Abstract
The biosynthesis of the tetracyclic diterpene ent-kaurene is a critical step in the general (primary) metabolism of gibberellin hormones. ent-Kaurene is formed by a two-step cyclization of geranylgeranyl diphosphate via the intermediate ent-copalyl diphosphate. In a lower land plant, the moss Physcomitrella patens, a single bifunctional diterpene synthase (diTPS) catalyzes both steps. In contrast, in angiosperms, the two consecutive cyclizations are catalyzed by two distinct monofunctional enzymes, ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS). The enzyme, or enzymes, responsible for ent-kaurene biosynthesis in gymnosperms has been elusive. However, several bifunctional diTPS of specialized (secondary) metabolism have previously been characterized in gymnosperms, and all known diTPSs for resin acid biosynthesis in conifers are bifunctional. To further understand the evolution of ent-kaurene biosynthesis as well as the evolution of general and specialized diterpenoid metabolisms in gymnosperms, we set out to determine whether conifers use a single bifunctional diTPS or two monofunctional diTPSs in the ent-kaurene pathway. Using a combination of expressed sequence tag, full-length cDNA, genomic DNA, and targeted bacterial artificial chromosome sequencing, we identified two candidate CPS and KS genes from white spruce (Picea glauca) and their orthologs in Sitka spruce (Picea sitchensis). Functional characterization of the recombinant enzymes established that ent-kaurene biosynthesis in white spruce is catalyzed by two monofunctional diTPSs, PgCPS and PgKS. Comparative analysis of gene structures and enzyme functions highlights the molecular evolution of these diTPSs as conserved between gymnosperms and angiosperms. In contrast, diTPSs for specialized metabolism have evolved differently in angiosperms and gymnosperms.
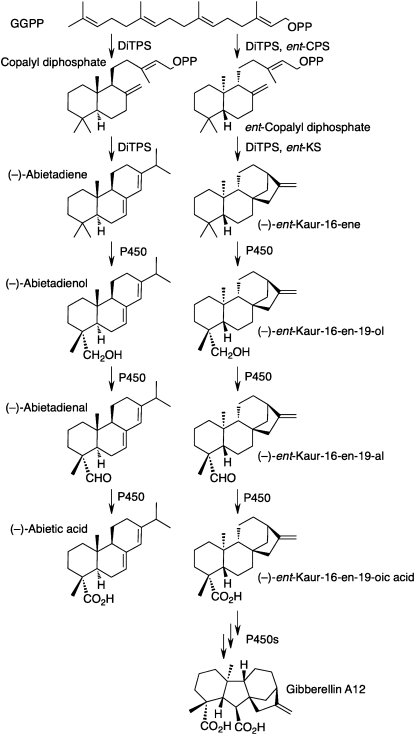
Conifers (Coniferophyta) are well known for producing an abundant and diverse assortment of oleoresin diterpenoids, predominantly in the form of diterpene resin acids from specialized (or secondary) metabolism, that play roles in conifer defense (Trapp and Croteau, 2001a; Keeling and Bohlmann, 2006a; Bohlmann, 2008) and are an important source of biomaterials (Bohlmann and Keeling, 2008). Several conifer diterpene synthases (diTPSs) that biosynthesize these compounds have been functionally characterized (Stofer Vogel et al., 1996; Peters et al., 2000; Martin et al., 2004; Keeling and Bohlmann, 2006b; Ro and Bohlmann, 2006). The formation of diterpene resin acids of conifer specialized metabolism parallels the formation of ent-kaurenoic acid in the biosynthesis of the gibberellin diterpenoid phytohormones (Fig. 1; Keeling and Bohlmann, 2006a; Yamaguchi, 2008). In gibberellin biosynthesis, geranylgeranyl diphosphate (GGPP) is cyclized by diTPS activity to ent-copalyl diphosphate (ent-CPP), and the ent-CPP is further cyclized by diTPS activity to ent-kaurene. A cytochrome P450 (P450)-dependent monooxygenase (CYP701) oxidizes ent-kaurene to ent-kaurenoic acid (Davidson et al., 2006), paralleling the activity of a P450 (CYP720B1) that oxidizes abietadiene to abietic acid in conifer diterpene resin acid biosynthesis (Ro et al., 2005). Other P450s further functionalize ent-kaurenoic acid to form the biologically active gibberellins. Surprisingly, no conifer diTPS involved in the general (or primary) metabolism of gibberellins has been reported to date, while metabolite profiles of gibberellins have been well characterized in conifers for their role in flowering (Moritz et al., 1990).
Figure 1.
Comparison of the biosynthesis of gibberellins, as it is known in angiosperm and lower plants, with the biosynthesis of diterpene resin acids in conifers, a large group of gymnosperm trees. In conifers, the formation of diterpene resin acids involves bifunctional diTPS (e.g. abietadiene synthase) for the stepwise cyclization of GGPP into diterpenes such as abietadiene via a copalyl diphosphate intermediate that moves between the two active sites of the bifunctional diTPS (Peters et al., 2001). The products of the diTPS are subsequently oxidized by P450 to the resin acids. In contrast, gibberellin biosynthesis in angiosperms requires two monofunctional diTPSs to convert GGPP into ent-kaurene, which is subsequently modified by P450s. The two monofunctional diTPSs in angiosperm gibberellin biosynthesis are CPS and KS. In the lower plant P. patens, the CPS and KS activities are combined in a bifunctional diTPS similar to the bifunctional diTPS in conifer diterpene resin acid biosynthesis. Prior to this work, to our knowledge, it was not known if the formation of gibberellins in a gymnosperm involves two monofunctional diTPSs, as in angiosperms, or a bifunctional diTPS, as in gymnosperm diterpene resin acid biosynthesis and in P. patens gibberellin biosynthesis. (Figure adapted from Keeling and Bohlmann [2006a].)
In the fungi Gibberella fujikuroi (Toyomasu et al., 2000) and Phaeosphaeria species L487 (Kawaide et al., 1997) and in the primitive land plant Physcomitrella patens (Bryophyta; Hayashi et al., 2006; Anterola and Shanle, 2008), the formation of ent-kaurene from GGPP is catalyzed by bifunctional diTPS enzymes. These enzymes contain two active sites. The N-terminal active site domain harbors a conserved DXDD motif and catalyzes the protonation-initiated cyclization of GGPP to ent-CPP (Prisic et al., 2007). In the C-terminal active site domain, a conserved DDXXD motif is essential for the diphosphate ionization-initiated cyclization of ent-CPP to ent-kaurene (Christianson, 2006). The presence of two active sites with their characteristic DXDD and DDXXD motifs resembles the structure of conifer bifunctional diTPSs in specialized metabolism of diterpene resin acid biosynthesis (Fig. 1), such as the grand fir (Abies grandis) abietadiene synthase (AgAS) and Norway spruce (Picea abies) levopimaradiene/abietadiene synthases (PaLAS; Peters et al., 2001; Martin et al., 2004; Keeling and Bohlmann, 2006a). In contrast, the formation of ent-kaurene from GGPP in angiosperms is catalyzed by two separate monofunctional enzymes, one with only the DXDD motif and having ent-copalyl diphosphate synthase (ent-CPS) activity and the other with only the DDXXD motif and having ent-kaurene synthase (ent-KS) activity (Yamaguchi, 2008).
