Abstract
Bromodomain and Extra Terminal domain (BET) proteins are characterized by the presence of two types of domains, the bromodomain and the extra terminal domain. They bind to acetylated lysines present on histone tails and control gene transcription. They are also well known to play an important role in cell cycle regulation. In Arabidopsis (Arabidopsis thaliana), there are 12 BET genes; however, only two of them, IMBIBITION INDUCIBLE1 and GENERAL TRANSCRIPTION FACTOR GROUP E6 (GTE6), were functionally analyzed. We characterized GTE4 and show that gte4 mutant plants have some characteristic features of cell cycle mutants. Their size is reduced, and they have jagged leaves and a reduced number of cells in most organs. Moreover, cell size is considerably increased in the root, and, interestingly, the root quiescent center identity seems to be partially lost. Cell cycle analyses revealed that there is a delay in activation of the cell cycle during germination and a premature arrest of cell proliferation, with a switch from mitosis to endocycling, leading to a statistically significant increase in ploidy levels in the differentiated organs of gte4 plants. Our results point to a role of GTE4 in cell cycle regulation and specifically in the maintenance of the mitotic cell cycle.
The histones H2A, H2B, H3, and H4 are the four core proteins that built nucleosomes (Hansen, 2002). They can all be modified by acetylation, methylation, phosphorylation, and ubiquitination, and these posttranslational modifications provide in a combinatorial or sequential fashion the histone code that can dictate specific cellular processes (Strahl and Allis, 2000; Turner, 2000; Zhang and Reinberg, 2001; Eberharter and Becker, 2002; Sun and Allis, 2002; Nowak and Corces, 2004; Bode and Dong, 2005). It has been demonstrated that some of the modifications, especially acetylations, of histone tails are able to relax the packing of the DNA, facilitating the access to DNA of many regulatory proteins involved in replication, transcription, repair, and recombination (Lorch et al., 1999; Wolffe, 2001; de la Cruz et al., 2005).
Proteins containing bromodomains have the important role of deciphering the histone acetylation codes, since bromodomains bind acetylated Lys residues on histone tails (Dhalluin et al., 1999; Jacobson et al., 2000; Strahl and Allis, 2000; Dey et al., 2003; Liu et al., 2008). The bromodomain was first discovered in the Drosophila Brahma protein (Kennison and Tamkun, 1988; Tamkun et al., 1992) and is present in a broad range of chromatin-modifying proteins (Haynes et al., 1992; Jeanmougin et al., 1997; Jacobson et al., 2000; Syntichaki et al., 2000; Dyson et al., 2001; Horn and Peterson, 2001; Schwanbeck et al., 2004; Yang, 2004). Bromodomain and Extra Terminal domain (BET) proteins form a separate group of bromodomain proteins that all share besides the N-terminal bromodomain(s) an extra terminal (ET) domain (Haynes et al., 1992; Lygerou et al., 1994; Pandey et al., 2002). The ET domain consists of three separate regions, of which only the N-terminal ET domain is conserved in all BET proteins. The ET domain was shown to have Ser kinase activity, and it functions as an interaction domain to recruit other proteins or complexes to acetylated histones (Platt et al., 1999).
The first BET family member that was functionally analyzed is female sterile homeotic of Drosophila (Huang and Dawid, 1990; Chang et al., 2007). This gene has been shown to activate the ULTRABITORAX gene, a homeotic gene involved in Drosophila embryo development. Other well-characterized members are the mammalian BET proteins BRD2 and BRD4 (formally RING3 and MCAP), for which it has been demonstrated that they bind to acetylated histones (Dey et al., 2003; Kanno et al., 2004). The same has been observed for their yeast homolog Bromodomain factor1 (Bdf1; Pamblanco et al., 2001). Brd2 and Brd4 are expressed in proliferating cells and are fundamental for cell cycle progression (Dey et al., 2000; Houzelstein et al., 2002; Maruyama et al., 2002; Shang et al., 2004; Wu and Chiang, 2007; Mochizuki et al., 2008; Yang et al., 2008). BET proteins identified in yeast are also involved in regulation of the cell cycle. The yeast bdf1 mutant is defective in meiosis and was also shown to be associated with mitotic chromosomes. Also, BRD2 and BRD4 bind to mitotic chromosomes and binding persists during mitosis (Platt et al., 1999; Dey et al., 2003). It has been hypothesized that these proteins contribute to the transmission of the transcriptional memory from one generation of cells to the next (Matangkasombut et al., 2000; Dey et al., 2003).
Although BET proteins were first identified and mostly studied in yeast, human, mouse, and Drosophila, BET proteins were also recently identified in plants. All plant BET proteins are different from those of yeast and animals, since they have only one bromodomain instead of two (Florence and Faller, 2001; Pandey et al., 2002). In animals, the presence of two bromodomains seems to be fundamental for their function. However, in yeast, genetic analysis suggests that the function of Bdf1 in sporulation is only dependent on one bromodomain (Chua and Roeder, 1995). The presence of only one bromodomain in the plant BET protein members was supposed to be compensated by a dimerization event, but there is still no evidence that can support this hypothesis (Florence and Faller, 2001). Twelve BET-encoding genes have been identified in the Arabidopsis (Arabidopsis thaliana) genome. Until now, only two of these have been functionally characterized, IMBIBITION INDUCIBLE1 (IMB1) and GENERAL TRANSCRIPTION FACTOR GROUP E6 (GTE6). IMB1 plays a role in the promotion of seed germination by regulating negatively the abscisic acid pathway and positively the phytochrome transduction pathway (Duque and Chua, 2003). Microarray analysis of the imb1 mutant showed that IMB1 acts predominantly as a transcriptional activator and is responsible for the activation of genes involved in cell wall metabolism and plastid-encoded genes. The other characterized plant BET-encoding gene, GTE6, is involved in the establishment of elliptical leaf shape in mature leaves (Chua et al., 2005). GTE6 was shown to positively regulate the myb domain gene ASYMMETRIC LEAF1 (AS1), which is involved in leaf axis specification in mature leaves. It is associated with the promoter and the start of the transcribed region of AS1 and up-regulates AS1 expression through the acetylation of histones H3 and H4.
Although both IMB1 and GTE6 seem to have a role in transcriptional regulation, for neither of the two is there evidence for a function in cell cycle regulation.
Here, we present a functional analysis of GTE4, a member of a so far uncharacterized group of Arabidopsis BET proteins. The gte4 mutant shows defects during root, leaf, and flower development. Cell numbers are significantly reduced in most of the organs. Cell cycle reactivation is delayed in germinating seeds, and a premature switch from mitosis to endoreduplication occurs. Furthermore, a partial loss of root quiescent center (QC) identity is observed. The results that we obtained show that GTE4 is involved in the activation and maintenance of cell division in the meristems and by this controls cell numbers in most organs.
RESULTS
GTE4 Encodes a BET Bromodomain Protein and Is Expressed in All Organs
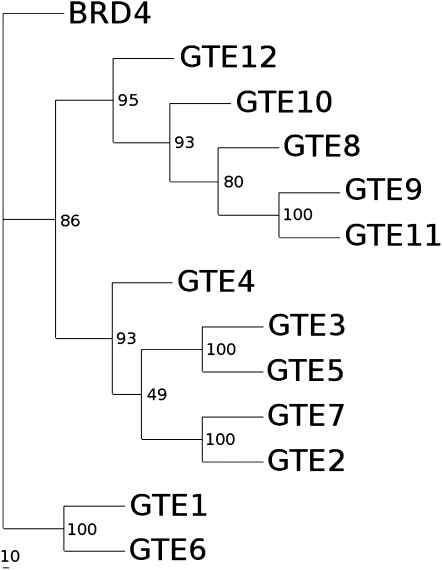
The analysis of the Arabidopsis genome sequence allowed the identification of 12 BET proteins. We produced a rooted tree for these Arabidopsis BET proteins using the bromodomain and the ET domain. This analysis supports the previous results obtained by Florence and Faller (2001), since it shows that Arabidopsis BET proteins can be divided into three groups that clearly reflect the differences in the entire protein sequences of these Arabidopsis BET proteins. Both IMB1 (GTE1) and GTE6, the first two bromodomain genes that were studied in detail, belong to the same group, whereas GTE4 belongs to a new group of uncharacterized genes (Fig. 1).
Figure 1.
Phylogenetic analysis of the Arabidopsis BET bromodomain family. The rooted tree was obtained with a ClustalX alignment and subsequent analysis with Phylyp protdist neighbor joining; the alignment was made considering only the bromodomain (of human BRD4, only the second bromodomain was considered). The numbers represent the branching reproducibility over 100 bootstraps.
To analyze the expression of GTE4, RNA was extracted from roots, rosette leaves, stems, flowers, and siliques and analyzed by reverse transcription (RT)-PCR. This analysis revealed that GTE4 is expressed in all tissues (Supplemental Fig. S1).
