Abstract
Histidine (His) plays a critical role in plant growth and development, both as one of the standard amino acids in proteins, and as a metal-binding ligand. While genes encoding seven of the eight enzymes in the pathway of His biosynthesis have been characterized from a number of plant species, the identity of the enzyme catalyzing the dephosphorylation of histidinol-phosphate to histidinol has remained elusive. Recently, members of a novel family of histidinol-phosphate phosphatase proteins, displaying significant sequence similarity to known myoinositol monophosphatases (IMPs) have been identified from several Actinobacteria. Here we demonstrate that a member of the IMP family from Arabidopsis (Arabidopsis thaliana), myoinositol monophosphatase-like2 (IMPL2; encoded by At4g39120), has histidinol-phosphate phosphatase activity. Heterologous expression of IMPL2, but not the related IMPL1 protein, was sufficient to rescue the His auxotrophy of a Streptomyces coelicolor hisN mutant. Homozygous null impl2 Arabidopsis mutants displayed embryonic lethality, which could be rescued by supplying plants heterozygous for null impl2 alleles with His. In common with the previously characterized HISN genes from Arabidopsis, IMPL2 was expressed in all plant tissues and throughout development, and an IMPL2:green fluorescent protein fusion protein was targeted to the plastid, where His biosynthesis occurs in plants. Our data demonstrate that IMPL2 is the HISN7 gene product, and suggest a lack of genetic redundancy at this metabolic step in Arabidopsis, which is characteristic of the His biosynthetic pathway.
His is one of the standard 20 amino acids found in proteins, and is required for plant growth and development (Muralla et al., 2007; Bikard et al., 2009). The occurrence of His in the active sites of numerous enzymes is attributable to the imidazole functional group (pKa approximately 6), which can alternate between the protonated and unprotonated states under physiologically relevant conditions, allowing its participation in acid-base catalysis (Fersht, 1999). His also plays important biochemical roles as a nucleophile in phosphoryl group transfer, and as a metal-binding ligand (Fraústo da Silva and Williams, 2001; Harding, 2004).
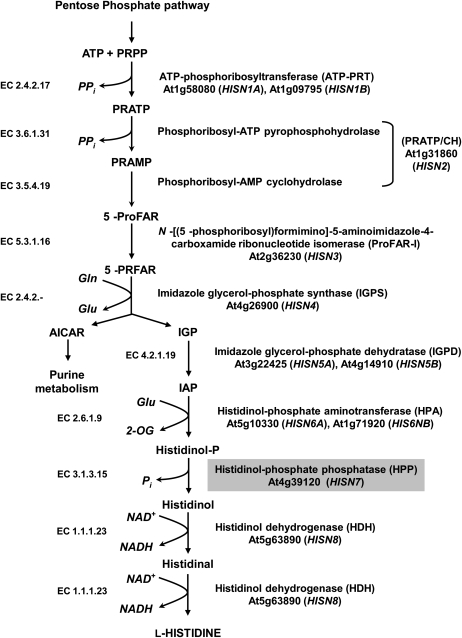
Genetic analysis of His biosynthesis has provided insights into key regulatory mechanisms in microorganisms, notably the discovery of operon structure and metabolic regulation through attenuation (Ames et al., 1960; Roth and Ames, 1966). However, this was the last amino acid biosynthetic pathway to be characterized in plants, and only recently has it been demonstrated that His biosynthesis follows the same route as that in microorganisms with 10 reactions catalyzed by eight enzymes (Fig. 1), beginning with the condensation of ATP and 5-phosphoribosyl-1-pyrophosphate catalyzed by the enzyme ATP-phosphoribosyl transferase (Ohta et al., 2000). His biosynthesis is linked to purine metabolism through its precursors 5-phosphoribosyl-1-pyrophosphate and ATP, and the release of an intermediate (5′-phosphoribosyl-4-carboxamide-5-aminoimidazole) at the branch point step catalyzed by imidazole-glycerol phosphate synthase, which enters the de novo purine synthesis pathway (Alifano et al., 1996; Ward and Ohta, 1999). While the regulation of His biosynthesis has not been as comprehensively investigated in plants as it has in microorganisms, analysis of transgenic Arabidopsis (Arabidopsis thaliana) plants overexpressing single HISN genes under the control of the cauliflower mosaic virus 35S promoter suggests that ATP-phosphoribosyl transferase activity controls the size of the pool of free His in plants (Ingle et al., 2005; Rees et al., 2009).
Figure 1.
The His biosynthetic pathway in plants. Abbreviations used for enzyme names in this work are shown in parentheses, and enzyme commission numbers and Arabidopsis gene loci indicated. Modified after Ward and Ohta (1999), Ingle et al. (2005), and Muralla et al. (2007).
Genes encoding seven of the eight enzymes in the His pathway were identified in plants during the late 1990s (Nagai et al., 1993; Mori et al., 1995; El Malki et al., 1998; Fujimori and Ohta, 1998a, 1998b; Fujimori et al., 1998; Ohta et al., 2000) and, in contrast to most other amino acid biosynthetic pathways, the majority were found to be encoded by single genes in the Arabidopsis genome (Stepansky and Leustek, 2006). The missing link in the pathway has been HISN7, histidinol-phosphate phosphatase (HPP; EC 3.1.3.15). While dephosphorylation of histidinol-P to histidinol was first detected in plant extracts almost 40 years ago (Wiater et al., 1971), the identity of the enzyme(s) catalyzing this reaction in plants is unknown. HPP proteins identified from microorganisms prior to 2006 can be grouped into one of two superfamilies—the DDDD (which contain four invariant Asp residues) and PHP (polymerase and histidinol phosphatase) superfamilies (Lee et al., 2008). The DDDD superfamily contains bifunctional enzymes from enteric bacteria (such as Escherichia coli) where dephosphorylation of histidinol-P is catalyzed by the N-terminal domain, while the C terminus displays imidazole glycerol-phosphate dehydratase (IGPD) activity (Alifano et al., 1996; Fani et al., 2007), and monofuctional HPP proteins such as that recently identified in Thermococcus onnurineus (Lee et al., 2008). In contrast, all members of the PHP family identified to date are monofunctional HPP enzymes, including those encoded by HIS2 in Saccharomyces cerevisiae and ytvP in Bacillus subtilis (Alifano et al., 1996; le Coq et al., 1999). Notably, neither the Arabidopsis nor rice (Oryza sativa) genomes contain any sequences with significant sequence identity to members of either the DDDD or PHP superfamilies (Stepansky and Leustek, 2006).
