Abstract
Although phenylpropanoid-polyamine conjugates (PPCs) occur ubiquitously in plants, their biological roles remain largely unexplored. The two major PPCs of Nicotiana attenuata plants, caffeoylputrescine (CP) and dicaffeoylspermidine, increase dramatically in local and systemic tissues after herbivore attack and simulations thereof. We identified NaMYB8, a homolog of NtMYBJS1, which in BY-2 cells regulates PPC biosynthesis, and silenced its expression by RNA interference in N. attenuata (ir-MYB8), to understand the ecological role(s) of PPCs. The regulatory role of NaMYB8 in PPC biosynthesis was validated by a microarray analysis, which revealed that transcripts of several key biosynthetic genes in shikimate and polyamine metabolism accumulated in a NaMYB8-dependent manner. Wild-type N. attenuata plants typically contain high levels of PPCs in their reproductive tissues; however, NaMYB8-silenced plants that completely lacked CP and dicaffeoylspermidine showed no changes in reproductive parameters of the plants. In contrast, a defensive role for PPCs was clear; both specialist (Manduca sexta) and generalist (Spodoptera littoralis) caterpillars feeding on systemically preinduced young stem leaves performed significantly better on ir-MYB8 plants lacking PPCs compared with wild-type plants expressing high levels of PPCs. Moreover, the growth of M. sexta caterpillars was significantly reduced when neonates were fed ir-MYB8 leaves sprayed with synthetic CP, corroborating the role of PPCs as direct plant defense. The spatiotemporal accumulation and function of PPCs in N. attenuata are consistent with the predictions of the optimal defense theory: plants preferentially protect their most fitness-enhancing and vulnerable parts, young tissues and reproductive organs, to maximize their fitness.
In nature, plants are frequently exposed to abiotic and biotic stress factors, including drought, extreme temperatures, high winds, UV light, pathogens, and herbivores. These selection pressures enabled plants to refine their constitutive and inducible defenses (Purrington, 2000; Zangerl, 2003; Howe and Jander, 2008; Walling, 2009). A shift from constitutive to inducible defense strategies can be considered a potential cost-saving mechanism whereby plants tune the production and accumulation of defenses with the need for the defenses and thereby forgo the production and opportunity costs they incur when they are not needed; for example, UV light induces secondary metabolite-flavonoid accumulation to intensify their UV light-protective screen (Li et al., 1993; Zhao et al., 2007; Jenkins, 2009; Wang et al., 2009).
Once recognized via specific receptors, stress factors activate phytohormone signaling networks that trigger downstream defense responses in plants. Jasmonic acid (JA) is known to mediate wound and herbivore stress signals in plants that activate local and systemic defenses and lead to the accumulation of antifeedants and/or ovipositioning deterrents against herbivores. These toxins largely impair insect growth and reduce their survivorship rates, helping plants to diminish further damage (Steppuhn and Baldwin, 2007; Chen, 2008). In a similar manner, salicylic acid coordinates the elicitation of defenses against invading pathogens, resulting in the accumulation of phytoalexins that limit the spread of pathogens in the plant tissues, but an increasing amount of evidence points to antagonism between salicylic acid and JA signaling (Loake and Grant, 2007; Diezel et al., 2009). Therefore, defense-related hormones in plants are engaged in a complex cross talk that still needs to be fully examined (Koornneef and Pieterse, 2008).
After herbivore attack, the wounds in plants often come in direct contact with herbivores’ oral secretions (OS), a potential carrier of herbivore-specific elicitors, which can be recognized by plant cells. Upon perception of these elicitors, such as fatty acid-amino acid conjugates (FACs), inceptins, and caeliferins, plants trigger herbivore-specific defense responses, which involve large-scale transcriptional, translational, and posttranslational changes in plants at both local and systemic levels (Halitschke et al., 2001; Gatehouse, 2002; Howe and Jander, 2008). In Nicotiana attenuata plants, FACs are known to rapidly activate the mitogen-activated protein kinase cascade, followed by JA accumulation and conjugation of JA to Ile, yielding the active signal molecule JA-Ile (Wu et al., 2007; Meldau et al., 2009). JA-Ile is required for the interaction of JAZ (for jasmonate-ZIM domain) repressor proteins with the SCFcoi1 (for Skp1-Cullin-F-box) E3-ubiquitin ligase complex that results in the degradation of JAZ repressors and the activation of MYC2 transcription factor (Santner and Estelle, 2007; Chico et al., 2008; Eckardt, 2008; Kazan and Manners, 2008; Browse, 2009; Chini et al., 2009a, 2009b; Chung et al., 2009; Fonseca et al., 2009; Santner and Estelle, 2009; Yan et al., 2009). MYC2 and a putative suite of MYC2-controlled secondary transcription factors like NAC domain-containing transcription factors ANAC019 and ANAC055 in Arabidopsis (Arabidopsis thaliana; Bu et al., 2008) then activate the transcription of genes encoding defense-related enzymes responsible for the biosynthesis of defense-related metabolites in plants (Kazan and Manners, 2008; Chini et al., 2009a). Notably, with the exception of two already mentioned NAC proteins, the identities of other putative MYC2-regulated secondary transcription factors involved in JA signaling cascade are unknown (Bu et al., 2008; Chini et al., 2009a).
Secondary metabolites constitute an important part of both inducible and constitutive plant defenses that target insects, plant pathogens, and other competitors, including plants (Sudha and Ravishankar, 2002; Zhao et al., 2005). One of the widespread secondary metabolic pathways in plants activated by stress is the biosynthesis of phenylpropanoid-polyamine conjugates (PPCs), which has been frequently found to be positively correlated with increased plant resistance to pathogens, viruses, and fungi (for review, see Edreva et al., 2007); however, the exact role and mode of action of these metabolites in plants remain unclear (Cowley and Walters, 2005; Edreva et al., 2007). In addition, the enzymes responsible for the conjugation of acetyl-CoA-activated phenylpropanoid moieties to polyamines have only started to be identified (Grienenberger et al., 2009; Luo et al., 2009).
The NtMYBJS1 gene, previously reported from tobacco (Nicotiana tabacum), is a JA-dependent R2R3-MYB transcription factor that regulates the accumulation of PPCs in BY-2 tobacco cell cultures (Gális et al., 2006); however, the physiological and ecological functions of this gene in intact plants are not known. Therefore, we cloned a functional homolog of NtMYBJS1 from the established ecological model N. attenuata (designated NaMYB8 in this study) and explored the potential role of this transcription factor in plant defense. The transcripts of NaMYB8 accumulated rapidly and transiently upon mechanical wounding of local N. attenuata leaves; however, a simultaneous application of Manduca sexta OS to the wounds amplified the wound-induced increase in NaMYB8 transcripts at 1 to 2 d after OS elicitation, suggesting that NaMYB8 could play an important role in plant-herbivore interactions, possibly by regulating PPC levels in response to herbivory.
Following the initial characterization of NaMYB8 expression and the accumulation of caffeoylputrescine (CP) in N. attenuata, we used the NaMYB8 transcription factor as a genetic tool to examine the ecological relevance of OS-induced PPC accumulation and the significance of PPC presence in N. attenuata. The plants specifically silenced in the expression of NaMYB8 were generated through RNA interference technology. As expected, the inverted repeat NaMYB8-silenced (referred to as ir-MYB8) plants were unable to accumulate two major PPCs (CP and dicaffeoylspermidine [DCS]) in their tissues. A microarray study conducted with ir-MYB8 and wild-type plants revealed that NaMYB8 protein is required for transcriptional activation of genes involved in phenylpropanoid and polyamine biosynthesis, in addition to several other genes with unknown functions. As both specialist (M. sexta) and generalist (Spodoptera littoralis) caterpillars performed better on ir-MYB8 plants compared with wild-type plants, we propose that PPCs and their regulation by NaMYB8 are vital parts of the direct defense mechanisms used by plants against attacking herbivores.
RESULTS
Isolation and Expression of an NtMYBJS1 Homolog in N. attenuata
The transcripts of the NtMYBJS1 gene accumulated 3 h after treatment of tobacco BY-2 cell cultures with methyl jasmonate (Gális et al., 2006). Therefore, we used a 1-h wounding+OS-elicited leaf cDNA pool, a time point known to be associated with the accumulation of many JA-responsive genes, to clone a homolog of NtMYBJS1 from N. attenuata. The cloned sequence, which was designated NaMYB8 in this study (Supplemental Fig. S1; GenBank accession no. GU451752), occurs as a single-copy gene in the N. attenuata genome (Supplemental Fig. S2C).
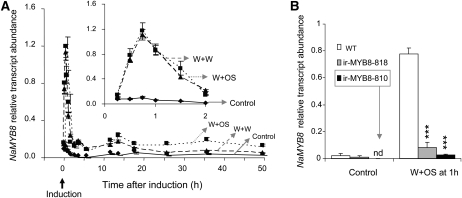
We then examined the transcriptional response of NaMYB8 to wounding and herbivore cues: a fully expanded rosette leaf was wounded with a pattern wheel, and either water (W+W), mimicking mechanical wounding, or M. sexta OS (W+OS), representing simulated herbivory, was applied to the wounds. Biotic stresses are known to trigger transcriptional responses that typically follow one of two expression profiles: (1) rapid and transient up-regulation of transcripts encoding primary regulators, and (2) steadily increasing but long-lasting transcriptional responses of genes encoding defensive metabolites (e.g. trypsin proteinase inhibitors) or enzymes involved in their biosynthesis (Zhao et al., 2005). Interestingly, NaMYB8 gene showed a mixed pattern of its transcript regulation in the local leaves, including rapid transient increase of transcripts 45 min after W+W and W+OS elicitations (Fig. 1A, inset), followed by stably elevated levels of NaMYB8 transcripts in the W+OS-elicited leaves for an additional 1 to 2 d (Fig. 1A). Because NaMYB8 transcripts responded differentially to the presence of M. sexta OS, we assumed that it could play an important role in the inducible defenses employed by plants against attacking herbivores. The NaMYB8 transcript levels in unwounded control leaves did not change significantly over 2 d (0–50 h; Fig. 1A), excluding the possibility that NaMYB8 transcript accumulation is controlled by circadian rhythm.
Figure 1.
