Abstract
Abscisic acid (ABA) is postulated to be a ubiquitous hormone that plays a central role in seed development and responses to environmental stresses of vascular plants. However, in liverworts (Marchantiophyta), which represent the oldest extant lineage of land plants, the role of ABA has been least emphasized; thus, very little information is available on the molecular mechanisms underlying ABA responses. In this study, we isolated and characterized MpABI1, an ortholog of ABSCISIC ACID INSENSITIVE1 (ABI1), from the liverwort Marchantia polymorpha. The MpABI1 cDNA encoded a 568-amino acid protein consisting of the carboxy-terminal protein phosphatase 2C (PP2C) domain and a novel amino-terminal regulatory domain. The MpABI1 transcript was detected in the gametophyte, and its expression level was increased by exogenous ABA treatment in the gemma, whose growth was strongly inhibited by ABA. Experiments using green fluorescent protein fusion constructs indicated that MpABI1 was mainly localized in the nucleus and that its nuclear localization was directed by the amino-terminal domain. Transient overexpression of MpABI1 in M. polymorpha and Physcomitrella patens cells resulted in suppression of ABA-induced expression of the wheat Em promoter fused to the β -glucuronidase gene. Transgenic P. patens expressing MpABI1 and its mutant construct, MpABI1-d2, lacking the amino-terminal domain, had reduced freezing and osmotic stress tolerance, and associated with reduced accumulation of ABA-induced late embryogenesis abundant-like boiling-soluble proteins. Furthermore, ABA-induced morphological changes leading to brood cells were not prominent in these transgenic plants. These results suggest that MpABI1 is a negative regulator of ABA signaling, providing unequivocal molecular evidence of PP2C-mediated ABA response mechanisms functioning in liverworts.
Land plants are repeatedly challenged by environmental stresses such as desiccation, salinity, and freezing, which can cause irreversible damage to intracellular structures and membranes by severe dehydration. Abscisic acid (ABA), which is known to be a phytohormone responsible for physiological control of seed maturation and germination, bud dormancy, and stomata closure, has been postulated to be involved in the development of cellular dehydration tolerance under stress conditions. In higher plants, water-stress conditions trigger a transient or sustained increase in endogenous ABA content in tissues. Increased ABA stimulates the expression of a number of stress-related genes such as those of late embryogenesis abundant (LEA) proteins, which are thought to protect cells from the damage caused by the dehydration stress (Chandler and Robertson, 1994). It has been demonstrated that various tissue-cultured cells of vascular plants, including those of angiosperms, gymnosperms, and ferns, develop desiccation and freezing tolerance upon exogenous ABA treatment, indicating that ABA plays a key role in stress tolerance at cellular levels (Reynolds and Bewley, 1993; Attree et al., 1995; Shiota et al., 1999; Gusta et al., 2005).
The role of ABA in desiccation and freezing stress tolerance has been implicated not only in vascular plants but also in nonvascular plants (i.e. bryophytes). In mosses (Bryophyta), it has been shown that desiccation tolerance of protonema cells of Funaria hygrometrica is increased by exogenous ABA treatment (Werner et al., 1991). Pretreatment with ABA has been shown to be effective for cryopreservation of several moss species (Pence, 1998). We previously reported that the moss Physcomitrella patens increases freezing tolerance upon ABA treatment with increased expression of stress-related genes (Minami et al., 2003, 2006). Analysis of endogenous ABA in P. patens revealed that hyperosmotic stress treatment of its protonemata increases the levels of endogenous ABA (Minami et al., 2005). The role of ABA in stress responses has been studied in another class of bryophytes, liverworts (Marchantiophyta). It has been shown that exogenous application of ABA induces desiccation tolerance of the xerophilic liverwort Exormotheca holstii (Hellwege et al., 1994). Exogenously applied ABA also induces changes in Riccia fluitans from the aqueous form to the desiccation-tolerant land form (Hellwege et al., 1996; Pence et al., 2005). These studies have suggested that the mechanisms for ABA responses are conserved in vascular and nonvascular plants.
In higher plants, genetic analysis of ABA-insensitive mutants of Arabidopsis (Arabidopsis thaliana) has resulted in the identification of genes involved in ABA-induced signaling processes. The ABSCISIC ACID INSENSITIVE1 (ABI1) gene has been cloned from Arabidopsis by molecular analysis of an ABA-insensitive mutant, abi1-1. The abi1-1 plants had seeds with reduced dormancy and vegetative tissues susceptible to wilting. Exogenous application of ABA had limited effects on the inhibition of seed germination and closure of stomata in the abi1-1 plants. Cloning of ABI1 revealed that the gene encoded a 434-amino acid polypeptide consisting of two distinct domains: the N-terminal domain with no sequence similarity to known protein sequences and the C-terminal type 2C protein phosphatase (PP2C) domain (Leung et al., 1994; Meyer et al., 1994). It was found that abi1-1 had a single Gly-to-Asp substitution at amino acid 180 in the PP2C domain, which caused a drastic reduction in the phosphatase activity in vitro but conferred a semidominant phenotype in vivo. Another ABA-insensitive locus, abi2-1, has also been analyzed, and it was shown that ABI2 encodes the ABI1-related PP2C (Leung et al., 1997; Rodriguez et al., 1998). ABI1 is thought to affect ABA-dependent signaling processes by interacting with the SNF1-like Ser/Thr protein kinase OST1/SnRK2.6 (Ohta et al., 2003; Yoshida et al., 2006a), which phosphorylates and regulates bZIP transcription factors such as AREB1, AREB2, and ABI5 (Furihata et al., 2006).
Analyses of transgenic plants and gene knockout lines have indicated that ABI1 is a negative regulator of ABA signaling. Overexpression of ABI1 resulted in an ABA-insensitive phenotype, while its disruption or antisense inhibition resulted in an ABA-hypersensitive phenotype (Sheen, 1998; Gosti et al., 1999; Tähtiharju and Palva, 2001). Later studies revealed that ABI1-like genes are conserved in vascular plants and play a role in the negative regulation of stress responses (González-García et al., 2003). Among seventy-six PP2Cs encoded in the Arabidopsis genome, ABI1, ABI2, HAB1, HAB2, and AtPP2CA are members of group A PP2C, which possibly functions in the regulation of ABA signaling (Schweighofer et al., 2004). Transgenic plants overexpressing these PP2Cs typically show ABA-insensitive phenotypes such as slow stomata closure and enhanced seed germination by exogenously applied ABA, while the knockout lines of these genes exhibit ABA-hypersensitive phenotypes (Yoshida et al., 2006b). Recent studies indicated that negative regulation by the ABI1-related PP2Cs plays a central role in ABA signaling through signal perception and transduction mediated by ABA receptors (Ma et al., 2009; Park et al., 2009).
In comparison with accumulated knowledge of signaling molecules involved in ABA responses in higher plants, very little is known about the ABA signaling processes leading to stress tolerance in bryophytes. Marella et al. (2006) identified PpABI3, which positively regulates ABA-induced expression of the wheat (Triticum aestivum) Em promoter fused to the GUS reporter gene (Em-GUS). This indicated that transcriptional machinery for ABA-induced gene expression is conserved in mosses. In this study, we focused on analysis of ABA responses in liverworts, which constitute the oldest extant land plant lineage among three bryophyte classes. Despite the experimental facts that show increased stress tolerance induced by exogenous ABA application, the physiological role of ABA in liverworts has been underestimated because it has been believed that liverworts do not have endogenous ABA. However, Li et al. (1994) reported that gametophytes of Marchantia polymorpha contain endogenous ABA at concentrations similar to those in higher plants. It was also reported that E. holstii and R. fluitans contain endogenous ABA (Hellwege et al., 1992, 1994). These results suggested that ABA acts as a hormone in liverworts. Considering the fact that liverworts are the earliest diverging branch of land plants, clarification of the mechanisms of ABA sensing and signal transduction will be key for understanding evolutionary aspects of the utilization of this substance by land plant ancestors as a stress hormone for their adaptation to the terrestrial environment.
In this study, we took a molecular approach to analyze the functional conservation of cellular processes involved in the regulation of ABA signaling in liverworts. A sequence search was carried out in the EST database of M. polymorpha to find orthologs of known signaling molecules involved in ABA-dependent processes. One of clones was found to have high similarity to ABI1 in the conserved PP2C domain. This clone, designated as MpABI1, was used for in vitro and in vivo analyses to clarify its physiological roles. Analysis of transgenic plants expressing MpABI1 revealed that it has a conserved function as a regulator of ABA-dependent signaling processes.