GTE4 Is Not Able to Form Homodimers in a Yeast Two-Hybrid Assay
Plant BET bromodomain proteins have only one bromodomain instead of two. The presence of only one bromodomain was supposed to be overcome by the capacity of plant BET proteins to form dimers (Florence and Faller, 2001). To address if GTE4 could form homodimers, we performed a yeast two-hybrid analysis. For this purpose, we cloned the GTE4 open reading frame in two different vectors, creating fusion proteins with the GAL4 activation domain and the binding domain. We tested at 21°C and 29°C for interaction by selecting yeast double transformants on medium lacking His to which we added increasing concentrations of 3-amino-1,2,4-triazole, and we also tested for interactions using the β -galactosidase assay. Whereas all positive controls using interacting MADS box proteins (de Folter et al., 2005) were positive, all the assays in which we tested homodimerization of GTE4 were negative, indicating that GTE4 is not forming homodimers. This assay obviously does not exclude the possibility that GTE4 forms heterodimers with other BET proteins.
The gte4 Mutant Presents Defects in Aerial Organ Size and Shape
To functionally analyze GTE4, we searched the Salk T-DNA collection (Alonso et al., 2003) for GTE4 insertion mutants. In this collection, we identified a line that has a T-DNA insertion 200 bp downstream of the bromodomain (Fig. 2A). Homozygous mutant lines were identified by PCR and confirmed by Southern-blot analysis (data not shown). Expression analysis by RT-PCR using RNA extracted from the homozygous gte4 mutant showed that the T-DNA insertion caused complete silencing of the GTE4 gene (Supplemental Fig. S2).
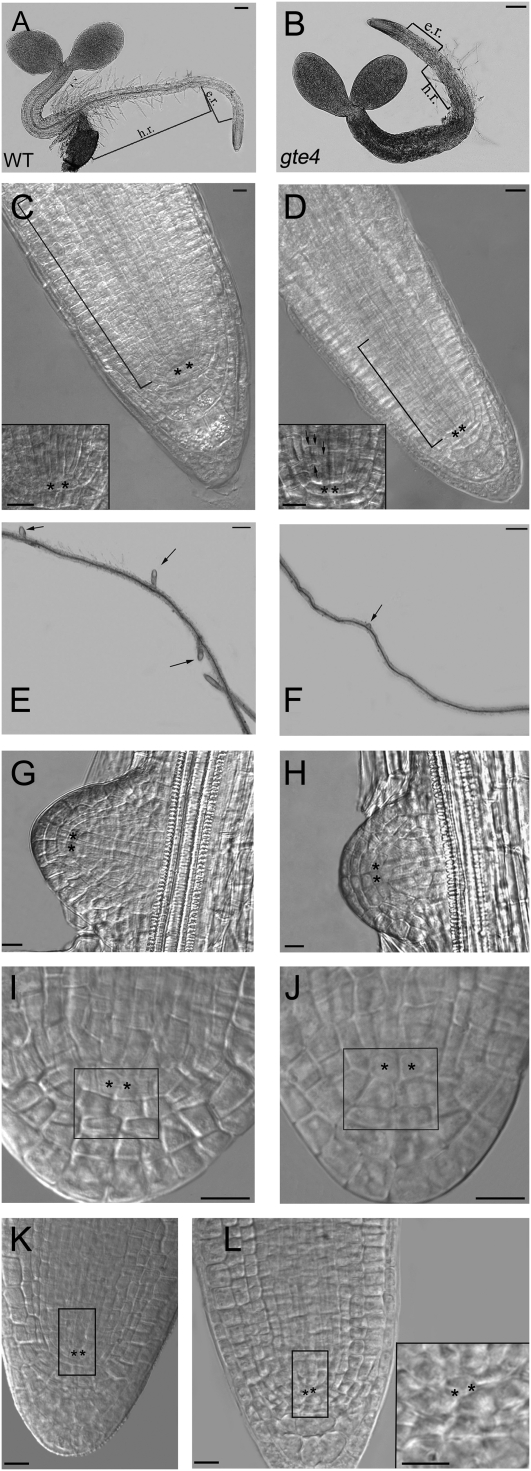
Figure 2.
Analysis of the gte4 mutant. A, Schematic representation of the T-DNA insertion in GTE4. B, Differences in inflorescence height in the wild type (WT) and the gte4 mutant. C, Rosettes of the gte4 mutant are smaller than those of wild-type plants. D, Comparison of leaf size and shape between the gte4 mutant and the wild type. gte4 leaves are visibly more jagged than those of the wild type. E, Transections of wild-type and gte4 stem internodes showing the reduced stem thickness in gte4 plants at 55 d after germination with unstained sections under light microscopy. Bars = 50 μ m. F, Wild-type and gte4 flowers at anthesis. The stamen number (arrows) in the gte4 flower is reduced in comparison with the wild type. Bars = 0.5 mm. [See online article for color version of this figure.]
Initial phenotypic analyses of the gte4 mutant showed that these plants were significantly reduced in size at all stages of plant development (Fig. 2, B and C). Comparing leaves of wild-type and gte4 mutant plants evidenced that those of the mutant were not only smaller but also had a slightly serrated shape (Fig. 2D). The production of rosette leaves was delayed in the gte4 mutant, with a significant (P < 0.01) reduction in leaf number up to day 14. At day 20, the mean number of rosette leaves had become the same (6.6 ± 0.3 for the wild type and 6.6 ± 0.4 for gte4), although differences in leaf morphology remained. The leaves of gte4 plants (e.g. the sixth leaf) were significantly (P < 0.05) smaller than those of the wild type (Table I). Histological analysis carried out on the sixth leaf showed a higher variability in the cellular dimensions of the mutant leaf in comparison with the wild-type leaf (as shown by se values in Table I); however, the mean areas of the subepidermal (Table I) and epidermal (data not shown) cells were not statistically different between the genotypes. Moreover, there was no significant change in the mean area of the intercellular spaces (data not shown). However, in the gte4 mutant, the number of cells in the subepidermis was about half with respect to the wild type (Table I). This suggests that in the gte4 mutant there is a general reduction in leaf cell number. During later developmental stages, the leaf phenotype of gte4 plants was not rescued; in fact, at day 35, the leaves remained significantly (P < 0.05) smaller than wild-type leaves (Table I).
Table I. Microscopic analysis of rosette leaves of 20- and 35-d-old gte4 and wild-type plants.
The mean area (±se) of the sixth rosette leaf, the mean area of the subepidermal cells (±se), and the mean number of subepidermal cells (±se) are shown at days 20 and 35 (70 cells randomly chosen in 10 specimens per genotype). Values followed by no letter or by the same letter are not significantly different.
| Parameter | Wild Type | gte4 | |
| Day 20 | Leaf area (mm2) | 31.4 ± 3.4ab | 17.7 ± 3.9c |
| Cell area (μ m2) | 516.5 ± 31.6c | 554.2 ± 130c | |
| Cell no. | 60,947.5 ± 6,044.6d | 34,375.1 ± 3,168.1 | |
| Day 35 | Leaf area (mm2) | 50 ± 4a | 37.4 ± 1.1 |
| Cell area (μ m2) | 1,031.8 ± 118 | 1,017 ± 129.3 | |
| Cell no. | 50,186.9 ± 4,218.3a | 36,792.6 ± 3,957.4 | |
Values of the wild type differ statistically from the gte4 mutant (P < 0.05).
Value of day 20 is statistically different with respect to the corresponding value of day 35 within the same genotype (P < 0.05).
Values of day 20 are statistically different with respect to the corresponding values of day 35 within the same genotype (P < 0.01).
Values of the wild type differ statistically from the gte4 mutant (P < 0.01).
The floral transition was not affected in the gte4 mutant and occurred under long-day conditions at the same time (day 16) as in wild-type plants. However, in gte4 plants, the height of the inflorescence stem remained significantly (P < 0.01) reduced up to the end of the life cycle (i.e. 23.3 ± 0.8 versus 27.9 ± 1.3 cm for the wild type; Fig. 2B). Stem thickness was also significantly (P < 0.01) reduced, which was due to a reduction in the mean area of the cortex and stelar regions (Fig. 2E; Supplemental Table S1). These differences were due to a reduced cell number, as exemplified for the cortex in Supplemental Table S1.
Analysis of gte4 flowers revealed no differences in floral organ size. The only difference detected in floral structures was a reduction in stamen number. In wild-type flowers, we always observed six stamens, whereas in the gte4 mutant, this number was reduced to 4.7 ± 0.2 (Fig. 2F). Some delay in silique growth also occurred; however, at the end of the reproductive phase, neither the length of the siliques nor the number of seeds per silique changed in comparison with the wild type (data not shown).
The gte4 Mutant Affects Root Development
Root development was clearly affected in gte4 mutant plants. Significant differences in primary root length were already observed at day 4 (Fig. 3, A and B). The gte4 mutant roots were about three times shorter than the wild-type ones (Table II). The mutant hairy region was about 25% of the length compared with the wild type, whereas the elongation region was about 50% reduced in length. The cell area in both the elongation region and in the hairy region was not significantly different until day 4, which shows that there is a clear reduction in cell number in both regions of the mutant (Table II). At day 9, the reduction in cell numbers was enhanced in the mutant (Table II). The roots of the mutant continued to be shorter than in the wild type after day 9, although the difference in length was progressively reduced (data not shown).
Figure 3.