NaMYB8 responds differentially to M. sexta OS elicitation in N. attenuata. A, Technical replicate means ± se of NaMYB8 transcript relative abundances quantified with RT-qPCR from pooled samples using five independent control, W+W-elicited, and W+OS-elicited plants. The inset shows a detail of rapid transient accumulation of transcripts between 15 and 120 min after elicitation. B, Mean ± se levels of NaMYB8 transcripts in unelicited and 1-h W+OS-elicited local leaves of two independent homozygous ir-MYB8 and wild-type (WT) plants (n = 3). Asterisks represent significantly different transcript abundances between genotypes within the same treatment group at P < 0.001. nd, Not detected.
Transient and Stable Silencing of NaMYB8 Expression in Planta
In order to confirm the role of the NaMYB8 transcription factor in plant-insect interactions, we used posttranscriptional gene silencing approaches to generate transiently silenced NaMYB8-VIGS (for virus-induced gene silencing; Saedler and Baldwin, 2004) as well as stably transformed ir-MYB8 plants. In the VIGS experiment, Agrobacterium tumefaciens cultures carrying vectors pTV-MYB8 (containing a fragment of the NaMYB8 sequence) and pTV-00 (empty vector control [EV]) were used to inoculate wild-type plants, generating NaMYB8-VIGS and EV-VIGS plants, respectively. The endogenous levels of NaMYB8 transcripts in NaMYB8-VIGS plants were reduced by approximately 70% relative to EV-VIGS plants (Supplemental Fig. S3A); however, no visible changes in plant morphology or growth were observed in these plants compared with the EV-VIGS plants (data not shown). This largely ruled out the possibility that NaMYB8 is involved in the vegetative or reproductive development of N. attenuata, allowing preparation of the stably silenced transgenic plants.
Stably silenced ir-MYB lines were produced by the A. tumefaciens-mediated transformation method (Krügel et al., 2002) using pSOL8 binary vector containing the ir fragment of the NaMYB8 sequence (Supplemental Fig. S2A). After screening for single T-DNA insertion lines (Supplemental Fig. S2B), all further experiments were performed with two independently transformed homozygous diploid lines, 810 and 818 (referred to as ir-MYB8-818 and ir-MYB8-810). The lines were analyzed for their endogenous NaMYB8 transcript levels before and 1 h after W+OS elicitation, showing significantly reduced levels of NaMYB8 transcripts (ir-MYB8-818, approximately 90% silenced; ir-MYB8-810, approximately 96% silenced; ANOVA, Tukey post-hoc test, F2,6 = 128.512, Pir-MYB8-818 < 0.001, Pir-MYB8-810 < 0.001) relative to similarly elicited wild-type plants (Fig. 1B).
In order to determine the position of NaMYB8 upstream or downstream of hormonal signals in plant defense against herbivores, we quantified JA, JA-Ile accumulation, and ethylene emission levels in ir-MYB8 and wild-type plants before elicitation and 60, 90, and 180 min after W+W and W+OS elicitations. No significant differences in maximal JA (Supplemental Fig. S4A; 60 min; ANOVA, Tukey post-hoc test, F2,12 = 2.547, Pir-MYB8-818 = 0.124, Pir-MYB8-810 = 0.904), JA-Ile accumulation (Supplemental Fig. S4B; 60 min; ANOVA, Tukey post-hoc test, F2,12 = 1.148, Pir-MYB8-818 = 0.487, Pir-MYB8-810 = 0.65), or ethylene emission (Supplemental Fig. S4C) were observed between the ir-MYB8 and wild-type plants, which was consistent with the previous phytohormone analyses conducted with NaMYB8-VIGS and EV-VIGS plants (Supplemental Fig. S4, D–F). Therefore, we concluded that NaMYB8 should function downstream of JA and ethylene signals in N. attenuata, most probably below or in parallel with the putative transcriptional activator MYC2.
NaMYB8 Regulates the Accumulation of PPCs in N. attenuata Leaves
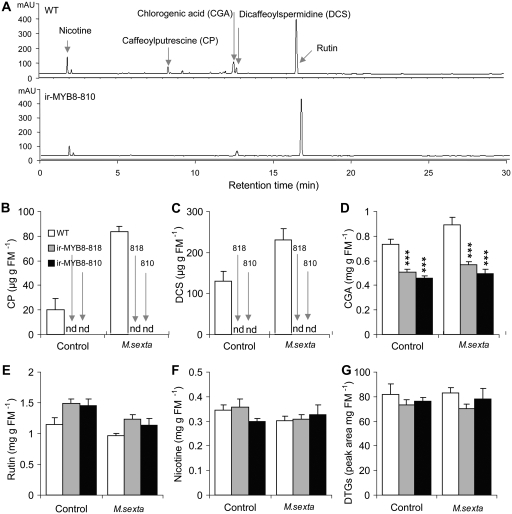
To obtain comprehensive information about the changes in herbivory-elicited secondary metabolite profiles in ir-MYB8 plants, we examined the content of secondary metabolites in (+1) rosette leaves of transgenic and wild-type plants that were fed by M. sexta caterpillars for 4 d. A simple comparison of the chromatograms, obtained from acidic methanolic plant extracts separated by HPLC, showed clear differences between ir-MYB8 and wild-type plants (Fig. 2A). In particular, CP (Fig. 2B) and DCS (Fig. 2C) could not be detected in ir-MYB8-818 and -810 leaves on which caterpillars fed, confirming previously observed trends observed in NaMYB8-VIGS leaves (Supplemental Fig. S3, B and C). Moreover, CP and DCS were not detected in unelicited leaves of ir-MYB8 plants, suggesting that both constitutive and inducible levels of CP and DCS in N. attenuata are dependent on the activity of the NaMYB8 transcription factor (Fig. 2, B and C). However, an expected overaccumulation of parental compounds, putrescine, spermidine, and caffeic acid, due to NaMYB8 silencing had no impact on the accumulation of several compounds derived from interconnected metabolic pathways, including nicotine (Fig. 2F; control, ANOVA, Tukey post-hoc test, F2,12 = 1.488, Pir-MYB8-818 = 0.939, Pir-MYB8-810 = 0.424; herbivory, ANOVA, Tukey post-hoc test, F2,12 = 0.219, Pir-MYB8-818 = 0.987, Pir-MYB8-810 = 0.804) and rutin (Fig. 2E; control, ANOVA, Tukey post-hoc test, F2, 12 = 3.746, Pir-MYB8-818 = 0.071, Pir-MYB8-810 = 0.101; herbivory, ANOVA, Tukey post-hoc test, F2,12 = 2.989, Pir-MYB8-818 = 0.077, Pir-MYB8-810 = 0.326). In contrast, a significant reduction in chlorogenic acid levels (CGA; Fig. 2D) was found in both ir-MYB8 lines (control, ANOVA, Tukey post-hoc test, F2,12 = 22.1, Pir-MYB8-818 < 0.001, Pir-MYB8-810 < 0.001; herbivory, ANOVA, Tukey post-hoc test, F2,12 = 23.792, Pir-MYB8-818 < 0.001, Pir-MYB8-810 < 0.001). Finally, no changes in the accumulation of diterpene glycosides (DTGs), unrelated but very abundant defensive metabolites (Jassbi et al., 2008), were observed in ir-MYB8 plants compared with the wild-type plants (Fig. 2G; control, ANOVA, Tukey post-hoc test, F2,12 = 0.654, Pir-MYB8-818 = 0.569, Pir-MYB8-810 = 0.994; herbivory, ANOVA, Tukey post-hoc test, F2,12 = 2.411, Pir-MYB8-818 = 0.130, Pir-MYB8-810 = 0.864). In summary, silencing NaMYB8 abolished the accumulation of CP and DCS, reduced CGA levels, and showed no statistically significant effects on other examined metabolites, namely nicotine, rutin, and DTGs, in stably silenced ir-MYB8 lines.
Figure 2.
NaMYB8 regulates the accumulation of CP and DCS in M. sexta-attacked leaves. M. sexta caterpillars were allowed to feed on N. attenuata wild-type (WT) and NaMYB8 transgenic plants for 4 d before harvesting samples for analysis. A, HPLC chromatograms obtained from wild-type and ir-MYB-810 M. sexta-fed rosette leaf methanolic extracts detected at 254-nm wavelength. B to G, Mean ± se concentrations of CP (B), DCS (C), CGA (D), rutin (E), nicotine (F), and DTGs (G) in wild-type, ir-MYB8-818, and ir-MYB8-810 leaves directly attacked by herbivores quantified by HPLC. Asterisks represent significant differences among the genotypes within the treatment group at P < 0.001 (n = 5). FM, Fresh mass; nd, not detected.
Transcriptional Targets of NaMYB8
In order to correlate the observed metabolic changes in NaMYB8-silenced lines with the NaMYB8 transcript accumulation in N. attenuata, we used a custom oligonucleotide microarray (Biochip version 4), spotted specifically with herbivory-activated genes from N. attenuata and other related species (1,421 gene probes; Meldau et al., 2009). Referring to the maximal accumulation of NaMYB8 transcripts at 45 min after W+OS treatment (Fig. 1A), we used leaf samples collected at 45 and 90 min after treatment with W+OS from each ir-MYB8-818 and wild-type plant for the microarray analysis. Three pairs of labeled cDNA probes (WT-Cy5/ir-MYB8-Cy3) were hybridized with the microarray chip to obtain differential expression data for each time point (45 min and 90 min).
Statistical analysis with three biological replications of the experiment revealed transcript abundances of eight and 44 genes to be significantly down-regulated in ir-MYB8 leaves after W+OS elicitation at 45 and 90 min, respectively (2-fold threshold; P ≤ 0.05; Supplemental Table S1). These genes predominantly included sequences from phenylpropanoid metabolism, such as Phe ammonia lyase and 4-coumaroyl-CoA ligase, and genes encoding enzymes involved in the synthesis of polyamines (Supplemental Table S1); we assume that these genes represent direct targets of NaMYB8 transcriptional activity in N. attenuata. The transcripts of the key biosynthetic gene involved in putrescine biosynthesis, Orn decarboxylase, were also less abundant in ir-MYB8 leaves, but only at 1.8-fold change level, and therefore below our arbitrarily selected threshold (2-fold). In contrast, NaMYB8 silencing significantly influenced the accumulation of spermidine synthase transcripts (approximately 3.9-fold reduction), which are required for DCS biosynthesis from spermidine.
Only a few genes were actually up-regulated in NaMYB8-silenced plants compared with the wild-type plants (Supplemental Table S1), demonstrating that NaMYB8 generally functions as a positive transcriptional regulator in N. attenuata.