A novel HPP protein, showing no sequence similarity to members of either the DDDD or PHP superfamilies, was recently isolated from Corynebacterium glutamicum (Mormann et al., 2006), and orthologs have subsequently been identified in other Actinobacteria, including Streptomyces coelicolor (Marineo et al., 2008). All members of this new family display significant sequence similarity to known myoinositol monophosphatases (IMPs; Mormann et al., 2006), which catalyze the hydrolysis of the ester bond of d-myoinositol 1(or 3)-P (d-Ins 1-P, d-Ins 3-P) to generate myoinositol, without the production of a phospho-enzyme intermediate (Leech et al., 1993). In eukaryotes, myoinositol plays a crucial role in a number of cellular processes including phosphatidylinositol-mediated signaling and the membrane anchoring of proteins (Boss et al., 2006; Fujita and Jigami, 2008). To date, three putative IMP encoding sequences have been identified in the Arabidopsis genome: VTC4 (At3g02870), myoinositol monophosphatase-like1 (IMPL1; At1g31190), and IMPL2 (At4g39120; Torabinejad et al., 2009). VTC4 was first reported as the l-Gal 1-P phosphatase in ascorbate biosynthesis in Arabidopsis (Laing et al., 2004; Conklin et al., 2006), but was recently demonstrated to be a bifunctional enzyme also able to catalyze the dephosphorylation of d-Ins 1-P and d-Ins 3-P to myoinositol in vitro (Torabinejad et al., 2009). vtc4 null mutants showed only a 30% reduction in myoinositol content, suggesting genetic redundancy in the capacity to generate myoinositol, and accordingly IMPL1 and IMPL2 were shown to have in vitro IMP activity (Torabinejad et al., 2009). Both enzymes were also able to utilize l-Gal 1-P as a substrate, suggesting some promiscuity in their substrate specificity. Here we demonstrate that the IMPL2 protein encoded by At4g39120 is also able to catalyze the dephosphorylation of histidinol-P to histidinol, completing the His biosynthetic pathway in plants.
RESULTS
Bioinformatic Analysis Identifies IMPL2 as a Candidate HPP in Arabidopsis
To identify putative homologs in Arabidopsis of the IMP-like proteins that catalyze the dephosphorylation of histidinol-P in the Actinobacteria, we carried out BLAST analysis of the Arabidopsis protein RefSeq database with the S. coelicolor HPP protein (NP_629355). This identified three proteins with approximately 30% sequence identity to NP_629355: VTC4 (At3g02870), IMPL1 (At1g31190), and IMPL2 (At4g39120; Supplemental Fig. S1), all members of the IMP superfamily. VTC4, IMPL1, and IMPL2 have recently been shown to catalyze the dephosphorylation of d-Ins 1-P, d-Ins 3-P, and l-Gal 1-P (Torabinejad et al., 2009). However, their ability to utilize histidinol-P as a substrate has not been reported.
All known plant HISN genes contain N-terminal plastid transit peptide encoding sequences, and it is thought that His biosynthesis is localized to the plastid (Stepansky and Leustek, 2006). It is thus highly probable that any HPP enzyme(s) in Arabidopsis would also be targeted to this organelle. Both IMPL1 and IMPL2 satisfy this criterion. These two proteins contain putative plastid transit peptide sequences as predicted by ChloroP analysis (Emanuelsson et al., 1999), and have been detected in mass spectrometry analyses of purified chloroplast preparations from Arabidopsis (Zybailov et al., 2008). In contrast, VTC4 does not contain a predicted plastid transit peptide, and is instead a membrane protein; HMMTOP analysis (Tusnady and Simon, 2001) predicts the presence of three transmembrane α -helices and VTC4 has been detected in purified plasma membrane preparations by mass spectrometry analysis (Marmagne et al., 2007).
In contrast to yeast, there is little evidence for the operation of a general control of nitrogen metabolism regulatory mechanism in plants. However, the genes encoding the first six enzymes in Arabidopsis have GCN4-like binding sites in their upstream regions (Stepansky and Leustek, 2006), which may be indicative of transcriptional coregulation. In addition, inhibition of IGPD activity has been shown to result in increased expression of histidinol dehydrogenase (HDH) in Arabidopsis (Guyer et al., 1995). We used the Arabidopsis Coexpression Data Mining tool (Manfield et al., 2006) to identify genes coexpressed with IMPL1 and IMPL2 across 322 publicly available Affymetrix ATH1 microarray datasets. This analysis identified two His biosynthetic genes (in the top five) as showing the most positive correlation in expression with IMPL2; ProFAR-I (At2g36230, HISN3) ranked first and HDH (At5g63890, HISN8) ranked fourth (Supplemental Table S1). In addition, HPA (At5g10330, HISN6A and At1g71920, HISN6B) was ranked 61st with an r value of 0.56. In contrast, none of the known HISN genes were returned in the top 200 genes most positively correlated with IMPL1 expression in Arabidopsis.
Finally, analysis of the IMPL1 and IMPL2 amino acid sequences revealed that a Gly residue required for HPP activity in the S. coelicolor enzyme (Marineo et al., 2008) was conserved in IMPL2 but not IMPL1 where it is replaced by Glu (Supplemental Fig. S1). On the basis of our bioinformatic analyses we investigated whether IMPL2 encodes a functional HPP enzyme in Arabidopsis.
Heterologous Expression of IMPL2, But Not IMPL1, Rescues the His Auxotrophy of a S. coelicolor HPP Mutant
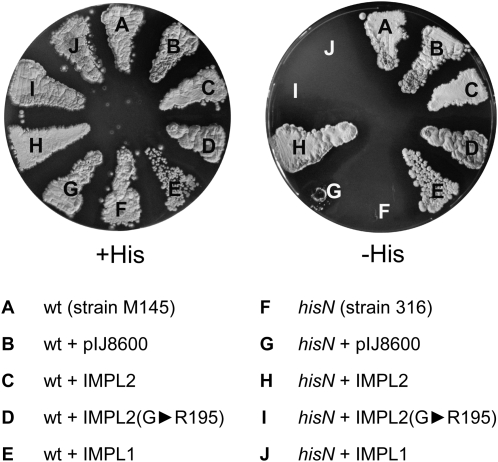
As histidinol-P is no longer commercially available to test for HPP activity of recombinant IMPL2 protein, we instead determined whether heterologous expression of IMPL2 was sufficient to complement the His auxotrophy of the S. coelicolor HPP mutant hisN (Puglia et al., 1982). hisN protoplasts transformed with the empty pIJ8600 expression vector were able to grow on minimal media (plus Phe) only if supplemented with 50 μ g mL−1 His, while transformation with pIJ8600:IMPL2 allowed the mutant to grow on minimal media without added His (Fig. 2), confirming that IMPL2 possesses HPP activity. In contrast, heterologous expression of IMPL1 was not sufficient to complement the His auxotrophy of the hisN strain (Fig. 2), suggesting that IMPL1 does not possess HPP activity.
Figure 2.
Functional complementation of the S. coelicolor hisN mutant. Bacteria transformed with pIJ8600:IMPL2 were able to grow in the absence of supplemental His (50 μ g mL−1), while those transformed with pIJ8600:IMPL1 or a mutant version of IMPL2 where Gly-195 was replaced with Arg (IMPL2G▸R195) were not. wt, Wild type.
