjugation systems (Atg5, Atg7, Atg10, and Atg12) have been characterized, and they function in a similar manner to their counterparts in yeast (Mizushima et al. 1998b; Mizushima et al. 2002; Tanida et al. 1999). There is also a mammalian Atg16-like protein, Atg16L, which forms an approximately 800 kDa protein complex with the Atg12—Atg5 conjugate, again mediated by the homo-oligomerization of Atg16L (Mizushima et al. 2003). There are at least four mammalian Atg8 homologs, MAP1LC3, GATE16, GABARAP and Atg8L. All proteins possess a conserved glycine residue near their C terminus and are conjugated to PE in the same manner as occurs in yeast via the catalysts Atg4, Atg7, and Atg3 (Hemelaar et al. 2003; Kabeya et al. 2000, 2004; Tanida et al. 2002, 2003, 2006). Among them, LC3 is most abundant in autophagosomal membranes and is well established as a marker to monitor the autophagosome and autophagic activity.
During autophagosome formation, both the Atg12—Atg5—Atg16 complex and the Atg8—PE conjugate localize at the PAS (Kim et al. 2002; Suzuki et al. 2001). Electron microscopy analysis clearly shows that these two conjugates decorate the expanding phagophore (Kabeya et al. 2000; Kirisako et al. 1999; Mizushima et al. 2001, 2003). The Atg12—Atg5—Atg16 complex is mainly localized on the outer side of the phagophore and released into the cytosol before or after autophagosome completion. These observations suggest that the Atg12—Atg5—Atg16 complex might serve as a coat component to drive the expansion and/or curvature of the membrane leaflet during autophagosome formation. Recent data, however, indicate that the Atg12—Atg5 conjugate might function as an E3, ubiquitin ligase, for Atg8—PE conjugation (Fig. 6), although it is not essential for the latter process to occur (Hanada et al. 2007). In contrast, Atg8—PE displays an apparently symmetrical distribution on both sides of the phagophore. The Atg8—PE that resides on the surface that becomes the outer face of the sequestering vesicle is released from the membrane by a second Atg4-dependent cleavage, whereas the inner population remains inside the vesicle and is delivered into the vacuole/lysosome, where it is degraded (Huang et al. 2000; Kabeya et al. 2000; Kirisako et al. 1999). Accordingly, Atg8—PE is another scaffold candidate to drive membrane expansion and vesicle completion. Upon autophagy induction, Atg8 protein levels increase, and this is needed to accommodate the larger-sized autophagosome relative to the Cvt vesicle. A quantitative correlation between the amount of Atg8 and the size of the sequestering vesicle has recently been determined (Xie et al. 2008). Atg8 is also suggested to act during the expansion of the autophagosomal membrane by mediating membrane tethering and hemifusion (Nakatogawa et al. 2007), although the physiological significance of this activity is not yet known.
2.5 Atg9 and Its Cycling Systems
One of the intriguing questions concerning autophagy is the source of the lipid that is used for autophagosome formation and the mechanism used for lipid movement to the site of autophagosome assembly. Atg9 is an integral membrane protein and is thought to be a “membrane carrier” during the assembly process (He et al. 2006; Noda et al. 2000). Unlike most other Atg proteins, which primarily display single punctate localization at the PAS, Atg9 localizes to multiple punctate structures, including the PAS (Reggiori et al. 2005b; Reggiori et al. 2004a). The cycling of Atg9 between the PAS and the non-PAS punctate structures is essential for autophagosome formation. Potentially, membrane could be delivered to the PAS through this shuttling process. In yeast, several Atg9 non-PAS puncta are found to localize adjacent to or at the surface of mitochondria (Reggiori et al. 2005b). It is still unclear, however, whether Atg9 is an integral component of the mitochondrial outer membrane or the membrane component of an organelle or other structure associated with mitochondria. Moreover, a population of the Atg9 peripheral pool does not colocalize with either the PAS or mitochondria, but rather is dispersed throughout the cytosol. This portion of Atg9 is thought to be associated with membranes in the process of trafficking between the PAS and the membrane donor sites.
