Abstract
The role of nitrogen metabolism in the survival of prolonged periods of waterlogging was investigated in highly flood-tolerant, nodulated Lotus japonicus plants. Alanine production revealed to be a critical hypoxic pathway. Alanine is the only amino acid whose biosynthesis is not inhibited by nitrogen deficiency resulting from RNA interference silencing of nodular leghemoglobin. The metabolic changes that were induced following waterlogging can be best explained by the activation of alanine metabolism in combination with the modular operation of a split tricarboxylic acid pathway. The sum result of this metabolic scenario is the accumulation of alanine and succinate and the production of extra ATP under hypoxia. The importance of alanine metabolism is discussed with respect to its ability to regulate the level of pyruvate, and this and all other changes are discussed in the context of current models concerning the regulation of plant metabolism.
Plants initiate several responses to alleviate the consequences of oxygen deprivation during flooding or waterlogging of the soil. The combination of these adaptive responses is known as the low oxygen escape syndrome (Bailey-Serres and Voesenek, 2008). Among those adaptations, various anatomical and morphological changes, such as aerenchyma formation (Justin and Armstrong, 1987; Kennedy et al., 1992), stem elongation (Voesenek and Blom, 1989; Armstrong et al., 1994), the presence of gas films around flooded leaves (Pederson et al., 2009), and increasing shoot biomass can be observed. The latter are supposed to help provide oxygen via photosynthesis (Mommer and Visser, 2005; Pederson et al., 2009).
Another class of hypoxic responses includes the down-regulation of a suite of energy-, and therefore, oxygen-consuming, metabolic pathways (Geigenberger, 2003). Examples of such metabolic adaptations to hypoxia include the down-regulation of storage metabolism (Geigenberger et al., 2000), the shift from invertase to Suc synthase routes of Suc hydrolysis (Bologa et al., 2003; Huang et al., 2008), and the inhibition of mitochondrial respiration (Gupta et al., 2009; Zabalza et al., 2009). These responses are already initiated before oxygen becomes limiting as a substrate for respiration. Therefore, it is suggested that these metabolic changes are important components of the survival strategy as they considerably extend the period of hypoxia that a plant can withstand.
When the oxygen availability decreases below the level at which oxygen becomes limiting for oxidative phosphorylation, plant cells will depend on alternative metabolic pathways to produce ATP. Under such anoxic circumstances, the major source for energy is the glycolytic pathway, which produces two ATP and two pyruvate molecules per unit of hexose while concomitantly reducing NAD+ to NADH. In order to maintain glycolysis under anoxic conditions, NAD+ must be continuously regenerated from NADH via fermentative reactions. Using pyruvate as substrate, fermentative metabolism either produces lactate via lactate dehydrogenase or ethanol via two subsequent reactions catalyzed by pyruvate decarboxylase and alcohol dehydrogenase (Tadege et al., 1999). However, these two pathways have clear drawbacks: lactate is toxic for the cells, and ethanol diffuses rapidly out of the cells, which leads to a considerable loss of carbon during hypoxia.
In addition to lactate and ethanol fermentation, many plant species accumulate Ala under anoxic conditions (de Sousa and Sodek, 2003; Miyashita et al., 2007). Ala can accumulate to high concentrations without harmful consequences. It is even suggested that Ala production would help to regulate the pH balance within an anoxic cell (Reggiani et al., 1988). Nevertheless, it remains unclear how exactly the production of Ala supports anoxic metabolism, as no NADH oxidation occurs during the production of Ala (de Sousa and Sodek, 2002). Accumulation of Ala without any alternative NADH oxidation would therefore lead to a decrease of the glycolytic flux due to NAD+ limitation.
Several metabolic pathways have been proposed to explain the accumulation of Ala upon anoxia. A rapid induction of the expression of AlaAT, a gene encoding for the enzyme Ala amino transferase, as well as an increase in the activity of the enzyme during flooding have previously been documented (Good and Crosby, 1989; Good and Muench, 1992; Muench and Good, 1994). These inductions could potentially explain the increased production of Ala. However, Ala accumulation was also observed in T-DNA knockout plants in which AlaAT activity was reduced to almost zero (Miyashita et al., 2007). Apparently, Ala production does not depend solely on the activity of AlaAT. An alternative reaction that is able to produce Ala is catalyzed by γ-aminobutyric acid transaminase (GABA-T) using pyruvate as cosubstrate. Similar to the AlaAT knockout plants, GABA-T null mutants accumulated only slightly less Ala upon hypoxia compared with wild-type plants (Miyashita and Good, 2008). These results suggest at least partial redundancy of AlaAT and GABA-T under hypoxic conditions.
To better understand the role of Ala accumulation during hypoxia, metabolic changes were analyzed following root waterlogging of plants suffering from nitrogen limitation. Nodulated Lotus japonicus was grown on nitrogen-free substrate and exposed to several days of waterlogging. Changes in the levels of metabolites of primary carbon and nitrogen metabolism were compared to changes that were observed in plants not able to fix nitrogen via symbiotic interaction with rhizobia. For this purpose, transgenic L. japonicus were used in which the genes encoding Leghemoglobin were silenced via an RNA interference (RNAi) approach (Ott et al., 2005, 2009). We preferred this transgenic approach to change the intrinsic nitrogen status of the plants to feeding experiments with various amounts of nitrate since it has been demonstrated that under anoxic conditions, nitrate is converted via nitrate reductase into nitrite and subsequently to nitric oxide (Rockel et al., 2002; Planchet et al., 2005). Nitric oxide is known to be a versatile signaling molecule that could induce multiple pleiotropic effects (Besson-Bard et al., 2008).
Here, we show that L. japonicus roots are very tolerant to waterlogging-induced hypoxia. However, their susceptibility to hypoxia increased upon nitrogen deprivation. Upon waterlogging, pronounced accumulation of succinate and Ala was observed that was independent of the nitrogen status of the plant, whereas the metabolic behavior during flooding of all other amino acids was strongly influenced by the nitrogen status of the plant. Based on our observations, a metabolic model is proposed that explains the role of Ala accumulation during waterlogging via reorganization of the tricarboxylic acid (TCA) cycle in a modular manner where ATP is produced from an oxidizing pathway leading from oxoglutarate to succinate, whereas a parallel reducing branch oxidizes NADH during hypoxia. According to this scheme, Ala metabolism would also prevent pyruvate accumulation, thereby facilitating the continued operation of glycolysis during waterlogging.
RESULTS
L. japonicus Is Highly Tolerant to Waterlogging
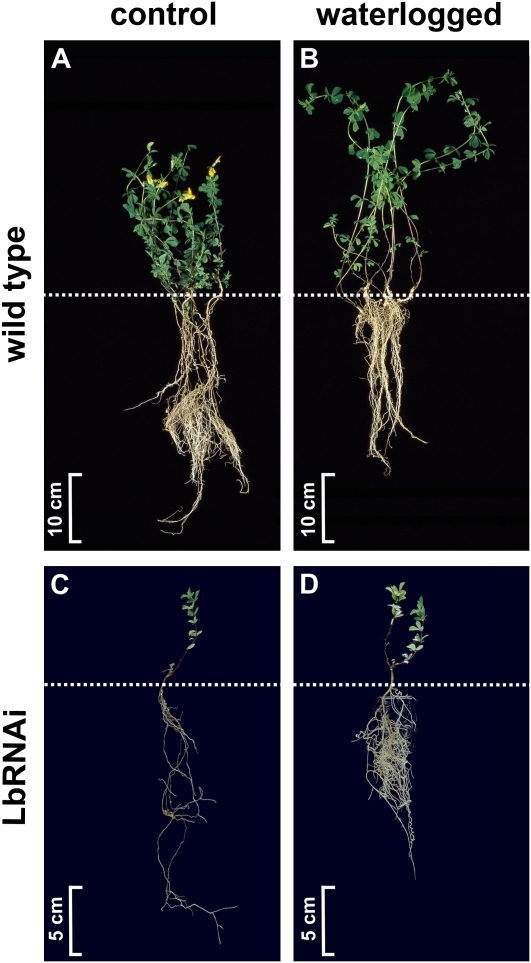
The effect of waterlogging on wild-type L. japonicus plants was determined by visual observation. Even after 4 weeks, none of the plants of our test population showed any sign of reduced vitality, though several morphological changes could be observed in the waterlogged plants (Fig. 1). These changes included the presence of etiolated stems, slightly yellowish leaves, delayed flowering and seed set, and stunted root growth in comparison to non-waterlogged plants. Apparently, L. japonicus wild-type plants are well able to adapt to prolonged periods of waterlogging, even though this occurs at the cost of seed production.