Effects of the gte4 mutation on root development in young seedlings. A and B, Young seedlings. C and D, Primary roots. E to L, Lateral root apices. A and B, Wild-type (WT; A) and gte4 (B) unstained sections under light microscopy of 4-d-old seedlings. The gte4 primary root is shorter than that of the wild type due to a reduction in length in the hairy region (h.r.) and the elongation region (e.r.). Bars = 100 μ m. C and D, Wild-type (C) and gte4 (D) apices of the primary root of seedlings at day 4 after germination showing the different lengths of the apical region (brackets). Bars = 10 μ m. Magnifications of the QC cells and surrounding initial cells are shown in the insets. The arrows in the inset of D show anomalous anticlinal divisions in the procambial cells. Bars = 10 μ m. E and F, Unstained sections under light microscopy showing details of the primary root of wild-type (E) and gte4 (F) 9-d-old seedlings revealing a lower density, and a reduced development, of lateral roots (arrows) in the mutant. Bars = 200 μ m. G and H, Wild-type (G) and gte4 (H) lateral root apices at early emergence from the primary root in 12-d-old seedlings showing apical initials and QC cells equally defined in both genotypes. Bars = 10 μ m. I and J, Details of wild-type (I) and gte4 (J) lateral root apices during the expansion phase of the emerged organ in 14-d-old seedlings showing that expansion in the QC cells and columella initials is higher in gte4 roots than in the wild type (squares). Bars = 10 μ m. K and L, Apices of protruded lateral roots in wild-type (K) and gte4 (L) plants. Rectangles show QC cells, columella, and procambial initials in the gte4 mutant in comparison with the wild type. A magnification of hardly visible QC cells and of irregularly sized surrounding cells is shown in the inset. Bars = 10 μ m. In C, D, and G to L, images are by Nomarski microscopy. QC cells are marked with asterisks.
Table II. Histological analysis of the hairy and elongation regions in the primary root of 4- and 9-d-old gte4 and wild-type seedlings.
Mean length, mean cell area, and mean cell number (±se) of cortical cells in the hairy region and in the elongation region are shown at days 4 and 9 (100 cells randomly chosen in 10 specimens per genotype). Values followed by no letter are not significantly different between genotypes in the same region on the same day.
| Parameter | Wild-Type Hairy Region | Wild-Type Elongation Region | gte4 Hairy Region | gte4 Elongation Region | |
| Day 4 | Length (μ m) | 2,342.3 ± 210.5a | 565 ± 45.5a | 594.8 ± 82.9 | 308.8 ± 39.2 |
| Cell area (μ m2) | 1,571.4 ± 151 | 587.6 ± 122.1 | 1,861.5 ± 131.5 | 369.9 ± 95.8 | |
| Cell no. per region | 202.4 ± 19a | 118.7 ± 15.3b | 66.6 ± 6.3 | 95.3 ± 20.6 | |
| Day 9 | Length (μ m) | 15,406.3 ± 1,181.4a | 509.7 ± 30a | 6,419.5 ± 1,023.9 | 342.5 ± 23.4 |
| Cell area (μ m2) | 1,467.8 ± 66.9 | 384.9 ± 11.7 | 1,654.7 ± 258 | 577.8 ± 113.2 | |
| Cell no. per region | 1,265.4 ± 109.9a | 155.3 ± 12a | 479.6 ± 145.7 | 68.7 ± 15.5 | |
Values statistically different (P < 0.01) with respect to the corresponding region in the gte4 genotype on the same day.
Values statistically different (P < 0.05) with respect to the corresponding region in the gte4 mutant on the same day.
At 4 d after germination, the primary root apical region was analyzed in detail, starting from the basal walls of the QC cells up the distal border of the elongation region (Fig. 3, C and D). The length and area of this region were significantly reduced in mutant roots in comparison with the wild-type ones (Table III). The cell size in the gte4 mutant was instead more than twice that of wild-type cells, resulting in a highly reduced cell number in gte4 apices (Table III). The length and area of the apex did not change during the following days in both genotypes, as shown for day 9 in Table III. The only difference was that in wild-type primary roots the mean cell size reached a value comparable to those measured in the gte4 mutant. However, since the region area in the wild type remained more than twice that in mutant roots, the cell number in the gte4 mutant continued to be highly reduced (Table III).
Table III. Mean length and area of the root apex of primary and lateral roots in 4- and 9-d-old gte4 and wild-type seedlings.
The mean cell area and cell number of the cortical derivative cells (±se) are shown (30 cells randomly chosen in 10 specimens per genotype and day for the primary root, and 10 cells randomly chosen in 10 specimens per genotype for the lateral root). Comparative values between the wild type and gte4 followed by no letter are not significantly different.
| Parameter | Wild Type | gte4 | |
| Primary roots | |||
| Day 4 | Length (μ m) | 163.2 ± 4.0a | 88.6 ± 9.3 |
| Region area (μ m2) | 9,671.9 ± 717.6a | 4,137.6 ± 797.2 | |
| Cell area (μ m2) | 46.1 ± 2.2a | 93.9 ± 15.7 | |
| Cell no. per region | 216.1 ± 16.2a | 42.6 ± 8.9 | |
| Day 9 | Length (μ m) | 188.8 ± 13a | 104.4 ± 9.8 |
| Region area (μ m2) | 11,052.4 ± 486.1a | 4,851.4 ± 563.5 | |
| Cell area (μ m2) | 95.1 ± 10 | 96.1 ± 10.1 | |
| Cell no. per region | 121.5 ± 12.7a | 51.0 ± 5.5 | |
| Lateral roots | |||
| Day 9 | Region area (μ m2) | 1,752.1 ± 28.7 | 1,732.9 ± 57.5 |
| Cell area (μ m2) | 33.1 ± 2.1b | 44.6 ± 4.5 | |
| Cell no. per region | 53.9 ± 3.7b | 40.7 ± 4.1 | |
Values statistically different (P < 0.01) with respect to the gte4 mutant.
Values statistically different (P < 0.05) with respect to the gte4 mutant.
Also, the development of the lateral roots and their contribution to the root apparatus were different in gte4 plants, considering both the timing of their macroscopic protrusion from the primary root and their number. In fact, at day 9, in comparison with wild-type roots, lateral root formation was still poor on gte4 plants. At day 20, the differences in lateral root formation continued to be observed, with a significantly lower number of lateral roots (P < 0.01) in the mutant (i.e. 1.6 ± 0.6 compared with 6.9 ± 1.3 in the wild type) and a reduced density (data not shown).
At day 9, the root apex of lateral roots was histologically examined. Besides the differences in cell number and density when compared with wild-type roots (see above), the lateral roots seemed to develop slower in the mutant. In gte4 plants, the lateral roots were still primordia, whereas in wild-type roots, they were already in a much more advanced stage of development (Fig. 3, E and F). The apical dome of the emerging lateral roots exhibited the same shape in the two genotypes (Fig. 3, G and H), and as a consequence, its area was similar (Table III). The mean cell area in the mutant was instead higher (P < 0.05), resulting in a reduced (P < 0.05) cell number in comparison with the wild type (Table III).
Loss of GTE4 Function Affects Seed Development and Germination
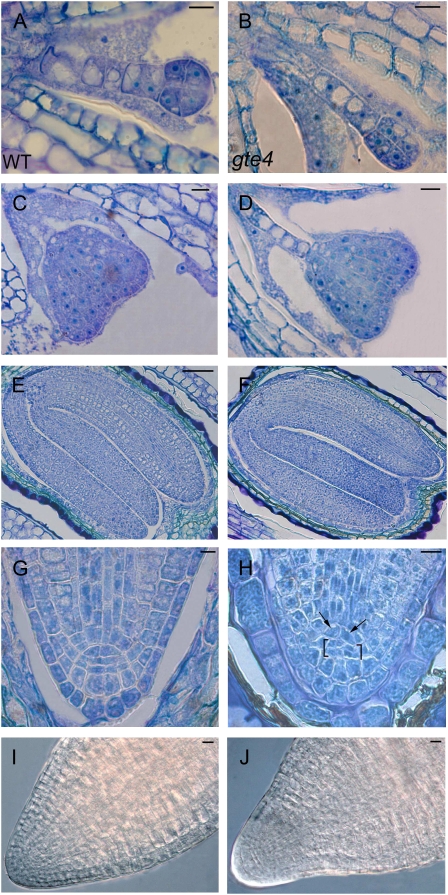
To investigate whether the gte4 mutant already demonstrated developmental defects at the embryonic stage, we analyzed the formation of the embryo and the suspensor during seed development. This analysis showed that no visible changes occurred in the mutant embryo from octant to heart stage (Fig. 4, A–D). However, the area of the suspensor cells was significantly (P < 0.05) increased in the mutant relative to the wild-type (i.e. 148.9 ± 15 and 109.2 ± 9.3 μ m2, respectively). This resulted into a more elongated suspensor in gte4 at the heart stage (Fig. 4, C and D). Moreover, at the cotyledonary stage, about half of the gte4 embryos showed anomalous root pole development (Fig. 4, E–H), with irregularly shaped QC cells and irregularly dividing columella initials (Fig. 4, G and H).
Figure 4.