Young Systemic Leaves Accumulate High Levels of CP after Simulated Herbivory
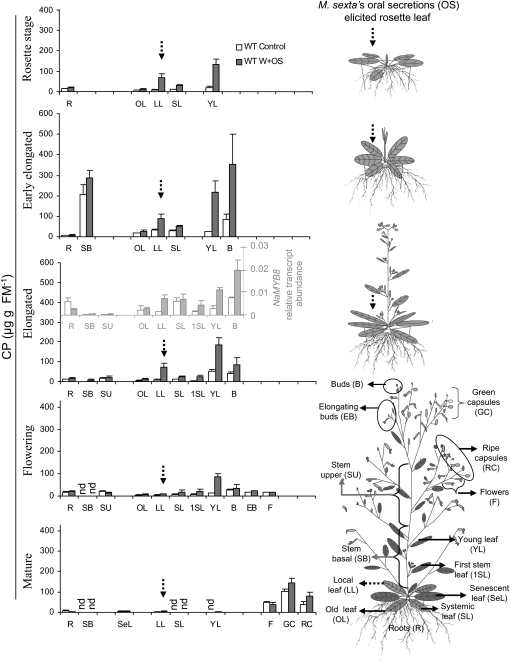
Resource allocation and spatiotemporal accumulation of defense-related metabolites vary within plant tissues and often reflect the degree and frequency of stresses that plants have to face during their development (Boege and Marquis, 2005; Boege et al., 2007). Therefore, we analyzed NaMYB8-dependent CP accumulation in plant ontogeny, namely at rosette, early elongated, elongated, flowering, and mature stages of the development, using 3-d W+OS-elicited (stressed) and unelicited (control) wild-type plants. Even though CP accumulation followed a complex developmental pattern (Fig. 3), several general trends could be recognized. For example, the high constitutive levels of CP in the vegetative tissues at rosette and early elongated stages clearly shifted toward reproductive tissues after flowering and capsule development (Figs. 3 and 4A). In the mature plants, almost no CP could be detected in the leaves. Interestingly, while CP levels always increased in the local leaves following W+OS elicitation, CP accumulated even more in the systemically induced young stem leaves of these plants, even at the flowering stage when it could be barely detected in the vegetative plant parts (Fig. 3).
Figure 3.
Young systemically induced N. attenuata leaves and reproductive tissues accumulate high levels of CP upon OS elicitation of rosette leaves. Wild-type (WT) plants were germinated in sand and supplemented with nutrients dissolved in water to the roots. At each developmental stage, a single fully expanded rosette leaf at (+1) position was OS elicited, and 3 d later the samples from representative plant parts were collected and analyzed by HPLC. The dotted arrow shows the position of the locally W+OS-elicited rosette leaf at different stages of development. NaMYB8 transcripts, shown in the gray inset, were analyzed from sample tissues used for the determination of CP at the elongated stage. R, Root; SB, stem basal; SU, stem upper; SeL, senescent leaf; OL, old leaf; LL, locally W+OS-induced rosette leaf; SL, systemic rosette leaf; 1SL, first stem leaf; YL, young stem leaf; B, flower buds; EB, elongating flower buds; F, open flowers; GC, green capsules with seeds; RC, ripe capsules with seeds; FM, fresh mass; nd, not detected.
Figure 4.
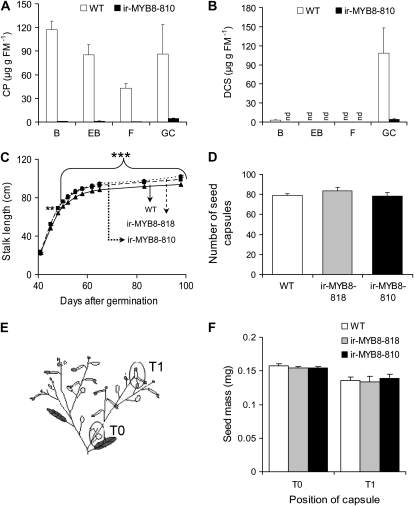
NaMYB8-silenced plants show normal growth and reproductive fitness. A and B, CP (A) and DCS (B) contents in reproductive tissues of wild-type (WT) and ir-MYB8-810 transgenic lines determined by HPLC. B, Flower buds; EB, elongating flower buds; F, open flowers; GC, green capsules with seeds; FM, fresh mass; nd, not detected. C, Mean ± se stalk lengths measured throughout plant development in the glasshouse, starting from day 41 after germination of plants. Asterisks represent significantly different growth parameters between wild-type plants and two ir-MYB8 homozygous lines at specific time points at P < 0.01 (**) and P < 0.001(***; n = 12). D, Mean ± se lifetime seed capsules produced by plants determined 98 d after germination. No statistically significant differences were observed. E, Schematic representation of the seed capsule position on the uppermost lateral branch used for seed mass determinations. T0, Seed capsule located nearest to the branching point on the top most lateral branch; T1, most distal seed capsule on the same branch. F, Mean ± se seed mass in representative seed capsules at T0 and T1 positions measured in 98-d-old plants. No statistically significant differences were observed.
We examined the transcript profile of NaMYB8 in wild-type plants at an elongated stage of development to address whether the systemic accumulation of CP could be due to metabolite mobilization to the systemic leaves or the transmission of systemic signal from the locally W+OS-induced leaves to systemic ones, which would require NaMYB8 to mediate the up-regulation of downstream biosynthetic genes. The accumulation of CP coarsely correlated with the NaMYB8 transcript abundances in both control and OS-elicited leaves (Fig. 3, inset), suggesting that CP accumulation in the distal leaves is most likely subject to systemic activation of NaMYB8 gene expression and, hence, transcriptional activity of the NaMYB8 protein.
As both local and systemic accumulation of CP and DCS was completely abolished in ir-MYB8 plants (Fig. 5A), we propose that the NaMYB8 gene serves as a universal master regulator for CP and DCS biosynthesis in N. attenuata plants. However, in contrast to a strong inducible character of CP accumulation in the leaves (Fig. 5A), no significant increase in DCS accumulation was observed in the systemically induced young stem leaves of wild-type plants (Fig. 5A), while locally W+OS-induced leaves still showed a small but significant increase in DCS levels (Fig. 5A). In addition, the constitutive levels of DCS were usually higher compared with the constitutive levels of CP. This suggests that other cis-acting regulatory elements most probably contribute to the regulation of CP and DCS synthase genes, which additionally modifies the rate-limiting NaMYB8 transcriptional activity. Alternatively, a variation in the substrate availability of putrescine and spermidine could be contributing to the differential spatiotemporal accumulation of CP and DCS in N. attenuata plants (Paschalidis and Roubelakis-Angelakis, 2005).
Figure 5.
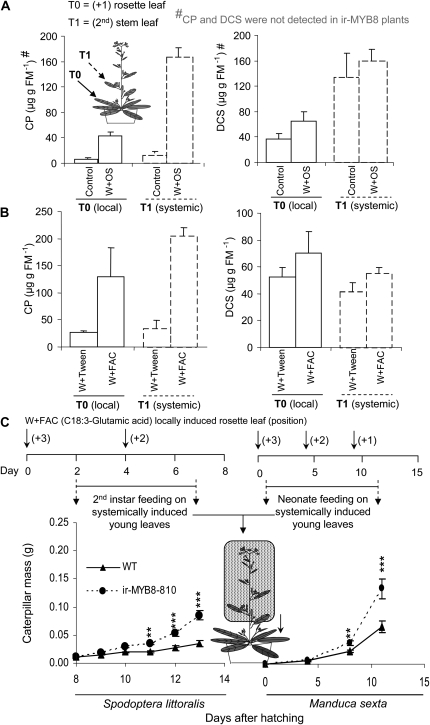
Silencing NaMYB8 in N. attenuata makes plants vulnerable to insect herbivores. A, Mean ± se CP and DCS accumulation in locally W+OS-induced rosette leaves and systemically induced young stem leaves in wild-type N. attenuata analyzed 3 d after induction (n = 4); control plants remained unelicited. Neither CP nor DCS was detected in ir-MYB8 plants. B, A pattern wheel-wounded rosette leaf of a wild-type plant was treated either with the OS-specific elicitor C18:3-Glu dissolved in 0.02% Tween or with 0.02% Tween (mock treatment), and both local and systemic accumulation of CP and DCS were analyzed as above. C, Mean ± se mass gained by N. attenuata generalist herbivore (S. littoralis) and specialist herbivore (M. sexta) caterpillars when fed on systemically preinduced young stem leaves of ir-MYB8-810 and wild-type (WT) plants, whose rosette leaves were elicited with C18:3-Glu (FAC). A single rosette leaf was elicited every 4th d to enhance the CP accumulation in the young stem leaves. Asterisks represent significantly different growth responses of herbivores that fed on wild-type and homozygous ir-MYB8-810 plants at specific time points at P < 0.01 (**) and P < 0.001 (***; n = 16). FM, Fresh mass.
Examination of Putative Roles of CP and DCS in Plant Development
The preferential accumulation of CP in the young leaves and reproductive tissues of N. attenuata suggested that CP might play multiple defensive and/or developmental roles in N. attenuata. Several previous reports have associated high CP levels with flower initiation and bud tissue development in tobacco (Martin-Tanguy, 1985, 1997; Balint et al., 1987), indirectly proposing a role for CP in flower development. Moreover, the accumulation of phenylpropanoids, polyamines, and their conjugates in the reproductive tissues has been previously associated with pollen fertility and floral initiation in plants (Wada et al., 1994; Imai et al., 2004; Kasukabe et al., 2004; Paschalidis and Roubelakis-Angelakis, 2005; Fellenberg et al., 2008, 2009; Grienenberger et al., 2009; Matsuno et al., 2009).
