Abstract
The ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif is a transcriptional regulatory motif identified in members of the ethylene-responsive element binding factor, C2H2, and auxin/indole-3-acetic acid families of transcriptional regulators. Sequence comparison of the core EAR motif sites from these proteins revealed two distinct conservation patterns: LxLxL and DLNxxP. Proteins containing these motifs play key roles in diverse biological functions by negatively regulating genes involved in developmental, hormonal, and stress signaling pathways. Through a genome-wide bioinformatics analysis, we have identified the complete repertoire of the EAR repressome in Arabidopsis (Arabidopsis thaliana) comprising 219 proteins belonging to 21 different transcriptional regulator families. Approximately 72% of these proteins contain a LxLxL type of EAR motif, 22% contain a DLNxxP type of EAR motif, and the remaining 6% have a motif where LxLxL and DLNxxP are overlapping. Published in vitro and in planta investigations support approximately 40% of these proteins functioning as negative regulators of gene expression. Comparative sequence analysis of EAR motif sites and adjoining regions has identified additional preferred residues and potential posttranslational modification sites that may influence the functionality of the EAR motif. Homology searches against protein databases of poplar (Populus trichocarpa), grapevine (Vitis vinifera), rice (Oryza sativa), and sorghum (Sorghum bicolor) revealed that the EAR motif is conserved across these diverse plant species. This genome-wide analysis represents the most extensive survey of EAR motif-containing proteins in Arabidopsis to date and provides a resource enabling investigations into their biological roles and the mechanism of EAR motif-mediated transcriptional regulation.
Plants respond to various developmental and environmental cues by regulating gene expression at the transcriptional and posttranscriptional levels. Gene regulation at the transcriptional level is orchestrated by a complex and coordinated network of activators, repressors, coactivators, and corepressors. In general, there has historically been greater research focus on transcriptional activation mechanisms and positive control of gene regulation. In comparison, relatively little is known about transcriptional repression and negative regulation of gene expression. Transcriptional repression mechanisms are currently under intense investigation, and results obtained in recent years have convincingly shown that transcriptional repression is a major regulatory mechanism and plays a key role in many biological processes (for review, see Thiel et al., 2004). Transcriptional repressors are basically classified as passive or active repressors (Hanna-Rose and Hansen, 1996). Passive repressors do not possess an intrinsic repression domain and inhibit activation of transcription by competing with transcriptional activators for cognate DNA-binding sites or by directly interacting with them to form inactive heterodimers. Conversely, active repressors generally contain a distinct, small, and portable repression domain(s) that inhibits the activation of transcription either by interacting with components of basal transcription machinery or positive transcriptional regulators and/or by recruiting histone deacetylases (HDACs), which modify chromatin structure and prevent other transcriptional activators from binding to their target cis-elements (Hanna-Rose and Hansen, 1996; Pazin and Kadonaga, 1997). A repression domain was initially identified almost two decades ago in the Drosophila active repressors Engrailed (Han and Manley, 1993) and Kruppel (Licht et al., 1994). Since then, numerous active repression domain-containing proteins have been reported in yeast, Drosophila, and animals (Hanna-Rose and Hansen, 1996), but in plants only a few active repression domains essential for transcriptional repression have been identified (Ohta et al., 2001; Matsui et al., 2008; Ikeda and Ohme-Takagi, 2009).
A subset of class II ethylene-responsive element binding factors (ERFs) and TFIIIA-type zinc finger proteins (ZFPs) in various plant species, including Arabidopsis (Arabidopsis thaliana), tobacco (Nicotiana tabacum), wheat (Triticum aestivum), petunia (Petunia hybrida), and soybean (Glycine max), have been identified as active repressors (Ohta et al., 2001; Zhang et al., 2010). The repression domain located in their C-terminal region is designated as the ERF-associated amphiphilic repression (EAR) motif and contains a conserved consensus sequence of L/FDLNL/F(x)P (Ohta et al., 2001). The complete deletion of the C-terminal EAR motif region or targeted mutation of Asp residue within the EAR motif of NtERF3, an ERF protein from tobacco, completely abolishes its capacity for repression of transcription (Ohta et al., 2001). Similarly, the deletion of C-terminal EAR motif regions of Zinc Finger of Arabidopsis10 (ZAT10) and ZAT11 results in complete loss of their repression activity (Ohta et al., 2001), confirming an essential role for the EAR motif in the repressor activity of these proteins. Several other ERFs and ZFPs containing an EAR motif function as transcriptional repressors and are known to play roles in hormone signaling, disease resistance, and abiotic stress resistance (Fujimoto et al., 2000; Sakamoto et al., 2004; McGrath et al., 2005; Song et al., 2005; Yang et al., 2005; Kazan, 2006; Jiang et al., 2008).
A sequence similar to the EAR motif in ERFs and ZFPs is also found in the C-terminal region of SUPERMAN (SUP), a negative regulator of transcription involved in flower development (Sakai et al., 1995). SUP contains a sequence (QDLDLELRLGFA) that when fused to the GAL4 DNA-binding domain converts it into a dominant repressor (Hiratsu et al., 2002, 2003). Transient expression assays demonstrated that the repression domain of SUP, when tethered to a GAL4 DNA-binding domain, had approximately five times stronger repressive activity than the EAR motif of ERFs (Hiratsu et al., 2002). The minimal functional unit responsible for the repressive activity of SUP was identified as an amphiphilic motif composed of six amino acids, DLELRL (Hiratsu et al., 2004), which is similar to the EAR motif found in ERFs and ZFPs. Mutational analysis within the DLELRL hexapeptide motif revealed that the Leu residues within this motif are essential and sufficient for strong repressive activity (Hiratsu et al., 2004).
The AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) proteins, another group of active repressors comprising 29 proteins in Arabidopsis, also contain a LxLxL type of motif that bears resemblance to the EAR motif from SUP, some ERFs, and ZFPs (Tiwari et al., 2001, 2004). The repression motifs within AUX/IAAs have been shown to confer repression of primary auxin-responsive genes, and the Leu residues within the hexapeptide motif were found to be crucial for their repressive activity (Tiwari et al., 2001, 2004). A recent study (Szemenyei et al., 2008) has further confirmed the role of the EAR motif in suppressing auxin-regulated gene expression during embryonic apical-basal cell fate determination. Perhaps the most interesting discovery of this study was demonstrating the direct interaction of the EAR motif of INDOLE-3-ACETIC ACID INDUCIBLE12 (IAA12), an AUX/IAA protein, with the CTLH (C-Terminal to LISH) domain of TOPLESS (TPL), a member of the Groucho/Tup1 family of corepressors. Genetic evidence suggests that TPL works in conjunction with HISTONE DEACETYLASE19 (HDA19) during the transition stage of embryogenesis (Long et al., 2006). Considering the general role of HDACs in suppressing gene expression by deacetylation of Lys residues on histones, it appears likely that TPL may facilitate EAR motif-mediated gene regulation through chromatin modification.
Apart from ERFs, ZFPs, and AUX/IAAs, several other repressor proteins, including Arabidopsis MYB DOMAIN PROTEIN4 (AtMYB4) and AtMYB32 belonging to the MYB family (Jin et al., 2000; Preston et al., 2004), HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE2 (HSI2) and related proteins of the ABSCISIC ACID-INSENSITIVE3/VIVIPAROUS1 (ABI3/VP1) family (Tsukagoshi et al., 2005), AGAMOUS-LIKE15 (AGL15) of the MADS (for MCM1, AGAMOUS, DEFICIENS, and SRF [serum response factor]) family (Hill et al., 2008), and NIM1-INTERACTING1 (NIMIN1; Weigel et al., 2005), have been reported to contain EAR-like motifs. Among these, AtMYB4 and AtMYB32 are required for normal pollen development (Preston et al., 2004). In addition, AtMYB4 also plays a role in the development of UV light-protecting sunscreens (Jin et al., 2000). The B3 domain and EAR motif-containing proteins such as HSI2 and related proteins are involved in the repression of seed maturation genes during the transition from seed maturation to seedling growth (Tsukagoshi et al., 2007). The MADS factor protein AGL15 along with AGL18 is also known to act as a repressor of floral transition in Arabidopsis (Adamczyk et al., 2007). NIMIN1 interacts with NONEXPRESSOR OF PR GENES1 (NPR1), a key regulator of systemic acquired resistance in Arabidopsis, and modulates the expression of pathogenesis-related (PR) genes (Weigel et al., 2005).
Considering the diverse set of biological traits that the currently known EAR motif-containing proteins influence and their distribution across multiple families, it is reasonable to hypothesize that the EAR motif is a distinct regulatory motif that is utilized by many additional transcriptional repressor proteins involved in different signaling pathways. From this perspective, a genome-wide survey in Arabidopsis would aid in the identification of the complete repertoire of EAR motif-containing proteins and thereby shed light on the transcriptional regulatory mechanisms influenced by them. In this study, we report the identification, classification, and structural analysis of 219 Arabidopsis EAR motif-containing proteins through a systematic genome-wide bioinformatics analysis. Alignment of the large data set of EAR motifs presented here enabled the identification of additional conserved amino acids proximal to the EAR motif, of which some residues may provide a means for posttranslational modification and regulation of the function of EAR motifs. Putative orthologs for each of these proteins are identified from other plant species, such as poplar (Populus trichocarpa), grapevine (Vitis vinifera), rice (Oryza sativa), and sorghum (Sorghum bicolor), and evidence supporting the broad evolutionary conservation of EAR motifs is provided. Finally, the mechanism of action of EAR motif-containing proteins and their use in generating dominant-negative chimeric repressors are discussed.
RESULTS
Identification of EAR Motif-Containing Proteins in Arabidopsis
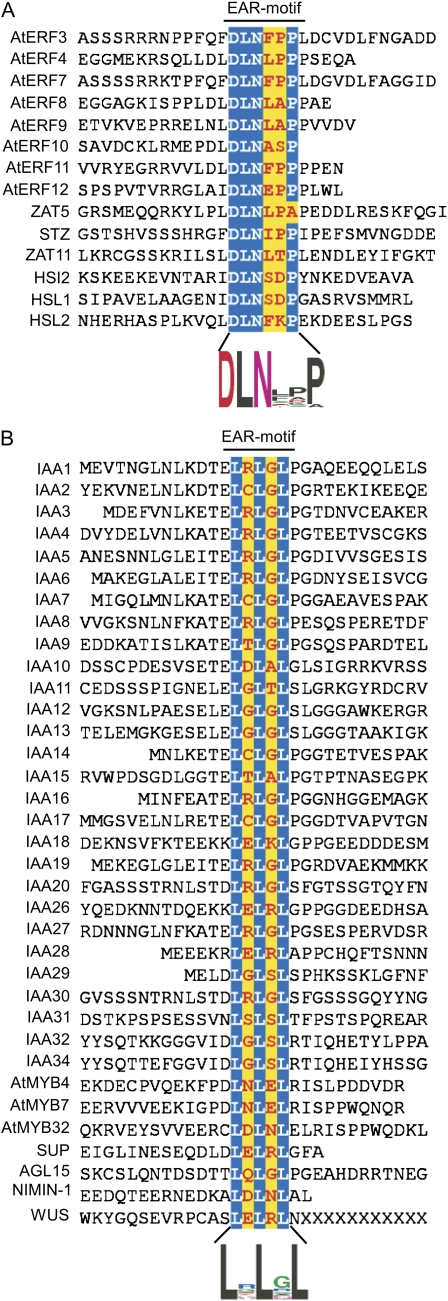
A survey of published literature identified 49 EAR motif-containing proteins (Supplemental Table S1) belonging to various transcription factor (TF) and other transcriptional regulator (OTR) families such as ERFs (eight proteins; Ohta et al., 2001; McGrath et al., 2005; Yang et al., 2005), ZFPs (four proteins; Ohta et al., 2001; Sakamoto et al., 2004; Mittler et al., 2006), ABI3/VP1 family (three proteins; Tsukagoshi et al., 2005, 2007), MYB family (five proteins; Jin et al., 2000; Preston et al., 2004), AUX/IAA family (28 proteins; Tiwari et al., 2004), Homeobox family (one protein; Kieffer et al., 2006), MADS family (one protein; Hill et al., 2008), and NPR family (one protein; Weigel et al., 2005). The protein sequence comparison of these proteins in the conserved EAR motif region (Fig. 1) revealed two distinct degenerate consensus sequence patterns: DLNxxP (where X can be one of the 20 common amino acids), consisting of a conserved DLN box and a Pro residue at the fifth or sixth position, wherein the Asp and Leu residues are implicated in repression activity (Ohta et al., 2001; Tsukagoshi et al., 2005); and LxLxL, consisting of three conserved Leu residues in alternate positions, which have been shown to be critical for both repressor activity and interaction with corepressors (Hiratsu et al., 2004; Tiwari et al., 2004; Szemenyei et al., 2008). To gain further insight into the potential utilization of the EAR motif in plant gene regulation, we conducted a genome-wide analysis of Arabidopsis to identify novel EAR motif-containing proteins. We generated an initial collection by performing a nonweighted degenerate pattern matching (DPM) analysis of the Arabidopsis proteome using sequence patterns DLNxxP or LxLxL as queries (Yan et al., 2005). This search identified 187 and 5,471 nonredundant proteins containing at least one occurrence of the DLNxxP or LxLxL type of motif, respectively, within their protein sequences (Supplemental Table S2). Only 73 out of the 187 group of proteins and 352 out of the 5,471 group of proteins were found to be potential transcriptional regulators based on The Arabidopsis Information Resource (TAIR) Gene Ontology (GO) annotations, providing an initial set of 398 (considering an overlap of 27 proteins between the 187 and 352 protein groups) candidate transcriptional regulators potentially employing an EAR motif to influence gene expression. Since the evidence from the current literature suggests a unique role for the EAR motif in transcriptional repression, identification of this motif in a large number of nontranscriptional regulator proteins (approximately 5,200 proteins) by DPM analysis reflects the possibility that the consensus sequence of EAR motifs identified in many of the proteins in the initial data sets may not function as transcriptional repressors. The relatively short and partially degenerate sequences of the LxLxL type of EAR motif might have contributed to the identification of false positives using the initial analysis parameters. To assess the incidence of false positives, we performed additional DPM searches using control sequence patterns derived from LxLxL by replacing Leu residues with one of the remaining 19 amino acids (e.g. VxVxV). Except for using SxSxS as a query employing the structurally dissimilar Ser, all other pattern searches returned substantially fewer hits compared with LxLxL (Supplemental Table S2); notably, the query patterns with similar hydrophobic residues Ile and Val returned a much lower number of hits (approximately 9% and 22%, respectively) than LxLxL. However, the recovery of hits with the alternate amino acid patterns suggested that the LxLxL pattern may occur to some degree unrelated to its function in regulating gene expression. Therefore, we concluded that the nonrandom and potentially functional EAR repressome in Arabidopsis is possibly smaller than the approximately 400 transcriptional regulators originally identified by DPM analysis.