Effects of the gte4 mutation on embryo development. Images on the left are from the wild type (WT) and those on the right are from the gte4 mutant. A to D, In the gte4 mutant, there is no visible effect on embryo development at the octant stage (A and B) and the heart stage (C and D), whereas the suspensor cells are enlarged. Bars = 10 μ m. E and F, Wild-type (E) and gte4 (F) embryos at the cotyledonary stage showing no significant alteration in the cotyledons. Bars = 50 μ m. G and H, Details of the root pole of the embryo at the cotyledonary stage showing irregularly shaped QC cells (arrows) and irregularly divided columella initials (brackets) in the mutant. Bars = 10 μ m. I and J, Details of wild-type and gte4 apices of mature embryos showing a different shape in the mutant. Bars = 10 μ m. In A to H, longitudinal toluidine blue-stained sections of developing seeds are observed under light microscopy; in I and J, images by Nomarski microscopy are shown after 5 h of imbibition on filter paper. [See online article for color version of this figure.]
The root pole of the mature embryo was also analyzed after 5 h of imbibition on filter paper (Fig. 4, I and J). Differences in the shape and size of the most apical region (50 μ m in length, starting from the basal walls of the QC) were observed (i.e. 2,305.1 ± 64.8 and 2,692.1 ± 71.5 μ m2 for gte4 and the wild type, respectively; P < 0.01). Since dimensions of the embryonic cells did not change significantly between the genotypes (i.e. 66 ± 1.9 μ m2 in gte4, 60.1 ± 2.3 μ m2 in the wild type), the cell number in this region was significantly (P < 0.01) reduced in the mutant embryos in comparison with that in the wild type (i.e. 35 ± 0.6 and 45.1 ± 1.3, respectively).
To test whether seed germination was affected, we plated wild-type and gte4 mutant seeds on filter paper for 7 d. This analysis showed that after 2 d, 75% of the wild-type seeds were germinated whereas only 8% of the mutant seeds were germinated. The delay in germination continued up to day 4 (67% of gte4 germinating seeds with respect to 98% of wild-type seeds; Table IV).
Table IV. Germination and cell cycle analysis for BrdU continuous labeling.
Values are means ± sd of the percentages obtained in three independent experiments. In each experiment, the percentage of BrdU-positive nuclei was determined in 20 randomly chosen root tips per genotype and developmental stage.
| Time from Start of Seed Imbibition | Percentage of Germination |
Percentage of BrdU-Positive Nuclei |
|||
| Wild Type | Mutant | Wild Type | Mutant |
||
| Germinated | Nongerminated | ||||
| 12 h | 0 | 0 | 0 | 0 | 0 |
| 24 h | 0 | 0 | 4 ± 1a | 0 | 0 |
| 48 h | 75 ± 2 | 8 ± 5a | 89 ± 12 | 87 ± 9 | 0 |
| 72 h | 95 ± 4 | 55 ± 7a | 98 ± 6 | 98 ± 11 | 0 |
| 96 h | 98 ± 7 | 67 ± 9a | 99 ± 12 | 98 ± 13 | 0 |
These values are statistically different (P < 0.01).
Finally, after 1 week, both wild-type and mutant seeds had all germinated, indicating that there is only a delay in germination, not a defect that completely abolishes germination of the mutant seeds.
The gte4 Mutant Shows Defects in the QC
Histological analysis of the gte4 mutant showed that QC cells were always present in the primary root apex (Fig. 3, C and D). QC cells were normally defined during embryogenesis; however, they became progressively smaller and, starting from day 4, confined with irregularly dividing cells, procambial ones in particular (Fig. 3, C and D, insets). During the formation of lateral roots in the gte4 mutant, QC cells were normally produced and exhibited a normal shape at the early emergence stage of the primordium (Fig. 3, G and H). At the following expansion phase (Malamy and Benfey, 1997), the QC cells appeared to be more expanded than in the wild type. The same was observed for some columella initials that surround the QC (Fig. 3, I and J). Subsequently, in the protruded lateral roots, QC cells became highly reduced in size (Fig. 3, K and L), whereas cells surrounding the QC were irregular in size (Fig. 3L, inset) and columella initials were expanded (Fig. 3, K and L, insets), resulting in an anomalous morphology of the apical meristem.
These observations suggest that QC cells are not functioning correctly in mature apices, and this might result in a lack of coordination in division activity of the surrounding initials, affecting macroscopic organ development.
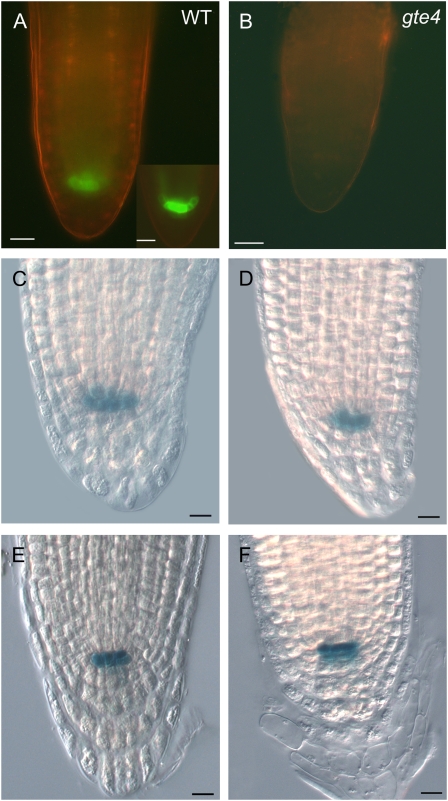
To investigate whether the loss of GTE4 function causes QC defects, we crossed the gte4 mutant with three QC marker lines: two QC-expressed promoter trap GUS lines, QC25 and QC46 (Sabatini et al., 2003), and the AGL42:GFP line (Nawy et al., 2005). This analysis showed that in the gte4 mutant, the QC25 and QC46 markers are normally expressed (Fig. 5, C–F). However, in the gte4 mutant containing the AGL42:GFP marker, no GFP signal could be detected in all stages of root development following QC definition (Fig. 5, A and B), thus indicating a partial, but not transient, loss of QC identity. This might explain the observed defects in the root meristem morphology.
Figure 5.
Expression of QC markers in the root apex of wild-type (A, C, and E) and gte4 (B, D, and F) seedlings. A and B, Expression of AGL42:GFP in the apex of the primary root. A, Wild-type (WT) root showing a strong GFP signal in the QC cells (higher magnification in the inset). B, Absence of GFP signal in the root apex of a gte4 mutant containing the same construct. Bars = 20 μ m. C to F, Expression of the QC46 (C and D) and QC25 (E and F) GUS reporter in the QC cells of wild-type and gte4 mutant roots. Bars = 10 μ m.
Loss of GTE4 Function Affects Cell Proliferation in the Root Meristem and Promotes Endoreduplication in Both Roots and Shoots
The onset of cell proliferation during seed germination was investigated by applying the bromodeoxyuridine (BrdU) incorporation/immunodetection method. BrdU labeling allows the detection of all the cells that enter S phase at least once during the period in which BrdU is present in the medium. Wild-type and gte4 mutant seeds were imbibed and grown for 96 h in the presence of BrdU. No labeled nuclei were detected up to 12 h in both wild-type and mutant roots (Table IV). At 24 h, a low percentage of proliferating cells (4%) was only revealed in wild-type roots. Such a percentage increased in the next hours, reaching a mean value of 89% after 48 h. At 96 h, nearly all wild-type root cells were BrdU positive. Similar kinetics for cell cycle activation was observed in gte4 mutant root as seeds germinated. However, most seeds had not yet germinated after 48 h. In these seeds, no cycling cells were detected (Table IV; Supplemental Fig. S3), suggesting that the delay in root emergence might be related to the delay in cell cycle activation.
Additional analyses to investigate for anomalies in cell cycle regulation were performed on roots and shoots applying the BrdU pulse/chase method along with the determination of mitotic index and flow cytometry (Galbraith et al., 1991). Three-day-old wild-type and mutant seedlings, incubated with BrdU during the last 2 h of growth, showed no significant difference in the percentage of root meristem cells that were running either through S phase or M phase (Table V). This indicated that the loss of GTE4 function did not affect the cell cycle progression in the root meristem of 3-d-old plants. Nevertheless, the analysis of 4- and 5-d-old seedlings showed that the proportion of cells that underwent mitosis was significantly reduced (P < 0.01) in mutant root meristem, whereas the percentage of S phase-traversing cells was similar to that observed in the wild type (Table V). This suggested that in the mutant, a substantial number of cells exit from the mitotic cell cycle earlier than the corresponding wild-type cells and switch from mitosis to endoreduplication. Accordingly, flow cytometry analysis of 4- to 5-d-old roots revealed a statistically (P < 0.05) higher percentage of 8C polyploid cells in the gte4 mutant, whereas no difference occurred in 3-d-old roots in comparison with the wild type (Table VI). The enhanced endoreduplication in gte4 roots was confirmed by the analysis of fully differentiated plant organs (e.g. differentiated portions of roots from 10- and 30-d-old seedlings) in which the percentage of polyploids was significantly higher (P < 0.01; Table VI). Similar results were obtained for the leaves. As in 3-d-old roots, in the first formed leaf primordia cell cycle progression was not affected (Table VI), whereas enhanced endoduplication occurred in the fully differentiated leaves and cotyledons as in roots (P < 0.01; Table VI).
Table V. Cell cycle analysis for BrdU pulse and chase labeling and mitotic index determination.
