Abstract
(1,3;1,4)-β-d-Glucan (β-glucan) accounts for 20% of the total cell walls in the starchy endosperm of wheat (Triticum aestivum) and is an important source of dietary fiber for human nutrition with potential health benefits. Bioinformatic and array analyses of gene expression profiles in developing caryopses identified the CELLULOSE SYNTHASE-LIKE F6 (CSLF6) gene as encoding a putative β-glucan synthase. RNA interference constructs were therefore designed to down-regulate CSLF6 gene expression and expressed in transgenic wheat under the control of a starchy endosperm-specific HMW subunit gene promoter. Analysis of wholemeal flours using an enzyme-based kit and by high-performance anion-exchange chromatography after digestion with lichenase showed decreases in total β-glucan of between 30% and 52% and between 36% and 53%, respectively, in five transgenic lines compared to three control lines. The content of water-extractable β-glucan was also reduced by about 50% in the transgenic lines, and the Mr distribution of the fraction was decreased from an average of 79 to 85 × 104 g/mol in the controls and 36 to 57 × 104 g/mol in the transgenics. Immunolocalization of β-glucan in semithin sections of mature and developing grains confirmed that the impact of the transgene was confined to the starchy endosperm with little or no effect on the aleurone or outer layers of the grain. The results confirm that the CSLF6 gene of wheat encodes a β-glucan synthase and indicate that transgenic manipulation can be used to enhance the health benefits of wheat products.
Cell wall polysaccharides account for about 10% of the dry weight of the mature wheat (Triticum aestivum) grain, and about 2% to 3% dry weight of the white flour fraction that is derived from the major storage tissue of the grain, the starchy endosperm (Stone, 1996). Although they are relatively minor components of white flour, the cell wall polysaccharides are immensely important in determining the properties of the flour for processing (Saulnier et al., 2007a) and in human nutrition, forming a major source of dietary fiber (Saulnier et al., 2007b; Topping, 2007).
Wheat endosperm cell walls comprise two major components, arabino-(1,4)-β-d-xylan (arabinoxylan [AX]) and (1,3;1,4)-β-d-glucan (β-glucan), which account for about 70% and 20% of the total, respectively (Bacic and Stone, 1980). In addition about 4% cellulose [(1,4)-β-d-glucan] and 7% (1,4)-β-d-glucomannans are present (Bacic and Stone, 1980). This contrasts with starchy endosperm tissues of oats (Avena sativa) and barley (Hordeum vulgare), in which the proportions of AX and β-glucan are reversed (Henry, 1987; Stone, 1996). This is of particular interest as soluble β-glucans from barley and oats have benefits in reducing coronary heart disease that have been accepted by the U.S. Food and Drug Administration for health claims on food products (Anonymous, 2008). It is not known whether these benefits are shared by β-glucan from wheat, which differs from barley and oat β-glucans in its distribution of 1,3 and 1,4 linkages (Lazaridou and Biliaderis, 2007). Differences in Mrs of β-glucans have also been reported, although it is difficult to conclude whether these result from true genetic differences, the effects of growth conditions, or the methods used for extraction and characterization (Lazaridou and Biliaderis, 2007). However, differences in linkage distribution are likely to affect β-glucan solubility and viscosity (Cui et al., 2000; Lazaridou and Biliaderis, 2007), which are considered to be key parameters determining health benefits (Wood, 2007).
Despite the biological importance of plant cell walls, we know relatively little about their biosynthesis. However, it is becoming clear that members of the cellulose synthase super family are responsible for the synthesis of the backbone of hexose polysaccharides, with cellulose itself being synthesized by enzymes encoded by the CELLULOSE SYNTHASE A (CESA) family (Richmond and Somerville, 2000) and other polysaccharides by enzymes encoded by genes in CELLULOSE SYNTHASE-LIKE (CSL) families designated A to H and J. The latter include genes encoding (1,4)-β-d-mannans (CSLA; Dhugga et al., 2004; Liepman et al., 2007) and the (1,4)-β-d-glucan backbone of xyloglucan (CSLC; Cocuron et al., 2007).
Burton et al. (2006) provided convincing evidence that the rice (Oryza sativa) CSLF genes encode β-glucan synthases, by expression in transgenic Arabidopsis (Arabidopsis thaliana) and detection of β-glucan (which is not normally synthesized in Arabidopsis) with a highly specific antibody. Furthermore, four of the corresponding CSLF genes of barley were mapped to a locus on chromosome 2H corresponding to a major quantitative trait locus (QTL) for grain β-glucan content, and two other CSLF genes to loci on chromosomes 1H and 7H that were also close to β-glucan QTLs. The seventh barley CSLF gene mapped to a locus on chromosome 5H and showed no linkage to β-glucan QTLs (Burton et al., 2008). However, more recently Doblin et al. (2009) have shown that the barley CSLH gene also leads to the synthesis of β-glucan when expressed in transgenic Arabidopsis. The authors concluded that both the CSLF and CSLH gene families contribute to β-glucan synthesis in this species, and suggested that they act independently, based on the fact that there was no significant correlation between their transcripts.
We have identified the CSLF6 gene of wheat as a putative β-glucan synthase and confirmed this function by RNA interference (RNAi) suppression in transgenic wheat. This demonstrates that transgenic manipulation can be used to modify the amounts and properties of β-glucan in wheat, to enhance the health benefits and improve the properties for processing and livestock feed.
RESULTS
Bioinformatic Identification of Candidate Genes
We hypothesized that candidate genes for β-glucan synthesis should be expressed in the developing caryopsis during the early (cell division and expansion) and mid (grain filling) stages of development, when cell wall synthesis is actively occurring. Furthermore, we proposed that such genes were likely to be members of the CSL gene family. Figure 1 therefore shows the abundances of transcripts for CSL genes represented on the wheat Affymetrix GeneChip, hybridized to cDNA isolated from whole caryopses of wheat throughout development (Wan et al., 2008; Fig. 1A) and from fractions enriched in starchy endosperm tissue taken at 14 to 21 DPA (Fig. 1B). Transcripts are labeled according to the most similar rice sequences following the naming convention for CSL genes (Hazen et al., 2002). Data are only shown for the six most abundant CSL transcripts in the endosperm-enriched samples; the complete list of CSL genes represented on the chip and their average expression levels are given in Supplemental Table S1. The most highly expressed transcripts in the endosperm-enriched fraction (Fig. 1B) are members of the CSLD family (D2) and the CSLA family (A7-1, A7-2, A12), the latter being implicated in mannan synthesis (Dhugga et al., 2004; Liepman et al., 2007). The molecular function of CSLD genes, which are the most similar to CESA, are unknown, but Arabidopsis knockout mutants for CSLD genes have defective root tip or pollen tube development (Bernal et al., 2008). The next most abundant transcripts in the endosperm-enriched fraction (Fig. 1B) are CSLF6 and CSLC1, implicated in β-glucan and xyloglucan synthesis, respectively (Burton et al., 2006; Cocuron et al., 2007), with both being particularly abundant early in grain development (Fig. 1A). These expression patterns are consistent with the report that β-glucan synthesis is initiated earlier and proceeds more actively during early grain development than the synthesis of AX in wheat (Philippe et al., 2006).
