Abstract
In both applied and basic research, Agrobacterium-mediated transformation is commonly used to introduce genes into plants. We investigated the effect of three Agrobacterium tumefaciens strains and five transferred (T)-DNA origins of replication on transformation frequency, transgene copy number, and the frequency of integration of non-T-DNA portions of the T-DNA-containing vector (backbone) into the genome of Arabidopsis (Arabidopsis thaliana) and maize (Zea mays). Launching T-DNA from the picA locus of the Agrobacterium chromosome increases the frequency of single transgene integration events and almost eliminates the presence of vector backbone sequences in transgenic plants. Along with novel Agrobacterium strains we have developed, our findings are useful for improving the quality of T-DNA integration events.
Since the generation of transgenic plants approximately 25 years ago, Agrobacterium tumefaciens has been widely used for introducing genes into plants for purposes of basic research as well as for generation of commercially used transgenic crops. For plant transformation, the gene of interest is placed between the left and right border repeats of Agrobacterium transferred (T)-DNA (Gelvin, 2003). The T-DNA region harboring the transgene is stably integrated into the plant genome by using an appropriate plant transformation protocol. T-DNA originates from the Agrobacterium tumor-inducing (Ti) plasmid. Because Ti plasmids are large and difficult to manipulate, smaller T-DNA binary vectors are currently predominately used for generation of transgenic plants (de Framond et al., 1983; Lee and Gelvin, 2008).
Although Agrobacterium has been used for plant transformation for more than two decades, problems using this bacterium remain. Agrobacterium-mediated transformation generally results in lower transgene copy numbers than do other transformation methods such as particle bombardment or polyethylene glycol-mediated transformation (Kohli et al., 1998; Shou et al., 2004). On the other hand, transformation frequently results in unwanted high copy number T-DNA integration events (Jorgensen et al., 1987; Deroles and Gardner, 1988; Shou et al., 2004; De Buck et al., 2009). Multiple integration events, often coupled with inverted repeat T-DNA integration patterns, may affect the stability of transgene expression by silencing mechanisms (Jorgensen et al., 1996). An additional problem with Agrobacterium-mediated transformation is the propensity for DNA sequences outside the T-DNA region to integrate into the plant genome (Kononov et al., 1997; Wenck et al., 1997; Shou et al., 2004). Integration of such vector backbone sequences can occur with high frequency. For example, Kononov et al. (1997) detected backbone sequences in 75% of tested transgenic tobacco (Nicotiana tabacum) plants, and very often the entire vector backbone is introduced into the plant genome (De Buck et al., 2000). T-DNA vector backbones usually harbor bacterial antibiotic resistance genes that can create governmental regulatory concerns.
Here we show that launching T-DNA from the A. tumefaciens chromosome reduces integrated transgene copy number and almost eliminates the presence of T-DNA backbone sequences. We describe several plasmids and bacterial strains to facilitate use of this methodology.
RESULTS
A. tumefaciens Strains and T-DNA Constructions
Our investigation utilized various combinations of the commonly used A. tumefaciens strains EHA101, GV3101, and LBA4404 with five different T-DNA binary systems. These Agrobacterium strains are nononcogenic (disarmed) and have been used for transformation of a large variety of plants. EHA101 (Hood et al., 1987) harbors a derivative of the agropine/l,l-succinamopine-type Ti-plasmid pTiBo542, GV3101 (Koncz and Schell, 1986), a derivative of the nopaline-type Ti-plasmid pTiC58, and LBA4404 (Ooms et al., 1981), a derivative of the octopine-type Ti-plasmid pTiAch5. The tested T-DNA vectors contain an identical T-DNA region plus an aadA gene for bacterial selection for spectinomycin resistance. However, they contain different origins of replication (ori): the pVS ori, the pSa ori, the RK2 ori, and the pRiA4b ori. We furthermore analyzed the effect of launching T-DNA from the Agrobacterium C58 chromosome at the picA locus in strains EHA101 and GV3101. Disruption of this locus does not affect transformation (Rong et al., 1990) and we have generated vectors specifically designed to integrate genes into this locus (Lee et al., 2001).
We analyzed 14 different A. tumefaciens strain by replication origin combinations. The T-DNA region, derived from the binary vector pTF101.1 (Paz et al., 2004), harbors a bar gene as a plant selectable marker under the control of a cauliflower mosaic virus (CaMV) double 35S promoter (Fig. 1). Utilizing identical T-DNA regions with the same plant selectable marker and the identical non-T-DNA sequence proximal to the T-DNA left border in all constructions enabled us directly to compare results obtained for transformation frequencies, integrated transgene copy numbers, and backbone integration for all strain by construct combinations. All plasmids and A. tumefaciens strains used in this study are listed in Supplemental Table S1.
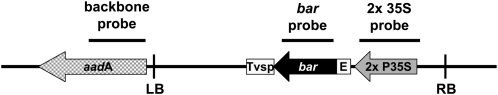
Figure 1.
Schematic map of the T-DNA and neighboring regions of the base vector pTF101.1 (Paz et al., 2004) used in constructing the various vectors. Black bars represent fragments used for DNA-blot hybridizations experiments. LB and RB, T-DNA left and right borders, respectively; aadA, gene encoding spectinomycin resistance; E, translational leader from Tobacco etch virus; Tvsp, VSP (soybean [Glycine max] vegetative storage protein) terminator sequence; bar, gene conferring resistance to Basta/Bialophos/phosphinothricin; 2x P35S, CaMV double 35S promoter sequence.
Effect of Binary Vector Replication Origin on Binary Vector Copy Number in Agrobacterium
We determined the copy number in A. tumefaciens of the four binary vectors used in our study. We placed a bar gene into the EHA105 chromosome and separately introduced each of the four T-DNA binary vectors into this strain. The resulting strains were incubated with or without acetosyringone to induce vir gene expression, and total bacterial DNA was extracted and subjected to DNA-blot analysis using the bar gene as a probe. T-DNA binary vector copy numbers were determined by comparison of the signal intensity of the chromosomal band (one per cell) to the T-DNA binary vector band. Figure 2 shows that plasmids containing the pSa origin are maintained at approximately four copies per cell. The copy numbers of plasmids containing the RK2 and pVS origins are seven to 10 per cell, and plasmids containing the pRi origin replicate to 15 to 20 copies per cell. No significant differences were seen when the strains were cultured under inducing or noninducing conditions.