Values are means ± sd of the percentages obtained in four independent experiments. In each experiment, at least 20 randomly chosen root tips per genotype and developmental stage were analyzed.
| Time from Start of Seed Imbibition | Percentage of S Phase-Traversing Cells (BrdU-Positive Cells) |
Percentage of M Phase-Traversing Cells (Mitotic Index) |
||
| Wild Type | Mutant | Wild Type | Mutant | |
| 72 h | 18.1 ± 1.1 | 16.7 ± 0.9 | 4.8 ± 0.2 | 4.5 ± 0.4 |
| 96 h | 15.3 ± 0.7 | 16.0 ± 1.1 | 4.2 ± 0.1 | 2.1 ± 0.3a |
| 120 h | 14.2 ± 1.3 | 13.5 ± 1.2 | 4.1 ± 0.4 | 1.8 ± 0.5a |
These values are statistically different (P < 0.01).
Table VI. Ploidy level distribution in wild-type and gte4 mutant plants.
Pooled plantlets or single mature plants were subjected to flow cytometric analysis according to organ type at successive times from the beginning of seed imbibition. The reported values are means ± sd of the percentages obtained in four independent experiments. In each experiment, at least five pools of about 100 plantlet leaf primordia, five pools of 10 cotyledons, and at least five pools of about 50 roots were analyzed (3-, 4-, 5-, and 10-d-old plantlets); also in each experiment, about 20 30-d-old plants were individually analyzed to determine ploidy in mature leaves and roots. Asterisks indicate statistical differences (P < 0.01). n.d., Not determined.
| 3-d Roots |
4-d Roots |
||||
| Wild Type | gte4 | Wild Type | gte4 | ||
| 2c | 44.3 ± 2.2 | 45.5 ± 2.9 | 2c | 46.1 ± 3.3 | 34.8 ± 2.1* |
| 4c | 43.8 ± 2.8 | 43.1 ± 3.3 | 4c | 42.5 ± 4.2 | 43.4 ± 1.4 |
| 8c | 11.9 ± 1.7 | 11.4 ± 1.9 | 8c | 11.4 ± 2.1 | 21.8 ± 3.8* |
| 16c | n.d. | n.d. | 16c | n.d. | n.d. |
| 32c | n.d. | n.d. | 32c | n.d. | n.d. |
| 5-d Roots |
10-d Roots |
||||
| Wild Type | gte4 | Wild Type | gte4 | ||
| 2c | 41.9 ± 2.7 | 33.3 ± 2.6* | 2c | 36.8 ± 2.7 | 22.7 ± 2.1* |
| 4c | 43.5 ± 1.9 | 45.2 ± 2.1 | 4c | 27.4 ± 1.9 | 31.3 ± 1.6* |
| 8c | 14.6 ± 1.9 | 21.5 ± 1.8* | 8c | 30.7 ± 1.7 | 36.6 ± 2.1* |
| 16c | n.d. | n.d. | 16c | 5.1 ± 1.4 | 9.4 ± 0.6* |
| 32c | n.d. | n.d. | 32c | n.d. | n.d. |
| 30-d Roots |
First Immature Leaves |
||||
| Wild Type | gte4 | Wild Type | gte4 | ||
| 2c | 32.3 ± 2.3 | 24.3 ± 2.6* | 2c | 74.2 ± 3.8 | 75.2 ± 3.4 |
| 4c | 24.9 ± 1.9 | 24.7 ± 2.3 | 4c | 25.1 ± 1.9 | 23.9 ± 1.6 |
| 8c | 34.2 ± 2.1 | 40.4 ± 2.1* | 8c | 0.7 ± 0.4 | 0.9 ± 0.6 |
| 16c | 8.4 ± 0.9 | 9.9 ± 0.9 | 16c | n.d. | n.d. |
| 32c | 0.1 ± 0.06 | 0.7 ± 0.1* | 32c | n.d. | n.d. |
| Differentiated Leaves |
Cotyledons |
||||
| Wild Type | gte4 | Wild Type | gte4 | ||
| 2c | 49.9 ± 1.8 | 44.2 ± 2.1* | 2c | 42.6 ± 2.1 | 30.2 ± 2.4* |
| 4c | 42.2 ± 1.6 | 36.5 ± 2.1* | 4c | 39.9 ± 2.7 | 36.3 ± 2.6 |
| 8c | 6.3 ± 1.7 | 13.8 ± 1.3* | 8c | 13.5 ± 1.7 | 26.3 ± 2.2* |
| 16c | 1.4 ± 0.8 | 5.2 ± 1.6* | 16c | 2.8 ± 1.0 | 5.2 ± 1.1* |
| 32c | 0.2 ± 0.1 | 0.3 ± 0.1 | 32c | 1.2 ± 0.6 | 2.0 ± 0.7 |
Based on these results, GTE4 plays a role in the control of cell proliferation and can be supposed to be involved in the retinoblastoma (RB)-E2F pathway, which is one of the most important pathways involved in the control and coupling of cell division and cell differentiation in both animals and plants (van den Heuvel and Dyson, 2008). To test this hypothesis, we analyzed by quantitative RT-PCR the expression of a few genes that are involved in the E2F pathway in 4-d-old plantlets.
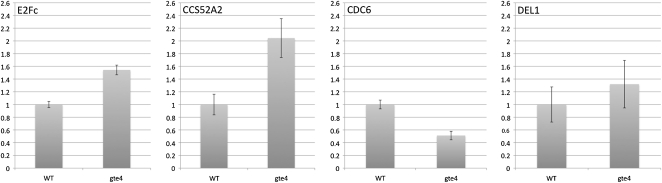
We tested E2Fc, E2Fe/DEL1, CDC6, and CCS52A2. EF2c was selected because it was shown to function at two stages during the cell cycle: at G0 (cell cycle enter), during which E2Fc actively represses E2F target genes, and later in the cell cycle, likely in G2/M, when E2Fc might participate in the decision to progress through mitosis or to switch to the endocycle and differentiation programs (del Pozo et al., 2006). E2Fe/DEL1 expression was tested because it is another important cell cycle regulator. It is an inhibitor of the endocycle and preserves the mitotic state of proliferating cells by suppressing the transcription of genes that are required for cells to enter the DNA endoreduplication cycle (Vlieghe et al., 2005). Finally, CDC6 and CCS52A2 were selected among the E2F target genes because CDC6 is critical for DNA replication and its mRNA level is reduced by overexpression of the nondegradable form of the E2Fc protein, whereas CCS52A2 is critical for the onset of endoreduplication and is a direct E2Fe/DEL1 target (Castellano et al., 2004; del Pozo et al., 2006; Lammens et al., 2008).
From this analysis emerges the idea that, except for E2Fe/DEL1, the expression of which seems to be not significantly changed, E2Fc and CCS52A2 expression is significantly up-regulated and that of CDC6 is down-regulated (Fig. 6). These changes in expression are in agreement with the possible involvement of GTE4 in the RB-E2F pathway for the control of cell proliferation.
Figure 6.
Quantitative RT-PCR analysis of genes that are involved in the E2F pathway. The graphs show the relative expression of each gene in the gte4 mutant compared with the wild type (WT). Error bars indicate se. RNA was extracted from 4-d-old Arabidopsis plantlets.
Rescue of the gte4 Mutant Phenotype by Complementation
To provide proof that the observed phenotypes of the gte4 mutant are due to the loss of GTE4 activity, a complementation experiment was performed. Plants homozygous for the gte4 allele were transformed using a binary vector carrying a genomic fragment that included sequences 1.6 kb upstream of the translation start site, the entire coding region, and 947 bp downstream of the stop codon. Transformants were identified by kanamycin selection, and the presence of the complementation construct was confirmed by PCR. We obtained 12 transformants (homozygous for the gte4 allele), of which seven had a normal wild-type phenotype (data not shown). This confirms that the introduced construct complements the gte4 phenotypes.
DISCUSSION
GTE4 is a member of the BET protein family, which is known to contain a bromodomain capable of interacting with acetylated Lys residues usually found in histones and an ET domain that is a protein-protein interaction motif (Denis et al., 2006). These sequence characteristics make BET proteins key players in the modulation of gene expression by epigenetic mechanisms (Florence and Faller, 2001; Yang et al., 2008). Epigenetic mechanisms regulate genome reprogramming during early embryogenesis and gametogenesis, cell differentiation, and maintenance of a committed lineage (Delcuve et al., 2009).
BET proteins have been studied in a variety of animal organisms, where they are mainly implicated in cell cycle progression by transmitting epigenetic memory through mitosis (Florence and Faller, 2001; Yang et al., 2008). In plants until now, only two members of the Arabidopsis BET gene family have been characterized, IMB1 and GTE6, with a role in the promotion of seed germination and leaf shape, respectively (Duque and Chua, 2003; Chua et al., 2005).
Here, we report the characterization of GTE4, which belongs to a different phylogenetic group of the Arabidopsis BET family of which no member, to our knowledge, was studied previously. Phenotypic characterization of the gte4 mutant shows a variety of developmental defects in roots, leaves, and flowers that might be caused by defective cell cycle regulation in meristematic cells, leading to a significant reduction in cell number and a significant increase in ploidy level in most organs. Interestingly, in this mutant, the functionality of the root QC also seems to be partially lost.