HPP Activity of IMPL2 Requires a Conserved Gly Residue at Position 195
A conserved Gly residue has previously been shown to be required for HPP activity in S. coelicolor (Marineo et al., 2008). This residue is present in all putative HPP homologs subsequently identified in the Actinobacteria, and is also present in IMPL2 but not IMPL1 (Supplemental Fig. S1). To determine whether Gly-195 was required for the HPP activity of IMPL2, we used PCR site-directed mutagenesis to generate a mutant protein where this residue was substituted with Arg. hisN cells expressing the mutant IMPL2 protein were unable to grow on minimal media (plus Phe) without supplementation with 50 μ g mL−1 His, demonstrating that Gly-195 is also required for HPP activity of IMPL2 (Fig. 2).
Homology Modeling Suggests That Substitution of Gly-195 with Arg Leads to Disruption of the IMPL2 Active Site
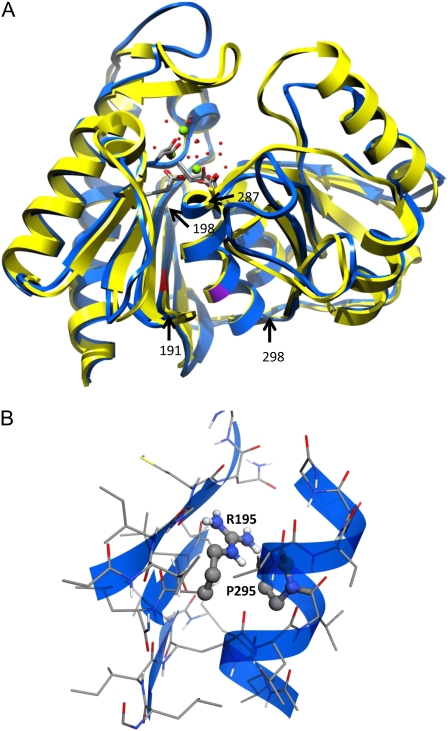
A search of the Protein Data Bank for homologs of IMPL2 produces 22 protein structures which, according to the criteria of pGenTHREADER (McGuffin and Jones, 2003), are certain to have similar folds. Structural alignment of several of these proteins using the Matchmaker tool of UCSF Chimera (Pettersen et al., 2004) demonstrated that there was considerable conservation of the location of most of the structural elements of the protein family. Variability in structure occurred mainly in the loops, especially in those residues corresponding to 100 to 118 in IMPL2. The two structures chosen as models were SuhB: the inositol monophosphatase and extragenic suppressor from E. coli (2qfl) and the inositol-1(or 4)-monophosphatase (A-chain) from Bos taurus (2bji). IMPL2 has sequence identities of 24.8% and 20.4% to 2qfl and 2bji, respectively. The pGenTHREADER generated alignment of IMPL2 with these two proteins was slightly modified to ensure that gaps were inserted in the most appropriate loops (Supplemental Fig. S2), and used to produce a homology model using MODELLER (Eswar et al., 2006; Fig. 3). The molecule has a α β α β α layered structure and a pronounced active site cleft in which are situated three Mg2+ ions held in position by carboxylate groups of Glu and Asp residues located on loops at the side of the six-stranded sheet, and on one of the central helices (Fig. 3).
Figure 3.
Homology modeling of IMPL2. A, Superposition of the homology model of IMPL2 (blue) and inositol-1(or 4)-monophosphatase (A-chain) from B. taurus (2bji; yellow). The location of Gly-195 is indicated in red and that of Pro-295 in purple. The active site carboxylates, Mg2+ ions (green spheres), and closely associated water molecules (red spheres) are also shown. The β -sheet at residues 191 to 198 and α -helix from 287 to 298 are indicated by black arrows. Residues 1 to 60 comprising the plastid transit peptide of IMPL2 were not included in the model. B, Close-up view of the best packing around Arg-195 achieved by SCWRL4. The two residues with the greatest overlap (Arg-195 and Pro-295) are represented as ball and stick models.
Gly-195 is located in the third β -strand (191–198) of the six-stranded sheet and is in close contact with one of the central α -helices (287–298; Fig. 3). The location of this helix determines the position of Asp-288, which forms a salt bridge to one of the active site Mg2+ ions. Gly-195 is strongly conserved in the IMP superfamily, and the only other residue tolerated at this position is Ala, which is found in 2qfl. Replacement of Gly-195 with an Arg using SCWRL4 (Shapovalov and Dunbrack, 2007) resulted in severe clashes with Pro-265. Since SCWRL4 searches for an optimum packing of side chains without moving the protein backbone we conclude that it is impossible to accommodate an Arg residue at this position without separating the β -strand (191–198) and the α -helix (287–298) by at least 3Å. Such a separation would alter the relative locations of the key active site residues: Asp-289, Glu-147, Asp-165, and Asp-168. The latter three amino acids are located on other strands in the β -sheet. This, in turn, would disrupt the active site and account for the observed inactivity of the mutant IMPL2 protein.
Analysis of impl2 Mutants Suggests a Lack of Genetic Redundancy in the Capacity for Histidinol Biosynthesis in Arabidopsis
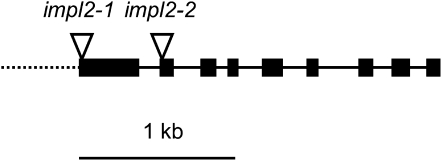
Homozygosity for null alleles of any of the four nonredundant genes in the His biosynthetic pathway (see Fig. 1) results in embryonic lethality in Arabidopsis (Muralla et al., 2007). If IMPL2 was the only protein with HPP activity in Arabidopsis, this phenotype should also be observed in impl2 T-DNA insertion lines. Two T-DNA insertion lines were obtained from the European Arabidopsis Stock Centre, SAIL_146_E09 (impl2-1) and SALK_076930 (impl2-2; Fig. 4). Both impl2-1 and impl2-2 were supplied as segregating lines and PCR genotyping of 115 plants failed to identify a single homozygous individual in either line, consistent with homozygous lethality for null mutations in IMPL2. Instead χ2 analysis revealed no significant difference from a ratio of two heterozygous to one wild-type plant in either line (Table I).
Figure 4.
Map of At4g39120 showing T-DNA insertion sites. Schematic of At4g39120 coding and upstream sequence, indicating T-DNA insertion sites in the lines used in this study. Blocks represent exons, solid lines represent introns, and dashed line 5 ′ upstream sequence.
Table I. PCR genotyping of segregating impl2-1 and impl2-2 populations.