Figure 1.
Transcript abundance of CSL genes determined on the wheat Affymetrix GeneChip. Labels indicate the closest rice CSL gene that the transcript corresponds to; there are two wheat paralogs matching the rice OsCSLA7 gene. A, Whole-grain samples isolated at 10 stages of development in cv Hereward (each point is the average of two replicates; ANOVA of stages × genes gives a lsd 5% of 30.1 and there are significant differences between points at P < 0.001). B, Endosperm-enriched samples isolated at 14 to 21 DPA in cv Cadenza (each bar is the average of four replicates; ANOVA of genes gives a lsd 5% of 99.3 and there are significant differences between points at P = 0.001).
The relative abundances of CSL transcripts from the array data are consistent with counts of wheat ESTs from grain libraries; the five transcripts identified as most abundant in Figure 1 being in the top six ranked by EST counts (Supplemental Table S2). These ESTs indicate the presence of at least seven CSLF genes in wheat, corresponding to the rice CSLF1, CSLF2, CSLF3, CSLF4, CSLF6, CSLF8, and CSLF9 genes. Of the CSLF transcripts in barley, the HvCSLF6 gene, an ortholog of TaCSLF6, is also by far the most abundant during the cell expansion phase in developing barley endosperm (Burton et al., 2008). Another barley transcript, HvCSLF9, was shown to be reasonably abundant early in endosperm development, but the wheat ortholog has only two associated wheat ESTs (Supplemental Table S2), compared with 30 barley ESTs, and is not represented on the wheat Affymetrix array.
Since the CSLA genes are implicated in the synthesis of mannans (Dhugga et al., 2004; Liepman et al., 2007) that account for only 7% of wheat endosperm cell walls (Stone and Morell, 2009), CSLD2 and CSLF6 transcripts were selected as targets for RNAi suppression to generate changes in endosperm cell wall composition. During the progress of this work, the rice CSLF genes were also implicated in the synthesis of β-glucan by expression in transgenic Arabidopsis (Burton et al., 2006), making the CSLF6 gene the stronger candidate. Similarly, a recent study of the β-d-glucan less (bgl) mutant of barley showed cosegregation of the mutation with polymorphism in the CSLF6 gene, suggesting that CSLF6 encodes a β-glucan synthase (Tonooka et al., 2009).
Construction of RNAi Cassettes
Supplemental Figure S1 shows the protein sequence corresponding to the TaCSLF6 gene cloned from cv Cadenza endosperm cDNA, compared with those of the rice OsCSLF2 gene that was shown by Burton et al. (2006) to confer the capacity to synthesize β-glucan by expression in transgenic Arabidopsis and the orthologous barley HvCSLF6 gene that is highly expressed in the endosperm (Burton et al., 2008). Supplemental Figure S2 compares the nucleotide sequence of the 3′ end of this gene with single nucleotide polymorphisms present in two other forms (determined by sequencing of 15 cDNA clones covering the whole region shown and a further 21 covering the region used for the RNAi construct and that are almost certainly homoeologs) present in hexaploid cv Cadenza cDNA. The fragments used for the RNAi constructs against CSLF6 (Supplemental Fig. S2) and against CSLD2/CSLD4 (Supplemental Fig. S3) are also shown; an RNAi construct is expected to repress transcripts that contain regions of 20 bp identity or more, so homoeologs and other closely related isoforms will be affected. Partial CSLF6 and CSLD4 cDNAs cloned from cv Cadenza were used as templates to PCR amplify fragments for RNAi. Two RNAi plasmids were constructed (Supplemental Fig. S4), one designed to silence only CSLF6 (pHMW-Adh-Nos-f6ri/ri) and the other designed to simultaneously silence both CSLD2 and CSLD4 (pHMW-Adh-Nos-d4ri/ri) by using regions of CSLD4 cDNA possessing high similarity to CSLD2. The RNAi cassettes were constructed with inverted repeats flanking the maize (Zea mays) Adh2 intron and under the control of the endosperm-specific high Mr glutenin subunit 1Dx5 promoter (Lamacchia et al., 2001).
Production and Initial Screening of RNAi Lines
Two RNAi cassettes were used, both together and separately, to generate transgenic lines of wheat (cv Cadenza). A total of 29 independent transgenic T0 lines were made, 13 containing the CSLF6 RNAi cassette, eight containing the CSLD2/4 RNAi cassette, and a further eight containing both constructs. Presence of the selectable marker gene and RNAi cassette in genomic DNA from each T0 plant was confirmed by PCR. Additional evidence for the presence of transgene cassettes in these plants was provided using TaqMan real-time PCR analysis performed on selected T1 plants by iDNA Genetics.
T1 seed from 16 lines (10 containing the CSLF6 RNAi cassette only, four containing the CSLD2/4 RNAi cassette only, and two containing both constructs) were germinated and the T1 progeny were screened by PCR of genomic DNA to determine the segregation ratios of the transgenes. The χ2 goodness of fit test indicated that the transgenes were segregating with a 3:1 ratio in 12 of these lines, including line 15T that possessed both CSLF6 and CSLD2/4 cassettes, indicative of a single transgene locus containing both cassettes (Supplemental Table S5). In two of the lines, the χ2 goodness of fit test predicted a 15:1 ratio, indicating that a CSLF6 cassette had inserted in two unlinked loci (Supplemental Table S5). Line 22T that also contained both CSLF6 and CSLD2/4 cassettes possessed a classic 9:3:3:1 ratio of two unlinked loci, with one containing at least one copy of CSLF6 and the other, at least one copy of CSLD2/4. The remaining line (10T) segregated in a non-Mendelian ratio.
Enzyme fingerprinting of T1 seed of transgenic wheat plants carrying only RNAi designed against the CSLD genes showed no detectable differences in grain cell wall composition from the controls. We therefore concluded that the CSLD genes do not control the synthesis of the major groups of cell wall polymers in wheat endosperm (β-glucan and AX). In contrast, enzyme fingerprinting of T1 seeds of 10 heterozygous transgenic lines carrying RNAi designed against CSLF6 showed a mean decrease in β-glucan content of over 20% (data not shown). It was therefore decided to focus attention on the lines expressing the CSLF6 RNAi constructs.
Five independent lines that displayed both 3:1 segregation of the transgene and significantly reduced β-glucan content in T1 seeds were subjected to transgene copy number analysis and zygosity estimation via quantitative PCR. From each of these lines, homozygous T1 sublines were propagated to collect T2 seed that were then sown to provide T3 seed for further analyses. Four of these lines possessed the CSLF6 RNAi cassette (lines 4T, 7T, 9T, and 21T) and one possessed both CSLF6 and CSLD2/4 cassettes (line 15T). Transgene copy number ranged from one to 14 copies per line (Supplemental Table S5). These five lines were used for β-glucan analysis. The 100 grain weights of the five transgenic lines ranged from 2.96 g to 4.21 g (mean 3.75 g), which did not differ significantly (P = 0.14) from the 100 grain weights of three control lines (range 2.91 g to 3.45 g, mean 3.22 g). This confirms that the expression of the transgene did not have any significant effect on grain filling.