The anterograde movement of Atg9 to the PAS involves several Atg proteins (Fig. 7). In the absence of Atg11, the transport of Atg9 to the PAS is blocked (He et al. 2006; Shintani and Klionsky 2004b). The efficient anterograde movement of Atg9 to the PAS also involves Atg23 and Atg27, which form a cycling unit with Atg9 (Legakis et al. 2007; Yen et al. 2007). Atg23 is a peripheral membrane protein, whereas Atg27 is a type I transmembrane protein. Both of these proteins are required for the Cvt pathway and efficient autophagy. Similar to Atg9, they localize to the PAS and several other punctate structures. Current evidence suggests that Atg9, Atg23 and Atg27 are in a heterotrimeric complex, and travel together to the PAS. Based on fluorescence microscopy, the anterograde transport of these three proteins is largely interdependent. In the absence of Atg23 or Atg27, Atg9 is at multiple punctate sites other than the PAS, whereas Atg23 is dispersed throughout the cytosol without either Atg9 or Atg27.
Fig. 7.
Cycling of Atg9. In yeast, Atg9 cycles between the PAS and non-PAS punctate structures (peripheral sites), some of which are found to localize adjacent to or at the surface of mitochondria. The efficient anterograde movement of Atg9 to the PAS requires Atg11, Atg23, Atg27, and the actin cytoskeletion. Atg9, Atg23, and Atg27 are in a heterotrimeric complex, and their movement to the PAS is interdependent. Atg11 acts as a potential adaptor between Atg9 and actin, and between Atg9 and the Arp2/3 complex, while the latter may provide the force to push the cargo (Atg9 and its associated membrane) away from the peripheral sites and toward the PAS. The retrograde transport of Atg9 from the PAS back to the peripheral sites depends on the Atg1–Atg13 kinase complex, Atg2, Atg18, and the PtdIns3K complex I. The Atg1–Atg13 complex promotes the association of Atg2 and the PtdIns(3)P binding protein Atg18 with Atg9, and the formation of this ternary complex initiates Atg9 retrieval for another round of membrane delivery
The actin cytoskeleton also participates in Atg9 anterograde movement (He et al. 2006). Disruption of the actin cytoskeleton prevents correct targeting of Atg9 to the PAS. Moreover, an actin-related protein, Arp2, interacts with Atg9 and directly regulates the dynamics of Atg9 PAS targeting (Monastyrska et al. 2008). Arp2 is one subunit of the Arp2/3 complex, the nucleation factor of branched actin filaments. Thus, one model is that Atg11 acts as an adaptor between the cargo (Atg9 and actin), while the Arp2/3 complex provides the force to push the cargo (Atg9 and its associated membrane) away from the membrane donor and toward the forming autophagosome (He et al. 2006; Monastyrska et al. 2008; Monastyrska et al. 2006).
The retrieval of Atg9 from the PAS back to the peripheral, non-PAS sites depends on the Atg1–Atg13 kinase complex, Atg2, Atg18, and the PtdIns3K complex I (Fig. 7); the absence of any of these proteins results in the accumulation of Atg9 at the PAS (Reggiori et al. 2004a). Similarly, the retrieval of Atg23 and Atg27 requires the Atg1–Atg13 complex; however, only Atg23 retrieval needs a high level of Atg1 kinase activity (Legakis et al. 2007; Yen et al. 2007). Atg2 and Atg18 are two interacting peripheral membrane proteins (Suzuki et al. 2007). They can both interact with Atg9, and the interaction of Atg18 with Atg9 requires Atg2 and Atg1 (Reggiori et al. 2004a; Wang et al. 2001a). The PAS localization of Atg2 and Atg18 depends on each other, Atg1, Atg13, Atg9, and the PtdIns3K complex I. Atg18 can bind two phosphoinositides, PtdIns(3)P, and PtdIns(3,5)P2, but only the former is essential for autophagy (Stromhaug et al. 2004). One model is that once the Atg1–Atg13 complex and Atg9 are recruited to the PAS separately, Atg1–Atg13 promotes Atg9 interaction with Atg2 and Atg18, and the formation of this ternary complex allows Atg9 to be released for another round of membrane delivery (Reggiori et al. 2004a).