Figure 1.
Arabidopsis EAR motif-containing proteins described in the literature. The 49 proteins are divided into two groups based on the sequence conservation pattern within the core EAR motif sites (highlighted in color). The alignment includes 12 amino acid residues upstream and downstream of the EAR motif, or up to where the nominal 12-amino acid sequence is abridged by encountering the first or last amino acid of the protein. A, The DLNxxP motif is conserved in some members of class II ERFs, TFIIIA-type ZFPs, and ABI3/VP1 family proteins. B, The LxLxL motif is conserved in AUX/IAAs and some members of the MYB and HD-Zip family proteins. Sequence logos (Crooks et al., 2004) illustrating the frequency of amino acids within the EAR motifs are presented below the respective alignments.
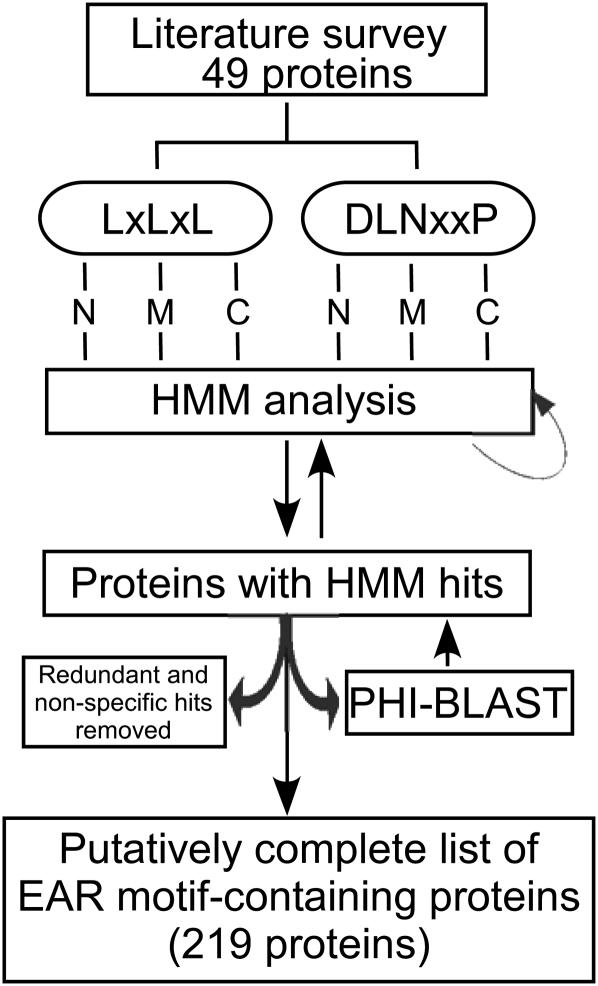
In an attempt to identify bona fide EAR motif-containing proteins in Arabidopsis, we employed a strategy containing hidden Markov model (HMM; Eddy, 1998) analysis combined with pattern hit-initiated BLAST (PHI-BLAST; Zhang et al., 1998; Fig. 2) in searching the Arabidopsis proteome. Because of sound statistical foundation, as well as modularity and flexibility to incorporate prior knowledge into the model architecture, HMM-based strategies have been routinely and reliably employed in genome-wide searches for target proteins, motifs, and domains. The protein sequences of the set of 49 known EAR motif-containing proteins derived from a literature survey (Fig. 1) were grouped into six subgroups based on motif type (DLNxxP or LxLxL) and location (N-terminal [N], middle [M], or C-terminal [C] region). To build HMM profiles, protein sequences were truncated to an approximately 30-amino acid region of each protein comprising the core EAR motif site and adjoining sequence of ± 12 amino acids. The subgrouping of proteins by location of the EAR motif was carried out to minimize sequence variations within the EAR motif region that may be due to the relative positions of EAR motif sites in their protein sequences. The rationale for including adjoining sequences in HMM analysis was based on the assumption that the sequences flanking the EAR motif may possess additional signals required for its proper function, which may be newly identified by our whole proteome analysis. A role for flanking sequences in transcriptional repression by the EAR motif has been demonstrated previously; for example, an Ala substitution for Thr or Glu residues at −2 or −3 positions of the EAR motif in IAA17 resulted in partial loss of its transcriptional repression activity (Tiwari et al., 2004). The individual HMM profiles generated from the six subgroups of sequences were used to scan the Arabidopsis proteome for putative EAR motif-containing proteins via HMMER (HMM software for biological sequence analysis). The HMM hits with a score of 5 or greater were manually inspected and included in further steps if they appeared to be bona fide EAR motif-containing proteins based on two criteria: (1) the presence of an amphiphilic pentapeptide (LxLxL) and/or hexapeptide (DLNxxP) EAR motif(s); and (2) the presence, based on GO annotation or literature evidence, of a well-defined DNA-binding domain or prior knowledge of interaction with proteins containing a DNA-binding domain, suggesting a role in transcriptional regulation. The first iteration of HMM analysis recovered 33 novel EAR motif-containing proteins in addition to the proteins used for training HMM profiles. In the next step, sequences of these novel proteins and the set of 49 EAR motif-containing proteins derived from our literature survey (Fig. 1) were used to perform PHI-BLAST analysis (Zhang et al., 1998). This step was carried out to help ensure that no distantly related homologs of EAR motif-containing proteins were missed. The PHI-BLAST hits with an E-value significantly below threshold were manually assessed for the presence of an EAR motif. The novel positive hits recovered by PHI-BLAST and the first iteration of HMM analysis were reincorporated into the six subgroups of EAR motif-containing proteins classified by type and location of EAR motif, and the HMM profiles were refined. Subsequent iterations of HMM/PHI-BLAST analysis were carried out until no more novel EAR motif-containing proteins were identified. Combined, the iterative HMM/PHI-BLAST strategy identified an additional 209 EAR motif-containing proteins, out of which 170 proteins have been previously annotated as transcriptional regulators and the remaining 39 proteins were either ambiguously annotated or lacked prior knowledge of their involvement in transcriptional regulation. Since the focus of our study was to identify transcriptional regulators containing bona fide EAR motifs, only 170 proteins were added to the list of 49 EAR motif-containing proteins that were previously validated in the literature. The remaining 39 proteins lacking transcription-related annotation may not function as transcriptional repressors and were hence considered as false positives. Unlike DPM analysis (false discovery rate [FDR] = 0.92), only a small proportion of the predictions made by HMM/PHI-BLAST analysis were found to be false positives (FDR = 0.15). Overall, this analysis identified 219 distinct, nonredundant Arabidopsis EAR motif-containing transcription-related proteins (Table I; Supplemental Table S3).
Figure 2.
Strategy for the identification of EAR motif-containing proteins in Arabidopsis. An iterative approach including literature survey, HMM analysis, and PHI-BLAST was used to search through the Arabidopsis protein database. Details of each step are given in the text. N, M, and C refer to locations of the EAR motifs as being in the N-terminal, middle, and C-terminal regions of the protein, respectively.
Table I. Arabidopsis transcriptional regulator proteins with conserved EAR motif sequence.
For each protein, AGI code, protein name, protein size (amino acids), and sequence and location of core EAR motif site(s) are listed. Proteins with a known role in negative regulation of gene expression are underlined. The location refers to the position of the Leu (L) residue underlined in the corresponding EAR motif sequence. Additional information about each protein is provided in Supplemental Table S3.
| AGI Code | Protein Name | Protein Size | Core EAR Motif Site |
|
| Sequence | Location | |||
| ABI3VP1 family | ||||
| AT2G30470abc | HSI2 | 790 | DLNSDP | 730 |
| AT4G21550a | HSL2 | 721 | DLNFKP | 653 |
| AT4G32010ac | HSL1 | 780 | DLNSDP | 710 |
| ERF family | ||||
| AT1G03800ab | AtERF10 | 245 | DLNASP | 241 |
| AT1G28360ab | AtERF12 | 189 | LSLDLNHLP | 73 |
| DLNEPP | 181 | |||
| AT1G28370ab | AtERF11 | 166 | DLNFPP | 158 |
| AT1G46768 | DEAR6 | 153 | DLNQIP | 144 |
| AT1G50640ab | AtERF3 | 225 | DLNFPP | 201 |
| AT1G53170ab | AtERF8 | 185 | LELSL | 117 |
| LDLNLAP | 178 | |||
| AT2G23340 | DEAR3 | 176 | DLNKLP | 156 |
| AT2G28550 | TOE1 | 449 | LDLNL | 4 |
| AT2G39250 | SNZ | 325 | LDLNL | 4 |
| LELSL | 252 | |||
| AT3G15210abc | AtERF4 | 222 | LELSL | 129 |
| LDLNLPP | 213 | |||
| AT3G20310ab | AtERF7 | 244 | DLNFPP | 219 |
| AT3G50260bc | DEAR1 | 153 | DLNKLP | 137 |
| AT3G54990 | SMZ | 346 | LDLNL | 4 |
| LELSL | 273 | |||
| AT4G36900 | DEAR4 | 196 | DLNKLP | 184 |
| AT4G36920c | AP2 | 432 | DLNDAP | 4 |
| LDLSL | 306 | |||
| AT5G44210ab | AtERF9 | 200 | LDLNLAP | 191 |
| AT5G67190 | DEAR2 | 184 | DLNKLP | 167 |
| AS2 family | ||||
| AT1G67100 | ASL37 | 233 | LELTLGL | 189 |
| AT1G68510 | ASL36 | 233 | LELRL | 230 |
| AT3G02550 | ASL38 | 263 | LDLTLRL | 224 |
| AT3G49940 | ASL40 | 247 | LDLSL | 186 |
| AT5G67420 | ASL39 | 250 | LDLSL | 189 |
| bZIP family | ||||
| AT1G06850 | AtBZIP52 | 337 | LKLRL | 213 |
| AT2G40620 | AtbZIP18 | 367 | LKLRL | 213 |
| AT4G37730 | AtBZIP7 | 305 | LRLVL | 243 |
| LRLRL | 267 | |||
| BZR family | ||||
| AT1G19350 | BES1 | 335 | LELTL | 326 |
| AT1G75080bc | BZR1 | 336 | LELTL | 327 |
| AT1G78700 | BEH4 | 325 | LELTL | 317 |
| AT3G50750 | BEH1 | 276 | LELTL | 266 |
| AT4G18890 | BEH3 | 284 | LELTL | 276 |
| C2C2-CO-like family | ||||
| AT2G21320 | 172 | DLNSNP | 142 | |
| AT4G38960 | 183 | DLNANP | 141 | |
| C2C2-Dof family | ||||
| AT1G69570 | 399 | DLNEPP | 82 | |
| C2H2 family | ||||
| AT1G02030 | 267 | LSLML | 132 | |
| DLNLPA | 251 | |||
| AT1G10480 | ZFP5 | 211 | LDLHL | 202 |
| AT1G13400 | NUB | 207 | DLNNLP | 11 |
| AT1G24625 | ZFP7 | 209 | LDLKL | 37 |
| LTLRL | 207 | |||
| AT1G26610 | 455 | DLNLPA | 447 | |
| AT1G27730ab | STZ | 227 | DLNIPP | 191 |
| AT1G34790 | TT1 | 303 | DLNPNP | 59 |
| AT1G49900 | ATZF1 | 917 | LDLKLSL | 340 |
| DLNNPP | 390 | |||
| AT1G66140 | ZFP4 | 260 | LNLSL | 38 |
| LTLPL | 59 | |||
| LTLKL | 258 | |||
| AT1G67030 | ZFP6 | 197 | LDLHLSL | 191 |
| AT1G68480b | JAG | 253 | DLNNLP | 10 |
| AT1G80730 | ZFP1 | 228 | LELGLTL | 24 |
| LTLKL | 156 | |||
| AT2G17180 | ZAT2 | 270 | DLNVPA | 248 |
| LDLRLGL | 266 | |||
| AT2G26940 | 286 | DLNQPP | 276 | |
| AT2G28200a | ZAT5 | 286 | LPLDLNLPA | 254 |
| AT2G28710 | ZAT17 | 156 | LCLDLNLTP | 135 |
| LKLEL | 149 | |||
| AT2G37430ab | ZAT11 | 178 | LSLDLNLTP | 149 |
| AT2G37740 | ZFP10 | 304 | LDLELRL | 292 |
| AT2G41940 | ZFP8 | 257 | LDLHL | 255 |
| AT2G42410 | ZFP11 | 214 | LDLELRL | 207 |
| AT2G45120 | ZAT4 | 314 | DLNLPA | 296 |
| AT3G09290 | TAC1 | 172 | LDLELRL | 158 |
| AT3G10470 | ZAT15 | 398 | LDLDLNLPA | 342 |
| AT3G19580b | AZF2 | 273 | LALCL | 73 |
| DLNLPA | 225 | |||
| AT3G23130abc | SUP | 204 | LDLELRL | 197 |