Figure 2.
Vector copy number in Agrobacterium under inducing and noninducing conditions. A. tumefaciens cells harboring a fragment of the bar gene in the chromosome and a T-DNA binary vector were grown under vir gene inducing and noninducing conditions. DNA-blot analysis was done to determine copy numbers of various T-DNA binary vectors. Membranes were probed with a bar gene fragment. The top band represents the chromosomal integrated bar gene fragment (copy no. per cell: 1) and the bottom band the T-DNA binary vector.
Effect of A. tumefaciens Strain and T-DNA ori on Transformation Frequency
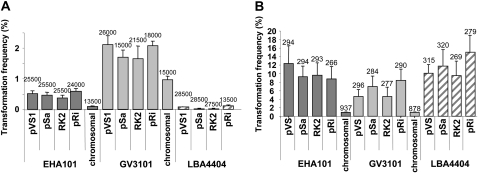
We determined the effect of 14 Agrobacterium strain-by-construct combinations on transformation frequency of Arabidopsis (Arabidopsis thaliana) and maize (Zea mays). Arabidopsis was transformed using a floral-dip protocol (Clough and Bent, 1998). At least four independent transformation experiments were conducted for each vector-by-strain combination, and transformation frequencies were determined by analyzing 1,500 to 4,500 seeds per experiment (Fig. 3A). Transformation frequency was highly dependent upon the A. tumefaciens strain utilized. GV3101 resulted in the highest transformation frequencies (0.97%–2.11%), whereas EHA101 and LBA4404 effected medium (0.09%–0.58%) and low (0.01%–0.12%) transformation frequencies, respectively. T-DNA replication origin had little effect on transformation frequency with one exception: Launching T-DNA from the Agrobacterium chromosome of EHA101 or GV3101 resulted in transformation frequencies lower than those of the other four T-DNA binary systems of the respective strain. Because the picA sequence of the Ach5 chromosome of LBA4404 does not share sufficient homology with the picA sequence of the C58-derived recombination vector we used (Lee et al., 2001), we were unable to integrate the T-DNA region into the picA locus of LBA4404.
Figure 3.
Transformation frequencies of Arabidopsis (A) and maize (B) with different Agrobacterium strain by origin of replication combinations. Arabidopsis was transformed by a floral-dip protocol and maize by embryo inoculation. Error bars represent the se among different transformation experiments. At least four independent transformation experiments were conducted for both Arabidopsis and maize. The total number of seeds screened (A) or embryos infected (B) to calculate transformation frequencies is indicated above each error bar. For maize, five independent experiments were conducted to establish relative transformation frequencies for each of 12 strain-by-replication origin combinations (except chromosomal). Embryos from each of the 22 ears dissected were shared 12 ways at infection. Subsequently, transformation frequencies for the two chromosomal replication origin combinations (in EHA101 and GV3101) were established by inoculating them beside the pTF101.1 replication origin vector in EHA101 and GV3101. In four independent experiments, embryos from 21 dissected ears were shared four ways at infection. Throughout the remainder of this study, additional maize transformation experiments using these 14 strain-by-replication origin combinations were conducted as needed to recover adequate numbers of transgenic events for molecular analysis.
Using an embryo inoculation protocol (Frame et al., 2002), we conducted at least four independent maize transformation experiments from which relative transformation frequencies for each of 12 strain-by-replication origin combinations (except chromosomal) were established (Fig. 3B). Four experiments were conducted to establish relative transformation frequencies for the two chromosomal replication origin combinations (in EHA101 and GV3101; Fig. 3B). As with Arabidopsis, launching T-DNA from the chromosomes of EHA101 and GV3101 resulted in low transformation frequencies (0.9%), whereas transformation frequencies were considerably higher (5%–15%) when T-DNA was placed on a plasmid binary vector.
Launching T-DNA from the Agrobacterium Chromosome Results in a High Percentage of Plants Containing a Single Integrated T-DNA Copy
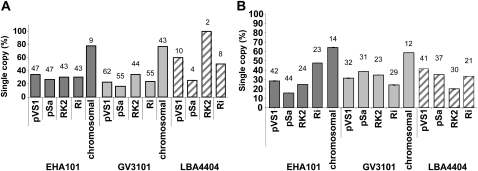
We investigated the number of copies of integrated T-DNA in transgenic events by DNA dot-blot hybridization. T1 generation (heterozygous for T-DNA) Arabidopsis leaf samples were analyzed using a bar gene-specific fragment (Fig. 1). Figure 4A shows, strikingly, that launching T-DNA from the Agrobacterium picA chromosomal locus resulted in 77% to 78% of the events containing a single transgene copy. The percentage of single transgene copy events resulting from use of “conventional” T-DNA binary vectors was much lower. Correspondingly, the average transgene copy number for events generated using conventional T-DNA binary vectors was higher (3.3–4.9 copies/genome; Table I) than was the integrated transgene copy number of events generated using strains with T-DNA launched from the bacterial chromosome (1.3–1.6 copies/genome; Table I).
Figure 4.
The percentage of events containing a single copy of integrated T-DNA following transformation by the 14 T-DNA replication origin-by-Agrobacterium strain combinations for Arabidopsis (A) and maize (B). Copy number reconstructions were done by DNA dot-blot experiments using T-DNA-specific probes (Fig. 1). Analysis was performed on heterozygous T1 generation Arabidopsis plants and heterozygous regenerated T0 generation maize plants. Numbers over the bars represent the number of independent events analyzed.
