Abstract
Transporters move hydrophilic substrates across hydrophobic biological membranes and play key roles in plant nutrition, metabolism, and signaling and, consequently, in plant growth, development, and responses to the environment. To initiate and support systematic characterization of transporters in the model legume Medicago truncatula, we identified 3,830 transporters and classified 2,673 of these into 113 families and 146 subfamilies. Analysis of gene expression data for 2,611 of these transporters identified 129 that are expressed in an organ-specific manner, including 50 that are nodule specific and 36 specific to mycorrhizal roots. Further analysis uncovered 196 transporters that are induced at least 5-fold during nodule development and 44 in roots during arbuscular mycorrhizal symbiosis. Among the nodule- and mycorrhiza-induced transporter genes are many candidates for known transport activities in these beneficial symbioses. The data presented here are a unique resource for the selection and functional characterization of legume transporters.
Transporters are membrane-spanning proteins that selectively transport hydrophilic solutes across hydrophobic membranes. They are present and required in all cellular membranes, including the cell or plasma membrane that separates cellular contents from the external environment and membranes of the various subcellular organelles. By transporting metabolites and nonmetabolites, such as inorganic ions, transporters play integral roles in cell metabolism, ion homeostasis, osmoregulation, signaling, and other processes. Transporters move solutes not only within cells but also between cells, tissues, and organs of complex, multicellular organisms such as higher plants. Therefore, they help to coordinate metabolic, physiological, and developmental processes in higher plants and other organisms.
Transporter proteins/complexes contain multiple membrane-spanning domains that form an aqueous pore in the membrane, which enables movement of selected solutes from one side of the membrane to the other. Membrane-spanning domains are hydrophobic in nature, or at least partially so, which enables them to interact with the phospholipid bilayer of membranes. Many transporters contain hydrophobic α -helical segments that span the membrane, while others contain β -barrel transmembrane domains (TMD). Computer programs have been developed to identify putative membrane-spanning α -helices (Hoffman and Stoffel, 1993; Hirokawa et al., 1998; Tusnady and Simon, 2001) and β -barrels (Koebnik et al., 2000; Valavanis et al., 2006), which facilitate de novo prediction of putative membrane proteins, including transporters. Databases of known, characterized transport proteins aid in the identification and classification of transporters in new species via sequence similarity. Perhaps the most comprehensive of these is the Transporter Classification Database (TCDB; Saier et al., 2006), which was created to serve as a repository of functionally characterized transporters. It also serves to categorize new transporters into families and subfamilies based on molecular, evolutionary, and functional properties. At present, it consists of approximately 3,000 transporters classified in more than 500 families (www.tcdb.org).
The legume family is second only to the grass family in importance to humans as a source of food, feed for livestock, and raw materials for industry (Graham and Vance, 2003). Legumes are the lynch pin of sustainable agriculture, because they supply their own nitrogen by “fixing” it (reducing N2 to NH3) in a symbiotic association with bacteria called rhizobia. This mutually beneficial association provides legumes and subsequent crops with a free and renewable source of usable nitrogen (Udvardi and Day, 1997). Legumes also establish symbiosis with mycorrhizal fungi that help the plant mine phosphorous and other nutrients from the soil (Smith and Read, 2008).
Symbiotic nitrogen fixation (SNF) in root nodule cells of legumes is carried out by rhizobia that are completely surrounded by a plant membrane called the symbiosome membrane (SM), which forms a nitrogen-fixing organelle, the symbiosome, within the plant cytoplasm. Infected cortical cells of nodules contain thousands of symbiosomes, each containing one or a few bacteria. Infected plant cells, interspersed with noninfected cells, constitute the central tissue of nodules, which is surrounded by uninfected tissue that restricts gas exchange with the soil, and phloem and xylem, which import and export nutrients from the nodule, respectively. In exchange for ammonium produced by bacterial nitrogenase and released to the plant, rhizobia receive reduced carbon (principally dicarboxylic acids such as malate) and every other nutrient required for bacterial cell growth and maintenance (Udvardi and Day, 1997). Exchange of nutrients between the plant cell cytoplasm and rhizobia is mediated by a variety of transporters in the SM, some of which are induced during nodule development (Benedito et al., 2008). Transporters perform many other important roles in nodules, such as short- and long-distance transport of nutrients between plant cells and tissues and between the nodule and other organs, processes facilitated by proteins of the plant cell plasma membrane. On the other hand, transporters on the membranes of organelles such as mitochondria, plastids, and peroxisomes facilitate the movement of metabolites between cellular compartments, which is crucial for nodule metabolism and SNF.
In the arbuscular mycorrhizal (AM) symbiosis, the fungal symbionts inhabit the root cortex, where they obtain carbon from the plant, and in exchange they deliver mineral nutrients, particularly phosphorus and nitrogen, to the root. Mineral nutrient transfer between symbionts occurs at a specialized symbiotic interface between branched hyphae, called arbuscules, and the cortical cells that they inhabit (Parniske, 2008). The interface is delimited by a plant-derived membrane called the periarbuscular membrane, which is continuous with the plasma membrane but contains some unique proteins, including novel inorganic phosphate (Pi) transporters (Harrison et al., 2002; Paszkowski et al., 2002). These transporters are required to transfer Pi that is released from the arbuscule into the cortical cell. It is assumed, but not yet shown directly, that nitrogen, and possibly other mineral nutrients such as zinc, is also transferred between the symbionts at this membrane interface (Smith and Read, 2008). However, the transport proteins involved are currently unknown. Likewise, transporters involved in carbon transfer to the fungal symbiont have not been identified. While it is expected that the periarbuscular membrane will contain additional transport activities, only a handful of transporters residing in this membrane have been identified to date.
Although inroads have been made in the characterization of individual transporters in a variety of legume species, no systematic work has been done to identify and characterize all the transporters in any one species. Three legume species, Medicago truncatula, Glycine max (soybean), and Lotus japonicus, have been the subject of extensive cDNA and genomic DNA sequencing over the past few years (Young et al., 2003, 2005; Sato et al., 2007, 2008), making them interesting model systems for whole-genome analysis of transporters. The genome sequence of M. truncatula is being annotated by the International Medicago Genome Annotation Group (IMGAG), which described 38,335 genes in its version 2.0 of the genome sequence (http://www.medicago.org/genome/downloads/Mt2/). Additional resources relevant to Medicago functional genomics include the Medicago Gene Expression Atlas (http://bioinfo.noble.org/gene-atlas/v2), which provides developmental expression data for the majority of Medicago genes (Benedito et al., 2008), and a Tnt1 transposon-insertion mutant population with insertions in the majority of genes, which enables efficient forward and reverse genetics (Tadege et al., 2005, 2008). To facilitate systematic functional analysis of transporters in Medicago, and especially those involved in nitrogen-fixing and AM symbioses, we have identified and categorized 2,673 transporter genes and analyzed the expression patterns of 2,604 of these. The results of this work are presented here.
RESULTS
Identification of Putative Transporters
Initially, M. truncatula proteins predicted from genome sequence (IMGAG sequence release version 2.0) were analyzed for the presence of potential TMD using three algorithms: HMMTOP 2.0 (Tusnady and Simon, 2001), TMPred (Hoffman and Stoffel, 1993), and SOSUI (Hirokawa et al., 1998). HMMTOP utilizes a machine-learning hidden Markov model (HMM) approach, whereas TMPred uses amino acid properties to identify hydrophobic stretches of amino acids that could interact with lipid membranes. SOSUI integrates multiple properties, including hydropathy, amphiphilicity, amino acid charges, and sequence length, to predict protein topology in the membrane.
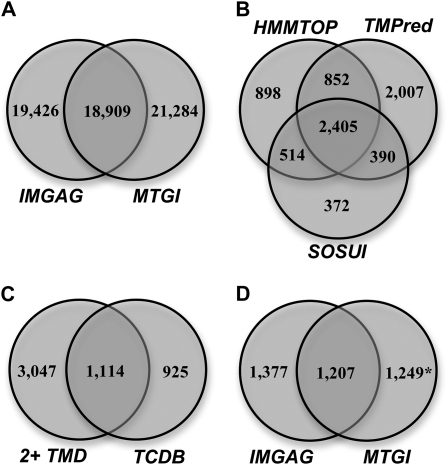
Among the 38,335 IMGAG-annotated gene products (Fig. 1A), 44% were predicted by TMPred to contain at least one TMD, while 32% and 21% were predicted to have one or more TMD by HMMTOP and SOSUI, respectively. In total, 18,684 proteins were predicted to contain at least one TMD by one or more of the three programs. A total of 7,438 proteins were predicted to contain two or more TMD by at least one program, of which 2,405 were identified by all three programs (Fig. 1B). Additionally, all 38,335 IMGAG proteins were compared by sequence homology with proteins of the TCDB (Saier et al., 2006). This approach was used to identify potential transporters not recognized by any of the TMD prediction algorithms as well as to guide transporter classification. Among the IMGAG-predicted proteins, 2,039 (5.3%) showed significant similarity to a TCDB sequence. Of these, 1,114 proteins were also found to contain at least two TMD by at least two prediction programs (Fig. 1C).
Figure 1.
Genomic analysis of M. truncatula membrane transporters. A, Number of genome-predicted genes (IMGAG version 2) and transcripts (MTGI) and overlap between databases. B, Number of IMGAG-predicted proteins with at least two TMD (2+ TMD) according to different algorithms. C, Overlap between the 2,039 IMGAG-predicted proteins with significant similarity (e-value < e-3) to TCDB members and the 4,161 proteins with at least two TMD predicted by at least two topology algorithms. D, Number of identified membrane transporters derived from IMGAG version 2.0 or MTGI version 8. *, Note that the total number of MTGI sequences is higher than shown in Table I due to gene redundancy or multiple probe set mapping.