Figure 1.
Phenotype of L. japonicus wild-type and LbRNAi plants before and during waterlogging. Ten-week-old wild-type (A and B) and LbRNAi (C and D) plants were photographed after a 4-week period of continuous waterlogging (B and D) or a similar growth period under control conditions (A and C). The photographs show representative plants of the entire population. The dotted lines indicate the transition from root to shoot.
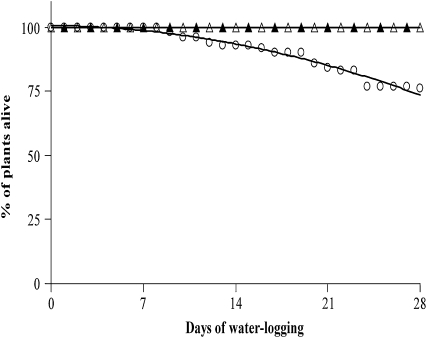
Tolerance to waterlogging was investigated in more detail by determining the amount of plants surviving the course of the experiment (Fig. 2). Notably, even after 4 weeks, no dead wild-type plants were observed. In contrast, plants in which the genes encoding nodular leghemoglobin were silenced by an RNAi approach already showed first signs of reduced vitality within the first week of waterlogging. These transgenic plants have been described in detail before (Ott et al., 2005, 2009), and various independent lines were shown to be identically affected by the RNAi silencing approach. Upon waterlogging, LbRNAi plants displayed a reduced but denser root system, and shoots produced more side branches (Fig. 1). After 4 weeks, about 25% of these plants were dead (Fig. 2). These data thus illustrate that the ability to fix nitrogen is required for maximum flooding resistance.
Figure 2.
Survival rate of L. japonicus wild-type and LbRNAi plants during an extended waterlogging period. The amount of healthy plants was determined throughout a period of 30 d for LbRNAi plants (circles) and for wild-type plants of the same age (black triangles) or the same size (white triangles) as the LbRNAi plants.
Waterlogging-Induced Changes in the Expression of Genes Encoding Enzymes of Fermentation and Nitrogen Fixation
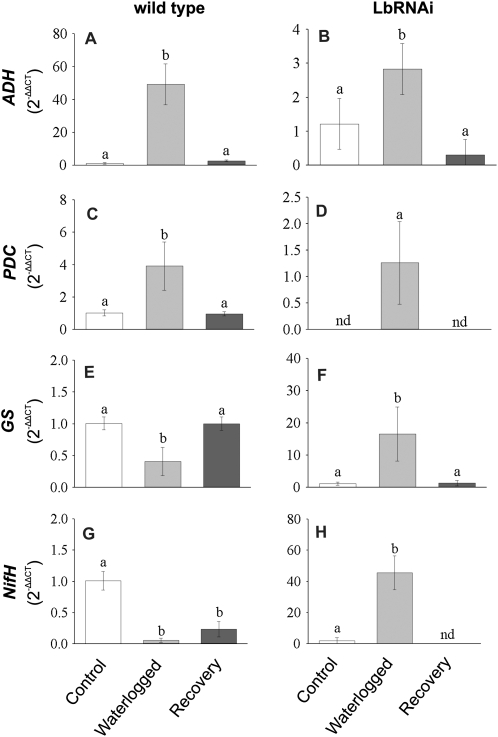
Waterlogging is generally assumed to reduce the availability of oxygen to the roots. The induction of the plant's response to hypoxia was verified by measuring the expression of alcohol dehydrogenase (ADH) and pyruvate decarboxylase (PDC; Fig. 3). These genes are all well known to be expressed upon hypoxia (Bologa et al., 2003; Ismond et al., 2003). As expected, the expression levels of ADH and PDC increased during waterlogging in both wild-type and the LbRNAi plants, and during recovery the expression levels returned to the same levels as detected prior to the waterlogging treatment.
Figure 3.
Changes in the transcript levels of hypoxia-responsive genes. Changes in the transcript levels of selected genes known to be induced by hypoxia. Samples from wild-type (A, C, E, and G) and LbRNAi (B, D, F, and H) plants were analyzed at normoxia, after 5 d of waterlogging, and after a 5-d recovery period. Values represent means ± sd of six replicates. Different letters denote significant differences among means judged by a one-way ANOVA in relation to the control (P < 0.05). nd, Not detected.
In addition to the expression of these hypoxia-induced genes, transcript levels of two additional genes encoding for Gln synthase (GS) and a subunit of the nitrogenase complex (NifH) were analyzed. These genes are known to be only expressed in nodules in which the oxygen concentration is so low that nitrogen fixation is possible. As expected, the expression level of both genes appeared to be very low in nodules of LbRNAi plants (Fig. 3). However, both genes were strongly induced during waterlogging of LbRNAi plants, again indicating that the oxygen concentration in these nodules had dropped. Expression levels of GS and NifH in wild-type nodules changed in a manner opposite to that observed in LbRNAi plants, being initially high, reduced during waterlogging, and increased again during the recovery phase.
Quantitative Changes of Total Amino Acids, Sugars, Protein, and Starch Induced by Waterlogging Differ between Roots and Shoot
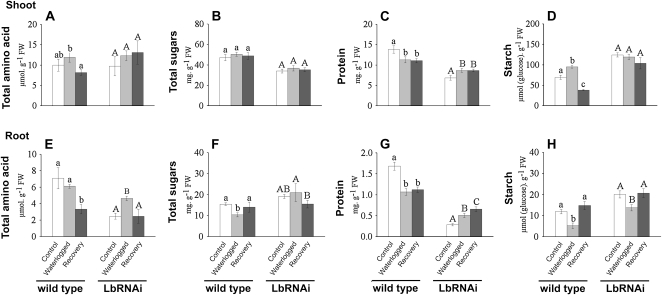
Changes in the amount of total amino acids, sugars, protein, and starch were determined before the start of the waterlogging treatment, as well as following 5 d of waterlogging and after 5 d of subsequent recovery from the treatment. In leaves of LbRNAi plants, the levels of total sugars and protein appeared to be slightly lower than in the wild type, whereas the amount of starch was about 45% higher when assessed on a per gram fresh weight (FW) basis (Fig. 4). The amount of free amino acids was similar between the genotypes. Waterlogging had hardly any effect on the quantity of the above-mentioned metabolites in leaves, with only the level of protein decreasing slightly in wild-type leaves and moderately increasing in LbRNAi plants. During the recovery phase, these changes were not reversed.
Figure 4.
Determination of changes in total amino acids, protein, total soluble sugar, and starch in L. japonicus wild-type and LbRNAi plants. Quantification of total soluble amino acids (A and E), total soluble sugars (B and F), total protein (C and G), and starch (E and H) measured in shoot (A–D) and roots (E–H) of wild-type and LbRNAi plants. Samples were taken prior to any treatment (control), after 5 d of waterlogging, and after 5 d of recovery. Values are presented as means ± sd of six replicates. Bars marked with dissimilar letters are significantly different from each other by one-way ANOVA (P < 0.05).