Table I. Average transgene copy numbers for T-DNA replication origins-by-A. tumefaciens strain combinations.
| A. tumefaciens Strain | T-DNA Vector ori (Incompatibility Group) | Average Transgene Copy No. | No. of Events Analyzed |
| Arabidopsis | |||
| EHA101 | pVS1 | 4.1 | 47 |
| EHA101 | pSa (incW) | 4.7 | 47 |
| EHA101 | RK2 (incP α) | 4.1 | 43 |
| EHA101 | pRiA4 | 3.3 | 43 |
| EHA101 | Chromosomal | 1.3 | 9 |
| GV3101 | pVS1 | 4.4 | 62 |
| GV3101 | pSa (incW) | 4.9 | 55 |
| GV3101 | RK2 (incP α) | 3.3 | 44 |
| GV3101 | pRiA4 | 4.3 | 55 |
| GV3101 | Chromosomal | 1.6 | 43 |
| LBA4404 | pVS1 | 1.7 | 10 |
| LBA4404 | pSa (incW) | 2.0 | 4 |
| LBA4404 | RK2 (incP α) | 1.0 | 2 |
| LBA4404 | pRiA4 | 1.5 | 8 |
| Maize | |||
| EHA101 | pVS1 | 3.7 | 42 |
| EHA101 | pSa (incW) | 3.5 | 44 |
| EHA101 | RK2 (incP α) | 4.2 | 24 |
| EHA101 | pRiA4 | 3.5 | 23 |
| EHA101 | Chromosomal | 1.7 | 14 |
| GV3101 | pVS1 | 3.1 | 32 |
| GV3101 | pSa (incW) | 2.6 | 31 |
| GV3101 | RK2 (incP α) | 3.3 | 23 |
| GV3101 | pRiA4 | 3.2 | 29 |
| GV3101 | Chromosomal | 1.7 | 12 |
| LBA4404 | pVS1 | 2.1 | 41 |
| LBA4404 | pSa (incW) | 2.9 | 37 |
| LBA4404 | RK2 (incP α) | 3.0 | 30 |
| LBA4404 | pRiA4 | 2.4 | 21 |
Maize T-DNA copy number determinations were made using heterozygous T0 generation plants (one plant per event). Because the bar gene resulted in background hybridization signals in maize (H. Oltmanns, unpublished data), we used the CaMV double 35S promoter fragment as the probe (Fig. 1). Figure 4B shows that for events generated using conventional T-DNA binary vectors, the percentage of events carrying a single transgene copy ranged from 16% to 48%. Average T-DNA copy numbers resulting from the use of binary vectors ranged from 2.1 to 4.2 copies per cell (Table I). As with Arabidopsis, use of Agrobacterium strains containing chromosomal integration of T-DNA resulted in a higher percentage (58%–64%) of single transgene copy number events; the average T-DNA copy number in these transgenic events was 1.7 copies per cell (Table I).
Although antibiotics were used to eradicate Agrobacterium after infection, bacterial cells might still contaminate selected transgenic Arabidopsis and maize plants. To eliminate the possibility of contaminating Agrobacterium DNA falsely increasing the apparent T-DNA copy number, we hybridized several membranes with the Agrobacterium chromosomal picA gene. We did not detect a hybridization signal using this probe (data not shown).
Launching T-DNA from the Agrobacterium Chromosome Mitigates Integration of T-DNA Backbone Sequences in Transgenic Plants
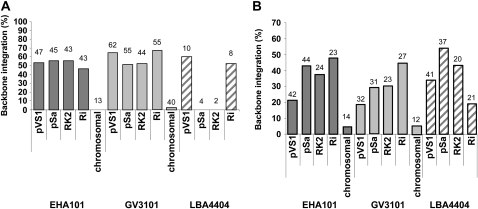
Integration of T-DNA backbone sequences into the genome of transgenic plants can present regulatory problems, especially when bacterial antibiotic resistance genes are transferred. We investigated whether the Agrobacterium strain or T-DNA replication origin affects the frequency of backbone integration events. We used the spectinomycin resistance (aadA) gene immediately outside the T-DNA left border (Fig. 1) as a hybridization probe to detect backbone sequences within the genome of transgenic plants.
Figure 5, A and B show the percentage of transgenic plants with backbone integration events for the 14 strain-by-origin combinations in Arabidopsis and maize, respectively. For Arabidopsis, no plants from 13 EHA101 events (0%) and one plant from 40 GV3101 events (3%), respectively, contained backbone sequences when they were generated by Agrobacterium strains in which the T-DNA was launched from the chromosome. For maize, one plant from 14 EHA101 events (7%) and one plant from 12 GV3101 events (8%), respectively, contained the vector backbone. On the other hand, the use of conventional T-DNA binary vectors resulted in a relatively high percentage of plants containing backbone sequences (47%–67% for Arabidopsis; 19%–55% for maize).
Figure 5.
The percentage of independent events containing vector backbone sequences following transformation by the 14 T-DNA replication origin-by-Agrobacterium strain combinations for Arabidopsis (A) and maize (B). Blots were hybridized with a backbone-specific probe, the aadA gene (Fig. 1). Numbers over the bars represent the number of independent events analyzed.
Vectors and A. tumefaciens Strains to Facilitate Integration of T-DNA into the Agrobacterium Chromosome
We generated two systems to facilitate integration of T-DNA into the Agrobacterium chromosome (Fig. 6). We first introduced a T-DNA region into the picA locus of the C58 chromosome. This T-DNA contains a CaMV 35S-bar gene as a plant selection marker, and a small region of pBluescript to provide homology for recombination with a variety of pBluescript-derived plasmids, such as the pSAT series of expression vectors (Chung et al., 2005). Because plasmids harboring the ColE1 ori cannot replicate in A. tumefaciens, ampicillin/carbenicillin-resistant bacteria can only be selected when the introduced plasmid cointegrates into the homologous T-DNA region (Fig. 6A). This bacterial strain contains an aadA gene directly outside the T-DNA left border to detect transfer of non-T-DNA sequences. In addition, the strain can be eliminated from cocultivation reactions by using β -lactam antibiotics containing clavulanate, such as Timentin (L.-Y. Lee, unpublished data).
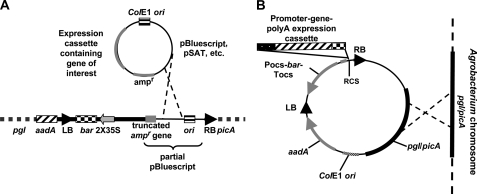
Figure 6.