Recent studies on mammalian Atg9 (mAtg9) have revealed that mAtg9 resides in a juxtanuclear region corresponding to the trans-Golgi network (TGN) and late endosomes (Young et al. 2006). Starvation triggers the distribution of mAtg9 from the TGN to a dispersed peripheral endosomal pool, and knockdown of Ulk1, the mammalian ortholog of Atg1, restricts mAtg9 to the TGN. These observations lead to the idea that mAtg9 traffics between the TGN and late endosomes, and that, potentially, membranes are delivered from the TGN to the forming autophagosomes.
2.6 De Novo Vesicle Formation
Unlike most other intracellular trafficking processes, autophagy undergoes de novo formation of double-membrane vesicles. This is a de novo process in that the sequestering vesicles do not bud from a pre-existing organelle. Instead, these vesicles are thought to form through the expansion of a membrane core of unknown origin, termed the phagophore (Mizushima et al. 2001; Noda et al. 2002; Seglen et al. 1990). Figure 8 shows a hypothetical model for de novo vesicle formation. The proposed site for vesicle formation is the phagophore assembly site (Kim et al. 2002; Suzuki et al. 2001). In yeast, the PAS is a perivacuolar site and is defined in part as the site where almost all of the Atg proteins reside, at least transiently (Suzuki et al. 2001). The PAS can be also defined as a hybrid of the phagophore and its associated Atg proteins (Xie and Klionsky 2007). In mammalian cells, colocalization of the Atg proteins has also been observed, although a comprehensive study has been lacking (Yamada et al. 2005; Young et al. 2006). In these observations, cells lack a single specialized site for autophagosome formation that is similar to the yeast PAS, and instead display multiple sites of Atg protein colocalization, possibly corresponding to multiple PAS.
Fig. 8.
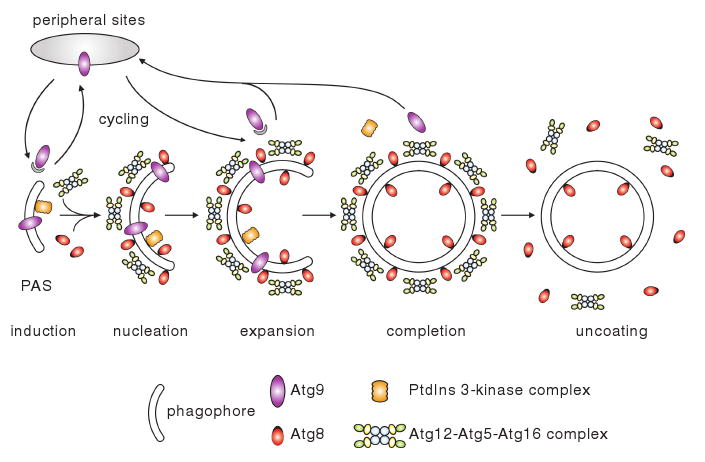
Schematic depiction of double-membrane vesicle formation. The PAS serves to facilitate the nucleation and/or expansion of the phagophore, the precursor of the autophagosome, through recruitment of Atg proteins. Atg9 and the PtdIns3K complex I are recruited relatively early to the PAS, and act in membrane nucleation. Atg9 cycles between peripheral sites and the PAS; potentially, Atg9 cycling delivers lipids to the expanding membrane. The Atg12—Atg5—Atg16 complex and the Atg8—PE conjugate, components of the vesicle-forming machinery, are subsequently recruited to the PAS, and mediate the expansion of the sequestering vesicle. The Atg12—Atg5—Atg16 complex may in part behave like a coat or may function as an E3 ubiquitin ligase-like enzyme, whereas Atg8—PE acts in the elongation of the vesicle as a structural component. Before or immediately after the autophagosome is completed, most of the Atg components, including the putative coat proteins, dissociate from the vesicle; the portion of Atg8—PE on the outer surface of the vesicle is normally cleaved off by the Atg4 protease. Finally, the sequestering vesicle can fuse with the vacuole
Understanding the nature of the PAS is a key to studying this novel type of membrane-forming process. However, the PAS is poorly characterized. Although the role of the PAS is not fully understood, one model suggests that the PAS serves to facilitate the nucleation and/or expansion of the phagophore, the precursor of the autophagosome, through recruitment of Atg proteins (Mizushima et al. 2001; Suzuki et al. 2007). In addition, membrane has to be delivered to the phagophore; although the origin of this membrane is also not clear, it appears to include the early secretory pathway and, in yeast, the mitochondria (Reggiori et al. 2004b, 2005b).