A previously published model for the evolution of plant diTPS (Trapp and Croteau, 2001b) suggests that genes encoding the monofunctional CPS and KS enzymes known in angiosperms originated by gene duplication and subfunctionalization (Lynch and Force, 2000) of an ancestral bifunctional CPS/KS gene that may have been similar to the gene for the CPS/KS enzyme of the moss P. patens. The same model also suggests that genes for diTPSs of gymnosperm specialized diterpene resin acid metabolism arose from duplication and subsequent neofunctionalization of an ancestral bifunctional diTPS of the gibberellin pathway (Trapp and Croteau, 2001b). The pathways to specialized oleoresin diterpenes existed in ancient plants prior to the differentiation of gymnosperms and angiosperms (Bray and Anderson, 2009). Vascular plants split from nonvascular plants approximately 500 million years ago, and angiosperms split from gymnosperms approximately 300 million years ago (Palmer et al., 2004). As there has been no report to date of genes involved in gibberellin biosynthesis in gymnosperms, it remains unresolved and cannot be predicted whether conifers have a bifunctional CPS/KS for the formation of ent-kaurene similar to the primitive land plant P. patens and paralleling the diTPSs for conifer specialized diterpene resin acid biosynthesis or whether they have separate monofunctional CPS and KS enzymes, as is the case in angiosperms.
In this study, we made use of the extensive EST resources for spruce species (Pavy et al., 2005; Ralph et al., 2008), combined with isolation and sequencing of full-length cDNAs, genomic (g)DNA, and targeted bacterial artificial chromosome (BAC) clones, as well as enzyme assays with recombinant proteins to search for, and functionally characterize, possible monofunctional or bifunctional diTPS for ent-kaurene biosynthesis in a gymnosperm. In summary, we successfully isolated and characterized monofunctional ent-CPS (PgCPS) and ent-KS (PgKS) from white spruce (Picea glauca) and isolated orthologous cDNAs from Sitka spruce (Picea sitchensis). Comparison of enzyme functions and gene structures support common ancestry but different routes of evolution of monofunctional and bifunctional diTPS in conifer general and specialized metabolism, respectively.
RESULTS
Initial EST, cDNA, and gDNA Discovery of Spruce Candidate CPS and KS Genes
Using tBLASTn and a selection of plant CPS and KS protein sequences to identify CPS-like and KS-like sequences in the available spruce ESTs, we identified two candidate cDNA clones from approximately 500,000 ESTs: cDNA clone WS0403_I07 (GenBank accession no. ES262766) originating from the young and mature roots of Sitka spruce and cDNA clone WS0074_G02 (GenBank accession no. CO237940) originating from the buds, young shoots, and mid shoots of white spruce. The best BLASTx (National Center for Biotechnology Information Nonredundant [NCBI nr] database) hits were to bifunctional CPS/KS from P. patens (BAF61135; E value of 2 × 10−69) and a predicted ent-KS from cottonwood (Populus trichocarpa; XP_002311286; E value of 1 × 10−35), respectively. Both cDNA clones contained only partial open reading frames (ORFs). WS0403_I07 was truncated at both ends but contained the conserved DXDD motif. The WS0074_G02 clone was too short at the 5 ′ end to observe either of the conserved DXDD and DDXXD motifs. However, by 5 ′ RACE, we isolated a white spruce cDNA sequence (PgKS) containing a full ORF representing the WS0074_G02 clone. This sequence contained the DDXXD motif but not the DXDD motif, consistent with putative monofunctional KS activity (Prisic et al., 2007). To isolate a full-length cDNA representing the WS0403_I07 clone, we screened a white spruce BAC library as described below to identify the gDNA of a CPS-like gene (PgCPS) prior to isolating the cDNA. We also sequenced targeted BAC clones to isolate the genomic sequence of PgKS and reisolate the corresponding full-length cDNA.
gDNA Sequences of PgCPS and PgKS Genes
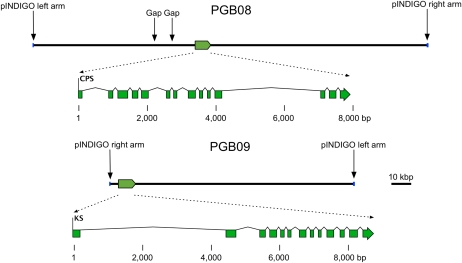
We used white spruce gDNA and GenomeWalker libraries, and primers based upon the white spruce cDNA sequence, to amplify genomic PgKS fragments that covered almost the complete gene except for a very large intron at the 5 ′ end (see below). To obtain the full genomic sequences for PgCPS and PgKS, we used the recently described approach for isolating targeted BAC clones and subsequent sequence assembly of insert DNA (Hamberger et al., 2009). PCR-based screening of a white spruce BAC library resulted in the isolation of an individual BAC clone for PgCPS (BAC clone PGB08) and PgKS (BAC clone PGB09; Fig. 2). BAC clones PGB08 and PGB09 contained gDNA inserts of approximately 195 and 160 kb, respectively, based upon their mobility in pulsed-field gel electrophoresis. The presence of the gene of interest in each BAC was confirmed by comparing the sequence of a PCR product of the insert with that of the available cDNA sequences. The complete gDNA inserts were excised, sheared into fragments of 700 to 2,000 bp, shotgun subcloned into plasmid libraries, and pair-end sequenced, and the sequences were assembled as described previously (Hamberger et al., 2009).
Figure 2.
Structure of white spruce genomic DNA of BAC clones PGB08 and PGB09. The positions of the target genes PgCPS and PgKS in the pIndigoBAC-5 vector inserts are indicated. The scale bar represents 10 kb in the BAC sequences. Blue bars indicate the left and right arms of the pIndigoBAC-5 vector. In PGB08, the positions of gaps in the sequence assembly are indicated. Detailed gene structures are shown to scale below each BAC sequence, with green rectangles representing exons separated by introns.
The initial sequence assemblies of PGB08 and PGB09 gDNA inserts yielded seven and four contigs after 1,536 and 1,152 plasmid clones were pair-end sequenced, respectively. Subsequent targeted DNA amplification and sequencing from the isolated BAC clones yielded additional contig-bridging sequences, allowing the full gDNA insert to be assembled for PGB09. As occurred previously with other white spruce BACs (Hamberger et al., 2009), regions with highly repetitive sequences in PGB08 prevented sequencing and assembling the full gDNA insert and resulted in three contigs and two gaps. The length of white spruce gDNA in these BAC inserts was 198,274 bp (including estimated gap sizes) and 122,148 bp for PGB08 and PGB09, respectively. Average sequence coverage was 12.3 × and 14.8 × , respectively. Results from the overall sequence analyses of the BAC clones PGB08 and PGB09, visualized using gbrowse, are available at http://gb2.treenomix3.msl.ubc.ca/cgi-bin/gbrowse/PGB08/ and http://gb2.treenomix3.msl.ubc.ca/cgi-bin/gbrowse/PGB09/, respectively. These descriptions include BLAST annotations (against NCBI nr, Repbase, Arabidopsis [Arabidopsis thaliana] library, version 13, issue 4 [Jurka et al., 2005], and spruce ESTs), GC content, gene predictions (Genemark Prediction [Eukaryotic HMM], FGENESH Prediction, Genescan Prediction), and putative cis-acting regulatory elements (from the PlantCARE database) in regions of 3,000 bp upstream from each ORF. PGB08 and PGB09 each contained a single functional gene identified by BLAST searches, which match the targeted genes PgCPS (PGB08) and PgKS (PGB09), respectively (Fig. 2), in addition to many transposons and repetitive elements that have previously been shown to be abundant in white spruce gDNA (Hamberger et al., 2009). PGB08 contained a full-length gene representing the targeted partial sequence from WS0403_I07, and this gene had the DXDD motif but not the DDXXD motif, suggestive of putative CPS activity (PgCPS). PGB09 contained the target gene consistent with the PgKS cDNA described above.