Transgene and Endogenous CSLF6 Expression
The expression of the CSLF6 RNAi transgene and its effect on the abundance of the endogenous CSLF6 transcript were determined in line 15T and controls by quantitative reverse transcription (qRT)-PCR (Fig. 2). In RNA samples isolated from dissected endosperm at 14 DPA, primers designed against the transgene as well as the native gene showed abundance 11-fold greater in 15T compared to null controls (P < 0.001), whereas primers designed exclusively to the endogenous CSLF6 transcript showed abundance decreased by 4-fold on average (P = 0.025).
Figure 2.
Comparison of transcript abundance in line 15 transgenic (15T) and null segregant (N) samples of TaCSLF6 endogenous gene plus RNAi transgene (A) and TaCSLF6 endogenous gene only (B), determined by qRT-PCR using endosperm samples at 14 DPA. Bars represent average ± 1 se of ratio from four biological replicate samples for 15T and either four (A) or seven (B) biological replicate samples for nulls. [See online article for color version of this figure.]
β-Glucan Contents of Mature Grain
The mean content of total β-glucan in wholemeal fractions of the five transgenic lines was reduced by 42.2% compared to that in the three control lines, with the greatest reduction of 52.2% occurring in line 21T (Table I).
Table I. Contents and properties of β-glucans in wholegrain samples of the transgenic and control lines.
Contents of total and hot-water-extractable (WE) β-glucans. Mrs of WE β-glucans determined by se-HPLC using Calcofluor for detection and percentiles describing the Mr at which 10%, 50%, and 90% of the distribution fall below that value (g/mol × 104).
| Line (n = 3)a | Total β-Glucan (g/100 g Dry Wt; Mean ± sd) P < 0.001 | WE β-Glucan (% Dry Wt; Mean ± sd)b | WE β-Glucan (% Total β-Glucan) |
 of β-Glucan (g/mol × 104) of β-Glucan (g/mol × 104) |
|||
| Averageb (Mean ± sd) | p10 | p50 | p90 | ||||
| 9T (transgenic) | 0.437 ± 0.091 | 0.05 ± 0.011 | 12.58 | 36.0 ± 4.51 | 2.61 | 16.31 | 89.71 |
| 7T (transgenic) | 0.577 ± 0.092 | 0.06 ± 0.011 | 9.71 | 43 ± 6.41,2 | 2.91 | 17.61 | 113.41,2 |
| 21T (transgenic) | 0.410 ± 0.091 | 0.06 ± 0.011 | 14.63 | 45.3 ± 4.21.2 | 2.91 | 18.11 | 116.81,2 |
| 4T (transgenic) | 0.461 ± 0.091 | 0.06 ± 0.011 | 13.67 | 47.5 ± 6.11,2 | 2.01 | 15.71 | 128.01.2 |
| 15T (transgenic) | 0.607 ± 0.92 | 0.07 ± 0.012,3 | 11.86 | 56.6 ± 1.72,3 | 3.11,2 | 21.31 | 160.32,3 |
| 20N (control)c | 0.872 ± 0.043 | 0.16 ± 0.023 | 18.24 | 84.8 ± 9.54 | 4.93 | 44.52 | 229.14 |
| 23C (control)c | 0.888 ± 0.043 | 0.14 ± 0.022,3 | 15.54 | 78 ± 5.14 | 5.23 | 41.62 | 212.83,4 |
| 15N (control)c | 0.825 ± 0.043 | 0.11 ± 0.021.2 | 13.57 | 83.9 ± 2.54 | 4.62,3 | 40.42 | 232.44 |
Biological replicates.
Figures with different superscripts within a column are significantly different from each other (P < 0.01).
15n and 20n are null segregants from transformed lines. 23C is a nontransformed control line that had been subjected to the same transformation and regeneration conditions as the transformed lines.
This decrease in total β-glucan was confirmed by enzyme fingerprinting, digesting with lichenase and endoxylanase to determine AX and β-glucan in a single analysis (Ordaz-Ortiz et al., 2005; Saulnier et al., 2009). Typical high-performance anion-exchange chromatography (HPAEC) separations of the digestion products from grain of control (null) and transgenic lines are shown in Figure 3.
Figure 3.
HPAEC analysis of oligosaccharides produced by enzyme mapping of transgenic and control lines. A trace from transgenic line 9 (green) is overlaid on a trace from the null control line 15 (red). Detector response is measured in nC, retention time in minutes. G3 and G4 are glucan fragments released from β-glucan by digestion with lichenase; X and XX are Xyl and xylobiose released from AX by digestion with endoxylanase. Peaks 1 to 7 are AXOS released from AX by digestion with endoxylanase: 1 = XA3X, 2 = XA3XX, 3 = XA2+3XX, 4 = XA3A3XX, 5 = XA3XA3XX, 6 = XA3A2+3XX, and 7 = XA3XA2+3XX. All identified peaks were integrated (Supplemental Table S4) and used for PCA analysis (Fig. 4; Supplemental Fig. S5).
Oligosaccharides released from AX by endoxylanase were Xyl (X), xylobiose (XX), and oligosaccharides of DP4 to 9 (XA3X, XA3XX, XA2+3XX, XA3A3XX, XA3A2+3XX, XA3XA3XX, and XA3XA2+3XX; Ordaz-Ortiz et al., 2005; Saulnier et al., 2009; see Fauré et al., 2009 for nomenclature). In addition, lichenase mainly released glucooligosaccharides of DP3 (3-O-β-cellobiosyl-d-Glc, G3) and DP4 (3-O-β-cellotriosyl-d-Glc, G4). These two fragments have been reported to account for 90% of all wheat β-glucan (Lazaridou and Biliaderis, 2007). Further small peaks corresponding to larger glucooligosaccharides (DP5, DP6) were also observed, but these accounted for small proportions of the total glucooligosaccharides and were not quantified in this study. The full analyses of the transgenic and control lines are reported in Supplemental Table S3.
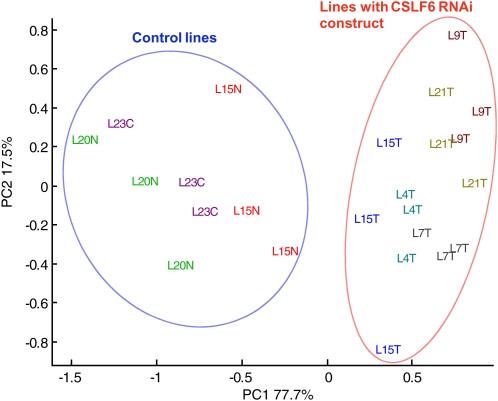
Principal component analysis (PCA) was applied to HPAEC peaks obtained from transgenic and control samples (five transgenic and three control lines, three biological replicates). This revealed variation in AX and β-glucan composition of endosperm cell walls and showed clear separation between the transgenic and control lines. Figure 4 shows the similarity map for the two first principal components that together accounted for 95.2% of the total variance. The loading plot (Supplemental Fig. S5) indicated that the first principal component that accounted for 77.7% of the total variance related to G3 and G4 and thus indicated variation in the proportions of β-glucan in the samples. Principal component 2 mainly related to xylobiose (peak XX) and accounted for 17.5% of the total variation. Principal component 2 revealed small differences between the lines irrespective of the presence or absence of the transgene, showing that AX structure was not affected by the presence of the transgene. Since β-glucan and AX are both synthesized from UDP, Glc compensatory increases in AX could occur if β-glucan synthesis is down-regulated. However, this was not the case, the HPAEC analysis (Fig. 2; Supplemental Table S3) showed that the amount of AXOS released by the enzyme was similar in transgenic and in control lines.