| AT3G23140 | 172 | LNLSLGL | 130 | |
| LDLDLRL | 162 | |||
| AT3G46090c | ZAT7 | 168 | LDLDL | 146 |
| AT3G49930 | ZAT13 | 215 | LALCL | 62 |
| DLNLPA | 189 | |||
| AT3G53600 | ZAT18 | 175 | LDLNLTP | 148 |
| AT3G53820 | 142 | LELRLGL | 126 | |
| AT3G58070 | GIS | 253 | LDLHL | 251 |
| AT3G60580 | ZAT9 | 288 | DLNLPA | 272 |
| AT3G62850 | 472 | DLNPPP | 101 | |
| AT4G16610 | 204 | DLNLPP | 187 | |
| AT4G17810b | SAZ | 204 | LDLELRL | 195 |
| AT4G35280 | ZAT3 | 284 | DLNVPP | 259 |
| LDLRLGL | 280 | |||
| AT4G35610 | 271 | DLNVEP | 234 | |
| DLNKSP | 256 | |||
| AT4G35700 | 275 | DLNADP | 236 | |
| DLNRSP | 258 | |||
| AT5G01860 | 215 | LSLKL | 213 | |
| AT5G03510 | ZAT14 | 292 | LGLGL | 112 |
| LQLDLNLPA | 251 | |||
| AT5G04340c | ZAT6 | 238 | LCLML | 56 |
| DLNIPP | 204 | |||
| AT5G04390 | 362 | LDLDLNLPA | 310 | |
| AT5G05120 | 201 | LDLCL | 161 | |
| LSLSLKL | 197 | |||
| AT5G06070c | RBE | 226 | LDLELRL | 214 |
| AT5G06650 | GIS2 | 191 | LDLRL | 189 |
| AT5G10970 | 272 | LDLML | 62 | |
| LDLSLKL | 268 | |||
| AT5G14010c | Knuckles | 161 | LDLDLSLRL | 155 |
| AT5G25160 | ZFP3 | 235 | LDLSLKL | 231 |
| AT5G27880 | 278 | LSLSL | 276 | |
| AT5G43170b | AZF3 | 193 | DLNIIP | 167 |
| AT5G43540 | 137 | LELRLGL | 127 | |
| AT5G48890 | 173 | LDLSLHL | 169 | |
| AT5G56200 | 493 | DLNELP | 481 | |
| AT5G57520 | ZFP2 | 150 | LRLSL | 11 |
| LNLELVL | 23 | |||
| LDLSLRL | 146 | |||
| AT5G59820bc | ZAT12 | 162 | LDLSL | 143 |
| LNLKLEL | 153 | |||
| AT5G67450b | AZF1 | 245 | LALCL | 66 |
| DLNLPA | 233 | |||
| C3H family | ||||
| AT1G53010 | 178 | LDLDL | 58 | |
| AT2G42030 | 425 | LDLNLGP | 13 | |
| AT3G58030 | 436 | LDLNLGP | 13 | |
| EIL family | ||||
| AT3G20770c | EIN3 | 628 | DLNIPN | 532 |
| AT5G21120 | EIL2 | 518 | DLNPSP | 473 |
| LGLVL | 486 | |||
| AT5G65100 | 557 | DLNQLP | 522 | |
| G2-like family | ||||
| AT1G14600 | 255 | LSLSLSL | 195 | |
| LNLNL | 246 | |||
| AT2G02060 | 256 | LSLSLSL | 202 | |
| AT2G40260 | 410 | LVLQL | 114 | |
| LDLSLSL | 336 | |||
| LSLSL | 372 | |||
| LDLTL | 408 | |||
| AT4G04580 | 166 | LSLELTL | 158 | |
| AT2G38300 | 340 | LDLSLKL | 277 | |
| LSLSL | 305 | |||
| LDLTL | 338 | |||
| Homeobox family | ||||
| AT1G19700 | BLH10 | 538 | LSLSL | 82 |
| AT1G28420 | HB-1 | 1,705 | LKLDL | 1,005 |
| AT1G34650 | HDG10 | 708 | LSLPL | 540 |
| AT1G62990 | KNAT7 | 291 | LKLEL | 197 |
| AT1G75410 | BLH3 | 524 | LSLSL | 83 |
| AT1G79840 | GL2 | 747 | LSLSL | 24 |
| AT2G17950ac | WUS | 292 | LELRL | 289 |
| AT2G22800 | HAT9 | 274 | LVLGL | 13 |
| LTLCL | 42 | |||
| AT2G23760c | BLH4 | 627 | LSLSL | 156 |
| LTLGL | 604 | |||
| AT2G27220 | BLH5 | 431 | LSLGL | 394 |
| AT2G27990 | BLH8 | 584 | LTLEL | 553 |
| AT2G32370 | HDG3 | 725 | LALNL | 283 |
| AT2G35940 | BLH1 | 680 | LSLTL | 141 |
| LTLGL | 598 | |||
| AT2G44910 | ATHB4 | 318 | LGLSLSL | 10 |
| LRLNL | 26 | |||
| AT3G11260 | WOX5 | 182 | LDLRL | 177 |
| AT3G60390 | HAT3 | 315 | LGLSLSLSL | 10 |
| AT3G61150 | HDG1 | 808 | LSLGL | 55 |
| LDLAL | 320 | |||
| AT4G00730 | ANL2 | 570 | LSLAL | 67 |
| LSLCL | 567 | |||
| AT4G04890 | PDF2 | 743 | DLNLEP | 99 |
| AT4G16780bc | HAT4 | 284 | LGLSLGL | 10 |
| AT4G17460 | HAT1 | 282 | LGLSLSL | 10 |
| LQLNL | 24 | |||
| AT4G32980 | ATH1 | 473 | LSLSL | 156 |
| AT4G34610 | BLH6 | 532 | LSLSL | 98 |
| LTLGL | 472 | |||
| AT4G36870c | BLH2 | 739 | LSLSL | 242 |
| AT4G37790 | HAT22 | 278 | LVLGLGL | 13 |
| LTLSL | 48 | |||
| AT5G02030c | BLR | 575 | LSLSL | 117 |
| LTLGL | 517 | |||
| AT5G06710 | HAT14 | 336 | LALSL | 5 |
| LQLQL | 92 | |||
| AT5G17320 | HDG9 | 718 | LSLPL | 549 |
| AT5G47370c | HAT2 | 283 | LGLSLSL | 10 |
| HSF family | ||||
| AT1G46264 | AtHSFB4 | 348 | LALNL | 340 |
| AT4G13980bc | AtHSFA5 | 466 | LRLEL | 258 |
| LNLTL | 353 | |||
| AT4G18880 | AtHSFA4A | 401 | LALNL | 195 |
| AT5G45710 | AtHSFA4C | 345 | LSLNL | 192 |
| MADS family | ||||
| AT1G24260c | SEP3 | 251 | LRLRL | 173 |
| AT1G26310c | CAL | 255 | LDLTL | 241 |
| AT1G31140 | AGL63 | 213 | LKLNLQL | 107 |
| AT1G65360 | AGL23 | 226 | LELNL | 140 |
| AT1G69120c | AP1 | 256 | LELTL | 241 |
| AT2G03060 | AGL30 | 386 | LPLTL | 314 |
| AT2G14210 | ANR1 | 234 | LGLQL | 231 |
| AT2G22540c | SVP | 240 | LELQL | 86 |
| LRLGL | 234 | |||
| AT3G57390c | AGL18 | 256 | LQLGL | 226 |
| AT4G24540c | AGL24 | 220 | LKLGL | 214 |
| AT5G06500 | AGL96 | 242 | DLNMEP | 215 |
| AT5G10140c | FLC | 196 | LMLKL | 140 |
| AT5G13790ac | AGL15 | 268 | LQLGL | 213 |
| AT5G20240c | PI | 208 | LQLEL | 103 |
| AT5G23260 | TT16 | 252 | LELRL | 105 |
| AT5G26870 | AGL26 | 121 | LSLFL | 62 |
| AT5G26950 | AGL93 | 289 | LSLFL | 106 |
| AT5G27050 | AGL101 | 120 | LSLFL | 62 |
| AT5G27070 | AGL53 | 287 | LSLFL | 106 |
| AT5G27090 | AGL54 | 187 | LSLFL | 105 |
| AT5G27580 | AGL89 | 223 | LSLFL | 106 |
| AT5G27960 | AGL90 | 320 | LNLNLNL | 247 |
| AT5G48670 | AGL80 | 321 | LNLNL | 236 |
| MYB family | ||||
| AT1G22640 | AtMYB3 | 257 | LNLEL | 191 |
| AT2G16720a | AtMYB7 | 269 | LNLEL | 192 |
| AT2G23290 | AtMYB70 | 309 | LRLSL | 217 |
| AT1G35515c | HOS10 | 212 | LNLDLTL | 194 |
| AT2G37630c | AS1 | 367 | LELQL | 284 |
| AT3G13540 | AtMYB5 | 249 | LILRL | 90 |
| AT3G50060 | MYB77 | 301 | LSLSL | 207 |
| AT4G09460 | AtMYB6 | 236 | LNLDLTL | 199 |
| AT4G34990ac | AtMYB32 | 274 | LDLNLEL | 185 |
| AT4G38620ac | AtMYB4 | 282 | LNLEL | 200 |
| AT5G67300c | AtMYB44 | 305 | LSLSL | 196 |
| NAC family | ||||
| AT2G33480 | ANAC041 | 268 | DLNLTP | 221 |
| AT5G13180 | ANAC083 | 252 | DLNLLP | 217 |
| PHD family | ||||
| AT4G23860 | 452 | DLNSCP | 325 | |
| AT5G12400 | 1,595 | DLNCGP | 121 | |
| AT5G16680 | 1,290 | LELAL | 1,210 | |
| LSLSL | 1,265 | |||
| SBP family | ||||
| AT1G20980 | SPL14 | 1,035 | LDLNL | 83 |
| LSLQL | 390 | |||
| LPLEL | 461 | |||
| AT1G27360 | SPL11 | 393 | LCLKL | 127 |
| AT1G27370 | SPL10 | 396 | LCLKL | 127 |
| WRKY family | ||||
| AT1G30650 | WRKY14 | 430 | DLNINP | 53 |
| AT1G68150 | WRKY9 | 374 | LSLKL | 33 |
| LSLSL | 177 | |||
| AT4G31800 | WRKY18 | 310 | DLNTNP | 13 |
| AUX/IAA family | ||||
| AT1G04100abc | IAA10 | 261 | LDLALGL | 47 |
| AT1G04240abc | IAA3 | 189 | LRLGL | 14 |
| AT1G04250abc | IAA17 | 229 | LCLGL | 16 |
| LKLNL | 42 | |||
| AT1G04550abc | IAA12 | 239 | LELGLGLSL | 22 |
| AT1G15050abc | IAA34 | 185 | LGLSL | 65 |
| AT1G15580abc | IAA5 | 163 | LRLGL | 17 |
| AT1G51950abc | IAA18 | 267 | LELKL | 44 |
| AT1G52830abc | IAA6 | 189 | LRLGL | 15 |
| AT1G80390abc | IAA15 | 179 | LTLAL | 23 |
| AT2G01200abc | IAA32 | 143 | LGLSL | 34 |
| LALQL | 127 | |||
| AT2G22670abc | IAA8 | 321 | LRLGL | 56 |
| AT2G33310abc | IAA13 | 246 | LELGLGLSL | 16 |
| AT2G46990abc | IAA20 | 175 | LRLGL | 36 |
| AT3G04730abc | IAA16 | 236 | LRLGL | 11 |
| LKLNL | 40 | |||
| AT3G15540abc | IAA19 | 197 | LRLGL | 15 |
| AT3G16500abc | IAA26 | 269 | LELRL | 40 |
| AT3G17600abc | IAA31 | 158 | LSLSL | 31 |
| AT3G23030abc | IAA2 | 174 | LCLGL | 18 |
| AT3G23050abc | IAA7 | 243 | LCLGL | 15 |
| LMLNL | 46 | |||
| AT3G62100abc | IAA30 | 172 | LRLGL | 37 |
| AT4G14550abc | IAA14 | 228 | LCLGL | 10 |
| LKLNL | 41 | |||
| AT4G14560abc | IAA1 | 168 | LRLGL | 16 |
| AT4G28640abc | IAA11 | 246 | LELGLTLSL | 36 |
| AT4G29080abc | IAA27 | 305 | LRLGL | 47 |
| AT4G32280abc | IAA29 | 251 | LDLGLSL | 5 |
| AT5G25890abc | IAA28 | 175 | LELRL | 9 |
| AT5G43700abc | IAA4 | 186 | LRLGL | 20 |
| AT5G65670abc | IAA9 | 338 | LTLGL | 70 |
| JAZ family | ||||
| AT1G17380c | JAZ5 | 274 | DLNEPT | 156 |
| LDLRL | 272 | |||
| AT1G72450c | JAZ6 | 269 | DLNEPT | 162 |
| LELKL | 267 | |||
| AT1G30135c | JAZ8 | 131 | LELRL | 10 |
| AT2G34600c | JAZ7 | 148 | LELRL | 28 |
| NPR family | ||||
| AT1G02450ac | NIMIN1 | 142 | LDLNLAL | 138 |
| AT1G09415c | NIMIN3 | 112 | LDLNLSL | 108 |
| AT3G25882c | NIMIN2 | 122 | LGLDLNCKP | 107 |
| AT4G19660 | NPR4 | 574 | DLNETP | 467 |
| AT4G26120c | NPR2 | 600 | DLNMAP | 479 |
| AT5G45110c | NPR3 | 586 | DLNETP | 474 |
EAR motif-containing proteins that are previously validated in the literature as transcriptional repressors.
Evidence from transient expression assays supports these proteins functioning as transcriptional repressors.
Genetic evidence supports these proteins functioning as negative regulators of gene expression.
A comparison between this set of 219 proteins and the protein set identified by DPM analysis revealed that the DPM search data set has 179 additional transcriptional regulator proteins. A majority of these proteins contain a LxLxL type of EAR motif (Supplemental Table S4). The high-stringency parameters included in our HMM/PHI-BLAST analysis may have excluded these proteins from being retrieved. To rule out this possibility, we did an additional search, using the multiple expectation maximization for motif elicitation (MEME) method (Bailey and Elkan, 1995), for significantly overrepresented motifs within the protein sequences of the set of 179 additional proteins discovered by DPM analysis. Interestingly, the most highly overrepresented sequence identified by MEME output was similar to the consensus sequence of the EAR motif; however, the combined E-value of the output was too high to convincingly define the consensus sequence as not being a potential artifact (data not shown). Owing to this lack of statistical evidence, these 179 proteins were not included in the final list of EAR motif-containing transcription-related proteins (Table I) but are reported in Supplemental Table S4 to provide a complete list of possible candidates. It is worth noting that the bHLH class of transcription factors with 23 proteins is one of the most prevalent TF families in this protein set.