χ2 tests were used to determine whether the ratio of heterozygous to wild-type individuals was significantly different from 2:1.
| Insertion Line | Wild Type | Heterozygote | Homozygote | χ2 Value | P Value |
| impl2-1 | 17 | 30 | 0 | 0.1702 | 0.68 |
| impl2-2 | 23 | 45 | 0 | 0.0075 | 0.93 |
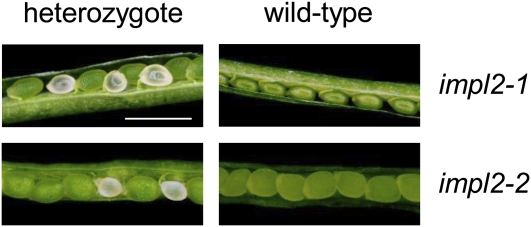
To determine the developmental stage at which lethality occurs, we examined the siliques of heterozygous plants following self-pollination. Approximately a quarter of the seeds in these siliques were observed to have aborted prior to the late globular stage of development (when embryos begin to turn green), in both the impl2-1 and impl2-2 insertion lines (Fig. 5; Table II). Thus lethality of the null impl2 mutants occurred during embryo development, as previously reported for other single copy HISN genes (Muralla et al., 2007). In contrast to several of the nonredundant HISN genes analyzed by Muralla et al. (2007), χ2 analysis revealed no significant difference from a ratio of three wild-type to one mutant seed, and thus no evidence of reduced transmission of impl2 mutant pollen tubes.
Figure 5.
Embryonic lethality of null impl2 mutants. Representative images of immature siliques from heterozygous and wild-type plants isolated from segregating impl2-1 and impl2-2 T-DNA insertion populations. Scale bar indicates a distance of 1 mm.
Table II. Analysis of siliques from plants heterozygous for null impl2 alleles.
Het, Heterozygote; Wt, wild type.
| Insertion Line | Genotype | Siliques Screened | Total No. of Seeds | Aborted Seeds |
| impl2-1 | Het | 41 | 1,811 | 440 (24.3%) |
| Wt | 10 | 429 | 9 (2.1%) | |
| impl2-2 | Het | 23 | 1,188 | 287 (24.2%) |
| Wt | 8 | 272 | 1 (0.4%) |
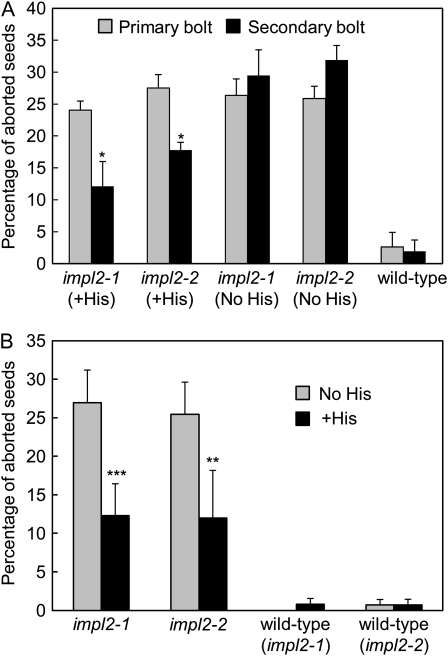
The supply of exogenous His to heterozygous plants can partially rescue the embryonic lethality of null hisn mutant seeds (Muralla et al., 2007). To determine whether His supplementation could rescue the embryonic lethality phenotype in the impl2 mutants, we first identified heterozygous plants by PCR, and determined the frequency of seed abortion in siliques from the primary bolt. The primary bolt was then removed and the plants supplied with 1 mm His daily (added to the roots). Application of His resulted in a reduced frequency of seed abortion in the secondary bolts (compared to that observed in the primary bolts prior to the supply of His); from 24.1% to 12.1% in impl2-1 and 27.5% to 17.8% in impl2-2. No such decrease was observed in plants supplied only with water (Fig. 6A). In an alternative experimental approach, 1 mm His was supplied to heterozygous plants identified by PCR prior to bolting, and siliques from the primary bolt analyzed 20 d after feeding began. The percentage of aborted seeds in siliques from plants supplied with His was significantly lower than that of control plants (Fig. 6B), and the majority of these plants contained several siliques with no aborted seeds.
Figure 6.
Exogenous His partially rescues the embryonic lethality of null impl2 mutants. A, The frequency of seed abortion in siliques from primary bolts was determined in plants heterozygous for impl2 mutant alleles and in wild-type plants isolated from the impl2-1 segregating population. Following removal of the primary bolt, plants were supplied daily with 1 mm His (+His) or water (No His). Analysis of siliques from secondary bolts was carried out 16 d after supplementation with His began. Five siliques were analyzed per plant, and values shown are the mean percentage of aborted seeds per plant +sd (n = 4 plants). Significant differences in the percentage of aborted seeds (P < 0.05) between primary and secondary bolts (determined by paired t test analysis) are indicated by an asterisk (*). B, Percentage seed abortion in primary bolts from heterozygous plants supplied with 1 mm His (+His) or water (No His) daily for 20 d prior to analysis of siliques. Wild-type plants isolated from the impl2-1 and impl2-2 segregating populations were used as controls. Five siliques were analyzed per plant, and values shown are the mean percentage frequency of seed abortion per plant +sd (n = 5 plants). Significant differences in the percentage of aborted seeds between plants supplied with His and control plants (determined by two-sample t test analysis) are indicated by asterisks: ** P < 0.01; *** P < 0.001.
Together, these data suggest that the embryonic lethality phenotype of impl2 null mutants is a consequence of their inability to synthesize His, and that there is no apparent genetic redundancy in the capacity for histidinol biosynthesis in Arabidopsis, at least in the developing embryo.
IMPL2 Is Expressed in All Plant Tissues and at All Developmental Stages
RNA gel-blot and microarray analyses of known HISN genes from Arabidopsis has demonstrated that they are constitutively expressed in all plant tissues, and at all developmental stages (Ohta et al., 2000; Muralla et al., 2007). We analyzed the temporal and spatial expression patterns of IMPL2 across multiple microarray expression profiling experiments using Genevestigator V3 (Hruz et al., 2008). IMPL2 displayed a similar pattern of expression to other known HISN genes (Supplemental Fig. S3, A and B); IMPL2 transcripts were detected in all tissues and throughout development, and mRNA levels were lowest in pollen, as previously reported for all other HISN genes by Muralla et al. (2007).