Since Medicago genome sequencing is not yet complete, we also analyzed Medicago EST data (Medicago truncatula Gene Index [MTGI] version 8.0; http://compbio.dfci.harvard.edu/tgi/) using tBLASTX. Among the 36,850 Tentative Consensus (TC) sequences and singletons in the EST database, 2,051 (5.6%) encoded proteins similar to TCDB transporters, of which 1,249 did not match transporters predicted from IMGAG (genomic) sequences (Table I; Fig. 1D). TMD prediction was avoided as a method to select putative transporters encoded by ESTs because many of these sequences are incomplete.
Table I. Genomic analysis of M. truncatula membrane transporters.
| Variable | IMGAG Version 2 | MTGI Version 8 | Totala |
| No. of sequences | 38,335 | 36,850 | 60,823 |
| Proteins with TMDb | 18,684 | – | – |
| Proteins with TCDB homologs | 2,039 | 2,051 | 3,830 |
| Classified transporters | 1,681 | 1,759 | 2,673 |
Number of nonredundant genes.
Proteins with TMD identified by at least one algorithm.
Transporter Classification
Medicago proteins with similarity to TCDB proteins were classified into families and, when sufficient evidence was available, into subfamilies. Many transporter families have distinctive characteristics, such as membrane topology (number of TMD), presence of conserved domains, and approximate protein size. These features were considered during transporter classification and used to determine a level of confidence (1–5) for our classification of each transporter. All putative membrane protein sequences (with two or more TMD or a match to TCDB proteins) were further analyzed with respect to predicted length, presence of conserved domains (Pfam and InterPro), Gene Ontology (GO) annotation, and predicted subcellular localization (Supplemental Table S1). Medicago proteins with significant similarity to TCDB transporters, original annotations indicative of transporter activity, or with conserved domains characteristic of transporters were collected for manual curation and classification into TCDB families. To avoid false positives resulting from “forced” matches to proteins in TCDB, which contains only 3,000 transporters, Medicago sequences were also compared with more comprehensive sequence databases: the well-annotated Swiss-Prot database (Boeckmann et al., 2003) and the comprehensive (Viridiplantae) Nonredundant National Center for Biotechnology Information (NR-NCBI) GenBank (Benson et al., 2008). Annotation based on TCDB analysis and other protein characteristics was checked against annotations of homologous proteins in the Swiss-Prot and NR-NCBI databases. Confidence level 1 in our classification indicates that all features of a protein are consistent with its membership in a particular TCDB transporter family/subfamily, while level 2 indicates some divergence from expected features. Level 3 means functionality is doubtful due to a lack of key expected features (such as protein size, TMD absence or an unexpected number, or dubious TCDB homologies) or that classification is loose due to conflicting or weak pieces of evidence. Level 4 indicates that the putative membrane protein did not match any TCDB transporter, although there was some evidence of transporter function, such as a characteristic transporter domain, original annotation from IMGAG or EST, or GO annotation. TCDB contains some proteins, such as heat shock protein HSP70 family (TCDB 1.A.33) and group translocators, which transfer chemical groups from one molecule to another (TCDB category 4) that do not fit our conception of a transporter as a pore-forming membrane protein. We assigned such proteins to level 5, although this does not necessarily indicate a lack of confidence in their classification.
A total of 2,039 proteins predicted from the genome sequence had homology to TCDB proteins, and an additional 543 proteins had features consistent with transporter function (such as the presence of conserved transporter domains). Of these, 1,681 proteins were classified into transporter families and subfamilies with confidence levels from 1 to 3, 389 putative membrane transporters with no significant TCDB hit were assigned level 4, and 514 were assigned to level 5 (Supplemental Table S1). Likewise, among the 2,051 EST-encoded proteins with homology to TCDB proteins, 1,759 sequences were classified into transporter families and subfamilies with confidence levels 1 to 3 (Tables I and II). A total of 2,673 proteins predicted from genomic and EST sequences were classified into TCDB families and subfamilies. The largest Medicago transporter families are listed in Table III, and a complete list is given in Supplemental Table S2.
Table II. Membrane transporter classification according to TCDB classes and mapping onto the Affymetrix Medicago GeneChip.
| Transporter Class | IMGAG Version 2 | MTGI Version 8 | Totala | Probe Sets |
| Class 1: channels and pores | 223 | 244 | 365 | 379 |
| Class 2: secondary transporters | 755 | 756 | 1,181 | 1,132 |
| Class 3: primary active transporters | 452 | 536 | 791 | 734 |
| Class 8: accessory factors | 35 | 20 | 47 | 41 |
| Class 9: incompletely characterized | 213 | 203 | 289 | 318 |
| Total (confidence levels 1–3) | 1,681 | 1,759 | 2,673 | 2,611 |
| Not classified (level 4)b | 389 | – | 389 | 275 |
| Excluded from analysis (level 5)c | 514 | 292 | 768 | 715 |
Number of nonredundant genes.
Genes with no significant TCDB homology but with indications of transport function.
TCDB families that were not considered further or potentially false positives (see “Discussion”).
Table III. The 30 most abundant membrane transporter families in M. truncatula.
The complete list is given in Supplemental Table S2.
| TCDB No. | Family Name (Acronym) | IMGAG Genes | MTGI Transcripts | Nonredundant Sequences | Affymetrix Probe Sets |
| 1.A.1 | Voltage-Gated Ion Channel (VIC) Superfamily | 30 | 13 | 33 | 23 |
| 1.A.4 | Transient Receptor Potential Ca2+ Channel (TRP-CC) | 73 | 65 | 116 | 113 |
| 1.A.8 | Major Intrinsic Protein (MIP) | 32 | 53 | 64 | 72 |
| 1.A.20 | gp91phox Phagocyte NADPH Oxidase-Associated Cytochrome b558 H+ Channel (CytB) | 15 | 34 | 34 | 43 |
| 1.A.31 | Annexin (Annexin) | 19 | 14 | 25 | 22 |
| 1.C.45 | Plant Defensin (PD) | 9 | 14 | 21 | 20 |
| 2.A.1 | Major Facilitator Superfamily (MFS) | 110 | 116 | 175 | 163 |
| 2.A.3 | Amino Acid-Polyamine-Organocation (APC) | 8 | 27 | 29 | 33 |
| 2.A.5 | Zinc (Zn2+)-Iron (Fe2+) Permease (ZIP) | 11 | 15 | 19 | 23 |
| 2.A.6 | Resistance-Nodulation-Cell Division (RND) Superfamily | 13 | 18 | 23 | 27 |
| 2.A.7 | Drug/Metabolite Transporter (DMT) Superfamily | 119 | 93 | 168 | 159 |
| 2.A.17 | H+-Dependent Oligopeptide Transporter (POT) | 71 | 79 | 111 | 101 |
| 2.A.18 | Amino Acid/Auxin Permease (AAAP) | 57 | 61 | 96 | 87 |
| 2.A.19 | Ca2+:Cation Antiporter (CaCA) | 11 | 14 | 18 | 26 |
| 2.A.29 | Mitochondrial Carrier (MC) | 91 | 86 | 134 | 119 |
| 2.A.37 | Monovalent Cation-Proton Antiporter-2 (CPA2) | 29 | 6 | 30 | 29 |
| 2.A.40 | Nucleobase:Cation Symporter-2 (NCS2) | 10 | 14 | 19 | 18 |
| 2.A.49 | Chloride Channel (CIC) | 26 | 14 | 32 | 38 |
| 2.A.53 | Sulfate Permease (SulP) | 12 | 29 | 33 | 34 |
| 2.A.66 | Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | 53 | 48 | 73 | 79 |
| 2.A.67 | Oligopeptide Transporter (OPT) | 23 | 17 | 30 | 26 |
| 2.A.69 | Auxin Efflux Carrier (AEC) | 22 | 9 | 27 | 23 |
| 2.A.72 | K+ Uptake Permease (KUP) | 15 | 22 | 30 | 25 |
| 3.A.1 | ATP-Binding Cassette (ABC) Superfamily | 160 | 160 | 254 | 219 |
| 3.A.2 | H+/Na+-Translocating F-, V-, and A-type ATPase (F-ATPase) Superfamily | 34 | 49 | 64 | 77 |
| 3.A.3 | P-Type ATPase (P-ATPase) Superfamily | 37 | 71 | 85 | 84 |
| 3.A.5 | General Secretory Pathway (Sec) | 63 | 82 | 108 | 114 |
| 3.A.8 | Mitochondrial Protein Translocase (MPT) | 11 | 26 | 30 | 34 |
| 3.A.9 | Chloroplast Envelope Protein Translocase (CEPT or Tic-Toc) | 40 | 51 | 73 | 75 |
| 3.A.16 | Endoplasmic Reticular Retrotranslocon (ER-RT) | 72 | 67 | 109 | 114 |
Our analysis of transporters encoded by genomic DNA was based on IMGAG version 2.0 annotation of the genome, which discarded seven IMGAG version 1.0 gene models encoding putative transporters represented by probe sets on the Affymetrix Medicago GeneChip, including a putative ammonium transporter gene known to be expressed during mycorrhizal symbiosis (Gomez et al., 2009). Therefore, we included these seven putative transporter genes in our subsequent analyses of gene expression.
Analysis of Transporter Gene Expression
The Affymetrix Medicago GeneChip contains 50,900 M. truncatula probe sets corresponding to most of the gene transcripts in this species. A script was written in Perl to map probe sets to IMGAG version 2.0 gene sequences (see “Materials and Methods”). In this way, Affymetrix probe sets were assigned to 18,909 IMGAG-annotated genes and 21,284 ESTs (TCs and singlets) not represented by genomic sequence (Fig. 1A). Of the 2,673 genes encoding TCDB-classified transporters, 2,604 were represented by probe sets on the Affymetrix Medicago GeneChip (Table II; Supplemental Table S3). Affymetrix probe sets matched seven additional transporter genes predicted by IMGAG version 1 but not IMGAG version 2. We included these in our analyses and assigned them confidence level “X” to make them easy to identify in the tables.