A recent systematic analysis demonstrated that the Atg proteins depend on each other for PAS recruitment (Cheong et al. 2008; Kawamata et al. 2008; Suzuki et al. 2007). In particular, Atg11 and Atg17 act as scaffold proteins for PAS assembly, meaning that they may be the initial factors responsible for subsequent recruitment of the remaining Atg proteins. Atg11 is essential for PAS organization under vegetative conditions, whereas Atg17 (and associated proteins) plays a critical role during starvation. In cells lacking both Atg11 and Atg17, there is a complete absence of PAS localization of other Atg proteins. Starvation-induced PAS assembly, however, requires more than Atg17. In addition to the Atg1–Atg13–Atg17 ternary complex, Atg17 also interacts with two autophagy-specific proteins, Atg29 and Atg31 (Kawamata et al. 2008). Cells lacking Atg11 and any of the components in the Atg17 complex display essentially the same phenotype as the atg11Δ atg17Δ double mutant, suggesting that these two complexes function as a PAS-organization center to induce the ordered recruitment of Atg proteins (Cheong et al. 2008; Kawamata et al. 2008). Although Atg1 kinase activity is not essential for PAS recruitment of other Atg proteins, it might play a role in disassembly of the PAS or the dissociation of Atg proteins from the PAS. A dynamic process of Atg protein cycling is thought to be critical to proper autophagosome expansion (Cheong et al. 2008). This concept fits with the idea that Atg1 kinase activity is related to the size of the sequestering vesicle (Noda et al. 2002).
Among the remaining Atg proteins, Atg9 is recruited relatively early to the PAS, and this also requires the function of the PtdIns3K complex I. Atg9 and the PtdIns3K complex I may play some role in membrane nucleation and facilitating the subsequent recruitment of certain Atg components, including the Atg12—Atg5—Atg16 complex and the Atg8—PE conjugate. In their absence, these two conjugates can be formed, but they become completely diffuse in the cytosol without any punctate localization. The PAS localization of Atg8—PE depends on the Atg12—Atg5–Atg16 complex (Suzuki et al. 2001).
As mentioned above, before or immediately after the autophagosome is completed, most of the Atg components dissociate from the vesicle. The sequestering vesicle must be completed before fusion with the vacuole. The Atg12—Atg5–Atg16 complex may in part behave like a coat to prevent premature fusion; the portion of Atg8—PE on the outer surface of the vesicle might also play such a role. Atg8—PE is normally cleaved off by the Atg4 protease prior to vesicle fusion. Furthermore, there may be certain unknown factors that can sense the completion of the double-membrane vesicle and trigger the disassembly of the vesicle-forming machinery. Atg1 is one possible candidate because it functions at later stages in the vesicle-forming process, such as Atg9 retrieval and Atg11 release from the PAS, and its kinase activity has been also suggested to play a role in the disassembly of the PAS or the dissociation of Atg proteins from the PAS (Cheong et al. 2008; Reggiori et al. 2004a; Yorimitsu and Klionsky 2005a).
2.7 Vesicle Docking and Fusion with the Vacuole
Once the double-membrane vesicle is formed, it is targeted to the vacuole for the fusion process. Molecular genetic studies have indicated that the machinery involved in homotypic vacuole fusion is also essential for the fusion of autophagosomes and Cvt vesicles with the vacuole (Fig. 9). This machinery includes the SNARE proteins Vam3, Vam7, Vti1 and Ykt6, the NSF Sec18, the α-SNAP Sec17, the Rab GTPase Ypt7 and the class C Vps/HOPS complex; the two recently characterized proteins Mon1 and Ccz1 are also part of the fusion machinery (Klionsky 2005; Wang and Klionsky 2003). Mon1 and Ccz1 form a complex and are critical for the Ypt7-dependent tethering/docking stage leading to the subsequent formation of the SNARE complex (Wang et al. 2003). The class C Vps/HOPS complex functions in concert with Ypt7 during the tethering/docking stage (Wang and Klionsky 2003). After fusion, the autophagosome inner single-membrane vesicle is released inside the vacuole lumen, which is termed the autophagic body.