Full-Length cDNAs of White Spruce and Sitka Spruce CPS and KS
Based upon the gDNA and cDNA sequences, full-length cDNA clones for both CPS- and KS-like genes were amplified and sequenced from white spruce (Pg) and Sitka spruce (Ps) cDNA template. The CPS-like cDNA sequences were 2,525 and 2,544 bp for PgCPS and PsCPS, respectively. With the exception of a 76-bp insertion at the 5 ′ untranslated region of PsCPS, the sequences are 99.7% identical between species. The KS-like cDNA sequences were 2,519 and 2,521 bp for PgKS and PsKS, respectively, and are 99.4% identical to each other. At the protein level, CPS-like sequences of both species were 761 amino acids and differed by only one amino acid (Fig. 3). They contained the conserved DXDD motif (DVDD) at the 5 ′ end but lacked the DDXXD motif at the 3 ′ end (Fig. 3). Both KS-like sequences were 757 amino acids and differed by six amino acids. They contained the DDXXD motif (DDFFD) but lacked the DXDD motif (Fig. 3). The CPS-like sequences were only 35% identical to the spruce KS-like sequences. The spruce CPS and KS proteins were shorter at the N terminus by 54 and 19 amino acids, respectively, than the corresponding proteins from Arabidopsis. BLASTp analyses against NCBI nr identified a putative CPS from Scoparia dulcis and the bifunctional CPS/KS from P. patens (PpCPS/KS) as the closest matches (66% and 68% sequence similarity and E values of < 1 × 10−200) for PgCPS and PgKS, respectively.
Figure 3.
Amino acid alignment of monofunctional and bifunctional diTPSs of general and specialized metabolism, generated by MUSCLE. AtCPS, A. thaliana ent-CPS (NCBI accession no. AAA53632); PgCPS, P. glauca ent-CPS; PsCPS, P. sitchensis ent-CPS; PpCPS/KS, P. patens ent-CPS/ent-KS (BAF61135); AgAS, A. grandis abietadiene synthase (Q38710); AtKS, A. thaliana ent-KS (AAC39443); PgKS, P. glauca ent-KS; PsKS, P. sitchensis ent-KS. Amino acids with gray and black backgrounds indicate highly and completely conserved residues, respectively. Asp-rich motifs are indicated by underlines; a single underline indicates the DXDD motif necessary for protonation-initiated cyclization of GGPP to CPP, and a double underline indicates the DDXXD motif necessary for diphosphate ionization-initiated cyclization of CPP to the final diterpene products such as abietadiene and ent-kaurene.
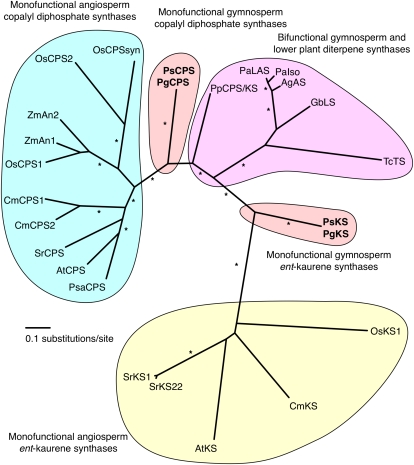
Phylogenetic analysis (Fig. 4) shows that the clades of monofunctional CPS and KS proteins from angiosperms are well separated from each other and from the clade of bifunctional diTPS enzymes found in gymnosperms and the moss P. patens. The newly identified spruce putative CPS proteins, PgCPS and PsCPS, are approximately equidistant between the clade of bifunctional PpCPS/KS and diTPSs of gymnosperm specialized (secondary) metabolism and the clade of monofunctional angiosperm CPS proteins. Similarly, the spruce putative KS proteins, PgKS and PsKS, are approximately equidistant between the clade of bifunctional PpCPS/KS and diTPSs of gymnosperm specialized metabolism and the clade of monofunctional angiosperm KS proteins.
Figure 4.
Unrooted phylogenetic tree of functionally characterized monofunctional and bifunctional diTPS proteins in general and specialized metabolism. AgAS, A. grandis abietadiene synthase (NCBI accession no. Q38710); AtCPS, A. thaliana ent-CPS (AAA53632); AtKS, A. thaliana ent-KS (AAC39443); CmCPS1, Cucurbita maxima ent-CPS1 (AAD04292); CmCPS2, C. maxima ent-CPS2 (AAD04293); CmKS, C. maxima ent-KS (AAB39482); GbLS, Ginkgo biloba levopimaradiene synthase (AAL09965); OsCPS1, O. sativa ent-CPS1 (BAD42449); OsCPS2, O. sativa ent-CPS2 (AAT11021); OsCPSsyn, O. sativa syn-CPS (AAS98158); OsKS1, O. sativa ent-KS (BAE72099); PaIso, P. abies isopimaradiene synthase (AAS47690); PaLAS, P. abies levopimaradiene/abietadiene synthase (AAS47691); PgCPS, P. glauca ent-CPS; PgKS, P. glauca ent-KS; PpCPS/KS, P. patens ent-CPS/ent-KS (BAF61135); PsCPS, P. sitchensis ent-CPS; PsKS, P. sitchensis ent-KS; PsaCPS, Pisum sativum ent-CPS (AAB58822); SrCPS, S. rebaudiana ent-CPS (AAB87091); SrKS1, S. rebaudiana ent-KS 1 (AAD34294); SrKS22, S. rebaudiana ent-KS 22 (AAD34295); TcTS, Taxus canadensis taxadiene synthase (AAR13860); ZmAn1, Z. mays ent-CPS1 (AAA73960); ZmAn2, Z. mays ent-CPS1 (AAT70083). The phylogenetic tree was prepared by protein alignment with MUSCLE, curation with Gblocks, phylogenetic analysis by PhyML (four rate substitution categories, gamma shape parameter optimized, Jones-Taylor-Thornton substitution model, and 100 bootstrap repetitions), and visualization with DrawTree. Asterisks indicate nodes supported by 80% or greater bootstrap values. The spruce CPS and KS proteins are positioned in the tree equidistant between the bifunctional diTPSs from gymnosperms and lower plants and the angiosperm monofunctional CPS and KS proteins, respectively. Without prior knowledge of the conserved DXDD and DDXXD motifs and functional characterization of these enzymes, it would not have been possible to predict whether the spruce enzymes were monofunctional or bifunctional.