Classification of EAR Motif-Containing Proteins
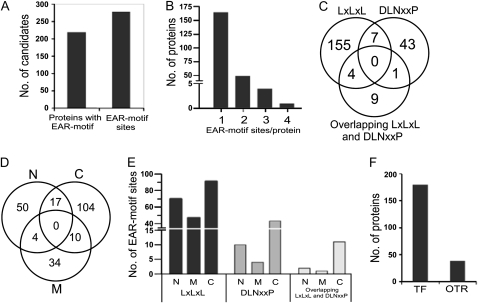
A list of Arabidopsis Genome Initiative (AGI) codes and the sequences of core EAR motif sites of all 219 proteins are presented in Table I. Additional information, such as protein name and description, protein identifiers, and location of EAR motif sites in each sequence, is listed in Supplemental Table S3. Upon careful inspection of the protein sequences of all 219 proteins, about 279 “high-confidence” (based on HMM/PHI-BLAST analysis) EAR motif sites were identified (Fig. 3A). The number of EAR motif sites found in each protein varies from one to four (Fig. 3B). Approximately 75% (165 out of 219) of the proteins contain only one EAR motif site, while the remaining 25% contain two (49 proteins), three (four proteins), or four (one protein) EAR motif sites. The EAR motif-containing proteins were further classified based on type of the motif: LxLxL (containing three or more Leu residues in alternate positions), DLNxxP, and an additional class containing overlapping (not mutually exclusive) LxLxL and DLNxxP motif sites. The LxLxL, DLNxxP, and overlapping LxLxL and DLNxxP type of EAR motif sites were found in 165, 51, and 14 proteins, respectively, of which 11 proteins contained at least two different types of EAR motifs (Fig. 3C). The EAR motif sites were mainly found in the C-terminal region (131 out of 219 proteins) and at slightly lower frequency in the N-terminal (71 proteins) and middle (48 proteins) regions (Fig. 3, D and E).
Figure 3.
Overview of the Arabidopsis EAR repressome. A, Total number of predicted EAR motif-containing proteins with an annotated transcription-related function, and incidence of EAR motif sites within this protein set. B, Distribution of EAR motif sites in transcription-related proteins. C, Incidence of EAR motif types in transcription-related proteins. The overlapping LxLxL and DLNxxP type includes mutually nonexclusive EAR motif sites. D, Distribution of EAR motif locations in transcription-related proteins (C, C terminal; M, middle; N, N terminal). E, Frequency of EAR motif sites in the N-terminal, C-terminal, or middle region of transcription-related proteins. F, Distribution of EAR motif-containing proteins as TF or OTR based on the presence or absence of annotated DNA-binding domains, respectively.
To obtain insight into the biological functions of the EAR motif-containing proteins, they were further classified according to families. A total of 180 out of 219 proteins contain distinct DNA-binding domains and were classified into 18 different TF families (Fig. 3F; Table I). The remaining 39 proteins do not possess a defined DNA-binding domain but are known in the literature to regulate transcription by interacting with TFs; hence, they were designated as OTRs and categorized into three distinct OTR families: AUX/IAA, JAZ (for Jasmonate; ZIM domain), and NPR. The families with the greatest number of EAR motif-containing proteins include C2H2 (56 proteins), HOMEOBOX (29 proteins), AUX/IAA (28 proteins), MADS (23 proteins), and ERF (17 proteins); the remaining families contain one to six proteins each (Table I). Proteins belonging to each of the TF families and OTR categories are discussed below as per the order of their appearance in Table I.
Transcription Factor Families
ABI3/VP1 Family
Three ABI3/VP1 family proteins, HSI2, HSI2-LIKE1 (HSL1), and HSL2, contain the DLNxxP type of EAR motif in the C-terminal region (Table I; Supplemental Table S3). These proteins contain plant-specific B3 DNA-binding domains similar to those in ABI3, LEAFY COTYLEDON2, and FUSCA3 and are believed to regulate the expression of seed maturation genes by binding to a Sph/RY element in their promoters and first introns (Suzuki et al., 2007). HSI2 and HSL1 are known to repress sugar-inducible expression of seed maturation genes during seedling growth (Tsukagoshi et al., 2005, 2007). Deletion or mutation of the EAR motif in HSI2 significantly reduces its in vitro transrepression activity, supporting that the EAR motif of HSI2 is an active repression domain (Tsukagoshi et al., 2005).
ERF Family
The EAR motif was first identified in members of class II ERFs such as AtERF3, AtERF4, and NtERF3 (Ohta et al., 2001). The ERF family in Arabidopsis contains 122 proteins that bind to the “ethylene-responsive element” or the GCC boxes found in the promoters of target genes. Protein sequences of eight members of this gene family, including AtERF3, AtERF4, and AtERF7 to AtERF12, which contain an EAR motif and act as active repressors (Fujimoto et al., 2000; Ohta et al., 2001; McGrath et al., 2005; Song et al., 2005; Yang et al., 2005), were included in the training set used to build the HMM profiles used in this study. The HMM analysis revealed an additional nine proteins belonging to the ERF family that contained EAR motif(s). In this collective set of 17 members of the ERF family, both the LxLxL and DLNxxP types of EAR motif are present at approximately the same frequency and located in the C-terminal region (Table I; Supplemental Table S3). Among the EAR motif-containing ERFs, AtERF4 is known to act as a negative regulator of jasmonate-ethylene-dependent expression of PLANT DEFENSIN1.2 (pDF1.2), possibly through EAR motif-mediated repression (Yang et al., 2005). Similarly, AtERF7 binds to GCC box-containing promoters and recruits AtSIN3 (Arabidopsis SIN3 homolog) and HDA19 to repress the expression of target genes such as pDF1.2 (Song et al., 2005). Another ERF family protein, DEAR1 (for dehydration-responsive element binding protein 1/C-repeat-binding factor), identified by our HMM analysis as containing a DLNxxP type of EAR motif, was recently shown to function as a repressor of dehydration-responsive element binding proteins that mediate biotic and abiotic stress responses (Tsutsui et al., 2009). Four other proteins, DEAR2, DEAR3, DEAR4, and DEAR5, structurally similar to DEAR1, also are predicted to contain a DLNxxP type of EAR motif in the C-terminal region (Table I).
AS2/LBD Family
Five proteins belonging to the ASYMMETRIC LEAVES2 (AS2) family (also known as LATERAL ORGAN BOUNDARIES DOMAIN [LBD] family) in Arabidopsis, namely AS2-Like36 (ASL36/LBD42), ASL37 (LBD40), ASL38 (LBD41), ASL39 (LBD37), and ASL40 (LBD38), were predicted to contain a LxLxL type of EAR motif in the C-terminal region (Table I; Supplemental Table S3). The AS2 family in Arabidopsis includes 42 proteins that are divided into two classes: class I consists of 35 proteins and class II consists of six proteins (Iwakawa et al., 2002). All five proteins that were predicted to contain an EAR motif belong to class II. The sixth member of class II, ASL41 (LBD39), lacks an EAR motif and is also weakly conserved in the AS2 domain compared with the other five proteins. Currently, there is no evidence supporting repressor activity for EAR motif-containing proteins of the AS2 family; however, similar to the ERF family EAR motif-containing proteins, these proteins are structurally very similar to each other but distinct from other members of the AS2 family (Iwakawa et al., 2002), suggesting that they may also have distinct roles, possibly in transcriptional repression.
bZIP Family
The basic leucine zipper (bZIP) is one of the largest and diverse dimerizing TF families present exclusively in eukaryotes (Riechmann et al., 2000; Fassler et al., 2002; Vinson et al., 2002; Deppmann et al., 2004). The bZIP TFs can bind to abscisic acid-responsive elements and regulate the expression of various genes involved in pathogen defense, light and stress signaling, seed maturation, and flower development (Jakoby et al., 2002). Arabidopsis contains 73 bZIP TFs, of which only three proteins (AtbZIP7, AtbZIP18, and AtbZIP52) were predicted to contain a LxLxL type of EAR motif occurring either in the C-terminal or middle region (Table I; Supplemental Table S3). Future studies are required to determine the role of the EAR motif in these proteins.
BZR Family
The BRASSINAZOLE-RESISTANT (BZR) is a small family of six proteins including BZR1, BES1 (for BRI1-EMS-SUPPRESSOR1), and BEH1 (for BES1 HOMOLOG1) to BEH4. Except for BEH2, all members of this family contain a LxLxL type of EAR motif (DLELTL) in the C-terminal region (Table I; Supplemental Table S3). BZR1, upon activation by brassinosteroids, acts as a transcriptional repressor of brassinosteroid biosynthesis genes to confer feedback inhibition of brassinosteroid biosynthesis (He et al., 2005). BZR1 shares close similarity with BES1 and the BEH proteins, and together these proteins are predicted to perform partially redundant functions in brassinosteroid signaling (Yin et al., 2005). Future studies aimed at understanding negative regulation of gene expression by BZR family proteins may identify a role for the EAR motif in brassinosteroid signaling.
C2C2 Family
The C2C2 family has four subfamilies: CO-like (for CONSTANS-like), DOF (for DNA binding with one finger), GATA, and YABBY. Two proteins, At2g21320 and At4g38960, belonging to the C2C2-CO-like subfamily, contain the DLNxxP type of EAR motif in the C-terminal region. One protein (At1g69570) of the C2C2-DOF subfamily contains a DLNxxP type of EAR motif in the N-terminal region (Table I; Supplemental Table S3). The potential of these proteins to act as repressors has not yet been reported.
C2H2 Family
The C2H2 family is the largest family of EAR motif-containing proteins, as a total of 56 proteins belonging to this family were predicted to contain EAR motif-like sequences (Table I; Supplemental Table S3). Some of these proteins contain more than one EAR motif site. As a result, 78 high-confidence EAR motif sites were identified in 56 proteins; 47 sites (60%) and 24 sites (31%) are LxLxL and DLNxxP type, respectively. The remaining seven sites (9%) are the overlapping LxLxL and DLNxxP type. The EAR motif sites are mainly located in the C-terminal region (59 sites) but are also identified in the N-terminal (14 sites) and middle (five sites) regions. For many of the C2H2 family EAR motif-containing proteins, results from in vitro and in planta assays support their function as transcriptional repressors (Ohta et al., 2001; Hiratsu et al., 2002, 2004; Payne et al., 2004; Sakamoto et al., 2004; Vogel et al., 2005; Ciftci-Yilmaz et al., 2007; Devaiah et al., 2007). The EAR motif of ZAT7, one of the EAR motif-containing C2H2-ZFP proteins, was shown to be directly involved in enhancing abiotic stress tolerance of Arabidopsis by mediating the physical interaction between ZAT7 and the stress response and defense-related TFs WRKY70 and HASTY (Ciftci-Yilmaz et al., 2007).
C3H Family
Three zinc finger (C3HC4-type RING finger) family proteins, At1g53010, At2g42030, and At3g58030, were identified as containing EAR motif-like sequences (Table I; Supplemental Table S3). At1g53010 contains a LxLxL type of EAR motif in the middle region, whereas At2g42030 and At5g58030 each contain overlapping LxLxL and DLNxxP type of EAR motifs in the N-terminal region. At present, these proteins have not been associated with any known repressor activity.
G2-Like Family
The G2 (for GOLDEN2)-like family proteins are members of the GARP (for GOLGI-ASSOCIATED RETROGRADE PROTEIN) superfamily of TFs. Out of 40 proteins belonging to this family, five proteins were identified as containing a LxLxL type of EAR motif sequence located in the C-terminal region (Table I; Supplemental Table S3). The repressor activity of these proteins is currently unknown.
EIL Family
The ETHYLENE-INSENSITIVE3-LIKE (EIL) family includes six proteins that are involved in the ethylene signal transduction pathway in Arabidopsis (Lee and Kim, 2003). Three of the proteins of this family (EIN3, EIL2, and At5g65100) contain the DLNxxP type of EAR motif in the C-terminal region (Table I; Supplemental Table S3). EIL2 contains an additional LxLxL type of motif in the C-terminal region. It has recently been shown that EIN3 represses the expression of SALICYLIC ACID INDUCTION DEFICIENT2 to negatively regulate innate plant immunity (Chen et al., 2009). The direct involvement of the EAR motif in mediation of this repression has not been reported.
Homeobox Family
Arabidopsis homeodomain-containing proteins can be classified into six families: homeodomain associated to a Leu zipper (HD-Zip), plant homeodomain associated to a finger domain (PHD finger), BEL1-like TALE homeodomain (BELL), zinc finger associated with a homeodomain, WUSCHEL-related homeobox (WOX), and Knotted-related homeobox (KNOX; Ariel et al., 2007). Our analysis identified 26 EAR motif-containing proteins that can be classified into four of these families/subfamilies, such as HD-Zip II (nine proteins), HD-Zip IV (seven proteins), WOX (two proteins), BELL (10 proteins), and KNOX (one protein; Table I; Supplemental Table S3). Except for PROTODERMAL FACTOR2 (PDF2), a member of the HD-Zip IV subfamily that contains a DLNxxP type EAR motif, all other proteins contain a LxLxL type of EAR motif (Table I; Supplemental Table S3). A total of 40 EAR motif sites, distributed in N-terminal (22 sites), C-terminal (11 sites), and middle (seven sites) regions, were identified in this group of proteins. The biological functions of several of these proteins are well known (Ariel et al., 2007). For example, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN2/HISTONE ACETYLTRANSFERASE4 (ATHB2/HAT4), one of the EAR motif-containing proteins belonging to the HD-Zip II family, has been described as a negative regulator of paralogous genes; it recognizes its own promoter region, and its endogenous expression is repressed when it is overexpressed in plants (Steindler et al., 1999; Ohgishi et al., 2001). Similarly, HAT2, another EAR motif-containing protein, also regulates its endogenous expression by negatively regulating its own promoter (Sawa et al., 2002). Two BELL family proteins, BEL1-LIKE HOMEODOMAIN2/SAWTOOTH1 (BLH2/SAW1) and BLH4/SAW2, contain an EAR motif and are involved in establishing leaf shape by repressing the expression of one or more KNOX genes (Kumar et al., 2007).