Fig. 9.

Vesicle docking and fusion with the vacuole. The SNARE proteins Vam3, Vam7, Vti1, and Ykt6 function in various membrane fusion events, including the process of autophagosome fusion with the vacuole. Also shown are the Mon1–Ccz1 complex and the class C Vps/HOPS complex, which function in concert at the Ypt7-dependent tethering/docking stage. This figure is modified from Fig. 7 of Klionsky (2005)
In mammalian cells, maturation of autophagosomes includes several fusion events with vesicles originating from early and late endosomes, as well as lysosomes. Fusion with endosomes to become amphisomes allows convergence of the endocytic and autophagic pathways; subsequent fusion of autophagosomes or amphisomes with lysosomes generates autolysosomes (Berg et al. 1998; Tooze et al. 1990). In some cases where it is not possible to distinguish the precise nature of the compartment, the term “autophagic vacuoles” is used to cover all three autophagic structures: autophagosomes, amphisomes, and autolysosomes. Mammalian Vtilb is involved in the fusion of autophagsomes with multivesicular endosomes (Atlashkin et al. 2003), and the Rab GTPase Rab7 plays a role in the fusion with lysosomes (Jager et al. 2004).
2.8 Vesicle Breakdown and Recycling of the Resulting Macromolecules
Upon release into the vacuole, the single-membrane subvacuolar vesicle, the autophagic or Cvt body, is broken down inside the vacuolar lumen. This process depends on proper vacuole function (including vacuolar acidification) and the activity of vacuolar resident hydrolases (including Pep4 and Prb1). In addition to these factors, Atg15, a putative lipase, is also implicated at this step and seems likely to function directly in the intravacuolar lysis of the autophagic/Cvt body (Epple et al. 2001; Teter et al. 2001). Atg15 contains a lipase active-site motif, and mutations in the corresponding active site eliminate its function. Atg15 is targeted to the vacuolar lumen via the multivesicular body (MVB) pathway (Epple et al. 2003). Atg15 seems to function as a general lipase because it is also involved in the disintegration of intravacuolar MVB vesicles.
The main purpose of autophagy is to degrade cytoplasm and recycle the resulting macromolecules for the synthesis of essential components to overcome various stress conditions. Accordingly, the resulting macromolecules must be released back to the cytosol for reuse; however, little is known about this process. Atg22, a putative amino acid effluxer on the vacuolar membrane, has been found to play such role in mediating the efflux of leucine and other amino acids resulting from autophagic degradation (Yang et al. 2006). In addition, Avt3 and Avt4 seem to be part of the same family of permeases (Russnak et al. 2001). Upon elimination of all three partially redundant vacuolar effluxers, cells rapidly lose viability under starvation conditions, whereas supplementation with leucine partially restores viability. Although a mammalian homolog of Atg22 has not been identified, homologs of Avt3 and Avt4 have been characterized as SLC36A1/LYAAT-1 (lysosomal amino acid transporter 1) (Sagne et al. 2001), and SLC36A4/LYAAT-2, respectively. How autophagy contributes to the recycling of other macromolecules, such as carbohydrate or lipids, remains unknown.
3 Conclusion
As a conserved cellular degradative pathway in eukaryotes, autophagy protects cells during various types of stress. Defects in autophagy have been linked to human diseases, indicating its crucial physiological significance. Autophagy involves dynamic membrane rearrangement for sequestration of cytoplasm and its delivery into the vacuole/lysosome. Significant breakthroughs in understanding the molecular mechanism of autophagy have been achieved from studies in yeast and other model systems. Currently, analyses of autophagy, pexophagy and the Cvt pathway in fungi have identified 31 ATG genes, corresponding to the unique molecular machinery that drives these pathways. However, the fundamental biochemical questions that concern the functions of Atg proteins still need to be resolved, especially those related to sequestering vesicle formation, such as the ordered vesicle assembly process and the origin of the lipid membrane. In addition, as more examples of selective types of autophagy emerge, continued studies on the specific nature of autophagy are becoming increasingly important. Yeast remains a powerful system to address these questions. Further studies of autophagy-related pathways will facilitate our understanding of the molecular mechanism and regulation of these pathways, and may allow the practical use of autophagy for therapeutic purposes.
