Abstract
Killer Ig-like receptors (KIRs) are implicated in protection from multiple pathogens including HIV, human papillomavirus, and malaria. Nonhuman primates such as rhesus and cynomolgus macaques are important models for the study of human pathogens; however, KIR genetics in nonhuman primates are poorly defined. Understanding KIR allelic diversity and genomic organization are essential prerequisites to evaluate NK cell responses in macaques. In this study, we present a complete characterization of KIRs in Mauritian cynomolgus macaques, a geographically isolated population. In this study we demonstrate that only eight KIR haplotypes are present in the entire population and characterize the gene content of each. Using the simplified genetics of this population, we construct a model for macaque KIR genomic organization, defining four putative KIR3DL loci, one KIR3DH, two KIR2DL, and one KIR1D. We further demonstrate that loci defined in Mauritian cynomolgus macaques can be applied to rhesus macaques. The findings from this study fundamentally advance our understanding of KIR genetics in nonhuman primates and establish a foundation from which to study KIR signaling in disease pathogenesis.
Killer Ig-like receptors (KIRs)3 are a multigeneic family of cell surface receptors expressed on NK cells and a subset of T lymphocytes (1–3). KIR signaling is an important component in the recognition of foreign cells and the antiviral response (4). KIRs can mediate either activating or inhibitory signals (5). The primary known KIR ligands are MHC class I molecules; however, ligands have not been identified for all KIRs (6, 7). Unlike the highly specific TCR-HLA interactions, each KIR generally interacts with a broad subset of HLA class I alleles (8, 9). When an NK cell encounters a normal host cell, inhibitory KIRs interact with self-HLA Ag on the target and the NK cell is inhibited (10). However, when HLA class I expression is altered, such as when HLA class I is down-regulated by a viral pathogen, inhibition does not occur and the target is lysed (11). The role of activating KIRs is less characterized and in most cases their ligands are unknown.
KIR genotypes are extremely polymorphic and have a major impact on NK activity (12, 13). The primary mechanism by which KIR polymorphism influences NK function is through variable KIR gene content (14). A total of 16 human KIR genes have been identified (3). Each chromosome encodes a subset of these genes, with the number and identity of KIRs varying between haplotypes (15, 16). To date, more than 37 human KIR haplotypes with distinct gene content have been described (16, 17). In general, the protein product of each KIR gene binds a unique set of ligands (18). Therefore, the identity of the KIR genes encoded in the genome dictates the potential KIR-HLA interactions that can occur. The HLA class I genotype of the individual also influences NK activity by controlling the set of ligands available for KIR interaction (19). It should be noted that although alleles of each KIR gene appear to have broadly similar specificity for HLA ligands, KIR allelic variability can result in slightly altered binding affinities, creating functional polymorphism (20). KIRs are also regulated at the level of gene expression, with the expression level of each KIR heavily influenced by the other KIR and HLA alleles encoded in an individual's genome (19, 21, 22).
Associations have been identified between KIR/HLA genotype and protection from infectious diseases including HIV, malaria, hepatitis B virus, and human papillomavirus (23–27). Although there have been significant advances in our understanding of KIR signaling, the mechanisms mediating control are elusive. This mystery is at least partially due to the complexity of KIR interactions. The entire KIR and HLA class I genotype of an individual influences NK activity, weaving a complex tapestry of interactions that are difficult to decipher (5, 28). This response is unlike the T cell, for which meaningful results can be obtained by studying individuals that share a single HLA class I allele (29, 30). Untangling the role of KIRs in NK activity requires controlling both KIR and HLA genetics; however, individuals identical at both HLA and KIR loci are exceptionally rare.
Macaques are important models for the study of many human pathogens including HIV (31). They offer advantages including access to samples from precisely defined time points following infection and the ability to control the dose and route of infection. They have provided important insights into the role of MHC genetics in HIV pathogenesis (32–34). For the study of KIR function, a significant advantage of macaques is the ability to selectively infect specific animals based on genotype. However, KIR genetics are poorly characterized in macaques. The sequence of one rhesus macaque KIR haplotype and a limited number of macaque KIR cDNA sequences have been published (35–37). These studies demonstrate an overall conservation of structure between human and macaque KIRs, but a distinct genomic organization (35). To conduct KIR functional studies, several basic questions about KIR biology need to be addressed. Foremost, we need a more comprehensive understanding of KIR allelic diversity and genomic organization. This understanding requires the description of the gene content of additional KIR haplotypes and cannot be accomplished using cDNA sequences alone.
In this study we used a novel combination of microsatellite analysis and cDNA sequencing to characterize the gene content from eight macaque KIR haplotypes. We performed these studies in Mauritian cynomolgus macaques (MCM) from the Indian Ocean island of Mauritius, a geographically isolated population with limited genetic diversity. Through comparison of the gene content of these haplotypes, we defined a model for MCM KIR genomic organization. The findings from this study serve as a framework to understand KIR genetics in macaques and provide a foundation for functional studies investigating the role of KIR genetics in infectious disease pathogenesis.
Materials and Methods
Animal selection
Blood samples from feral MCM were purchased directly for genetic analyses (Charles River Breeding Facilities). This cohort has been used previously for MHC studies (38). Genomic DNA and total RNA was purified from whole blood using the MagNA LC DNA and RNA isolation kits (Roche) according to the manufacturer's instructions. For microsatellite analysis, samples from all 274 animals were used. For cDNA and gDNA cloning, animals homozygous throughout the KIR region were selected, with a minimum of three per haplotype. In the case of K6 and K8, homozygotes were not available, so heterozygotes were used.