In roots, waterlogging-induced differences in the quantity of sugars, starch, amino acids, and protein were pronounced. Even prior to waterlogging, the levels of free amino acids and total protein were significantly lower in the LbRNAi plants than in the wild type. In wild-type plants, waterlogging induced a decrease in the amount of total protein. This level did not recover within 5 d after cessation of the treatment. Also, the amount of soluble amino acids was reduced to about half of the level that was present before the waterlogging treatment throughout the recovery period. In contrast, the amount of amino acids in the LbRNAi plants increased during waterlogging and returned to its original value after waterlogging was ceased. However, the amount of total protein increased during waterlogging and during the subsequent recovery period.
In contrast to the nitrogen-containing compounds, the level of sugars and starch were significantly higher in the LbRNAi plant than in the wild type. Upon waterlogging, the levels of starch in both wild-type and LbRNAi roots decreased, whereas the levels returned again to their initial levels during the recovery period. The amount of sugars decreased during waterlogging in wild-type roots but not in those of LbRNAi plants.
In general, it is shown here that the hypoxic stress imposed on the plants by waterlogging induces strongest metabolic changes in roots, whereas the well-aerated parts of the plant remained rather unaffected. Furthermore, whereas nitrogen assimilation is inhibited by waterlogging in wild-type plants, the opposite is observed in LbRNAi plants.
Metabolite Profiling
A comprehensive profiling of primary metabolism was obtained by gas chromatography-mass spectrometry (Fernie et al., 2004; Lisec et al., 2006) on samples taken daily during the 5-d period of waterlogging and the five subsequent days of the recovery phase. More than 40 metabolites were reproducibly identified in both genotypes across the three treatments, but not all metabolites were detected in each instance. An overview of the level of all metabolites determined across the experiment is provided in Supplemental Table S1. In addition, the production of ethanol was determined using an enzymatic essay.
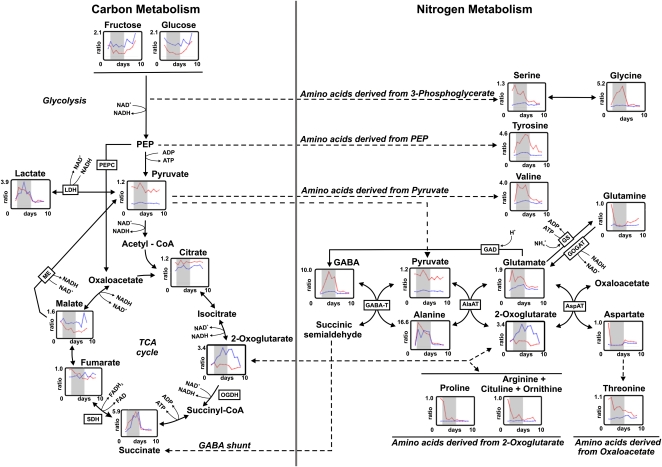
In order to investigate correlations between changes in the levels of metabolic intermediates of primary carbon and nitrogen metabolism, the levels of selected metabolites were plotted within the context of the metabolic pathways to which they belong. Visualizations of the data obtained for roots and nodules are shown in Figures 5and 6, respectively, whereas the relative changes (±se) of all these metabolites compared to the control value measured for wild-type tissue before waterlogging are given in Supplemental Tables S2 and S3, respectively. In roots, changes in the level of almost all nitrogen-containing metabolites were detected in wild-type plants, whereas only moderate changes were observed in the LbRNAi plants. In general, amino acids that are derived from precursors from the glycolytic pathway increased during waterlogging, whereas the levels declined for most amino acids that are linked to the TCA cycle. However, Ala, GABA, and Glu behaved differently, as these amino acids increased during waterlogging.
Figure 5.
Visualization of changes in selected metabolites of primary carbon and nitrogen metabolism in roots of L. japonicus wild-type and LbRNAi plants. Relative changes in the levels of metabolites as detected by GC-TOF-MS. Metabolite levels are calculated relative to the data obtained from wild-type tissue before waterlogging (day 0). The red line represents data obtained from wild-type plants, and the blue line shows values measured in LbRNAi plants. The gray area within each graph indicates the 5-d period during which the plants were waterlogged. Solid arrows indicate enzymatic reactions, and the dashed arrow link the same metabolite when this is present in two linked pathways. All data (±se) depicted in this figure are also listed in Supplemental Table S2. A full list of the levels of all metabolites that were detected can be found in Supplemental Table S1. GAD, Glu decarboxylase; GS, Gln synthetase; PEP, phosphoenolpyruvate; PEPC, phosphoenolpyruvate carboxylase.
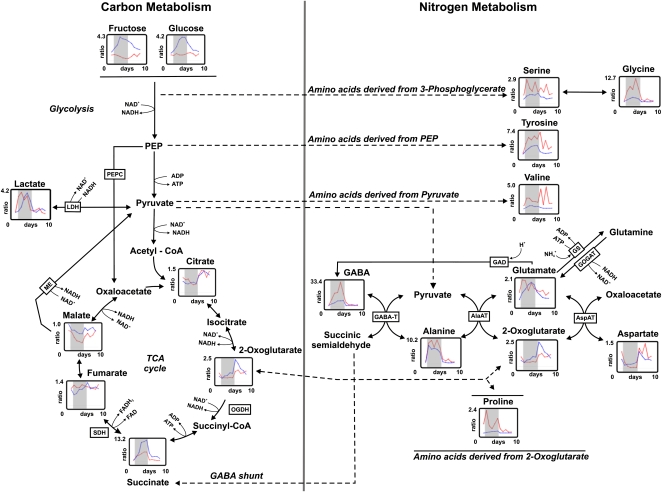
Figure 6.
Visualization of changes in selected metabolites of primary carbon and nitrogen metabolism in nodules of L. japonicus wild-type and LbRNAi plants. For an explanation of the experiment and the symbols and abbreviations used in the figure, see legend to Figure 5. The data (±se) that are shown in this figure are also listed in Supplemental Table S3.
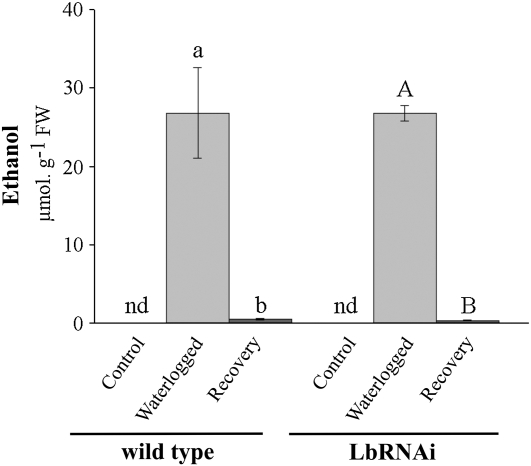
In contrast to the levels of amino acids, intermediates of glycolysis and the TCA cycle behaved rather similarly in both genotypes. A most pronounced increase was observed for the level of succinate. In nodules, the levels of amino acids behaved similarly in both genotypes, but marked differences were observed in the metabolic behavior of Fru and Glc (a strong increase was observed during waterlogging in the LbRNAi plants, but no obvious change occurred in the wild type), as well as succinate, which increased in both genotypes, though in LbRNAi plants, the increase was twice as strong. Also, the production of the fermentative end products lactate (Figs. 5 and 6) and ethanol (Fig. 7) was similar in both genotypes.
Figure 7.