Expression data for putative transporter genes were retrieved from the Medicago Gene Expression Atlas (Benedito et al., 2008), including data from all major organ systems (Supplemental Table S4), a nodule developmental series, and mycorrhizal roots (Supplemental Table S5). Based on presence/absence calls (at least two present calls within three biological replicates) obtained from chip analysis, 94% of transporter genes were expressed in at least one organ, with each organ expressing about 60% of all transporter genes. Only 4.5% of transporter genes (129) were organ specific (i.e. active in a single organ type), while 29% of genes (823) were expressed in more than one, but not all, organ types. The majority of transporter genes (61%) were expressed in all organs analyzed, although at different levels (Supplemental Table S4).
Transporter Gene Expression during Root Symbioses
Regulation of transporter gene expression during development of two different root symbioses was investigated: nitrogen-fixing and AM symbioses.
Gene expression data for developing and nitrogen-fixing root nodules were obtained from two sets of experiments, one in which plants were grown in solid substrate (turface) and one in which plants were grown aeroponically (Benedito et al. 2008). In the first set of experiments, mature, nitrogen-fixing nodules were harvested 28 d after inoculation with Sinorhizobium meliloti and compared with noninoculated, control roots of the same age. In the second set of experiments, plants were grown aeroponically and nodules were harvested at 4, 10, and 14 d after inoculation and compared with control, noninoculated roots harvested immediately prior to inoculation of symbiotic plants. In both sets of experiments, three biological replicates were performed for both treated and control samples. Using a Bonferroni-corrected P value cutoff of 1.14e-6 (Benedito et al., 2008) in pair-wise comparisons of nodules against control roots, 49% to 65% of transporter genes exhibited differential expression during nodule development. A total of 196 genes showed more than 5-fold increase in expression in nodules compared with root controls, with 25 genes showing greater than 100-fold change in expression (Supplemental Table S5). Table IV shows 37 genes that were induced more than 50-fold during nodule development. Figure 2 shows membrane transporters highly induced in, or specific to, nodules.
Table IV. Membrane transporters induced in nodules in comparison with nonnodulating roots (> 50-fold change).
Data were retrieved from the Medicago Gene Expression Atlas (Benedito et al., 2008).
| Probe Sets | Family | Certainty | Superfamily/Family Name | IMGAG Locus | MTGI | Nod0a | Nod4 | Nod10 | Nod14 | Rootb | Nod28 | Maximum Ratio |
| Mtr.2246.1.S1_at | 1.A.8 | 1 | Major Intrinsic Protein (MIP) | BG582951 | 13 | 103 | 1,830 | 1,866 | 13 | 529 | 149 | |
| Mtr.37525.1.S1_at | 1.A.8 | 1 | Major Intrinsic Protein (MIP) | AC168148_28.4 | TC100851 | 175 | 1,701 | 16,032 | 14,866 | 187 | 9,459 | 92 |
| Mtr.32104.1.S1_s_at | 1.A.20 | 3 | gp91phox Phagocyte NADPH Oxidase-Associated Cytochrome b558 (CytB) H+ Channel | AC161241_8.5 | AW329243 TC105131 | 12 | 28 | 1,447 | 1,051 | 276 | 379 | 121 |
| Mtr.39812.1.S1_s_at | 1.A.20 | 3 | gp91phox Phagocyte NADPH Oxidase-Associated Cytochrome b558 (CytB) H+ Channel | AC161241_8.5 | TC105783 | 26 | 39 | 2,028 | 1,966 | 275 | 519 | 78 |
| Mtr.52057.1.S1_at | 1.A.20 | 3 | gp91phox Phagocyte NADPH Oxidase-Associated Cytochrome b558 (CytB) H+ Channel | AC161241_18.5 | 17 | 24 | 1,476 | 1,341 | 163 | 368 | 85 | |
| Mtr.9266.1.S1_at | 1.C.45 | 1 | Plant Defensin (PD) | CT963132_15.4 | TC102850 | 14 | 16 | 4,896 | 3,661 | 13 | 2,971 | 340 |
| Mtr.433.1.S1_at | 2.A.1 | 1 | Major Facilitator Superfamily (MFS) | AC137838_17.4 | TC107287 | 131 | 1,424 | 7,902 | 7,662 | 180 | 6,272 | 60 |
| Mtr.21610.1.S1_at | 2.A.1 | 1 | Major Facilitator Superfamily (MFS) | AC147178_32.4 | 10 | 10 | 2,868 | 1,753 | 10 | 928 | 277 | |
| Mtr.37205.1.S1_at | 2.A.1 | 1 | Major Facilitator Superfamily (MFS) | AC167034_32.4 | TC100124 | 11 | 133 | 4,620 | 2,789 | 11 | 3,197 | 404 |
| Mtr.4676.1.S1_at | 2.A.3 | 1 | Amino Acid-Polyamine-Organocation (APC) | AC154036_21.4 | AL375006 | 10 | 2,288 | 4,790 | 4,052 | 11 | 3,099 | 457 |
| Mtr.42325.1.S1_at | 2.A.3 | 1 | Amino Acid-Polyamine-Organocation (APC) | AC154036_21.4 | TC111233 | 14 | 1,203 | 2,952 | 2,168 | 15 | 2,149 | 205 |
| Mtr.12845.1.S1_at | 2.A.7 | 1 | Drug/Metabolite Transporter (DMT) Superfamily | TC96179 | 8 | 1,080 | 881 | 965 | 8 | 610 | 137 | |
| Mtr.28141.1.S1_at | 2.A.7 | 1 | Drug/Metabolite Transporter (DMT) Superfamily | BG583743 | 15 | 41 | 1,173 | 760 | 15 | 458 | 79 | |
| Mtr.28851.1.S1_at | 2.A.7 | 1 | Drug/Metabolite Transporter (DMT) Superfamily | AC150779_27.4 | BQ124404 | 26 | 2,004 | 597 | 1,225 | 46 | 2,684 | 59 |
| Mtr.37389.1.S1_at | 2.A.7 | 1 | Drug/Metabolite Transporter (DMT) Superfamily | TC100560 | 21 | 1,202 | 1,950 | 1,712 | 35 | 2,498 | 71 | |
| Mtr.40650.1.S1_at | 2.A.7 | 1 | Drug/Metabolite Transporter (DMT) Superfamily | TC107753 | 15 | 133 | 9,430 | 8,067 | 18 | 7,357 | 629 | |
| Mtr.41092.1.S1_at | 2.A.7 | 1 | Drug/Metabolite Transporter (DMT) Superfamily | TC108704 | 8 | 712 | 2,387 | 1,667 | 9 | 2,106 | 291 | |
| Mtr.27727.1.S1_at | 2.A.17 | 1 | H+-Dependent Oligopeptide Transporter (POT) | AC149471_42.5 | BE997589 | 12 | 124 | 439 | 332 | 13 | 724 | 58 |
| Mtr.31737.1.S1_at | 2.A.17 | 1 | H+-Dependent Oligopeptide Transporter (POT) | AC175049_14.4 | AL377677 | 8 | 766 | 1,866 | 1,650 | 11 | 1,450 | 225 |
| Mtr.50491.1.S1_at | 2.A.18 | 1 | Amino Acid/Auxin Permease (AAAP) | AC140027_4.5 | 20 | 1039 | 401 | 808 | 16 | 1,472 | 95 | |
| Mtr.1587.1.S1_at | 2.A.29 | 3 | Mitochondrial Carrier (MC) | AW573662 | 9 | 405 | 1,792 | 1,259 | 19 | 1,200 | 194 | |
| Mtr.43719.1.S1_at | 2.A.29 | 3 | Mitochondrial Carrier (MC) | CT573353_28.4 | TC95911 | 9 | 9 | 1,359 | 1,070 | 9 | 689 | 154 |
| Mtr.19388.1.S1_at | 2.A.49 | 1 | Chloride Channel (CIC) | AC140549_27.5 | 14 | 14 | 866 | 403 | 12 | 83 | 63 | |
| Mtr.37708.1.S1_at | 2.A.53 | 1 | Sulfate Permease (SulP) | AC139842_4.5 | TC101252 | 12 | 730 | 8,943 | 6,309 | 237 | 4,474 | 776 |
| Mtr.49248.1.S1_at | 2.A.53 | 1 | Sulfate Permease (SulP) | TC100150 | 36 | 14,336 | 8,140 | 8,895 | 35 | 10,186 | 397 | |
| TC100173 | ||||||||||||
| Mtr.27423.1.S1_at | 2.A.66 | 1 | Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | AW980583 | 20 | 280 | 423 | 651 | 20 | 1,801 | 89 | |
| Mtr.28858.1.S1_at | 2.A.66 | 1 | Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | AC165438_46.5 | BQ124842 | 14 | 1,586 | 219 | 575 | 16 | 1,783 | 108 |
| Mtr.49406.1.S1_at | 2.A.66 | 1 | Multidrug/Oligosaccharidyl-lipid/Polysaccharide (MOP) Flippase Superfamily | AC144375_31.4 | 10 | 32 | 3,064 | 2,463 | 12 | 1,947 | 303 | |
| Mtr.49182.1.S1_x_at | 2.A.67 | 1 | Oligopeptide Transporter (OPT) | AC143341_26.4 | 21 | 1,149 | 430 | 649 | 17 | 1,061 | 55 | |
| Mtr.20006.1.S1_at | 2.A.85 | 1 | Aromatic Acid Exporter (ArAE) | AC151522_26.4 | 12 | 22 | 828 | 1,162 | 11 | 972 | 95 | |
| Mtr.49882.1.S1_at | 3.A.1 | 1 | ATP-Binding Cassette (ABC) Superfamily | AC146548_55.5 | 21 | 85 | 428 | 625 | 15 | 1,191 | 81 | |
| Mtr.3724.1.S1_at | 3.A.2 | 1 | H+- or Na+-Translocating F-, V-, and A-type ATPase (F-ATPase) Superfamily | BG583840 | 14 | 31 | 5,652 | 4,130 | 14 | 1,608 | 403 | |
| Mtr.42937.1.S1_s_at | 3.A.2 | 2 | H+- or Na+-Translocating F-, V-, and A-type ATPase (F-ATPase) Superfamily | TC93945 | 17 | 40 | 7,945 | 7,575 | 20 | 8,828 | 451 | |
| TC94148 | ||||||||||||
| Mtr.42017.1.S1_at | 3.A.3 | 1 | P-Type ATPase (P-ATPase) Superfamily | TC110571 | 13 | 12 | 2,003 | 1,390 | 10 | 1,427 | 152 | |
| Mtr.40309.1.S1_s_at | 3.A.8 | 3 | Mitochondrial Protein Translocase (MPT) | CT573353_23.4 | TC98311 | 14 | 7,079 | 10,753 | 11,381 | 69 | 10,337 | 803 |
| TC107014 | ||||||||||||
| TC100596 | ||||||||||||
| Mtr.9479.1.S1_at | 9.B.24 | 3 | Testis-Enhanced Gene Transfer (TEGT) | TC103475 | 20 | 245 | 2,814 | 1,845 | 22 | 2,077 | 139 | |
| Mtr.636.1.S1_at | Unclassified | 4 | Not defined | AC148995_45.5 | 15 | 13 | 741 | 470 | 11 | 546 | 51 |
Nod0 to Nod14 constitute a time course of nodule development, with numbers indicating days after inoculation of rhizobia.