HSF Family
Arabidopsis contains 21 heat stress TFs (HSFs), which are key regulators of heat stress response (Nover et al., 2001). Our analysis revealed that four HSFs (AtHSFB4, AtHSFA5, AtHSFA4A, and AtHSFA4C) contain a LxLxL type of EAR motif in the middle or C-terminal region (Table I; Supplemental Table S3). One of these HSFs, AtHSFA5, specifically represses the transcriptional activation potential of AtHSFA4A and AtHSFA4C (Baniwal et al., 2007).
MADS Family
The MADS domain proteins form a large and diverse family of regulatory factors involved in various biological processes, including floral transition and floral organ identity determination. The MADS box gene family includes both transcriptional activators and repressors, which are known to form complex interaction networks among themselves as well as with various other regulatory factors such as corepressors (Sridhar et al., 2006), RNA-binding proteins (Pelaz et al., 2001), and posttranslation modifying factors (Fujita et al., 2003). A total of 23 proteins belonging to this family were identified as containing a LxLxL type of EAR motif in their sequences (Table I; Supplemental Table S3). Only one family member, AGL96, contained a DLNxxP type of motif. The EAR motifs in these proteins were found in either the middle or C-terminal region at about the same frequency. Many EAR motif-containing proteins belonging to this category have been implicated in negative regulation of floral transition (AGL15 and AGL18; Adamczyk et al., 2007) and also floral organ identity determination (Gregis et al., 2006). FLOWERING LOCUS C (FLC), the central regulator of flowering (Michaels et al., 2005), also contains an EAR motif in the middle region of the protein. The functions of the EAR motif in several of these proteins are not known except for AGL15, in which the EAR motif has been shown to mediate the recruitment of an HDAC complex to confer repression of target genes (Hill et al., 2008).
MYB Family
R2R3 MYB proteins are characterized by a highly conserved DNA-binding domain and a variable C-terminal domain that is thought to be responsible for the activation or repression of specific target genes (Jin and Martin, 1999). Our analysis has identified 11 members of this protein family containing a LxLxL type of EAR motif in the C-terminal region (Table I; Supplemental Table S3). One of these 11 proteins, AtMYB4, has been reported to act as a negative regulator of hydroxycinnamic acid metabolism, mainly by repressing genes involved in the phenylpropanoid pathway (Jin et al., 2000). Interestingly, the repression effect of AtMYB4 is attributed to the C-terminal region (Jin et al., 2000), which is where we detected an EAR motif, thus supporting the possibility that the repressor activity of AtMYB4 is mediated by the EAR motif. AtMYB4 belongs to subgroup IV of the R2R3 MYB family, which also includes the other EAR motif-containing proteins AtMYB3, AtMYB6, AtMYB7, and AtMYB32 (Kranz et al., 1998). Conservation of the EAR motif in these latter proteins supports the possibility that they may act similarly to AtMYB4 in negative regulation of phenylpropanoid pathway genes. Another EAR motif-containing R2R3 MYB protein, AS1, is known to form a protein complex with AS2 and repress important developmental genes, such as BREVIPEDICELLUS, KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 (KNAT2), KNAT6, FILAMENTOUS FLOWER, KANADI, PETAL LOSS, CUP-SHAPED COTYLEDON1 (CUC1), and CUC2 (Semiarti et al., 2001; Byrne et al., 2002; Lin et al., 2003; Xu et al., 2008). AtMYB8, also known as HOS10 (for HIGH RESPONSE TO OSMOTIC STRESS10), contains an EAR motif and is known to act as a negative regulator of stress-responsive genes (Zhu et al., 2005).
NAC, PHD, SBP, and WRKY Families
Two proteins belonging to the NAC (for NAM, ATAF1, and CUC2) family and three proteins each of the PHD (for plant homeodomain), SBP (for squamosa promoter binding), and WRKY families were identified as containing an EAR motif-like sequence (Table I; Supplemental Table S3). Although various members of the NAC, PHD, and WRKY families have been implicated in plant defense responses, the biological functions of proteins in these families containing an EAR motif are not known.
Other Transcriptional Regulator Families
The AUX/IAA, JAZ, and NPR families of proteins do not bind to DNA directly but interact with TF proteins such as ARFs (for auxin response factors), MYC2, and TGAs (for TGACG motif-binding factors; Guilfoyle et al., 1998; Despres et al., 2003; Chini et al., 2007), respectively, to mediate gene regulation; hence, they are grouped into a separate category as OTRs.
AUX/IAA Family
AUX/IAA proteins are auxin-responsive nuclear proteins involved in the repression of early auxin-responsive genes by dimerizing with ARF proteins, transcriptional activators of auxin-responsive genes (Tiwari et al., 2001, 2004). Arabidopsis contains 29 AUX/IAA proteins, such as IAA1 to IAA20 and IAA26 to IAA34 (Liscum and Reed, 2002). Except for IAA33, which does not contain an EAR motif, the remaining 28 proteins contain a LxLxL type of EAR motif in the N-terminal region (Table I; Supplemental Table S3; Tiwari et al., 2004). Most AUX/IAA proteins contain four conserved domains referred to as domains I, II, III, and IV. The EAR-repression motif in all AUX/IAA proteins is located in domain I (Tiwari et al., 2004), which has been shown to function as an active repression domain (Ulmasov et al., 1997; Tiwari et al., 2001). The Leu residues within the EAR motif in domain I were identified to be important for the repression activity (Tiwari et al., 2004), consistent with the involvement of the EAR motifs in the repression activity of AUX/IAAs. A recent study has demonstrated that the EAR motif in IAA12 mediates physical interaction between IAA12 and TPL, a corepressor that functions in auxin-dependent transcriptional repression during Arabidopsis embryogenesis, thus confirming an in planta role of the EAR motif in transcriptional repression (Szemenyei et al., 2008).
Interestingly, the AUX/IAA proteins in Physcomitrella patens (a moss) lack the typical LxLxL type of EAR motif in domain I; instead, they are proposed to contain a similar LxLxPP motif (Paponov et al., 2009). Notably, the LxLxPP motif also occurs, either exclusively or in addition to the LxLxL type of EAR motif, in three Arabidopsis AUX/IAA proteins (IAA18, IAA26, and IAA28), in AUX/IAAs from other flowering plants such as poplar (six), grapevine (two), rice (three), and sorghum (three), as well as in an annotated AUX/IAA from a primitive nonflowering plant (Selaginella moellendorffii; Paponov et al., 2009). The function of the LxLxPP motif is currently unknown. It has been proposed to function as a putative repression domain similar to the LxLxL type of EAR motif (Paponov et al., 2009); however, experimental evidence is currently lacking.
JAZ Family
JAZ family proteins are a new group of regulators involved in jasmonic acid (JA) signaling and are known to repress the transcription of JA-responsive genes by directly binding to their transcriptional activator, MYC2 (Chini et al., 2007; Chico et al., 2008). There are 12 members of the JAZ family in Arabidopsis (JAZ1–JAZ12; Katsir et al., 2008). Our analysis indicated that four of these 12 proteins, namely JAZ5, JAZ6, JAZ7, and JAZ8, contain EAR motif sequences (Table I; Supplemental Table S3). JAZ5 and JAZ6 contain both LxLxL and DLNxxP types of EAR motif; the LxLxL motif is located in the C-terminal region, whereas the DLNxxP motif is located in the middle region. JAZ7 and JAZ8 contain a LxLxL type of EAR motif in the N-terminal region (Table I; Supplemental Table S3). The repressor activity of the EAR motifs found in JAZ proteins is currently not known; however, the functional similarity between JAZ proteins and AUX/IAAs supports the possibility that the EAR motif present in a subset of JAZ proteins is responsible for their repression activity. Further research is required to test this possibility.
NPR Family
The Arabidopsis NPR1 protein is an essential regulator of salicylic acid signaling. There are six NPR1-related proteins in the Arabidopsis genome, NPR1 to NPR6. Two of these proteins, NPR3 and NPR4, have been proposed to function as negative regulators of PR gene expression by directly interacting with transcriptional activators of PR1, such as TGA2, TGA3, TGA5, and TGA6 (Zhang et al., 2006). Interestingly, both NPR3 and NPR4 contain a DLNxxP type of EAR motif in their C-terminal regions (Table I; Supplemental Table S3). Whether the repression activity of NPR3 and NPR4 is facilitated by the EAR motif is not yet known.
Another group of pathogen response proteins known as NIMIN proteins (NIMIN1, NIMIN2, and NIMIN3) is involved in negative regulation of NPR1-mediated PR gene expression (Weigel et al., 2001, 2005). Our analysis indicates that all three of these proteins contain a LxLxL type of EAR motif in the C-terminal region (Table I; Supplemental Table S3). It has been proposed that NIMIN1 decreases transcriptional activation of the PR1 gene, possibly through its EAR motif (Weigel et al., 2005). Further studies are required to evaluate a link between the EAR motif and negative regulatory mechanisms of the NPR family of proteins.
Protein Sequence Context around EAR Motifs
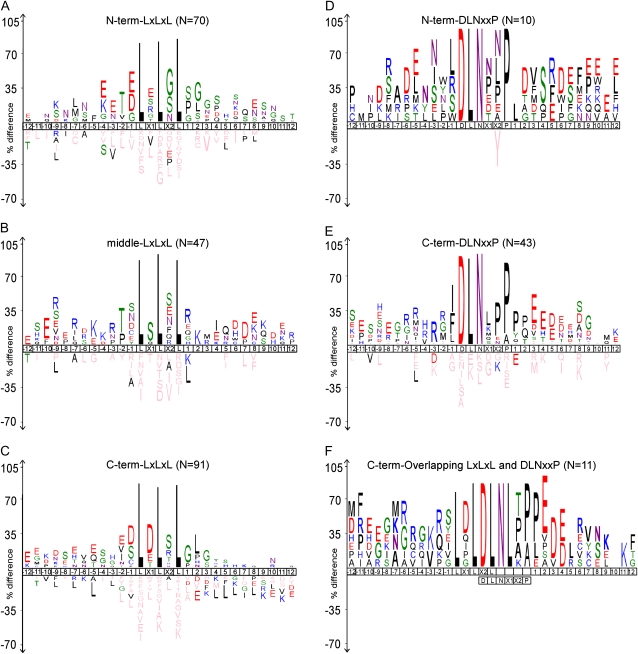
From the current literature, it is evident that the EAR motif functions as a “potential repressor” by physically interacting with other corepressors (Hill et al., 2008; Szemenyei et al., 2008). The protein sequence context in which the EAR motif appears may influence its ability to interact with potential interacting proteins. We analyzed the local sequence context within and around the EAR motif sites by comparing a protein sequence region of approximately 30 amino acids, comprising the core EAR motif sites ± 12 amino acids, in all 219 proteins of our data set, which was divided into nine groups based on type (LxLxL, DLNxxP, or overlapping LxLxL and DLNxxP) and location (N-terminal, C-terminal, or middle region) of the EAR motif(s). As shown in Figure 4A, among the 70 N-terminal LxLxL types of EAR motif sites, acidic amino acids are more enriched at X1, −1, −3, −4, and +8 positions. Similarly, Gly or polar residues such as Ser and Gln are more common at the X2 position and in sequences downstream of the core EAR motif site. Interestingly, Thr is a preferred residue at the −2 position both in N-terminal and middle region LxLxL type of EAR motif sites. Notably, mutation of Thr at the −2 position to Ala in IAA17 results in the reduction of its repression activity (Tiwari et al., 2004), supporting the influence of flanking residues on the function of the EAR motif.
Figure 4.
Amino acid sequence context of EAR motif-containing transcriptional regulator proteins in Arabidopsis. The core EAR motif site and flanking ± 12 amino acid sequences of N-terminal LxLxL (A), middle LxLxL (B), C-terminal LxLxL (C), N-terminal DLNxxP (D), C-terminal DLNxxP (E), and C-terminal overlapping LxLxL and DLNxxP (F) types of EAR motif-containing proteins were used for generating IceLogos using the IceLogo program (Colaert et al., 2009). The number of sequences containing the EAR motif site (N) in each category is indicated. The IceLogos for middle DLNxxP and N- and C-terminal overlapping LxLxL and DLNxxP categories of EAR motifs were not generated, as the number of sequences in each of these categories is too small (less than four) for IceLogo determination of statistical significance. Only significant amino acids (P < 0.05) are shown. The P value of each amino acid at every position was calculated by testing its frequency in the experimental set comprising the 219 high-confidence EAR motif-containing proteins against a reference set comprising the same number of random Arabidopsis proteins. Single letter codes for each amino acid are color coded according to their chemical properties as follows: black (hydrophobic residues), green and pink (polar residues), blue (residues with positive charge), red (residues with negative charge), and light pink (residues that do not occur in both experimental and reference sets). The height of the letter representing an amino acid in each position reflects the difference in the frequency of its occurrence in the experimental and reference sets.
The X1, X2, and −1 positions in the middle region LxLxL type of EAR motif sites are occupied more frequently by Ser (Fig. 4B). Immediately downstream of the EAR motif is a preference for the positively charged amino acids Arg and Lys at +1 and +2 positions, with subsequent positions principally occupied by acidic residues. In the region upstream of this class of EAR motif site can be seen a preference for charged residues (Arg at −3, −7, and −9 positions and Lys at −4 and −5), with other sites occupied by acidic (Asp or Glu) or polar (Ser) residues (Fig. 4B).
Approximately 45% of the LxLxL types of EAR motif sites occur in the C-terminal region of proteins. Among these, a preference for acidic residues in many positions within and around the core EAR motif sites can be observed (Fig. 4C). For example, while Asp residue is more abundant in X1, −1, and −9 positions, Glu residues are enriched in −2, −5, −7, −11, and −12 positions. Ser residues appear more frequently at X2, −4, and −8 positions.
The DLNxxP type of EAR motif sites are mainly found in the C-terminal (43 out of total 57 sites; Fig. 4E) and N-terminal (10 out of total 57 sites; Fig. 4D) regions, with only four instances in the middle region (data not shown). Leu and Pro are the most preferred residues within and in close vicinity of these EAR motif sites. Adjoining sequences again show a preference for charged amino acids (Glu, Asp, and Arg) or polar residues (Ser; Fig. 4, D and E). The overlapping LxLxL and DLNxxP type of EAR motif sites are mainly found in the C-terminal region and also exhibit an enrichment of acidic, polar, or charged residues within and around the EAR motif sites (Fig. 4F).