IMPL2 Localizes to the Chloroplast in Planta
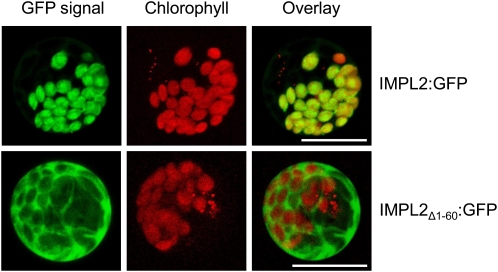
All of the His biosynthetic enzymes identified to date from plants contain a putative N-terminal plastid transit peptide (Stepansky and Leustek, 2006), and two proteins (IGPD and HDH) have been immunolocalized to this organelle (Nagai et al., 1993; Tada et al., 1995). To demonstrate conclusively that the Arabidopsis HPP (IMPL2) is also targeted to the plastid in planta, we generated two C-terminal GFP fusion proteins; the first a full-length IMPL2 protein fused to GFP (IMPL2:GFP) and the second a truncated IMPL2 lacking the first 60 amino acids predicted to act as the plastid transit peptide by ChloroP (Emanuelsson et al., 1999) fused to GFP (IMPL2Δ 1–60:GFP). Confocal imaging of maize (Zea mays) leaf protoplasts transiently expressing these proteins revealed that IMPL2:GFP was targeted to the chloroplast, as evidenced by the colocalization of GFP signal and chlorophyll autofluoresence (Fig. 7). In contrast, the truncated IMPL2Δ 1–60:GFP protein remained in the cytoplasm (Fig. 7). Thus the 60 amino acid N-terminal sequence serves as a functional plastid transit peptide, and is required for transport into the chloroplast.
Figure 7.
Subcellular localization of IMPL2. Confocal images of maize protoplasts transiently expressing either full-length IMPL2 protein fused to GFP (IMPL2:GFP) or a truncated IMPL2 protein lacking the first 60 amino acids predicted to act as the plastid transit peptide fused to GFP (IMPL2Δ 1–60:GFP). GFP emission at 500 to 520 nm (GFP signal), chlorophyll autofluoresence at 575 to 650 nm (chlorophyll), and a merged image (overlay) are shown. Scale bars indicate a distance of 20 μ m.
Phylogenetic Analysis of Plant IMP Sequences
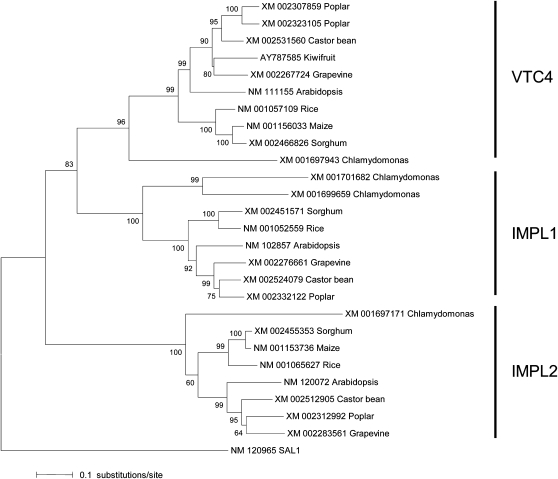
Querying the National Center for Biotechnology Information mRNA RefSeq database with the Arabidopsis VTC4, IMPL1, and IMPL2 cDNA sequences identified putative homologs of each gene in a range of monocot and dicot plants for which full genome sequence is available, as well as in the chlorophyte green alga Chlamydomonas reinhardtii. Phylogenetic analysis of the aligned IMP cDNA sequences, using the neighbor-joining method (rooted with the Arabidopsis SAL1 gene that encodes a 3 ′ ,5′-bisphosphate nucleotidase), revealed the existence of three well-supported clades (Fig. 8), with the orthologs from the various species showing greater sequence similarity than paralogs within the same species. The phylogenetic tree is consistent with the occurrence of two gene duplication events, with subsequent sequence divergence; the first duplication event giving rise to the IMPL2 clade, and the second to the IMPL1 and VTC4 clades. These gene duplication events apparently occurred prior to the divergence of the plant and green algal lineages from their last common ancestor, which has been dated at 1,000 million years ago (Heckman et al., 2001).
Figure 8.
Neighbor-joining tree depicting phylogenetic relationships between IMP cDNA sequences in plants. cDNA sequences obtained from the National Center for Biotechnology Information RefSeq database (accession nos. indicated) were aligned using ClustalW. The alignment was refined manually, including the removal of sequences encoding plastid transit peptides. Phylogenetic analysis was performed with Mega 4.0 (Tamura et al., 2007) using the neighbor-joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) is indicated above the nodes subtending the branches. Evolutionary distances were computed using the Tajima-Nei method. The tree is rooted on the SAL1 gene from Arabidopsis (At5g63980), which encodes a 3 ′ ,5′-bisphosphate nucleotidase.
DISCUSSION
The biochemical pathway of His biosynthesis (Fig. 1) was elucidated in plants during the late 1990s largely as a result of efforts to identify novel targets for herbicides (Mori et al., 1995), and genes encoding seven of the eight enzymes identified through functional complementation of bacterial or yeast His biosynthetic mutants. The identity of the enzyme(s) catalyzing the dephosphorylation of histidinol-P to histidinol was unknown, and analysis of the Arabidopsis genome sequence revealed no likely homologs of either DDDD or PHP-type HPP proteins. Ward and Ohta (1999) questioned whether plants possessed a specific HPP enzyme, and suggested instead that additional phosphatase activities might function adequately to catalyze the dephosphorylation of histidinol-P. Our data suggest that this is the case in Arabidopsis; functional complementation of the S. coelicolor hisN mutant by IMPL2 (Fig. 2) demonstrated that in addition to Gal-P and inositol-P substrates, this enzyme can also hydrolyze histidinol-P to histidinol. While the majority of His enzymes from plants have not been biochemically characterized, they are thought to function solely in His biosynthesis, as is the case in microorganisms (Alifano et al., 1996). IMPL2 is thus unusual, as its previously reported in vitro activity against d-Ins 1-P and l-Gal 1-P (Torabinejad et al., 2009) is suggestive of additional roles for IMPL2 in myoinositol and ascorbate metabolism, though this has yet to be demonstrated in vivo.
While the substrate specificity of the IMPL2 enzyme may be relatively broad, we found no evidence for genetic redundancy in histidinol biosynthesis in Arabidopsis. Null impl2 mutants displayed embryonic lethality (Fig. 5) that could be partially rescued by supplementation of plants heterozygous for impl2 mutant alleles with 1 mm His (Fig. 6, A and B). This indicates that IMPL2 was required for His production in the developing embryo, and that endogenous His levels in the surrounding maternal tissue were insufficient to rescue the His auxotrophy of the null impl2 embryos, as previously observed for mutations in four other nonredundant HisN genes (Muralla et al., 2007). VTC4 and IMPL1, although expressed in embryonic tissue (Supplemental Fig. S3C), were not sufficient to provide histidinol for the developing embryo.