Production of ethanol by nodulated roots from wild-type and LBRNAi plants during hypoxia. The level of ethanol was determined in the nutrient solution of the plants prior to the hypoxic treatment (control), after 5 d of hypoxia as induced by blowing nitrogen gas through the nutrient solution for 15 min, and 5 d after the plants were transferred to fresh, well-aerated nutrient solution to allow the tissue to recover from the hypoxic treatment. Values are presented as means ± sd. Bars that are marked with a dissimilar letters are judged to be significantly different from each other (one-way ANOVA, P < 0.05). nd, Not detected.
TCA Cycle Enzyme Activities
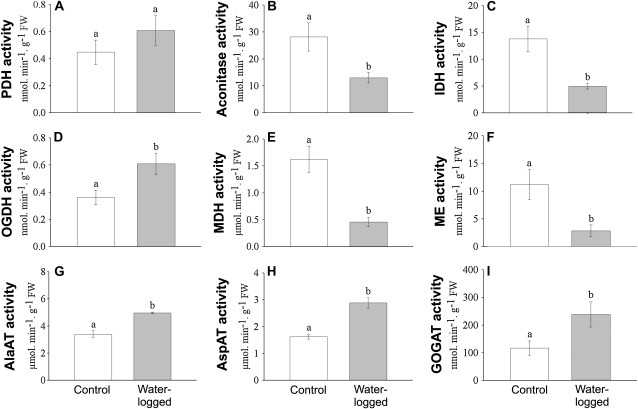
The effect of flooding on the activities of pyruvate dehydrogenase (PDH; EC 1.2.4.1), aconitase (EC 4.2.1.3), NAD+-dependent isocitrate dehydrogenase (IDH; EC 1.1.1.41), 2-oxoglutarate dehydrogenase (OGDH; EC 1.2.4.2), NAD+-dependent malate dehydrogenase (MDH; EC 1.1.1.37), NAD+-dependent malic enzyme (ME; EC 1.1.1.39), AlaAT (EC 2.6.1.2), Asp aminotransferase (AspAT; EC 2.6.1.1), and NADH-dependent Glu synthase (GOGAT; EC 1.4.1.14) was determined in desalted enzyme extracts from roots of wild-type plants (Fig. 8). The enzyme activities as measured under normoxic control conditions were as follows: PDH, 0.74 nmol min−1 g−1 FW; aconitase, 28.18 nmol min−1 g−1 FW; IDH, 13.78 nmol min−1 g−1 FW; OGDH, 0.36 nmol min−1 g−1 FW; MDH, 1.62 μmol min−1 g−1 FW; ME, 11.18 nmol min−1 g−1 FW; AlaAT, 3.39 μmol min−1 g−1 FW; AspAT, 1.62 μmol min−1 g−1 FW; and GOGAT, 116.29 nmol min−1 g−1 FW. After 5 d of waterlogging, the activity of PDH did not change significantly, whereas aconitase, IDH, MDH, and ME were significantly reduced to 12.99 nmol min−1 g−1 FW, 4.97 nmol min−1 g−1 FW, 0.46 μmol min−1 g−1 FW, and 2.84 nmol min−1 g−1 FW, respectively. In contrast, the activity of OGDH, AlaAT, AspAT, and GOGAT increased to 0.60 nmol min−1 g−1 FW, 4.96 μmol min−1 g−1 FW, 2.88 μmol min−1 g−1 FW, and 238.25 nmol min−1 g−1 FW, respectively. Of course, in vivo these enzymes are not necessarily active at maximum activity, but these data show unequivocally that various enzymes of the TCA cycle are differentially regulated upon flooding.
Figure 8.
Changes in enzyme activities from roots as induced by waterlogging of wild-type L. japonicus plants. The maximum activity of PDH (A), aconitase (B), IDH (C), OGDH (D), MDH (E), ME (F), AlaAT (G), AspAT (H), and Glu synthase (I) were determined in root material that was collected from wild-type plants grown under normoxic control conditions or after 5 d of waterlogging. The bars indicate mean values ± sd of three independent biological replicate experiments. Means that differ significantly according to a one-way ANOVA (P < 0.05) are marked with different letters.
DISCUSSION
Tolerance to Waterlogging Is Dependent on Cellular Nitrogen Status
Plants can initiate several adaptive responses in order to alleviate the consequences of oxygen deprivation during flooding or waterlogging of the soil. In this study, nodulated L. japonicus was shown to be tolerant to prolonged waterlogging (Fig. 2). Waterlogged plants were characterized by reduced root growth and extensive stem elongation (Fig. 1), as well as the induction of genes encoding for the fermentative enzymes ADH and PDC (Fig. 3). These adaptations are described to be part of the so-called low oxygen escape syndrome (Bailey-Serres and Voesenek, 2008) that delineates a characteristic response to hypoxia of flooding-tolerant plant species.
In addition to the morphological changes that were observed during waterlogging, both nitrogen and carbon metabolism are clearly affected in roots of wild-type plants during this treatment. At the end of the 5-d period of waterlogging, the amounts of protein and starch were reduced in roots of wild-type plants (Fig. 4, G and H). Similar observations were made in other studies in which various plant species, including Arabidopsis (Arabidopsis thaliana), barley (Hordeum vulgare), wheat (Triticum aestivum), soybean (Glycine max), and potato (Solanum tuberosum) were exposed to hypoxia (Thorne, 1982; Geigenberger et al., 2000; Gibon et al., 2002; Rolletschek et al., 2003; van Dongen et al., 2003, 2004). These changes were best explained as part of the adaptive response of plants to hypoxia: the increased demand for carbon to drive glycolysis during hypoxia can lead to the decrease of the starch pool. Additionally, plants limit their energy demand and, therefore, respiratory oxygen consumption by down-regulating the synthesis of storage products like starch and protein (Geigenberger, 2003; Gupta et al., 2009).
The depletion in protein and amino acids during hypoxia proceeds even beyond the waterlogging treatment, whereas the amounts of sugar and starch retained their original levels during the recovery period (Fig. 4). Apparently, amino acid metabolism is not only inhibited as an adaptive response to hypoxia, but also by a reduction in the rate of nitrogen fixation due to hypoxia-induced energy deprivation leading to a prolonged depletion of cellular nitrogen levels. This hypothesis is further supported by the observation that the genes NifH and GS, which encode enzymes known to play essential roles in nodule nitrogen metabolism, are strongly down-regulated during waterlogging (Fig. 3). Also in soybean, it was shown that nitrogen fixation dropped severely during the first days of flooding (Thomas et al., 2005). Only after acclimation over a period of 10 d, during which aerenchyma developed, partial recovery of the nitrogen fixation capacity was observed in that study.
The importance of nitrogen metabolism for hypoxia tolerance was further investigated using L. japonicus plants in which the expression of nodular leghemoglobin (Lb) was silenced via an RNAi approach. The function of Lb in these transgenic plants has been elaborately characterized in various independent lines before (Ott et al., 2005, 2009), and we performed our extensive biochemical investigations here on the strongest stable LbRNAi line. LbRNAi plants are unable to decrease the oxygen concentration inside nodules to the level that is required for nitrogen fixation by nitrogenase. Indeed, the levels of amino acids and protein were lower in LbRNAi plants compared to the wild type, with this difference being particularly prominent in roots. Moreover, LbRNAi plants were less resistant against waterlogging than wild-type plants (Fig. 2). However, during waterlogging of LbRNAi plants, the level of amino acids and protein in both root and shoot increased (Fig. 4). Probably, this increase is explained by the activation of nitrogenase due to the decrease in the concentration of oxygen within the nodules during waterlogging. Indeed, when LbRNAi plants were waterlogged, not only the genes encoding fermentative enzymes were induced but also the expression levels of NifH and GS increased (Fig. 3). However, it should be noted that even during waterlogging the levels of amino acids and protein documented in the transgenics still remained below the values measured in the wild type, indicating a low level of nitrogen fixation only.