A prediction of the above hypothesis would be that CP (and DCS) deficiency should strongly influence flower development and seed set of ir-MYB8 N. attenuata plants. At first, the contents of CP and DCS were examined in reproductive tissues of the ir-MYB8-810 transgenic line; the contents of CP (Fig. 4A) and DCS (Fig. 4B) were heavily reduced in mature MYB8-silenced plants compared with corresponding wild-type tissues. We then closely examined the reproductive fitness-associated traits of ir-MYB8 and wild-type plants and found that ir-MYB8 plants grew better during their vegetative developmental phase, having slightly but significantly taller stalks compared with the wild-type plants (Fig. 4C; repeated-measures ANOVA, F2,33 = 55.609, Pir-MYB8-818 < 0.001, Pir-MYB8-810 < 0.001). In addition, their lifetime seed capsule production, which is considered as an important fitness measure, was comparable in wild-type and ir-MYB8 plants (Fig. 4D; ANOVA, Tukey post-hoc test, F2,33 = 0.864, Pir-MYB8-818 = 0.510, Pir-MYB8-810 = 0.999). We further quantified the seed mass in two seed capsules located on the uppermost lateral branch, one located nearest (T0) and one farthest (T1) from the branching point (Fig. 4E); the seed mass was not statistically different between ir-MYB8 and wild-type plants (Fig. 4F; T0, ANOVA, Tukey post-hoc test, F2,33 = 0.793, Pir-MYB8-818 = 0.469, Pir-MYB8-810 = 0.609; T1, ANOVA, Tukey post-hoc test, F2,33 = 0.188, Pir-MYB8-818 = 0.986, Pir-MYB8-810 = 0.902). The lack of any observable statistically significant differences between ir-MYB8 and wild-type plants in terms of reduced growth and/or reproductive fitness supports the idea that flower and seed development in N. attenuata, despite being positively correlated with the occurrence of high levels of PPCs, are independent of CP and DCS accumulation. We next focused on an alternative hypothesis that CP and DCS could be involved in direct defenses targeted against attacking herbivores, accounting for the high levels of PPC accumulation in reproductive plant parts.
Lack of CP and DCS Accumulation Makes Plants Susceptible to Herbivores
To test whether CP accumulation is part of plant defense activated against attacking herbivores, we assessed the performance of M. sexta (N. attenuata specialist) and S. littoralis (N. attenuata generalist) caterpillars on wild-type and NaMYB8-silenced plants. It is known that the elicitors present in herbivore OS, for instance, the FAC N-linolenoyl-l-Glu (C18:3-Glu), are responsible for the activation of defense mechanisms targeted against herbivores in N. attenuata (Halitschke et al., 2003; Giri et al., 2006); C18:3-Glu elicitor was found predominantly in OS of both N. attenuata generalist and specialist herbivores (Diezel et al., 2009). Therefore, we analyzed the CP and DCS accumulation in both W+C18:3-Glu-induced local rosette and systemically induced young stem leaves of N. attenuata. This treatment enhanced both local and systemic accumulation of CP in the plants, while the levels of DCS did not change significantly (Fig. 5B). Because the overall pattern of CP accumulation in W+FAC-treated leaves was comparable to W+OS-elicited leaves, we used the standardized FAC treatment to maximize the accumulation of CP in the systemically induced young leaves (Fig. 3) in the following herbivory bioassays.
To precondition the plants, we treated rosette leaves of early elongated wild-type and ir-MYB8-810 N. attenuata, one at the time, with W+C18:3-Glu 1 to 2 d before placing the caterpillars onto the young, systemically preinduced leaves. This experimental setup was specifically designed to mimic the initial feeding of the rosette leaves by neonates just hatched from oviposited eggs, before they increased size and mobility of first instar larvae migrating to other plant parts (Stork et al., 2009). Both generalist and specialist herbivores were allowed to feed only on systemically induced young stem leaves (Fig. 5C) to highlight the effect of systemic accumulation of CP in combination with the constitutive levels of DCS (Fig. 5, A and B); in the course of the experiment, additional rosette leaves were elicited with W+C18:3-Glu every 4th d to maintain the elicitation effect.
To conduct the actual bioassays, freshly hatched M. sexta (specialist herbivore) neonates were placed directly onto the FAC preinduced stem leaves. Due to the high sensitivity of S. littoralis (generalist herbivore) to N. attenuata’s defenses, hatched neonates had to be first prereared on artificial diet for 6 d. To eliminate the effect of artificial diet present in a caterpillar's gut, a short feeding on wild-type or ir-MYB8-810 leaves was then carried out with the larvae, and subsequently, preweighed caterpillars were transferred to wild-type and ir-MYB8-810 young stem leaves and their body mass gain was recorded every day. Both M. sexta (unpaired t test, P = 0.004) and S. littoralis (unpaired t test, P < 0.001) caterpillars performed better on ir-MYB8-810 plants compared with the wild-type plants (Fig. 5C). A similar trend in caterpillar performance was previously observed in M. sexta neonates that were allowed to feed directly on NaMYB8-VIGS and EV-VIGS plants (Supplemental Fig. S5; repeated-measures ANOVA, F1,28 = 6.265, P = 0.018). These results provided strong evidence that PPCs function as indispensable defensive metabolites targeted against leaf-chewing herbivores in N. attenuata plants.
Exogenous Application of Synthetic CP Impairs the Growth of M. sexta Caterpillars
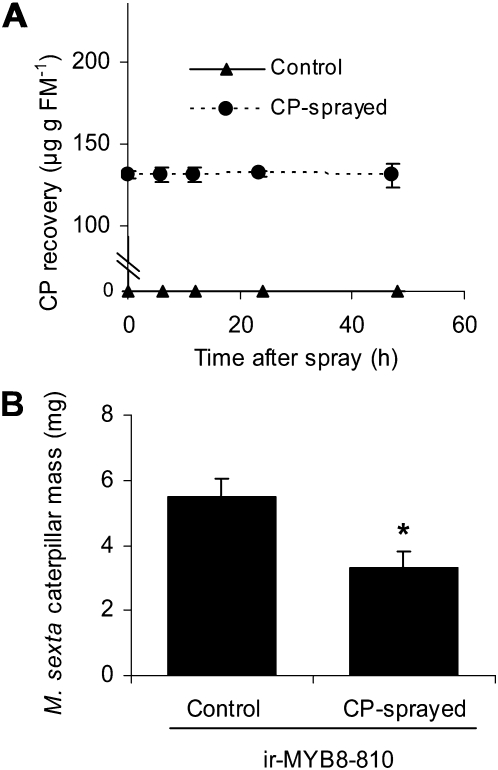
We further tested the role of CP as a plant-specific defensive metabolite against herbivores by spraying physiologically relevant concentrations of synthetic CP on ir-MYB8 leaves, which are known to be deficient in the accumulation of CP and DCS. We first examined the turnover and stability of the exogenously applied CP on the leaf surface and found that CP sprayed on ir-MYB8-810 leaves remained stable for at least 2 d after application. This shows that CP was neither degraded nor mobilized to other plant parts. After the spray, we recovered about 130 μ g CP g−1 fresh mass (Fig. 6A), which was just below the CP concentrations accumulated in the leaves after W+OS elicitation (approximately 170 μ g CP g−1 fresh mass; Fig. 5A). In the control treatment, no CP was found in the water-sprayed leaves of ir-MYB8-810 plants (Fig. 6A). We then clip caged two neonates on CP- or water-sprayed ir-MYB8-810 leaves and examined the growth of the caterpillars. The caterpillars fed for 4 d on CP-sprayed leaves showed less mass gain compared with caterpillars fed on water-sprayed (control) ir-MYB8-810 leaves (Fig. 6B; unpaired t test, P = 0.011), further highlighting the role of CP as a direct defense metabolite in response to herbivory in N. attenuata plants.
Figure 6.
CP functions as an indispensable part of direct defense against M. sexta in N. attenuata. A, Mean ± se concentrations of CP sprayed on ir-MYB8-810 leaves over a period of 48 h. B, Mean ± se M. sexta caterpillar mass gain 4 d after feeding on CP-sprayed or water-sprayed ir-MYB8-810 leaves. The asterisk represents significantly different M. sexta caterpillar growth responses between the treatments at P < 0.05 (n = 16). FM, Fresh mass.
DISCUSSION
PPCs belong to a group of ubiquitously occurring secondary metabolites in plants (Edreva et al., 2007). While their chemistry and distribution in plants are relatively well documented, much less is known about their biological functions. PPCs predominantly accumulate after pathogen and viral infections, during tuberization, and/or in response to fungal elicitors (Martin-Tanguy, 1985, 2001; Rabiti et al., 1998; Facchini et al., 2002; Walters, 2003; Cowley and Walters, 2005; Rodríguez-Kessler et al., 2008; Muroi et al., 2009), suggesting that PPCs might be involved in plant defense. Previously, the accumulation of several PPCs in N. attenuata leaves in response to herbivory has been reported (Kessler and Baldwin, 2004; Paschold et al., 2007), but no conclusive experimental evidence exists for the role of PPCs in plant-herbivore interactions. Here, we show that CP and DCS accumulation in N. attenuata, regulated by the NaMYB8 transcription factor (Fig. 7), plays an important role in plant defense against leaf-chewing herbivores.
Figure 7.
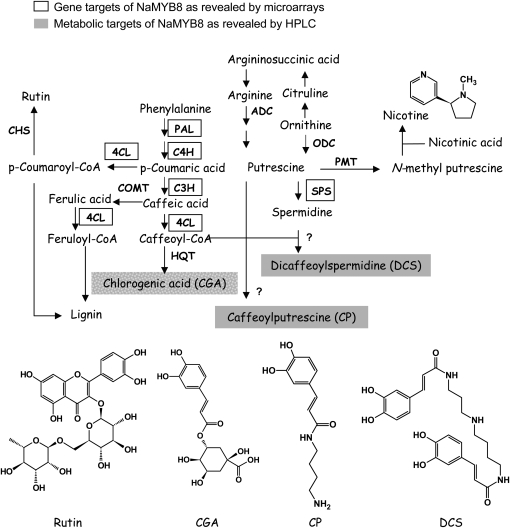
NaMYB8-regulated metabolic pathways in N. attenuata plants. The NaMYB8 transcription factor transcriptionally controls the accumulation of CP and DCS metabolites in N. attenuata plants. NaMYB8-dependent genes determined by microarray analysis are enclosed in rectangles. ADC, Arg decarboxylase; C3H, cinnamic acid-3-hydroxylase; C4H, cinnamic acid-4-hydroxylase; CHS, chalcone synthase; 4CL, 4-coumaroyl-CoA:ligase; COMT, caffeic acid O-methyltransferase; HQT, hydroxycinnamoyl-CoA quinate transferase; ODC, Orn decarboxylase; PAL, Phe ammonia lyase; PMT, putrescine methyltransferase; SPS, spermidine synthase.