RESULTS
Response of M. polymorpha to ABA
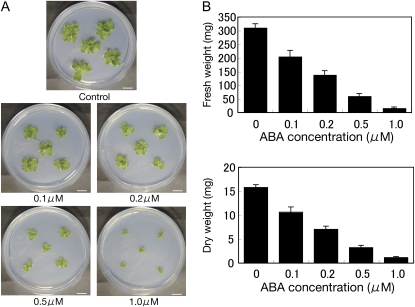
M. polymorpha has been reported as a liverwort species that shows little ABA responses (Pence et al., 2005). Thus, we first examined ABA responses of M. polymorpha gametophytes by using axenic culture of thalli and gemmae. When a piece of mature thallus cultured for 3 weeks was planted in the medium containing 10 μm ABA, it did not show significant changes in growth in comparison with that grown in normal medium (data not shown). However, when a gemma was planted in the ABA-containing medium, we observed that its growth was effectively inhibited by submicromolar concentrations of ABA (Fig. 1A). An inhibitory effect of ABA on gemmae growth was confirmed by measurement of their fresh and dry weights (Fig. 1B). These results indicated that M. polymorpha gametophytes have sensitivity to ABA, which convinced us that M. polymorpha has mechanisms for ABA signaling.
Figure 1.
Effect of ABA on growth of the M. polymorpha gametophyte. Gemmae of M. polymorpha gametophytes were grown on control M51C agar medium or medium containing 0.1, 0.2, 0.5, or 1.0 μm ABA. After photographs had been taken, the gametophyte tissues were used for measurement of fresh weight. The tissues were then dried at 90°C for 24 h for measurement of dry weight. A, Photographs of representative gametophytes grown for 4 weeks. Bars = 10 mm. B, Fresh weight and dry weight of gametophytes grown for 4 weeks (n = 5).
Isolation of a cDNA Clone of MpABI1
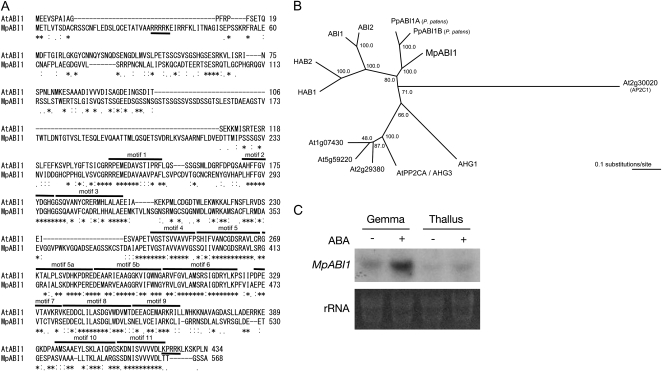
A BLAST search against published and unpublished M. polymorpha ESTs was carried out to find sequences with similarity to genes involved in ABA-dependent processes. The search resulted in the identification of one assembled sequence that has amino acid sequence similarity to ABI1. Individual ESTs belonging to the same sequence were derived from young and mature thalli. On the basis of the sequences identified, the cDNA containing the longest open reading frame was amplified by PCR using first-strand cDNA synthesized from the mRNA isolated from male mature thalli. The amplified clone encoding a protein with 568 amino acids was designated as MpABI1 (GenBank accession no. GQ504039). Sequence analysis revealed that the MpABI1 polypeptide contains the PP2C domain in its C-terminal half domain (amino acid residues 253–568), with characteristic conserved amino acid motifs (Bork et al., 1996; Fig. 2A). Sequence comparison with known protein sequences revealed that MpABI1 had sequence similarity to ABI1, ABI2, HAB1, HAB2, and PP2CA of Arabidopsis in the C-terminal half PP2C domain. The PP2C domain of MpABI1 had the highest sequence similarity to ABI1 orthologs of P. patens, PpABI1A and PpABI1B (Komatsu et al., 2009). Phylogenetic analysis indicated that MpABI1 is clustered with group A PP2Cs of Arabidopsis and PpABI1A and PpABI1B of P. patens (Fig. 2B). In contrast, the sequence of the N-terminal domain (1–252) had low similarity to that of ABI1. However, sequence analysis revealed that the N-terminal half domain contained a putative nuclear localization signal (NLS) with a stretch of basic amino acid residues: pat4 motifs in amino acids 31 to 35 (RRRRK; Hicks and Raikhel, 1995).
Figure 2.
Molecular characteristics of MpABI1. A, Sequence alignment of MpABI1 and Arabidopsis ABI1. The alignment was made by the T-COFFEE program. Identical amino acids are marked with asterisks, very similar ones are marked with colons, and weakly similar ones are marked with dots. Conserved motifs 1 to 11 of PP2Cs assigned by Bork et al. (1996) are indicated by overlines. Putative NLSs are underlined. B, Phylogenetic representations of MpABI1, PpABI1A, PpABI1B, and Arabidopsis ABI1-related PP2Cs. The phylogenetic tree was built on the alignment of amino acid sequences of the PP2C domains using the ClustalW program. The Arabidopsis Ap2C1 phosphatase sequence was used as the outgroup. The bar indicates 0.1 substitutions per site. C, Northern analysis of M. polymorpha mRNA for examination of MpABI1 expression. Total RNAs isolated from ABA-treated (+) or nontreated (−) gemma and thallus were electrophoresed, blotted onto a nylon membrane, and hybridized with a 32P-labeled MpABI1 cDNA probe. Ethidium bromide-stained rRNA of each sample is shown to verify equal loading.
In order to examine the expression of MpABI1 transcripts in M. polymorpha tissues, total RNA was isolated from mature thallus and gemma and used for northern-blot analysis. It has been reported that ABI1 and ABI2 transcripts are induced by exogenous ABA treatment in Arabidopsis (Leung et al., 1997). Thus, ABA-induced expression of MpABI1 in M. polymorpha thallus and gemma was examined. The results showed that transcripts of MpABI1 were expressed both in the thallus and gemma. Furthermore, the expression was remarkably increased by exogenous ABA treatment in the gemma (Fig. 2C).
MpABI1 Is a Protein Phosphatase with Divalent Cation-Dependent Activity
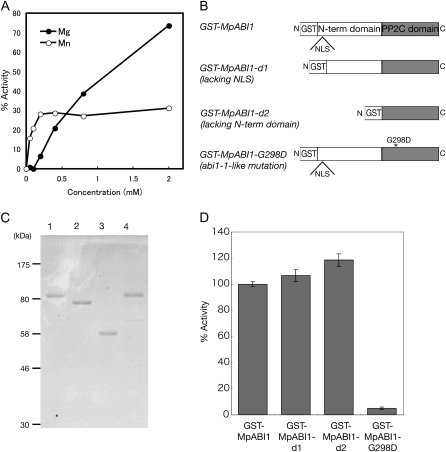
To determine phosphatase activity of MpABI1, glutathione-S-transferase fusion proteins of MpABI1 (GST-MpABI1) were expressed in Escherichia coli and purified by glutathione-Sepharose affinity chromatography. An isolated protein with an approximate molecular mass of 85 kD was used for in vitro phosphatase assays using 32P-labeled myelin basic protein (MBP) phosphorylated by a protein Ser/Thr kinase as a substrate (Takezawa, 2003). The enzyme showed no activity in the presence of 0.1 mm EDTA. Divalent cations, Mg2+ and Mn2+, increased phosphatase activity of GST-MpABI1 in a concentration-dependent manner (Fig. 3A). The enzyme activity was differentially affected by the two cations: Mn2+ activated the enzyme at a much lower concentration than did Mg2+. The concentration of Mn2+ required for half-maximal activity was approximately 50 μm, and the activity was nearly saturated at 200 μm. In contrast, there was no enzyme activation by Mg2+ up to a concentration of 100 μm. The enzyme was activated by Mg2+ at concentrations greater than 200 μm, and the activity was not saturated even by 2 mm Mg2+. These results indicated that MpABI1 has higher affinity to Mn2+ than to Mg2+ but that Mg2+ is required for its maximal activity.
Figure 3.
Protein phosphatase assays of GST-MpABI1 fusion proteins. A, Mg2+- and Mn2+-dependent activities of GST-MpABI1 toward the 32P-labeled MBP. B, Schematic representation of various GST-MpABI1 constructs used for protein phosphatase assays. C, SDS-PAGE of GST fusion proteins of MpABI1 and its mutant proteins. The proteins were stained with Coomassie Brilliant Blue after electrophoresis. Positions of the molecular mass markers are shown on the left. Lane 1, GST-MpABI1; lane 2, GST-MpABI1-d1; lane 3, GST-MpABI1-d2; lane 4, GST-MpABI1-G298D. D, Protein phosphatase activity of wild-type and mutant GST fusion proteins of MpABI1 toward the 32P-labeled MBP (n = 3). Phosphatase reactions were carried out at 30°C for 10 min and terminated by the addition of TCA. Activity is represented as a percentage of [32P]orthophosphate released by GST-MpABI1 in the presence of 5 mm Mg2+.
A previous study showed that deletion of the N-terminal domain of ABI1 increases its phosphatase activity, suggesting that it may serve as an intramolecular negative regulator (Sheen, 1998). To analyze the role of the N-terminal domain of MpABI1, we made two deletion constructs: GST-MpABI1-d1, lacking the putative NLS in the N-terminal domain, and GST-MpABI1-d2, lacking most of the N-terminal domain (Fig. 3B). E. coli-expressed and purified GST-MpABI1-d1 and GST-MpABI1-d2, with molecular masses of 79 and 58 kD, respectively (Fig. 3C), were used for in vitro phosphatase assays. The results indicated that both deletion mutant proteins showed activity similar to that of GST-MpABI1, indicating that deletion of the N-terminal domain or impairment of the NLS motif therein has little effect on its PP2C activity in vitro (Fig. 3D). We also used the GST-MpABI1-G298D protein with an abi1-1-like mutation for phosphatase assays (Fig. 3B). As has been reported for the abi1-1 protein, substitution by Asp of the Gly residue located C terminally to the conserved DGH stretch in motif 2 in the reaction center was thought to reduce the phosphatase activity (Bertauche et al., 1996). Phosphatase assays using the recombinant GST-MpABI1-G298D protein (Fig. 3C) revealed that a Gly-to-Asp substitution in this position dramatically reduced the phosphatase activity (Fig. 3D).