Control for the Nod28 sample (mature nodule belonging to the organ series).
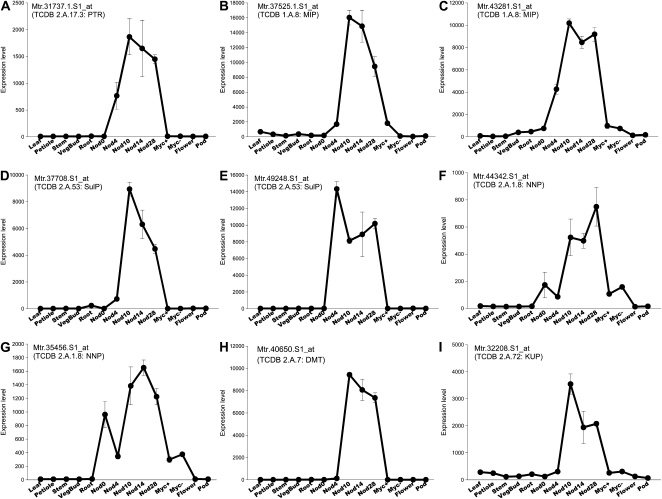
Figure 2.
Nodule-enhanced and nodule-specific membrane transporters in M. truncatula. Gene expression represents signal strength of Affymetrix Medicago GeneChip identifiers from the Medicago Gene Expression Atlas (Benedito et al., 2008) and Gomez et al. (2009). Samples for the mature organ series were taken from noninoculated 28-d-old plants (except nodules) growing at optimal conditions (Benedito et al., 2008). For nodule samples (Nod), numbers indicate days postinoculation with rhizobia. Myc indicate root samples with (+) or without (−) mycorrhizal association. Error bars indicate se of three biological replicates. Affymetrix probe sets and their respective TC (Saier et al., 2006) numbers and family acronyms are shown for each transcript. A, Member of the H+-dependent Oligopeptide Transporter family (POT/PTR). B and C, Members of the Major Intrinsic Protein family (MIP; aquaporins). D and E, Members of the Sulfate Permease family (SulP/SULTR). F and G, Members of the Nitrate/Nitrite Porter family (NNP), which belong to the Major Facilitator Superfamily (MFS). H, Member of the Drug/Metabolite Transporter superfamily (DMT). I, A member of the K+ Uptake Permease (KUP) family, homolog to LjKUP1 from L. japonicus.
Gene expression data for AM symbiosis were obtained from Medicago roots harvested 30 d after inoculation with Glomus intraradices and compared with data from noninoculated control roots, with three biological replicates in both cases (Gomez et al., 2009). Changes in transporter gene expression during AM symbiosis were more subtle than during nodule development and nitrogen-fixing symbiosis, as might be expected given the absence of new organ development during AM symbiosis. Nonetheless, 886 genes showed significantly altered expression during AM symbiosis, of which 44 genes were induced more than 5-fold compared with noninoculated roots (Table V; Supplemental Table S5).
Table V. Membrane transporters induced in mycorrhizal roots in comparison with control roots (> 10-fold change).
Data were retrieved from Gomez et al. (2009) and are publicly available in the Medicago Gene Expression Atlas version 2 (http://bioinfo.noble.org/gene-atlas/v2/).
| Probe Sets | Family | Certainty | Superfamily/Family Name | IMGAG Locus | MTGI | myc−a | myc+ | Ratio |
| Mtr.7596.1.S1_at | 1.A.8. | 1 | Major Intrinsic Protein (MIP) | AC174468_10.4 | AJ499650 | 11 | 318 | 30 |
| Mtr.37525.1.S1_at | 1.A.8 | 1 | Major Intrinsic Protein (MIP)-MtNIP I | AC168148_28.4 | TC100851 | 93b | 1,822 | 20 |
| Mtr.7210.1.S1_at | 1.C.45 | 1 | Plant Defensin (PD) | TC101060 | 13b | 1,863 | 146 | |
| Mtr.31214.1.S1_s_at | 1.C.45 | 1 | Plant Defensin (PD) | AL385826 | 8 | 164 | 20 | |
| AJ499425 | ||||||||
| Mtr.35854.1.S1_at | 1.C.45 | 1 | Plant Defensin (PD) | TC98064 | 12 | 495 | 40 | |
| Mtr.35484.1.S1_at | 1.C.45 | 1 | Plant Defensin (PD) | TC104515 | 10 | 147 | 14 | |
| Mtr.43062.1.S1_at | 2.A.1.9 | 1 | Phosphate:H+ Symporter Family (PHS)-MtPT4 | TC94453 | 14 | 1,902 | 136 | |
| Mtr.36985.1.S1_at | 2.A.17 | 1 | H+-Dependent Oligopeptide Transporter (POT) | AC140547_5.4 | TC105446 | 7 | 490 | 68 |
| Mtr.36985.1.S1_s_at | 2.A.17 | 1 | H+-Dependent Oligopeptide Transporter (POT) | AC140547_5.4 | TC105446 | 13 | 531 | 40 |
| Mtr.50194.1.S1_at | 2.A.17 | 1 | H+-Dependent Oligopeptide Transporter (POT) | AC140547_5.4 | 11 | 516 | 45 | |
| Mtr.39705.1.S1_at | 2.A.29 | 1 | Mitochondrial Carrier (MC) | TC105531 | 11 | 454 | 42 | |
| Mtr.1103.1.S1_at | 3.A.1 | 1 | ATP-Binding Cassette (ABC) Superfamily | AC141435_35.5 | 10 | 214 | 22 | |
| Mtr.44070.1.S1_at | 3.A.1 | 1 | ATP-Binding Cassette (ABC) Superfamily | AC202319_10.4 | TC96634 | 20 | 544 | 28 |
| Mtr.46524.1.S1_at | 3.A.1 | 1 | ATP-Binding Cassette (ABC) Superfamily | AC152057_1.5 | 21 | 888 | 42 | |
| Mtr.51195.1.S1_at | 3.A.1 | 1 | ATP-Binding Cassette (ABC) Superfamily | AC126010_13.4 | 10 | 249 | 24 | |
| Mtr.43470.1.S1_at | 3.A.3 | 1 | P-Type ATPase (P-ATPase) Superfamily | TC95400 | 13 | 610 | 48 | |
| Mtr.31910.1.S1_at | 3.A.16 | 2 | Endoplasmic Reticular Retrotranslocon (ER-RT) | AL385223 | 9 | 99 | 11 | |
| Mtr.4771.1.S1_atc | 9.A.10 | 1 | Iron/Lead Transporter (ILT) Superfamily | AL382013 | 11 | 369 | 33 | |
| Mtr.37110.1.S1_at | 9.A.12 | 1 | Copper Transporter (Ctr) | TC97522 | 12b | 552 | 47 |
Expression level in control roots (myc−) and mycorrhizal roots (myc+) as mean values of three biological replicates of Affymetrix chip expression.
Expression induction in root cells with arbuscules was confirmed by RT-PCR (Gomez et al., 2009).
This transcript is probably derived from AM fungal RNA. cDNA libraries generated from mycorrhizal roots contain transcripts from both plant and fungal symbionts.
DISCUSSION
Genome-wide identification and classification of transporters is an important first step in the systematic analysis of transporters in model organisms. Manual curation of information collected for all Medicago proteins, including predictions of the number of TMD and homology to transporters in the TCDB, resulted in the identification and classification of 2,673 distinct transporters. This represents 4.4% of all predicted proteins in Medicago and is in line with what has been found in other plant species. For example, Arabidopsis has 1,269 transporter genes (4.6% of all genes; Bock et al., 2006), while transporter genes account for approximately 5% of all rice (Oryza sativa) genes (Amrutha et al., 2007).
Classification into a specific family/subfamily was given a confidence score from 1 to 3, based on whether or not additional information supported the results of TCDB analysis, as described in “Results.” Proteins with two or more TMD that did not match proteins in the TCDB, but for which additional information pointed to possible transporter function, were given a confidence score of 4. Such additional information included the presence of conserved domains typical of transporters or annotation of homologous proteins in more extensive databases (Plant NR-NCBI and Swiss-Prot). Automatization of the classification process is under way (Li et al., 2008, 2009). A total of 389 putative transporters received a score of 4. Medicago proteins with similarity to TCDB proteins that we consider not to be involved in the transport of solutes across lipid bilayers were given a score of 5 and not subjected to gene expression analysis. There were 768 such proteins. All putative Medicago transporters predicted from genomic and nonredundant cDNA/EST sequences are linked to Affymetrix probe sets listed in the Medicago Gene Expression Atlas database (http://bioinfo.noble.org/gene-atlas), which includes further links to gene expression and analysis tools. This database will be updated following the release of IMGAG version 3 annotation of the Medicago genome.