Systems for introducing T-DNA harboring transgene expression cassettes into the pgl/picA region of the A. tumefaciens chromosome. A, T-DNA containing a CaMV 35S promoter-bar gene plant selection marker and a fragment of pBluescript, and an aadA gene to the left of the T-DNA left border, has been inserted into the A. tumefaciens C58 chromosome. When a pUC-derived plasmid containing an expression cassette is introduced into this strain, carbenicillin-resistant colonies can only be obtained if homologous recombination occurs, resulting in a cointegrate between this plasmid and the pBluescript region of the T-DNA. B, In this system, a transgene expression cassette is first cloned into a T-DNA region located on a plasmid harboring a ColE1 ori. The plasmid also contains the pgl/picA region of the A. tumefaciens C58 chromosome. When this plasmid is introduced into A. tumefaciens C58-derived strains, carbenicillin-resistant colonies can only be obtained if homologous recombination occurs between the pgl/picA regions of the plasmid and the chromosome. LB, T-DNA left border; RB, T-DNA right border; ori, origin of replication; ampr, β -lactamase gene conferring ampicillin/carbenicillin resistance upon the bacterium; aadA, spectinomycin resistance gene; 2X35S, double CaMV 35S promoter; bar, gene conferring resistance to Basta/Bialophos/phosphinothricin; Pocs, octopine synthase promoter; Tocs, octopine synthase terminator; RCS, multiple rare cutting sites flanking various pSAT vectors (AscI, I-PpoI, I-SceI, I-CeuI, PI-PspI, and PI-TilI). Broken crossed lines indicate homologous recombination events.
Although the strain described in Figure 6A is easy to use, it has the potential regulatory disadvantage of introducing a β -lactamase antibiotic resistance gene into the plant when T-DNA integrates. To eliminate such potential regulatory problems for plants destined for field release, we generated a second system to launch T-DNA from the Agrobacterium chromosome. A region homologous to the Agrobacterium pgl/picA locus was cloned into a plasmid containing a T-DNA. Gene expression cassettes from, for example, pSAT vectors can be cloned into the rare-cutting multiple cloning site of this vector, and the entire vector can be introduced into A. tumefaciens. Spectinomycin resistance conferred by this plasmid results from homologous recombination with the picA/pgl region of the A. tumefaciens C58 chromosome (Fig. 6B). Thus, integration of T-DNA into the A. tumefaciens chromosome occurs without introduction of an antibiotic resistance gene into T-DNA.
DISCUSSION
We studied the effect of three commonly used A. tumefaciens strains and five T-DNA replication origins on transformation frequency and the quality of T-DNA integration events in Arabidopsis and maize. Within a factor of approximately three, there was little difference among tested Agrobacterium strains harboring various T-DNA binary vectors with respect to maize transformation frequency. These results contrast with Arabidopsis flower-dip transformation, where we observed a strong dependency upon the Agrobacterium strain, but not the T-DNA binary vector, used. Interestingly, efficient transformation of Brassica juncea (Pandian et al., 2006), Brassica napus, and Brassica oleracea (De Block et al., 1989; Bhalla and Singh, 2008) was obtained using a variety of Agrobacterium strains, including GV3101, LBA4404, and AGL1 (which is similar to EHA101; Lee and Gelvin, 2008). However, none of these studies utilized a flower-dip transformation protocol. It may be that flower-dip transformation is more sensitive to the Agrobacterium strain used than is somatic cell transformation.
We decided to integrate T-DNA into the Agrobacterium chromosome at the picA locus because previous work had indicated that disruption of this site in the bacterial chromosome does not negatively affect transformation frequency (Rong et al., 1990). In addition, we had previously developed plasmids to facilitate integration of DNA into this locus (Lee et al., 2001). Miranda et al. (1992) had previously launched T-DNA from the picA locus. It is possible that launching T-DNA from other positions of the Agrobacterium chromosome would also be suitable. However, we have not investigated this possibility. We note, however, that Hoekema et al. (1984) launched T-DNA from an uncharacterized position in the Agrobacterium chromosome, suggesting that other chromosomal sites could function as T-DNA launching pads. Launching T-DNA from the Agrobacterium chromosome results in fewer integrated transgene copies and almost eliminates the presence of T-DNA binary backbone sequences in recovered transgenic events. However, these two advantageous aspects of plant transformation are accompanied by decreased transformation frequency (Fig. 3). In Arabidopsis, this decrease is slight (2- to 4-fold) but it can be greater (approximately 10-fold) in maize. Whether scientists are willing to compensate decreased transformation frequency with a higher quality transformation event will depend upon the ease, time, and cost in generation of multiple transgenic events for different plant species.
Stable and predictable transgene expression is a major objective for both basic and applied research. Multiple integrated T-DNA copies, especially when combined with complex T-DNA integration patterns, can trigger transgene silencing (Jorgensen et al., 1987, 1996; Stam et al., 1997). The routine generation of single-copy transgenic events is therefore a major goal for agricultural biotechnology. Launching T-DNA from the Agrobacterium chromosome may provide one approach for achieving this goal.
Several studies have analyzed T-DNA locus and/or copy numbers in transgenic Arabidopsis. Feldmann (1991) concluded that the average number of independently segregating, active transgene loci in his initial library of T-DNA-tagged plants is 1.4. This value is similar to that of other T-DNA-tagged collections in Arabidopsis (McElver et al., 2001; Alonso et al., 2003) and rice (Oryza sativa; Barakat et al., 2000; Jeon et al., 2000). However, the number of active loci in these plants is generally less than the number of integrated T-DNA molecules. T-DNA insertions frequently occur as partial or complete multimers in direct or inverted repeat orientation (Jorgensen et al., 1987; Feldmann, 1991). Bechtold et al. (1993) showed that 70% of tested Arabidopsis transformants generated by a vacuum infiltration protocol carried direct or indirect tandem repeat copies of T-DNA. In this study the average T-DNA copy number in Arabidopsis ranged from 1.0 to 4.9, and in maize from 1.3 to 3.9 per diploid genome (Fig. 4; Table I). Our results agree with those of Galbiati et al. (2000) who investigated 38,000 transgenic Arabidopsis plants generated by a floral-dip method. Interestingly, transformation using A. tumefaciens LBA4404 resulted in an average transgene copy number (1.0–1.7 in Arabidopsis and 2.1–3.0 in maize; Table I) lower than that resulting from transformation using the other tested strains. Grevelding et al. (1993) investigated whether the transformation method affected transgene copy number in Arabidopsis. Most transgenic plants produced by a leaf-disc inoculation method contained multiple T-DNA insertions, whereas root transformation resulted mostly in single T-DNA insertions. Therefore, the Agrobacterium strain, transformation method, and plant target tissue may influence the number of integrated T-DNA molecules (De Buck et al., 2009).