Cellular Localization of MpABI1
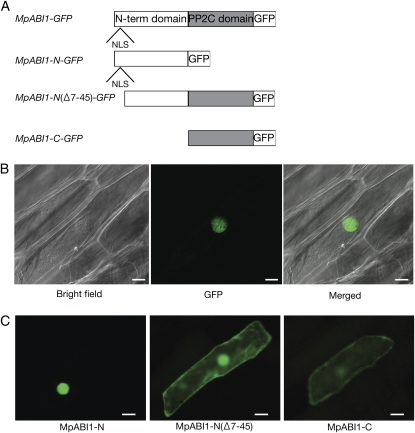
To analyze the cellular localization of MpABI1, gene constructs for MpABI1-GFP fusion proteins were generated (Fig. 4A). The constructs were introduced into epidermal cells of onion (Allium cepa) by particle bombardment, and GFP fluorescence was observed. Transient expression of the full-length MpABI1 fused to GFP (MpABI1-GFP) resulted in localization of the GFP fluorescent signals mainly in the nucleus (Fig. 4B). To identify domains responsible for the nuclear localization, we separately generated GFP constructs for the N-terminal domain fusion (MpABI1-N-GFP) and the C-terminal domain fusion (MpABI1-C-GFP) and transiently expressed them in onion epidermal cells. The results indicated that the MpABI1-N-GFP signal was mostly localized in the nucleus, with slight fluorescence in the cytoplasm. When the amino acid region containing the putative NLS was deleted from the N-terminal domain [MpABI1-N(Δ 7–45)-GFP], the GFP signal was found in the nucleus and cytoplasm. Fluorescence of MpABI1-C-GFP was found in the nucleus and cytoplasm (Fig. 4C). These results indicated that MpABI1 is mainly localized in the nucleus and that the putative NLS in the N-terminal domain is likely to be involved in the nuclear localization.
Figure 4.
Localization study using GFP fusion proteins. A, Schematic representation of GFP fusion constructs of MpABI1 and its mutant constructs used for analysis. B, Cellular localization of MpABI1-GFP. The DNA construct for the MpABI1-GFP fusion protein was bombarded into epidermal cells of onion, and the cells were observed after 1 d of incubation at 25°C in the dark. Bright-field, GFP fluorescence, and merged images are shown. Bars = 30 μ m. C, N-terminal domain-directed nuclear localization of MpABI1. Fluorescent images show onion epidermal cells bombarded with DNA constructs for the MpABI1 N-terminal domain (MpABI1-N; amino acids 1–225), MpABI1-N (Δ 7–45) lacking NLS, and the C-terminal PP2C domain (MpABI1-C; amino acids 213–568) fused to the upstream region of the GFP coding sequence. Bars = 50 μ m.
Effect of Transient Expression of MpABI1 on ABA-Induced Gene Expression
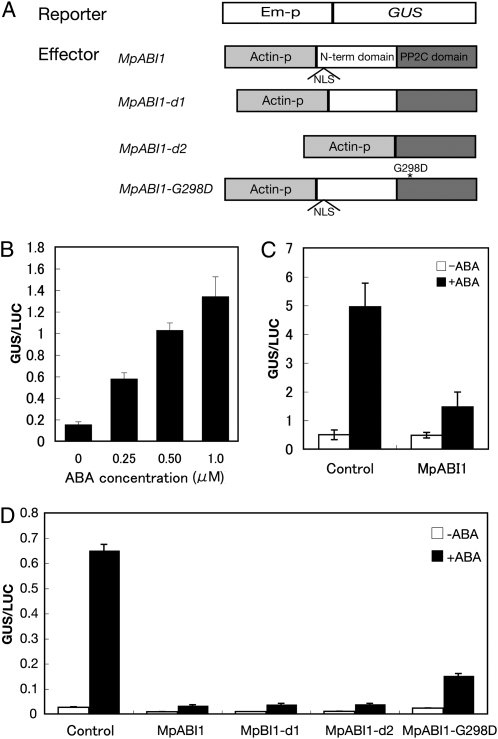
In order to test the effect of MpABI1 on ABA-induced gene expression, transient assays using reporter genes were carried out. The Em-GUS construct has been used for studies of ABA-responsive gene expression in transient assays of higher and lower plants (Marcotte et al., 1989; Marella et al., 2006). To examine the effect of MpABI1 overexpression on GUS activity, MpABI1 cDNA was fused to the downstream region of the rice (Oryza sativa) actin promoter (Fig. 5A) and cointroduced with the Em-GUS construct into M. polymorpha cells by particle bombardment.
Figure 5.
Effect of transient overexpression of MpABI1 on ABA-induced gene expression. A, Constructs used for transient assays. The wheat Em promoter (Em-p) fused to the GUS reporter gene was used as a reporter construct for the detection of ABA-induced gene expression (Marcotte et al., 1989). The rice actin promoter (Actin-p) fused to MpABI1, MpABI-d1, MpABI1-d2, or MpABI1-G298D was used as an effector construct to see their effects on ABA-induced GUS gene expression. The rice ubiquitin promoter fused to the luciferase gene was used as a control construct (data not shown). Plasmid DNAs with these constructs were cobombarded into M. polymorpha or P. patens cells using the PDS-1000He particle delivery system, and the cells were then incubated with or without ABA (for details, see “Materials and Methods”). Protein fractions prepared from the cells were used for GUS and LUC assays, and the relative GUS activity-to-LUC activity ratios (GUS/LUC) are represented in B to D. B, Effect of different concentrations of ABA on Em-GUS expression in M. polymorpha cells. C, Effect of MpABI1 overexpression on 1- μm ABA-induced Em-GUS expression in M. polymorpha cells. D, Effect of overexpression of wild-type MpABI1 and deletion mutants MpABI1-d1, MpABI1-d2, and MpABI1-G298D on ABA-induced Em-GUS expression in P. patens.
In liverworts, transient assay systems for the study of ABA-induced gene expression have not been established. When the thallus of M. polymorpha was used for particle bombardment of Em-GUS, the level of GUS activity was too low to detect even after treatment with ABA. The gemmae that showed ABA sensitivity (Fig. 1) were too small to obtain enough material for consistent experiments. Thus, we used suspension-cultured cells of M. polymorpha for the transient assays. In order to increase the signal-to-noise ratio in the experiments, the wheat ubiquitin promoter fused to the GFP gene (Ubi-GFP) was cointroduced with the effector and reporter constructs into the cells, and the GFP-positive cells were manually enriched with a fluorescence microscope after 1 d of incubation. GUS assays using GFP-positive cell-enriched samples gave a higher signal-to-noise ratio than did samples without enrichment (data not shown), and the Em-GUS-bombarded cells showed a clear ABA response. Figure 5B shows that Em-GUS expression was increased by submicromolar concentrations of ABA (Fig. 5B). Coexpression of MpABI1 and Em-GUS resulted in suppression of the ABA-induced gene expression (Fig. 5C). These results indicate that MpABI1 has a negative effect on ABA-induced gene expression in M. polymorpha cells. We also carried out transient assays in protonema cells of the moss P. patens. Overexpression of MpABI1 effectively suppressed the ABA-induced Em-GUS expression in P. patens (Fig. 5D). In subsequent experiments, P. patens cells were used for assays because the cells gave a higher signal-to-noise ratio without enrichment of GFP-transfected cells than did the cells of M. polymorpha.
To determine which domain is important for the suppression of ABA-induced gene expression, mutant constructs were used for transient assays. Constructs for MpABI1-d1, without a putative NLS in the N-terminal domain, and MpABI1-d2, lacking most of the N-terminal domain, were generated (Fig. 5A) and introduced into P. patens cells to see their effect on ABA-induced Em-GUS expression. The results indicated that transient expression of these constructs suppressed expression of the Em-GUS gene to an extent similar to that achieved by intact MpABI1 (Fig. 5D). This indicated that the N-terminal domain of MpABI1 is not necessary for its activity in ABA-induced gene expression. A construct for a mutant with the abi1-1-like substitution (MpABI1-G298D) was also used for the transient assay, because previous studies indicated that the abi1-1-like mutation causes a hypermorphic ABA-insensitive phenotype in higher plant cells. The results of the bombardment of MpABI1-G298D, however, indicated that the Gly-to-Asp substitution had a reduced effect on the suppression of ABA-induced Em-GUS expression in comparison with the wild-type MpABI1 (Fig. 5D).