Ala Metabolism Appears Indispensible for Lotus during Hypoxia
The observations described above indicate a correlation between the nitrogen status of L. japonicus roots and the ability of this species to survive prolonged periods of waterlogging. Since the activation of nitrogen metabolism and primary carbon metabolism were shown to be tightly coregulated (Scheible et al.,1997; Stitt et al., 2002), we performed a detailed analysis of changes in both primary carbon and nitrogen metabolism following waterlogging. For this purpose, we carried out gas chromatography time-of-flight mass spectrometry (GC-TOF-MS)-based metabolite profiling on samples taken daily during 5 d of waterlogging and five subsequent days of the recovery phase (Supplemental Table S1). The levels of various metabolites detected in roots (Fig. 5) and nodules (Fig. 6) were plotted, within their metabolic context, onto a pathway map describing glycolysis, TCA cycle, and amino acid biosynthesis.
In roots of wild-type plants, the majority of amino acids that are derived from intermediates of the TCA cycle decrease during waterlogging. In LbRNAi plants, the amount of these amino acids is already very low initially; therefore, no further decrease can be observed. Three amino acids behave differently from this general trend: in roots, Ala increases to a similar extent in both wild-type and LbRNAi plants, whereas the levels of Glu and GABA, both direct cosubstrates for Ala synthesis, increase strongly during flooding in the wild type but only moderately in roots of the LbRNAi line. The situation in nodules is slightly different. Especially striking is that the amount of Glu in LbRNAi plants increases during waterlogging to the same extent as in the wild type, which is probably explained by the slight activation of nitrogen fixation in the LbRNAi due to the decreased cellular oxygen concentration during flooding.
It is most intriguing that the production of Ala under hypoxia appears to occur at the cost of nearly all other amino acids, especially when nitrogen availability is limited. Even LbRNAi plants accumulate Ala to quantities that are indistinguishable from those of the wild type. Apparently, Ala biosynthesis upon hypoxia is important for L. japonicus in the survival of waterlogging. The change in the level of Ala, as well as other metabolically linked amino acids like Gln and GABA, was also observed in Arabidopsis when the respiratory activity was artificially inhibited either by using rotenone, a chemical inhibitor of complex I of the mitochondrial electron chain (Garmier et al., 2008), or by a genetic knockout approach that lead to the dissociation and functional impairment of complex I (Meyer et al., 2009). Apparently, the changes induced in Ala metabolism are characteristic for a general adaptive metabolic response to situations in which respiration is hampered.
The important role of Ala biosynthesisis during hypoxia is further sustained by the observation that AlaAT activity increases significantly during waterlogging (Fig. 8). The induction of AlaAT and the concomitant accumulation of Ala have been shown previously for many plant species; however, its significance was discussed. de Sousa and Sodek (2002) argued that the benefit of Ala production as a fermentation pathway can be questioned because no NAD+ is regenerated during operation of the pathway, which would be required to drive glycolysis at hypoxia. In this study, the significance of Ala accumulation is demonstrated since Ala accumulation does not decline even if plants are suffering from very low nitrogen availability. It appears likely that Ala fermentation primarily functions to regulate the level of pyruvate. Pyruvate is not only a known activator of the alternative oxidase (Vanlerberghe et al., 1999) but has also recently been shown to interfere with the hypoxia-induced inhibition of respiration (Gupta et al., 2009; Zabalza et al., 2009). Therefore, in order to control the rate of respiratory oxygen consumption when the oxygen availability is low, it is important to prevent pyruvate accumulation. Ala fermentation would be able to do just this, with the additional advantage that Ala can accumulate to high concentrations without the detrimental side effects that go along with the lactate or ethanol fermentation pathways.
Modular Reorganization of the TCA Cycle into Oxidative and Reductive Reactions
Amino acid metabolism cannot be regarded independently of primary carbon metabolism. Therefore, we linked the changes in amino acid levels to the carbohydrates we determined from glycolysis and the TCA cycle (Figs. 5 and 6). The increase in the production of lactate (Fig. 5) and ethanol (Fig. 7) in roots of both wild-type and LbRNAi plants indicates the activation of fermentation during waterlogging. The decrease of both Glc and Fru in wild-type roots confirms the observation described for total sugars and is also indicative for an increased glycolytic flux, such as that which characterizes the fermentative Pasteur effect (Summers et al., 2000).
The levels of most intermediates of the TCA cycle that we were able to determine did not change strongly during waterlogging, with the clear exception of succinate. In both roots and nodules of wild-type and LbRNAi plants, the amount of succinate increased strongly during waterlogging. Succinate accumulation appears to be a general phenomenon in plants exposed to hypoxia, as similar observations were made for many other plants species (Menegus et al., 1989; Good and Muench, 1993; Narsai et al., 2009; van Dongen et al., 2009). It has recently been discussed that succinate accumulation upon various biotic and abiotic stresses could potentially be explained via activation of the GABA shunt (Fait et al., 2008). Indeed, GABA accumulated during waterlogging in Lotus too. The accumulation of GABA during hypoxia has previously been explained by the activation of Glu decarboxylase when the cytosolic pH decreases (Carroll et al., 1994; Crawford et al., 1994). The reaction from Glu to GABA, catalyzed by Glu decarboxylase, consumes protons and has been shown to buffer cellular acidification during hypoxia. However, the other enzymes of the GABA shunt that are required to produce succinate exhibit pH optima between 8 and 10 (Satya Narayan and Nair, 1989; Shelp et al., 1995). It thus seems highly unlikely that a major role of the GABA shunt would be to catalyze reactions under hypoxia. Therefore, we suggest that the GABA shunt is not responsible for the accumulation of succinate upon hypoxia, but another pathway must exist to explain the simultaneous accumulation of succinate and GABA during waterlogging of L. japonicus.
The accumulation of succinate as observed in Figures 5 and 6 is more likely to be explained by an inhibition of succinate dehydrogenase (SDH) under hypoxia. Since SDH participates in both the TCA cycle and the mitochondrial electron transport chain, the succinate oxidation activity of SDH will decrease when the ubiquinone pool is electron saturated. Concomitantly, the activity of aconitase, IDH, and MDH also decreased after 5 d of waterlogging to between 28% and 46% of the activities as determined in normoxic roots (Fig. 8, B, C, and E). In contrast, the capacity of OGDH increased by approximately 33% (Fig. 8D). Probably, this might help to drive the production of ATP by succinate CoA ligase (also known as succinyl CoA synthetase) in the subsequent step of the TCA cycle and that would of course be most valuable under waterlogged conditions.
OGDH requires NAD+ as cosubstrate. Therefore, NADH must continuously be oxidized within the mitochondria to keep this reaction going, even when the mitochondrial electron transport chain is strongly inhibited due to the hypoxia. However, much evidence exists that regeneration of NAD+ could occur via MDH by catalyzing the reverse reaction leading to malate formation. First, metabolic flux analysis using 14C-labeled carbohydrates in Selenastrum minutum revealed that under anoxia, the TCA cycle can split in into two independent branches, both commencing from oxaloacetate. One branch proceeds in the common oxidative direction, whereas the second branch follows the opposite reductive direction (Vanlerberghe et al., 1989, 1990). Second, flux balance analysis of a metabolic model of barley seeds subjected to various oxygen concentrations (Grafahrend-Belau et al., 2009) revealed that upon anoxia, the TCA cycle breaks up in parts. It was predicted that the initial oxidizing reactions of the cycle are reversed to citrate production from oxoglutarate. Furthermore, it was suggested that the reaction catalyzed by MDH is reversed such that malate is produced and NADH is oxidized.