The 3,062 putative transporters assigned confidence scores from 1 to 4 were represented by 2,886 nonredundant probe sets on the Affymetrix Medicago GeneChip. Therefore, we were able to query published gene expression data (Benedito et al., 2008; Gomez et al., 2009) for 94% of all predicted Medicago transporters. While the majority of transporter genes were expressed in two or more organs, approximately 4.5% (129) were expressed in an organ-specific manner (Supplemental Table S4). Presumably, these play specialized roles in organ development, differentiation, and/or function, and they represent interesting targets for future functional analysis. However, because of our interest in beneficial plant-microbe interactions, we focused most of our attention on genes induced during nodule development and SNF or during AM symbiosis. Eighty-seven percent of all transporter genes were differentially expressed during nodule development and SNF, of which 196 were induced more than 5-fold compared with noninoculated roots and 25 were induced more than 100-fold (Supplemental Table S5). A total of 886 genes were differentially expressed during AM symbiosis, of which 44 were induced more than 5-fold compared with noninoculated roots. In the following paragraphs, we discuss some of these genes in the context of what is known about transport and transporters in the two types of symbiosis.
Transporters Involved in N2 Fixation Symbiosis
A variety of complementary data (for review, see Udvardi and Day, 1997) indicate that Suc translocated into nodules from the shoot is converted to dicarboxylic acids such as malate before being transported from the cytoplasm of infected cells to nitrogen-fixing bacteroids. The high-affinity, energy-dependent bacterial transporter DctA is mainly responsible for dicarboxylate uptake by nitrogen-fixing bacteroids and is indispensable for SNF (Ronson et al., 1981; Udvardi and Day, 1997). The plant counterpart of DctA on the SM is a lower affinity transporter that has been characterized biochemically (Udvardi et al., 1988) but not yet at the molecular level. However, the discovery of a dicarboxylate transporter, AgDCAT1, located on the SM in actinorrhizal nodules of the nonlegume Alnus glutinosa (Jeong et al., 2004) suggests that related H+-dependent oligopeptide transporter family (POT/PTR; TC 2.A.17) members may transport dicarboxylates across the SM of legume nodules. Interestingly, two POT/PTR genes are strongly induced during nodule development in M. truncatula (Table IV; Fig. 2A) and L. japonicus (Colebatch et al., 2004).
Ammonia produced by nitrogen-fixing bacteroids appears to be transported out across the bacteroid membranes by simple diffusion before being transported across the SM as either NH4+, via a cation channel that also transports K+ (Tyerman et al., 1995; Roberts and Tyerman, 2002; Obermeyer and Tyerman, 2005), or NH3 (Niemietz and Tyerman, 2000), possibly via aquaglyceroporins of the NIP (for Nodulin-like Intrinsic Protein) family (TC 1.A.8.12). The archetype of the NIP family is soybean nodulin 26, a nodule-specific protein of the SM. In L. japonicus, nodule-induced LIMP1 (a Tonoplast Intrinsic Protein family member) and LIMP2 (possibly an ortholog of Nod26) have been characterized, although their roles during SNF remain unclear (Guenther and Roberts, 2000). Two homologs of these proteins are also expressed more or less specifically in Medicago nodules (Table IV; Fig. 2, B and C). The molecular identity of the SM NH4+/K+ channel is unknown. However, it is unlikely to be a member of the Ammonium Transporter (AMT) family of NH4+ transporters, which are relatively specific for NH4+, do not transport K+, and seem to be located in the plasma membrane, where they are likely to be involved in the recovery of ammonia lost from nodule cells by diffusion (Simon-Rosin et al., 2003; D'Apuzzo et al., 2004; Rogato et al., 2008). We identified nine AMT family (TC 1.A.11) members in Medicago, none of which were induced more than 5-fold during nodule development.
A range of inorganic nutrients, including phosphorus, sulfur, potassium, sodium, calcium, vanadium, iron, molybdenum, nickel, and cobalt, are required by rhizobia for multiplication and maintenance (Rosendahl et al., 1991), but little is known about how most of these nutrients are obtained from the plant. Although sulfate transport into symbiosomes has not been studied directly, map-based cloning of LjSST1 identified a nodule-induced sulfate transporter gene in L. japonicus that is essential for nodule function (Krusell et al., 2005). Its reported location on the SM (Wienkoop and Saalbach, 2003) suggests that SST1 is essential for sulfur supply to the bacteroids. Thirteen homologs of LjSST1 in the SulP/SULTR family (TC2.A.53) were induced more than 2-fold during nodule development in Medicago, and two were essentially nodule specific (Table IV; Fig. 2, D and E). Some members of the SulP family transport substrates other than sulfate, including nitrate, bicarbonate, chloride, and molybdate (Tejada-Jimenez et al., 2007; Tomatsu et al., 2007), so it will be interesting to determine the substrates of the various nodule-induced SulP transporters in Medicago.
Lotus LjN70 and soybean GmN70, two nodulins of the Nitrate/Nitrite Porter family (NPP; TC 2.A.1.8), were shown to be anion channels with ion selectivity similar to a soybean SM transporter characterized biochemically earlier (Udvardi et al., 1991; Vincill et al., 2005). Two homologs of these NPP transporters, together with 32 other members of the Major Facilitator Superfamily (MFS; TC 2.A.1), were induced during nodule development in Medicago (Table IV; Fig. 2, F and G).
Iron transport across the SM and bacteroid membranes of soybean nodules has been characterized biochemically (Moreau et al., 1995, 1998; LeVier et al., 1996), and a nodule-induced divalent metal transporter, GmDMT1, capable of ferrous iron transport has been cloned and characterized (Kaiser et al., 2003). GmDMT1 was localized to the SM and also appears to transport zinc, copper, and manganese, so it may play a role in supplying a variety of metal ions to bacteroids (Kaiser et al., 2003). A homolog of GmDMT1 (NRAMP; TC 2.A.55 [not to be confused with the DMT superfamily; TC 2.A.7]) is expressed in a nodule-specific manner in Medicago (Table IV). An unrelated, nodule-specific SM protein of soybean called GmZIP1 (TC 2.A.5), which transports zinc, has also been characterized (Moreau et al., 2002). Inhibition of zinc transport into isolated symbiosomes by an antibody to GmZIP1 implicates the transporter in zinc supply to the bacteroids (Moreau et al., 2002). We identified 23 ZIP family members in Medicago, six of which were induced more than 2-fold during nodule development (Supplemental Table S5).
Potassium is transported across the SM of soybean, broad bean (Vicia faba), and Lotus (Tyerman et al., 1995; Roberts and Tyerman, 2002; Andreev et al., 2005) via the NH4+/K+ channel described above, and possibly others, but the identity of these proteins is unknown. A nodule-induced potassium transporter of the KUP family (TC 2.A.72), LjKUP1 from Lotus, was cloned and characterized, but its location on the plasma membrane of plant cells suggests that it plays a role in plant rather than bacteroid potassium nutrition/homeostasis (Desbrosses et al., 2004). Of the 25 putative potassium transporters identified in Medicago, one gene (represented by two probe sets) showed a massive induction during the onset of nitrogen fixation and in mature nodules (Table IV; Fig. 2I).
Calcium uptake into symbiosomes has been documented for yellow lupin (Lupinus luteus; Andreev et al., 1998) and broad bean (Andreev et al., 1999), driven by a Ca2+-ATPase (TC 3.A.3.2) in the latter case, but the corresponding proteins remain to be discovered. Interestingly, the NH4+/K+ channel of the Lotus SM also appears to transport Ca2+ (Roberts and Tyerman, 2002). Among the 15 Ca2+-ATPases found in Medicago, one EST showed greater than 150-fold change during late states of nodule development and nitrogen fixation (Table IV).
A P-type H+-ATPase (TC 3.A.3) and possibly other proton pumps energize the SM, which drives many secondary transport processes on this membrane (Udvardi and Day, 1989; Fedorova et al., 1999). However, the specific isoforms responsible remain to be identified. We found a nodule-specific P-type ATPase in Medicago, which is an interesting candidate for this activity (Table IV).
ATP for plant membrane energization and metabolism in nodule cells is provided by mitochondria, which undergo both morphological (Werner and Mörschel, 1978) and biochemical (Suganuma and Yamamoto, 1987) differentiation during nodule development. We found that several mitochondrial transporter genes are induced during Medicago nodule development, including a Mitochondrial Protein Translocase (MPT; TC 3.A.8; Table IV), which generally transports preproteins into mitochondria (Lister et al., 2007), and two Mitochondrial Carriers (MC; 2.A.29), which may transport tricarboxylic acid cycle intermediates like α -oxoglutarate to the cytoplasm for ammonium assimilation/amino acid biosynthesis (Picault et al., 2002; Palmieri et al., 2008).
The ultimate source of energy and carbon skeletons for nodule metabolism and ammonium assimilation is Suc, which is imported into nodules from photosynthetic organs (Vance et al., 1997; Gordon et al., 1999). LjSUT4 is a Suc transporter in L. japonicus that is up-regulated during nodule development and presumably plays a role in Suc uptake into nodule cells (Flemetakis et al., 2003). Three members of the Suc/proton symporter subfamily (TC 2.A.2.4) of the Glycoside-Pentoside-Hexuronide:Cation Symporter family (TC 2.A.2) were identified in our analysis of Medicago transporters, but none of them was induced during nodulation.
Nodules of temperate legumes, such as Medicago, typically export fixed nitrogen to the root and shoot in the form of amides (Gln and Asn; Temple et al., 1998; Lodwig and Poole, 2003). Putative amino acid transporters from different families were identified in our analysis as strongly induced during nodule development (more than 5-fold change over control roots): five members of the Amino Acid-Polyamine-Organocation family (APC; TCDB 2.A.3), 17 Drug/Metabolite Transporters (DMT; 2.A.7), 12 Multidrug/Oligosaccharidyl-lipid/Polysaccharide Flippases (MOP/MATE; 2.A.66), four Oligopeptide Transporters (OPT; 2.A.67), and one Aromatic Acid Exporter (ArAE; 2.A.85; Supplemental Table S5), making them interesting targets for future work aimed at identifying transporters with key roles in amino acid export from nodules.