Analysis of Transgenic Plants Expressing MpABI1
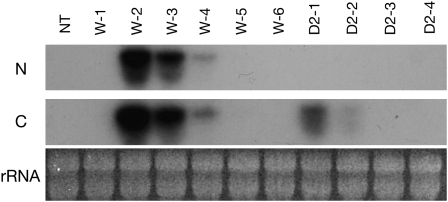
To analyze the function of MpABI1 in ABA-induced stress tolerance, transgenic P. patens plants expressing MpABI1 were generated. The MpABI1 cDNA fused to the rice actin promoter was introduced into protoplasts of P. patens protonema with the hygromycin phosphotransferase gene cassette, and hygromycin-resistant cells were selected. We also generated transgenic plants expressing MpABI-d2, which lacks most of the N-terminal domain. Six hygromycin-resistant lines, W-1 to W-6, expressing MpABI1 and four lines, D2-1 to D2-4, expressing MpABI1-d2 were isolated and used for northern analysis to determine expression levels of the transcripts.
The cDNA fragments of the N-terminal domain and the C-terminal domain of MpABI1 cDNA were separately labeled and used as probes for detection of the transcripts in the transgenic plants (Fig. 6). Transcripts of MpABI1 were detected by both the N-terminal and C-terminal domain probes, and those of MpABI1-d2 were detected only by the C-terminal domain probe. Of the six transgenic lines expressing MpABI1, the W-2 line showed the highest level of transcript accumulation. Four lines expressing MpABI-d2 showed transcript accumulation with relatively low abundance, among which the D2-1 line showed the highest transcript accumulation.
Figure 6.
Northern-blot analysis of transgenic P. patens plants expressing MpABI1 and MpABI1-d2. Total RNAs extracted from nontransgenic (NT) and independent transgenic lines expressing intact MpABI1 (W-1 to W-6) and MpABI1-d2 (D2-1 to D2-4) were electrophoresed and blotted onto a nylon membrane. The blotted RNAs were hybridized with 32P-labeled probes for either the N-terminal domain region (N) or the C-terminal domain region (C) of MpABI1 cDNA. Ethidium bromide-stained rRNA for each sample is also shown.
Changes in Osmotic and Freezing Stress Tolerance in Transgenic Plants
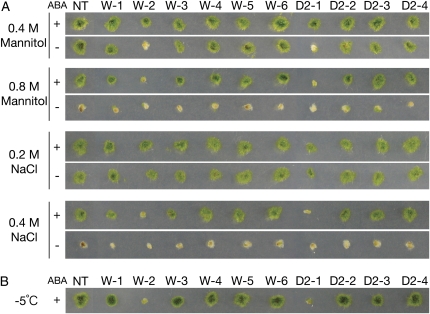
To analyze changes in stress tolerance by expression of MpABI1 and MpABI1-d2, sensitivities to osmotic stress of nontransgenic and transgenic moss lines (W-1 to W-6 and D2-1 to D2-4) were compared. We previously reported that cellular osmotic concentrations of nontreated and ABA-treated cells were approximately 0.3 and 0.5 osmol L−1, respectively, in protonemata of nontransgenic P. patens (Nagao et al., 2005). Treatment of protonema cells with 0.4 and 0.8 m mannitol or with 0.2 and 0.4 m NaCl, followed by regrowth on normal agar medium, revealed that transgenic lines W-2 and D2-1 are hypersensitive to osmotic stress (Fig. 7A). The W-3 plants showed moderate sensitivity to osmotic stress such as 0.8 m mannitol and 0.4 m NaCl. Protonemata of the transgenic plants were also used for freezing tests. We previously showed that P. patens protonema cells responded to ABA and acquired tolerance to freezing. One-day treatment with 10 μm ABA typically alters the temperature at which 50% of the cells are damaged by freezing, from −2°C to −10°C in protonemata of P. patens (Minami et al., 2003; Nagao et al., 2005). Thus, ABA-treated protonemata of nontransgenic and transgenic moss lines were subjected to freezing to −5°C, and after thawing, survival was determined by regrowth on agar medium. As shown in Figure 7B, transgenic mosses, W-2 and D2-1, expressing the largest amount of MpABI1 and MpABI1-d2 transcripts showed sensitivity to freezing. Freezing tolerance of P. patens protonema cells has been shown to be associated with their tolerance to freeze-induced dehydration (Nagao et al., 2005). Thus, our results suggested that transgenic plants expressing high levels of MpABI1 and MpABI1-d2 are susceptible to freezing due to their increased sensitivity to dehydration stress.
Figure 7.
Effect of ABA treatment on osmotic and freezing stress-induced damage of control and transgenic P. patens plants. Protonema cells of nontransgenic control (NT) and independent transgenic lines expressing wild-type MpABI1 (W-1 to W-6) and MpABI1-d2 (D2-1 to D2-4) were incubated for 1 d with or without 10 μm ABA and used for osmotic stress and freezing treatments. A, Representative results of growth of cells that had been either pretreated with 10 μm ABA (+) or nontreated (−) prior to the osmotic stress treatment by the indicated concentrations of mannitol or NaCl. After 15 min of osmotic stress treatment at room temperature, a piece of protonemata was planted on fresh BCD agar medium and grown at 25°C for 1 week. B, Representative results of growth after freezing of nontransgenic and transgenic plants that had been pretreated with 10 μm ABA for 1 d. All samples were ice inoculated at −1°C and subjected to equilibrium freezing to −5°C. After thawing, a piece of protonemata was planted on fresh BCD agar medium and grown at 25°C for 1 week.
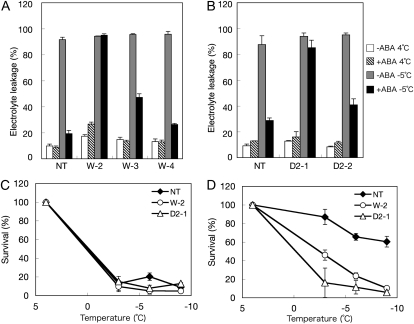
Freezing survival of representative transgenic lines was quantitatively determined by measurement of electrolyte leakage after freezing. We previously showed that the percentage of electrolyte leakage after freezing is in fair correlation with cell death (Nagao et al., 2005). Results of freezing to −5°C indicated that protonemata of nontransgenic plants acquired freezing tolerance upon ABA treatment, but the transgenic plants W-2 and D2-1 failed to acquire freezing tolerance by ABA treatment (Fig. 8, A and B). These transgenic plants were also subjected to freezing at different temperatures, and the effect of ABA on their freezing survival was examined (Fig. 8, C and D). The D2-1 plants did not show ABA-induced freezing tolerance, whereas tolerance of W-2 plants to freezing at −3°C was slightly increased by ABA treatment but that at −6°C was not affected by ABA (Fig. 8D).
Figure 8.
Freezing damage of nontransgenic (NT) and representative transgenic lines expressing different levels of transcripts of MpABI1 (W2, W3, and W4) and MpABI1-d2 (D2-1 and D2-2). Protonema cells were incubated for 1 d with or without 10 μm ABA and subjected to freezing. The cells were then ice inoculated at −1°C and subjected to equilibrium freezing at a rate of −2.4°C h−1 to desired temperatures. After thawing, electrolyte leakage was measured and survival was estimated as described previously (Nagao et al., 2005). A and B, Comparison of electrolyte leakage (%) of unfrozen and frozen plants that had been pretreated with 10 μm ABA for 1 d prior to freezing. A, Comparison of nontransgenic, W2, W3, and W4 plants. B, Comparison of nontransgenic, D2-1, and D2-2 plants. C and D, Nontreated (C) or 10 μm ABA-treated (D) protonema cells of nontransgenic, W-2, and D2-1 plants were frozen to −3°C, −6°C, and −9°C, and survival (%) after thawing was estimated by electrolyte leakage measurement.
Levels of ABA-Induced Proteins in Transgenic Plants
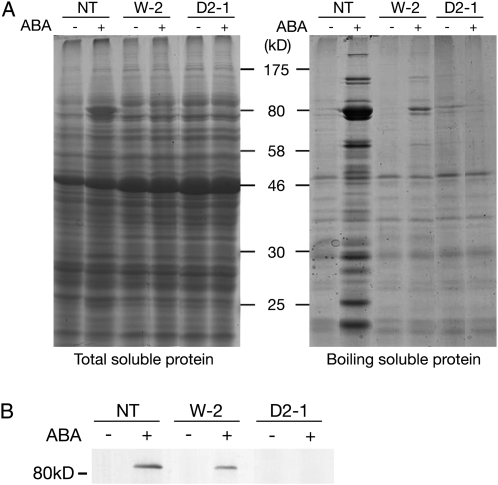
Next, we examined levels of ABA-induced soluble proteins in transgenic plants. In higher plants, cold stress, osmotic stress, and exogenous ABA treatment increase the accumulation of specific proteins, especially LEA-like proteins, many of which are rich in hydrophilic amino acid residues and remain soluble even after heat treatment. Such LEA-like proteins have been shown to accumulate in response to ABA and dehydration stress in P. patens (Knight et al., 1995; Minami et al., 2005, 2006). Analysis of the total soluble protein fraction by SDS-PAGE revealed that ABA treatment increases levels of specific proteins, especially those with molecular mass slightly greater than the 80-kD protein marker, in nontransgenic P. patens protonema cells. However, levels of these proteins were significantly decreased in transgenic W-2 and D2-1 plants (Fig. 9A, left). We also analyzed proteins of boiling-soluble fractions in which LEA-like hydrophilic proteins are enriched. Total soluble proteins were boiled for 3 min, and the fractions that remained soluble after centrifugation were analyzed. The results indicated that the nontransgenic plants accumulated a number of boiling-soluble proteins upon ABA treatment, but the levels of these proteins were decreased in the W-2 plant. Furthermore, the levels of the boiling-soluble proteins were unchanged by ABA treatment in the D2-1 plant (Fig. 9A, right). Immunoblot analysis using antiserum raised against the LEA-like protein encoded by the 17B9 gene, which had been isolated as one of most abundantly expressed genes upon ABA treatment (Minami et al., 2006), revealed that ABA-induced accumulation of the 17B9 protein was decreased in W-2 plants and not detected in D2-1 plants (Fig. 9B).