In the Arabidopsis root meristem, the initial cells produce mitotically active derivative cells that further differentiate, giving rise to all cell types of the mature root. The initial cells surround a small group of mitotically less active cells that is called the QC. The QC cells and the initials form the stem cell niche (Sabatini et al., 2003). Loss of QC identity causes the loss of the stem cell niche and prevents the root from growing (Aida et al., 2004; Tucker and Laux, 2007).
The gte4 mutation affects the root stem cell niche and derivative cells from the very first stages of development. This is demonstrated by the fact that the embryo root apex shows after 5 h of imbibition a reduced number of cells, and the morphology of the QC and surrounding cells is abnormal in a large proportion of embryos at the cotyledonary stage.
The aberrant morphology of the gte4 QC and surrounding cells is likely correlated to the partial loss of QC identity, shown by the lack of the QC-specific pAGL42:GFP reporter expression in these mutant plants and the presence of QC25 and QC46 promoter trap expression. Partial loss of QC identity has also been observed in the scarecrow (scr) mutant. In this mutant, the QC184 marker maintains its expression in the QC, whereas the QC25 and QC46 lose their expression (Sabatini et al., 2003). Interestingly, cells in the scr QC region are aberrant in shape and roots ultimately cease growth (Scheres et al., 1995; DiLaurenzio et al., 1996). gte4 roots are less affected than scr roots, since root growth is not completely abolished. However, gte4 QC cells, and surrounding ones, have an aberrant morphology and the root meristem is disorganized, showing altered pattern formation.
The analysis of the cell cycle in developing seedlings revealed a premature arrest of the cycling cells, which switch from mitosis to endoreduplication. No anomalies in cell cycle progression through the cell cycle phases were detected. These results indicate that gte4 meristematic cells can normally proliferate, but they are incapable of balancing the number of differentiated cells with the maintenance of an adequate pool of self-renewing cells. This failure to properly coordinate differentiation with the permanent exit from the cell cycle should explain the phenotypic characteristics of the gte4 mutant (i.e. small organs composed of a reduced number of cells).
An additional feature of gte4 mutant plants is that the differentiated organs are composed of a statistically higher number of polyploid cells. As mentioned above, cell cycle analysis showed that the premature arrest of the mitotic cell cycle is coupled with the onset of endoreduplication, which leads to an increase in ploidy level. The link between mitotic cell cycle and endoreduplication has been widely investigated, and these data suggest that endoreduplication is achieved by a modification of the mitotic cell cycle. For instance, Arabidopsis plants with a reduced CDKB1;1 activity prematurely exit the mitotic cell cycle and have elevated ploidy levels (Boudolf et al., 2004). Similarly, the Arabidopsis SIAMESE gene is required to suppress mitosis as part of the switch to endoreduplication in trichomes (Churchman et al., 2006). Thus, GTE4 seems to be involved in the maintenance of the mitotic cell cycle, and the endoreduplication observed in the gte4 mutant seems directly linked to premature cell cycle exit and might be positively related to the cell size increase observed in some organs. An increase in cell size in response to the inhibition of cell division, and induction of endopolyploidy, have also been observed previously and can be attributed to an uncoupling of cell division and cell expansion (De Veylder et al., 2001; Sugimoto-Shirasu and Roberts, 2003). This hypothesis has already been formulated for other mutants with a reduced cell number, such as the Arabidopsis struwwelpeter (swp) mutant (Autran et al., 2002). Similar to GTE4, SWP plays a role in pattern formation in the meristem and is important for defining the duration of cell proliferation.
Finally, the analysis of the onset of the cell cycle in gte4 germinating seeds showed a delay in cell cycle reactivation in comparison with the wild type. This delay might explain the slower protrusion of the embryonic root from the seed coat registered for the mutant. Moreover, a delay in cell cycle activation in the pericycle founder cells could also explain the drastic delay in lateral root formation that we observed in the gte4 mutant. Many studies both in animals and plants suggest a tight relationship among the onset of the cell cycle, the maintenance of cell proliferation, and differentiation. For instance, Dewitte et al. (2003) demonstrated that Arabidopsis CYCD3 promotes the mitotic cell cycle and affects differentiation by inhibiting mitotic exit and/or endocycles, either independently or through its regulation of RB function. In mammalian development, in particular, the RB-E2F pathway is of central importance and represents a link between the activation of cell proliferation and cell cycle exit, leading to terminal differentiation (Kirshenbaum, 2001).
Thus, GTE4 seems to regulate not only the maintenance of meristem cell proliferation but also the onset of the cell cycle. The pathway(s) that is affected by the gte4 mutation is currently not defined, although on the basis of the literature it is possible to give some hypotheses. It has been shown that the reentrance into the cell cycle is preceded by a change in chromatin condensation. Heterochromatin starts to unpack and new euchromatic regions, compatible with transcriptional activity, are formed (Zhao et al., 2001; Williams et al., 2003). These epigenetic changes might be mediated by GTE4 through its ability, as a BET protein, to interact with acetylated histone tails. Therefore, we can hypothesize that GTE4 may play a role in translating the histone acetylation marks into cell cycle gene activity, allowing the entrance into the cell cycle. GTE4 is also necessary to maintain cell proliferation in the root meristem, since its role does not seem to be limited to reentry into the cell cycle. E2F transcription factors have also been shown to regulate these two processes. E2F is responsible for stem cell maintenance in the root, regulating the transition from cell proliferation to differentiation, and is involved in the reactivation of cell proliferation in the QC (Wildwater et al., 2005; Lammens et al., 2008). In addition, when a cell is reentering the cell cycle, the chromatin around E2F target genes becomes decondensed, allowing E2F to regulate target gene transcription (Williams et al., 2003). Moreover, in mammals, the decision of cells to continue or stop dividing depends largely on the activity of the E2F transcription factors (van den Heuvel and Dyson, 2008). In Arabidopsis, six E2F (E2Fa to -f) and two DP (DPa and -b) proteins were identified, and some of them were demonstrated to be key regulators of cell proliferation and endoreduplication (De Veylder et al., 2002). For instance, E2Fc-DPb restricts cell division and is one of the components in the coordination between cell proliferation and endoreduplication during Arabidopsis development (del Pozo et al., 2006). In agreement with the involvement of GTE4 in the RB-E2F pathway, 4-d-old gte4 mutant plantlets showed changes in the expression of E2Fc, CDC6, and CCS52A2 genes. Specifically, E2Fc was significantly up-regulated and its target gene, CDC6, was down-regulated. Interestingly, like plants overexpressing E2Fc/DPb, gte4 mutant plants are characterized by a reduced number of mitotic cells and an increased DNA content. Moreover, also the expression of the APC/C activator gene CCS52A2 was significantly enhanced in 4-d-old gte4 plantlets, suggesting a transcription deregulation of genes important for correct cell proliferation/endoreduplication and related to the RB-E2F pathway.
Due to these analogies, we can speculate that GTE4 might be involved in E2F-related pathways controlling gene transcription. This hypothesis is also supported by the facts that the BRD2 (RING3) BET bromodomain protein in animals binds to E2F and, together, they regulate gene transcription and cell cycle activity (Denis et al., 2000).
Although our analyses suggest a link between GTE4 and E2F-related pathways, some caution has to be taken, since the number of genes that we analyzed is limited and we do not know if the observed changes in expression levels are physiologically relevant and if the effects on the expression of these genes are direct or indirect. Future studies directed to identify target genes that are under the control of GTE4 will be needed to draw a clearer picture of the regulatory pathways that are controlled by this plant BET protein and to get a better understanding of its role in the epigenetic control of plant development.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) gte4 mutant (ecotype Columbia) was obtained from the Salk collection (SALK_113292 code N613292). Seeds were vernalized for 2 d at 4°C under continuous darkness, sterilized for 10 min in 10% sodium hypochlorite, rinsed with three changes of sterile distilled water, and then sown on petri plates on half-strength Murashige and Skoog medium supplemented with 0.55 mm myoinositol, 0.3 μm thiamine-HCl, 2% (w/v) Glc, and 0.8% (w/v) agar (Sigma). Seeds were incubated under long-day conditions (16/8-h light/dark) at a fluence rate of 150 μ E m−2 s−1 and 22°C ± 2°C. The plates used for the root apparatus analyses were oriented vertically to ease the observation and removal of roots (Malamy and Benfey, 1997). For observation of germination, vernalized seeds were sterilized and put on filter paper for 7 d, whereas for the observation of aerial organs, the seeds were imbibed on filter paper for 2 d at 4°C and sown on commercial soil (Universal Soil). The same growth chamber was used as described above.
QC46 and QC25 promoter trap GUS lines were kindly provided by S. Sabatini (Sabatini et al., 2003), and the AGL42:GFP line was provided by P. Benfey (Nawy et al., 2005).
Phylogenetic Analysis
We produced a rooted tree for these Arabidopsis BET proteins using the bromodomain and the ET domain. The protein sequences have been analyzed using the program ClustalX for protein sequence alignments (Jeanmougin et al., 1998). The tree was obtained using Phylip (Retief, 2000).