AlaAT Links Glycolysis and the TCA Cycle during Hypoxia
The observations described above can be summarized in a metabolic model from which the role of AlaAT during waterlogging of Lotus becomes clear. Pyruvate reacts with Glu to form Ala and oxoglutarate via AlaAT. This prevents pyruvate from accumulating, and simultaneously oxoglutarate is produced that can react in the mitochondria to form succinate via OGDH and succinate CoA ligase to produce ATP. The NAD+ that is required for this reaction is regenerated from NADH via MDH catalyzing the reaction from oxaloacetate to malate. The oxaloacetate that is required as substrate for this reaction is produced by AspAT. Concomitantly, Glu is produced, which is the cosubstrate for Ala synthesis. To keep this cycle running, the pools of Asp and Gln deplete. Malate either reacts with NAD+ via ME to form pyruvate, which then can be used for Ala synthesis, or malate reacts via fumarate to succinate. Both pathways might function in parallel.
This pathway explains the role of Ala accumulation during hypoxia as well as the strong decline in most other TCA cycle-related amino acids. All other changes in metabolite levels (as indicated by the font size in Fig. 9) observed in roots and nodules of waterlogged L. japonicus are in agreement with the metabolic equilibriums that are expected to drive the metabolic flux from glycolysis, via Ala synthesis and oxoglutarate to succinate. Since all enzymes that are required for these reactions are known to have isoforms located within the mitochondria, the cycle can take place entirely within the mitochondrion. The amount of ATP gained via this hypoxic pathway is twice as much as that produced by glycolysis alone. The importance of Ala accumulation for surviving waterlogging of L. japonicus was shown here. The relevance of this metabolic pathway in other plant species now has to be investigated.
Figure 9.
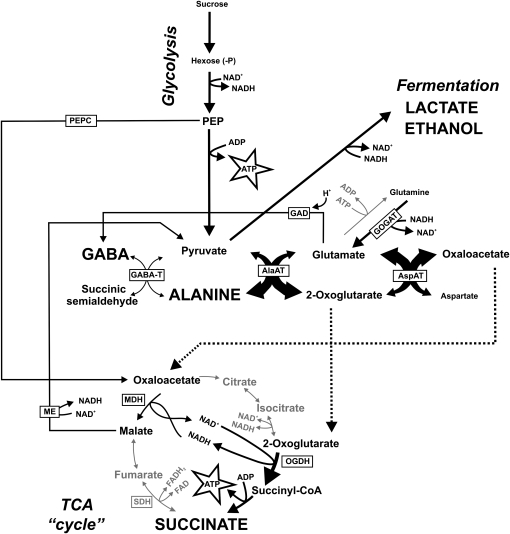
Metabolic model of primary metabolism under hypoxia in L. japonicus. When respiratory energy production is inhibited by decreased oxygen availability to a plant, drastic changes are observed in the steady-state levels of primary metabolites. Here, a model is described that agrees with the changes in metabolite levels and enzyme activities observed in L. japonicus during waterlogging. Pyruvate production is enhanced due to the activation of glycolysis (Pasteur effect). To keep glycolysis going, cytosolic NAD+ must be continuously regenerated from NADH via fermentation reactions that lead to the accumulation of lactate and ethanol. By producing pyruvate, ATP is formed. Accumulation of pyruvate should be prevented during hypoxia as this activates respiratory oxygen consumption (Zabalza et al., 2009). Via AlaAT, the amount of pyruvate can be reduced. Moreover, this reaction produces 2-oxoglutarate, which can be used by OGDH and succinate CoA ligase to produce another ATP. Succinate will accumulate as the TCA cycle will be further blocked due to the oxygen limitation at the reaction catalyzed by SDH. The mitochondrial NAD+ that is required to oxidize 2-oxoglutarate can be recycled via the enzyme MDH, which obtains its substrate either via phosphoenolpyruvate carboxylase (PEPC) or via AspAT. The latter enzyme is known to be activated upon anoxia and provides Glu as a substrate for AlaAT. All reactions leading to the production of pyruvate can be catalyzed by enzymes that are located within the mitochondrion. The model explains why Ala and succinate accumulate during anoxia in plants and provides a pathway that improves the ATP production during anoxia.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type Lotus japonicus ecotype GIFU (B-129) and transgenic plants (LbRNAi) in which the expression of all three homologous genes encoding leghemoglobin in nodules were reduced to <3% of the wild type via an RNAi approach (Ott et al., 2005) were grown from seeds in a controlled greenhouse environment (16 h day, 60% relative humidity, and 21°C/17°C day/night temperate regime). Seeds were sterilized in 2% sodium hypochlorite for 10 min, rinsed five times with sterile water, and germinated on moist, sterile filter paper for 3 d. Germinated seedlings were transferred to pots filled with 2- to 4-mm-diameter hydroponic culture substrate granules (Lecaton Original; Fibo Exclay Deutschland). Plants were watered daily, and twice per week, a nitrogen-free nutrient solution was supplied (0.5 mm CaCl2, 0.5 mm KCl, 0.25 mm KH2PO4, 0.25 mm K2HPO4, 1.0 mm MgSO4, 0.05 mm FeEDTA, and trace elements 9.1 μm MnCl2, 0.046 mm H3BO3, 0.765 μm ZnCl2, 0.56 μm NaMoO4, and 0.32 μm CuCl2). Seven days after transferring seedlings to the pots, plants were inoculated with Mesorhizobium loti strain R7A.
Waterlogging of the plants was performed by placing the pots inside a second pot of similar size that was filled with nitrogen-free nutrient solution at one-third of normal strength sufficient to bring its level just above the substrate. Subsequently, nitrogen gas was bubbled through the solution until the concentration of dissolved oxygen decreased to 55 μm, which is the equivalent of 20% of air saturation. The oxygen concentration of the solution was controlled throughout the course of the experiment and was in the range of 15 to 50 μm.
Plant organs (root, nodule, and leaves) were harvested from 12-week-old plants directly into liquid nitrogen and stored at −80°C. Sampling was on consecutive days within 15 min at 12 am, with sunrise at approximately 6 am. Six biological replicates were taken for each sample.
Plant survival during waterlogging was determined by counting the number of dead plants each day during a 30-d time period. The survival rate of wild-type and LbRNAi plants was compared between nonflowering plants of the same age (4–5 weeks, wild-type shoots were approximately 6–8 cm, and LbRNAi shoots were 2.5–3.0 cm long), as well as between plants of the same size (wild-type plants were 2 to 3 weeks old, and LbRNAi plants were 4 to 5 weeks old).
Metabolic Analyses
Nodulated roots and leaves were sampled during the course of the experiment at the time points indicated in the text and immediately frozen in liquid nitrogen. Nodules were subsequently separated from the roots. Aliquots of 50 to 100 mg tissue FW were ground in 2-mL Eppendorf tubes each containing a clean stainless steel metal ball (5 mm diameter) using a mixer-mill grinder (MM200; Retsch) for 2 min with a frequency of 30 Hz. Grinding components of the mill were cooled with liquid nitrogen to keep the samples frozen at all times. Metabolites were extracted as described by Lisec et al. (2006) by thoroughly shaking for 15 min at 70°C in a mixture of 1400 μL methanol supplemented with 60 μL 1.3 μm ribitol in water. Water-soluble metabolites were separated by adding 750 μL chloroform and 400 μL water to the extract. Subsequently, the aqueous phase was transferred into to clean Eppendorf tubes. Samples were dried at room temperature by vacuum centrifugation (Concentrator 5301; Eppendorf).
Derivatization of the metabolites was performed by a methoxyamination reaction prior to GC-TOF-MS analysis. After baseline correction (ChromaTOF software version 1.00, Pegasus driver 1.61; LECO), peak heights of the mass (m/z) fragments were normalized to the internal standard (ribitol) and FW of the samples. Annotation of the mass fragments was manually supervised using TagFinder (Luedemann et al., 2008). Statistical analysis by two-way ANOVA of the data was done using MeV (Multi Experiment Viewer) software (Saeed et al., 2003).