Figure 9.
Analysis of ABA-induced soluble proteins in control (nontransgenic [NT]) and transgenic plants, W-2 and D2-1. Total soluble proteins (left) and boiling-soluble proteins (right) were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue. Positions of molecular mass markers are shown in kD. B, The boiling-soluble proteins were used for immunoblot analysis after electroblotting onto an Immobilon-P membrane. The blotted proteins were reacted with antiserum raised against the recombinant 17B9 protein. Signals were detected using an alkaline phosphatase-conjugated secondary antibody and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate color-developing reagent.
ABA-Induced Morphological Changes in Transgenic Plants
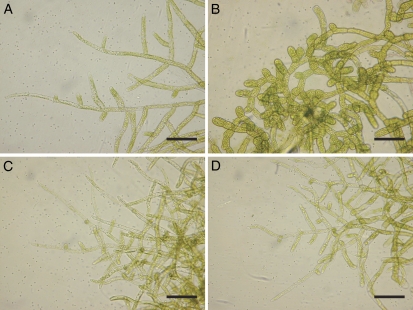
It is known that ABA not only affects stress tolerance but also induces specific morphological changes in protonema cells of mosses. ABA treatment of protonema cells of various moss species for several days has been shown to induce structural changes leading to the formation of spherical, thick-walled brood cells (Goode et al., 1992). Thus, we observed ABA-induced changes of protonema cells in transgenic P. patens expressing MpABI1 and MpABI1-d2. Comparison of cellular appearance after treatment with 10 μm ABA for 7 d revealed that typical morphological changes observed in protonema cells of nontransgenic control plants (Fig. 10, A and B) did not occur in W-2 and D2-1 plants (Fig. 10, C and D). These results indicated that expression of MpABI1 and MpABI1-d2 had a negative effect on the ABA-induced brood cell formation as well as on stress tolerance in the protonemata of the transgenic plants.
Figure 10.
ABA-induced morphological changes of protonema cells in control and transgenic plants. The protonemata of control, W-2, and D2-1 plants were incubated in normal BCD medium or the medium containing 10 μm ABA for 7 d. A, Nontreated control protonemata. B, ABA-treated control protonemata. C, ABA-treated W-2 protonemata. D, ABA-treated D2-1 protonemata. Bars = 100 μ m. [See online article for color version of this figure.]
DISCUSSION
In contrast to the accumulated knowledge on molecular machinery controlling ABA-dependent processes in angiosperms, very little information is available on that in nonflowering plants, especially in bryophytes. Among bryophytes, liverworts are the most basal lineage of extant land plants, and the role of ABA is especially of interest, because they are the only group of plants that do not have stomata, not to mention the fact that they also lack seeds, lateral roots, and inflorescences, on which many studies for ABA-signaling processes have been carried out. Furthermore, since liverworts are the key group of organisms sister to all other plant lineages (Qiu et al., 2006; Bowman et al., 2007), determination of ABA response in liverworts is critical for understanding the conserved mechanisms for stress tolerance in land plants. However, there are a number of unsolved issues in liverworts regarding ABA, such as its synthetic pathway, metabolism, cellular and tissue distribution, and storage as well as perception and signal transduction mechanisms.
Recent studies have indicated that ABA might play a role in stress tolerance of liverworts (Hellwege et al., 1994; Pence et al., 2005). Nevertheless, physiological roles of ABA in liverworts have been controversial for years because of unsuccessful detection of endogenous ABA (Weiler, 1979). In addition, lunularic acid, a substance structurally similar to ABA, is found in a wide range of liverwort species and has been suspected to be substituting for the role of ABA (Pryce, 1971). However, the results of ABA measurement by ELISA using monoclonal antibodies against ABA indicated that its levels are increased by slow desiccation and decreased by rehydration in E. holstii (Hellwege et al., 1994). Levels of endogenous ABA have also been estimated in R. fluitans using a similar method (Hellwege et al., 1992, 1996). Li et al. (1994) reported on the detection of ABA in M. polymorpha thalli by measurements using gas chromatography with mass spectrometry and estimated its endogenous levels to be 4 to 16 ng g−1 fresh weight, similar to those of higher plants. Our estimation by ELISA indicated that thalli of M. polymorpha cultured in normal medium for 4 weeks (Fig. 1A) contain 16.5 ± 3.9 ng g−1 fresh weight endogenous ABA, comparable to the levels reported by Li et al. (1994). Previous reports indicated that thalli of M. polymorpha show little ABA responses and desiccation stress tolerance (Hellwege et al., 1994; Pence et al., 2005). However, we observed that submicromolar concentrations of exogenous ABA retarded the growth of its gemma, known as a stress-tolerant form (Fig. 1). These results convinced us of the role of ABA in growth regulation and stress tolerance in M. polymorpha, at least in particular developmental stages.
In this study, we took an alternative approach for analyzing ABA-dependent responses in liverworts by searching for genes that function as regulators of ABA signaling. By a search of the M. polymorpha EST database, we identified MpABI1, with significant similarity to Arabidopsis ABI1, known as a negative regulator of ABA-dependent signaling processes. We considered that analyses of MpABI1 by biochemical and transgenic approaches would be important for understanding the built-in regulatory systems for ABA responses in liverworts.
MpABI1 Is a PP2C with Divalent Cation-Dependent Activity
Sequence analysis of MpABI1 revealed that it encodes a protein containing the PP2C domain in its C-terminal half region. Phylogenetic analysis revealed that MpABI1 is closely associated with the group A PP2Cs of Arabidopsis (Fig. 2). Among 76 PP2C genes encoded in the Arabidopsis genome, the group A PP2Cs, including ABI1, ABI2, HAB1, and PP2CA, have been shown to play a role in the negative regulation of ABA signaling (Schweighofer et al., 2004). The close association of MpABI1 with these group A PP2Cs of Arabidopsis and PpABI1A and PpABI1B of P. patens (Komatsu et al., 2009) indicates that ABI1-related PP2Cs are conserved in higher and lower land plants (Fig. 2).
Results of protein phosphatase assays using recombinant GST fusion proteins indicated that MpABI1 is a functional protein phosphatase with divalent cation-dependent activity (Fig. 3). GST-MpABI1 showed higher affinity for Mn2+ than for Mg2+ for the phosphatase activity, but Mg2+ appeared to be required for its maximal activity. ABI1, ABI2, and HAB1 of Arabidopsis have been shown to exhibit similar divalent cation-dependent activity (Bertauche et al., 1996; Leung et al., 1997; Robert et al., 2006), although their preference for divalent cations has not been studied. The abi1-1-like G298D substitution of MpABI1 dramatically reduced its phosphatase activity (Fig. 3D), indicating that disturbance of divalent cation association might have resulted in reduced enzyme activity, similar to group A PP2Cs of Arabidopsis (Bertauche et al., 1996; Leung et al., 1997; Robert et al., 2006). From these results, we conclude that biochemical characteristics of MpABI1 are typical of PP2Cs and similar to those of ABI1 and ABI1-related PP2Cs.
Roles of the N-Terminal Domain of MpABI1
Sequence analysis of MpABI1 resulted in identification of the putative NLS in the N-terminal domain. This suggests the role of the N-terminal domain in its localization to the nucleus. Analysis of localization using GFP fusion proteins of MpABI1 revealed that the protein is localized mainly in the nucleus and that its localization is directed by the NLS sequence in the N-terminal domain (Fig. 4). Arabidopsis members of group A PP2Cs have various N-terminal domains, but their functions have not been elucidated. Among nine group A PP2Cs, only the gene products of AHG1, At2g29380, and At5g59220 have the putative NLS in the N-terminal domain. Interestingly, a sequence around NLS of At5g59220 (amino acids 37–49: AARRRRMEIRRFK) has a significant similarity to that of MpABI1 (amino acids 29–41: AARRRRKEIRRFK), implicating functional significance to these amino acids in the N-terminal domain. In contrast, gene products of At1g07430, AtPP2CA/AHG3, and ABI1 have NLS in the C terminus of the PP2C domain, while ABI2, HAB1, and HAB2 gene products do not have obvious NLS. The role of nuclear localization of group A PP2Cs is not clearly understood, but, recently, it has been shown that the basic amino acid cluster found in the C terminus of ABI1 directs nuclear localization when the abi1-1 mutation has been introduced, suggesting that its nuclear localization is responsible for causing a hypermorphic phenotype (Moes et al., 2008). The mode of regulation of MpABI1 by NLS in the N-terminal domain would be different from that of ABI1 with C-terminal NLS. The role of the N-terminal domain of MpABI1 could be to regulate its activity by facilitating its compartmentalization to the nucleus. Our results of transient assays using MpABI1-d2 suggested that nuclear localization is at least not necessary for negative regulation of ABA-induced gene expression (Fig. 5). Examination of how ABA treatment affects cellular localization of MpABI1 in future experiments will be important for elucidation of the role of the N-terminal domain.