While it is possible that phosphatase enzymes other than IMPL2 may also be capable of producing histidinol in other plant tissues, or at different developmental stages, it is unlikely that either VTC4 or IMPL1 play such a role in Arabidopsis. Lethality of impl1 null mutants has recently been reported (Torabinejad et al., 2009), however this phenotype does not appear to be linked to His biosynthesis, as heterologous expression of IMPL1 was not able to complement the His auxotrophy of the S. coelicolor hisN mutant (Fig. 3), indicating that IMPL1 does not possess HPP activity. While we did not test the ability of VTC4 to complement the hisN mutant in this study, this protein is localized to the plasma membrane (Marmagne et al., 2007), and is thus unlikely to play a role in His biosynthesis. All known plant His proteins contain putative plastid transit peptides, and two (IGPD and HDH) have been immunolocalized to the chloroplast (Nagai et al., 1993; Tada et al., 1995). The observation that full-length IMPL2:GFP fusion protein was targeted to the chloroplast (Fig. 7) provides further evidence that His biosynthesis in plants is confined to this organelle.
Phylogenetic analysis of plant IMP cDNA sequences (Fig. 8) suggested that the gene duplication events giving rise to the three IMP clades in plants occurred prior to the split of lineages giving rise to chlorophyta algae and plants (Fig. 8), which has been dated through analysis of 41 protein coding sequences at around 1,000 million years ago (Heckman et al., 2001). In light of this ancient evolutionary divergence, there is a remarkable degree of conservation in substrate specificity between the three Arabidopsis enzymes. VTC4, IMPL1, and IMPL2 are all capable of catalyzing the dephosphorylation of d-Ins 1-P, d-Ins 3-P, and l-Gal 1-P in vitro (Torabinejad et al., 2009), although IMPL1 displays 10-fold reduced activity with d-Ins 3-P and l-Gal 1-P compared to d-Ins 1-P. vtc4 null mutants are viable, suggesting genetic redundancy in all the enzymatic reactions catalyzed by VTC4 (Torabinejad et al., 2009). In contrast, impl1 null mutants are not viable, suggesting that IMPL1 may perform additional uncharacterized reaction(s), while the lethality of the impl2 null mutants can be explained by the role of IMPL2 in His biosynthesis. The substrate specificities of the majority of putative plant IMP proteins have not been determined, though an IMP from kiwifruit (Actinidia deliciosa; that falls in the VTC4 clade, Fig. 8) has also been shown to dephosphorylate both l-Gal 1-P and d-Ins 1-P (Laing et al., 2004), and this bifunctionality is also found in IMP proteins from mammals (Parthasarathy et al., 1997). Homology modeling of the IMPL2 protein against solved structures for IMP enzymes from Escherichia coli and B. taurus also revealed a very high degree of structural conservation, despite the relatively low overall sequence identity (Fig. 3). The ability to utilize both Gal-P and inositol-P substrates may thus be common to all IMP proteins in plants (and beyond). Secondary substrate specificity (e.g. histidinol-P in IMPL2) may have arisen subsequent to gene duplication, with the cooption of the duplicated gene into alternative metabolic functions. Alternatively the ability to dephosphorylate histidinol-P may be an ancestral state, subsequently lost from the VTC4/IMPL1 clade. We suggest that proteins within the IMPL2 clade (Fig. 8) are likely HISN7 candidates.
In summary, we have demonstrated that IMPL2 functions as an HPP enzyme in Arabidopsis, identifying the missing link in this important primary metabolic pathway. IMPL2 is thus the HISN7 gene according to the nomenclature of Muralla et al. (2007). While the enzyme has a broad substrate range in vitro, our data suggest that it is the only protein with HPP activity in Arabidopsis, at least in the developing embryo. This lack of genetic redundancy is characteristic of the His biosynthetic pathway in Arabidopsis, with five of the eight enzymes now known to be encoded by single copy genes, in contrast to the extensive redundancy observed in other amino acid biosynthetic pathways.
MATERIALS AND METHODS
Plant Growth Conditions
Plants were grown on a 1:1 mix of peat (Jiffy Products International) and vermiculite in a controlled environment room with a photosynthetic flux of 60 to 80 μ mol m−2 s−1 under a 16-h-light/8-h-dark cycle at 22°C and 55% relative humidity. Plants were fertilized with 0.14% (w/v) Phostrogen plant food (Bayer CropScience) 7 and 21 d postgermination.
Genotypic Analysis of T-DNA Insertion Lines
Two segregating T-DNA insertion knockout lines for At4g39120, SAIL_146_E09 (impl2-1), and SALK_076930 (impl2-2) were obtained from the European Arabidopsis Stock Centre. Plants were genotyped using gene-specific primers in combination with the LB3 primer (5′-GAATTTCATAACCAATCTCGATACAC-3 ′) for impl2-1, or the LBb1.3 primer (5′-ATTTTGCCGATTTCGGAAC-3 ′) for impl2-2. Gene-specific primers were as follows, impl2-1: left (5′-TGCTGTGAAGAAACAGAGCAG-3 ′) and right (5′-AGGAGAATGGCGAAGGAGTAG-3 ′) and impl2-2: left (5′-ATTATCAAACCGAACCGAACC-3 ′) and right (5′-AGCAATCAGCGTACCAAACAC-3 ′). Two PCR reactions were set up for each DNA sample, one with the gene-specific left and right primers, and a second with the left border primer and right primer, both at an annealing temperature of 60°C, and an extension time of 1 min. The exact position of the T-DNA insertions in these lines was determined by DNA sequencing of the PCR product generated using the left border and right primers, and was found to lie at the 5 ′ boundary of exon I (impl2-1) and 5 ′ boundary of exon II (impl2-2), respectively.
Phenotypic Analysis of T-DNA Insertion Lines
The analysis of siliques for the presence of aborted seeds in heterozygous individuals was carried out as described in the tutorial section of the Seed Genes Project Web site (www.seedgenes.org). Rescue experiments were carried out by supplying 1 mm His (dissolved in water) daily to the roots of heterozygous plants prior to bolting. Siliques were analyzed 14 to 20 d after supplementation with His began.
Complementation of the Streptomyces coelicolor hisN Mutant
Following an initial round of PCR site-directed mutagenesis (Ho et al., 1989) to remove an internal NdeI site, the IMPL2 open reading frame (ORF; minus putative plastid transit peptide sequence) was amplified by PCR using the primers IMPL2-F (5′-GCCATATGGCTTCAAACTCAAAAC-3 ′) and IMPL2-R (5′-GCTCTAGATCAATGCCACTCAAGTGAC-3 ′), and cloned into the conjugative and integrative expression vector pIJ8600 (Sun et al., 1999) via the XbaI and NdeI sites in the multiple cloning site. A second construct containing the IMPL2 ORF (minus plastid transit peptide) with a Gly-195 to Arg-195 substitution was also generated by PCR site-directed mutagenesis. Two overlapping PCR products were amplified using IMPL2-F in combination with Arg-R (5′-TGATCAATCAGTCGTAGAATCG-3 ′) and IMPL2-R with Arg-F (5′-CGATTCTACGACTGATTGATCA-3 ′). The resulting products were used as the template in a second round of PCR with the IMPL2-F and IMPL2-R primers and the 760 bp PCR product cloned into pIJ8600 via the XbaI and NdeI sites. The IMPL1 ORF (minus putative plastid transit peptide sequence) was amplified by PCR using the primers 5′-GCCATATGCTGTCTGAGGTTTCTGATCAG-3 ′ and 5′-GCGGATCCTTAAAGCTCTGTATGATAATC-3 ′ , and cloned into pIJ8600 via the BamHI and NdeI sites in the multiple cloning site. The pIJ8600 vectors were transformed into the methylation-deficient Escherichia coli ET12567 strain containing pUZ8002 that supplies transacting functions for the mobilization of ori-T containing plasmids (Flett et al., 1997), and introduced into Streptomyces coelicolor strains by conjugation according to Kieser et al. (2000). The S. coelicolor strains used in this study were wild type (strain M145) and the HPP mutant (strain 316; pheA1, hisN, strA1, SCP2+) that requires supplementation with 50 μ g mL−1 His and 37 μ g mL−1 Phe for growth on minimal media (Puglia et al., 1982).