The amount of total amino acids was determined from methanol extracts using the ninhydrin method of Yemm and Cocking (1955) using a concentration series of Leu for quantification. Total soluble sugars were determined with the anthrone method of Graham and Smydzuk (1965) using Glc for the calibration curve. Starch was determined from the insoluble pellet as described by Hendriks et al. (2003) using the Starch UV-method kit (Roche), and total protein was measured as described by Bradford (1976) after solubilization of the insoluble pellet with 0.1 m NaOH. Absorbancies were read in a Spectra Max Plus microplate and cuvette spectrophotometer (Molecular Devices).
Ethanol production was determined by measuring the accumulation of ethanol in the nutrient solution as described by Zabalza et al. (2009).
Enzyme Analysis
Enzyme extracts were prepared as described previously (Gibon et al., 2004), except Triton X-100 was used at a concentration of 1% and glycerol at 20%. The maximum activity of OGDH (EC 1.2.4.2) was determined exactly as described by Araújo et al. (2008). Aconitase (EC 4.2.1.3) activity was determined as detailed by Carrari et al. (2003), PDH (EC 1.2.4.1) was assayed as described by Randall and Miernyk (1990), IDH (EC 1.1.1.41) as described by Jenner et al. (2001), MDH (EC 1.1.1.37) as described by Jenner et al. (2001), ME (EC 1.1.1.39) as detailed by Kulichikhin et al. (2009), AlaAT (EC 2.6.1.2) as described by Good and Muench (1992), AspAT (EC 2.6.1.1) as described by Griffith and Vance (1989), and GOGAT (EC 1.4.1.14) as described by Groat and Vance (1981).
Analysis of Gene Expression
Total RNA was extracted from nodulated roots using the RNeasy kit (Qiagen) according to the manufacturer's instructions. DNase treatment was done with the TURBO DNA-free kit (Ambion). cDNA was prepared from 5 μg RNA using the Superscript III reverse transcriptase kit (Invitrogen). Real-time PCR amplification was carried out with the ABI Prism 7900 sequence detection system (Applied Biosystems), using power Sybr-green master mix (Applied Biosystems) and the primers described in Supplemental Table S4. Transcript levels of most genes were normalized against ubiquitin mRNA as described previously by Colebatch et al. (2004) except for the levels of bacterial nifH transcript that was normalized against the constitutively expressed gene sigA as described by Ott et al. (2005). Relative quantification of gene expression was performed using the comparative threshold cycle method, as described in the ABI PRISM 7900 Sequence Detection System User Bulletin 2 (Applied Biosystems).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Overview of all metabolites determined in roots and nodules of Lotus japonicus wild-type and LbRNAi plants during a 5-d period of waterlogging and 5 d of subsequent recovery.
Supplemental Table S2. Overview of the relative changes change (±se) in metabolite levels extracted from roots of wild-type and LbRNAi Lotus japonicus plants before, during, and after waterlogging.
Supplemental Table S3. Overview of the relative changes change (±se) in metabolite levels extracted from nodules of wild-type LbRNAi Lotus japonicus plants before, during, and after waterlogging.
Supplemental Table S4. List of primers used for quantitative RT-PCR.
Acknowledgments
The Deutscher Akademischer Austausch Dienst and Deutsch Forschungsgemeinschaft (SFB 429) are kindly acknowledged for their financial support.
References
- Araújo WL, Nunes-Nesi A, Trenkamp S, Bunik VI, Fernie AR. (2008) Inhibition of 2-oxoglutarate dehydrogenase in potato tuber suggests the enzyme is limiting for respiration and confirms its importance in nitrogen assimilation. Plant Physiol 148: 1782–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong W, Brändle R, Jackson MB. (1994) Mechanisms of flood tolerance in plants. Acta Bot Neerl 43: 307–358 [Google Scholar]
- Bailey-Serres J, Voesenek LACJ. (2008) Flooding stress: acclimations and genetic diversity. Annu Rev Plant Biol 59: 313–339 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A, Pugin A, Wendehenne D. (2008) New insights into nitric oxide signaling in plants. Annu Rev Plant Biol 59: 21–39 [DOI] [PubMed] [Google Scholar]
- Bologa KL, Fernie AR, Leisse A, Loureiro ME, Geigenberger P. (2003) A bypass of sucrose synthase leads to low internal oxygen and impaired metabolic performance in growing potato tubers. Plant Physiol 132: 2058–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of proteindye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Carrari F, Nunes-Nesi A, Gibon Y, Lytovchenko A, Loureiro ME, Fernie AR. (2003) Reduced expression of aconitase results in an enhanced rate of photosynthesis and marked shifts in carbon partitioning in illuminated leaves of wild species tomato. Plant Physiol 133: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AD, Fox GG, Laurie S, Phillips R, Ratcliffe RG, Stewart GR. (1994) Ammonium assimilation and the role of γ-aminobutyric acid in pH homeostasis in carrot cell suspensions. Plant Physiol 106: 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebatch G, Desbrosses G, Ott T, Krusell T, Montanari O, Kloska S, Kopka J, Udvardi MK. (2004) Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J 39: 487–512 [DOI] [PubMed] [Google Scholar]
- Crawford LA, Bown AW, Breitkreuz KE, Guinel FC. (1994) The synthesis of γ-aminobutyric acid in response to treatments reducing cytosolic pH. Plant Physiol 104: 865–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sousa CAF, Sodek L. (2002) The metabolic response of plants to oxygen deficiency. Braz J Plant Physiol 14: 83–94 [Google Scholar]
- de Sousa CAF, Sodek L. (2003) Alanine metabolism and alanine aminotransferase activity in soybean (Glycine max) during hypoxia of the root system and subsequent return to normoxia. Environ Exp Bot 50: 1–8 [Google Scholar]
- Fait A, Fromm H, Walter D, Galili G, Fernie AR. (2008) Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci 13: 14–19 [DOI] [PubMed] [Google Scholar]
- Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. (2004) Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5: 763–769 [DOI] [PubMed] [Google Scholar]
- Garmier M, Carroll AJ, Delannoy E, Vallet C, Day DA, Small ID, Millar AH. (2008) Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol 148: 1324–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P. (2003) Response of plant metabolism to too little oxygen. Curr Opin Plant Biol 6: 247–256 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Fernie AR, Gibon Y, Christ M, Stitt M. (2000) Metabolic activity decreases as an adaptive response to low internal oxygen in growing potato tubers. Biol Chem 381: 723–740 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M. (2004) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes in enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16: 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Vigeolas H, Tiessen A, Geigenberger P, Stitt M. (2002) Sensitive and high throughput metabolite assays for inorganic pyrophosphate, ADPGlc, nucleotide phosphates, and glycolytic intermediates based on a novel enzymic cycling system. Plant J 30: 221–235 [DOI] [PubMed] [Google Scholar]
- Good AG, Crosby WL. (1989) Anaerobic induction of alanine aminotransferase in barley root tissue. Plant Physiol 90: 1305–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AG, Muench DG. (1992) Purification and characterization of an anaerobically induced alanine aminotransferase from barley roots. Plant Physiol 99: 1520–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good AG, Muench DG. (1993) Long-term anaerobic metabolism in root tissue (metabolic products of pyruvate metabolism metabolism). Plant Physiol 101: 1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafahrend-Belau E, Schreiber F, Koschutzki D, Junker BH. (2009) Flux balance analysis of barley seeds: a computational approach to study systemic properties of central metabolism. Plant Physiol 149: 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D, Smydzuk J. (1965) Use of anthrone in the quantitative determination of hexose phosphates. Anal Biochem 11: 246–255 [DOI] [PubMed] [Google Scholar]