MpABI1 Serves as a Negative Regulator of ABA Signaling
To show molecular responses to ABA in M. polymorpha, we conducted transient assays using the Em-GUS construct, which had been successfully used for studies on ABA signaling (Marella et al., 2006). Specific sequence elements such as the ABA-responsive element ABRE found in the promoter of Em are thought to function in both higher plant and moss cells (Knight et al., 1995; Kamisugi and Cuming, 2005). Since transient assay systems for detection of ABA-induced gene expression in liverwort cells have not been reported previously, we tested different tissues of M. polymorpha for detection of ABA-induced Em-GUS expression. Little ABA response was observed when the thallus of M. polymorpha was used for the assay, as expected from our observation that thallus growth was not significantly inhibited by ABA. Instead, we used suspension-cultured cells of M. polymorpha and detected an ABA-induced increase in Em-GUS expression (Fig. 5B). Establishment of this transient assay system has made it possible to study ABA-induced gene expression processes in the liverwort. We found that transient overexpression of MpABI1 resulted in suppression of ABA-induced expression of Em-GUS (Fig. 5C). These results provided, to our knowledge, the first molecular evidence showing conservation of a regulatory system for an ABA-dependent signaling process in liverworts. Similar results were obtained when P. patens cells were used for transient assays (Fig. 5D), indicating that the function of ABI1-like PP2C as a negative regulator is conserved in bryophytes. A model presented in recent reports highlighted the role of the ABA receptor(s) in direct regulation of group A PP2Cs (Ma et al., 2009; Park et al., 2009). In this model, the receptor binds and inhibits ABI1 and ABI2 in the presence of ABA, which leads to activation of SnRKs and downstream signaling molecules (Park et al., 2009; Nishimura et al., 2010). Whether or not similar ABA receptor molecule(s) bind to PP2Cs in mosses or liverworts is not clear. The differences in the mode of action in higher and lower plants must depend on the types of receptors and signaling molecules functioning in ABA-dependent pathways, which remain to be clarified.
Analysis of Transgenic Mosses Expressing MpABI1 and MpABI1-d2
In this study, we analyzed stress tolerance and morphological changes of transgenic P. patens expressing MpABI1 and MpABI1-d2 to examine their effect on ABA-induced signaling processes (Figs. 7 and 8). Transgenic W-2 and D2-1 plants expressing the highest levels of MpABI1 and MpABI1-d2 transcripts showed the most severe phenotypes, and ABA had little effect on their freezing and osmotic stress tolerance (Fig. 7). The levels of tolerance were associated with reduced accumulation of ABA-induced boiling-soluble proteins, including that of the LEA-like 17B9 protein (Fig. 9). LEA-like proteins have been postulated to be involved in desiccation tolerance of seeds during late embryogenesis and freezing/dehydration stress tolerance in vegetative tissues (Wise and Tunnacliffe, 2003). Our results indicated that accumulation of LEA-like proteins is essential for cellular stress tolerance in P. patens cells. It should be noted that, even without ABA pretreatment, W-2 and D2-1 plants showed increased sensitivity to osmotic stress imposed by 0.4 m mannitol or 0.2 m NaCl. Since the amounts of boiling-soluble proteins in nontreated cells of nontransgenic, W-2, and D2-1 plants were similar (Fig. 9A), it is likely that other cellular factors are involved in their stress tolerance. Hypersensitivity of these lines to osmotic stress could be in part due to reduced levels of soluble sugars that contribute to both cellular osmotic adjustment and membrane protection. Analysis of total soluble sugars indicated that their levels in W-2 and D2-1 plants were lower than that of the nontransgenic control, with or without ABA treatment (data not shown). Our previous results indicated that stress tolerance of P. patens cells was associated with morphological changes such as vacuolar fragmentation, cell wall thickening, and chloroplast flattening, with concomitant accumulation of soluble sugars and boiling-soluble proteins (Nagao et al., 2005; Minami et al., 2006). Physiological aspects of W-2 and D2-1 are being studied in detail, but it is likely that their reduced tolerance is due not only to reduced levels of LEA-like proteins or soluble sugars but also to changes in factors controlling cellular morphology necessary for being adaptive to stress conditions.
CONCLUSION
Although ABA is a widely distributed compound detected in plants, algae, and pathogenic fungi, its utilization as a hormonal signal for development and stress tolerance appears to be a feature commonly found in land plants. It is not clear when and how the land plant ancestors integrated this compound in their stress-response processes as a signaling molecule, but it is possible that establishment of perception and signaling transduction mechanisms for ABA was one of the keys for their successful colonization of land, where plants are readily exposed to dehydration conditions. Our results for ABA-induced Em-GUS expression and the finding that MpABI1 is a regulator of ABA-induced stress tolerance suggest that liverworts, positioned at the basal lineage of land plant evolution, have mechanisms to control ABA signals both positively and negatively, which are likely to be conserved in all extant land plants. Physiological roles of ABA in liverworts have yet to be clarified. Since our results indicated that the gemma, a dormant stress-tolerant reproductive form, showed a clear ABA response, we are currently investigating the role of ABA in the gemma of transgenic M. polymorpha overexpressing MpABI1. Functional analyses of signaling molecules are indispensable for the identification of their role in ABA-induced signal transduction. M. polymorpha could be an ideal model for studies of ABA response and environmental adaptation of nonvascular plants. High-efficiency transformation protocols (Ishizaki et al., 2008) and the transient assay system established in this study will be powerful tools for molecular studies using M. polymorpha as a model organism.
MATERIALS AND METHODS
Plant Materials
Male gametophyte of Marchantia polymorpha, accession Takaragaike-1, was cultured at 22°C on the M51C agar medium containing 2% Suc under continuous light as described by Ishizaki et al. (2008). Suspension culture of M. polymorpha was maintained in the M51C liquid medium containing 2% Suc and 1 mg L−1 2,4-dichlorophenoxyacetic acid at 22°C with shaking at 130 rpm (Ono et al., 1979). Protonemata of Physcomitrella patens were cultured using the modified BCD agar medium as described by Nagao et al. (2005).
Extraction of Total RNA
M. polymorpha and P. patens tissues were ground in liquid nitrogen using mortars and pestles and transferred to a plastic tube. The tissue powder was quickly suspended in three volumes of 2 × CTAB buffer (2% cetyl trimethyl ammonium bromide, 1.4 m NaCl, 100 mm Tris-Cl, pH 8.0, 20 mm EDTA, and 10 mm β -mercaptoethanol) at 65°C. The suspension was mixed well with an equal volume of chloroform and centrifuged at 10,000g at room temperature to remove insoluble materials. The upper aqueous phase was transferred to a new tube, mixed with 0.75 volume of isopropyl alcohol, and then centrifuged at 10,000g at 4°C. The pellet was dissolved in 0.2% diethyl pyrocarbonate (DEPC)-treated water. The sample was mixed with one-third volume of 8 m LiCl and kept at 4°C to precipitate RNA. After centrifugation at 10,000g at 4°C, the pellet was dissolved in 0.2% DEPC-treated water. Following extraction with phenol:chloroform (1:1, v/v) and precipitation with ethanol, the RNA was finally dissolved in 0.2% DEPC-treated water and stored at −80°C.
Amplification of MpABI1 cDNA
Total RNA of M. polymorpha was used as a template for synthesis of the first-strand cDNA using the oligo(dT)15 primer and ReverTraAce reverse transcriptase (Toyobo). PCR was conducted for amplification of the cDNA encompassing the entire MpABI1 open reading frame using the first-strand cDNA and specific oligonucleotide primers, 5′-GAACTGCCGGAAGTGGATTG-3 ′ and 5′-GCGACATATACCTTTCTCAC-3 ′ , designed on the basis of the EST sequences of M. polymorpha. The amplified PCR fragment was cloned into pGEM-T Easy vector (Promega) and used for dideoxy nucleotide sequencing.
Northern-Blot Analysis
Electrophoresis of RNAs and transfer to a nylon membrane were carried out according to the method described by Minami et al. (2005). The RNAs blotted onto the membrane were hybridized with the 32P-labeled probe of either the N-terminal or C-terminal half region of the MpABI1 cDNA. Probe labeling was carried out using the DNA labeling kit (Takara-Shuzou) and [ α -32P]dCTP. Hybridization was carried out using Ultrahyb hybridization buffer (Ambion) at 42°C overnight, and washing was carried out at 60°C for 1 h in a buffer containing 0.2 × SSC and 0.2% SDS.