Construction of GFP Fusion Protein Vectors
The full-length IMPL2 ORF was amplified by PCR using the primers GFP-F (5′-ATGCGTCGACATGTTAGCTCAGTCGCACTTC-3 ′) and GFP-R (5′-ATTTGCGGCCGCGAATGCCACTCAAGTGACTCCA-3 ′), and the ORF minus putative plastid transit peptide sequence (IMPL2Δ 1–60) with the primers GFP-CTP (5′-ATGCGTCGACATGGCTTCAAACTCAAAACG-3 ′) and GFP-R. The amplified sequences were cloned into the Gateway pENTR1A vector (Invitrogen, www.invitrogen.com) via the SalI and NotI sites, and then recombined into binary GATEWAY destination vector pK7FWG2.0 (Karimi et al., 2002).
Localization of IMPL2:GFP Fusion Proteins
Protoplasts were isolated from 0.3 g of leaf tissue from maize (Zea mays) plants grown under standard growth conditions for 9 d, followed by 2 d in continuous darkness according to the protocol of Yoo et al. (2007). Polyethylene glycol calcium-mediated transfection of 1 × 104 protoplasts with 10 μ g of plasmid DNA was carried out as described by Yoo et al. (2007). Imaging was carried out using a Zeiss LSM 510 Meta confocal microscope, with an LD C-Apochromat 40 × /1.1 W M27 objective. GFP was excited at 488 nm and emission detected in the 500 to 520 nm range. Chlorophyll autoflouresence was detected in the 575 to 600 nm range.
Molecular Modeling
Homologs to IMPL2 (excluding the N-terminal 60 amino acid plastid transit peptide sequence) were found in the Protein Data Bank using pGenTHREADER (McGuffin and Jones, 2003). The alignment generated by pGenTHREADER was used as the basis for the construction of a homology model using MODELLER (Eswar et al., 2006). Several iterations were necessary to ensure that insertions and deletions were appropriately placed in the loops connecting the secondary structural elements (Supplemental Fig. S2). Side chains were placed using SCWRL4 (Shapovalov and Dunbrack, 2007). Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (Pettersen et al., 2004).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_001118558, NP_195623, and NP_564376.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence alignment of Arabidopsis IMPL1, IMPL2, and VTC4 proteins with known HPP enzymes from Corynebacterium glutamicum and S. coelicolor.
Supplemental Figure S2. pGenTHREADER alignment used to generate homology model of IMPL2.
Supplemental Figure S3. Expression patterns of VTC4, IMPL1, and VTC4 in Arabidopsis.
Supplemental Table S1. Top 10 genes showing the strongest positive correlation with IMPL2 expression across 322 publicly available microarray datasets.
Acknowledgments
We thank Dirk Lang (University of Cape Town [UCT]) for confocal imaging of protoplasts, Nicci Illing (UCT) for donation of cellulase and macerozyme, Jacqueline Bishop (UCT) for advice on phylogenetic analyses, Marc and Heather Knight (University of Durham, UK) for assistance with Gateway cloning, and Dave Woods (UCT) for critical reading of the manuscript.
References
- Alifano P, Fani R, Liò P, Lazcano A, Bazzicalupo M, Carlomagno MS, Bruni CB. (1996) Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev 60: 44–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN, Garry B, Herzenberg LA. (1960) The genetic control of the enzymes of histidine biosynthesis in Salmonella typhimurium. J Mol Biol 33: 533–546 [DOI] [PubMed] [Google Scholar]
- Bikard D, Patel D, Le Mette C, Giorgi V, Camilleri C, Bennett MJ, Loudet O. (2009) Divergent evolution of duplicate genes leads to genetic incompatibilities within Arabidopsis thaliana. Science 323: 623–626 [DOI] [PubMed] [Google Scholar]
- Boss WF, Davis AJ, Im YJ, Galvão RM, Perera IY. (2006) Phosphoinositide metabolism: towards an understanding of subcellular signaling. Subcell Biochem 39: 181–205 [DOI] [PubMed] [Google Scholar]
- Conklin PL, Gatzek S, Wheeler GL, Dowdle J, Raymond MJ, Rolinski S, Isupov M, Littlechild JA, Smirnoff N. (2006) Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J Biol Chem 281: 15662–15670 [DOI] [PubMed] [Google Scholar]
- El Malki F, Frankard V, Jacobs M. (1998) Molecular cloning and expression of a cDNA sequence encoding histidinol phosphate aminotransferase from Nicotiana tabacum. Plant Mol Biol 37: 1013–1022 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Marti-Renom MA, Webb B, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A. (2006) Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics Chapter 5: Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani R, Brilli M, Fondi M, Lió P. (2007) The role of gene fusions in the evolution of metabolic pathways: the histidine biosynthesis case. BMC Evol Biol 7: S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. (1999) Structure and Mechanism in Protein Science. W.H. Freeman and Company, New York [Google Scholar]
- Flett F, Mersinias V, Smith CP. (1997) High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett 155: 223–229 [DOI] [PubMed] [Google Scholar]
- Fraústo da Silva JJR, Williams RJP. (2001) The Biological Chemistry of the Elements: The Inorganic Chemistry of Life, Ed 2. Oxford University Press, Oxford [Google Scholar]
- Fujimori K, Ohta D. (1998a) An Arabidopsis cDNA encoding a bifunctional glutamine amidotransferase/cyclase suppresses the histidine auxotrophy of a Saccharomyces cerevisiae his7 mutant. FEBS Lett 428: 229–234 [DOI] [PubMed] [Google Scholar]
- Fujimori K, Ohta D. (1998b) Isolation and characterization of a histidine biosynthetic gene in Arabidopsis encoding a polypeptide with two separate domains for phosphoribosyl-ATP pyrophosphohydrolase and phosphoribosyl-AMP cyclohydrolase. Plant Physiol 118: 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori K, Tada S, Kanai S, Ohta D. (1998) Molecular cloning and characterization of the gene encoding N′-[(5′-phosphoribosyl)-formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (BBM II) isomerase from Arabidopsis thaliana. Mol Gen Genet 259: 216–223 [DOI] [PubMed] [Google Scholar]
- Fujita M, Jigami Y. (2008) Lipid remodeling of GPI-anchored proteins and its function. Biochim Biophys Acta 1780: 410–420 [DOI] [PubMed] [Google Scholar]