While no absolute pattern of amino acid residue conservation could be distinguished between the regions where the EAR motif was located (N-terminal, C-terminal, or middle region) or between the motif types, it can be observed that there is an overall high incidence of charged and polar residues surrounding the motif. Given the protein-protein interactions known for EAR motif-containing proteins (Ciftci-Yilmaz et al., 2007; Hill et al., 2008; Szemenyei et al., 2008), this sequence context may be important for exposure of the EAR motif and possible physical contact with interacting partners. Additionally, residues with known roles in posttranslational modifications, such as Ser, Thr, and Lys (for review, see Seo and Lee, 2004), also occur more commonly proximal or integral with EAR motifs, suggesting a potential role for these residues in regulating the repressor activity of the EAR motif via phosphorylation, acetylation, methylation, or glycosylation. The possible influence of phosphorylation on the function of the EAR motif is alluded to by the essentiality of an adjoining Thr residue (Tiwari et al., 2004). This possibility is further supported by results from a large-scale analysis of the Arabidopsis phosphoproteome (PhosPhAt 3.0 database; Heazlewood et al., 2008). We used this data set to analyze the phosphorylation status of the Ser and Thr residues within and around the EAR motif sites. Interestingly, Ser and Thr residues within the EAR motif sites and flanking regions of five different proteins, IAA9 (At5G65670), ERF10 (At1g03800), BEH4 (At1g78700), DEAR4 (At4g36900), and a C2H2 family member (At5g05120), were identified by mass spectrometry as being phosphorylated (Table II). These proteins include examples of both LxLxL and DLNxxP types of EAR motifs, with four examples of TFs and one OTR (IAA9) that does not encode a DNA-binding domain. Thus, it appears that phosphorylation may have a role in regulating the function of EAR motifs and may possibly provide another level of regulation controlling gene repression.
Table II. Phosphorylation status of Ser and Thr residues integral and adjacent to the EAR motif in Arabidopsis EAR motif-containing proteins.
The phosphorylated peptide sequences are annotated using the modification nomenclature outlined in the publication guidelines of Molecular & Cellular Proteomics. An experimentally confirmed phosphorylation site is indicated in boldface letters with a lowercase p and is contained in parentheses: (pS). pST sites wherein which residue(s) within the peptide is phosphorylated remains ambiguous are presented as boldface lowercase letters, and each potential candidate (not necessarily all S and T residues) is contained in parentheses: (s) or (t). The sequence of the core EAR motif site is underlined.
| Protein Name | Position of the EAR Motif | Modified Peptide Sequence |
| IAA9 (At5g65670) | N terminal | A(t)EL(t)LGLPG(s)Q(s)PAR |
| ERF10 (At1g03800) | N terminal | MEPDLDLNA(pS)P |
| BEH4 (At1g78700) | C terminal | IHEE(pS)GSDDLELTLGN(s)(s)(t)R |
| DEAR4 (At4g36900) | C terminal | VDLNKLPDPET(pS)DDD |
| C2H2 family protein (At5g05120) | C terminal | ENDG(pS)(pS)LSLSLK |
Comparative Analysis of Putative Orthologs of Arabidopsis EAR Motif-Containing Proteins
The comparison of sequences of novel functional domains/motifs across multiple orthologous proteins is an approach validated in the literature for predicting protein functions based on evolutionary conservation. With the availability of genome sequences of many plant species, comparative genomics/proteomics approaches have become more feasible. The rice genome annotation project team (Yuan et al., 2003) has compiled a list of putative orthologous groups from five different plant species for which genome sequences are available: Arabidopsis, rice, poplar, grapevine, and sorghum. These lists are made available through a Web resource (http://rice.plantbiology.msu.edu/ortholog/index.shtml; Yuan et al., 2003). We utilized this resource to search for putative orthologs of Arabidopsis EAR motif-containing proteins in the other four plant species in order to develop additional evidence supporting the authenticity of the EAR motifs we identified based on their evolutionary conservation.
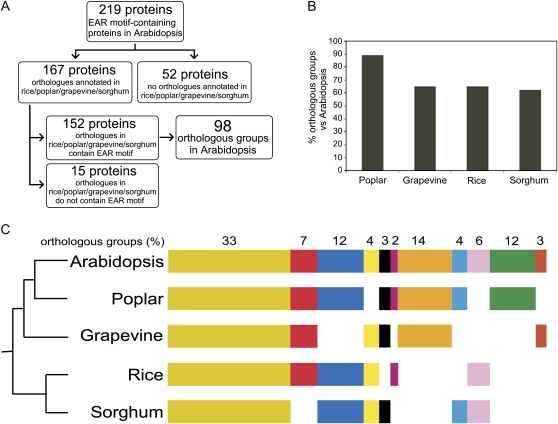
Putative orthologs for 167 of the 219 Arabidopsis EAR motif-containing proteins were identified in rice, poplar, grapevine, and/or sorghum (Fig. 5A; Supplemental Table S5). For the remaining 52 proteins, no orthologs have yet been annotated in these plant species; however, more complete proteome annotation in the future may enable this number to be reduced. The orthologs of 152 out of the 167 Arabidopsis EAR motif-containing proteins contain conserved sequences that are similar to the EAR motif in the corresponding Arabidopsis counterparts at relatively similar positions within the protein (Fig. 5A; Supplemental Table S5). We aligned the EAR motifs of orthologous proteins of each orthologous group and found the composition of LxLxL and DLNxxP to be highly conserved in the majority of orthologous sets (Supplemental Table S5). The adjacent residues (x) in most cases were found to be either fully conserved or replaced by residues with similar properties. The orthologs of the remaining 15 proteins from the initial set of 167 do not contain an EAR motif-like sequence. The 152 protein set from Arabidopsis and their corresponding orthologs in rice, poplar, grapevine, and/or sorghum were grouped into 98 orthologous groups (Supplemental Table S5) as per the orthoMCL algorithm (Li et al., 2003). Each orthologous group contains one to many paralogs (within each plant species) and orthologs (between plant species).
Figure 5.
Conservation of Arabidopsis EAR motif-containing proteins in rice, poplar, grapevine, and sorghum. A, Flow chart showing the classification of Arabidopsis EAR motif-containing proteins based on the presence or absence of EAR motif-containing orthologs in rice, poplar, grapevine, and sorghum. The EAR motif-containing proteins from Arabidopsis and their putative orthologs in other plant species were grouped into orthologous groups using the plant orthologs database of the rice genome annotation project (Yuan et al., 2003). B, Similarity between Arabidopsis and other plant species as indicated in terms of orthologous groups shared by their EAR motif-containing proteins. C, Clustering of orthologous groups. The orthologous groups were clustered based on the presence or absence of orthologs to Arabidopsis EAR motif-containing proteins in rice, poplar, grapevine, and sorghum. Each cluster is assigned a color. For example, the dark yellow cluster indicates the presence of EAR motif-containing orthologs to Arabidopsis EAR motif-containing proteins in all four plant species. Similarly, the blue cluster indicates the presence of EAR motif-containing Arabidopsis orthologs only in rice, poplar, and sorghum but not in grapevine. The pictogram representing the evolutionary level of genome conservation among all five plant species was adapted from Yang et al. (2009). Additional information about clustering of orthologous groups and locus identifiers of proteins in each orthologous group is provided in Supplemental Table S5.
The similarity between Arabidopsis and other plant species in terms of orthologous groups shared by their EAR motif-containing proteins was assessed. There is approximately 90% similarity between Arabidopsis and poplar, and approximately 60% similarity between Arabidopsis and grapevine, rice, and sorghum (Fig. 5B). The higher similarity between Arabidopsis and poplar may be due to their closer genomic relatedness compared with other plant species (Yang et al., 2009). In 33% of the orthologous groups, the EAR motif is conserved across all five plant species (Fig. 5C), whereas conservation in only four or only three plant species is observed in 26% of the orthologous groups, and conservation in only two plant species is observed in 15% of the orthologous groups (Fig. 5C; Supplemental Table S5). To assess the validity of the observed evolutionary conservation of EAR motifs, we conducted a mock analysis following the same approach but using a “mock” data set of 219 Arabidopsis proteins randomly picked from the set of false positives identified by DPM analysis; the mock analysis was performed using the rice proteome. Rice orthologs have been annotated for 141 of the Arabidopsis mock set and for 116 of the 219 Arabidopsis high-confidence EAR motif-containing proteins set. We analyzed the protein sequences of the orthologs and determined that only 33 of 141 rice orthologs in the mock set versus 102 of 116 rice orthologs of the Arabidopsis high-confidence set contained conserved EAR motif sequences. This mock analysis supports that the EAR motifs identified in rice orthologs of the Arabidopsis high-confidence set represent authentic evolutionary conservation of protein sequence. These results collectively demonstrate that the EAR motif is highly conserved across diverse plant species and support that the EAR motif identified in these proteins possesses functional relevance retained during evolution.
Functional Categorization of EAR Motif-Containing Proteins
A GO term enrichment analysis was performed to characterize the predicted functions of the EAR motif-containing proteins. As expected, the proteins associated with DNA binding, transcription factor activity, and transcriptional repressor activity were significantly enriched in the protein set representing the Arabidopsis EAR repressome (Table III). The proteins associated with plant developmental processes including tissue, organ, reproductive, and postembryonic development, abiotic stresses including osmotic and salt stress, and hormonal stimuli including auxin, abscisic acid, gibberellic acid, ethylene, salicylic acid, and JA also occurred significantly more frequently in the EAR motif-containing proteins set (Table III), suggesting a functional role for EAR motif-containing proteins in these biological processes.
Table III. GO term enrichment in EAR motif-containing proteins of Arabidopsis.
| GO Identifiera | Biological Process or Molecular Function | Percentage of Total Query Proteins (n = 208)b | Percentage of Total Reference Proteins (n = 22,393)c | FDR P Valued |
| GO:0003677 | DNA binding | 90.9 | 9.9 | 5.85E−161 |
| GO:0003700 | Transcription factor activity | 88.9 | 7.4 | 1.03E−174 |
| GO:0016564 | Transcription repressor activity | 6.7 | 0.2 | 1.98E−15 |
| GO:0032502 | Developmental processes | 25.5 | 8.2 | 1.78E−12 |
| GO:0048513 | Organ development | 17.8 | 2.8 | 1.38E−17 |
| GO:0009888 | Tissue development | 7.7 | 1.1 | 6.29E−08 |
| GO:0003006 | Reproductive developmental processes | 11.1 | 3.3 | 7.45E−06 |
| GO:0048869 | Cellular developmental processes | 8.7 | 1.9 | 2.36E−06 |
| GO:0009791 | Postembryonic development | 14.4 | 4.0 | 3.19E−08 |
| GO:0009628 | Response to abiotic stimulus | 15.4 | 5.1 | 4.45E−07 |
| GO:0006970 | Response to osmotic stress | 6.7 | 1.7 | 0.000206 |
| GO:0009651 | Response to salt stress | 6.7 | 1.6 | 0.000104 |
| GO:0009725 | Response to hormone stimulus | 26.9 | 3.3 | 2.27E−32 |
| GO:0009733 | Response to auxin stimulus | 17.8 | 1.3 | 1.43E−28 |
| GO:0009737 | Response to abscisic acid stimulus | 5.3 | 1.2 | 0.000382 |
| GO:0009739 | Response to gibberellin stimulus | 4.8 | 0.5 | 3.94E−06 |
| GO:0009723 | Response to ethylene stimulus | 6.7 | 0.6 | 3.85E−09 |
| GO:0009751 | Response to salicylic acid stimulus | 4.8 | 0.6 | 7.45E−06 |
| GO:0009753 | Response to JA stimulus | 5.8 | 0.7 | 4.45E−07 |
GO categories significantly enriched in the set of EAR motif-containing proteins as identified using the hypergeometric P value method are presented.
Total number of GO-annotated proteins from the set of 219 EAR motif-containing proteins identified by HMM/PHI-BLAST analysis.
Total number of GO-annotated proteins in the reference list.
FDR for multiple comparisons to detect overrepresented GO terms-adjusted P values obtained by Fisher's exact test.
To gain further insight into the possible influence of EAR motif-containing proteins on gene expression, an in silico transcriptomic analysis was performed; the results of the analysis are summarized in Supplemental Figures S1, S2, and S3, and the differential expression patterns of individual genes encoding the EAR motif-containing proteins are presented in Supplemental Tables S7, S8, and S9. The in silico analysis revealed tissue-, stress-, hormone-, and time-specific expression patterns of genes encoding EAR motif-containing proteins. For example, a majority of the genes encoding EAR motif-containing proteins belonging to the ERF and C2H2 families were induced in roots, hypocotyls, and the late stages of seed development (Supplemental Table S7). In contrast, a majority of genes encoding EAR motif-containing proteins belonging to the AUX/IAA, homeobox, NPR, and ERF families were repressed in shoot apices during transition to flowering and inflorescence stages as well as in stamens, sepals, petals, and several seed developmental stages. Consistent with their roles in stress and defense responses (McGrath et al., 2005; Yang et al., 2005; Ciftci-Yilmaz and Mittler, 2008), the genes encoding EAR motif-containing proteins of the ERF and C2H2 families responded to various abiotic stresses including cold, osmotic, salt, and drought and the plant stress hormone abscisic acid (Supplemental Tables S8 and S9). Many genes encoding EAR motif-containing AUX/IAA and JAZ proteins were induced by auxin and JA, respectively. Overall, this analysis is supportive of a role for EAR motif-containing proteins participating in the regulation of various plant growth and developmental processes as well as plant responses to various environmental stresses and hormonal signals.
DISCUSSION
In this study, we have shown through genome-wide analysis that the EAR motif is conserved in a large number of transcriptional regulator proteins in plants. The repertoire of EAR motif-containing transcriptional regulators in Arabidopsis comprises 219 proteins belonging to 18 different TF families and three OTR families (Table I; Supplemental Table S3). These proteins represent approximately 12% (219 of 1,770; AGRIS AtTFDB database) of all predicted transcriptional regulator proteins in Arabidopsis. TFs in Arabidopsis are grouped into 41 different families (Davuluri et al., 2003). The distribution of EAR motif-containing proteins across 21 (18 TF plus three OTR) transcriptional regulator families indicates the vast diversity of functions the EAR motif may have in regulating gene expression. Approximately 23% of the 219 EAR motif-containing proteins have previously been reported to function as transcriptional repressors in transient expression assays. Similarly, genetic evidence suggests that approximately 31% of the 219 proteins function as negative regulators of gene expression in various biological processes (individual references are listed in Supplemental Table S3). In addition, evidence for direct involvement of the EAR motif in conferring repression ability to transcriptional regulators also exists in the literature (Hill et al., 2008; Szemenyei et al., 2008).