Microsatellite marker design and analysis
Short tandem repeats were identified using eTandem software (39). Microsatellite primers flanking these repeats were designed with the assistance of Primer3Plus (40). Primer sequences are listed in supplemental Table I.4 We performed multiplexed PCR using the following touchdown PCR conditions: 98°C for 30 s; 3 cycles of 98°C for 5 s, 64°C for 5 s, 72°C for 20 s; 3 cycles of 98°C for 5 s, 62°C for 5 s, 72°C for 20 s; 3 cycles of 98°C for 5 s, 60°C for 5 s, 72°C for 20 s; 6 cycles of 98°C for 5 s, 58°C for 5 s, 72°C for 20 s; 25 cycles of 98°C for 5 s, 50°C for 5 s, 72°C for 20 s; and a final extension at 72°C for 5 min. Fluorescently labeled PCR products were resolved by using capillary gel electrophoresis on an ABI 3730×l (Applied Biosystems). Fragment analysis was performed using DAx software (Van Mierlo).
KIR cDNA cloning and sequencing
First strand cDNA was generated using the Superscript III RT kit (Invitrogen) according to the manufacturer's instructions. KIR cDNAs were amplified by PCR using Phusion high-fidelity polymerase (New England Bio-labs) and the following PCR primers: 5′-CAGCACCATGTCGCTCAT-3′ and 5′-GGGGTCAAGTGAAGTGGAGA-3′. In all cases, PCR conditions were as follows: 98°C for 30 s; 28 cycles of 98°C for 5 s, 63°C for 1 s, 72°C for 20 s; and a final extension of 72°C for 5 min. Products were cloned into pCR-Blunt II TOPO (Invitrogen) and bidirectionally sequenced using DYEnamic ET Terminator cycle sequencing kit (GE Healthcare). In addition to vector-specific primers, internal primers 5′-AACCTTCCCTCCTGGCC-3′ and 5′-TTGGTTCAGTGGGTGAAGGCCAA-3′ were used. Sequences were analyzed using CodonCode Aligner software. To minimize artifacts inherent in the PCR process, a sequence was only considered to be a novel allele when three or more identical cDNA clones were observed.
KIR genomic DNA cloning and sequencing
Amplicons containing intron 4 along with portions of the flanking exons were generated using the following primers: 5′-GTCCCCTGGTGAAAT CAGGA-3′ and 5′-AGGTCACGTTCTCTCCTGC-3′. In addition to vector-specific primers, the following internal primers were used for sequencing: 5′-GAGAGACTGAGAGGCAGAG-3′, 5′-TCTTCCTTTTTCTTTAATTCTGAG-3′, 5′-TCTATGACCTAATGCTCTCT-3′, and 5′-TGTCACAGCTCCCCTCAC-3′. PCR conditions, cloning, sequencing and analysis were otherwise identical to the methods described for KIR cDNA cloning.
Phylogenetic analysis
Sequences were aligned by the CLUSTAL W program (41). Phylogenetic trees were constructed on the basis of the JTT amino acid sequence distance (42) by the neighbor-joining method (43). The reliability of clustering patterns in phylogenetic trees was assessed by bootstrapping (44); 1000 bootstrap samples were used.
Results
Microsatellite design and analysis
To characterize the KIR region in macaques, we used microsatellite analysis. This technique takes advantage of short tandem repeats interspersed at regular intervals throughout the region of interest. These repeats are inherently unstable in length and evolve quickly. Primers are designed to flank each repeat, and the size of the resulting PCR product is determined. Because microsatellite repeats are physically linked to the surrounding genes, microsatellite allele sizes can be used to infer genotype. The closer the microsatellite is to the gene of interest, the higher the resolution of genotyping.
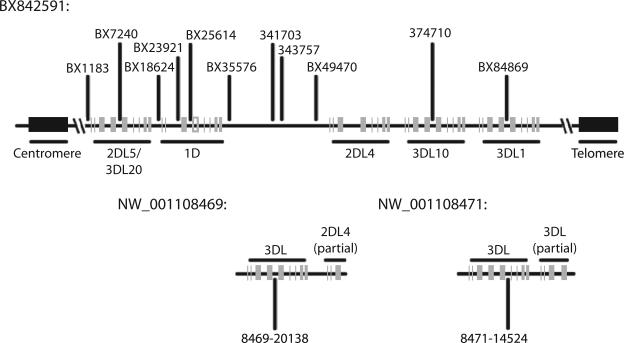
To design the microsatellite panel, we compiled all published macaque KIR genomic sequence. Although no KIR genomic data has been published from cynomolgus macaques, one complete genomic rhesus macaque KIR haplotype has been sequenced (accession no. BX842591) (35). In this haplotype, the KIR region spans 86 kb and contains five KIR genes, organized into telomeric and centromeric clusters separated by a 21-kb unique region. Using this genomic sequence, we identified 11 microsatellite markers distributed throughout the KIR region (Fig. 1). Five of these markers are located within KIR introns, primarily within intron 4. Because these repeats are located within genes themselves, they are tightly linked to the surrounding gene and have the potential to provide very high resolution genotyping. In addition to these microsatellites, six other markers were identified within the intergenic regions or the unique region between the centromeric and telomeric ends.
FIGURE 1.
Diagram of microsatellite markers. The diagram displays the position of microsatellite markers relative to published rhesus macaque KIR genes. Markers were designed using a complete haplotype sequenced by Sambrook et al. (35) (accession no. BX842591) and two genomic sequence contigs from the rhesus genome project (NW_001108469 and NW_001108471).
A challenging characteristic of the KIR region is that the number and identity of KIR genes varies between haplotypes. Therefore, although the single sequenced haplotype can serve as a framework, it is unlikely to contain all macaque KIR genes. We also identified two rhesus macaque genomic sequence contigs containing KIR genes that have been deposited as part of the rhesus genome project (accession nos. NW_001108469 and NW_001108471). Each genomic contig contains an intact KIR3DL gene that is divergent in sequence from either of the KIR3DL genes present on the intact haplotype. Presumably these represent distinct macaque KIR genes that are not present on the haplotype that has been sequenced. These sequences were used to design additional microsatellite primer pairs located within KIR introns. We also identified a primer pair, 374710-20138, which amplifies the microsatellite located in intron 4 from all sequenced KIR3DL loci except KIR3DL1. The position of these markers and genes relative to those present on the intact KIR haplotype cannot be determined from sequence alone.