- Guyer D, Patton D, Ward E. (1995) Evidence for cross-pathway regulation of metabolic gene expression in plants. Proc Natl Acad Sci USA 92: 4997–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding MM. (2004) The architecture of metal coordination groups in proteins. Acta Crystallogr D Biol Crystallogr 60: 849–859 [DOI] [PubMed] [Google Scholar]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB. (2001) Molecular evidence for the early colonization of land by fungi and plants. Science 293: 1129–1133 [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle RA, Mugford ST, Rees JD, Campbell MM, Smith JAC. (2005) Constitutively high expression of the histidine biosynthetic pathway contributes to nickel tolerance in hyperaccumulator plants. Plant Cell 17: 2089–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. (2000) Practical Streptomyces Genetics. John Innes Centre, Norwich, UK [Google Scholar]
- Laing WA, Bulley S, Wright M, Cooney J, Jensen D, Barraclough D, MacRae E. (2004) A highly specific l-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. Proc Natl Acad Sci USA 101: 16976–16981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Coq D, Fillinger S, Aymerich S. (1999) Histidinol phosphate phosphatase, catalyzing the penultimate step of the histidine biosynthesis pathway, is encoded by ytvP (hisJ) in Bacillus subtilis. J Bacteriol 181: 3277–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Cho Y, Lee JH, Kang SG. (2008) Novel monofunctional histidinol-phosphate phosphatase of the DDDD superfamily of phosphohydrolases. J Bacteriol 190: 2629–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech AP, Baker GR, Shute JK, Cohen MA, Gani D. (1993) Chemical and kinetic mechanism of the inositol monophosphatase reaction and its inhibition by Li+. Eur J Biochem 212: 693–704 [DOI] [PubMed] [Google Scholar]
- Manfield IJ, Jen CH, Pinney JW, Michalopoulos I, Bradford JR, Gilmartin PM, Westhead DR. (2006) Arabidopsis Co-expression Tool (ACT): web server tools for microarray-based gene expression analysis. Nucleic Acids Res 34: W504–W509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marineo S, Cusimano MG, Limauro D, Coticchio G, Puglia AM. (2008) The histidinol phosphate phosphatase involved in histidine biosynthetic pathway is encoded by SCO5208 (hisN) in Streptomyces coelicolor A3(2). Curr Microbiol 56: 6–13 [DOI] [PubMed] [Google Scholar]
- Marmagne A, Ferro M, Meinnel T, Bruley C, Kuhn L, Garin J, Barbier-Brygoo H, Ephritikhine G. (2007) A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Mol Cell Proteomics 6: 1980–1996 [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Jones DT. (2003) Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics 19: 874–881 [DOI] [PubMed] [Google Scholar]
- Mori I, Fonnepfister R, Matsunaga S, Tada S, Kimura Y, Iwasaki G, Mano J, Hatano M, Nakano T, Koizumi S, et al. (1995) A novel class of herbicides—specific inhibitors of imidazoleglycerol phosphate dehydratase. Plant Physiol 107: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mormann S, Lömker A, Rückert C, Gaigalat L, Tauch A, Pühler A, Kalinowski J. (2006) Random mutagenesis in Corynebacterium glutamicum ATCC 13032 using an IS6100-based transposon vector identified the last unknown gene in the histidine biosynthesis pathway. BMC Genomics 7: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralla R, Sweeney C, Stepansky A, Leustek T, Meinke D. (2007) Genetic dissection of histidine biosynthesis in Arabidopsis. Plant Physiol 144: 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai A, Suzuki K, Ward E, Moyer M, Mano J, Beck J, Tada S, Hashimoto M, Chang J, Ryals J, et al. (1993) Histidinol dehydrogenase in higher plants. Purification, cloning and expression. Murata N, , Research in Photosynthesis, Vol 4. Kluwer Academic Publishers, Dordrecht, The Netherlands, 95–98 [Google Scholar]
- Ohta D, Fujimori K, Mizutani M, Nakayama Y, Kunpaisal-Hashimoto R, Munzer S, Kozaki A. (2000) Molecular cloning and characterization of ATP-phosphoribosyl transferase from Arabidopsis, a key enzyme in the histidine biosynthetic pathway. Plant Physiol 122: 907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy R, Parthasarathy L, Vadnal R. (1997) Brain inositol monophosphatase identified as a galactose-1-phosphatase. Brain Res 778: 99–106 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Puglia AM, Misuraca F, Randazzo R, Sciandrello G, Sermonti G. (1982) Plasmid-dependent co-mutation in Streptomyces coelicolor A3(2). Curr Genet 5: 89–92 [DOI] [PubMed] [Google Scholar]
- Rees JD, Ingle RA, Smith JAC. (2009) Relative contributions of nine genes in the pathway of histidine biosynthesis to control of free histidine concentrations in Arabidopsis thaliana. Plant Biotechnol J 7: 499–511 [DOI] [PubMed] [Google Scholar]
- Roth JR, Ames BN. (1966) Histidine regulatory mutants having altered histidyl-tRNA synthetase. J Mol Biol 22: 325–334 [DOI] [PubMed] [Google Scholar]
- Shapovalov MV, Dunbrack RL., Jr (2007) Statistical and conformational analysis of the electron density of protein side chains. Proteins 66: 279–303 [DOI] [PubMed] [Google Scholar]
- Stepansky A, Leustek T. (2006) Histidine biosynthesis in plants. Amino Acids 30: 127–142 [DOI] [PubMed] [Google Scholar]
- Sun J, Kelemen GH, Fernández-Abalos JM, Bibb MJ. (1999) Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145: 2221–2227 [DOI] [PubMed] [Google Scholar]
- Tada S, Hatano M, Nakayama Y, Volrath S, Guyer D, Ward E, Ohta D. (1995) Insect cell expression of recombinant imidazoleglycerolphosphate dehydratase of Arabidopsis and wheat and inhibition by triazole herbicides. Plant Physiol 109: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabinejad J, Donahue JL, Gunesekera BN, Allen-Daniels MJ, Gillaspy GE. (2009) VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol 150: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17: 849–850 [DOI] [PubMed] [Google Scholar]
- Ward E, Ohta D. (1999) Histidine biosynthesis. Singh BK, , Plant Amino Acids: Biochemistry and Biotechnology. Marcel Dekker, New York, 293–303 [Google Scholar]
- Wiater A, Krajewska-Grynkiewicz K, Kloptowski T. (1971) Histidine biosynthesis and its regulation in plants. Acta Biochim Pol 18: 299–307 [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]