Our analysis has helped identify several sets of proteins that are thought to play crucial roles in plant development as well as plant responses to environmental and hormonal stimuli. An impressive number of novel EAR motif-containing proteins identified in this study belong to the homeobox (29 proteins) and MADS (23 proteins) families (Table I), which are known to be exclusively involved in plant developmental processes (for review, see Nam et al., 2003; Ariel et al., 2007). Interestingly, four MADS box proteins, APETALA1 (AP1), SEPALLATA3 (SEP3), AGL24, and SHORT VEGETATIVE PHASE (SVP), and one homeobox protein, BELLRINGER (BLR), which are known to function in outer whorl-specific transcriptional repression of AGAMOUS (AG; Bao et al., 2004; Gregis et al., 2006; Sridhar et al., 2006), contain an EAR motif sequence (Table I). AG is a floral homeotic gene involved in specification of the inner two whorls of a flower (Yanofsky et al., 1990; Bowman et al., 1991; Drews et al., 1991). The MADS family repressor proteins of AG are implicated in the recruitment of SEUSS, a corepressor known to form a repression complex with LEUNIG and HDA19, to regulatory regions of AG (Sridhar et al., 2004, 2006; Gregis et al., 2006; Gonzalez et al., 2007). It remains to be seen if the EAR motif in these MADS proteins are directly or indirectly involved in recruiting the SEUSS-LEUNIG-HDA19 complex to AG.
Several other novel EAR motif-containing candidate transcriptional regulators identified in this study belonging to various families are known to play key roles in various developmental processes such as petal development (RABBIT EARS; Krizek et al., 2006), cellular proliferation and development of basal patterns in gynoecium (KNUCKLES; Payne et al., 2004), shoot maturation (GLABROUS INFLORESCENCE STEMS [GIS]; Gan et al., 2006), regulation of epidermal cell fate (GIS, GIS2, and ZFP8; Gan et al., 2007), floral transition (AGL15 and AGL18; Adamczyk et al., 2007), stem cell fate determination in shoot apical meristem (WUSCHEL; Leibfried et al., 2005), and leaf tissue, stamen, and carpel development (JAGGED and NUBBIN; Ohno et al., 2004; Dinneny et al., 2006). The identification of a function for the EAR motifs in these proteins will enhance our understanding of the possible role of EAR motif-mediated gene repression in developmental processes.
The spatial and temporal regulation of gene expression in response to stress and hormonal signals is achieved through a multidimensional network of transcriptional activation and repression mechanisms. In nature, plants encounter a combination of abiotic and biotic stresses to which they respond by modulating their gene expression. A dynamic system using antagonistic mechanisms should exist in plants to induce the expression of defense genes following the onset of stress and to completely or partially suppress their expression in the absence of stress. Interestingly, about 33% of the Arabidopsis EAR repressome is composed of class II ERFs and C2H2-ZFPs (Table I). The class II ERFs and C2H2-ZFPs function as key transcriptional repressors involved in stress acclimation responses of plants (Ciftci-Yilmaz and Mittler, 2008), thus indicating a broad role for EAR-mediated modulation of the expression of abiotic and biotic stress response genes. Hormonal responses in plants are regulated by negative feedback loops to avoid harmful runaway responses (Kazan, 2006). The EAR motif-containing AUX/IAAs act as negative regulators of auxin signaling in an auxin concentration-dependent manner (Tiwari et al., 2004). Auxin and JA signaling modules are strikingly similar. In JA signaling, a set of JAZ proteins act as active repressors of JA-responsive genes (Chini et al., 2007). Interestingly, four of the JAZ proteins, JAZ5 to JAZ8, contain an EAR motif sequence. Thus, it is possible that the EAR motif in JAZ proteins functions in their repression activity similar to the EAR motif in AUX/IAAs. BZR family proteins also contain an EAR motif (Table I). At least one of these proteins, BZR1, is known to play a role in BR signaling by negatively regulating BR biosynthesis genes (He et al., 2005). These observations collectively support an essential role for the EAR motif in regulating hormonal responses and maintenance of their homeostasis in plants.
Collectively, the published literature on EAR motif-containing proteins supports this class of proteins as having transcriptional repression functions that are utilized by plants to respond to various developmental, stress, and hormonal signals. The specific as well as overlapping expression patterns of the EAR motif-containing proteins at one or more stages of development, and also in response to one or more stress and hormonal signals elucidated in our in silico transcriptomic analysis (Supplemental Tables S7–S9), further support a role for EAR motif-containing proteins in diverse developmental and physiological functions, possibly by acting as transcriptional repressors. From a developmental perspective, the spatiotemporal expression patterns of EAR motif-containing proteins may be important in the demarcation of expression boundaries for the potential target genes to allow proper organ development and facilitate smooth transitioning to the next developmental stage. For example, genetic and molecular evidence indicates that SUP, an authentic EAR motif-containing repressor (Hiratsu et al., 2002, 2004), functions in establishing a boundary between stamen and carpel development by repressing the activity of AP3 and PISTILLATA in the carpel (Sakai et al., 1995). In addition, a role for EAR motif-containing proteins in physiological responses is supported by studies wherein ectopic constitutive expression of C2H2 family EAR motif-containing proteins, such as ZAT7, ZAT10, and ZAT12, resulted in enhanced tolerance of transgenic plants to abiotic stresses (Rizhsky et al., 2004; Sakamoto et al., 2004; Davletova et al., 2005; Vogel et al., 2005; Mittler et al., 2006). Similarly, the involvement of EAR motif-containing proteins in hormone signaling is supported by the involvement of ERF proteins, AtERF4 and AtERF7, in negative regulation of ethylene, JA, and abscisic acid responses (McGrath et al., 2005; Yang et al., 2005). The differential expression of EAR motif-containing proteins in response to stress and hormone signals may, at least in part, influence the effectiveness by which plants can respond to these signals.
Despite a pivotal role in several biological processes, the precise mechanism by which EAR motifs function has not yet been elucidated. Several recent publications provide insight into possible repression mechanisms utilized by EAR repressors. The recent discovery of a direct physical interaction between the EAR motif of IAA12 and TPL (Szemenyei et al., 2008) and the genetic interaction between the mutants of TPL and HDA19 (Long et al., 2006) suggests a possible link between the EAR motif and chromatin-remodeling factors. Negative regulation of gene expression by AGL15 is also achieved by a similar mechanism wherein AGL15 recruits the SIN3/HDAC complex by interacting with an adapter protein known as SIN3-ASSOCIATED POLYPEPTIDE P18 (SAP18; Hill et al., 2008). The EAR motif in AGL15 facilitates its interaction with SAP18, thus providing additional evidence for the involvement of an HDAC complex in the EAR-mediated repression of genes. AtERF7 is also known to recruit the SIN3/HDAC complex by directly interacting with SIN3; however, the direct involvement of the EAR motif of AtERF7 in mediating this interaction has not been determined (Song et al., 2005). These findings collectively support a model for EAR motif-mediated repression via chromatin modification through the recruitment and actions of chromatin-remodeling factors such as histone deacetylases. Indeed, transcriptional repression by chromatin modification is a frequently used mechanism by many active repressors in other eukaryotes (Thiel et al., 2004).
Although the minimal repression unit of the EAR motif is defined as a short peptide of five to six amino acids, the sequence context in the region surrounding the EAR motif may influence its ability to interact with a corepressor or other potential interactors. The analysis of sequences adjoining the EAR motif sites of all EAR motif-containing proteins in Arabidopsis revealed an enrichment of charged and polar residues (Fig. 4). Similarly, amino acids known to be involved in posttranslational modification were also more abundant (Fig. 4). These observations suggest two possibilities: (1) the high incidence of charged and polar residues in the flanking region may aid in exposing the EAR motif to potential interactors such as corepressors; and (2) the residues with known roles in posttranslational modifications may potentially undergo phosphorylation, acetylation, methylation, or glycosylation, which may in turn lead to either enhancement or relief of the suppression effects of the EAR motif, depending upon whether the actions of the EAR motifs are required or not. Interestingly, residues adjacent and integral to the EAR motif have been detected as being phosphorylated in five proteins (Table II; Heazlewood et al., 2008), supporting the possibility of posttranslational regulation of EAR motif function. Future research in this area will provide insight to a novel element of gene regulation networks.
A novel gene-silencing technology exploiting the EAR motif has been developed for the conversion of transcriptional activators into dominant repressors by fusing them with a slightly modified EAR motif repression domain of SUP (LDLDLELRLGFA). This simple yet effective repressor technology has a unique capability to overcome the challenges of gene redundancy in plants and is designated CRES-T (for Chimeric Repressor Silencing Technology; Hiratsu et al., 2003). This system has been successfully employed in the conversion of approximately 25 transcriptional activators into dominant repressors (Supplemental Table S10) and in the generation of a CRES-T mutant library for 178 TFs in the families of AP2, ARF, HD, MADS, MYB, NAC, and TCP (Kunieda et al., 2008). Interestingly, transcription factors engineered to encode an EAR motif and expressed in heterologous transgenic plants have the ability to repress the expression of the target orthologous genes across multiple plant species (Shikata and Ohme-Takagi, 2008), consistent with EAR motif-mediated repression mechanisms being evolutionarily conserved. The results of our analysis of putative orthologs of EAR motif-containing proteins across diverse plant species, including a dicotyledonous annual (Arabidopsis), two monocotyledonous annuals (rice and sorghum), and two dicotyledonous perennial woody plants (poplar and grapevine), illustrate conservation of the EAR motif across these evolutionarily diverse plant species (Fig. 5). Thus, it is possible that EAR-mediated repression of gene expression is a general regulatory mechanism functional across multiple gene families and diverse plant species. Future elucidation of the precise mechanisms of EAR motif-mediated transcriptional repression, the phenotypic effects of proteins identified in this study, and the influence of posttranslational modifications of EAR motifs will not only shed light on the effect of the EAR motif on physiological processes but also significantly advance our understanding of negative regulatory mechanisms of gene expression in plants.
MATERIALS AND METHODS
Protein Sequences
The entire proteome data sets or protein sequences of individual EAR motif-containing proteins in Arabidopsis (Arabidopsis thaliana) and their corresponding orthologs in rice (Oryza sativa), poplar (Populus trichocarpa), grapevine (Vitis vinifera), and sorghum (Sorghum bicolor) were obtained from the following databases: Arabidopsis (release 8; http://www.arabidopsis.org/tools/bulk/sequences/index.jsp), rice (release 6.1; http://rice.plantbiology.msu.edu/data_download.shtml), poplar (release 1.1; http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.download.ftp.html), grapevine (release 1; http://www.geno scope.cns.fr/spip/Vitis-vinifera-e.html), and sorghum (release 1.4; http://genome.jgi-psf.org/Sorbi1/Sorbi1.download.html).
Database Search for EAR Motif-Containing Proteins in Arabidopsis
The seed sequence used for database searches was a list of 49 EAR motif-containing proteins described in the literature. Full-length or truncated sequences of these proteins were used to iteratively search for novel EAR motif-containing candidates in the Arabidopsis proteome using various approaches, including DPM and sequence-based HMM and PHI-BLAST strategies (Fig. 2).
DPM
A Web-based pattern-matching tool at TAIR (PatMatch tool; http://www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl; Yan et al., 2005) was used to establish an initial data set of novel EAR motif-containing proteins. Using DLNxxP or LxLxL as query sequence patterns and the PatMatch tool set to its default configuration, the Arabidopsis protein database was searched to identify proteins with sequences similar to query sequence patterns. Additional DPM searches with control query variants of the LxLxL pattern derived by replacing the Leu residues with one of the remaining 19 common amino acids were carried out to assess the frequency of nonauthentic incidence of the above pattern. From the individual PatMatch outputs, a list of protein identifiers of all hits was extracted using an in-house PERL script and was submitted to the Batch Entrez server at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/sites/batchentrez) to obtain corresponding unique AGI codes, which were used for further classification using GO terms at TAIR to identify a subset of transcription-related proteins with sequence patterns similar to the query.
HMM Analysis
Proteins identified through the literature survey were grouped into six subgroups based on type (DLNxxP or LxLxL) and location (N-terminal, middle, or C-terminal region) of the EAR motif site. The first 25% of the protein sequence adjoining the initial amino acid residue and the last 25% of the protein sequence adjoining the final amino acid residue were defined as N- and C-terminal regions, respectively, and the remaining part of the protein was called the middle region. The truncated protein sequences, comprising the core EAR motif sites and adjoining ± 12 amino acids, of multiple proteins in each category were then aligned by ClustalW (Thompson et al., 1994) using default parameters, and the resulting multiple sequence alignments were used to generate HMM models using hmmbuild (Eddy, 1998). These models were then calibrated via hmmcalibrate and used to search through the Arabidopsis proteome (TAIR release 8) via hmmsearch (Eddy, 1998).
BLAST Search
The sequence of each protein identified through literature survey and HMM analysis was iteratively compared by PHI-BLAST (Zhang et al., 1998) with the Arabidopsis protein database at the National Center for Biotechnology Information BLAST server. PHI-BLAST searches the specified database for other protein sequences containing a user-specified pattern and having significant similarity to the query sequence in the vicinity of the pattern occurrence (Zhang et al., 1998) and thus provides an opportunity to explore variations of a known pattern among homologous sequences. For all PHI-BLAST searches, the expected threshold cutoff was set to 0.005, and hits with an E-value significantly better than the threshold were manually examined for the presence of a bona fide EAR motif.
MEME Analysis
The MEME analysis (Bailey and Elkan, 1995) on 179 proteins that were identified by DPM analysis but not by HMM/PHI-BLAST analysis was carried out using the following parameters: (1) occurrences of a single motif distributed per protein sequence were set to any number of repetitions; (2) the maximum number of motifs to find was 10; and (3) the optimum width of each motif was from six to 12 amino acids.