Functional Characterization of Recombinant PgCPS and PgKS Enzymes
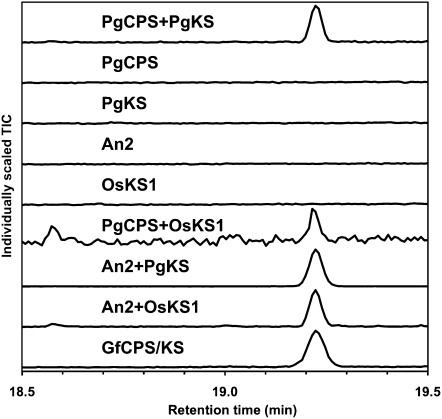
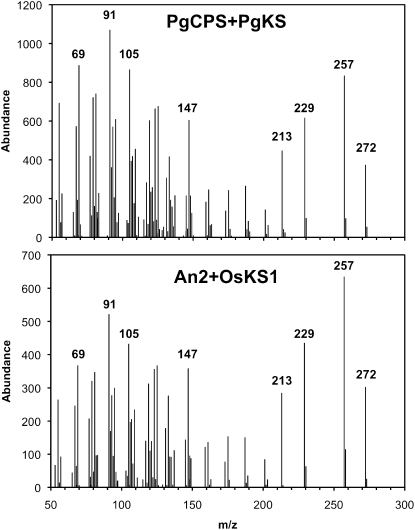
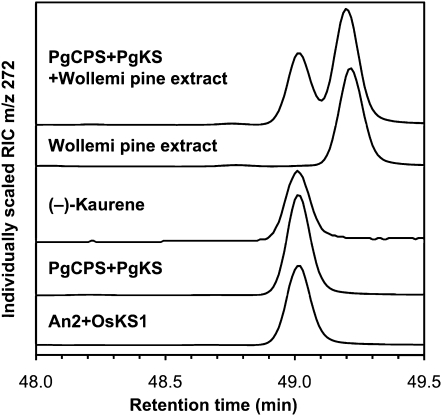
We performed a series of assays with recombinant PgCPS and PgKS proteins to test for the possibility of bifunctional or monofunctional enzyme activities. Enzyme assays were conducted with PgCPS or PgKS alone and with their possible functional complement so that the final ent-kaurene product could be observed by gas chromatography-mass spectrometry (GC-MS; Fig. 5). For coupled enzyme assays and as relevant controls, we used the corn (Zea mays) An2 protein encoding a monofunctional ent-CPS, the rice (Oryza sativa) OsKS1 encoding a monofunctional ent-KS, and the bifunctional fungal GfCPS/KS. PgCPS, PgKS, the bifunctional fungal GfCPS/KS, and the monofunctional angiosperm enzymes An2 and OsKS1 were expressed in Escherichia coli and nickel-affinity purified. For in vitro assays, GGPP was incubated with each purified recombinant protein individually (PgCPS, PgKS, An2, OsKS1, and GfCPS/KS), and the products were analyzed for the presence of ent-kaurene by GC-MS. GGPP was also incubated with functionally complementary enzyme pairs (An2+OsKS1, An2+PgKS, PgCPS+OsKS1, and PgCPS+PgKS). The combined results from these assays established that the white spruce PgCPS and PgKS enzymes are monofunctional for the formation of ent-CPP and ent-kaurene, respectively (Figs. 5 and 6). While neither PgCPS nor PgKS alone yielded ent-kaurene in assays with GGPP, as would be expected of a bifunctional enzyme (see GfCPS/KS in Fig. 5), combination of these two proteins catalyzed the complete series of cyclizations from GGPP to ent-kaurene. Similarly, PgCPS successfully complemented OsKS1, and PgKS successfully complemented An2, in the formation of ent-kaurene from GGPP. Although both PgCPS and OsKS1 did not express well in E. coli, the yield of these purified enzymes was sufficient to observe activity even when this pair was combined. Unlike the bifunctional moss PpCPS/KS, which produces predominantly ent-16 α -hydroxykaurene and some ent-kaurene (Hayashi et al., 2006), only ent-kaurene was detected when PgKS was incubated with GGPP and either An2 or PgCPS. Stereochemical analysis of enzyme assay products and comparison with authentic standards identified the product of PgCPS+PgKS as (–)-ent-kaurene, identical to that produced by the combination of angiosperm enzymes An2+OsKS1 (Fig. 7).
Figure 5.
GC-MS analysis on a DB-WAX column of in vitro assays with purified recombinant proteins incubated with GGPP. TIC, Total ion current. To identify whether the white spruce PgCPS and PgKS enzymes were monofunctional or bifunctional enzymes, they were assayed with GGPP, alone or in combination with other enzymes. Neither PgCPS nor PgKS produced ent-kaurene when incubated alone with GGPP. However, when incubated with GGPP together or with complementary angiosperm monofunctional enzymes, ent-kaurene was produced, with identical mass spectral and elution characteristics to the product of bifunctional GfCPS/KS.
Figure 6.
Mass spectra of recombinant enzyme assay products. When incubated with GGPP, recombinant PgCPS+PgKS produced a product with identical elution (see Fig. 5) and mass spectral fragmentation patterns as the ent-kaurene produced by An2+OsKS1.
Figure 7.
Stereochemical analysis of enzyme assay products on a Cyclodex- β GC column by GC-MS. RIC, Reconstructed ion current of the molecular ion of kaurene. The assay product of PgCPS+PgKS incubated with GGPP eluted at the same retention time as an authentic standard of (–)-kaurene as well as the assay product of An2+OsKS1, which is known to produce (–)-kaurene when incubated with GGPP. When mixed with Wollemi pine extract, the assay product of PgCPS+PgKS did not coelute with the (+)-kaurene from Wollemi pine, confirming that PgCPS+PgKS produced (–)-kaurene when incubated with GGPP.
Sequence alignment of PgKS showed a DIVS motif in place of the DXDD motif found in CPS and in the bifunctional diTPS of PpCPS/KS and AgAS, the latter representing a bifunctional conifer diTPS of specialized metabolism (Fig. 3). To determine whether the presence of the DXDD motif was sufficient to restore bifunctional activity to PgKS, we used site-directed mutagenesis to modify the DIVS of PgKS (DIVSTSI to DIDDTSI and DIDDTAM). Neither mutation resulted in a bifunctional enzyme with CPS and KS activities when incubated with GGPP alone, although both mutants still retained monofunctional KS activity when incubated with An2 and GGPP (data not shown).
Analysis of gDNA Sequences of PgCPS and PgKS
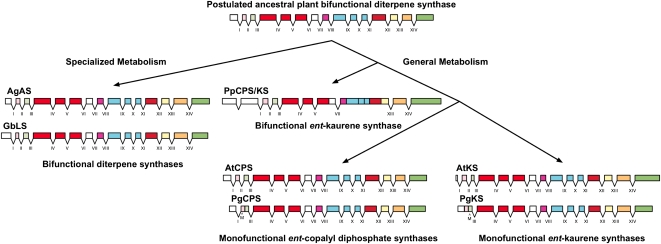
Trapp and Croteau (2001b) had previously shown conservation of gene structure between the genes encoding monofunctional CPS and KS enzymes of angiosperm gibberellin formation and a gene for a bifunctional diTPS (AgAS) in conifer specialized metabolism. Based on this finding, they proposed that AgAS resembles a putative ancestral bifunctional diTPS from which the monofunctional CPS and KS descended through gene duplication and subsequent specialization of each of the duplicated genes for only one of the two ancestral activities (Fig. 8). This model of an ancestral bifunctional diTPS was corroborated with the discovery of a bifunctional CPS/KS of similarly conserved gene structure in the lower land plant P. patens (Hayashi et al., 2006; Anterola and Shanle, 2008). The identification of complete genomic sequences for the monofunctional PgCPS and PgKS from a gymnosperm plant allowed us to further the analysis of Trapp and Croteau (2001b). To this end, we compared the genomic structures of PgCPS and PgKS with representative monofunctional and bifunctional diTPS genes from angiosperms (AtCPS and AtKS), gymnosperms (e.g. AgAS), and the moss P. patens (PpCPS/KS; Fig. 8).