JA Signaling Is Required for CP Accumulation
The accumulation of CP was previously shown to be strongly induced by JA in N. attenuata and tomato (Solanum lycopersicum) plants (Keinänen et al., 2001; Chen et al., 2006), and CP accumulated in a dose-dependent manner in JA-treated sweet pepper (Capsicum annuum) cotyledons (Tebayashi et al., 2007). The dependence of CP accumulation on JA was further highlighted using N. attenuata transgenic plants silenced in their expression of lipoxygenase 3 (antisense-LOX3), an enzyme essential for JA biosynthesis, which resulted in strongly reduced CP levels in M. sexta OS-elicited leaves (Paschold et al., 2007). The accumulation of CP in antisense-LOX3 plants could be rescued by exogenously applied JA, suggesting that JA is one of the major limiting factors in CP accumulation. The role of JA and its active metabolite JA-Ile in CP biosynthesis was further examined in ir-COI1 N. attenuata plants impaired in JA-Ile perception due to a nonfunctional SCFcoi1 protein complex. ir-COI1 plants showed strongly reduced CP levels (Paschold et al., 2007), similar to tomato jai-1 mutants defective in JA perception, which failed to accumulate CP in the flowers and in the methyl jasmonate-treated leaves (Chen et al., 2006). Altogether, these observations provide a strong link between JA signaling and CP accumulation; however, the actual regulatory mechanisms initiated after JA perception, particularly those downstream of putative MYC2 protein regulation in N. attenuata, leading to PPC biosynthesis in plants, and activation of enzymes such as Phe ammonia lyase (phenylpropanoid biosynthesis) or Orn decarboxylase/Arg decarboxylase (polyamines), remain elusive.
Transcriptional Regulation of the Phenylpropanoid Pathway
The phenylpropanoid biosynthesis in plants is often controlled by specific transcription factors, including the members of the MYB gene family (Arabidopsis [Stracke et al., 2001] and Populus trichocarpa [Wilkins et al., 2009]). The most abundant subgroup of MYB transcriptional regulators, R2R3-MYBs, is frequently activated by biotic and abiotic stress factors, such as UV light exposure, wounding, osmotic stress, anaerobic conditions, herbivory, and pathogen infections, which further regulate the accumulation of secondary metabolites in stressed plants (Jin et al., 2000; Vailleau et al., 2002; Mengiste et al., 2003; De Vos et al., 2006; Gigolashvili et al., 2007; Sønderby et al., 2007; Tong Geon et al., 2007; Lippold et al., 2009; Mellway et al., 2009). In contrast, several MYB genes have been shown to regulate processes different from secondary metabolism, including cellular morphogenesis, cell cycle, meristem formation, fiber and trichome development, and lignification in plants (Baumann et al., 2007; Legay et al., 2007; Bomal et al., 2008; Petroni et al., 2008; Cominelli and Tonelli, 2009; Machado et al., 2009; Zhang et al., 2009).
The direct connection between MYB transcriptional activity and the regulation of PPC biosynthesis was first reported by Gális et al. (2006) and Shinya et al. (2007). Ectopically expressed R2R3-MYB transcription factors, such as NtMYBJS1 (methyl jasmonate responsive) and NtMYBGR1 (glucan elicitor responsive), increased the accumulation of CP and feruloylputrescine (another PPC) in unelicited BY-2 tobacco cell cultures. Both transcription factors selectively responded to their own elicitors; however, both proteins converged in the regulation of a similar group of genes that were involved in phenylpropanoid biosynthesis and controlled the accumulation of PPCs in transformed plant cells (Gális et al., 2006; Shinya et al., 2007). The relatively large spectrum of elicitors that could induce various MYB transcription factors, and enhance the accumulation of PPCs, suggests that these metabolites could be mediating a broad range of resistances to fungi, necrotrophic pathogens, and herbivores.
NaMYB8: OS-Responsive Regulator of CP and DCS Accumulation
NaMYB8 transcripts accumulated rapidly in N. attenuata rosette leaves after wounding and returned to the preelicited levels within 3 h after treatment (Fig. 1A); however, the M. sexta OS-elicited rosette leaves showed delayed reinstatement of transcripts to their basal levels at late hours after induction, reflecting that NaMYB8 expression discriminates between herbivory-associated damage and simple mechanical wounding. Similar trends in transcript accumulation are typically found in genes involved in defense triggered by M. sexta OS elicitation, for instance, genes encoding trypsin proteinase inhibitors (Halitschke et al., 2001). Previously, we showed that the NaMYB8 gene is also induced by UV-B light in the glasshouse and by cumulative stress conditions in the natural environment of N. attenuata, both following an RdR2-dependent induction pattern (Pandey and Baldwin, 2008). This suggests that, apart from regulating N. attenuata’s response to herbivores, the NaMYB8 gene may also be regulated by various abiotic stresses, namely high levels of UV-B light in the natural environment.
We examined the transcription regulatory activity of NaMYB8 by performing microarray analysis with OS-elicited ir-MYB8-818 leaves hybridized against identically elicited wild-type leaves. This analysis confirmed that NaMYB8 specifically activates the transcription of genes involved in phenylpropanoid and polyamine biosynthesis (Fig. 7; Supplemental Table S1). A similar transcriptional regulatory pattern of genes involved in the phenylpropanoid pathway was observed in BY-2 tobacco cell cultures ectopically expressing the NtMYBJS1 gene using an independent tobacco microarray system (Gális et al., 2006). Interestingly, several novel NaMYB8-controlled genes with a potential role in PPC biosynthesis have been identified in the current microarray experiment, and the functional characterization of these genes is in progress. Remarkably, one of the NaMYB8-regulated genes encodes a functional DCS synthase in N. attenuata (N. Onkokesung, unpublished data).
The accumulation of CP and DCS in wild-type plants correlated well with the pattern of NaMYB8 transcript abundance. The absence of both metabolites in NaMYB8-silenced lines, regardless of plant treatment, provided final conclusive proof that the NaMYB8 transcription factor is essential for CP and DCS biosynthesis and accumulation in N. attenuata plants (Fig. 7), similar to the role of the PAP1 gene, an AtMYB75 transcriptional regulator that governs anthocyanin biosynthesis in Arabidopsis (Teng et al., 2005; Tohge et al., 2005).
CP Accumulates in the Young Vegetative and Reproductive Tissues
The ecological roles of metabolites can often be deduced from their spatiotemporal accumulation patterns. CP and DCS accumulated in the shoot apices, young leaves, and female reproductive organs during flower induction and development in tobacco (Cabanne et al., 1981; Martin-Tanguy, 1985; Edreva et al., 2007), and a similar pattern of PPC accumulation was also observed in some Araceae species (Ponchet et al., 1982). The accumulation of CP in reproductive organs is widely reported in the literature, which is generally linked to plant development. However, when the distribution of PPCs was examined across seven species belonging to the Solanaceae family, CP accumulated predominantly in the pistils of tobacco flowers but this pattern was absent in several other species, indicating that PPCs may not be the universal regulators of flower initiation in solanaceous plants (Leubner-Metzger and Amrhein, 1993). Moreover, the use of specific inhibitors that can reduce the accumulation of feruloylputrescine and CP in in vitro tobacco cultures did not affect the growth and floral bud formation in tobacco stem explants (Wyss-Benz et al., 1990), further supporting the alternative role of PPCs in plants.
In the search for ecologically meaningful interpretations of CP accumulation patterns, we decided to treat the plants with simulated herbivory (Halitschke et al., 2001) at five developmental stages and compared the CP accumulation with untreated plants. Interestingly, both constitutive and inducible levels of CP were higher in the young stem leaves compared with the mature rosette leaves (Figs. 3 and 5A). In agreement with previous studies (Edreva et al., 2007), CP was most abundant in the buds at the early elongated stage, but these levels progressively declined as plants aged (Fig. 3). At the mature stage, plants retained high levels of CP almost exclusively in the reproductive organs, flowers, and capsules. However, when ectopically silencing the ability of plants to accumulate PPCs, we found no aberrant morphological changes associated with this novel trait. In summary, the CP accumulation shifted primarily from photosynthetically active young leaves during vegetative growth to flower buds, flowers, and seed capsules at maturity, and it was significantly stimulated by simulated herbivory treatments (Fig. 3).
CP and DSC Are Indispensable for Plant Defense against Herbivores
In our follow-up hypothesis, CP could be a direct defensive metabolite that accumulates upon herbivory and against herbivores. According to the optimal defense theory, plants tend to allocate more defense-associated metabolites to the valuable plant parts during development, such as photosynthetically active tissues, meristems, and reproductive tissues (flowers and seeds), to protect these organs from stress factors, including herbivores (McKey, 1974; Ohnmeiss and Baldwin, 2000; Stamp, 2003). A shift in CP accumulation, consistent with the theory, was indeed observed in N. attenuata plants during development and in response to herbivore-associated cues, suggesting that N. attenuata plants allocate PPCs to the young leaves in an attempt to protect them from attack by herbivores and/or pathogens. Defensive roles of several PPCs in plant resistance to pathogens have already been demonstrated, including preferential accumulation of PPCs in systemically induced tissues: systemic leaves of barley (Hordeum vulgare) accumulated higher concentrations of PPCs compared with methyl jasmonate-induced local leaves, and this higher accumulation of PPCs in systemic leaves was correlated with stronger resistance to powdery mildew infection (Walters et al., 2002).
Previously, CP was tested as an ovipositioning deterrent in cotyledons of sweet pepper by dipping leaves in CP solutions (Tebayashi et al., 2007). Treated leaves showed fewer oviposition-associated punctures from Liriomyza trifolii compared with nontreated leaves, demonstrating for the first time a defensive function of CP against herbivores in plants. PPCs, in particular N1-coumaroylspermine, have been shown to inhibit mammalian and crayfish neuroreceptors in vitro, being structurally similar to acylated polyamines found in spider and wasp toxins. However, when the shorter naturally occurring polyamine conjugates in plants (N1- and N8-coumaroylspermidine) were supplied in artificial diet to European corn borer, tobacco budworm, and the oblique banded leaf roller, no toxic effects of these metabolites on insects were found (Fixon-Owoo et al., 2003). This suggests that certain leaf-derived metabolites and/or enzymatic activities may be required for demonstrated toxicity of PPCs against herbivores in natural situations.
Here, we made a targeted attempt to investigate the role of CP and DCS in plant-insect interaction, using a reverse genetic approach and plants silenced in the expression of the master transcriptional regulator responsible for the accumulation of CP and DCS in native tobacco plants. In accordance with the proposed defensive role against herbivores, both generalist and specialist caterpillars gained more body mass when allowed to feed on the systemically induced leaves of ir-MYB8 plants compared with the wild-type plants. The last piece of supporting evidence resulted from experiments using direct application of synthetic CP at physiological concentrations to the ir-MYB8 leaves: ir-MYB8 leaves, when sprayed with synthetic CP, supported less M. sexta larval growth than did the water-sprayed ir-MYB8 leaves. This experiment further showed that PPCs should be tested in a natural context, together with plant leaves, rather than with an artificial diet to show their toxic effects.