Figure 4.
PCA of the enzyme fingerprinting (HPAEC) results of mature seeds of five transgenic and three control lines. The loading plot (see Supplemental Fig. S5) shows that the first principal component PC1 is mainly related to variation in G3 and G4 peaks and thus indicated variation in the proportions of β-glucans among samples. Samples on the right part of the plot are depleted in β-glucans compared to samples on the left part of the plot.
The combined proportions of G3 + G4 decreased by a mean of 45.9% (range from 35.5% to 53.2%) in the transgenic lines (Fig. 5A), showing excellent agreement with the levels of suppression determined by total β-glucan analysis. The ratio of G3 to G4 fragments was also slightly higher (P < 0.001), with the mean in the control lines being 2.33 and that in the transgenic lines being 2.58. These G3 to G4 ratios are lower than those reported for β-glucan prepared from wheat bran (3.7–4.5; Cui et al., 2000; Li et al., 2006; Lazaridou and Biliaderis, 2007) and flour (3.04–3.84; Wood et al., 1991), but similar to those determined using the same method for developing and mature caryopses of wheat cv Hereward (ranging from 2.31–2.53 during the period from 10 DPA to maturity; authors’ unpublished results on the developmental series described by Shewry et al., 2009). The values are also within the range reported for β-glucan from a range of barley samples (1.8–3.5; for review, see Lazaridou and Biliaderis, 2007).
Figure 5.
Areas of HPAEC peaks corresponding to G3 + G4 fragments in mature grain (A) and developing caryopses (B) of transgenic (4T, 7T, 9T, 15T, and 21T) and control (15N, 20N, and 23C) lines. Values in bars show G3 to G4 ratios in the different lines. daa, Days after anthesis. [See online article for color version of this figure.]
These analyses were carried out on whole-grain samples, whereas the RNAi constructs were expressed under the control of the HMW subunit 1Dx5 gene promoter that is expressed only in the starchy endosperm and starting at about 10 to 12 DPA (Lamacchia et al., 2001), with significant levels of the encoded protein being detected from 14 DPA (Shewry et al., 2009). Hence, the true impact of the down-regulation on β-glucan content and properties would have been diluted by the presence of the β-glucan-rich aleurone but also by β-glucan synthesized in the young developing starchy endosperm.
β-Glucan in Developing Grain
The HMW subunit 1Dx5 promoter is only expressed from about 10 to 12 DPA (Lamacchia et al., 2001), whereas CSLF6 expression begins earlier (Fig. 1). Enzyme fingerprinting was therefore also carried out on developing caryopses of transgenic line 15 (chosen as the only line with a single copy of the transgene), the corresponding null segregant control line (15N), and the wild-type control line 23 at 10 DPA, 14 DPA, and 21 DPA (Fig. 5B). The results were consistent with the expression pattern of the HMW subunit promoter, with no differences being observed between the G3 + G4 contents of the three lines at 10 DPA but with significant (P = 0.002; P = 0.005) decreases in the proportions of G3 + G4 fragments in the transgenic line at 14 DPA (24.06% decrease) and 21 DPA (16.79% decrease). The difference in G3 + G4 between the 15T and 15n lines was therefore less than half as great at 21 DPA (Fig. 5B) as in mature seed (Fig. 5A). A possible explanation for this observation is that although the abundance of the CSLF6 transcript was low after 21 DPA the encoded enzyme was still active, leading to greater cumulative differences between β-glucan amounts in the mature 15T and 15n grain.
No statistically significant differences were observed in the ratios of G3 to G4 fragments in the three lines at 10 DPA and 14 DPA (P = 0.210 and 0.135, respectively) while at 21 DPA the transgenic line was not significantly different from the wild-type control line 23 (lowest value) but the null segregant control line 15n (highest value) was significantly different from the other two lines (P = 0.015). The ratios in the three lines at 10 DPA were consistent with those in the mature grain samples (2.01–2.65 compared with 2.25–2.62) whereas the ratios at 14 DPA and 21 DPA were lower (1.62–1.74 and 2.13–2.08, respectively). However, it is clear that these differences were not related to the presence of the transgene.
Effect on β-Glucan Molecular Weight
The content of hot-water-extractable β-glucan in wholemeal flour was 0.11% to 0.16% of dry matter in the control lines, with the highest content in line 20N. The content was significantly lower (P < 0.001) in the transgenic samples (0.05%–0.07% of dry matter), with the highest content being in sample 15T. These values correspond to between 9.7% and 14.6% of the total β-glucan in the transgenic lines and between 13.6% and 18.2% of the total β-glucan in the control lines. However, the differences between the transgenic and control lines were not significantly different (P = 0.069).
The Mr distribution of the hot-water-extractable β-glucan was determined by high-performance size-exclusion chromatography (Fig. 6). The Mr distribution in the control samples (lines 20N, 23C, and 15N) was either bimodal or polymodal (i.e. with two or three populations of β-glucan being present). The transgenic samples showed clear differences in Mr distribution, with the high Mr population of β-glucan being reduced to a small shoulder (lines 9T, 15T, and 21T are shown in Fig. 6; lines 4T and 7T had similar patterns and are therefore not shown).
Figure 6.
se-HPLC profiles showing the Mr distribution of water-extractable β-glucan from wholemeal samples of mature grain of transgenic (9T, 21T, and 15T) and control (20N, 23C, and 15N) lines of wheat. The dotted lines represent the Calcofluor average Mrs.
The Calcofluor average Mr ( ) of the hot-water-extractable β-glucan was 79 to 85 × 104 g/mol in the control samples, but was significantly lower in the transgenic samples (36–57 × 104 g/mol; Table I). Within the transgenic lines the highest
) of the hot-water-extractable β-glucan was 79 to 85 × 104 g/mol in the control samples, but was significantly lower in the transgenic samples (36–57 × 104 g/mol; Table I). Within the transgenic lines the highest  was observed in line 15T that had the smallest decrease in β-glucan compared to the control plants, and the
was observed in line 15T that had the smallest decrease in β-glucan compared to the control plants, and the  decreased with increasing reduction in β-glucan. The Mr percentiles at 10%, 50%, and 90% were also significantly lower for the transgenic lines (2.0–3.1 × 104 g/mol, 16.3–21.3 × 104 g/mol, and 89.7–160.3 × 104 g/mol, respectively) than for the control samples (4.6–5.2 × 104 g/mol, 40.4–44.5 × 104 g/mol, and 213–232 × 104 g/mol, respectively), with 15T again having the highest value of the transgenic lines. The Mr percentile at 90% also decreased with increasing down-regulation for the transgenic samples.
decreased with increasing reduction in β-glucan. The Mr percentiles at 10%, 50%, and 90% were also significantly lower for the transgenic lines (2.0–3.1 × 104 g/mol, 16.3–21.3 × 104 g/mol, and 89.7–160.3 × 104 g/mol, respectively) than for the control samples (4.6–5.2 × 104 g/mol, 40.4–44.5 × 104 g/mol, and 213–232 × 104 g/mol, respectively), with 15T again having the highest value of the transgenic lines. The Mr percentile at 90% also decreased with increasing down-regulation for the transgenic samples.