Figure 8.
Gene structures and schematic of proposed evolution of diTPS in plants. Roman numerals indicate intron numbers. Colored bars indicate exon sequences. Schematic, intron numbers, and exon coloring scheme are based upon Trapp and Croteau (2001b). Genomic DNA sequences compared are as follows: AgAS, A. grandis abietadiene synthase (NCBI accession no. AF326516); AtCPS, A. thaliana ent-CPS (AT4G02780); AtKS, A. thaliana ent-KS (AT1G79460.1); GbLS, G. biloba levopimaradiene synthase (AY574248); PgCPS, PgKS, PpCPS/KS, P. patens CPS/KS (scaffold_130:28184..34739, www.phytozome.org). The positions of the start codons in the white spruce genes PgCPS and PgKS are indicated by M. An ancestral plant bifunctional diTPS gene is postulated to have duplicated to give rise to the diTPSs of general and specialized metabolism. Neofunctionalization has given rise to the bifunctional diTPS of gymnosperm specialized metabolism (e.g. AgAS) while conserving the gene structure of the ancestral gene. In general metabolism, two paths have occurred. In the moss P. patens, a diTPS (PpCPS/KS) has remained bifunctional but has fewer conserved introns and one different intron from the ancestral gene. In the case of angiosperms and gymnosperms, further duplication and subfunctionalization of the ancestral gene have resulted in the monofunctional CPS and KS genes, although the gene structure has been well conserved except for the loss of one or two introns at the 5 ′ end of the gymnosperm genes.
Apart from the 5 ′ end, which generally shows considerable variation in gene structure and sequence among TPS (Aubourg et al., 2002), there was strong conservation of both intron number and nearly identical intron locations among all of the angiosperm and gymnosperm diTPSs of general and specialized metabolism (Fig. 8). The PpCPS/KS gene had fewer introns, but the intron locations were consistent with those in the angiosperm and gymnosperm genes. The PpCPS/KS gene also has an intron at the 5 ′ end not found in the angiosperm and gymnosperm genes. PgCPS is lacking intron II and PgKS is lacking both intron I and II. Where intron phase numbers 0, 1, and 2 refer to the intron insertion before the first nucleotide or after the first or second nucleotide of a codon, respectively, the intron phases for both PgCPS and PgKS were 0, 1, 2, 1, 2, 1, 0, 2, 2, 2, 0, and 0 for introns III to XIV, identical to those previously described for other plant diTPS genes (Trapp and Croteau, 2001b). The first intron of PgKS was very large (4,212 bp). Recovering this intron sequence was not successful by primer walking and amplification from GenomeWalker libraries but was successfully sequenced from the BAC clone PGB09. The 11th intron of PgCPS also was large (2,869 bp). Although conifer bifunctional diTPS of specialized metabolism (e.g. AgAS) and conifer monofunctional CPS and KS of gibberellin biosynthesis represent two distinct branches of diTPS evolution, their conserved gene structure is strong evidence for a common ancestry of diTPS of general and specialized metabolism (Fig. 8).
Analysis of Upstream Promoter Regions of PgCPS and PgKS
The sequencing of BAC clones for conifer CPS and KS genes allowed for initial sequence characterization of upstream promoter regions. Putative cis-acting regulatory elements were identified in the 3,000 bp upstream of the start codon of each gene using the PlantCARE database (Lescot et al., 2002) and are shown as supplementary annotation information at http://gb2.treenomix3.msl.ubc.ca/cgi-bin/gbrowse/PGB08/ and http://gb2.treenomix3.msl.ubc.ca/cgi-bin/gbrowse/PGB09/. Many of the putative cis-acting regulatory elements identified were common to both genes, including those associated with light, heat, MYB binding, meristem expression, endosperm expression, defense and stress, circadian rhythm, and anaerobic induction. The promoter region of PgCPS additionally contained putative response elements associated with auxin, methyl jasmonate, gibberellin, ethylene, cell cycle, and wounding. The promoter region of PgKS additionally contained putative response elements associated with zein metabolism, low temperature, and salicylic acid. The promoter regions of both genes were particularly abundant in putative regulatory elements for light and heat response, consistent with recent research in light response in angiosperms (Seo et al., 2009).
DISCUSSION
The major goal of this study was to fill a considerable gap in our knowledge of genes of ent-kaurene diterpene biosynthesis in gymnosperms and to further our understanding of the evolution of diTPSs in the general (i.e. primary) and specialized (i.e. secondary) metabolism of conifers. We used a combination of spruce EST mining, targeted BAC isolation, gDNA sequencing, full-length cDNA cloning, protein expression, and enzyme assays to identify the genes encoding CPS and KS in a gymnosperm. The functional characterization of the PgCPS and PgKS gene products showed that both are monofunctional diTPS enzymes in white spruce. Monofunctional diTPSs of general metabolism in spruce are in contrast with the previously characterized diTPSs of conifer specialized diterpene resin acid biosynthesis, which are all bifunctional enzymes in a suite of species including grand fir (Stofer Vogel et al., 1996; Peters et al., 2000), Norway spruce (Martin et al., 2004; Keeling et al., 2008), loblolly pine (Pinus taeda; Ro and Bohlmann, 2006), and Sitka spruce (C.I. Keeling and J. Bohlmann, unpublished data).
The first indication of monofunctional CPS and KS in spruce came from comparisons of the deduced protein sequences that revealed the presence of only one of two conserved Asp-rich motifs in each protein, the DXDD motif in PgCPS and PsCPS and the DDXXD motif in PgKS and PsKS. These Asp-rich motifs function separately in the active site binding of GGPP (DXDD) or CPP (DDXXD), but they occur together in bifunctional diTPSs (Fig. 3). Monofunctionality of spruce CPS and KS was then experimentally proven in a series of enzyme assays testing recombinant proteins individually or in combinations (Fig. 5). Without knowledge of the roles of each of the Asp-rich motifs and without functional characterization, it would not have been possible to predict whether the conifer PgCPS and PgKS genes were likely to have monofunctional or bifunctional activities, as both appear phylogenetically approximately equidistant between previously characterized monofunctional and bifunctional diTPSs in a phylogeny with monofunctional angiosperm CPS and KS, bifunctional moss CPS/KS, and bifunctional conifer diTPS for diterpene resin acid biosynthesis (Fig. 4).
We show that both monofunctional and bifunctional diTPSs exist in conifers, a large group of gymnosperm trees, where the monofunctional diTPSs catalyze cyclizations in general diterpenoid metabolism and the bifunctional diTPSs function in specialized diterpene resin acid biosynthesis. If indeed the bifunctional and monofunctional diTPSs share a common ancestor, as was previously suggested (Bohlmann et al., 1998; Trapp and Croteau, 2001b) and as is supported by the analysis of gene structure presented here (Fig. 8), then the conifer diTPSs of general and specialized metabolisms represent two different branches of diTPS gene evolution. On one branch, diTPSs have retained two active site functions on a single protein, as for example with AgAS (Stofer Vogel et al., 1996; Peters et al., 2000) or PaLAS and PaIso (Martin et al., 2004), and on the other branch, gene duplication followed by subfunctionalization has led to the monofunctional spruce CPS and KS enzymes.