Acknowledgments
This work was supported by National Institutes of Health Public Health Service grant GM53396 to D.J.K.
References
- Abeliovich H, Zhang C, Dunn WA, Jr, Shokat KM, Klinosy DJ. Chemical genetic analysis of Apgl reveals a non-kinase role in the induction of autophagy. Mol Biol Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J Biol Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- Atlashkin V, Kreykenbohm V, Eskelinen EL, Wenzel D, Fayyazi A, Fischer von Mollard G. Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8. Mol Cell Biol. 2003;23:5198–5207. doi: 10.1128/MCB.23.15.5198-5207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellu AR, Komori M, van der Klei IJ, Kiel JAKW, Veenhuis M. Peroxisome biogenesis and selective degradation converge at Pex14p. J Biol Chem. 2001;276:44570–44574. doi: 10.1074/jbc.M107599200. [DOI] [PubMed] [Google Scholar]
- Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Øvervatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera AC. TOR signaling in mammals. J Cell Sci. 2004;117:4615–4616. doi: 10.1242/jcs.01311. [DOI] [PubMed] [Google Scholar]
- Carroll AS, O'Shea EK. Pho85 and signaling environmental conditions. Trends Biochem Sci. 2002;27:87–93. doi: 10.1016/s0968-0004(01)02040-0. [DOI] [PubMed] [Google Scholar]
- Chan EYW, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- Chang CY, Huang WP. Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol Biol Cell. 2007;18:919–929. doi: 10.1091/mbc.E06-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R. Cell growth control: little eukaryotes make big contributions. Oncogene. 2006;25:6392–6415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]
- Ellson CD, Andrews S, Stephens LR, Hawkins PT. The PX domain: a new phosphoinositide-binding module. J Cell Sci. 2002;115:1099–1105. doi: 10.1242/jcs.115.6.1099. [DOI] [PubMed] [Google Scholar]
- Epple UD, Suriapranata I, Eskelinen EL, Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J Bacteriol. 2001;183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple UD, Eskelinen EL, Thumm M. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J Biol Chem. 2003;278:7810–7821. doi: 10.1074/jbc.M209309200. [DOI] [PubMed] [Google Scholar]
- Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–376. doi: 10.1016/j.devcel.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrance biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23:3898–3904. doi: 10.1038/sj.onc.1207539. [DOI] [PubMed] [Google Scholar]
- Guan J, Stromhaug PE, George MD, Habibzadegah-Tari P, Bevan A, Dunn WA, Jr, Klionsky DJ. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol Biol Cell. 2001;12:3821–3838. doi: 10.1091/mbc.12.12.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunkel K, van der Klei IJ, Barth G, Veenhuis M. Selective peroxisome degradation in Yarrowia lipolytica after a shift of cells from acetate/oleate/ethylamine into glucose/ammonium sulfate-containing media. FEBS Lett. 1999;451:1–4. doi: 10.1016/s0014-5793(99)00513-x. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12–Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 2002;129:1181–1193. doi: 10.1104/pp.011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003;278:51841–51850. doi: 10.1074/jbc.M308762200. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Høyer-Hansen M, Jäättelä M. AMP-activated protein kinase: a universal regulator of autophagy. Autophagy. 2007;3:381–383. doi: 10.4161/auto.4240. [DOI] [PubMed] [Google Scholar]
- Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- Huang WP, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;112(Pt 22):4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004;279:40584–40592. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- Iwata J, Ezaki J, Komatsu M, Yokota S, Ueno T, Tanida I, Chiba T, Tanaka K, Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Hall MN. Tor signalling in bugs, brain and brawn. Nat Rev Mol Cell Biol. 2003;4:117–126. doi: 10.1038/nrm1018. [DOI] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Jeffries TR, Dove SK, Michell RH, Parker PJ. PtdIns-specific MPR pathway association of a novel WD40 repeat protein, WIPI49. Mol Biol Cell. 2004;15:2652–2663. doi: 10.1091/mbc.E03-10-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–2553. doi: 10.1091/mbc.E04-08-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–2050. doi: 10.1091/mbc.E07-10-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang WP, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Meijer AJ, Codogno P. Autophagy and p70S6 kinase. Autophagy. 2005;1:59–61. doi: 10.4161/auto.1.1.1536. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Ueno T, Waguri S, Uchiyama Y, Kominami E, Tanaka K. Constitutive autophagy: vital role in clearance of unfavorable proteins in neurons. Cell Death Differ. 