KIR haplotypes in MCM
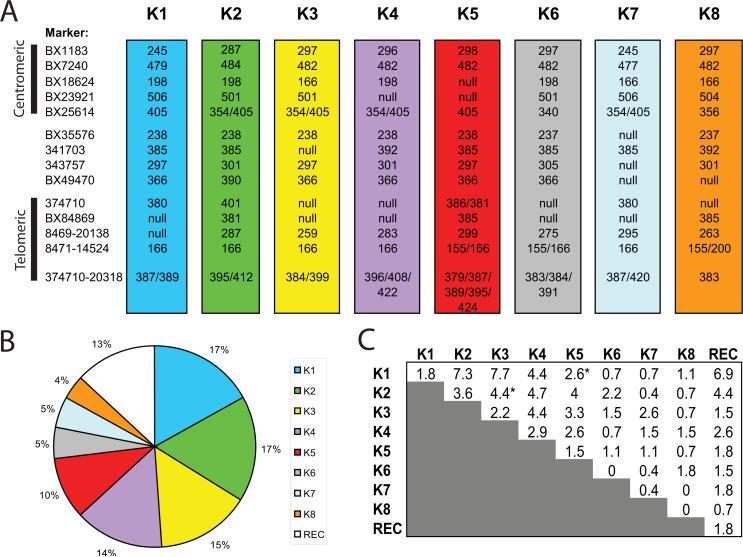
Cynomolgus macaques are believed to have been introduced to the island of Mauritius nearly 500 years ago (45). Although the population currently is greater than 25,000, evidence suggests it arose from a small founder population with as few as four animals (38, 46). Because of this limited genetic diversity, MCM provide an ideal macaque population in which to study the otherwise complex and polymorphic KIR region. Using the microsatellite panel, we screened a cohort of 274 MCM. We were able to identify 34 animals homozygous for the marker panel and use these to infer haplotypes. Analysis of the entire cohort revealed that eight common KIR haplotypes are present in MCM (Fig. 2A). The relative frequency of each haplotype varied, with the four most common haplotypes (K1, K2, K3, and K4) each present at a frequency of ~15%, followed by 10% for haplotypes K5 and ~5% each for K6, K7, and K8 (Fig. 2B).
FIGURE 2.
KIR haplotypes in MCM. A, The microsatellite profiles from the eight KIR haplotypes identified in MCM are shown. Marker name is indicated (left). Each column displays the size of the corresponding microsatellite repeat in base pairs. Null indicates lack of PCR amplification from a marker, suggestive of deletion within that haplotype. Several haplotypes contain multiple values for a microsatellite marker, suggesting duplication. Marker 374710-20138 was designed to amplify multiple microsatellites and therefore multiple allele sizes are expected. The order of markers was inferred using genomic sequence and segregation analysis. Markers within the centromeric and telomeric regions are indicated. B, The frequency of each haplotype within the MCM cohort is shown (n = 548 chromosomes). Recombinant haplotype (REC) is indicated. C, Shows the frequency of each KIR genotype within the population, expressed as the percentage within the 274 animal cohort. *, p < 0.05 for KIR genotypes with frequencies significantly lower than expected.
Next, we examined the cohort for associations between KIR or MHC haplotypes. First, we calculated the frequency of each KIR genotype (Fig. 2C). Within the population, the most frequent KIR genotypes were K1/K3 and K1/K2 heterozygotes, comprising 7.7% and 7.3% of the population, respectively. The MHC genotypes have been previously published for this cohort (38, 47, 48). We used Fisher's exact test to examine the association between each MHC haplotype and KIR haplotype; however, no tests were significant (p > 0.05 in all cases, whether corrected for multiple testing by the Holmes procedure or not corrected). In contrast, we were able to identify statistically significant associations among the KIR haplotypes themselves. There was a significant deficiency of K1/K5 heterozygotes (p < 0.05, corrected for multiple testing the by Holmes procedure). There was also a significant deficiency of K2/K3 heterozygotes (p < 0.05, corrected for multiple testing). It is possible that our sample size was not large enough to identify associations between KIR and MHC haplotypes. However, the fact that we were able to identify associations among the KIR haplotypes suggests that unless the associations were very weak, we should have been able to detect them. The evolution of associations between KIR and MHC haplotypes requires population subdivision as seen in humans (49). The 500 years following introduction of MCM to the island of Mauritius has been an insufficient time for population subdivision to occur, and genetic diversity may have been reduced due to a founder effect.
Within the MCM population, 13% of KIR haplotypes were recombinant. Of these, 86% could be explained by a single crossover event between two of the eight common haplotypes. We used the segregation patterns of microsatellite alleles in these recombinant haplotypes to infer their linear order on the chromosome. Virtually all of these simple recombination events occurred in the region between the centromeric and telomeric gene clusters, resulting in the centromeric end of one haplotype paired to the telomeric end of another. This finding is consistent with human data suggesting this region is a recombination hot spot (16). In these recombinants, the microsatellite alleles derived from genomic contigs NW_001108469 and NW_001108471 segregate with the telomeric end of the KIR region. Any genes physically linked to these microsatellite markers should also reside in the telomeric end. Unfortunately, an insufficient number of recombination events were detected within the telomeric end of the KIR region to more finely map the relative position of these markers.
KIR gene content by haplotype
Next, we sought to characterize the gene content of each KIR haplotype. We PCR amplified, cloned, and sequenced full-length KIR cDNA transcripts from a minimum of three representative animals possessing each haplotype. For six haplotypes, K1 through K5 and K7, we were able to identify and clone KIR sequences from homozygous animals. For haplotypes K6 and K8, homozygotes were not available, so we selected heterozygotes and inferred the gene content of these haplotypes.