Although T-DNA integration into the plant genome was experimentally shown almost 30 years ago, little is known about how many T-DNA strands are produced in Agrobacterium and transferred to the plant cell. It is likely that considerably more T-strands are transferred than are integrated (Virts and Gelvin, 1985; Atmakuri et al., 2007). T-DNA copy number of the chromosomal integration construction in the bacterial cell is one (1; except during replication before cell partition). Low integrated transgene copy numbers in plants may result from a limited number of T-strands transferred to the plant cell. We might therefore have expected to see a correlation between bacterial and plant T-DNA copy number using the different T-DNA replication origins because they replicate to different extents in the bacteria. However, we did not find such a correlation.
Integration of binary vector backbone sequences in transgenic plants is a common phenomenon (Kononov et al., 1997; Wenck et al., 1997; De Buck et al., 2000). Launching T-DNA from the Agrobacterium chromosome almost eliminates the presence of integrated T-DNA backbone sequences (Fig. 5). In contrast, 47% to 67% of the Arabidopsis plants generated by Agrobacterium strains harboring a T-DNA binary vector contained integrated vector backbone sequences. Although elimination of these sequences from transgenic plants is a major goal for agricultural biotechnology, only one previous report described a methodology to effect this result. By incorporating a lethal barnase gene into the non-T-DNA region of the binary vector, Hanson et al. (1999) reduced the number of plants harboring backbone sequences. However, up to 18% of the transgenic plants still carried backbone sequences.
Transfer of binary vector backbone sequences can occur when the T-DNA left border repeat is not recognized by the VirD2 endonuclease during processing of the T-DNA strand. It can also occur as a result of VirD2 linkage to the 5′ end of the vector DNA directly outside the T-DNA left border, followed by transfer of the backbone in a manner analogous to that of T-DNA transfer (Durrenberger et al., 1989; Kononov et al., 1997; De Buck et al., 2000). If the T-DNA strand were derived from a binary plasmid and during T-DNA processing the left border repeat is skipped, T-DNA processing will either end at a sequence in the backbone that resembles a T-DNA border or, due to the circular nature of binary vectors, when the right border repeat is reached. However, if T-DNA were integrated into the bacterial chromosome, read through at the T-DNA left border repeat could result in very long T-DNAs, theoretically as long as the Agrobacterium chromosome itself if no adequate termination site were present. Although transfer of long T-DNA molecules is possible (Miranda et al., 1992; Hamilton et al., 1996), it is less frequent than transfer of small T-DNAs. The observation that large T-DNAs only integrate into the plant genome very rarely offers a possible explanation for why chromosomal integration of T-DNA results in transgenic plants lacking backbone sequences. If the T-DNA left border were skipped during T-DNA strand processing (or if DNA transfer initiates from sequences directly to the left of the left T-DNA border), the resulting T-DNA would be too long for efficient transfer to the plant or integration into the plant genome. Although there might be concern that sequences from the bacterial chromosome next to the T-DNA right border could be integrated into the plant genome, probing of the DNA membranes with an Agrobacterium picA fragment, located immediately to the right of the right T-DNA border, failed to detect its presence (data not shown).
It is tempting to speculate whether integration of vector backbone sequences into plants is a consequence of simplifying Agrobacterium-mediated plant transformation by using small T-DNA binary vectors. Transfer of non-T-DNA portions of a large Ti plasmid to plants is possible but rare: On average only one out of 80 transgenic tobacco calli contained a nptII gene positioned outside the T-DNA left border (Ramanathan and Veluthambi, 1995). In contrast, Kononov et al. (1997) detected vector backbone sequences in approximately 75% of transgenic tobacco plants generated using an Agrobacterium strain carrying a small T-DNA binary vector. These results suggest that backbone integration occurs more frequently when a small T-DNA binary vector is used.
Although T-DNA binary vectors are ubiquitously used because of their ease of handling, we present here two vector systems to simplify launching T-DNA from the Agrobacterium chromosome. The first vector system (Fig. 6A) is easier to use because it requires only one cloning step. This is followed by introduction of the resulting expression plasmid into Agrobacterium for homologous recombination into a T-DNA already positioned at the picA locus. However, this system has the potential regulatory disadvantage of transferring a β -lactamase antibiotic resistance gene into the plant genome. We thus developed a second system (Fig. 6B) to integrate a T-DNA, containing a gene expression cassette but lacking an antibiotic resistance gene, into the A. tumefaciens picA locus. This latter system will not transfer an antibiotic resistance gene to plants. However, it requires two cloning steps before introducing the final plasmid into Agrobacterium for recombination into the picA locus. Together, these two vector systems will facilitate generation of single T-DNA copy number transgenic plants lacking vector backbone sequences.
MATERIALS AND METHODS
Agrobacterium tumefaciens Growth Conditions
Agrobacterium tumefaciens strains were grown on solidified or liquid AB Suc or yeast extract peptone medium (Lichtenstein and Draper, 1986) supplemented with appropriate antibiotics (rifampicin, 10 μ g/mL; spectinomycin, 100 μ g/mL; kanamycin, 25 μ g/mL; gentamicin, 25 μ g/mL).