CONCLUSION
We used a well-established ecological model plant, N. attenuata, combined with state-of-the-art molecular tools to construct transgenic plants deficient in the expression of a master transcriptional activator required for CP and DCS biosynthesis both at constitutive and elicited levels. The use of a master regulator (NaMYB8) allowed us to specifically down-regulate the biosynthetic pathway leading to CP and DCS accumulation without disturbing other defense-related mechanisms in plants. The functional analysis of ir-MYB8 plants then highlighted the important ecological roles of CP and DCS in plant defense operating against herbivorous insects. Further studies with ir-MYB8 plants will enable us to understand the roles of CP and DCS in defense against pathogens, and the results from our microarray analysis shall be used to identify novel genes responsible for CP and DCS biosynthesis.
MATERIALS AND METHODS
Plant Growth Conditions in the Glasshouse
Nicotiana attenuata (22nd inbred generation) seeds, originally collected from a native population from a field site located in Utah in the United States, were used for all described experiments, including transformation and generation of transgenic lines. The seeds were germinated on sterile Gamborg B5 medium (Sigma; http://www.sigmaaldrich.com) after 1 h of treatment with diluted smoke (House of Herbs) and 1 μm GA3 (www.carl-roth.de). Ten days after germination, seedlings were transferred into Teku pots containing peat-based substrate, and after an additional 10 to 12 d, the plantlets were transplanted into individual 1-L pots with the same substrate. In the glasshouse, plants were grown at 24°C to 26°C, relative humidity approximately 55%, and supplemented with light from 400- and 600-W sodium lamps (Philips Sun-T Agro; http://www.nam.lighting.philips.com) for 16 h.
VIGS
A VIGS system based on the Tobacco mosaic rattle virus was used as described by Saedler and Baldwin (2004). Three-week-old N. attenuata plants were inoculated with Agrobacterium tumefaciens and pTVMYB8 plasmid, carrying a fragment of NaMYB8 (NaMYB8-VIGS) or pTV00, carrying empty vector construct as a control (EV-VIGS).
Generation and Characterization of ir-MYB8 Transgenic Lines
Stably transformed ir-MYB8 lines were generated by introducing an ir construct containing a 309-bp NaMYB8 gene fragment and a gene for hygromycin resistance (hptII) as a screening marker in a pSOL8 transformation vector (Supplemental Fig. S2A) into N. attenuata plants as described by Bubner et al. (2006). A. tumefaciens-mediated plant transformation was followed as described by Krügel et al. (2002). Transformed lines, each containing a single insertion of the hptII marker gene as determined by Southern hybridization, were further screened by segregation analysis of T2 seedlings for their hygromycin resistance to obtain homozygous transformed lines. Quantitative real-time (RT-q)PCR was used to quantify the transcript accumulation of the NaMYB8 gene, and two independently transformed homozygous diploid lines with efficiently silenced expression of the NaMYB8 gene, ir-MYB8-810 and ir-MYB8-818, were selected for all subsequent experiments.
Southern-Blot Hybridizations
A modified cetyltrimethylammonium bromide method as described by Paschold et al. (2007) was followed for genomic DNA extraction from the fully expanded rosette leaves of wild-type, ir-MYB8-810, and ir-MYB8-818 N. attenuata plants (Rogers and Bendich, 1985). For Southern-blot hybridizations, 15 μ g of the DNA was digested overnight with two different restriction enzymes (New England Biolabs; http://www.neb.com) at 37°C, size fractionated on a 0.8% (w/v) agarose gel, and blotted onto a nylon membrane (GeneScreenPlus; Perkin-Elmer; http://www.perkinelmer.com). Fragments of hptII (forward primer 5′-CGTCTGTCGAGAAGTTTCTG-3′; reverse primer 3′-CCGGATCGGACGATTGCG-5′) were PCR amplified and used as probe for Southern hybridizations to confirm the single insertion of the hptII gene in ir-MYB8-transformed lines. The DNA probes were labeled with α -32P using the Rediprime II DNA labeling system (Amersham Biosciences; http://www.amershambiosciences.com).
Expression Analysis by RT-qPCR
To analyze NaMYB8 gene expression, total RNA was extracted from fully expanded control and W+OS-elicited rosette leaves of wild-type and ir-MYB8 plants, following the Trizol method as recommended by the manufacturer (Invitrogen; http://www.invitrogen.com). Thirty micrograms of RNA was given DNase treatment by adding 10 units of RQ1 RNase-free DNase in 1 × enzyme buffer following the manufacturer's instructions (Promega; http://www.promega.com). Each sample was incubated at 65°C for 20 min to inactivate DNase enzyme, extracted in phenol:chloroform:isoamyl alcohol (25:24:1), and precipitated in the presence of 0.1 volume of 3 m sodium acetate, pH 5.2, and 3 volumes of ice-cold pure ethanol at −20°C. Two micrograms of total RNA was reverse transcribed using oligo(dT) (Fermentas; http://www.fermentas.com) as primer and SuperScript II reverse transcriptase (Invitrogen). All RT-qPCR assays were performed with cDNA corresponding to 100 ng of RNA before reverse transcription and gene-specific primers using the qPCR core kit for SYBR Green I (Eurogentec; http://www.eurogentec.com), following the manufacturer's instructions.
To determine the NaMYB8 gene transcript levels in the ir-MYB8 lines, gene-specific primers were designed outside the region used for making the ir silencing construct. All gene-specific primers were designed with Primer3 online available software (http://frodo.wi.mit.edu/primer3). For all RT-qPCR analyses, unless stated differently, cDNA from five replicate biological samples was used and the assay was carried out on a Stratagene Mx3005P real-time PCR system (http://www.stratagene.com). Relative gene expression was calculated using a 10-fold dilution series of cDNA containing NaMYB8 as well as the elongation factor-1 α housekeeping gene from Nicotiana tabacum (accession no. D63396) as an endogenous reference.
To determine whether herbivore attack elicits NaMYB8 transcription, five fully expanded leaves at rosette stage were either W+W or W+OS elicited or left untreated as a control. All five leaves were pooled, and cDNA was prepared from each time point and treatment over a 48-h elicitation period. The transcript abundance of the NaMYB8 gene was quantified with RT-qPCR in three technical replicates of the pooled samples.
Microarray Analysis
We used a custom oligonucleotide DNA microarray selectively enriched with herbivore-activated genes from N. attenuata and related species (Biochip version 4; 1,421 gene probes; Meldau et al., 2009). The hybridizations were carried out with cDNA synthesized from pooled leaf tissues representing 15 plants in three biological replicates from wild-type and ir-MYB8-818 OS-elicited rosette leaves, which were harvested 45 and 90 min after elicitation. The cDNA samples from ir-MYB8-818 and the wild type were labeled with Cy3 and Cy5 fluorescent dyes, respectively, and three pairs of labeled probes were hybridized with the microarray (WT-Cy5/ir-MYB8-Cy3) to obtain differential expression data for each time point. Hybridization and analysis were carried out essentially as described by Wang et al. (2008).
Phytohormone Analysis
A (+1) ir-MYB8/WT or EV/NaMYB8-VIGS fully expanded N. attenuata rosette leaf was either W+W or W+OS elicited or left unelicited; the leaves were then collected, snap frozen in liquid nitrogen, and stored at −80°C until phytohormone analysis. The phytohormones JA and JA-Ile were extracted following the procedure described by Wu et al. (2007). In brief, 150 mg of leaf tissue was extracted with 1 mL of ethyl acetate spiked with 200 ng of JA-13C2, and JA-Ile-13C6 as internal standard, in FastPrep tubes containing 0.9 g of FastPrep Matrix (Sili; http://www.sigmund-lindner.com). The tissue was homogenized, and samples were centrifuged at 13,000 rpm for 15 min at 4°C before the supernatant was transferred to clean 2-mL Eppendorf tubes. Each pellet was reextracted with 1 mL of ethyl acetate and centrifuged; supernatants were combined and then dried in a vacuum concentrator to near dryness (Eppendorf; http://www.eppendorf.com). The residue in the tubes was resuspended in 0.5 mL of 70% methanol (v/v). A 10- μ L extract was subjected to reverse-phase HPLC coupled to tandem mass spectrometry, and the peaks were identified with a 1200LC MS/MS/MS system (Varian; http://www.varianinc.com) after negative ionization with parent ion/daughter ion selections as follows: 209/59 (JA), 211/61 (JA-13C2), 322/130 (JA-Ile), and 328/136 (JA-Ile-13C6). JA and JA-Ile were quantified with respect to each compound's labeled internal standard (Wu et al. 2007).
Secondary Metabolite Analysis
The secondary metabolite analysis was carried out on (+1) leaves after Manduca sexta caterpillar feeding on WT/ir-MYB8 and EV/NaMYB8-VIGS plants for 4 d, using HPLC as described by Keinänen et al. (2001). For analysis of the locally and systemically induced responses in WT/ir-MYB8-810/ir-MYB8-818, a (+1) leaf was either W+W or W+OS elicited or left unelicited. The 3-d OS-elicited rosette leaf served as the local leaf, and the second stem leaf was used for systemic response analysis.
To examine the spatiotemporal distribution of CP, a study was conducted on N. attenuata wild-type plants at five different stages of development (rosette, early elongated, elongated, flowering, and mature). At each stage, a (+1) leaf was either OS elicited or left unelicited, and 3 d after elicitation, representative tissue samples were collected and analyzed by HPLC. A 200-mg aliquot of tissue in FastPrep tubes containing 0.9 g of FastPrep matrix (Sili) was extracted with 1 mL of 40% methanol prepared with 0.5% acetic acid water. The sample in FastPrep tubes was homogenized on a FastPrep homogenizer (Thermo Electron) for 45 s and then centrifuged for 12 min at 13,000 rpm. The supernatant was transferred into 1.5-mL Eppendorf tubes, centrifuged, and finally transferred to a glass vial, where it was analyzed by an Agilent-HPLC 1100 series (http://www.chem.agilent.com). The ODS Inertsil C18 column (3 μ m, 150 × 4.6 mm i.d.) was attached to a Phenomenex Security Guard C18 precolumn (http://www.phenomenex.com). The solvents were 0.25% H3PO4 in water (A) and acetonitrile (B). The elution system was as follows: 0 to 6 min, 0% to 12% B; 6 to 10 min, 12% to 18% B; 10 to 30 min, 18% to 58% B. The flow rate was 1 mL min−1, the injection volume was 10 μ L, and the column oven was set at 24°C. The nicotine eluted at retention time of 1.83 min was detected at 254 nm; CP, CGA, and DCS eluted at retention times of 8.3, 12.3, and 12.7 min, respectively, were detected at 320 nm; rutin eluted at 16.4 min was detected at 360 nm; and diterpene glycosides eluting at 24 to 26 min were detected at 210 nm.