The decrease in β-glucan content determined by enzyme fingerprinting of the transgenic lines was significantly correlated with the decrease in the average Mr (r = 0.902, P = 0.036) and with p90 (r = 0.925, P = 0.024).
Immunolocalization of β-Glucan
Immunolocalization of β-glucan was carried out in semithin sections of mature grain (Fig. 7, A–D) and developing caryopses (Fig. 7, E–J). Strong labeling of the aleurone layer was observed in all samples (which is consistent with the presence of thick cell walls comprising about 29% β-glucan; Bacic and Stone, 1981) and of the pericarp and nucellar epidermis in the developing caryopses. No differences were observed between the control and transgenic lines in labeling of the starchy endosperm cell walls at 10 DPA (Fig. 7, I and J) and little difference at 14 DPA (Fig. 7, G and H). However, clear differences were observed at 21 DPA (Fig. 7, E and F) and maturity (Fig. 7, A–D) with line 9T (Fig. 7, A and E) showing reduced labeling throughout the starchy endosperm when compared with the control lines and line T15 showing a less uniform pattern of reduction in labeling (Fig. 7, C and F; being greater in the outer cells of the tissue). These outer endosperm cells correspond to those that are added most recently during grain development by division of the aleurone cells (Bechtel and Wilson, 2003). The labeling differences are consistent with the control of RNAi expression by the glutenin promoter and with the amounts of β-glucan determined using the Megazyme kit and enzyme fingerprinting.
Figure 7.
Immunolabeling of β-glucan in mature grain (A–D) and developing caryopses (E–J) of transgenic (sections on left) and control (sections on right) lines of wheat. Bars = 100 μm. al, Aleurone layer; en, starchy endosperm; ne, nucellar epidermis; pe, pericarp.
DISCUSSION
The amount and properties of β-glucan have important impacts on the utilization of both wheat and barley. Both cereals are widely used as feed for monogastric livestock, notably pigs and poultry. In this case the high viscosity conferred by β-glucan has negative effects on feed intake, feed conversion rate, and weight gain, and may result in sticky feces when used to feed chickens (Hesselman et al., 1981). Although this problem may be alleviated by adding β-glucanase to degrade β-glucan in the feed and thus lower gut viscosity (Hesselman and Åman, 1986), reducing the amount and viscosity of the β-glucan in the grain is a more attractive option. β-Glucan content and Mr are also important in malt and beer production, where high viscosity causes problems with haze formation and wort filtration (Bamforth, 1982), and in starch production where a high content of β-glucan may reduce the yield of starch. In contrast, high contents of soluble β-glucan are favored for food products as they may reduce serum cholesterol levels (McIntosh et al., 1991; Bourdon et al., 1999) as well as regulate blood Glc and insulin (Cavallero et al., 2002; Wood, 2007).
We have shown that down-regulation of the TaCSLF6 gene is sufficient to reduce the amount of β-glucan in the starchy endosperm of wheat to less than half of that in control lines. Expression of the transgene did not result in any consistent effects on the ratio of G3 to G4 fragments released on digestion with lichenase, suggesting that the synthesis of both 1,3 and 1,4 linkages was affected. This does not necessarily imply the encoded enzyme is capable of forming both linkages, as its removal could affect a multienzyme complex or stop the production of a substrate. However, the Mr of the polymer was reduced, which could result from the deposition of incompletely synthesized, and therefore shorter, polysaccharide chains into the wall due to limiting activity of the glucan synthase. Only one form of CSLF was expressed in the developing grain at significant levels, which is consistent with the same enzyme catalyzing β-glucan synthesis in the developing starchy endosperm (the tissue studied here) and other grain tissues.
The TaCSLF6 gene reported here is a close ortholog of the barley HvCSLF6 gene (Supplemental Fig. S1) that was shown by Burton et al. (2008) to be similarly highly expressed in barley endosperm. A role of this gene in β-glucan synthesis in barley grain is also implied by analysis of the bgl mutant line (Tonooka et al., 2009).
Rice CSLF genes were shown to mediate the synthesis of β-glucan in transgenic Arabidopsis (Burton et al., 2006). However, subsequent work from the same group showed that similar activity was displayed by transformation with a barley CSLH gene (Doblin et al., 2009). An ortholog of this gene is present in wheat but, as was the case in barley, the transcript levels in developing grain were very low (too low to show in Fig. 1A), which implies that it has little or no role in the synthesis of wheat grain β-glucan. Doblin et al. (2009) also suggested that the major role of the HvCSLH1 gene is the synthesis of β-glucan in secondary cell walls, based on in situ PCR localization in leaf tissues.
We therefore conclude that TaCSLF6 plays a major role in mediating β-glucan synthesis in developing grain of wheat and hence a key target for manipulation to increase or decrease total β-glucan. Furthermore, the effects of RNAi suppression on β-glucan Mr indicate that indirect effects on Mr and viscosity may occur by altering the relationship between enzyme activity and substrate availability.
MATERIALS AND METHODS
Array Analysis
Array analysis procedures were as previously reported (Wan et al., 2008). Wheat (Triticum aestivum) ESTs were assigned to rice (Oryza sativa) genes and assembled into contigs using the WhETS tool (Mitchell et al., 2007). PCR primers were designed to these sequences as the first step in cloning the CSL transcripts from grain cDNA.
cDNA Cloning and Construction of RNAi Expression Cassettes
Partial (CSLF6 and CSLD4) and full-length (CSLF6) cDNA was amplified by RT-PCR from RNA isolated from 14-DPA endosperm of cv Cadenza using PCR primers SP for CK201603, ASP1 for RACE/F6longer, SP_CSLF6 and ASP_CSLF6 (CSLF6), and SP_CSLD4 and ASP_CSLD4 (CSLD4) (Supplemental Table S4). cDNAs were A tailed and cloned in pGEM-T Easy (Promega) and sequenced.
For construction of RNAi expression cassettes DNA fragments corresponding to CSLF6 and CSLD4 were amplified using the PCR primers indicated in Supplemental Table S4 using partial cDNA clones as the templates. The RNAi fragment sequences were used to search all public domain wheat sequences and the only sequences that had stretches of identity of 20 bp or more were the target genes; namely CSLF6 for the CSLF6 fragment and CSLD2 and CSLD4 for the CSLD4 fragment. The reaction products were cloned, excised using BamHI and BglII, and ligated as inverted repeats on either side of the maize (Zea mays) Adh2 intron in the plasmid pHMW-Adh-Nos, an RNAi expression cassette constructed in pGEM-3Zf(+), under the control of the high Mr glutenin subunit 1Dx5 promoter (Supplemental Fig. S4; Lamacchia et al., 2001). The desired orientation of the inserts was confirmed by BamHI and BglII restriction digestion, PCR (using primers Dx5Pfwd with Adh5'rev and Adh3'fwd with Nos5'rev; Supplemental Table S4), and sequencing.