KIRs are classified by domain structure. In humans, KIRs contain either two Ig-like domains (KIR2D) or three Ig-like domains (KIR3D). In addition, KIRs contain either a long cytoplasmic tail (KIR3DL) or short cytoplasmic tail (KIR3DS). Previous work in macaques has identified the presence of KIR2DL and KIR3DL molecules (35, 36). Although neither KIR2DS nor KIR3DS molecules have been identified in macaques, macaques do express a hydrid (KIR3DH). These molecules contain a cytoplasmic tail resembling human KIR2DL4 (36). Macaques also express a single Ig domain (KIR1D) or KIR1D molecules. To differentiate species, cynomolgus macaque (Macaca fascicularis) KIRs are designated mfKIR, whereas rhesus macaque (Macaca mulatta) KIRs are designated mmKIR. Each novel MCM sequence has been assigned a temporary name. The KIR Nomenclature Committee is in the process of establishing a formal system for macaque KIR nomenclature. Once formal names have been assigned, this information will be available online through the Immuno Polymorphism Database (50).
We found significant variability in the number of KIRs per haplotype, with each haplotype expressing between 3 and 8 KIR sequences (Table I). In MCM, the most prevalent and diverse KIRs are the mfKIR3DL and mfKIR3DH molecules. Each MCM haplotype contains a minimum of two mfKIR3DL sequences, with K2 expressing six distinct mfKIR3DL sequences. KIR3DH sequences were present on six of the haplotypes, with a maximum of three per haplotype.
Table I.
MCM KIR gene content by haplotypea
| Haplotype | KIR1D | KIR2DL | KIR3DL | KIR3DH | Total Sequences |
|---|---|---|---|---|---|
| K1 | mfKIRnew37 (EU419110) | mfKIRnew38 (EU419111) | mfKIRnew24 (EU419101) | mfKIRnew40 (EU419113) | 5 |
| mfKIRnew35 (EU419109) | |||||
| K2 | mfKIRnew57 (EU419124) | mfKIRnew04 (EU419078) | mfKIRnew14 (EU419092) | 8 | |
| mfKIRnew05 (EU419079) | |||||
| mfKIRnew08 (EU419083) | |||||
| mfKIRnew12 (EU419089) | |||||
| mfKIRnew13 (EU419090) | |||||
| mfKIRnew02 (EU419076) | |||||
| K3 | mfKIRnew47 (EU419120) | mfKIRnew46 (EU419119) | 3 | ||
| mfKIRnew55 (EU419123) | |||||
| K4 | mfKIRnew42 (EU419114) | mfKIRnew44 (EU419115) | mfKIRnew48 (EU419121) | 4 | |
| mfKIRnew45 (EU419118) | |||||
| K5 | mfKIRnew01 (EU419075) | mfKIRnew20 (EU419097) | 8 | ||
| mfKIRnew22 (EU419099) | mfKIRnew21 (EU419098) | ||||
| mfKIRnew23 (EU419100) | mfKIRnew59 (EU419126) | ||||
| mfKIRnew25 (EU419103) | mfKIRnew61 (EU688994) | ||||
| mfKIRnew28 (EU419105) | mfKIRnew10 (EU419086) | ||||
| K6 | mfKIRnew60 (EU688993) | mfKIRnew49 (EU419122) | 3 | ||
| mfKIRnew62 (EU688995) | |||||
| K7 | mfKIRnew34 (EU419108) | mfKIRnew07 (EU419080) | 6 | ||
| mfKIRnew11 (EU419087) | |||||
| mfKIRnew15 (EU419093) | |||||
| mfKIRnew29 (EU419107) | |||||
| K8 | mfKIRnew03 (EU419077) | 4 | |||
| mfKIRnew09 (EU419084) | |||||
| mfKIRnew17 (EU419094) |
Cynomolgus macaque KIRs are designated mfKIR. Each novel MCM sequence has been assigned a GenBank ID and temporary name.
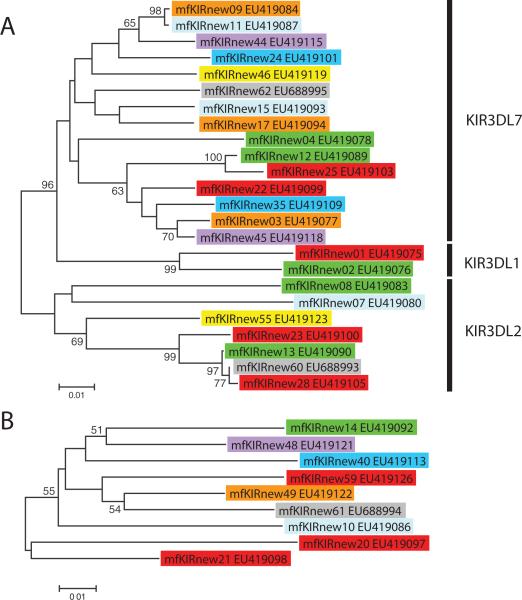
Fig. 3 shows a phylogenetic tree constructed using full-length amino acid sequences from all MCM KIR3DL (Fig. 3A) and KIR3DH (Fig. 3B) molecules. The mfKIR3DL molecules fell into three lineages (Fig. 3). These lineages have been named based on the closest published rhesus macaque homolog. The first group resembled mmKIR3DL1 (AF334616) and contains KIR3DL sequences from haplotypes K2 and K5. The second was most similar to mmKIR3DL2 (NM_001104553). KIRs from the second group were present on all MCM haplotypes except K1, K4, and K8 and there were two members of this group present in K5. The largest group was most similar to mmKIR3DL7 (AF334622). Most haplotypes contained two members of group 3, with the exception of K8, which contained three members and K3 and K6, each of which contained just one member of this group. Because most haplotypes contain multiple members from this group, we believe that this phylogenetic group represents two distinct loci. In haplotype K2, we detected a KIR3D sequence (EU419079) containing a 7nt deletion in the third Ig domain that results in a truncated molecule lacking both transmembrane and intracellular domains. Its extra-cellular domain shared 96.4% nucleotide identity with mmKIR3DL7 and we hypothesize it is a member of this group.
FIGURE 3.
Phylogenetic analysis of MCM KIR3D sequences. A, The tree contains KIR3DL sequences. B, Shows KIR3DH sequences. Vertical bars indicate phylogenetic groups. The colors highlighting sequence names correspond to their haplotype.