Nodule development is subject to hormonal regulation (Mabood et al., 2006; Prayitno et al., 2006; Murray et al., 2007; Ding et al., 2008; Oldroyd and Downie, 2008), which in the case of auxin appears to involve changes in auxin transport (de Billy et al., 2001; Wasson et al., 2006). An auxin influx carrier, CgAUX1 (Amino Acid/Auxin Permease Family [AAAP]; TC 2.A.18), was proposed to be active during actinorrhizal nodule formation in Casuarina glauca (Péret et al., 2007). We found one AAAP transporter that is essentially nodule specific in Medicago (Table IV), although it is not the most similar in sequence to the CgAUX1.
Apart from interesting candidates for known transport functions in nodules, our analysis of Medicago transporter gene expression identified many nodule-induced or nodule-specific transporters that presumably carry out novel transport functions in nodules. Clearly, both sets of transporters warrant further work in the future to characterize their biochemical and physiological roles in nodules.
Transporters Involved in Mycorrhizal Symbiosis
The number of differentially induced transporters in mycorrhizal roots in comparison with control roots was much smaller than that observed during nodulation, with only 33 transporters showing more than 5-fold change induction in mycorrhizal roots. This was probably due to a dilution effect, since whole root systems were sampled, in contrast to nodule samples, which were excised from nodulated roots and compared with nonnodulated control roots. Nonetheless, some transporters showed a massive induction (greater than 100-fold change) in mycorrhizal roots. Statistical tests revealed 658 transporters differentially expressed in mycorrhizal roots (Supplemental Table S5).
One of the greatest benefits of mycorrhizal symbiosis for the plants is an increased phosphate uptake, mediated by the AM fungi, which deliver Pi directly to the root cortex. Plant Pi transporters responsible for acquiring Pi delivered by the fungus have been identified previously, and MtPT4, a transporter belonging to the Phosphate:H+ Symporter Family (PHS; 2.A.1.9), is expressed specifically in mycorrhizal roots (Table IV; Harrison et al., 2002; Javot et al., 2007; Pumplin and Harrison, 2009). We found one member of the Sulfate Permease family (SP; 2.A.53) up-regulated 4.5-fold in mycorrhizal roots, indicating a possible symbiotic role in mycorrhizal roots. Moreover, this sulfate permease is different from the ones up-regulated in nodules. It is important to emphasize that members of this family have also been implicated in the transport of other anions as well, such as molybdate (Fitzpatrick et al., 2008), bicarbonate, and heavy metals. Likewise, the solute flippase (MOP/MATE family; 2.A.66), aforementioned as nodule induced, also showed induction (6-fold change) in mycorrhizal roots. Three members of the POT/PTR family (2.A.17), also within the MFS, were found to be induced in mycorrhizal roots. Although most members of this family transport peptides, some of them transport other substrates, such as nitrate and dicarboxylates; therefore, their roles in mycorrhizal roots remain uncertain.
The high energetic requirement of AM symbiosis likely influences mitochondrial metabolism. Our data show two putative Mitochondrial Carriers (2.A.29), one of them induced more than 100 times in mycorrhizal roots, indicating a possible function in this symbiosis. Many members of this family are ATP/ADP carriers, whereas others transport TCA-derived compounds. There was also high expression of a proton ATPase (3.A.3; 50-fold change), which polarizes the plasma membrane through proton extrusion at the expense of ATP hydrolysis. Interestingly, the same gene was also induced in nodules, although to a lesser extent. Proton ATPases induced in AM symbiosis have been reported previously (Rosewarne et al., 2007), including this particular gene, which was named Mtha1 (Krajinski et al., 2002). Mtha1 was shown to be expressed exclusively in cortical cells containing arbuscules, where it is assumed to maintain the proton gradient across the periarbuscular membrane (Krajinski et al., 2002). Ultimately, this provides the energy to drive proton-coupled symport activities such as that of MtPT4. A second gene classified in this family was found to be induced 9-fold in mycorrhizal roots (and not expressed in nodules) and may play a complementary role.
Many eukaryotic ATP-binding Cassette Transporters (ABC; 3.A.1) function as extruders of diverse solutes, including organic acids and secondary metabolites, at the expense of ATP. We identified four ABC transporters induced more than 20 times in mycorrhizal roots. This classification cannot predict whether these transporters import or export solutes, or the nature of their substrates, although the expression levels indicate an important function during AM symbiosis.
Nitrogen transfer from fungi to plants, potentially as ammonium, can occur in both endomycorrhizal and ectomycorrhizal symbioses (Govindarajulu et al., 2005; Chalot et al., 2006). In poplar (Populus species), among 14 AMT genes identified in the genome, PtrAMT1;2 was induced in ectomycorrhizal roots, whereas no expression was detected in mock roots (Couturier et al., 2007). Its ortholog, PttAMT1;2, was also induced in ectomycorrhizal roots (Selle et al., 2005). However, among 10 Medicago AMT genes, only one was found up-regulated more than 5-fold in mycorrhizal roots, and this was recently shown to be expressed in cortical cells containing arbuscules (Gomez et al., 2009). LjAMT2;2 was recently identified in Lotus and found to be expressed exclusively in mycorrhizal roots (Guether et al., 2009).
Surprisingly, a Voltage-Dependent Anion-Selective Channel (VDAC; 1.B.8.1) was induced almost 10-fold in mycorrhizal roots. VDAC porins belong to the Mitochondrial and Plastid Porin family (1.B.8) and have been implicated in organellar Ca2+-regulated homeostasis of ATP and other small molecules (Bathori et al., 2006). Additionally, a Transient Receptor Potential Ca2+ Channel (TRP-CC, 1.A.4) was induced more than 3-fold in mycorrhizal roots. The same gene was also induced (greater than 20-fold) in nodules. So far, this TCDB family of channels and sensors does not include any plant proteins, and its members are mostly from animals, which resulted in low confidence scores for putative members of this family during manual curation. Many members of this family have an ankyrin-repeat domain and consequently are classified in the Ankyrin family (8.A.28). Functional analysis of these transporters would help better resolve this classification as well as their symbiotic roles.
Plant defensins (PD; 1.C.45) are Cys-rich polypeptides proposed to transport small molecules, such as ions, by forming channels in the membrane (Kagan et al., 1990). Isolated proteins presented antimicrobial properties (Thomma et al., 2002; Finkina et al., 2008), and in vivo they may regulate microsymbiont differentiation. We noticed four mycorrhiza-induced genes classified in this family, with transcription induction ranging from 14- to 145-fold. Whether they have a defensive function or alternatively control development of the fungal symbiont remains to be determined. With four very similar proteins, the potential for functional redundancy is high, and this makes analysis of their roles in symbiosis a challenge.
Three aquaporins (Major Intrinsic Proteins [MIP]; 1.A.8) were induced in mycorrhizal roots (up to 30 times) relative to nonmycorrhizal controls. Interestingly, the most up-regulated aquaporin did not show differential expression during nodulation, indicating a specific role in mycorrhizal symbiosis, whereas other members induced in nodules did not show variation in mycorrhizal roots.
Gomez et al. (2009) reported 49 Medicago EST-derived probe sets on the Affymetrix GeneChip of probable fungal origin. The mycorrhizal root cDNA libraries have the potential to contain transcripts from both plant and AM fungal symbionts. Five of these were classified as putative transporters (Supplemental Tables S1–S5) and were induced between 3- and 10-fold in mycorrhizal roots (Supplemental Table S5). In our study, we found evidence for an additional fungus-derived transporter belonging to the Iron/Lead Transporter superfamily (ILT; 9.A.10) that is induced 33-fold in mycorrhizal roots (Table IV).
Perspectives
Our search for transporters in Medicago uncovered and classified 2,673 putative transporter genes, many of which are induced during symbiosis with nitrogen-fixing rhizobia or AM fungi. Given the importance of these symbioses to plant nutrition and sustainable agriculture, it will be interesting to characterize the function of many of the symbiosis-induced transporters. Systematic analysis of transporters in Medicago will be aided not only by the results presented here but also by the availability of Tnt1 insertion lines of Medicago (Tadege et al., 2008).
MATERIALS AND METHODS
Transporter Identification and Sequence Analyses
Predicted protein sequences of Medicago truncatula from IMGAG version 2 were retrieved (ftp://ftpmips.gsf.de/plants/medicago/MT_2_0/) and analyzed for the presence of TMD using two algorithms: HMMTOP 2.0 (Tusnady and Simon, 2001) and TMPred (Hoffman and Stoffel, 1993). The resulting putative TMD proteins were analyzed further using a more conservative TMD identification algorithm, SOSUI (Hirokawa et al., 1998). In parallel, all IMGAG version 2 predicted proteins were analyzed for sequence similarity to transporters of the TCDB (http://www.tcdb.org/index.php) using BLASTP with an e-value cutoff of e-3 or less. All sequences with two or more predicted TMD or with significant similarity to TCDB proteins were selected for manual curation.
ESTs (tentative consensuses and singlets) of the MTGI version 8 (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/gimain.pl?gudb=medicago) were retrieved and compared with TCDB proteins through tBLASTX with e-values of e-3 or less. Since most ESTs are not full-length cDNA sequences, TMD analysis, although performed, was not taken into account for selection. Sequences with significant similarity to TCDB proteins were selected for further analyses.
Medicago proteins predicted from genomic DNA and cDNA (mostly EST) sequences were screened for conserved domains by Pfam (http://pfam.sanger.ac.uk/) and InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/) with e-values of e-3 or less and submitted to sequence similarity analysis against broader databases: the curated Swiss-Prot database (e-value ≤ e-3; http://expasy.org/sprot/) and the Viridiplantae subset of the comprehensive NR-NCBI GenBank (e-value ≤ e-3; http://www.ncbi.nlm.nih.gov/). GO classification was carried out through protein homology to subterms of GO: 0022857 (transmembrane transporter activity; http://www.geneontology.org/). Additionally, TMD proteins without TCDB homologs but with annotation (IMGAG, Medicago Gene Index, GO, Swiss-Prot, or NCBI) indicating transporter functions (such as transpor*, *porter, carrier, channel, translocase, permease, ATPase, extrusion, and exchanger) were retrieved for further analyses.