Production of Transgenic Wheat Lines and Analysis of Transgene Segregation and Copy Number
Transformation was carried out by particle bombardment (PDS1000, Bio-Rad) of immature scutella of cv Cadenza (Sparks and Jones, 2009) with RNAi constructs being cobombarded with plasmid pCalNeo containing NptII for plant selection. PCR of genomic DNA was used to confirm the presence of the transgenes and the segregation in T1 plants: using primers Neo1 with Neo2 for the NptII gene and SP_CSLF6_BglII or SP_CSLD4_BglII with Nos5'rev for the CSLF6 and CSLD4 genes, respectively (Supplemental Table S4). The χ2 goodness of fit test (with 1 degree of freedom for 3:1 ratios and 3 degrees of freedom for 9:3:3:1 ratios) was used to separate plants into genetic classes. Genomic DNA was subjected to quantitative PCR using TaqMan probes to determine transgene copy number and zygosity of individual plants (Supplemental Table S5; analysis performed by iDNA Genetics).
Plant Material and Growth Conditions
Plant material for transformation was produced according to Sparks and Jones (2009). All other plant material was grown in the glasshouse with 18°C to 20°C day and 14°C to 16°C night temperatures with a 16-h photoperiod provided by natural light supplemented with banks of Son-T 400 W sodium lamps (Osram, Ltd.) giving 400 to 1,000 μmol m−2 s−1 photosynthetically active radiation.
T2 grain from five homozygous transgenic wheat lines carrying RNAi designed against CSLF6 (lines 9T, 7T, 21T, 4T, and 15T) and three controls (lines 20N, 15N, and 23C) were sown to provide mature T3 grain for analyses. For each line three 25-cm pots, with four plants per pot, were randomized and grown in the same glasshouse compartment at the same time. Grain weight was determined using four replicate samples (except for line 7T for which only two replicate samples were available) of grain from the main tillers of individual plants for each of five transgenic and three control lines.
qRT-PCR
Seeds of line 15T and segregating nulls, 14 DPA, were frozen in liquid nitrogen and stored at −80deg;C. For RNA extraction seeds were freeze dried and the endosperm extracted by peeling off the outer layers. After homogenization in a TissueLizer (Qiagen) total RNA was extracted according to Wan et al. (2008). RT and qRT-PCR was performed as described in Pellny et al. (2008). The expression of the native gene as well as the transgene was determined with the primers SP_CSLF6_RNAi_2739 and ASP_CSLF6_RNAi_2828 that give an amplicon within the RNAi construct. To test for the down-regulation of the native gene the sense primer was chosen to be just outside of the construct (SP_CSLF6_RNAi_2669) with the same antisense primer amplifying an extra 30 bp at the 5′ end of the RNAi construct and all but 3 bp at the 3′ end. The expression of these amplicons was normalized against Ta2526 (prTYW19_Ta2526_qPCR_F and prTYW20_Ta2526_qPCR_R), an EST that was shown to be stably expressed in wheat grain development (Wan et al., 2008). Results are displayed as relative expression of four transgenics compared to between four and seven nulls and significant differences were calculated by ANOVA after data transformations as proposed by Rieu and Powers (2009).
β-Glucan Analysis
For total β-glucan analysis, enzyme fingerprinting and determination of β-glucan Mr distribution finely ground freeze-dried developing caryopses (enzyme fingerprinting only) or wholegrain flour (all methods) was used. Three biological replicates consisting of 30 seeds each from the main tillers of three individual plants (a total of 90 seeds per replicate) were analyzed.
Total β-glucan was determined using a Megazyme kit. Endoxylanase (EC 3.2.1.8) from Trichoderma viride was purchased from Megazyme (Xylanase M1). The activity of the enzyme preparation determined by the supplier on WE-AX (40°C, pH 4.5) was 1,670 units/mL. pH optimum is 4.5 to 5.0. Lichenase [endo-1,3(4)-β-d-glucanase, EC 3.2.1.73] from Bacillus subtilis was purchased from Megazyme. The activity of the enzyme preparation on barley (Hordeum vulgare) β-glucan (40°C, pH 6.5) determined by the supplier was 1,000 units/mL. pH optimum is 6.5 to 7.0.
Enzyme fingerprinting samples (100 mg) weighed in an Eppendorf tube (2 mL) were treated with 1.5 mL ethanol solution (80%, v/v) in a boiling water bath for 5 min. The supernatant was removed and the residue successively washed with 1.5 mL ethanol (80%, v/v) and 1.5 mL ethanol (95%, v/v) solutions. The supernatant was then discarded and the residue dried for 2 h in an oven at 40°C. Samples were then digested overnight (16 h) with 2 units lichenase and 16 units endoxylanase (Ordaz-Ortiz et al., 2005; Saulnier et al., 2009). The degradation was carried out in water at a pH 6.0 compatible with the use of both enzymes in one run. After centrifugation, supernatant of the digested samples was diluted with water (1/40) and 10-μL aliquots were injected on a Carbopac PA-1 (5 × 250 mm) analytical column (Dionex) run at 25°C with a flow rate of 1 mL/min. AXOS and GOS were separated using the following elution conditions with Ultrapure water (A), 1 m NaOAc (B), and 0.5 m NaOH (C): 0 min (A: 80%, C: 20%), 35 min (A: 60%, B: 20%, C: 20%); 36 min (A: 80%, C: 20%), hold up to 60 min. Detection was realized with a TSP EC2000 pulse amperometric detector (Thermo Separation Products) using the pulse potentials E1 = +0.05 V, E2 = +0.6 V, and E3 = −0.6 V. Typical chromatograms are shown on Figure 2. Peak identification was based on the retention times of reference compounds previously isolated (Ordaz-Ortiz et al., 2004, 2005; Saulnier et al., 2009). It was checked that in the conditions used, GOS and especially G3 and G4 were not coeluted with peaks generated by the endoxylanase (Saulnier et al., 2009). Overlay of chromatogram was realized using raw data and adjustment of the baseline to the same level without peak normalization.
For PCA (Fig. 3; Supplemental Fig. S5) peak areas (Supplemental Table S3) were normalized for each chromatogram as follows:  , with SNPeaki being the normalized area of peak i and SPeaki the area of peak i (Ordaz-Ortiz et al., 2005; Saulnier et al., 2009).
, with SNPeaki being the normalized area of peak i and SPeaki the area of peak i (Ordaz-Ortiz et al., 2005; Saulnier et al., 2009).