RT-PCR
RT-PCR was performed on cDNA obtained as described previously (Lago et al., 2004) using the following primers specific for GTE4: Atp 388 (5′-GATCAGCTTAACGTAGTCAGAG-3 ′) and Atp 389 (5′-CGTCTACTGGAGCATTGAACAC-3 ′). PCR was performed for 30 cycles using an annealing temperature of 54°C.
Quantitative Real-Time RT-PCR
RNA was extracted using the Qiagen RNasy Plant Mini Kit (catalog no. 74904), followed by DNase treatment performed as described (Lago et al., 2004). Invitrogen SuperScriptII was used to reverse transcribe the RNA following the manufacturer's instructions. Real-time PCR was performed with a Bio-Rad IQ5 machine using IQ Bio-Rad SYBR Green Supermix (catalog no. 170-8882) and the primers listed below. Primer annealing was set at 59°C for 40 cycles, and ACTIN was used as a reference gene. Two separate real-time PCRs were performed with three replicas for each sample. The melting and standard curves were determined for each real-time PCR.
Real-time PCR primers were as follows: ACTIN2_f, 5′-GCTCCTCTTAACCCAAAGGC-3 ′ ; ACTIN2_r, 5′-ACACCATCACCAGAATCCAGC-3 ′ ; CCS52A2_f, 5′-ACTCGTACCGCGTTCTGTAC-3 ′ ; CCS52A2_r, 5′-CTCCCTGCTCTGAGATTTCG-3 ′ ; CDC6_f, 5′-GCCGGACCTAGTTCTTCC-3 ′ ; CDC6_r, 5′-GAAACCTCCGCAGCCGAATC-3 ′ ; E2Fc_f, 5′-GAGTCTCCCACGGTTTCAG-3 ′ ; E2FC_r, 5′-TCACCATCCGGTACTGTTGC-3 ′ ; DEL1_f, 5′-GTCCCAAGAAAACGCTACAGAG-3 ′ ; DEL1_r, 5′-AGTGCCTGGTGCAAAAGGTC-3 ′ .
Mutant Analysis
The position of the T-DNA insertion in the GTE4 gene was identified by PCR and subsequent sequencing using T-DNA-specific primers Atp 388 and Atp 58 (5′-TGGTTCACGTAGTGGGCCATCG-3 ′).
Histological Analysis
The histological analysis was carried out on leaves, stems, roots, siliques, and seeds.
Leaves were fixed in 70% ethanol, dehydrated, embedded in Technovit 7100 (Heraeus Kulzer), sectioned at 4 μ m with an automatic microtome (Microm HM 350 SV), stained with 0.05% (w/v) toluidine blue, and examined with a DAS Leica DMRB microscope (Leica). Alternatively, the leaves were treated with chloral hydrate solution (chloral hydrate:distilled water:glycerol, 8:1:2 [w/v/v]) for the observations with Nomarski optics applied to the same microscope.
To evaluate stem diameter, cortex area, stelar area, cortex cell area, and number, the median stem internodes of 55-d-old wild-type and gte4 plants were embedded in 5% (w/v) agar, sectioned at 30 μ m with a vibratome (Vibratome Series 1000), and observed by Nomarski optics. Embryos were analyzed in siliques and mature seeds. The siliques were fixed, dehydrated, embedded, sectioned, and stained as previously described, and the seeds were imbibed on filter paper for 5 h and treated for observation by Nomarski optics.
Primary and lateral root morphology, and QC and surrounding cells, were analyzed by Nomarski optics. In the primary root, the length of the hairy region was measured from the most proximal (i.e. toward the shoot apex) to the most distal (i.e. toward the root apex) trichoblast, that of the elongation region was measured from the most distal trichoblast to the most proximal cortical cell showing elongation, and that of the apical region was measured from this latter cell to the basal walls of the QC cells. To calculate the area of the apical region of the primary root, it was assimilated to an isosceles triangle, whose height was the length of the region, measured as described above, and the base was the width of the region, measured, including the root protoderm, at the distal border of the elongation region. To calculate the area of the apical region in the lateral root primordium, it was assimilated to an isosceles triangle, whose height was 50 μ m, measured proximally from the basal walls of QC cells, and the base was the width of the primordium at the proximal end of the height.
Histochemical Analysis of GUS Activity and GFP Analysis
gte4 mutant plants were crossed with the QC25, QC46, and pAGL42:GFP reporter lines, and from the F2 population gte4 homozygous plants containing the reporter constructs were selected and used to analyze the QC through GUS activity and monitoring of GFP fluorescence with a Leica DMRB microscope equipped with a double wavelength filter set (BP 490/20 and BP 575/30) with dichroic filters RKPs 505 and 600 and emission filters BPs 525/20 and 635/40. All the histological images were acquired with a DC500 video camera applied to the DMRB microscope and then analyzed with a personal computer (Opti-Xex GX 240 MT) using the Leica IM1000 image-analysis software (Leica).
Statistical Analysis
Differences between percentages were evaluated using the χ2 test, and differences between the means were evaluated by Student's t test, using GraphPad InStat3 software.
Complementation of the gte4 Mutant
The bacterial artificial chromosome clone F9P14 was used to obtain the GTE4 genomic region for the complementation experiment. The bacterial artificial chromosome clone was digested with SalI and KpnI, and a band of 3,900 bp was isolated containing a part of the At1G06230 locus and inserted into pCAMBIA 1300. To add the 3 ′ untranslated region, we performed PCR with Atp 597 (5′-AACCATATGGTACCAATGTCTG-3 ′) and Atp 598 (5′-GGGAGCTCACTGCACTTGCTTCCACAC-3 ′). The amplified fragment was cloned into the pGEM-T-Easy vector (Promega). Subsequently, this fragment was inserted as a KpnI-SacI fragment into the binary vector pCAMBIA 1300 already containing part of the At1G06230 locus. This binary vector was used to transform Agrobacterium tumefaciens C58C1/pMP90 (Koncz et al., 1984). Arabidopsis plants were transformed using the floral dip method described by Clough and Bent (1998).
Cell Cycle Analysis
Flow Cytometric Analyses
Nuclear suspensions were obtained from Arabidopsis plants at different development stages following the protocol of Galbraith et al. (1983). Chicken erythrocytes were added as a reference internal standard to each sample. The mixed nuclei were stained with the DNA-binding fluorochrome 4 ′ ,6-diamidino-2-phenylindole (DAPI) at a final concentration of 5.5 μm. The fluorescence intensity of the nuclei was measured with an arc lamp-based flow cytometer (Bryte-HS; Bio-Rad). Four independent experiments were carried out. In each experiment, 450 pooled plantlets were subjected to flow cytometric analysis according to organ type: at least five pools of about 100 plantlet leaf primordia, five pools of 10 cotyledons, and at least five pools of about 50 whole roots were analyzed in 3-, 4- , 5-, and 10-d-old plantlets. In 3-, 4-, 5-, and 10-d-old wild-type plantlets, the analyzed roots were 2.5 ± 0.2, 3.1 ± 0.2, 5.7 ± 0.9, and 17 ± 0.9 mm long, respectively. Smaller lengths (0.73 ± 0.1, 0.97 ± 0.2, 1.5 ± 0.5, and 7.4 ± 1.0 mm) were recorded for the analyzed gte4 roots at the same growth stages. About 20 plants were individually analyzed to determine ploidy in mature leaves and roots excised from 30-d-old plants. All the rosette leaves (about 9 ± 0.3 leaves for both the wild type and gte4) and the whole root were analyzed for each plant. The significant differences between gte4 and wild-type mean percentages were statistically analyzed by the Statgraphics plus program for Windows (version 4.0; Manugistic). ANOVA and Dunnet tests were applied when normality and homogeneity of variance were satisfied; data that did not conform to the assumptions were alternatively transformed into logarithms or were analyzed by Kruskal-Wallis nonparametric procedures.
BrdU Incorporation and Detection
In order to study the kinetics of cell cycle reactivation in root meristem during germination, plantlets were germinated from surface-sterilized seeds at 25°C on filter paper imbibed with 10 μm BrdU solution (Sigma-Aldrich). Root tips were excised after 72 h from the start of imbibition and were fixed in 4% (w/v) paraformaldehyde (Polysciences; 10% solution, methanol free) in Tris buffer [10 mm tris(hydroxy-methyl) aminomethane, 10 mm Na-EDTA, and 100 mm NaC1, pH 7.4] for 16 h at 4°C. Root tips were also collected and fixed from 3-, 4-, and 5-d-old plantlets grown on filter paper imbibed with distilled water and pulsed with 30 μm BrdU only during the last 2 h of growth to detect the percentage of meristematic cycling cells. After fixation, all the samples were embedded in London Resin Gold (Polysciences Europe). Sections were obtained with a Reichert Jung Ultracut E microtome and were collected on poly-l-Lys-coated slides. Selected sections were used for immunochemical detection of BrdU according to standard protocols. A negative control sample, without BrdU but with the rabbit primary antibody, was also included in the experiment. Slides were examined with a Zeiss Axioplan microscope equipped with a video camera (Media Cybernetics). The acquired digital images were analyzed by the Image-Pro Plus program (Media Cybernetics). The experiments were all repeated at least three times. In each experiment, at least 20 randomly chosen roots were analyzed per genotype and developmental stage. The figures show the results of one representative experiment. The tables report means ± sd of the data collected in all the experiments and their statistical analysis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_179270.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. GTE4 expression analysis by RT-PCR.