IceLogo Generation
Enrichment of amino acid residues within and adjoining EAR motifs was visualized using the IceLogo tool (Colaert et al., 2009). This tool provides a robust, probability-based analysis and improved visualization of consensus sequences in multiple sequence alignments by comparing the consensus with a background set of sequences tailored to the origins of sample and protein features. For generating the IceLogos in this analysis, we used the default percentage difference scoring method, which uses the difference in the frequency for an amino acid in the experimental set and the reference set as a measure of the height of the letter in the amino acid stack in each position. The positional background reference sets used in this analysis were created by random sampling of approximately 30-amino acid peptide sequences from the N-terminal, middle, and C-terminal regions of 219 random Arabidopsis proteins. The P value (cutoff = 0.05) of each amino acid at every position was calculated by testing the experimental frequency against the frequency of each amino acid in the reference set.
Analysis of the Phosphorylation Status of Ser and Thr Residues within EAR Motif-Containing Peptides
The phosphorylation status of Ser and Thr residues within EAR motif sites and adjoining regions of EAR motif-containing proteins was determined by searching the PhosPhAt database (Heazlewood et al., 2008) using default parameters.
Search for Orthologs in Other Plant Species
Putative orthologs of Arabidopsis EAR motif-containing proteins in rice, poplar, grapevine, and sorghum were identified by searching the plant orthologs database created by the rice genome annotation project team (http://rice.plantbiology.msu.edu/ortholog/index.shtml; Yuan et al., 2003). This Web-based resource contains information on putative orthologous groups of protein-coding genes from rice (release 6.1), Arabidopsis (release 8), poplar (release 1.1), grapevine (release 1), and sorghum (release 1.4) identified using OrthoMCL (Li et al., 2003) with default parameters. Protein sequences of putative orthologs in each plant species were manually assessed to confirm the presence or absence of an EAR motif in a relatively similar position corresponding to their Arabidopsis counterparts.
GO Analysis
GO enrichment analysis was performed using the EasyGO gene ontology enrichment analysis tool (http://bioinformatics.cau.edu.cn/easygo/; Zhou and Su, 2007). The GO term enrichment was calculated using hypergeometric distribution with a P value cutoff of 0.01 and a term-mapping count cutoff of 5. P values obtained by Fisher's exact test were adjusted with FDR for multiple comparisons to detect overrepresented GO terms.
Analysis of Gene Expression
In silico transcriptomic analysis was performed by utilizing the microarray data sets (Supplemental Table S6) generated by the AtGenExpress consortium (Schmid et al., 2005; Kilian et al., 2007; Goda et al., 2008) accessed through the expression browser tool of the Bio-Array Resource for Arabidopsis functional genomics database (Toufighi et al., 2005). Detailed description of the electronic northern analysis can be found in Supplemental Materials and Methods S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Differential expression of genes encoding EAR motif-containing transcriptional regulator proteins in various developmental stages of Arabidopsis.
Supplemental Figure S2. Differential expression of genes encoding EAR motif-containing transcriptional regulator proteins under abiotic stress conditions.
Supplemental Figure S3. Differential expression of genes encoding EAR motif-containing transcriptional regulator proteins in response to various hormonal signals.
Supplemental Table S1. AGI locus identifiers of the 49 EAR motif-containing proteins identified by literature survey.
Supplemental Table S2. DPM analysis of the Arabidopsis proteome.
Supplemental Table S3. List of protein names, descriptions, and identifiers of Arabidopsis EAR motif-containing transcriptional regulators.
Supplemental Table S4. List of transcription-related EAR motif-like sequence-containing proteins identified by DPM analysis but not by HMM/PHI-BLAST analysis.
Supplemental Table S5. Orthologs of Arabidopsis EAR motif-containing proteins from rice, poplar, grapevine, and sorghum.
Supplemental Table S6. An overview of microarray data sets included in this study.
Supplemental Table S7. Expression profiles of the Arabidopsis genes encoding EAR motif-containing transcriptional regulator proteins in various developmental stages.
Supplemental Table S8. Expression profiles of the Arabidopsis genes encoding EAR motif-containing transcriptional regulator proteins under abiotic stress conditions.
Supplemental Table S9. Expression profiles of the Arabidopsis genes encoding EAR motif-containing transcriptional regulator proteins in response to hormonal stimuli.
Supplemental Table S10. Application of CRES-T for the conversion of transcription factors into dominant repressors.
Supplemental Materials and Methods S1.
Acknowledgments
We thank Sifa Arafi for technical assistance. Dwayne Hegedus and Derek Lydiate are thanked for helpful discussions on gene regulation and the application of this research.
References
- Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J 50: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. (2007) The true story of the HD-Zip family. Trends Plant Sci 12: 419–426 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. (1995) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3: 21–29 [PubMed] [Google Scholar]
- Baniwal SK, Chan KY, Scharf KD, Nover L. (2007) Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. J Biol Chem 282: 3605–3613 [DOI] [PubMed] [Google Scholar]
- Bao X, Franks RG, Levin JZ, Liu Z. (2004) Repression of AGAMOUS by BELLRINGER in floral and inflorescence meristems. Plant Cell 16: 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Drews GN, Meyerowitz EM. (1991) Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3: 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. (2002) ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129: 1957–1965 [DOI] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico JM, Chini A, Fonseca S, Solano R. (2008) JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol 11: 486–494 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Mittler R. (2008) The zinc finger network of plants. Cell Mol Life Sci 65: 1150–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S, Morsy MR, Song L, Coutu A, Krizek BA, Lewis MW, Warren D, Cushman J, Connolly EL, Mittler R. (2007) The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J Biol Chem 282: 9260–9268 [DOI] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. (2009) Improved visualization of protein consensus sequences by IceLogo. Nat Methods 6: 786–787 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. (2005) The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, Grotewold E. (2003) AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deppmann CD, Acharya A, Rishi V, Wobbes B, Smeekens S, Taparowsky EJ, Vinson C. (2004) Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res 32: 3435–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, Fobert PR. (2003) The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15: 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. (2007) Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol 145: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF. (2006) NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis. Development 133: 1645–1655 [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. (1991) Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002 [DOI] [PubMed] [Google Scholar]
- Eddy SR. (1998) Profile hidden Markov models. Bioinformatics 14: 755–763 [DOI] [PubMed] [Google Scholar]
- Fassler J, Landsman D, Acharya A, Moll JR, Bonovich M, Vinson C. (2002) B-ZIP proteins encoded by the Drosophila genome: evaluation of potential dimerization partners. Genome Res 12: 1190–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Takemura M, Tani E, Nemoto K, Yokota A, Kohchi T. (2003) An Arabidopsis MADS-box protein, AGL24, is specifically bound to and phosphorylated by meristematic receptor-like kinase (MRLK). Plant Cell Physiol 44: 735–742 [DOI] [PubMed] [Google Scholar]
- Gan Y, Kumimoto R, Liu C, Ratcliffe O, Yu H, Broun P. (2006) GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 18: 1383–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Liu C, Yu H, Broun P. (2007) Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Gonzalez D, Bowen AJ, Carroll TS, Conlan RS. (2007) The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol 27: 5306–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. (2006) AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18: 1373–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T, Hagen G, Ulmasov T, Murfett J. (1998) How does auxin turn on genes?. Plant Physiol 118: 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Manley JL. (1993) Functional domains of the Drosophila Engrailed protein. EMBO J 12: 2723–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Rose W, Hansen U. (1996) Active repression mechanisms of eukaryotic transcription repressors. Trends Genet 12: 229–234 [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Durek P, Hummel J, Selbig J, Weckwerth W, Walther D, Schulze WX. (2008) PhosPhAt: a database of phosphorylation sites in Arabidopsis thaliana and a plant-specific phosphorylation site predictor. Nucleic Acids Res 36: D1015–D1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Wang H, Perry SE. (2008) A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J 53: 172–185 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Mitsuda N, Matsui K, Ohme-Takagi M. (2004) Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem Biophys Res Commun 321: 172–178 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. (2002) The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett 514: 351–354 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Ohme-Takagi M. (2009) A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol 50: 970–975 [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, et al. (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol 43: 467–478 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Jiang CJ, Aono M, Tamaoki M, Maeda S, Sugano S, Mori M, Takatsuji H. (2008) SAZ, a new SUPERMAN-like protein, negatively regulates a subset of ABA-responsive genes in Arabidopsis. Mol Genet Genomics 279: 183–192 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41: 577–585 [DOI] [PubMed] [Google Scholar]
- Katsir L, Chung HS, Koo AJ, Howe GA. (2008) Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol 11: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. (2006) Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci 11: 109–112 [DOI] [PubMed] [Google Scholar]
- Kieffer M, Stern Y, Cook H, Clerici E, Maulbetsch C, Laux T, Davies B. (2006) Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18: 560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C, et al. (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J 16: 263–276 [DOI] [PubMed] [Google Scholar]
- Krizek BA, Lewis MW, Fletcher JC. (2006) RABBIT EARS is a second-whorl repressor of AGAMOUS that maintains spatial boundaries in Arabidopsis flowers. Plant J 45: 369–383 [DOI] [PubMed] [Google Scholar]
- Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW. (2007) The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. Plant Cell 19: 2719–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunieda T, Mitsuda N, Ohme-Takagi M, Takeda S, Aida M, Tasaka M, Kondo M, Nishimura M, Hara-Nishimura I. (2008) NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell 20: 2631–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim WT. (2003) Molecular and biochemical characterization of VR-EILs encoding mung bean ETHYLENE INSENSITIVE3-LIKE proteins. Plant Physiol 132: 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht JD, Hanna-Rose W, Reddy JC, English MA, Ro M, Grossel M, Shaknovich R, Hansen U. (1994) Mapping and mutagenesis of the amino-terminal transcriptional repression domain of the Drosophila Kruppel protein. Mol Cell Biol 14: 4057–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. (2003) The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW. (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M. (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55: 954–967 [DOI] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM. (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu JK. (2006) Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett 580: 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J, dePamphilis CW, Ma H, Nei M. (2003) Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol Biol Evol 20: 1435–1447 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Doring P, Mishra SK, Ganguli A, Scharf KD. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need?. Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgishi M, Oka A, Morelli G, Ruberti I, Aoyama T. (2001) Negative autoregulation of the Arabidopsis homeobox gene ATHB-2. Plant J 25: 389–398 [DOI] [PubMed] [Google Scholar]
- Ohno CK, Reddy GV, Heisler MG, Meyerowitz EM. (2004) The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131: 1111–1122 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Teale W, Lang D, Paponov M, Reski R, Rensing SA, Palme K. (2009) The evolution of nuclear auxin signalling. BMC Evol Biol 9: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne T, Johnson SD, Koltunow AM. (2004) KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131: 3737–3749 [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. (1997) What's up and down with histone deacetylation and transcription?. Cell 89: 325–328 [DOI] [PubMed] [Google Scholar]
- Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF. (2001) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26: 385–394 [DOI] [PubMed] [Google Scholar]
- Preston J, Wheeler J, Heazlewood J, Li SF, Parish RW. (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J 40: 979–995 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279: 11736–11743 [DOI] [PubMed] [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. (1995) Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378: 199–203 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. (2004) Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol 136: 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T. (2002) The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 32: 1011–1022 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Seo J, Lee KJ. (2004) Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J Biochem Mol Biol 37: 35–44 [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. (2005) Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17: 2384–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata M, Ohme-Takagi M. (2008) The utility of transcription factors for manipulation of floral traits. Plant Biotechnol 25: 31–36 [Google Scholar]
- Sridhar VV, Surendrarao A, Gonzalez D, Conlan RS, Liu Z. (2004) Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc Natl Acad Sci USA 101: 11494–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar VV, Surendrarao A, Liu Z. (2006) APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133: 3159–3166 [DOI] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Wang HH, McCarty DR. (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol 143: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Thiel G, Lietz M, Hohl M. (2004) How mammalian transcriptional repressors work. Eur J Biochem 271: 2855–2862 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Morikami A, Nakamura K. (2007) Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc Natl Acad Sci USA 104: 2543–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Saijo T, Shibata D, Morikami A, Nakamura K. (2005) Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol 138: 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T, Kato W, Asada Y, Sako K, Sato T, Sonoda Y, Kidokoro S, Yamaguchi-Shinozaki K, Tamaoki M, Arakawa K, et al. (2009) DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J Plant Res 122: 633–643 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. (2002) Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol 22: 6321–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Weigel RR, Bauscher C, Pfitzner AJ, Pfitzner UM. (2001) NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Mol Biol 46: 143–160 [DOI] [PubMed] [Google Scholar]
- Weigel RR, Pfitzner UM, Gatz C. (2005) Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Li Z, Zhu Y, Wang H, Ma H, Dong A, Huang H. (2008) Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol 146: 566–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, Cherry JM, Rhee SY. (2005) PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res 33: W262–W266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Kalluri UC, Difazio SP, Wullschleger SD, Tschaplinski TJ, Cheng Z-M, Tuskan GA. (2009) Poplar genomics: state of the science. Crit Rev Plant Sci 28: 285–308 [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. (2005) Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol 58: 585–596 [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yuan Q, Ouyang S, Liu J, Suh B, Cheung F, Sultana R, Lee D, Quackenbush J, Buell CR. (2003) The TIGR rice genome annotation resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res 31: 229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Chen M, Chen X, Xu Z, Li L, Guo J, Ma Y. (2010) Isolation and characterization of a novel EAR-motif-containing gene GmERF4 from soybean (Glycine max L.). Mol Biol Rep 37: 809–818 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cheng YT, Qu N, Zhao Q, Bi D, Li X. (2006) Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J 48: 647–656 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schaffer AA, Miller W, Madden TL, Lipman DJ, Koonin EV, Altschul SF. (1998) Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res 26: 3986–3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Su Z. (2007) EasyGO: Gene Ontology-based annotation and functional enrichment analysis tool for agronomical species. BMC Genomics 8: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Verslues PE, Zheng X, Lee BH, Zhan X, Manabe Y, Sokolchik I, Zhu Y, Dong CH, Zhu JK, et al. (2005) HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc Natl Acad Sci USA 102: 9966–9971 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]