The situation in conifers, where both bifunctional and monofunctional diTPSs are found together but with separate roles in specialized and general metabolism, is different from any angiosperm species that has been characterized for parallel general and specialized diterpenoid biosynthesis. For example, in rice, one of the best characterized angiosperm systems for diTPSs of general and specialized metabolism, there are several OsCPS- and OsKS-like enzymes for specialized metabolism in antimicrobial defense in addition to the OsCPS and OsKS for gibberellin formation (Peters, 2006; Xu et al., 2007; Toyomasu, 2008). All of these enzymes are monofunctional. A similar pattern of all monofunctional diTPSs is found in general and specialized metabolism of other members of the grass (Poacea) family (Toyomasu et al., 2009) and in Stevia rebaudina, a dicot species that produces several specialized diterpene glycosides (Richman et al., 1999). In contrast to the conifer system described here, and to the best of our knowledge, there is no report of both monofunctional and bifunctional diTPS in any angiosperm system.
Restoring the DXDD motif in PgKS by site-directed mutagenesis did not restore bifunctional activity, as is found in the bifunctional conifer diTPSs of specialized metabolism as well as in the CPS/KS in fungi and P. patens. This result suggests a greater level of sequence divergence between monofunctional and bifunctional conifer diTPSs, as is also reflected in the diTPS phylogeny (Fig. 4). The existence of two monofunctional diTPSs for gibberellin biosynthesis in spruce suggests that the duplication and subfunctionalization of the ancestral bifunctional plant CPS/KS for gibberellin biosynthesis occurred before the divergence of angiosperms and gymnosperms.
Future work will have to explore a greater variety of lower plants for bifunctional CPS/KS, such as is found in the nonvascular plant P. patens, or for monofunctional CPS and KS, as we now know do not only occur in the angiosperms but also in gymnosperms. Lycophytes diverged from other vascular plants approximately 400 million years ago (Palmer et al., 2004), halfway between the split of vascular plants from nonvascular plants, and angiosperms from gymnosperms, 500 and 300 million years ago, respectively. The genome of the spike moss Selaginella moellendorffii has recently been sequenced as a reference genome for the lycophytes (http://genome.jgi-psf.org/Selmo1/Selmo1.home.html). Our preliminary examination of the draft genome assembly identified more than a dozen gene models with significant similarity to PpCPS/KS, PgCPS, and PgKS. These gene models contained either the DXDD motif, the DDXXD motif, or both motifs and had gene structures consistent with the ancestral plant diTPS gene structure rather than PpCPS/KS, which has fewer introns (Fig. 8). Without functional characterization of these genes, it is not yet possible to predict which of these genes in Selaginella are important for ent-kaurene biosynthesis and thus whether one bifunctional or two monofunctional enzymes function in ent-kaurene biosynthesis in this lycophyte.
MATERIALS AND METHODS
Identification of Putative ent-CPS and ent-KS ESTs from Spruce
Protein sequences of the bifunctional Physcomitrella patens CPS/KS and a set of known CPS and KS proteins from angiosperms were used to query Sitka spruce (Picea sitchensis), white spruce (Picea glauca), and interior spruce (Picea engelmannii × glauca) EST collections (Pavy et al., 2005; Ralph et al., 2008) by tBLASTn for candidate cDNA sequences. Candidate cDNA clones were fully sequenced. This sequence information was used to isolate and sequence full-length cDNAs and gDNA sequences as described below.
Identification and Sequencing of gDNA and BAC Clones for PgCPS and PgKS
White spruce (genotype Pg-653) GenomeWalker libraries (Clontech) and white spruce (genotype PG29) gDNA were used to amplify DNA fragments of PgCPS and PgKS genes. BAC clones of white spruce genotype PG29 gDNA in pIndigoBAC-5 containing the PgCPS and PgKS candidate genes were isolated, and their insert was sequenced and assembled as described previously (Hamberger et al., 2009). BAC clones were identified by PCR screening using primers (Supplemental Table S1) designed from the available partial PgCPS and PgKS cDNA sequences. Where the initial sequence assembly based upon shotgun sequencing did not result in one continuous sequence (i.e. contig) for a particular BAC insert, outward-facing primers near the ends of each contig were designed to amplify and sequence contig-bridging gDNA regions, using isolated BAC DNA as template, and the assembly was rerun.
Isolation of CPS and KS Full-Length cDNAs from White Spruce and Sitka Spruce
Prior to obtaining the above BAC sequences, we prepared cDNA from RNA from white spruce (genotype PG29) flushing bud tissue using the FirstChoice RLM-RACE kit (Ambion). Using primers designed from the white spruce WS0074_G02 clone with 5 ′ RACE and cDNA templates, we cloned the full ORF of a PgKS from white spruce cDNA with SuperTaq Plus (Ambion) into pCR-2.1-TOPO (Invitrogen). After the BAC sequences were completed, cDNA was prepared from RNA from white spruce (genotype Pg-653) plantlets and Sitka spruce (genotype FB3-425) flushing buds and interwhorl bark tissue using the SMART cDNA Synthesis Kit (Clontech). Using primers (Supplemental Table S1) designed from the white spruce gDNA and the available cDNA sequences, the full ORFs of CPS and KS from both species were amplified from cDNA, cloned into pJet1.2 (Fermentas), and sequenced.
Functional Characterization of PgCPS and PgKS
To determine whether the isolated white spruce genes encoded monofunctional or bifunctional enzymes, we used coupled enzyme assays with the recombinant spruce enzymes and monofunctional angiosperm CPS and KS enzymes so that the final ent-kaurene product could be detected by GC-MS. Authentic ent-kaurene was also produced via a bifunctional fungal enzyme. PgCPS full-length cDNA was cloned into NdeI/HindIII-digested pET28b(+) (Novagen, Merck) by InFusion cloning (Clontech). PgKS full-length cDNA was cloned into NheI/SpeI-digested pET28b(+) using standard protocols (Supplemental Table S1). Zea mays CPS (An2; Harris et al., 2005), Oryza sativa ent-KS (OsKS1; Xu et al., 2007), and bifunctional Gibberella fujikuroi CPS/KS (GfCPS/KS; Toyomasu et al., 2000) cDNAs were all cloned into NdeI/HindIII-digested pET28b(+) using a sticky-end PCR protocol (Zeng, 1998). All resulting recombinant proteins expressed from the plasmids described above were N-terminally His tagged.
Plasmids were transformed into C41 Escherichia coli cells (www.overexpress.com) containing the pRARE2 plasmid (coding for seven rare tRNAs in E. coli) prepared from Novagen Rosetta 2 cells (EMD Biosciences). Luria-Bertani medium containing kanamycin (50 mg L−1) and chloramphenicol (50 mg L−1) was inoculated with three individual colonies and cultured overnight at 37°C and 220 rpm. Terrific broth medium containing kanamycin (50 mg L−1) and chloramphenicol (50 mg L−1) was then inoculated with a 1:100 dilution of the overnight culture and grown at 37°C and 220 rpm until an optical density at 600 nm of at least 0.7 was reached. Cultures were then cooled to 16°C, induced with 0.2 mm isopropylthio- β -galactoside, and cultured for approximately 16 h at 16°C and 220 rpm before pelleting and freezing.