- Griffith SM, Vance CP. (1989) Aspartate aminotransferase in alfafa root nodules. I. Purification and partial characterization. Plant Physiol 90: 1622–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groat RG, Vance CP. (1981) Root nodule enzymes of ammonia assimilation in alfalfa (Medicago sativa L.): developmental patterns and response to applied nitrogen. Plant Physiol 67: 1198–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KJ, Zabalza A, Van Dongen JT. (2009) Regulation of respiration when the oxygen availability changes. Physiol Plant 137: 383–391 [DOI] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P. (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Colmer TD, Millar AH. (2008) Does anoxia tolerance involve altering the energy currency towards PPi?. Trends Plant Sci 13: 221–227 [DOI] [PubMed] [Google Scholar]
- Ismond KP, Dolferus R, De Pauw M, Dennis ES, Good AG. (2003) Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol 132: 1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner HL, Winning BM, Millar AH, Tomlinson KM, Leaver CJ, Hill SA. (2001) NAD Malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol 126: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justin SFW, Armstrong W. (1987) The anatomical characteristics of roots and plant response to soil flooding. New Phytol 106: 465–495 [Google Scholar]
- Kennedy RA, Rumpho ME, Fox TC. (1992) Anaerobic metabolism in plants. Plant Physiol 100: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulichikhin KY, Chirkova TV, Fagerstedt KV. (2009) Activity of biochemical pH-stat enzymes in cereal root tips under oxygen deficiency. Russ J Plant Physiol 56: 377–388 [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J. (2008) TagFinder for the quantitative analysis of gas chromatography–mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Menegus F, Cattaruzza L, Chersi A, Fronza G. (1989) Differences in the anaerobic lactate-succinate production and in the changes of cell sap pH for plants with high and low resistance to anoxia. Plant Physiol 90: 29–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, Tanz SK, Small ID, Pogson BJ, Millar AH. (2009) Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol 151: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y, Dolferus R, Ismond KP, Good AG. (2007) Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J 49: 1108–1121 [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Good AG. (2008) Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant Cell Physiol 49: 92–102 [DOI] [PubMed] [Google Scholar]
- Mommer L, Visser EJ. (2005) Underwater photosynthesis in flooded terrestrial plants: a matter of leaf plasticity. Ann Bot (Lond) 96: 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench DG, Good AG. (1994) Hypoxically inducible barley alanine aminotransferase: cDNA cloning and expression analysis. Plant Mol Biol 24: 417–427 [DOI] [PubMed] [Google Scholar]
- Narsai R, Howell KA, Carroll A, Ivanova A, Harvey Millar A, Whelan J. (2009) Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol 151: 306–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott T, Sullivan J, James EK, Flemetakis E, Günther C, Gibon Y, Ronson C, Udvardi MK. (2009) Absence of symbiotic leghemoglobins alters bacteroid and plant cell differentiation during development of Lotus japonicus root nodules. Mol Plant Microbe Interact 22: 800–808 [DOI] [PubMed] [Google Scholar]
- Ott T, van Dongen JT, Günther C, Krusell L, Desbrosses G, Vigeolas H, Bock V, Czechowski T, Geigenberger P, Udvardi MK. (2005) Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol 15: 531–535 [DOI] [PubMed] [Google Scholar]
- Pederson O, Rich SM, Colmer TD. (2009) Surviving floods: leaf gas films improve O2 and CO2 exchange, root aeration, and growth of completely submerged rice. Plant J 58: 147–156 [DOI] [PubMed] [Google Scholar]
- Planchet E, Gupta KJ, Sonoda M, Kaiser WM. (2005) Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J 41: 732–743 [DOI] [PubMed] [Google Scholar]
- Randall DD, Miernyk JA. (1990) The mitochondrial pyruvate dehydrogenase complex. Lea PJ, , Methods in Plant Biochemistry: Enzymes of Primary Metabolism, Vol 3. Academic Press, London, 175–199 [Google Scholar]
- Reggiani R, Cantú CA, Brambilla I, Bertani A. (1988) Accumulation and interconversion of amino acids in rice roots under anoxia. Plant Cell Physiol 29: 981–987 [Google Scholar]
- Rockel P, Strube F, Rockel A, Wildt J, Kaiser WM. (2002) Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot 53: 103–110 [PubMed] [Google Scholar]
- Rolletschek H, Weber H, Borisjuk L. (2003) Energy status and its control on embryogenesis of legumes. Embryo photosynthesis contributes to oxygen supply and is coupled to biosynthetic fluxes. Plant Physiol 132: 1196–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Satya Narayan V, Nair PM. (1989) Potato tuber succinate semialdehyde dehydrogenase: purification and characterization. Arch Biochem Biophys 275: 469–477 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M. (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp BJ, Walton CS, Snedden WA, Tuin LG, Oresnik IJ, Layzell DB. (1995) Gaba shunt in developing soybean seeds is associated with hypoxia. Physiol Plant 94: 219–228 [Google Scholar]
- Stitt M, Müller C, Matt P, Gibon Y, Carillo P, Morcuende R, Scheible WR, Krapp A. (2002) Steps towards an integrated view of nitrogen metabolism. J Exp Bot 53: 959–970 [DOI] [PubMed] [Google Scholar]
- Summers JE, Ratcliffe RG, Jackson MB. (2000) Anoxia tolerance in the aquatic monocot Potamogeton pectinatus: absence of oxygen stimulates elongation in association with an unusually large pasteur effect. J Exp Bot 51: 1413–1422 [PubMed] [Google Scholar]
- Tadege M, Dupuis II, Kuhlemeier C. (1999) Ethanolic fermentation: new functions for an old pathway. Trends Plant Sci 4: 320–325 [DOI] [PubMed] [Google Scholar]
- Thomas AL, Guerreiro SMC, Sodek L. (2005) Aerenchyma formation and recovery from hypoxia of the flooded root of system of nodulated soybean. Ann Bot (Lond) 96: 1191–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne JH. (1982) Temperature and oxygen effects on C14-photosynthate unloading and accumulation in developing soyabean seeds. Plant Physiol 69: 48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Fröhlich A, Ramírez-Aguilar SJ, Schauer N, Fernie AR, Erban A, Kopka J, Clark J, Langer A, Geigenberger P. (2009) Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann Bot (Lond) 103: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Roeb GW, Dautzenberg M, Froehlich A, Vigeolas H, Minchin PE, Geigenberger P. (2004) Phloem import and storage metabolism are highly coordinated by the low oxygen concentrations within developing wheat seeds. Plant Physiol 135: 1809–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen JT, Schurr U, Pfister M, Geigenberger P. (2003) Phloem metabolism and function have to cope with low internal oxygen. Plant Physiol 131: 1529–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Feil R, Turpin DH. (1990) Anaerobic metabolism in the N-limited green alga Selenastrum minutum: I. Regulation of carbon metabolism and succinate as a fermentation product. Plant Physiol 94: 1116–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Horsey AK, Weger HG, Turpin DH. (1989) Anaerobic carbon metabolism by the tricarboxylic acid cycle: evidence for partial oxidative and reductive pathways during dark ammonium assimilation. Plant Physiol 91: 1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Yip JY, Parsons HL. (1999) In organello and in vivo evidence of the importance of the regulatory sulfhydryl/disulfide system and pyruvate for alternative oxidase activity in tobacco. Plant Physiol 121: 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek LACJ, Blom CWPM. (1989) Growth responses of Rumex species in relation to submergence and ethylene. Plant Cell Environ 12: 433–439 [Google Scholar]
- Yemm EW, Cocking EC. (1955) The determination of amino acids with ninhydrin. Analyst (Lond) 80: 209–213 [Google Scholar]
- Zabalza A, van Dongen JT, Froehlich A, Oliver SN, Faix B, Gupta KJ, Schmäzlin E, Igal M, Orcaray L, Royuela M, et al. (2009) Regulation of respiration and fermentation to control the plant internal oxygen concentration. Plant Physiol 149: 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]