In the phylogenetic analysis of KIR3DH sequences, there were no clearly differentiated groups, making it difficult to identify potential distinct loci. All haplotypes except K3 contained at least one KIR3DH sequence, with three KIR3DH sequences expressed by haplotype K5. Two of the KIR3DH sequences from K5, mfKIRnew20 (EU419097) and mfKIRnew21 (EU419098), were significantly divergent from the other mfKIR3DH sequences. Of these, mfKIRnew20 shares greater similarity (95.4%) within its extracellular domain to mfKIR3DL1 molecules than to other mfKIR3DH molecules.
As compared with mfKIR3DL/H molecules, mfKIR1D and mfKIR2D were less polymorphic and prevalent within MCM haplotypes. mfKIR1D sequences were detected in haplotypes K1, K2, and K3. The sequences from K1 and K2 are >97% similar on the amino acid level to one another. These sequences are highly similar to rhesus macaque mmKIR1D molecules and each share >96% amino acid identity with mmKIR1D splice variant 6 (AF334640). A novel KIR lacking any Ig-like domains was detected in haplotype K3. Although it lacks Ig domains, it shares high sequence similarity to mfKIR1D and is presumably yet another mfKIR1D splice variant.
We identified KIR2DL molecules on three haplotypes: K1, K4, and K7. Based on amino acid sequence, these KIRs resemble either mmKIR2DL4 (K1 and K4) or mmKIR2DL5 (K7). The mfKIR2DL4 molecules are >98% identical to the amino acid level with one another and share >98% amino acid similarity to the rhesus macaque sequence mmKIR2DL4.2 (AF334645). A KIR2DL5 molecule was detected in haplotype K7. This sequence was 97% identical to the amino acid level to the rhesus macaque sequence mmKIR2DL5.1 (AF334646). In the same haplotype, we detected a KIR3D sequence identical to mfKIR2DL5 with the exception that it contained an additional Ig domain (accession no. EU419107). Similar findings have been reported in rhesus macaques with mmKIR2DL5/mmKIR3DL20 and it is likely these are alternatively spliced transcripts from the same locus. The KIR2DL5/KIR3DL20 locus has been speculated to represent an evolutionary intermediate between KIR2DL and KIR3DL genes (35). Because the KIR3DL version of this transcript lacks a stop codon with the region sequenced, we hypothesize that this variant is nonfunctional and have classified the pair as a KIR2DL molecule.
Order of KIR loci
Using segregation analysis, we previously defined the relative order of microsatellite markers. The majority of these microsatellites are located within KIR introns, primarily intron 4. By determining the KIR sequences physically linked to these microsatellite alleles, the segregation patterns of microsatellite alleles in recombinant haplotypes can be used to determine the relative positions of KIR genes. To accomplish this, we cloned and sequenced genomic DNA amplicons containing KIR intron 4 as well as the flanking exons, which encode Ig domains D1 and D2. For each genomic DNA amplicon, we used the sequence of the exons to identify the corresponding cDNA transcript. Next, we examined the intron sequence for sites recognized by any of the microsatellite primers and calculated the size of the predicted PCR product. These values were compared with the microsatellite alleles we determined experimentally. A complete summary of the microsatellite alleles and their linked KIR transcripts is shown in supplemental Table II.4 Although the corresponding microsatellite was not identified for all KIR sequences, in most cases we were able to determine whether the sequence was linked to a microsatellite located at either the telomeric or centromeric end of the region. Of the KIR3D sequences we detected, only KIR2DL5/KIR3DL20 (EU419108/EU419107) was linked to a marker located at the centromeric end (Table II). All other detected KIR3D sequences were linked to a microsatellite in the telomeric region.
Table II.
Marker of KIR3DL and KIR3DH alleles located at end of region
| KIR3DL | KIR3DH | |
|---|---|---|
| Centromeric | mfKIRnew29 (EU419107) | |
| Telomeric | mfKIRnew55 (EU419123) | mfKIRnew05 (EU419079) |
| mfKIRnew01 (EU419075) | mfKIRnew14 (EU419092) | |
| mfKIRnew03 (EU419077) | mfKIRnew20 (EU419097) | |
| mfKIRnew04 (EU419078) | mfKIRnew40 (EU419113) | |
| mfKIRnew09 (EU419084) | mfKIRnew48 (EU419121) | |
| mfKIRnew11 (EU419087) | mfKIRnew42 (EU419122) | |
| mfKIRnew12 (EU419089) | mfKIRnew61 (EU688994) | |
| mfKIRnew15 (EU419093) | ||
| mfKIRnew17 (EU419094) | ||
| mfKIRnew23 (EU419100) | ||
| mfKIRnew25 (EU419103) | ||
| mfKIRnew28 (EU419105) | ||
| mfKIRnew35 (EU419109) | ||
| mfKIRnew44 (EU419115) | ||
| mfKIRnew45 (EU419118) | ||
| mfKIRnew46 (EU419119) | ||
| Undetermined | mfKIRnew13 (EU419090) | mfKIRnew10 (EU419086) |
| mfKIRnew02 (EU419076) | mfKIRnew14 (EU419092) | |
| mfKIRnew07 (EU419080) | mfKIRnew21 (EU419098) | |
| mfKIRnew08 (EU419083) | ||
| mfKIRnew22 (EU419099) | ||
| mfKIRnew23 (EU419100) | ||
| mfKIRnew24 (EU419101) | ||
| mfKIRnew25 (EU419103) | ||
| mfKIRnew28 (EU419105) | ||
| mfKIRnew60 (EU688993) | ||
| mfKIRnew62 (EU688995) |
In several cases, we identified a genomic DNA fragment with exons that did not match any transcripts identified in that haplotype. We believe these represent pseudogenes, although it is possible that the corresponding transcripts were not detected due to low expression or primer selectivity.