T-DNA Constructions
The T-DNA region and the bacterial aadA (spectinomycin resistance) gene (Fig. 1) used in all T-DNA binary constructions derives from pTF101.1 (Paz et al., 2004). pTF101.1 contains a pVS1 origin of replication. To generate the various binary vectors, we replaced the pVS1 replication origin (ori) with those from other plasmids. For introducing the RK2 ori, we removed the pVS1 origin from pTF101.1 using ScaI and NotI and replaced it with a NotI/NruI fragment from pBIN19 (Bevan, 1984), generating pTF::Bin19. For introducing the pSa ori, we removed the pVS1 ori from pTF101.1 using ScaI and NsiI and replaced it with a PstI/SacII fragment from pUCD2 (Close et al., 1984). All overhanging ends were made blunt using T4 DNA polymerase (New England Biolabs) to enable ligation. The resulting plasmid was designated pTF::UCD2. The pRiA4b origin isolated from Agrobacterium rhizogenes A4 (Jouanin et al., 1986) was cloned as a BamHI-HindIII fragment into pBluescriptII KS+, generating pBluescript::Ri. To confirm that the cloned pRi replication origin effects replication in Agrobacterium, pBluescript::Ri was transformed into A. tumefaciens by electroporation and plasmid DNA was isolated from carbenicillin-resistant colonies. pBluescript::pRi was digested with ClaI and the overhanging ends were made blunt using T4 DNA polymerase. The product was subsequently digested with NotI and cloned into pTF101.1 prior digested with ScaI and NotI to remove the pVS1 replication origin. The resulting plasmid was designated pTF::Ri.
The four T-DNA vectors pTF101.1, pTF::Bin19, pTF::UCD2, and pTF::Ri (Supplemental Fig. S1) were separately transformed into A. tumefaciens EHA101 (Hood et al., 1987), GV3101 (Koncz and Schell, 1986), and LBA4404 (Ooms et al., 1981) by electroporation.
Construction of Agrobacterium Strains Containing T-DNA Integrated into the pgl/picA Locus of the C58 Chromosome
A 4.2 kb ScaI-NsiI fragment containing the T-DNA region plus the aadA gene of pTF101.1 was inserted into the blunted SpeI and PstI sites of the integration vector pE1931, generating pE2759 (Supplemental Fig. S1). pE2759 was separately introduced into A. tumefaciens EHA101 and GV3101, generating A. tumefaciens At1586 and At1588, respectively. The eviction plasmid pPH1JI or pVK102 was introduced into A. tumefaciens At1586 and At1588, respectively, and colonies were selected on gentamicin and carbenicillin. Tetracycline-sensitive colonies (that had lost pE2759) were selected, generating A. tumefaciens At1589 and At1591, respectively, and recombination of the T-DNA region into the pgl/picA locus of the Agrobacterium chromosome was confirmed by DNA-blot hybridization (Lee et al., 2001).
Construction of an Agrobacterium Strain to Facilitate Integration of Expression Cassettes into the T-DNA Region on the C58 Chromosome
A 1.549 kbp PvuII-ScaI fragment from pBluescript was cloned into the SmaI site of pTF101.1, generating pE3265. A 5.8 kbp ScaI-NsiI fragment from pE3265 containing T-DNA, part of pBluescript, and the aadA gene was cloned into blunted SpeI and PstI sites of pE1770, generating pE3349. pE3349 was introduced into A. tumefaciens EHA105, generating A. tumefaciens At1687. Escherichia coli strains containing pVK102 and pRK230, respectively, were used to conjugate with A. tumefaciens At1687. The recombinant Agrobacterium strain containing the T-DNA borders, aadA gene, and a portion of pBluescript sequence was named A. tumefaciens At1702.
Construction of an Integration Binary Vector to Facilitate Launching T-DNA from the A. tumefaciens C58 Chromosome
A blunted EcoRI fragment containing the pgl/picA locus was cloned into the blunted NdeI site of pE3055. Removal of the NdeI fragment from pE3055 resulted in the loss of the pVS ori to create pE3361, a binary vector containing rare cloning sites (AscI, I-PpoI, I-SceI, I-CeuI, PI-PspI, and PI-TilI), a plant selection marker (bar gene), a bacterial selection marker (aadA gene), and a ColE1 ori. This plasmid cannot replicate in Agrobacterium.
Determination of Binary Vector Copy Number in Agrobacterium
The four T-DNA vectors pTF101.1, pTF::Bin19, pTF::UCD2, and pTF::Ri were separately transformed into A. tumefaciens EHA105 (Hood et al., 1993) containing a bar gene introduced into the picA locus, grown under vir gene inducing or noninducing conditions (Gelvin, 2006), and total DNA isolated by phenol:chloroform extraction. Equal amounts of DNA were digested with BglII and EcoRI and separated by electrophroesis through 0.9% agarose gels. DNA was transferred to a Hybond N+ nylon membrane (Amersham Pharmacia Biotech) and hybridized with a bar gene PCR fragment using conditions described below. Hybridization signal intensity was determined by scanning autoradiograms and using Labworks 4.6 Image Acquisition and Analysis Software (UVP).
Agrobacterium-Mediated Transformation of Arabidopsis and Maize
Arabidopsis (Arabidopsis thaliana; ecotype Wassilewskija-2) was transformed by a floral-dip protocol (Clough and Bent, 1998). Phosphinothricin-resistant plants were selected on Gamborg's B5 medium (Caisson Laboratories) supplemented with 10 μ g/mL phosphinothricin and 100 μ g/mL Timentin. Transformation frequencies were calculated as follows: (no. of phosphinothricin-resistant plants/no. of seeds tested) × 100. Ten milligrams of seeds correspond to approximately 500 seeds. Maize (Zea mays) transformation was as previously described (Frame et al., 2002).
Genomic DNA Extraction and DNA Dot-Blot Hybridization
Genomic DNA was extracted from three to five leaves of 3- to 4-week-old Arabidopsis plants or approximately 100 mg maize leaf tissue according to Murray and Thompson (1980). Genomic DNA was quantified using a Gemini XPS microplate spectrofluorometer (Molecular Devices, excitation: 488 nm, emission: 525 nm) using a Quant-iT PicoGreen dsDNA assay kit (Invitrogen).