2007a;14:887–894. doi: 10.1038/sj.cdd.4402120. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007b;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Kubota H, Obata T, Ota K, Sasaki T, Ito T. Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2α kinase GCN2. J Biol Chem. 2003;278:20457–20460. doi: 10.1074/jbc.C300133200. [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12–Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legakis JE, Yen WL, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy. 2007;3:422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Levine B, Yuan J. Autophagy in cell death: an innocent convict. J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Shao SH, Xu ZX, Hennessy B, Ding Z, Larrea M, Kondo S, Dumont DJ, Gutterman JU, Walker CL, Slingerland JM, Mills GB. The energy sensing LKB1–AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007;9:218–224. doi: 10.1038/ncb1537. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Martinez E, Jimenez MA, Segui-Real B, Vandekerckhove J, Sandoval IV. Folding of the presequence of yeast pAPI into an amphipathic helix determines transport of the protein from the cytosol to the vacuole. J Mol Biol. 1997;267:1124–1138. doi: 10.1006/jmbi.1997.0925. [DOI] [PubMed] [Google Scholar]
- Massey A, Kiffin R, Cuervo AM. Pathophysiology of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2420–2434. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Mavrakis M, Lippincott-Schwartz J, Stratakis CA, Bossis I. Depletion of type IA regulatory subunit (RIα) of protein kinase A (PKA) in mammalian cells and tissues activates mTOR and causes autophagic deficiency. Hum Mol Genet. 2006;15:2962–2971. doi: 10.1093/hmg/ddl239. [DOI] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol Aspects Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Meley D, Bauvy C, Houben-Weerts JH, Dubbelhuis PF, Helmond MT, Codogno P, Meijer AJ. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998a;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998b;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. Mouse Apg10 as an Apg12-conjugating enzyme: analysis by the conjugation-mediated yeast two-hybrid method. FEBS Lett. 2002;532:450–454. doi: 10.1016/s0014-5793(02)03739-0. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I, Shintani T, Klionsky DJ, Reggiori F. Atg11 directs autophagosome cargoes to the PAS along actin cables. Autophagy. 2006;2:119–121. doi: 10.4161/auto.2.2.2298. [DOI] [PubMed] [Google Scholar]
- Monastyrska I, He C, Geng J, Hoppe AD, Li Z, Klionsky DJ. Arp2 links autophagic machinery with the actin cytoskeleton. Mol Biol Cell. 2008;19:1962–1975. doi: 10.1091/mbc.E07-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci USA. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Suzuki K, Ohsumi Y. Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 2002;12:231–235. doi: 10.1016/s0962-8924(02)02278-x. [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes: Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda MN, Scott SV, Hefner-Gravink A, Caffarelli AD, Klionsky DJ. Identification of a cytoplasm to vacuole targeting determinant in aminopeptidase I. J Cell Biol. 1996;132:999–1010. doi: 10.1083/jcb.132.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J Biol Chem. 2004;279:16071–16076. doi: 10.1074/jbc.M312706200. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Levine B. Viral evasion of autophagy. Autophagy. 2008;4:280–285. doi: 10.4161/auto.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J Biol Chem. 2004;279:15621–15629. doi: 10.1074/jbc.M311139200. [DOI] [PubMed] [Google Scholar]
- Panaretou C, Domin J, Cockcroft S, Waterfield MD. Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150·Ptdins 3-kinase complex. J Biol Chem. 1997;272:2477–2485. doi: 10.1074/jbc.272.4.2477. [DOI] [PubMed] [Google Scholar]
- Paz Y, Elazar Z, Fass D. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J Biol Chem. 2000;275:25445–25450. doi: 10.1074/jbc.C000307200. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005a;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005b;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004a;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Wang CW, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2004b;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russnak R, Konczal D, McIntire SL. A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J Biol Chem. 2001;276:23849–23857. doi: 10.1074/jbc.M008028200. [DOI] [PubMed] [Google Scholar]