A model for MCM KIR haplotypes
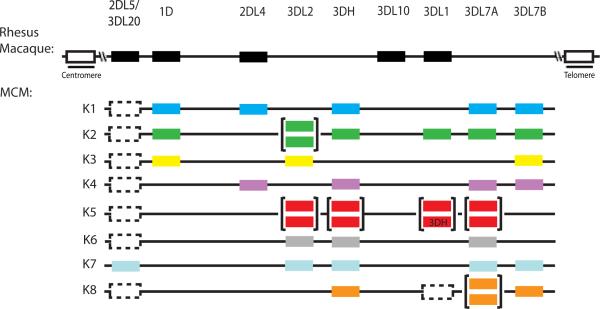
By comparing the gene content of the MCM KIR haplotypes, we constructed a model of MCM KIR genomic organization (Fig. 4). Unlike human KIRs, it appears the majority of macaque KIRs are located in the telomeric end of the region, where significant expansion of KIR3D loci has occurred. We defined four putative KIR3DL loci: KIR3DL1, KIR3DL2, KIR3DL7A, and KIR3DL7B along with one putative KIR3DH locus. These loci have been named based on the closest rhesus macaque homolog. Interestingly, haplotype K5 appears to have undergone a major duplication and contains two copies of every locus at the telomeric end of the region, with the exception that it lacks KIR3DL7B. The centromeric end of the region is comparatively gene poor, with only two loci detected: KIR2DL5/3DL20 and KIR1D.
FIGURE 4.
Model for MCM KIR genomic organization. The gene organization is illustrated for the published rhesus macaque haplotype (top) and MCM haplotypes (bottom). MCM organization has been inferred using phylogenetic analysis. Loci have been named based on the closest rhesus macaque homolog. Where identified, pseudogenes are indicated (dotted box). Brackets indicate duplications of a locus. The linear order of loci within the telomeric region is arbitrary.
Comparison with rhesus macaque KIRs
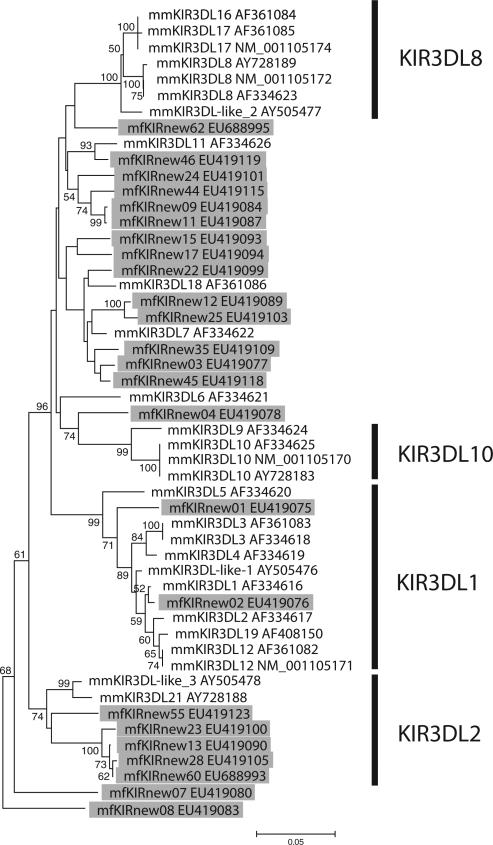
Using the framework defined in MCM, we analyzed all published rhesus macaque KIR3DL sequences. A phylogenetic tree constructed using full-length amino acid sequence is shown in Fig. 5. MCM sequences are highlighted in gray. Rhesus macaque sequences are labeled with both GenBank ID and sequence name under which they were published. There is significant redundancy among these names, which highlights the need to establish a formal system of classification for macaque KIRs.
FIGURE 5.
Phylogenetic tree of MCM and rhesus macaque KIR3DL sequences. MCM sequences are highlighted in gray; rhesus sequences are not highlighted. Phylogenetic groups are indicated (vertical bar).
There were four well-defined phylogenetic groups (Fig. 5, vertical bar). Two corresponded to groups defined in MCM, KIR3DL1 and KIR3DL2, and contained both MCM and rhesus sequences. There were also two groups present in rhesus macaques, but absent from MCM: KIR3DL8 and KIR3DL10. The absence of these groups in MCM could be due to species differences or result from the founder effect in the Mauritian population. The remaining macaque KIR3DL molecules did not form a well-defined phylogenetic group. These include those sequence defined as KIR3DL7 in MCM. It should be noted that KIR3DL7 was the most diverse of MCM KIR3DL groups. Given the data from MCM, we speculate that these sequences represent at least two loci. However, it is difficult to draw conclusions because only a single rhesus macaque haplotype has been characterized.
Discussion
Our results fundamentally advance understanding of KIR genetics in macaques. In this study, we characterize eight complete KIR haplotypes from MCM. The limited genetic diversity of this population creates several advantages for the study of KIRs. Unlike normal outbred populations, it is feasible to identify subjects homozygous throughout the KIR region. This identification provides a simplified system in which to describe KIR genetics, which can then be applied to broader macaque populations. MCM are also an extremely powerful model for the study of genetics in disease pathogenesis.
We demonstrate that eight KIR haplotypes account for virtually all KIR genetic diversity in MCM. We characterized the gene content of each haplotype, identifying a total of 40 novel KIR cDNA sequences. This nearly doubles the number of published macaque KIRs. A key feature of this study is that we characterized the gene content of complete haplotypes, rather than isolated cDNA sequences. By comparing the gene content of these haplotypes, we created a model for MCM KIR genomic organization, defining four presumptive KIR3DL loci, one KIR3DH, one KIR1D, and two KIR2DL. Using this framework, we analyzed KIR3DL sequences from rhesus macaques. Rhesus macaque KIR3DL molecules fall into four well-defined groups, two of which are not present in MCM. The remaining sequences do not form a highly similar phylogenetic group. This is unlike human KIR3DL organization, in which three loci exist, and the alleles of each locus are highly similar to one another (11). The distinct HLA/MHC class I organization between humans and macaques may at least partially explain these differences. Humans encode three classical HLA class I loci: HLA-A, HLA-B, and HLA-C. Macaques have a significantly more complex MHC organization, containing multiple duplications of the MHC class IA and MHC class IB loci (51, 52). In humans, KIR3DL molecules interact with HLA-A and HLA-B molecules. Given the evidence for co-evolution of KIR with MHC class I, the expansion of macaque KIR3DL loci would be expected.