For determination of Mr distribution, samples were extracted with boiling water containing thermostable α-amylase and analyzed by high-performance size-exclusion chromatography with fluorescence detection as described by Rimsten et al. (2003), except that 0.0025% Calcofluor was used and that the pH of the Calcofluor solution was not adjusted. This detection method is selective for β-glucan in extracts and the fluorescence response is independent of Mr. It will, however, exclude Mrs below 104 (Munck, 1989). The Mr is therefore given as Calcofluor average Mr ( ), which is the same as weight average Mr for samples that only contain high Mr
β-glucan.
), which is the same as weight average Mr for samples that only contain high Mr
β-glucan.  and percentiles (p10, p50, and p90) describing the Mr at which 10%, 50%, and 90% of the distribution fall below were calculated by using a calibration curve established with narrow Mr
β-glucan standards (Rimsten et al., 2003). The content of β-glucan in the extracts (content of water-extractable β-glucan) was also calculated by using the area of the peak as described by Rimsten et al. (2003). The repeatability was <15% for the three biological replicates analyzed. The results were evaluated statistically with ANOVA, generalized linear model procedure, and Tukey's pairwise comparison (Minitab 15).
and percentiles (p10, p50, and p90) describing the Mr at which 10%, 50%, and 90% of the distribution fall below were calculated by using a calibration curve established with narrow Mr
β-glucan standards (Rimsten et al., 2003). The content of β-glucan in the extracts (content of water-extractable β-glucan) was also calculated by using the area of the peak as described by Rimsten et al. (2003). The repeatability was <15% for the three biological replicates analyzed. The results were evaluated statistically with ANOVA, generalized linear model procedure, and Tukey's pairwise comparison (Minitab 15).
Immunomicroscopy
One-millimeter sections were fixed in 3% paraformaldehyde and 2% glutaraldehyde in phosphate buffer (0.1 m, pH 7.2), dehydrated in a graded aqueous ethanol series, and infiltrated with London Resin White acrylic resin. One-micrometer sections were mounted onto multiwell glass sides and preincubated in phosphate-buffered saline (PBS) containing 3% (w/v) bovine serum albumin (BSA) for 30 min and then incubated for 1 h with PBS containing 1% (w/v) BSA, 0.01% Tween 20 (PBS-BSA), and 1:1,000 dilution of mAb anti-β-glucan (Meikle et al., 1994; Biosupplies Pty Ltd.). After extensive washing in PBS they were incubated for 1 h in the dark with marker-conjugated goat anti-mouse antibody labeled with Alexa Fluor 546 (Molecular Probes, distributed by Interchim) diluted 100-fold in PBS 0.3% BSA. Nonspecific staining was identified using controls omitting the primary antibody. Sections were examined with a LEICA DMRD microscope equipped with epifluorescence using a band-pass filter at 515 to 560 nm for excitation filters and fluorescence detected at >570 nm.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AM743080.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of wheat, barley, and rice CSLF6 protein sequences, with that encoded by the OsCSLF2.
Supplemental Figure S2. Sequence of 3′ end of TaCSLF6 gene coding region (accession AM743080.2).
Supplemental Figure S3. Sequence of 3′ end of TaCSLD4 gene coding region and CSLD2/4 RNAi fragment.
Supplemental Figure S4. Schematic representation of construction of RNAi expression vectors.
Supplemental Figure S5. Loading plot for PCA of the enzyme fingerprints of mature seeds of five control and three transgenic lines.
Supplemental Table S1. Transcript abundance for all CSL genes represented on wheat Affymetrix GeneChip.
Supplemental Table S2. Wheat EST counts for sequences mapped to rice CSL genes.
Supplemental Table S3. Enzyme mapping of cell wall polysaccharides in mature grain, determined by HPAEC.
Supplemental Table S4. DNA sequences and targets for PCR primers used in this work.
Supplemental Table S5. Segregation and copy number analysis of 16 transgenic wheat lines.
Acknowledgments
This publication reflects the authors’ views and the European Union is not liable for any use that may be made of the information contained in this publication.
References
- Anonymous (2008) Final Rule on Soluble Fiber from Certain Foods and Risk of Coronary Heart Disease (73 FR 47828). U.S. Food and Drug Administration, College Park, MD: [PubMed] [Google Scholar]
- Bacic A, Stone BA. (1980) A (1→3)- and (1→4)-linked β-D-glucan in the endosperm cell walls of wheat. Carbohydr Res 82: 372–387 [Google Scholar]
- Bacic A, Stone BA. (1981) Chemistry and organisation of aleurone cell wall components from wheat and barley. Aust J Plant Physiol 8: 475–495 [Google Scholar]
- Bamforth CW. (1982) Barley β-glucans: their role in malting and brewing. Brewers Digest 57: 22–27 [Google Scholar]
- Bechtel D, Wilson JD. (2003) Amyloplast formation and starch granule development in hard red winter wheat. Cereal Chem 80: 175–183 [Google Scholar]
- Bernal AJ, Yoo CM, Mutwil M, Jensen JK, Hou G, Blaukopf C, Sorensen I, Blancaflor EB, Scheller HV, Willats WGT. (2008) Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol 148: 1238–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon I, Yokoyama W, Davis P, Hudson C, Backus R, Richter D, Knuckles B, Schneeman BO. (1999) Postprandial lipid, glucose, insulin and cholecystokinin responses in men fed barley pasta enriched with β-glucan. Am J Clin Nutr 69: 55–63 [DOI] [PubMed] [Google Scholar]
- Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, Fincher GB. (2008) The genetics and transcriptional profiles of the cellulose synthase-like HvCSLF gene family in barley. Plant Physiol 146: 1821–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. (2006) Cellulose synthase-like CSLF genes mediate the synthesis of cell wall (1,3;1,4)-β-D-glucans. Science 311: 1940–1942 [DOI] [PubMed] [Google Scholar]
- Cavallero A, Empilli S, Brighenti F, Stanca AM. (2002) High (1→3, 1→4)-β-glucan barley fractions in bread making and their effect on human glycemic response. J Cereal Sci 36: 59–66 [Google Scholar]
- Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. (2007) A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc Natl Acad Sci USA 104: 8550–8555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Wood PJ, Blackwell B, Nikiforuk J. (2000) Physicochemical properties and structural characterization by two-dimensional NMR spectroscopy of wheat beta-D-glucan—comparison with other cereal beta-D-glucans. Carbohydr Polym 41: 249–258 [Google Scholar]
- Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S, et al. (2004) Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science 303: 363–366 [DOI] [PubMed] [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A. (2009) A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci USA 106: 5996–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauré R, Courtin CM, Delcour JA, Dumon C, Faulds CB, Fincher GB, Fort S, Fry SC, Halila S, Kabel MA, et al. (2009) A brief and informationally rich naming system for oligosaccharide motifs of heteroxylans found in plant cell walls. Aust J Chem 62: 1–5 [Google Scholar]