Observation of the GFP-MpABI1 Fluorescence
The GFP fusion constructs for MpABI1 were generated by PCR using the high-fidelity KODplus polymerase (Toyobo) and MpABI1, MpABI1-d1, and MpABI1-d2 constructs as templates. A universal sense primer, 5′-GATTGGTCGACTATGGAGACGCTGGTCA-3 ′ , was used for the creation of the SalI site at the 5 ′ region. For MpABI1-N-GFP, an NcoI (CCATGG) site was created at Met-225 using an antisense primer, 5′-GGAACCATGGTGGTATCCTCCACATCGAG-3 ′ . For MpABI1-C-GFP, an NcoI site was created at the C terminus (Ala-568) of MpABI1 using the antisense primer 5′-GCAACCATGGCCGAAGATCCCGTCGTTAAG-3 ′ . The amplified fragments were used for ligation to the NcoI site of the GFP gene. Epidermal peels of onion (Allium cepa) bulbs were prepared and placed onto Murashige and Skoog agar medium containing 3% (w/v) Suc. The peels were bombarded with 1- μ m gold particles coated by DNA of the wild-type or mutant GFP-MpABI1 construct using the PDS-1000He particle delivery system (Bio-Rad) with a bombardment pressure of 1,100 p.s.i. under vacuum. The tissues were incubated for 16 h at 25°C in the dark and observed by the Olympus FV1000 confocal laser scanning microscope or Nikon TA300 inverted fluorescence microscope.
Preparation of GST-Fusion Proteins and Protein Phosphatase Assays
Constructs for GST fusion proteins of MpABI1 were generated using the pGEX-5X-3 vector (GE Healthcare). An oligonucleotide primer, 5′-CGCGAGATCTCTATGGAGACGCTGGTCACT-3 ′ , was used for the creation of the BglII site at the N terminus of each construct for in-frame fusions with the BamHI site of the pGEX-5X-3 vector. MpABI1, MpABI1-d1, MpABI1-d2, and MpABI1-G298D constructs used for transient assays were used as templates for PCR amplification for generation of GST-MpABI1, GST-MpABI1-d1, GST-MpABI1-d2, and GST-MpABI1-G298D, respectively.
Escherichia coli strain DH5 α containing each GST fusion protein construct was cultured in Luria-Bertani medium supplemented with 50 mg L−1 ampicillin at 37°C. Expression of fusion proteins was induced by addition of isopropyl-thio-galactoside, followed by overnight cultivation at 18°C. Bacterial extracts were prepared by sonication, and GST fusion proteins were purified by glutathione-Sepharose affinity chromatography. The affinity-purified fusion proteins were analyzed by SDS-PAGE, and the amounts were estimated by scanning the Coomassie Brilliant Blue-stained protein bands using bovine serum albumin as a standard.
Protein phosphatase assays of the GST fusion proteins were carried out as essentially described by Takezawa (2003). The [32P]MBP was prepared using the catalytic subunit of protein Ser/Thr kinase (P2645; Sigma) and [ γ -32P]ATP. The standard reaction mixture for the phosphatase assays contained 0.14 μ g of the GST fusion proteins and 4 μ g of [32P]MBP in a buffer of Tris-Cl (pH 7.5), 5 mm MgCl2, and 2 mm dithiothreitol. The reaction was carried out at 30°C and terminated by mixing with one-fourth volume of 100% (w/v) TCA. The mixture was kept on ice for 10 min and centrifuged at 4°C, 14,000g to remove precipitated proteins. The amount of 32P in the supernatant was quantified by scintillation counting.
Transient Assays of P. patens and M. polymorpha
MpABI1 cDNA was fused downstream of the rice (Oryza sativa) actin promoter and used for both transient assays and stable transformation. Deletion constructs of MpABI1 were made using the KODplus mutagenesis kit (Toyobo), with an antisense primer, 5′-GGAAGTGACCAGCGTCTCCA-3 ′ , and the following sense primers: 5′-GCTGGTATCTCGGAGCCATC-3 ′ for MpABI1-d1 and 5′-GCTGCGAGGAACTTTCTCGA-3 ′ for MpABI1-d2. The MpABI1-G298D construct was made using the same kit with an antisense primer, 5′-GATCCCAGGCAGCTGTGTTTTGTGCA-3 ′ , and a sense primer, 5′-CGTCATGTCCATCATATACTCCA-3 ′ .
Transient assays of P. patens protonema cells were carried out according to the protocol described by Marella et al. (2006). In brief, 6-d-old P. patens cells were bombarded with Em-GUS as a reporter construct, Ubi-LUC as an internal control construct, and the wild-type and mutant MpABI1 genes as effecter constructs using the PDS-1000He particle delivery system (Bio-Rad). The bombarded cells were cultured at 25°C under continuous light for 48 h on BCD medium with or without 10 μm ABA. Proteins extracted from the protonemata were then used for GUS and LUC assays to determine GUS-LUC ratio as described previously (Komatsu et al., 2009).
Transient assays of M. polymorpha cells were carried out using 7- to 10-d-old suspension culture. The cells were collected on a filter paper by filtration and cultured on M51C agar medium containing 2% Suc and 1 mg mL−1 2,4-dichlorophenoxyacetic acid. The DNAs of Em-GUS, Ubi-LUC, Ubi-GFP, and MpABI1 constructs were bound to 1.1- μ m tungsten particles and bombarded twice into the cells from a distance of 12 cm using the 1,100-p.s.i. rapture disc under a chamber vacuum of 28 mm of mercury. The cells were incubated on agar medium with or without ABA at 22°C for 24 h. After incubation, the cells were observed with a fluorescence stereomicroscope to collect GFP-positive cell clusters. The collected cell clusters were used for protein extraction for GUS and LUC reporter gene assays.
Generation of Transgenic P. patens Expressing MpABI1
Transgenic P. patens plants were generated by polyethylene glycol (PEG)-mediated transformation essentially as described previously (Nishiyama et al., 2000). In brief, protoplasts were prepared from 5-d-old protonemata of P. patens by incubation with 1.25% (w/v) driselase (Kyowa Hakko) in 8% (w/v) mannitol solution. After washing three times with 8% mannitol, the protoplasts were suspended in MMM solution containing 8% mannitol, 15 mm MgCl2, and 5 mm MES (pH 5.6). Linearized DNA (20 μ g in 20 μ L) was mixed with 200 μ L of the protoplast suspension and 200 μ L of the PEG solution [8% mannitol, 2% PEG-6000, 0.1 m Ca(NO3)2, and 10 mm Tris-Cl, pH 8.0]. The mixture was incubated at 45°C for 5 min and then kept at 20°C for 10 min. The protoplasts were gradually resuspended in the protoplast medium [6.6% mannitol, 0.5% Glc, 5 mm ammonium tartrate, 0.184 mm potassium phosphate, pH 5.6, 5 mm Ca(NO3)2, 1 mm MgSO4, and 0.045 mm FeSO4] and incubated overnight at 25°C in the dark. The protoplasts were collected by brief centrifugation and resuspended in the melted and 45°C-prewarmed BCD agar medium containing 8% mannitol and 10 mm CaCl2. The suspension was immediately spread over the cellophane-overlaid BCD agar medium containing 6% (w/v) mannitol and 10 mm CaCl2. After 3 d of incubation at 25°C under continuous light, the cells were transferred onto BCD agar medium containing 3% mannitol and appropriate concentrations of antibiotics. The cells were further incubated at 25°C under continuous light until the colonies of transformants appeared.
Measurement of Endogenous ABA
Extraction of ABA from tissues of M. polymorpha thalli cultured for 4 weeks was carried out as described by Hellwege et al. (1994), and the samples were finally dissolved in 50 mm Tris-Cl (pH 7.8), 150 mm NaCl, and 1 mm MgCl2. ABA content in the samples was determined by competitive ELISA using the Phytodetek ABA kit (Agdia).
Estimation of Freezing and Osmotic Stress Tolerance
Freezing tolerance tests were carried out essentially as described by Nagao et al. (2005). In brief, 7-d-old P. patens protonema cells were placed in a glass test tube containing 0.5 mL of water and kept at −1°C for ice inoculation. The ice-inoculated cells were cooled at a rate of −2.4°C h−1 to desired temperatures and then thawed at 4°C overnight. Electrolyte leakage from damaged cells was measured using a conductivity meter, and percentage survival was determined by comparison with the leakage of unfrozen control cells. Osmotic stress treatment was carried out using different concentrations of mannitol and NaCl solutions. Seven-day-old P. patens cells were soaked for 15 min in these solutions and then planted on BCD agar medium. The cells were cultured for 1 week under continuous light at 25°C to determine their survival.
Analysis of Soluble Proteins in Transgenic Mosses
Analyses of soluble proteins and boiling-soluble proteins by SDS-PAGE were carried out as described previously (Minami et al., 2005). For immunoblot analysis, the boiling-soluble proteins were electroblotted onto Immobilon-P membranes (Millipore) using the Transblot apparatus (Bio-Rad). Reactions with primary antiserum for the 17B9 protein and detection by nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate reagent were carried out as described previously (Minami et al., 2006).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GQ504039.