Supplemental Figure S2. RT-PCR expression analysis of GTE4 in wild-type and gte4 mutant plants.
Supplemental Figure S3. DAPI and BrdU staining analysis on wild-type and gte4 mutant roots.
Acknowledgments
We thank Drs. S. Sabatini and P. Benfey for providing the root QC marker lines, S. Berri for helping with the phylogenetic analysis of Arabidopsis BET family members, and A. Schnittger for helpful suggestions for the analysis of the E2F-related gene network.
References
- Aida K, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Autran D, Jonak C, Belcram K, Beemster GT, Kronenberger J, Grandjean O, Inzé D, Traas J. (2002) Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J 15: 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode AM, Dong Z. (2005) Inducible covalent posttranslational modification of histone H3. Sci STKE 26: re4. [DOI] [PubMed] [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inzé D, De Veylder L. (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano M M, Boniotti MB, Caro E, Schnittger A, Gutierrez C. (2004) DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16: 2380–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YL, King B, Lin SC, Kennison JA, Huang DH. (2007) A double-bromodomain protein, FSH-S, activates the homeotic gene ultrabithorax through a critical promoter-proximal region. Mol Cell Biol 27: 5486–5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua P, Roeder GS. (1995) Bdf1, a yeast chromosomal protein required for sporulation. Mol Cell Biol 15: 3685–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua YL, Channeliere S, Mott E, Gray JC. (2005) The bromodomain protein GTE6 controls leaf development in Arabidopsis by histone acetylation at ASYMMETRIC LEAVES1. Genes Dev 19: 2245–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Brown ML, Kato N, Kirik V, Hülskamp M, Inzé D, De Veylder L, Walker JD, Zheng Z, Oppenheimer DG, et al. (2006) SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18: 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicova L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz X, Lois S, Sánchez-Molina S, Martínez-Balbás MA. (2005) Do protein motifs read the histone code?. Bioessays 27: 164–175 [DOI] [PubMed] [Google Scholar]
- Delcuve GP, Rastegar M, Davie JR. (2009) Epigenetic control. J Cell Physiol 219: 243–250 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C. (2006) The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. (2006) Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res 5: 502–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis GV, Vaziri C, Guo N, Faller DV. (2000) RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ 11: 417–424 [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, et al. (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA. (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. (2003) The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA 100: 8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. (2000) A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol 20: 6537–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491–496 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Duque P, Chua NH. (2003) IMB1, a bromodomain protein induced during seed imbibition, regulates ABA- and phyA-mediated responses of germination in Arabidopsis. Plant J 35: 787–799 [DOI] [PubMed] [Google Scholar]
- Dyson MH, Rose S, Mahadevan LC. (2001) Acetyllysine-binding and function of bromodomain-containing proteins in chromatin. Front Biosci 6: D853–D865 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. (2002) Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep 3: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florence B, Faller DV. (2001) You bet-cha: a novel family of transcriptional regulators. Front Biosci 6: D1008–D1018 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Galbraith W, Wagner MC, Chao J, Abaza M, Ernst LA, Nederlof MA, Hartsock RJ, Taylor DL, Waggoner AS. (1991) Imaging cytometry by multiparameter fluorescence. Cytometry 12: 579–596 [DOI] [PubMed] [Google Scholar]
- Hansen JC. (2002) Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct 31: 361–392 [DOI] [PubMed] [Google Scholar]
- Haynes SR, Dollard C, Winston F, Beck S, Trowsdale J, Dawid IB. (1992) The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res 20: 2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Peterson CL. (2001) The bromodomain: a regulator of ATP-dependent chromatin remodeling?. Front Biosci 6: D1019–D1023 [DOI] [PubMed] [Google Scholar]
- Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. (2002) Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol 22: 3794–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DH, Dawid IB. (1990) The maternal-effect gene fsh is essential for the specification of the central region of the Drosophila embryo. New Biol 2: 163–170 [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science 288: 1422–1425 [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23: 403–405 [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Wurtz JM, Le Douarin B, Chambon P, Losson R. (1997) The bromodomain revisited. Trends Biochem Sci 22: 151–153 [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. (2004) Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol Cell 13: 33–43 [DOI] [PubMed] [Google Scholar]
- Kennison JA, Tamkun JW. (1988) Dosage-dependent modifiers of polycomb and antennapedia mutations in Drosophila. Proc Natl Acad Sci USA 85: 8136–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshenbaum LA. (2001) Death-defying pathways linking cell cycle and apoptosis. Circ Res 25: 978–980 [DOI] [PubMed] [Google Scholar]
- Koncz C, Kreuzaler F, Kalman Z, Schell J. (1984) A simple method to transfer, integrate and study expression of foreign genes, such as chicken ovalbumin and alpha-actin in plant tumors. EMBO J 3: 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago C, Clerici E, Mizzi L, Colombo L, Kater MM. (2004) TBP-associated factors in Arabidopsis. Gene 342: 231–241 [DOI] [PubMed] [Google Scholar]
- Lammens T, Boudolf V, Kheibarshekan L, Zalmas LP, Gaamouche T, Maes S, Vanstraelen M, Kondorosi E, La Thangue NB, Govaerts W, et al. (2008) Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc Natl Acad Sci USA 105: 14721–14726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wang X, Zhang J, Huang H, Ding B, Wu J, Shi Y. (2008) Structural basis and binding properties of the second bromodomain of Brd4 with acetylated histone tails. Biochemistry 47: 6403–6417 [DOI] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg RD. (1999) Histone octamer transfer by a chromatin-remodeling complex. Cell 96: 389–392 [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Conesa C, Lesage P, Swanson RN, Ruet A, Carlson M, Sentenac A, Seraphin B. (1994) The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res 22: 5332–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124: 33–44 [DOI] [PubMed] [Google Scholar]
- Maruyama T, Farina A, Dey A, Cheong J, Bermudez VP, Tamura T, Sciortino S, Shuman J, Hurwitz J, Ozato K. (2002) A mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol Cell Biol 22: 6509–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. (2000) Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev 14: 951–962 [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Nishiyama A, Jang MK, Dey A, Ghosh A, Tamura T, Natsume H, Yao H, Ozato K. (2008) The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J Biol Chem 283: 9040–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. (2005) Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak SJ, Corces VG. (2004) Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet 20: 214–220 [DOI] [PubMed] [Google Scholar]
- Pamblanco M, Poveda A, Sendra R, Rodriguez-Navarro S, Perez-Ortin JE, Tordera V. (2001) Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett 496: 31–35 [DOI] [PubMed] [Google Scholar]
- Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt GM, Simpson GR, Mittnacht S, Schulz TF. (1999) Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J Virol 73: 9789–9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retief JD. (2000) Phylogenetic analysis using PHYLIP. Methods Mol Biol 132: 243–258 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, DiLaurenzio L, Willemsen V, Hauser M-T, Janmaat K, Weisbeek P, Benfey P. (1995) Mutations affecting the radial organisation of the Arabidopsis root display specific defects throughout the embryonic axis. Development 121: 53–62 [Google Scholar]
- Schwanbeck R, Xiao H, Wu C. (2004) Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem 279: 39933–39941 [DOI] [PubMed] [Google Scholar]
- Shang E, Salazar G, Crowley TE, Wang X, Lopez RA, Wang X, Wolgemuth DJ. (2004) Identification of unique, differentiation stage-specific patterns of expression of the bromodomain-containing genes Brd2, Brd3, Brd4, and Brdt in the mouse testis. Gene Expr Patterns 4: 513–519 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts K. (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. (2002) Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Syntichaki P, Topalidou I, Thireos G. (2000) The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404: 414–417 [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. (1992) Brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68: 561–572 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Laux T. (2007) Connecting the paths in plant stem cell regulation. Trends Cell Biol 17: 403–410 [DOI] [PubMed] [Google Scholar]
- Turner BM. (2000) Histone acetylation and an epigenetic code. Bioessays 22: 836–845 [DOI] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ. (2008) Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GT, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé D, De Veylder L. (2005) The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15: 59–63 [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B. (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Williams L, Zhao J, Morozova N, Li Y, Avivi Y, Grafi G. (2003) Chromatin reorganization accompanying cellular dedifferentiation is associated with modifications of histone H3, redistribution of HP1, and activation of E2F-target genes. Dev Dyn 228: 113–120 [DOI] [PubMed] [Google Scholar]
- Wolffe AP. (2001) Transcriptional regulation in the context of chromatin structure. Essays Biochem 37: 45–57 [DOI] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. (2007) The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 282: 13141–13145 [DOI] [PubMed] [Google Scholar]
- Yang XJ. (2004) Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 26: 1076–1087 [DOI] [PubMed] [Google Scholar]
- Yang Z, He N, Zhou Q. (2008) Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol 28: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343–2360 [DOI] [PubMed] [Google Scholar]
- Zhao J, Morozova N, Williams L, Libs L, Avivi Y, Grafi G. (2001) Two phases of chromatin decondensation during dedifferentiation of plant cells: distinction between competence for cell fate switch and a commitment for S phase. J Biol Chem 276: 22772–22778 [DOI] [PubMed] [Google Scholar]