Curation and Classification of Transporters
Selected sequences were analyzed with respect to the presence of TMD, predicted protein size, presence of typical conserved domains, annotation of best matches in comprehensive databases, and homology to classified TCDB transporters. Medicago proteins were classified according to TCDB transporter homology at the family or subfamily level, and a confidence level was assigned to each categorization according to a variety of evidence. Confidence level 1 indicates that all features of a protein are consistent with its membership in a particular TCDB transporter family/subfamily, while level 2 indicates some divergence from expected features. Level 3 means functionality is doubtful due to a lack of key expected features (such as protein size, TMD absence or an unexpected number, or dubious TCDB homologies). Level 4 designates proteins with no TCDB homology but with some indications of transporter activity (such as conserved transporter domain, IMGAG annotation, transporter as best hit in broader databases). Level 5 indicates proteins classified into TCDB families that do not conform to our strict definition of a transporter, such as molecular chaperones and plasmodesmata proteins (potentially kinases). Transporter classification, confidence levels, and additional features are provided in Supplemental Table S1. The M. truncatula transporter classification resulting from this study has been incorporated into the Medicago Gene Expression Atlas (http://bioinfo.noble.org/gene-atlas/v2; Benedito et al., 2008).
Mapping of IMGAG Version 2-Predicted Genes onto the Affymetrix Medicago GeneChip
Affymetrix GeneChip Array (http://www.affymetrix.com) probe sets comprise 11 perfect-match 25-mer antisense probes designed to hybridize to each gene transcript. Because the current Affymetrix Medicago GeneChip was designed, in part, on a previous version of IMGAG gene annotations, we reanalyzed probe sets by mapping them onto the current IMGAG sequence release. Predicted coding sequences derived from IMGAG version 2 were matched to probe sets using ProbeMatch from the NetAffx package of Affymetrix (https://www.affymetrix.com/analysis/netaffx/probematch/probe_match.affx). This software postprocesses BLASTn results among probes and transcript sequences and scores the alignments using the position mismatch penalty matrix (1 1 1 1 1 2 2 2 2 2 3 3 3 3 3 2 2 2 2 2 1 1 1 1 1), which corresponds to the 25 nucleotides of each probe on a chip. Thus, the maximum score is 45, and a score of at least 43 implies two mismatches in the margin or one mismatch in the middle locations but no mismatches in the central five positions. Each probe set on the Affymetrix Medicago GeneChip was designed to have 11 perfect-match probes (as well as 11 mismatched probes, disregarded in this analysis). We set a threshold of eight out the 11 probes with a score of 43 or greater as the minimum requirement to match a gene to a probe set. Probe set mapping information for all identified transporters is provided in Supplemental Table S2. The complete mapping (all probe sets on the Affymetrix Medicago GeneChip to all IMGAG version 2 genes and MTGI version 8 transcripts) can be downloaded at http://bioinfo.noble.org/gateway/index.php?option=com_wrapper&Itemid=65.
Expression Analyses of Identified Medicago Transporters
The expression of IMGAG genes and MTGI transcripts mapped onto the Affymetrix GeneChip was analyzed for all organs of mature (4-week-old) plants, during nodule development, and in mycorrhizal roots. Expression data were retrieved from the Medicago Gene Expression Atlas version 1 (http://bioinfo.noble.org/gene-atlas/), normalized, and analyzed according to Benedito et al. (2008). Probe sets of the identified transporters are shown in Supplemental Tables S4 and S5. Statistical analyses for differential expression between control roots and nodules or mycorrhizal roots, and hierarchical cluster analyses, were also carried out as described by Benedito et al. (2008).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Classification of M. truncatula membrane transporters.
Supplemental Table S2. Number of members of each Medicago membrane transporter family.
Supplemental Table S3. Correspondence of Affymetrix Medicago Gene Chip membrane transporter probe sets to IMGAG version 2 coding sequences and MTGI transcripts.
Supplemental Table S4. Expression profile of each identified M. truncatula membrane transporter across vegetative and reproductive organs.
Supplemental Table S5. Differential gene expression and statistical analyses of M. truncatula membrane transporters during nodule development and in mycorrhizal roots.
References
- Amrutha RN, Sekhar PN, Varshney RK, Kishor PBK. (2007) Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci 172: 708–721 [Google Scholar]
- Andreev I, Krylova V, Dubrovo P, Izmailov S. (2005) Passive potassium transport by symbiosomes from broad bean root nodules. Plant Sci 168: 1005–1010 [Google Scholar]
- Andreev IM, Dubrovo PN, Krylova VV, Izmailov SF. (1998) Calcium uptake by symbiosomes and the peribacteroid membrane vesicles isolated from yellow lupin root nodules. J Plant Physiol 153: 610–614 [Google Scholar]
- Andreev IM, Dubrovo PN, Krylova VV, Izmailov SF. (1999) Functional identification of ATP-driven Ca2+ pump in the peribacteroid membrane of broad bean root nodules. FEBS Lett 447: 49–52 [DOI] [PubMed] [Google Scholar]
- Bathori G, Csordas G, Garcia-Perez C, Davies E, Hajnoczky G. (2006) Ca2+-dependent control of the permeability properties of the mitochondrial outer membrane and voltage-dependent anion-selective channel (VDAC). J Biol Chem 281: 17347–17358 [DOI] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. (2008) GenBank. Nucleic Acids Res 36: D25–D30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock KW, Honys D, Ward JM, Padmanaban S, Nawrocki EP, Hirschi KD, Twell D, Sze H. (2006) Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiol 140: 1151–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, et al. (2003) The SWISS-PROT protein knowledge base and its supplement TrEMBL in 2003. Nucleic Acids Res 31: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalot M, Blaudez D, Brun A. (2006) Ammonia: as candidate for nitrogen transfer at the mycorrhizal interface. Trends Plant Sci 11: 263–266 [DOI] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell L, Montanari O, Kloska S, Kopka J, Udvardi MK. (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39: 487–512 [DOI] [PubMed] [Google Scholar]
- Couturier J, Montanini B, Martin F, Brun A, Blaudez D, Chalot M. (2007) The expanded family of ammonium transporters in the perennial poplar plant. New Phytol 174: 137–150 [DOI] [PubMed] [Google Scholar]
- D'Apuzzo E, Rogato A, Simon-Rosin U, El-Alaoui H, Barbulova A, Betti M, Dimou M, Katinakis P, Marquez A, Marini AM, et al. (2004) Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol 134: 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Billy F, Grosjean C, May S, Bennett M, Cullimore JV. (2001) Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol Plant Microbe Interact 14: 267–277 [DOI] [PubMed] [Google Scholar]
- Desbrosses G, Kopka C, Ott T, Udvardi MK. (2004) Lotus japonicus LjKUP is induced late during nodule development and encodes a potassium transporter of the plasma membrane. Mol Plant Microbe Interact 17: 789–797 [DOI] [PubMed] [Google Scholar]
- Ding Y, Kalo P, Yendrek C, Sun J, Liang Y, Marsh JF, Harris JM, Oldroyd GE. (2008) Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20: 2681–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova E, Thomson R, Whitehead LF, Maudoux O, Udvardi MK, Day DA. (1999) Localization of H+-ATPases in root nodules. Planta 209: 25–32 [DOI] [PubMed] [Google Scholar]
- Finkina EI, Shramova EI, Tagaev AA, Ovchinnikova TV. (2008) A novel defensin from the lentil Lens culinaris seeds. Biochem Biophys Res Commun 371: 860–865 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick KL, Tyerman SD, Kaiser BN. (2008) Molybdate transport through the plant sulfate transporter SHST1. FEBS Lett 582: 1508–1513 [DOI] [PubMed] [Google Scholar]
- Flemetakis E, Dimou M, Cotzur D, Efrose RC, Aivalakis G, Colebatch G, Udvardi M, Katinakis P. (2003) A sucrose transporter, LjSUT4, is up-regulated during Lotus japonicus nodule development. J Exp Bot 54: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Gomez SK, Javot H, Deewatthanawong P, Torres-Jerez I, Tang Y, Blancaflor EB, Udvardi MK, Harrison MJ. (2009) Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AJ, Minchin FR, James CL, Komina O. (1999) Sucrose synthase in legume nodules is essential for nitrogen fixation. Plant Physiol 120: 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajulu M, Pfeffer PE, Jin H, Abubaker J, Douds DD, Allen JW, Bücking H, Lammers PJ, Shachar-Hill Y. (2005) Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435: 819–823 [DOI] [PubMed] [Google Scholar]
- Graham PH, Vance CP. (2003) Legumes: importance and constraints to greater use. Plant Physiol 131: 872–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P. (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182: 200–212 [DOI] [PubMed] [Google Scholar]
- Guenther JF, Roberts DM. (2000) Water-selective and multifunctional aquaporins from Lotus japonicus nodules. Planta 210: 741–748 [DOI] [PubMed] [Google Scholar]
- Harrison MJ, Dewbre GR, Liu J. (2002) A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14: 2413–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379 [DOI] [PubMed] [Google Scholar]
- Hoffman K, Stoffel W. (1993) Tmbase: a database of membrane spanning protein segments. Biol Chem Hoppe Seyler 374: 166 [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Suh S, Guan C, Tsay YF, Moran N, Oh CJ, An CS, Demchenko KN, Pawlowski K, Lee Y. (2004) A nodule-specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiol 134: 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan BL, Selsted ME, Ganz T, Lehrer RI. (1990) Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc Natl Acad Sci USA 87: 210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser BN, Moreau S, Castelli J, Thomson R, Lambert A, Bogliolo S, Puppo A, Day DA. (2003) The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. Plant J 35: 295–304 [DOI] [PubMed] [Google Scholar]