Measuring Ethylene Accumulation
At least three replicate measurements were used to quantify ethylene production in wild-type and NaMYB8 transgenic N. attenuata plants. Three leaves were treated either with W+W or W+OS in the case of EV/NaMYB8-VIGS or with W+OS in the case of WT/ir-MYB8-818/ir-MYB8-810, or plants were left untreated (controls). Leaves were cut from stems, immediately sealed in a three-neck 250-mL round-bottom flask, and kept in the glasshouse under light conditions for 5 h. The head space of the flasks was flushed into a photoacoustic laser spectrometer with hydrocarbon-free clean air, and the ethylene concentration was quantified by comparing ethylene peak areas with peak areas generated by a standard ethylene gas as described previously by Von Dahl et al. (2007).
Herbivore Performance
The growth performance of M. sexta (N. attenuata specialist) and Spodoptera littoralis (N. attenuata generalist) caterpillars was examined using wild-type and ir-MYB8-silenced plants. Due to the high sensitivity of the generalist herbivore to N. attenuata defenses, S. littoralis neonates were first reared on artificial diet for 6 d and then placed on wild-type or ir-MYB8 leaves for 1 d to get rid of artificial diet present in the caterpillars’ guts; subsequently, the preweighed S. littoralis caterpillars were placed on the plants. Freshly hatched N. attenuata specialist M. sexta neonates were placed directly on WT/ir-MYB8-810 stem leaves. Both generalist and specialist herbivores were allowed to feed only on systemically preinduced stem leaves of plants, which had their single rosette leaf elicited with FAC (C18:3-Glu; 0.07 nmol μ L−1 in 0.02% [v/v] Tween 20/water; 20 μ L per leaf) every 4th d from the start of the experiment. S. littoralis caterpillar mass was recorded daily over 5 d, while the M. sexta caterpillar mass was recorded on 4, 7, and 11 d of feeding.
CP Synthesis
CP was synthesized in the laboratory as described by Hu and Hesse (1996).
Statistical Analysis
All statistical analyses were performed with SPSS software (http://www.spss.com).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number GU451752.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Deduced NaMYB8 protein sequence aligned with its homolog from N. tabacum (NtMYBJS1).
Supplemental Figure S2. The map of NaMYB8 transformation vector pSOL8MYB8, and Southern-blot analysis of the two independently transformed ir-MYB8 lines.
Supplemental Figure S3. NaMYB8 is required for the accumulation of CP and DCS in M. sexta-attacked leaves.
Supplemental Figure S4. Silencing NaMYB8 does not change W+OS-elicited phytohormone concentrations in the leaves.
Supplemental Figure S5. M. sexta caterpillars perform better on NaMYB8-VIGS plants relative to EV-VIGS plants.
Supplemental Table S1. Silencing NaMYB8 suppresses the accumulation of CP and DCS by down-regulating the expression of genes involved in phenylpropanoid and polyamine metabolism.
Acknowledgments
We thank Thomas Hahn for sequencing; Klaus Gase and Antje Wissgott for vector construction; Susan Kutschbach and Wibke Kröber for generating the stably transformed plants; E. Rothe for technical assistance with HPLC; Tamara Krügel, Andreas Weber, and Andreas Schüenzel for growing the plants in the glasshouse; and Emily Wheeler for editorial assistance.
References
- Balint R, Cooper G, Staebell M, Filner P. (1987) N-Caffeoyl-4-amino-n-butyric acid, a new flower-specific metabolite in cultured tobacco cells and tobacco plants. J Biol Chem 262: 11026–11031 [PubMed] [Google Scholar]
- Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C. (2007) Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development 134: 1691–1701 [DOI] [PubMed] [Google Scholar]
- Boege K, Dirzo R, Siemens D, Brown P. (2007) Ontogenetic switches from plant resistance to tolerance: minimizing costs with age?. Ecol Lett 10: 177–187 [DOI] [PubMed] [Google Scholar]
- Boege K, Marquis RJ. (2005) Facing herbivory as you grow up: the ontogeny of resistance in plants. Trends Ecol Evol 20: 441–448 [DOI] [PubMed] [Google Scholar]
- Bomal C, Bedon F, Caron S, Mansfield SD, Levasseur C, Cooke JEK, Blais S, Tremblay L, Morency MJ, Pavy N, et al. (2008) Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. J Exp Bot 59: 3925–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C. (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18: 756–767 [DOI] [PubMed] [Google Scholar]
- Bubner B, Gase K, Berger B, Link D, Baldwin I. (2006) Occurrence of tetraploidy in Nicotiana attenuata plants after Agrobacterium-mediated transformation is genotype specific but independent of polysomaty of explant tissue. Plant Cell Rep 25: 668–675 [DOI] [PubMed] [Google Scholar]
- Cabanne F, Dalebroux MA, Martin-Tanguy J, Martin C. (1981) Hydroxycinnamic acid amides and ripening to flower of Nicotiana tabacum var. xanthi n.c. Physiol Plant 53: 399–404 [Google Scholar]
- Chen H, Jones AD, Howe GA. (2006) Constitutive activation of the jasmonate signaling pathway enhances the production of secondary metabolites in tomato. FEBS Lett 580: 2540–2546 [DOI] [PubMed] [Google Scholar]
- Chen MS. (2008) Inducible direct plant defense against insect herbivores: a review. Insect Sci 15: 101–114 [DOI] [PubMed] [Google Scholar]
- Chico JM, Chini A, Fonseca S, Solano R. (2008) JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol 11: 486–494 [DOI] [PubMed] [Google Scholar]
- Chini A, Boter M, Solano R. (2009a) Plant oxylipins: COI1JAZsMYC2 as the core jasmonic acid-signalling module. FEBS J 276: 4682–4692 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Chico JM, Fernández-Calvo P, Solano R. (2009b) The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J 59: 77–87 [DOI] [PubMed] [Google Scholar]
- Chung HS, Niu Y, Browse J, Howe GA. (2009) Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry 70: 1547–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Tonelli C. (2009) A new role for plant R2R3-MYB transcription factors in cell cycle regulation. Cell Res 19: 1231–1232 [DOI] [PubMed] [Google Scholar]
- Cowley T, Walters DR. (2005) Local and systemic changes in arginine decarboxylase activity, putrescine levels and putrescine catabolism in wounded oilseed rape. New Phytol 165: 807–811 [DOI] [PubMed] [Google Scholar]
- De Vos M, Denekamp M, Dicke M, Vuylsteke M, Van Loon LC, Smeekens SCM, Pieterse CMJ. (2006) The Arabidopsis thaliana transcription factor AtMYB102 functions in defense against the insect herbivore Pieris rapae. Plant Signal Behav 1: 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezel C, von Dahl CC, Gaquerel E, Baldwin IT. (2009) Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol 150: 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt NA. (2008) Oxylipin signaling in plant stress responses. Plant Cell 20: 495–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edreva AM, Velikova VB, Tsonev TD. (2007) Phenylamides in plants. Russ J Plant Physiol 54: 287–301 [Google Scholar]
- Facchini PJ, Hagel J, Zulak KG. (2002) Hydroxycinnamic acid amide metabolism: physiology and biochemistry. Can J Bot 80: 577–589 [Google Scholar]
- Fellenberg C, Böttcherb C, Vogt T. (2009) Phenylpropanoid polyamine conjugate biosynthesis in Arabidopsis thaliana flower buds. Phytochemistry 70: 1392–1400 [DOI] [PubMed] [Google Scholar]
- Fellenberg C, Milkowski C, Hause B, Lange PR, Böttcher C, Schmidt J, Vogt T. (2008) Tapetum-specific location of a cation-dependent O-methyltransferase in Arabidopsis thaliana. Plant J 56: 132–145 [DOI] [PubMed] [Google Scholar]
- Fixon-Owoo S, Levasseur F, Williams K, Sabado TN, Lowe M, Klose M, Mercier AJ, Fields P, Atkinson J. (2003) Preparation and biological assessment of hydroxycinnamic acid amides of polyamines. Phytochemistry 63: 315–334 [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chico JM, Solano R. (2009) The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Gális I, Simek P, Narisawa T, Sasaki M, Horiguchi T, Fukuda H, Matsuoka K. (2006) A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant J 46: 573–592 [DOI] [PubMed] [Google Scholar]
- Gatehouse JA. (2002) Plant resistance towards insect herbivores: a dynamic interaction. New Phytol 156: 145–169 [DOI] [PubMed] [Google Scholar]
- Gigolashvili T, Yatusevich R, Berger B, Müller C, Flügge UI. (2007) The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 51: 247–261 [DOI] [PubMed] [Google Scholar]
- Giri AP, Wunsche H, Mitra S, Zavala JA, Muck A, Svatos A, Baldwin IT. (2006) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. VII. Changes in the plant's proteome. Plant Physiol 142: 1621–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E, Besseau S, Geoffroy P, Debayle D, Heintz D, Lapierre C, Pollet B, Heitz T, Legrand M. (2009) A BAHD acyltransferase is expressed in the tapetum of Arabidopsis anthers and is involved in the synthesis of hydroxycinnamoyl spermidines. Plant J 58: 246–259 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Gase K, Hui DQ, Schmidt DD, Baldwin IT. (2003) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. VI. Microarray analysis reveals that most herbivore-specific transcriptional changes are mediated by fatty acid-amino acid conjugates. Plant Physiol 131: 1894–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT. (2001) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol 125: 711–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Hu W, Hesse M. (1996) Synthese der p-Cumaroylspermidine. Helv Chim Acta 79: 548–559 [Google Scholar]
- Imai A, Matsuyama T, Hanzawa Y, Akiyama T, Tamaoki M, Saji H, Shirano Y, Kato T, Hayashi H, Shibata D, et al. (2004) Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol 135: 1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassbi AR, Gase K, Hettenhausen C, Schmidt A, Baldwin IT. (2008) Silencing geranylgeranyl diphosphate synthase in Nicotiana attenuata dramatically impairs resistance to tobacco hornworm. Plant Physiol 146: 974–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. (2009) Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol 60: 407–431 [DOI] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasukabe Y, He L, Nada K, Misawa S, Ihara I, Tachibana S. (2004) Over-expression of spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana. Plant Cell Physiol 45: 712–722 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2008) Jasmonate signaling: toward an integrated view. Plant Physiol 146: 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinänen M, Oldham NJ, Baldwin IT. (2001) Rapid HPLC screening of jasmonate-induced increases in tobacco alkaloids, phenolics, and diterpene glycosides in Nicotiana attenuata. J Agric Food Chem 49: 3553–3558 [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. (2004) Herbivore-induced plant vaccination. Part I. The orchestration of plant defenses in nature and their fitness consequences in the wild tobacco Nicotiana attenuata. Plant J 38: 639–649 [DOI] [PubMed] [Google Scholar]