There are still many questions about macaque KIR biology that need to be addressed. We have presented a reasonable model for macaque genomic organization with the data available. As additional sequence data becomes available, this model may be refined. Although we demonstrate that our findings offer insight into rhesus macaque KIR organization, it is important to note that due to the rapid evolution of the KIR region, it is likely there will be significant differences between macaque species. This evolution should be taken into consideration when applying any genotyping technique across species. For these reasons, it will be essential to characterize additional KIR haplotypes and alleles from multiple species, either by sequencing or segregation analysis. Because rhesus macaques are commonly used in biomedical research, this species should be a priority. However, studies should be conducted in additional species such as pigtail macaques and cynomolgus macaques from outbred populations. In addition to genetic characterization, it will be essential to identify the ligands of macaque KIRs.
The KIR genetics of outbred macaque populations will require additional studies to unravel. Because disease models such as SIV/AIDS have significant investments in rhesus macaques, work should be conducted to understand KIR genetics in these animals. However, KIR biologists may consider alternate macaque populations. We have identified virtually all KIR and MHC class I molecules expressed within the entire MCM population. By virtue of their reduced genetic complexity, MCM provide a powerful model to study the role of genetics in disease pathogenesis. KIR signaling is influenced by the entire KIR/MHC genotype of the host. Subtle differences in KIR genotype including allelic variability and differences in regulatory elements can also modulate KIR function (20). Unlike virtually any other primate model, cohorts of MCM can be created that are completely KIR or MHC identical. This reduction in genetic variability creates significant advantages for the study of KIRs in disease pathogenesis. For example, NK activity can be compared between MCM with distinct KIR genotypes but identical MHC class I genetics. The KIR/MHC haplotypes could be selected to address questions such as whether an increased number of activating KIRs promotes disease protection or to test the protective capacity of specific KIR/MHC pairs. This novel model removes a major obstacle hindering the study of KIR function and creates opportunity for significant advancement in the understanding of KIRs in disease.
Supplementary Material
Acknowledgments
We thank Dr. T. Friedrich and A. Blasky for helpful comments and technical support. This publication's contents are solely the responsibility of the authors and do not necessarily represent the official views of the research sponsors.
Footnotes
This work was supported by Grant R24 RR021745-01A1 from National Institutes of Health (NIH), Contract number HHSN266200400088C/N01-AI-40088 from the National Institute of Allergy and Infectious Diseases, and by the International AIDS Vaccine Initiative. B.N.B. received support from NIH National Research Service Award T32 GM007215 from the National Institute of General Medical Sciences. A.L.H. received support by Grant GM43940 from the NIH. This research was conducted in part at a facility constructed with support by Grants RR15459-01 and RR020141-01 from the Research Facilities Improvement Program.
Abbreviations used in this paper: KIR, killer Ig-like receptor; MCM, Mauritian cynomolgus macaques.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat. Rev. Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter G, Rihn S, Streeck H, Teigen N, Piechocka-Trocha A, Moss K, Cohen K, Meier A, Pereyra F, Walker B, Altfeld M. Ligand independent exhaustion of killer immunoglobulin-like receptor-positive CD8+ T cells in human immunodeficiency virus type 1 infection. J. Virol. 2008;82:9668–9677. doi: 10.1128/JVI.00341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardiner CM. Killer cell immunoglobulin-like receptors on NK cells: the how, where and why. Int. J. Immunogenet. 2008;35:1–8. doi: 10.1111/j.1744-313X.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- 4.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parham P. Taking license with natural killer cell maturation and repertoire development. Immunol. Rev. 2006;214:155–160. doi: 10.1111/j.1600-065X.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 6.Kim S, Poursine-Laurent J, Truscott S, Lybarger L, Song Y, Yang L, French A, Sunwoo J, Lemieux S, Hansen T, Yokoyama W. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 7.Moretta L, Moretta A. Killer immunoglobulin-like receptors. Curr. Opin. Immunol. 2004;16:626–633. doi: 10.1016/j.coi.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Long E, Rajagopalan S. HLA class I recognition by killer cell Ig-like receptors. Semin. Immunol. 2000;12:101–108. doi: 10.1006/smim.2000.0212. [DOI] [PubMed] [Google Scholar]
- 9.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 10.Parham P. Killer cell immunoglobulin-like receptor diversity: balancing signals in the natural killer cell response. Immunol. Lett. 2004;92:11–13. doi: 10.1016/j.imlet.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Bashirova A, Martin M, McVicar D, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu. Rev. Genomics Hum. Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 12.Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J. Clin. Invest. 2008;118:1017–1026. doi: 10.1172/JCI32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yawata M, Yawata N, Draghi M, Little A, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrington M, Martin M. The impact of variation at the KIR gene cluster on human disease. Curr. Top. Microbiol. Immunol. 2006;298:225–257. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- 15.Wilson M, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu K, Chida S, Geraghty D, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol. Rev. 2002;190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 17.Witt C, Dewing C, Sayer D, Uhrberg M, Parham P, Christiansen F. Population frequencies and putative haplotypes of the killer cell immunoglobulin-like receptor sequences and evidence for recombination. Transplantation. 1999;68:1784–1789. doi: 10.1097/00007890-199912150-00024. [DOI] [PubMed] [Google Scholar]