- Koebnik R, Locher KP, Van Gelder P. (2000) Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol 37: 239–253 [DOI] [PubMed] [Google Scholar]
- Krajinski F, Hause B, Gianinazzi-Pearson V, Franken P. (2002) Mtha1, a plasma membrane H+-ATPase gene from Medicago truncatula, shows arbuscule-specific induced expression in mycorrhizal tissue. Plant Biol 4: 754–761 [Google Scholar]
- Krusell L, Krause K, Ott T, Desbrosses G, Krämer U, Sato S, Nakamura Y, Tabata S, James EK, Sandal N, et al. (2005) The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17: 1625–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVier K, Day DA, Guerinot ML. (1996) Iron uptake by symbiosomes from soybean root nodules. Plant Physiol 111: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Benedito VA, Udvardi MK, Zhao PX. (2009) TransportTP: a two-phase classification approach for membrane transporter prediction and characterization. BMC Bioinformatics 10: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Dai X, Zhao X. (2008) A nearest neighbor approach for automated transporter prediction and categorization from protein sequences. Bioinformatics 24: 1129–1136 [DOI] [PubMed] [Google Scholar]
- Lister R, Carrie C, Duncan O, Ho LHM, Howell KA, Murcha MW, Whelan J. (2007) Functional definition of outer membrane proteins involved in preprotein import into mitochondria. Plant Cell 19: 3739–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodwig E, Poole P. (2003) Metabolism of Rhizobium bacteroids. CRC Crit Rev Plant Sci 22: 37–78 [Google Scholar]
- Mabood F, Souleimanov A, Khan W, Smith DL. (2006) Jasmonates induce Nod factor production by Bradyrhizobium japonicum. Plant Physiol Biochem 44: 759–765 [DOI] [PubMed] [Google Scholar]
- Moreau S, Day DA, Puppo A. (1998) Ferrous iron is transported across the peribacteroid membrane of soybean nodules. Planta 207: 83–87 [Google Scholar]
- Moreau S, Meyer JM, Puppo A. (1995) Uptake of iron by symbiosomes and bacteroids from soybean nodules. FEBS Lett 361: 225–228 [DOI] [PubMed] [Google Scholar]
- Moreau S, Thomson RM, Kaiser BN, Trevaskis B, Guerinot ML, Udvardi MK, Puppo A, Day DA. (2002) GmZIP1 encodes a symbiosis-specific zinc transporter in soybean. J Biol Chem 277: 4738–4746 [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104 [DOI] [PubMed] [Google Scholar]
- Niemietz CM, Tyerman SD. (2000) Channel-mediated permeation of ammonia gas through the peribacteroid membrane of soybean nodules. FEBS Lett 465: 110–114 [DOI] [PubMed] [Google Scholar]
- Obermeyer G, Tyerman SD. (2005) NH4+ currents across the peribacteroid membrane of soybean: macroscopic and microscopic properties, inhibition by Mg2+, and temperature dependence indicate a subpicoSiemens channel finely regulated by divalent cations. Plant Physiol 139: 1015–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Palmieri L, Picault N, Arrigoni R, Besin E, Palmieri F, Hodges M. (2008) Molecular identification of three Arabidopsis thaliana mitochondrial dicarboxylate carrier isoforms: organ distribution, bacterial expression, reconstitution into liposomes and functional characterization. Biochem J 410: 621–629 [DOI] [PubMed] [Google Scholar]
- Parniske M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Paszkowski U, Kroken S, Roux C, Briggs SP. (2002) Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Swarup R, Jansen L, Devos G, Auguy F, Collin M, Santi C, Hocher V, Franche C, Bogusz D, et al. (2007) Auxin influx activity is associated with Frankia infection during actinorhizal nodule formation in Casuarina glauca. Plant Physiol 144: 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picault N, Palmieri L, Pisano I, Hodges M, Palmieri F. (2002) Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria: bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem 277: 24204–24211 [DOI] [PubMed] [Google Scholar]
- Prayitno J, Rolfe BG, Mathesius U. (2006) The ethylene-insensitive sickle mutant of Medicago truncatula shows altered auxin transport regulation during nodulation. Plant Physiol 142: 168–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N, Harrison MJ. (2009) Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol 151: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DM, Tyerman SD. (2002) Voltage-dependent cation channels permeable to NH4+, K+, and Ca2+ in the symbiosome membrane of the model legume Lotus japonicus. Plant Physiol 128: 370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogato A, D'Apuzzo E, Barbulova A, Omrane S, Stedel C, Simon-Rosin U, Katinakis P, Flemetakis M, Udvardi M, Chiurazzi M. (2008) Tissue-specific down-regulation of LjAMT1;1 compromises nodule function and enhances nodulation in Lotus japonicus. Plant Mol Biol 68: 585–595 [DOI] [PubMed] [Google Scholar]
- Ronson CW, Lyttleton P, Robertson JG. (1981) C4-dicarboxylate transport mutants of Rhizobium trifoli form ineffective nodules on Trifolium repens. Proc Natl Acad Sci USA 78: 4284–4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendahl L, Glenn AR, Dilworth MJ. (1991) Organic and inorganic inputs into legume root nodule nitrogen fixation. Dilworth MJ, Glenn AR, , Biology and Biochemistry of Nitrogen Fixation. Elsevier Science Publishers, Amsterdam, pp 259–291 [Google Scholar]
- Rosewarne GM, Smith FA, Schachtman DP, Smith SE. (2007) Localization of proton-ATPase genes expressed in arbuscular mycorrhizal tomato plants. Mycorrhiza 17: 249–258 [DOI] [PubMed] [Google Scholar]
- Saier MJ, Tran C, Barabote R. (2006) TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res 1: D181–D186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Asamizu E, Isobe S, Tabata S. (2007) Genome sequencing and genome resources in model legumes. Plant Physiol 144: 588–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15: 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selle A, Willmann M, Grunze N, Gessler A, Weiss M, Nehls U. (2005) The high-affinity poplar ammonium importer PttAMT1.2 and its role in ectomycorrhizal symbiosis. New Phytol 168: 697–706 [DOI] [PubMed] [Google Scholar]
- Simon-Rosin U, Wood C, Udvardi MK. (2003) Molecular and cellular characterisation of LjAMT2;1, an ammonium transporter from the model legume Lotus japonicus. Plant Mol Biol 51: 99–108 [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. (2008) Mycorrhizal Symbiosis. Academic Press, San Diego [Google Scholar]
- Suganuma N, Yamamoto Y. (1987) Respiratory metabolism of mitochondria in soybean root nodules. Soil Sci Plant Nutr 33: 93–101 [Google Scholar]
- Tadege M, Ratet P, Mysore KS. (2005) Insertional mutagenesis: a Swiss army knife for functional genomics of Medicago truncatula. Trends Plant Sci 10: 229–235 [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen JQ, He J, Tu HD, Kwak Y, Eschstruth A, Cayrel A, Endre G, Zhao PX, Chabaud M, et al. (2008) Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J 54: 335–347 [DOI] [PubMed] [Google Scholar]
- Tejada-Jimenez M, Llamas A, Sanz-Luque E, Galvan A, Fernandez E. (2007) A high-affinity molybdate transporter in eukaryotes. Proc Natl Acad Sci USA 104: 20126–20130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple SJ, Vance CP, Gantt JS. (1998) Glutamate synthase and nitrogen assimilation. Trends Plant Sci 3: 51–56 [Google Scholar]
- Thomma BP, Cammue BP, Thevissen K. (2002) Plant defensins. Planta 216: 193–202 [DOI] [PubMed] [Google Scholar]
- Tomatsu H, Takano J, Takahashi H, Watanabe-Takahashi A, Shibagaki N, Fujiwara T. (2007) An Arabidopsis thaliana high-affinity molybdate transporter required for efficient uptake of molybdate from soil. Proc Natl Acad Sci USA 104: 18807–18812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17: 849–850 [DOI] [PubMed] [Google Scholar]
- Tyerman SD, Whitehead LF, Day DA. (1995) A channel-like transporter for NH4+ on the symbiotic interface of N2-fixing plants. Nature 378: 629–632 [Google Scholar]
- Udvardi MK, Day DA. (1989) Electrogenic ATPase activity on the peribacteroid membrane of soybean (Glycine max L.) root nodules. Plant Physiol 90: 982–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi MK, Day DA. (1997) Metabolite transport across symbiotic membranes of legume nodules. Annu Rev Plant Physiol Plant Mol Biol 48: 493–523 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Lister DL, Day DA. (1991) ATPase activity and anion transport across the peribacteroid membrane of isolated soybean symbiosomes. Arch Microbiol 156: 362–366 [Google Scholar]
- Udvardi MK, Price GD, Gresshoff PM, Day DA. (1988) A dicarboxylate transporter on the peribacteroid membrane of soybean nodules. FEBS Lett 231: 36–40 [Google Scholar]
- Valavanis IK, Bagos PG, Emiris IZ. (2006) β -Barrel transmembrane proteins: geometric modelling, detection of transmembrane region, and structural properties. Comput Biol Chem 30: 416–424 [DOI] [PubMed] [Google Scholar]
- Vance CP, Miller SS, Driscoll BT, Robinson DL, Trepp G, Gantt JS, Samas DA. (1997) Nodule carbon metabolism: organic acids for N2 fixation. Elmerich CE, Kondorosi A, Newton WE, eds, Biological Nitrogen Fixation for the 21st Century. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 443–448 [Google Scholar]
- Vincill ED, Szczyglowski K, Roberts DM. (2005) GmN70 and LjN70: anion transporters of the symbiosome membrane of nodules with a transport preference for nitrate. Plant Physiol 137: 1435–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U. (2006) Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner D, Mörschel E. (1978) Differentiation of nodules of Glycine max. Planta 141: 169–177 [DOI] [PubMed] [Google Scholar]
- Wienkoop S, Saalbach G. (2003) Proteome analysis: novel proteins identified at the peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol 131: 1080–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Cannon SB, Sato S, Kim D, Cook DR, Town CD, Roe BA, Tabata S. (2005) Sequencing the genespaces of Medicago truncatula and Lotus japonicus. Plant Physiol 137: 1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Mudge J, Ellis THN. (2003) Legume genomes: more than peas in a pod. Curr Opin Plant Biol 6: 199–204 [DOI] [PubMed] [Google Scholar]