- Koornneef A, Pieterse CMJ. (2008) Cross talk in defense signaling. Plant Physiol 146: 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. (2002) Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12: 177–183 [Google Scholar]
- Legay S, Lacombe E, Goicoechea M, Briere C, Seguin A, MacKay J, Grima-Pettenati J. (2007) Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci 173: 542–549 [Google Scholar]
- Leubner-Metzger G, Amrhein N. (1993) The distribution of hydroxycinnamoyl-putrescines in different organs of Solanum tuberosum and other solanaceous species. Phytochemistry 32: 551–556 [Google Scholar]
- Li J, Ou-Lee TM, Raba R, Amundson RG, Last RL. (1993) Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK, Udvardi MK. (2009) AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol 149: 1761–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loake G, Grant M. (2007) Salicylic acid in plant defence: the players and protagonists. Curr Opin Plant Biol 10: 466–472 [DOI] [PubMed] [Google Scholar]
- Luo J, Fuell C, Parr A, Hill L, Bailey P, Elliott K, Fairhurst SA, Martin C, Michael AJ. (2009) A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell 21: 318–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES. (2009) The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J 59: 52–62 [DOI] [PubMed] [Google Scholar]
- Martin-Tanguy J. (1985) The occurrence and possible function of hydroxycinnamoyl acid amides in plants. J Plant Growth Regul 3: 381–399 [Google Scholar]
- Martin-Tanguy J. (1997) Conjugated polyamines and reproductive development: biochemical, molecular and physiological approaches. Physiol Plant 100: 675–688 [Google Scholar]
- Martin-Tanguy J. (2001) Metabolism and function of polyamines in plants: recent development (new approaches). J Plant Growth Regul 34: 135–148 [Google Scholar]
- Matsuno M, Compagnon V, Schoch GA, Schmitt M, Debayle D, Bassard JE, Pollet B, Hehn A, Heintz D, Ullmann P, et al. (2009) Evolution of a novel phenolic pathway for pollen development. Science 325: 1688–1692 [DOI] [PubMed] [Google Scholar]
- McKey D. (1974) Adaptive patterns in alkaloid physiology. Am Nat 108: 305–320 [Google Scholar]
- Meldau S, Wu J, Baldwin IT. (2009) Silencing two herbivory-activated MAP kinases, SIPK and WIPK, does not increase Nicotiana attenuata’s susceptibility to herbivores in the glasshouse and in nature. New Phytol 181: 161–173 [DOI] [PubMed] [Google Scholar]
- Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP. (2009) The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol 150: 924–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T, Chen X, Salmeron J, Dietrich R. (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15: 2551–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi A, Ishihara A, Tanaka C, Ishizuka A, Takabayashi J, Miyoshi H, Nishi T. (2009) Accumulation of hydroxycinnamic acid amides induced by pathogen infection and identification of agmatine coumaroyltransferase in Arabidopsis thaliana. Planta 230: 517–527 [DOI] [PubMed] [Google Scholar]
- Ohnmeiss TE, Baldwin IT. (2000) Optimal defense theory predicts the ontogeny of an induced nicotine defense. Ecology 81: 1765–1783 [Google Scholar]
- Pandey SP, Baldwin IT. (2008) Silencing RNA-directed RNA polymerase 2 increases the susceptibility of Nicotiana attenuata to UV in the field and in the glasshouse. Plant J 54: 845–862 [DOI] [PubMed] [Google Scholar]
- Paschalidis KA, Roubelakis-Angelakis KA. (2005) Spatial and temporal distribution of polyamine levels and polyamine anabolism in different organs/tissues of the tobacco plant: correlations with age, cell division/expansion, and differentiation. Plant Physiol 138: 142–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT. (2007) Co(i)-ordinating defenses: NaCOI1 mediates herbivore-induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J 51: 79–91 [DOI] [PubMed] [Google Scholar]
- Petroni K, Falasca G, Calvenzani V, Allegra D, Stolfi C, Fabrizi L, Altamura MM, Tonelli C. (2008) The AtMYB11 gene from Arabidopsis is expressed in meristematic cells and modulates growth in planta and organogenesis in vitro. J Exp Bot 59: 1201–1213 [DOI] [PubMed] [Google Scholar]
- Ponchet M, Martintanguy J, Poupet A, Marais A, Beck D. (1982) Separation and quantification of basic hydroxycinnamic amides and hydroxycinnamic acids by reversed-phase high-performance liquid-chromatography. J Chromatogr 240: 397–404 [Google Scholar]
- Purrington CB. (2000) Costs of resistance. Curr Opin Plant Biol 3: 305–308 [DOI] [PubMed] [Google Scholar]
- Rabiti AL, Betti L, Bortolotti C, Marini F, Canova A, Bagni N, Torrigiani P. (1998) Short-term polyamine response in TMV-inoculated hypersensitive and susceptible tobacco plants. New Phytol 139: 549–553 [Google Scholar]
- Rodríguez-Kessler M, Ruiz OA, Maiale S, Ruiz-Herrerac J, Jiménez-Bremonta JF. (2008) Polyamine metabolism in maize tumors induced by Ustilago maydis. Plant Physiol Biochem 46: 805–814 [DOI] [PubMed] [Google Scholar]
- Rogers SO, Bendich AJ. (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 5: 69–76 [DOI] [PubMed] [Google Scholar]
- Saedler R, Baldwin IT. (2004) Virus-induced gene silencing of jasmonate-induced direct defences, nicotine and trypsin proteinase-inhibitors in Nicotiana attenuata. J Exp Bot 55: 151–157 [DOI] [PubMed] [Google Scholar]
- Santner A, Estelle M. (2007) The JAZ proteins link jasmonate perception with transcriptional changes. Plant Cell 19: 3839–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078 [DOI] [PubMed] [Google Scholar]
- Shinya T, Gális I, Narisawa T, Sasaki M, Fukuda H, Matsuoka H, Saito M, Matsuoka K. (2007) Comprehensive analysis of glucan elicitor-regulated gene expression in tobacco BY-2 cells reveals a novel MYB transcription factor involved in the regulation of phenylpropanoid metabolism. Plant Cell Physiol 48: 1404–1413 [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Hansen BG, Bjarnholt N, Ticconi C, Halkier BA, Kliebenstein DJ. (2007) A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS One 2: e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamp N. (2003) Out of the quagmire of plant defense hypotheses. Q Rev Biol 78: 23–55 [DOI] [PubMed] [Google Scholar]
- Steppuhn A, Baldwin IT. (2007) Resistance management in a native plant: nicotine prevents herbivores from compensating for plant protease inhibitors. Ecol Lett 10: 499–511 [DOI] [PubMed] [Google Scholar]
- Stork W, Diezel C, Halitschke R, Gális I, Baldwin IT. (2009) An ecological analysis of the herbivory-elicited JA burst and its metabolism: plant memory processes and predictions of the moving target model. PLoS One 4: e4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Sudha G, Ravishankar GA. (2002) Involvement and interaction of various signaling compounds on the plant metabolic events during defense response, resistance to stress factors, formation of secondary metabolites and their molecular aspects. Plant Cell Tissue Organ Cult 71: 181–212 [Google Scholar]
- Tebayashi SI, Horibata Y, Mikagi E, Kashiwagi T, Mekuria DB, Dekebo A, Ishihara A, Kim CS. (2007) Induction of resistance against the leaf miner, Liriomyza trifolii, by jasmonic acid in sweet pepper. Biosci Biotechnol Biochem 71: 1521–1526 [DOI] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J-i, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Tong Geon L, Cheol Seong J, Jae Yoon K, Dong Sub K, Jae Han P, Dae Yeon K, Yong Weon S. (2007) A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol Plant 129: 375–385 [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylidès C, Roby D. (2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99: 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Dahl CC, Winz RA, Halitschke R, Kuhnemann F, Gase K, Baldwin IT. (2007) Tuning the herbivore-induced ethylene burst: the role of transcript accumulation and ethylene perception in Nicotiana attenuata. Plant J 51: 293–307 [DOI] [PubMed] [Google Scholar]
- Wada N, Shinozaki M, Iwamura H. (1994) Flower induction by polyamines and related compounds in seedlings of morning glory (Pharbitis nil cv. Kidachi). Plant Cell Physiol 35: 469–472 [Google Scholar]
- Walling LL. (2009) Adaptive defense responses to pathogens and insects. Van Loon LC, , Advances in Botanical Research: Plant Innate Immunity, Vol 51 Elsevier, London, pp 551–612 [Google Scholar]
- Walters D, Cowley T, Mitchell A. (2002) Methyl jasmonate alters polyamine metabolism and induces systemic protection against powdery mildew infection in barley seedlings. J Exp Bot 53: 747–756 [DOI] [PubMed] [Google Scholar]
- Walters DR. (2003) Polyamines and plant disease. Phytochemistry 64: 97–107 [DOI] [PubMed] [Google Scholar]
- Wang CY, Chen CT, Wang SY. (2009) Changes of flavonoid content and antioxidant capacity in blueberries after illumination with UV-C. Food Chem 117: 426–431 [Google Scholar]
- Wang L, Allmann S, Wu J, Baldwin IT. (2008) Comparisons of LIPOXYGENASE3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol 146: 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149: 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT. (2007) Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19: 1096–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Benz M, Streit L, Ebert E. (1990) Feruloylputrescine and caffeoylputrescine are not involved in growth and floral bud formation of stem explants from Nicotiana tabacum L. var Xanthi nc. Plant Physiol 92: 924–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. (2009) The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangerl AR. (2003) Evolution of induced plant responses to herbivores. Basic Appl Ecol 4: 91–103 [Google Scholar]
- Zhang Y, Cao G, Qu LJ, Gu H. (2009) Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis. Plant Cell Rep 28: 337–346 [DOI] [PubMed] [Google Scholar]
- Zhao J, Davis LC, Verpoorte R. (2005) Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol Adv 23: 283–333 [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, Wang X, Gong Z, Schumaker KS, Guo Y. (2007) SAD2, an importin-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19: 3805–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]