Cell pellets (typically 17 g L−1 culture) were resuspended in ice-cold lysis buffer (2 mL g−1 wet pellet, containing 20 mm HEPES, pH 7.5, 350 mm NaCl, 20 mm imidazole, 5 mm dithiothreitol [DTT], 5 mm phenylmethylsulfonyl fluoride or a protease inhibitor cocktail, 0.1 mg mL−1 DNase [Sigma], 0.1 mg mL−1 RNase [Sigma], and 1.0 mg mL−1 lysozyme). After 30 min of gentle mixing at 4°C, the lysate was disrupted by sonication (Bronson Sonifier S-250A). The lysate was then clarified by centrifugation (30 min, 10,000g, 4°C) and applied to a 1-mL HisTrap HP nickel-affinity column using an ÄKTApurifier 10 FPLC (GE Healthcare). The column was washed with 60 mL of wash buffer (20 mm HEPES, 350 mm NaCl, and 20 mm imidazole, pH 7.5) before eluting the His-tagged protein with elution buffer (20 mm HEPES, 350 mm NaCl, and 350 mm imidazole, pH 7.5). Buffer composition of eluate was exchanged to 20 mm HEPES and 350 mm NaCl, pH 7.5, with an Econo-Pac 10DG desalting column (Bio-Rad). Protein was quantified by direct measurement of A280 with a NanoDrop spectrophotometer (NanoDrop Technologies). Fresh DTT was added to 5 mm final concentration, and then the purified enzymes were divided into aliquots, flash frozen in liquid N2, and stored at –80°C.
Assays for each enzyme were conducted alone and with their likely functional complement so that the final ent-kaurene product could be observed by GC-MS. Single-vial enzyme assays were completed in 2-mL amber glass GC sample vials as described previously (Keeling et al., 2008). Buffer consisted of 50 mm HEPES, pH 7.2, 100 mm KCl, 7.5 mm MgCl2, 5 mm fresh DTT, 0.1 mg mL−1 bovine serum albumin, and 5% glycerol. Unless otherwise specified, GGPP (Sigma) was added to 50 μm in 500- μ L assays. Approximately 100 μ g of each purified protein was used in the assays. Assays were overlaid with 500 μ L of pentane and incubated at 30°C for 1 h, after which they were vortexed for 20 s to denature the proteins and stop the reaction. To completely separate the phases, vials were centrifuged for 30 min at 1,000g at 4°C.
Pentane overlays from the assays were analyzed on an Agilent HP5ms column (5% phenyl methyl siloxane, 30 m × 250 μ m i.d., 0.25- μ m film) at 1 mL min−1 helium with an Agilent 6890N gas chromatograph, 7683B series autosampler (vertical syringe position of 8 for single-vial assays), and 5975 Inert XL MS Detector. The GC temperature program was as follows: 40°C, hold 1 min; 7.5°C min−1 to 250°C; hold 2 min; pulsed splitless injector held at 250°C. Samples were also analyzed similarly on an Agilent DB-WAX column (polyethylene glycol, 30 m × 250 μ m i.d., 0.25- μ m film) with the following temperature program: 40°C, hold 3 min; 10°C min−1 to 240°C; hold 15 min; pulsed splitless injector held at 240°C. Compounds were identified by comparison with authentic standards, products generated from the previously characterized enzymes, retention indices, and MS from Adams (2007) and MS library searches (Hochmuth, 2007).
Stereochemistry of the enzyme assay products was analyzed on a J(W Cyclodex-β column (30 m × 250 μ m i.d., 0.25- μ m film) at 1 mL min−1 helium. The GC temperature program was as follows: 100°C, hold 2 min; 2°C min−1 to 230°C; hold 10 min; pulsed splitless injector held at 230°C. Assay products were compared with an authentic sample of (–)-kaurene. To obtain a sample of (+)-kaurene, we extracted the terpenoids from Wollemi pine (Wollemia nobilis) needle and stem tissue by shaking overnight in methyl tert-butyl ether, as this ancient conifer is known to contain abundant (+)-kaurene in its needles (Brophy et al., 2000).
Site-Directed Mutagenesis of PgKS
Site-directed mutagenesis of the DXDD motif of PgKS (DIVSTSI to DIDDTSI and DIDDTSI to DIDDTAM) was performed stepwise on the pET28b(+) construct using primers with multiple mutations (Supplemental Table S1) and the QuikChange Multi site-directed mutagenesis kit (Stratagene) following the manufacturer's protocols. Expression and functional characterization of these mutants were as described above.
Phylogenetic Analysis
Phylogenetic trees were constructed with the online tools at Phylogeny.fr (Dereeper et al., 2008). The workflow consisted of protein alignment with MUSCLE (Edgar, 2004), curation with Gblocks (Castresana, 2000), phylogenetic analysis by PhyML (four rate substitution categories, gamma shape parameter optimized, Jones-Taylor-Thornton substitution model, and 100 bootstrap repetitions; Guindon et al., 2005), and visualization with DrawTree (Felsenstein, 1989).
Analysis of Gene Structure
Gene structures were manually analyzed based upon published cDNA and gDNA sequences as well as those generated within this study. Exons were drawn to scale using the OmniGraffle Professional drawing program. Promoter analysis was conducted on the gDNA sequences 3,000 bp upstream of the start sites using the PlantCARE database (Lescot et al., 2002).
Sequence data from this article have been deposited in NCBI GenBank with the following accession numbers: PgCPS cDNA (genotype Pg-653), GU045755; PgKS cDNA (genotype Pg-653), GU045756; PgKS cDNA (genotype PG29), GU144565; PsCPS cDNA, GU045757; PsKS cDNA, GU045758; PGB08, GU059904; and PGB09, GU059905.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Oligonucleotide primers used for BAC screening, cloning, and site-directed mutagenesis.
Acknowledgments
We thank Reuben Peters (Iowa State University) for providing plasmids of OsKS1 and An2 cDNA, Bettina Tudzynskiof (Westfälische Wilhelms-Universität Münster) for G. fujikuroi cDNA, Armand Séguin (Natural Resources Canada, Canadian Forest Service, Laurentian Forestry Centre) for white spruce GenomeWalker libraries, C. Liewlaksanyanawin (University of British Columbia Forest Sciences) for white spruce gDNA, Ingrid Hoff (University of British Columbia Botanical Garden) and Susan Murch (University of British Columbia-Okanagan) for Wollemi pine samples, and Takeshi Sassa (Yamagata University) and Tomonobu Toyomasu (Yamagata University) for the (–)-kaurene sample. We thank Tristan Gillan, Björn Hamberger, Britta Hamberger, and Lina Madilao (University of British Columbia Michael Smith Laboratories) for technical assistance and Karen Reid for excellent laboratory management.