DNA dot blots were prepared as follows: Arabidopsis (75 ng) or maize (1.5 μ g) genomic DNA samples were denatured by adding NaOH and EDTA to final concentrations of 0.4 m and 10 mm, respectively, followed by 10 min incubation in boiling water. A Hybond N+ nylon membrane (Amersham Pharmacia Biotech) was prewetted with water and placed between the layers of the dot-blot apparatus. Samples were applied to the wells of the dot-blot apparatus, incubated for 30 min, and then drawn onto the nylon membrane using a gentle vacuum. DNA was cross-linked to the membrane by using a CL-1000 UV crosslinker (UVP). The membranes were incubated in 2 × SSC for 10 min and dried.
Probes for dot-blot hybridizations were generated using random prime Ready-To-Go DNA labeling beads and 32P-dCTP (both Amersham Pharmacia Biosciences). Unincorporated radioactive nucleotides were removed by Sephadex G-100 gel filtration. Membranes were prehybridized in 7% (w/v) SDS, 0.5 m sodium phosphate (pH 7.2), and 10 mm EDTA (pH 8.0) at 65°C for 2 h. Hybridization was conducted overnight at 65°C. After hybridization, membranes were washed two times with 2 × SSC, 0.1% (w/v) SDS, 10 mm EDTA (pH 8.0), then two times with 1 × SSC, 0.1% (w/v) SDS, 10 mm EDTA, and finally two times with 0.1 × SSC, 0.1% (w/v) SDS, 10 mm EDTA at 65°C. Membranes were exposed at −80°C for autoradiography. Integrated dot density was determined using Labworks 4.6 Image Acquisition and Analysis Software (UVP). For reprobing, membranes were stripped using boiling 0.1% (w/v) SDS twice. Calculations for T-DNA copy number standards were based on an Arabidopsis genome size of 125 Mb (Arabidopsis Genome Initiative, 2000) and a maize genome size of 2,500 Mb (Arumuganathan and Earle, 1991). The size of pTF101.1 is 9,189 bp (Paz et al., 2004).
DNA dot-blot membranes were hybridized sequentially with various probes. For determining T-DNA copy numbers of Arabidopsis plants, dot-blot membranes were hybridized with a bar gene PCR fragment (Fig. 1). To determine T-DNA copy numbers of maize plants, membranes were probed with a 759 bp PstI/XhoI fragment from pTF101.1 harboring the double CaMV 35S promoter (Fig. 1). To detect vector backbone sequences in both Arabidopsis and maize DNA, a 656 bp PCR fragment derived from the non-T-DNA region next to the T-DNA left border of pTF101.1 (Fig. 1) was used as a probe. The fragment was amplified using pTF101.1 as a template and 5′ -TCACCGTAACCAGCAAATCA-3′ and 5′-CTCGGCACAAAATCACCACT-3′ as primers. A 3.1 kb EcoRI fragment containing the pgl/picA locus (Rong et al., 1990) was used as a probe to check for the presence of contaminating Agrobacterium DNA in plant genomic DNA. All DNA fragments were gel purified prior to labeling using a QIAEX II gel extraction kit (Qiagen). To normalize amounts of DNA in each dot, membranes were hybridized with genomic Arabidopsis or maize DNA.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Schematic representation of the various T-DNA binary vectors and integration plasmid used in this study.
Supplemental Table S1. Plasmids and strains.
Acknowledgments
We thank Marcy Main, Jennifer McMurray, and Tina Paque for their assistance in maize transformation experiments.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. (1991) Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep 9: 229–233 [Google Scholar]
- Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. (2007) Agrobacterium ParA/Mind-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J 26: 2540–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A, Gallois P, Raynal M, Mestre-Ortega D, Sallaud C, Guiderdoni E, Delseny M, Bernardi G. (2000) The distribution of T-DNA in the genomes of transgenic Arabidopsis and rice. FEBS Lett 471: 161–164 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci III 316: 1194–1199 [Google Scholar]
- Bevan M. (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 12: 8711–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla PL, Singh MB. (2008) Agrobacterium-mediated transformation of Brassica napus and Brassica oleracea. Nat Protoc 3: 181–189 [DOI] [PubMed] [Google Scholar]
- Chung SM, Frankman EL, Tzfira T. (2005) A versatile vector system for multiple gene expression in plants. Trends Plant Sci 10: 357–361 [DOI] [PubMed] [Google Scholar]
- Close TJ, Zaitlin C, Kado CI. (1984) Design and development of amplifiable broad-host-range cloning vectors: analysis of the vir region of Agrobacterium tumefaciens plasmid pTiC58. Plasmid 12: 111–118 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- De Block M, De Brouwer D, Tenning P. (1989) Transformation of Brassica napus and Brassica oleracea using Agrobacterium tumefaciens and the expression of the bar and new genes in the transgenic plants. Plant Physiol 91: 694–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Buck S, de Wilde C, van Montagu M, Depicker A. (2000) T-DNA vector backbone sequences are frequently integrated into the genome of transgenic plants obtained by Agrobacterium-mediated transformation. Mol Breed 6: 459–468 [Google Scholar]
- De Buck S, Podevin N, Nolf J, Jacobs A, Depicker A. (2009) The T-DNA integration pattern in Arabidopsis transformants is highly determined by the transformed target cell. Plant J 60: 134–145 [DOI] [PubMed] [Google Scholar]
- de Framond AJ, Barton KA, Chilton MD. (1983) Mini-Ti: a new vector strategy for plant genetic engineering. Biotechnology (N Y) 1: 262–269 [Google Scholar]
- Deroles SC, Gardner RC. (1988) Analysis of the T-DNA structure in a large number of transgenic petunias generated by Agrobacterium-mediated transformation. Plant Mol Biol 11: 365–377 [DOI] [PubMed] [Google Scholar]
- Durrenberger F, Crameri A, Hohn B, Koukolikova-Nicola Z. (1989) Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc Natl Acad Sci USA 86: 9154–9158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann KA. (1991) T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J 1: 71–82 [Google Scholar]