- Sagne C, Agulhon C, Ravassard P, Darmon M, Hamon M, El Mestikawy S, Gasnier B, Giros B. Identification and characterization of a lysosomal transporter for small neutral amino acids. Proc Natl Acad Sci USA. 2001;98:7206–7211. doi: 10.1073/pnas.121183498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Oku M, van der Klei IJ, Kiel JAKW. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Beck T, Martin DE, Hall MN. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol. 2004;24:338–351. doi: 10.1128/MCB.24.1.338-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB, Holen I. Non-selective autophagy. Semin Cell Biol. 1990;1:441–448. [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004a;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004b;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Aasland R, Driscoll PC. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 2002;513:77–84. doi: 10.1016/s0014-5793(01)03308-7. [DOI] [PubMed] [Google Scholar]
- Stromhaug PE, Reggiori F, Guan J, Wang CW, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of plant Atg12 and its biological implication in autophagy. Autophagy. 2005;1:119–126. doi: 10.4161/auto.1.2.1859. [DOI] [PubMed] [Google Scholar]
- Tallóczy Z, Jiang W, Virgin HW, IV, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc Natl Acad Sci USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- Tanida I, Komatsu M, Ueno T, Kominami E. GATE-16 and GABARAP are authentic modifiers mediated by Apg7 and Apg3. Biochem Biophys Res Commun. 2003;300:637–644. doi: 10.1016/s0006-291x(02)02907-8. [DOI] [PubMed] [Google Scholar]
- Tanida I, Sou YS, Minematsu-Ikeguchi N, Ueno T, Kominami E. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. FEBS J. 2006;273:2553–2562. doi: 10.1111/j.1742-4658.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM, Klionsky DJ. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J Biol Chem. 2001;276:2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, de Winde JH. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–918. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- Tooze J, Hollinshead M, Ludwig T, Howell K, Hoflack B, Kern H. In exocrine pancreas, the basolateral endocytic pathway converges with the autophagic pathway immediately after the early endosome. J Cell Biol. 1990;111:329–345. doi: 10.1083/jcb.111.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle DL, Lewin AS, Dunn WA., Jr Selective autophagy of peroxisomes in methylotrophic yeasts. Eur J Cell Biol. 1993;60:283–290. [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Veenhuis M, Douma A, Harder W, Osumi M. Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch Microbiol. 1983;134:193–203. doi: 10.1007/BF00407757. [DOI] [PubMed] [Google Scholar]
- Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Kim J, Huang WP, Abeliovich H, Stromhaug PE, Dunn WA, Jr, Klionsky DJ. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem. 2001a;276:30442–30451. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Stromhaug PE, Kauffman EJ, Weisman LS, Klionsky DJ. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J Cell Biol. 2003;163:973–985. doi: 10.1083/jcb.200308071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wilson WA, Fujino MA, Roach PJ. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol. 2001b;21:5742–5752. doi: 10.1128/MCB.21.17.5742-5752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem. 2005;280:18283–18290. doi: 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- Yan J, Kuroyanagi H, Kuroiwa A, Matsuda Y, Tokumitsu H, Tomoda T, Shirasawa T, Muramatsu M. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem Biophys Res Commun. 1998;246:222–227. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]
- Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y, Suzuki Y, Ohshima Y, Mitani S, Masuho Y, Shirasawa T, Muramatsu M. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene. 1999;18:5850–5859. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006;17:5094–5104. doi: 10.1091/mbc.E06-06-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen WL, Legakis JE, Nair U, Klionsky DJ. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell. 2007;18:581–593. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005a;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005b;12 2:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, He C, Wang K, Klionsky DJ. Tap42-associated protein phosphatase type 2A negatively regulates induction of autophagy. Autophagy. 2009;5 doi: 10.4161/auto.5.5.8091. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ARJ, Chan EYW, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- Zurita-Martinez SA, Cardenas ME. Tor and cyclic AMP-protein kinase A: two parallel pathways regulating expression of genes required for cell growth. Eukaryot Cell. 2005;4:63–71. doi: 10.1128/EC.4.1.63-71.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]