- Hazen S, Scott-Craig SJ, Walton J. (2002) Cellulose synthase-like genes of rice. Plant Physiol 128: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RJ. (1987) Pentosan and (1→3),(1→4)-β-glucan concentrations in endosperm and wholegrain of wheat, barley, oats and rye. J Cereal Sci 6: 252–258 [Google Scholar]
- Hesselman K, Åman P. (1986) The effect of β-glucanase on the utilization of starch and nitrogen by broiler chickens fed on barley of low- or high-viscosity. Anim Feed Sci Technol 15: 83–93 [Google Scholar]
- Hesselman K, Elwinger K, Nilsson M, Thomke S. (1981) The effects of beta-glucanase supplementation, stage of ripeness, and storage treatment of barley in diets fed to broiler chickens. Poult Sci 60: 2664–2671 [Google Scholar]
- Lamacchia C, Shewry PR, Di Fonzo N, Forsyth J, Harris N, Lazzeri PA, Napier JA, Halford NG, Barcelo P. (2001) Endosperm-specific activity of a storage protein gene promoter in transgenic wheat seed. J Exp Bot 52: 243–250 [PubMed] [Google Scholar]
- Lazaridou A, Biliaderis CG. (2007) Molecular aspects of cereal β-glucan functionality: physical properties, technological applications and physiological effects. J Cereal Sci 46: 101–118 [Google Scholar]
- Li W, Cui SW, Kakuda Y. (2006) Extraction, fractionation, structural and physical characterization of wheat β-D-glucans. Carbohydr Polym 63: 408–416 [Google Scholar]
- Liepman AH, Nairn CJ, Willats WG, Sørensen I, Roberts AW, Keegstra K. (2007) Functional genomic analysis supports conservation of function among cellulose synthase-like a gene family members and suggests diverse roles of mannans in plants. Plant Physiol 143: 1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh GH, Whyte J, McArthur R, Nestel PJ. (1991) Barley and wheat foods: influence on plasma cholesterol concentration in hypercholesterolemic men. Am J Clin Nutr 53: 1205–1209 [DOI] [PubMed] [Google Scholar]
- Meikle PJ, Hoogenraad NJ, Bonig I, Clarke AE, Stone BA. (1994) A (1-3,1-4)-beta-glucan-specific monoclonal-antibody and its use in the quantitation and immunocyto-chemical location of (1-3,1-4)-beta-glucans. Plant J 5: 1–9 [DOI] [PubMed] [Google Scholar]
- Mitchell RAC, Castells-Brooke N, Taubert J, Verrier PJ, Leader DJ, Rawlings CJ. (2007) Wheat Estimated Transcript Server (WhETS): a tool to provide best estimate of hexaploid wheat transcript sequence. Nucleic Acids Res 35: W148– 151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munck L. (1989) Practical experiences in development of fluorescence analyses in an applied food research laboratory. Munck L, , Fluorescence Analyses in Foods. Longman Scientific and Technical, Harlow, UK, 1–32 [Google Scholar]
- Ordaz-Ortiz JJ, Devaux MF, Saulnier L. (2005) Classification of wheat varieties based on structural features of arabinoxylans as revealed by endoxylanase treatment of flour and grain. J Agric Food Chem 53: 8349–8356 [DOI] [PubMed] [Google Scholar]
- Ordaz-Ortiz JJ, Guillon F, Tranquet O, Dervilly-Pinel G, Tran V, Saulnier L. (2004) Specificity of monoclonal antibodies generated against arabinoxylans of cereal grains. Carbohydr Polym 57: 425–433 [Google Scholar]
- Pellny TK, Van Aken O, Dutilleul C, Wolff T, Groten K, Bor M, De Paepe R, Reyss A, Van Breusegem F, Noctor G, et al. (2008) Mitochondrial respiratory pathways modulate nitrate sensing and nitrogen-dependent regulation of plant architecture in Nicotiana sylvestris. Plant J 54: 976–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe S, Saulnier L, Guillon F. (2006) Arabinoxylan and (1 → 3),(1 → 4)-beta-glucan deposition in cell walls during wheat endosperm development. Planta 224: 449–461 [DOI] [PubMed] [Google Scholar]
- Richmond T, Somerville C. (2000) The cellulose synthase superfamily. Plant Physiol 124: 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I, Powers SJ. (2009) Real-time quantitative RT-PCR: design, calculations and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimsten L, Stenberg T, Andersson R, Andersson A, Åman P. (2003) Extraction of cereal β-glucan and determination of molecular weight using SEC with calcofluor detection. Cereal Chem 80: 485–490 [Google Scholar]
- Saulnier L, Guillon F, Sado PE. (2007a) Plant cell wall polysaccharides in storage organs: xylans (food applications). Kamerling J, Boons G-J, Lee Y, Suzuki A, Taniguchi N, Voragen AGJ, , Comprehensive Glycoscience. Elsevier, Amsterdam, 653–689 [Google Scholar]
- Saulnier L, Robert P, Grintchenko M, Jamme F, Bouchet B, Guillon F. (2009) Wheat endosperm cell walls: spatial heterogeneity of polysaccharide structure and composition using micro-scale enzymatic fingerprinting and FT-IR microspectroscopy. J Cereal Sci 50: 312–317 [Google Scholar]
- Saulnier L, Sado PE, Branlard G, Charmet G, Guillon F. (2007b) Wheat arabinoxylans: exploiting variation in amount and composition to develop enhanced varieties. J Cereal Sci 46: 261–281 [Google Scholar]
- Shewry PR, Underwood C, Wan Y, Lovegrove A, Bhandari D, Toole G, Mills ENC, Denyer K, Mitchell RAC. (2009) Storage product synthesis and accumulation in developing grains of wheat. J Cereal Sci 50: 106–112 [Google Scholar]
- Sparks CA, Jones HD. (2009) Biolistics transformation of wheat. Jones HD, Shewry PR, Walker J, , Methods in Biotechnology. Transgenic Wheat, Barley and Oats: Production and Characterization. Humana Press, Totowa, NJ, 71–92 [Google Scholar]
- Stone B, Morell MK. (2009) Carbohydrates. Khan K, Shewry PR, , Wheat: Chemistry and Technology, Ed 4. AACC, St. Paul, 299–362 [Google Scholar]
- Stone BA. (1996) Cereal grain carbohydrates. Henry RJ, Kettlewell PS, , Cereal Grain Quality. Chapman and Hall, London, 250–288 [Google Scholar]
- Tonooka T, Aoki E, Yoshioka T, Taketa S. (2009) A novel mutant gene for (1-3,1-4)-β-D-glucanless grain on barley (Hordeum vulgare L.) chromosome 7H. Breed Sci 59: 47–54 [Google Scholar]
- Topping D. (2007) Cereal complex carbohydrates and their contribution to human health. J Cereal Sci 46: 220–229 [Google Scholar]
- Wan Y, Poole RL, Huttly AK, Toscano-Underwood C, Feeney K, Welham S, Gooding MJ, Mills ENC, Edwards KJ, Shewry PR, et al. (2008) Transcriptome analysis of grain development in hexaploid wheat. BMC Genomics 9: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. (2007) Cereal β-glucans in diet and health. J Cereal Sci 46: 230–238 [Google Scholar]
- Wood PJ, Weisz J, Blackwell BA. (1991) Molecular characterization of cereal β-D-glucans: structural analysis of oat β-D-glucan and rapid structural evaluation of β-d-glucans from different sources by high-performance liquid chromatography of oligosaccharides released by lichenase. Cereal Chem 68: 31–39 [Google Scholar]