- Frame BR, Shou H, Chikwamba RK, Zhang Z, Xiang C, Fonger TM, Pegg SEK, Li B, Nettleton DS, Pei D, et al. (2002) Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol 129: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati M, Moreno MA, Nadzan G, Zourelidou M, Dellaporta SL. (2000) Large-scale T-DNA mutagenesis in Arabidopsis for functional genomic analysis. Funct Integr Genomics 1: 25–34 [DOI] [PubMed] [Google Scholar]
- Gelvin SB. (2003) Agrobacterium and plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67: 16–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin SB. (2006) Agrobacterium virulence gene induction. Wang K, , Methods in Molecular Biology: Agrobacterium Protocols, Vol 44 Humana Press, Totowa, NJ, pp 77–84 [DOI] [PubMed] [Google Scholar]
- Grevelding C, Fantes V, Kemper E, Schell J, Masterson R. (1993) Single-copy T-DNA insertions in Arabidopsis are the predominant form of integration in root-derived transgenics, whereas multiple insertions are found in leaf discs. Plant Mol Biol 23: 847–860 [DOI] [PubMed] [Google Scholar]
- Hamilton CM, Frary A, Lewis C, Tanksley SD. (1996) Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc Natl Acad Sci USA 93: 9975–9979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B, Engler D, Moy Y, Newman B, Ralston E, Gutterson N. (1999) A simple method to enrich an Agrobacterium-transformed population for plants containing only T-DNA sequences. Plant J 19: 727–734 [DOI] [PubMed] [Google Scholar]
- Hoekema A, Roelvink PW, Hooykaas PJJ, Schilperoort RA. (1984) Delivery of T-DNA from the Agrobacterium tumefaciens chromosome into plant cells. EMBO J 3: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood E, Gelvin SB, Melchers LS, Hoekema A. (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2: 208–218 [Google Scholar]
- Hood EE, Fraley RT, Chilton MD. (1987) Virulence of Agrobacterium tumefaciens strain A281 on legumes. Plant Physiol 83: 529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Lee S, Yang K, et al. (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22: 561–570 [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Snyder C, Jones JDG. (1987) T-DNA is organized predominantly in inverted repeat structures in plants transformed with Agrobacterium tumefaciens C58 derivatives. Mol Gen Genet 207: 471–477 [Google Scholar]
- Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA. (1996) Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol Biol 31: 957–973 [DOI] [PubMed] [Google Scholar]
- Jouanin L, Tourneur J, Tourneur C, Casse-Delbart F. (1986) Restriction maps and homologies of the three plasmids of Agrobacterium rhizogenes strain A4. Plasmid 16: 124–134 [DOI] [PubMed] [Google Scholar]
- Kohli A, Leech M, Vain P, Laurie DA, Christou P. (1998) Transgene organization in rice engineered through direct DNA transfer supports a two-phase integration mechanism mediated by the establishment of integration hot spots. Proc Natl Acad Sci USA 95: 7203–7208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204: 383–396 [Google Scholar]
- Kononov ME, Bassuner B, Gelvin SB. (1997) Integration of T-DNA binary vector “backbone” sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J 11: 945–957 [DOI] [PubMed] [Google Scholar]
- Lee LY, Gelvin SB. (2008) T-DNA binary vectors and systems. Plant Physiol 146: 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Humara JM, Gelvin SB. (2001) Novel constructions to enable the integration of genes into the Agrobacterium tumefaciens C58 chromosome. Mol Plant Microbe Interact 14: 577–579 [DOI] [PubMed] [Google Scholar]
- Lichtenstein C, Draper J. (1986) DNA cloning: a practical approach. Glover DM, , Genetic Engineering of Plants, Vol 2 IRL Press, Washington, DC, pp 67–119 [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda A, Janssen G, Hodges L, Peralta EG, Ream W. (1992) Agrobacterium tumefaciens transfers extremely long T-DNAs by a unidirectional mechanism. J Bacteriol 174: 2288–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. (1980) Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms G, Hooykaas PJJ, Moolenaar G, Schilperoort RA. (1981) Crown gall plant tumors of abnormal morphology, induced by Agrobacterium tumefaciens carrying mutated octopine Ti plasmids; analysis of T-DNA functions. Gene 14: 33–50 [DOI] [PubMed] [Google Scholar]
- Pandian A, Hurlstone C, Liu Q, Singh S, Salisbury P, Green A. (2006) Agrobacterium-mediated transformation protocol to overcome necrosis in elite Australian Brassica juncea lines. Plant Mol Biol Rep 24: 103a–103i [Google Scholar]
- Paz MM, Shou H, Guo Z, Zhang Z, Banerjee AK, Wang K. (2004) Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explant. Euphytica 136: 167–179 [Google Scholar]
- Ramanathan V, Veluthambi K. (1995) Transfer of non-T-DNA portions of the Agrobacterium tumefaciens Ti plasmid pTiA6 from the left terminus of TL-DNA. Plant Mol Biol 28: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Rong L, Karcher SJ, O'Neal K, Hawes MC, Yerkes CD, Jayaswal RK, Hallberg CA, Gelvin SB. (1990) picA, a novel plant-inducible locus on the Agrobacterium tumefaciens chromosome. J Bacteriol 172: 5828–5836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou H, Frame BR, Whitham SA, Wang K. (2004) Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Mol Breed 13: 201–208 [Google Scholar]
- Stam M, de Bruijn R, Kenter S, van der Hoorn RAL, van Blokland R, Mol JNM, Kooter JM. (1997) Post-transcriptional silencing of chalcone synthase in Petunia by inverted transgene repeats. Plant J 12: 63–82 [Google Scholar]
- Virts EL, Gelvin SB. (1985) Analysis of transfer of tumor-inducing plasmids from Agrobacterium tumefaciens to Petunia protoplasts. J Bacteriol 162: 1030–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenck A, Czako M, Kanevski I, Marton L. (1997) Frequent collinear long transfer of DNA inclusive of the whole binary vector during Agrobacterium-mediated transformation. Plant Mol Biol 34: 913–922 [DOI] [PubMed] [Google Scholar]