- 18.Anfossi N, André P, Guia S, Falk C, Roetynck S, Stewart C, Breso V, Frassati C, Reviron D, Middleton D, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Shilling HG, Young N, Guethlein LA, Cheng NW, Gardiner CM, Tyan D, Parham P. Genetic control of human NK cell repertoire. J. Immunol. 2002;169:239–247. doi: 10.4049/jimmunol.169.1.239. [DOI] [PubMed] [Google Scholar]
- 20.O'Connor G, Guinan K, Cunningham R, Middleton D, Parham P, Gardiner C. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J. Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 21.Yu J, Heller G, Chewning J, Kim S, Yokoyama WM, Hsu KC. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J. Immunol. 2007;179:5977–5989. doi: 10.4049/jimmunol.179.9.5977. [DOI] [PubMed] [Google Scholar]
- 22.Uhrberg M. Shaping the human NK cell repertoire: an epigenetic glance at KIR gene regulation. Mol. Immunol. 2005;42:471–475. doi: 10.1016/j.molimm.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 23.Khakoo S, Thio C, Martin M, Brooks C, Gao X, Astemborski J, Cheng J, Goedert J, Vlahov D, Hilgartner M, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 24.Carrington M, Wang S, Martin M, Gao X, Schiffman M, Cheng J, Herrero R, Rodriguez A, Kurman R, Mortel R, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J. Exp. Med. 2005;201:1069–1075. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin M, Qi Y, Gao X, Yamada E, Martin J, Pereyra F, Colombo S, Brown E, Shupert W, Phair J, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin M, Gao X, Lee J, Nelson G, Detels R, Goedert J, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 27.Hansen D, D'Ombrain M, Schofield L. The role of leukocytes bearing natural killer complex receptors and killer immunoglobulin-like receptors in the immunology of malaria. Curr. Opin. Immunol. 2007;19:416–423. doi: 10.1016/j.coi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, Vlahiotis A, Piccirillo JF, Cella M, Colonna M, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc. Natl. Acad. Sci. USA. 2008;105:3053–3058. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goulder P, Brander C, Tang Y, Tremblay C, Colbert R, Addo M, Rosenberg E, Nguyen T, Allen R, Trocha A, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- 30.Loffredo J, Burwitz B, Rakasz E, Spencer S, Stephany J, Vela J, Martin S, Reed J, Piaskowski S, Furlott J, et al. The antiviral efficacy of simian immunodeficiency virus-specific CD8+ T cells is unrelated to epitope specificity and is abrogated by viral escape. J. Virol. 2007;81:2624–2634. doi: 10.1128/JVI.01912-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bontrop RE, Watkins DI. MHC polymorphism: AIDS susceptibility in non-human primates. Trends Immunol. 2005;26:227–233. doi: 10.1016/j.it.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Loffredo J, Maxwell J, Qi Y, Glidden C, Borchardt G, Soma T, Bean A, Beal D, Wilson N, Rehrauer W, et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedrich T, Dodds E, Yant L, Vojnov L, Rudersdorf R, Cullen C, Evans D, Desrosiers R, Mothé B, Sidney J, et al. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 2004;10:275–281. doi: 10.1038/nm998. [DOI] [PubMed] [Google Scholar]
- 34.Wojcechowskyj J, Yant L, Wiseman R, O'Connor S, O'Connor D. Control of simian immunodeficiency virus SIVmac239 is not predicted by inheritance of Mamu-B*17-containing haplotypes. J. Virol. 2007;81:406–410. doi: 10.1128/JVI.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, Parham P, Carrington M, Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hershberger K, Shyam R, Miura A, Letvin N. Diversity of the killer cell Ig-like receptors of rhesus monkeys. J. Immunol. 2001;166:4380–4390. doi: 10.4049/jimmunol.166.7.4380. [DOI] [PubMed] [Google Scholar]
- 37.Grendell RL, Hughes AL, Golos TG. Cloning of rhesus monkey killer-cell Ig-like receptors (KIRs) from early pregnancy decidua. Tissue Antigen. 2001;58:329–334. doi: 10.1034/j.1399-0039.2001.580507.x. [DOI] [PubMed] [Google Scholar]
- 38.Wiseman R, Wojcechowskyj J, Greene J, Blasky A, Gopon T, Soma T, Friedrich T, O'Connor S, O'Connor D. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J. Virol. 2007;81:349–361. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 40.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 44.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 45.Sussman RW, Tattersall I. Distribution, abundance, and putative. Folia Primatol. 1986;46:28–43. [Google Scholar]
- 46.Lawler SH, Sussman RW, Taylor LL. Mitochondrial DNA of the Mauritian macaques (Macaca fascicularis): an example of the founder effect. Am. J. Phys. Anthropol. 1995;96:133–141. doi: 10.1002/ajpa.1330960203. [DOI] [PubMed] [Google Scholar]
- 47.O'Connor S, Blasky A, Pendley C, Becker E, Wiseman R, Karl J, Hughes A, O'Connor D. Comprehensive characterization of MHC class II haplotypes in Mauritian cynomolgus macaques. Immunogenetics. 2007;59:449–462. doi: 10.1007/s00251-007-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Florese RH, Wiseman RW, Venzon D, Karl JA, Demberg T, Larsen K, Flanary L, Kalyanaraman VS, Pal R, Titti F, et al. Comparative study of Tat vaccine regimens in Mauritian cynomolgus and Indian rhesus macaques: influence of Mauritian MHC haplotypes on susceptibility/resistance to SHIV(89.6P) infection. Vaccine. 2008;26:3312–3321. doi: 10.1016/j.vaccine.2008.03.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR, Kidd KK, Carrington M. Global diversity and evidence for coevolution of KIR and HLA. Nat. Genet. 2007;39:1114–1119. doi: 10.1038/ng2077. [DOI] [PubMed] [Google Scholar]
- 50.Robinson J, Marsh SG. IPD: the Immuno Polymorphism Database. Methods Mol. Biol. 2007;409:61–74. doi: 10.1007/978-1-60327-118-9_4. [DOI] [PubMed] [Google Scholar]
- 51.Otting N, de Vos-Rouweler AJ, Heijmans CM, de Groot NG, Doxiadis GG, Bontrop RE. MHC class I A region diversity and polymorphism in macaque species. Immunogenetics. 2007;59:367–375. doi: 10.1007/s00251-007-0201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc. Natl. Acad. Sci. USA. 2005;102:1626–1631. doi: 10.1073/pnas.0409084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.