References
- Adams RP. (2007) Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry, Ed 4. Allured Publishing, Carol Stream, IL [Google Scholar]
- Anterola A, Shanle E. (2008) Genomic insights in moss gibberellin biosynthesis. Bryologist 111: 218–230 [Google Scholar]
- Aubourg S, Lecharny A, Bohlmann J. (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267: 730–745 [DOI] [PubMed] [Google Scholar]
- Bohlmann J. (2008) Insect-induced terpenoid defenses in spruce. Schaller A, , Induced Plant Resistance to Herbivory. Springer Science, New York, 173–187 [Google Scholar]
- Bohlmann J, Keeling CI. (2008) Terpenoid biomaterials. Plant J 54: 656–669 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Meyer-Gauen G, Croteau R. (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray PS, Anderson KB. (2009) Identification of carboniferous (320 million years old) class Ic amber. Science 326: 132–134 [DOI] [PubMed] [Google Scholar]
- Brophy JJ, Goldsack RJ, Wu MZ, Fookes CJ, Forster PI. (2000) The steam volatile oil of Wollemia nobilis and its comparison with other members of the Araucariaceae (Agathis and Araucaria). Biochem Syst Ecol 28: 563–578 [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552 [DOI] [PubMed] [Google Scholar]
- Christianson DW. (2006) Structural biology and chemistry of the terpenoid cyclases. Chem Rev 106: 3412–3442 [DOI] [PubMed] [Google Scholar]
- Davidson SE, Reid JB, Helliwell CA. (2006) Cytochromes P450 in gibberellin biosynthesis. Phytochem Rev 5: 405–419 [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1989) PHYLIP: phylogeny inference package (version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Guindon S, Lethiec F, Duroux P, Gascuel O. (2005) PHYML Online: a Web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33: W557–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, Hall D, Yuen M, Oddy C, Hamberger B, Keeling CI, Ritland C, Ritland K, Bohlmann J. (2009) Targeted isolation, sequence assembly and characterization of two white spruce (Picea glauca) BAC clones for terpenoid synthase and cytochrome P450 genes involved in conifer defence reveal insights into a conifer genome. BMC Plant Biol 9: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T, Peters RJ. (2005) The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol Biol 59: 881–894 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsuo A, Nozaki H. (2006) Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett 580: 6175–6181 [DOI] [PubMed] [Google Scholar]
- Hochmuth DH. (2007) MassFinder 3. Hochmuth Scientific Consulting, Hamburg, Germany [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110: 462–467 [DOI] [PubMed] [Google Scholar]
- Kawaide H, Imai R, Sassa T, Kamiya Y. (1997) ent-Kaurene synthase from the fungus Phaeosphaeria sp. L487: cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J Biol Chem 272: 21706–21712 [DOI] [PubMed] [Google Scholar]
- Keeling CI, Bohlmann J. (2006a) Diterpene resin acids in conifers. Phytochemistry 67: 2415–2423 [DOI] [PubMed] [Google Scholar]
- Keeling CI, Bohlmann J. (2006b) Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol 170: 657–675 [DOI] [PubMed] [Google Scholar]
- Keeling CI, Weisshaar S, Lin RPC, Bohlmann J. (2008) Functional plasticity of paralogous diterpene synthases involved in conifer defence. Proc Natl Acad Sci USA 105: 1085–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. (2000) The probability of duplicate gene preservation by subfunctionalization. Genetics 154: 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Fäldt J, Bohlmann J. (2004) Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135: 1908–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz T, Philipson JJ, Oden PC. (1990) Quantitation of gibberellins A(1), A(3), A(4), A(9) and a putative A(9)-conjugate in grafts of Sitka spruce (Picea sitchensis) during the period of shoot elongation. Plant Physiol 93: 1476–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Soltis DE, Chase MW. (2004) The plant tree of life: an overview and some points of view. Am J Bot 91: 1437–1445 [DOI] [PubMed] [Google Scholar]
- Pavy N, Paule C, Parsons L, Crow JA, Morency MJ, Cooke J, Johnson JE, Noumen E, Guillet-Claude C, Butterfield Y, et al. (2005) Generation, annotation, analysis and database integration of 16,500 white spruce EST clusters. BMC Genomics 6: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RJ. (2006) Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67: 2307–2317 [DOI] [PubMed] [Google Scholar]
- Peters RJ, Flory JE, Jetter R, Ravn MM, Lee HJ, Coates RM, Croteau RB. (2000) Abietadiene synthase from grand fir (Abies grandis): characterization and mechanism of action of the “pseudomature” recombinant enzyme. Biochemistry 39: 15592–15602 [DOI] [PubMed] [Google Scholar]
- Peters RJ, Ravn MM, Coates RM, Croteau RB. (2001) Bifunctional abietadiene synthase: free diffusive transfer of the (+)-copalyl diphosphate intermediate between two distinct active sites. J Am Chem Soc 123: 8974–8978 [DOI] [PubMed] [Google Scholar]
- Prisic S, Xu J, Coates RM, Peters RJ. (2007) Probing the role of the DXDD motif in class II diterpene cyclases. ChemBioChem 8: 869–874 [DOI] [PubMed] [Google Scholar]
- Ralph SG, Chun HJ, Kolosova N, Cooper D, Oddy C, Ritland CE, Kirkpatrick R, Moore R, Barber S, Holt RA, et al. (2008) A conifer genomics resource of 200,000 spruce (Picea spp.) ESTs and 6,464 high-quality, sequence-finished full-length cDNAs for Sitka spruce (Picea sitchensis). BMC Genomics 9: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman AS, Gijzen M, Starratt AN, Yang Z, Brandle JE. (1999) Diterpene synthesis in Stevia rebaudiana: recruitment and up-regulation of key enzymes from the gibberellin biosynthetic pathway. Plant J 19: 411–421 [DOI] [PubMed] [Google Scholar]
- Ro D-K, Arimura G, Lau SY, Piers E, Bohlmann J. (2005) Loblolly pine abietadienol/abietadienal oxidase PtAO (CYP720B1) is a multifunctional, multisubstrate cytochrome P450 monooxygenase. Proc Natl Acad Sci USA 102: 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro DK, Bohlmann J. (2006) Diterpene resin acid biosynthesis in loblolly pine (Pinus taeda): functional characterization of abietadiene/levopimaradiene synthase (PtTPS-LAS) cDNA and subcellular targeting of PtTPS-LAS and abietadienol/abietadienal oxidase (PtAO, CYP720B1). Phytochemistry 67: 1572–1578 [DOI] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G, Yamaguchi S. (2009) Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69: 463–472 [DOI] [PubMed] [Google Scholar]
- Stofer Vogel B, Wildung MR, Vogel G, Croteau R. (1996) Abietadiene synthase from grand fir (Abies grandis): cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase involved in resin acid biosynthesis. J Biol Chem 271: 23262–23268 [DOI] [PubMed] [Google Scholar]
- Toyomasu T. (2008) Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosci Biotechnol Biochem 72: 1168–1175 [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Kagahara T, Hirose Y, Usui M, Abe S, Okada K, Koga J, Mitsuhashi W, Yamane H. (2009) Cloning and characterization of cDNAs encoding ent-copalyl diphosphate synthases in wheat: insight into the evolution of rice phytoalexin biosynthetic genes. Biosci Biotechnol Biochem 73: 772–775 [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Ishizaki A, Shinoda S, Otsuka M, Mitsuhashi W, Sassa T. (2000) Cloning of a full-length cDNA encoding ent-kaurene synthase from Gibberella fujikuroi: functional analysis of a bifunctional diterpene cyclase. Biosci Biotechnol Biochem 64: 660–664 [DOI] [PubMed] [Google Scholar]
- Trapp S, Croteau R. (2001a) Defensive resin biosynthesis in conifers. Annu Rev Plant Physiol Plant Mol Biol 52: 689–724 [DOI] [PubMed] [Google Scholar]
- Trapp SC, Croteau RB. (2001b) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158: 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya NM, Coates RM, Peters RJ. (2007) Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry 68: 312–326 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Zeng G. (1998) Sticky-end PCR: new method for subcloning. Biotechniques 25: 206–208 [DOI] [PubMed] [Google Scholar]