Acknowledgments
We thank Dr. Yasuo Niwa (University of Shizuoka) for a GFP construct and Dr. Ralph Quatrano (Washington University) for the Em-GUS and Ubi-LUC constructs. We also thank Masato Yahata (Saitama University) for technical assistance.
References
- Attree SM, Pomeroy MK, Fowke LC. (1995) Development of white spruce (Picea glauca (Moench.) Voss) somatic embryos during culture with abscisic acid and osmoticum, and their tolerance to drying and frozen storage. J Exp Bot 46: 433–439 [Google Scholar]
- Bertauche N, Leung J, Giraudat J. (1996) Protein phosphatase activity of abscisic acid insensitive I (ABII) protein from Arabidopsis thaliana. Eur J Biochem 241: 193–200 [DOI] [PubMed] [Google Scholar]
- Bork P, Brown NP, Hegyi H, Schultz J. (1996) The protein phosphatase 2C (PP2C) superfamily: detection of bacterial homologues. Protein Sci 5: 1421–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK, Sakakibara K. (2007) Green genes: comparative genomics of the green branch of life. Cell 129: 229–234 [DOI] [PubMed] [Google Scholar]
- Chandler PM, Robertson M. (1994) Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu Rev Plant Physiol Plant Mol Biol 45: 113–141 [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103: 1988–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García MP, Rodríguez D, Nicolás C, Rodríguez PL, Nicolás G, Lorenzo O. (2003) Negative regulation of abscisic acid signaling by the Fagus sylvatica FsPP2C1 plays a role in seed dormancy regulation and promotion of seed germination. Plant Physiol 133: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode JA, Stead AD, Duckett JG. (1992) Re-differentiation of moss protonemata: an experimental and immuno-fluorescence study of brood cell formation. Can J Bot 71: 1510–1519 [Google Scholar]
- Gosti F, Bequdoin N, Serizet C, Web AAR, Vartanian N, Giraudat J. (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusta LV, Trischuk R, Weiser CJ. (2005) Plant cold acclimation: the role of abscisic acid. J Plant Growth Regul 24: 308–318 [Google Scholar]
- Hellwege EM, Dietz KJ, Hartung W. (1996) Abscisic acid causes changes in gene expression involved in the induction of the landform of the liverwort Riccia fluitans L. Planta 198: 423–432 [DOI] [PubMed] [Google Scholar]
- Hellwege EM, Dietz KJ, Volk OH, Hartung W. (1994) Abscisic acid and the induction of desiccation tolerance in the extremely xerophilic liverwort Exormotheca holstii. Planta 194: 525–531 [Google Scholar]
- Hellwege EM, Volk OH, Hartung W. (1992) A physiological role of abscisic acid in the liverwort Riccia fluitans L. J Plant Physiol 140: 553–556 [Google Scholar]
- Hicks GR, Raikhel NV. (1995) Protein import into the nucleus: an integrated view. Annu Rev Cell Dev Biol 11: 155–188 [DOI] [PubMed] [Google Scholar]
- Ishizaki K, Chiyoda S, Yamato KT, Kohchi T. (2008) Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol 49: 1084–1091 [DOI] [PubMed] [Google Scholar]
- Kamisugi Y, Cuming AC. (2005) The evolution of the abscisic acid responses in land plants: comparative analysis of group 1 LEA gene expression in moss and cereals. Plant Mol Biol 59: 723–737 [DOI] [PubMed] [Google Scholar]
- Knight CD, Sehgal A, Atwal K, Wallace JC, Cove DJ, Coates D, Quatrano RS, Bahadur S, Stockley PG, Cuming AC. (1995) Molecular responses to abscisic acid and stress are conserved between moss and cereals. Plant Cell 7: 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K, Nishikawa Y, Ohtsuka T, Taji T, Quatrano RS, Tanaka S, Sakata Y. (2009) Functional analyses of the ABI1-related protein phosphatase type 2C reveal evolutionarily conserved regulation of abscisic acid signaling between Arabidopsis and the moss Physcomitrella patens. Plant Mol Biol 70: 327–340 [DOI] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J. (1997) The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatase 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wurtele ES, LaMotte CE. (1994) Abscisic acid is present in liverworts. Phytochemistry 37: 625–627 [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. (2009) Regulator of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Marcotte WR, Jr, Russel SH, Quatrano RS. (1989) Abscisic acid-responsive sequences from the Em gene of wheat. Plant Cell 1: 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella HH, Sakata Y, Quatrano RS. (2006) Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J 46: 1032–1044 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E. (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Minami A, Nagao M, Arakawa K, Fujikawa S, Takezawa D. (2003) Abscisic acid-induced freezing tolerance in the moss Physcomitrella patens is accompanied by increased expression of stress-related genes. J Plant Physiol 160: 475–483 [DOI] [PubMed] [Google Scholar]
- Minami A, Nagao M, Arakawa K, Fujikawa S, Takezawa D. (2006) Physiological and morphological alteration associated with freezing tolerance in the moss Physcomitrella patens. Chen T, Uemura M, eds, Plant Cold Hardiness CAB International, Wallingford, UK, 138–152 [Google Scholar]
- Minami A, Nagao M, Ikegami K, Koshiba T, Arakawa K, Fujikawa S, Takezawa D. (2005) Cold acclimation in bryophytes: low-temperature-induced freezing tolerance in Physcomitrella patens is associated with increases in expression levels of stress-related genes but not with increase in level of endogenous abscisic acid. Planta 220: 414–423 [DOI] [PubMed] [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E. (2008) Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. Plant J 54: 806–819 [DOI] [PubMed] [Google Scholar]
- Nagao M, Minami A, Arakawa K, Fujikawa S, Takezawa D. (2005) Rapid degradation of starch in chloroplasts and concomitant accumulation of soluble sugars associated with ABA-induced freezing tolerance in the moss Physcomitrella patens. J Plant Physiol 162: 169–180 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, et al. (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Hiwatashi Y, Sakakibara K, Kato M, Hasebe M. (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7: 9–17 [DOI] [PubMed] [Google Scholar]
- Ohta M, Guo Y, Halfter U, Zhu JK. (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci USA 100: 11771–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Ohyama K, Gamborg O. (1979) Regeneration of the liverwort Marchantia polymorpha L. from protoplasts isolated from cell suspension culture. Plant Sci Lett 14: 225–229 [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence V. (1998) Cryopreservation of bryophytes: the effects of ABA and encapsulation dehydration. Bryologist 101: 278–281 [Google Scholar]
- Pence V, Dundord SS, Redella S. (2005) Differential effects of abscisic acid on desiccation tolerance and carbohydrates in the three species of liverworts. J Plant Physiol 162: 1331–1337 [DOI] [PubMed] [Google Scholar]
- Pryce RJ. (1971) Lunularic acid, a common endogenous growth inhibitor of liverworts. Planta 97: 354–357 [DOI] [PubMed] [Google Scholar]
- Qiu YL, Li L, Wang B, Chen Z, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J, et al. (2006) The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci USA 103: 15511–15516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds TL, Bewley DJ. (1993) Abscisic acid enhances the ability of the desiccation-tolerant fern Polypodium virginianum to withstand drying. J Exp Bot 44: 1771–1779 [Google Scholar]
- Robert N, Merlot S, N'Guyen V, Boisson-Dernier A, Schroeder JI. (2006) A hypermorphic mutation in the protein phosphatase 2C HAB1 strongly affects ABA signaling in Arabidopsis. FEBS Lett 580: 4691–4696 [DOI] [PubMed] [Google Scholar]
- Rodriguez PL, Benning G, Grill E. (1998) ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185–190 [DOI] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. (2004) Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci 9: 236–243 [DOI] [PubMed] [Google Scholar]
- Sheen J. (1998) Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA 95: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota H, Tachibana K, Watabe K, Kamada H. (1999) Successful long-term preservation of abscisic-acid-treated and desiccated carrot somatic embryos. Plant Cell Rep 18: 749–753 [Google Scholar]
- Tähtiharju S, Palva T. (2001) Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J 26: 461–470 [DOI] [PubMed] [Google Scholar]
- Takezawa D. (2003) Characterization of a novel plant PP2C-like protein Ser/Thr phosphatase as a calmodulin-binding protein. J Biol Chem 278: 38076–38083 [DOI] [PubMed] [Google Scholar]
- Weiler EW. (1979) Radioimmunoassay for the determination of free and conjugated abscisic acid. Planta 144: 255–263 [DOI] [PubMed] [Google Scholar]
- Werner O, Espin RMR, Bopp M, Atzorn R. (1991) Abscisic-acid-induced drought tolerance in Funaria hygrometrica Hedw. Planta 186: 99–103 [DOI] [PubMed] [Google Scholar]
- Wise MJ, Tunnacliffe A. (2003) POPP the question: What do LEA proteins do?. Trends Plant Sci 9: 13–17 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Umezawa T, Mizoguchi T, Takahashi S, Takahashi F, Shinozaki K. (2006a) The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J Biol Chem 281: 5310–5318 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Nishiyama N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. (2006b) ABA-Hypersensitive Germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]