Abstract
To study the influence of PINFORMED1 (PIN1)-mediated auxin transport during embryogenesis and endosperm development in monocots, the expression pattern of the three identified ZmPIN1 genes was determined at the transcript level. Localization of the corresponding proteins was also analyzed during maize (Zea mays) kernel development. An anti-indole-3-acetic acid (IAA) monoclonal antibody was used to visualize IAA distribution and correlate the direction of auxin active transport, mediated by ZmPIN1 proteins, with the actual amount of auxin present in maize kernels at different developmental stages. ZmPIN1 genes are expressed in the endosperm soon after double fertilization occurs; however, unlike other tissues, the ZmPIN1 proteins were never polarly localized in the plasma membrane of endosperm cells. ZmPIN1 transcripts and proteins also colocalize in developing embryos, and the ZmPIN1 proteins are polarly localized in the embryo cell plasma membrane from the first developmental stages, indicating the existence of ZmPIN1-mediated auxin fluxes. Auxin distribution visualization indicates that the aleurone, the basal endosperm transfer layer, and the embryo-surrounding region accumulate free auxin, which also has a maximum in the kernel maternal chalaza. During embryogenesis, polar auxin transport always correlates with the differentiation of embryo tissues and the definition of the embryo organs. On the basis of these reports and of the observations on tissue differentiation and IAA distribution in defective endosperm-B18 mutant and in N-1-naphthylphthalamic acid-treated kernels, a model for ZmPIN1-mediated transport of auxin and the related auxin fluxes during maize kernel development is proposed. Common features between this model and the model previously proposed for Arabidopsis (Arabidopsis thaliana) are discussed.
In maize (Zea mays), the first step of endosperm development is the formation of a syncytium surrounding a large central vacuole, due to the first mitotic divisions. Subsequently, cellularization forms anticlinal cell walls. Cellularization continues centripetally until the entire endosperm cavity is contained within discrete cells. The aleurone is an epidermal layer that covers the endosperm, secretes hydrolytic enzymes, and mobilizes storage reserves from the starchy endosperm during seed germination (Becraft, 2001; Olsen, 2001). The basal endosperm transfer layer (BETL) is a highly specialized tissue with deep cell ingrowths at the basal surface that facilitate nutrient uptake from the apoplastic space in the placentochalazal region (Thompson et al., 2001) and is characterized by the BETL class of genes (Hueros et al., 1995; Muniz et al., 2006). Among the BETL genes, ZmTCI-1, a Myb-related transcription factor, has been shown to be a key regulator of transfer cell differentiation and function (Barrero et al., 2009). The establishment of the starchy endosperm and embryo-surrounding region (ESR), which is characterized by cells with dense cytoplasm and extensive rough endoplasmic reticulum, starts 4 to 6 d after pollination (DAP; Opsahl-Ferstad et al., 1997; Becraft, 2001). The ESR is also characterized by the expression of specific ESR genes (Bonello et al., 2000; Balandin et al., 2005). Interestingly, no ESR gene transcripts are detected in spontaneous embryoless kernels, suggesting that correct gene expression needs signaling from the embryo to the endosperm (Opsahl-Ferstad et al., 1997). During the maturation phase starting at 10 to 12 DAP, the starchy or internal endosperm cells accumulate starch and storage protein and cell divisions cease; however, nonselective endoreduplication cycles occur in the genome of central endosperm cells (Slocombe et al., 1999).
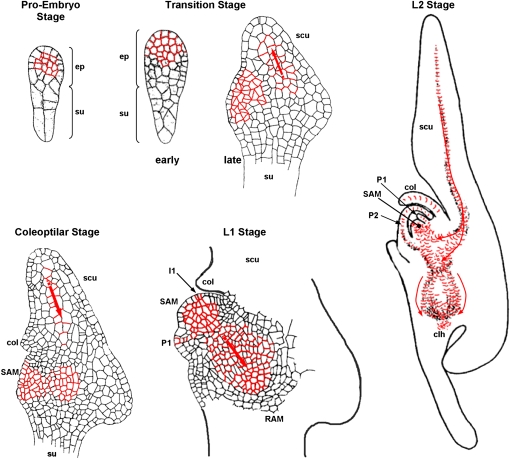
In maize, the first asymmetric divisions of the zygote determine the apical and basal poles of the embryo, as is the case with most angiosperms. However, unlike Arabidopsis (Arabidopsis thaliana), where apical/basal polarity and radial organization are established by a fixed program of cell division planes, only the plane of the first division is predictable in the maize zygote (Randolph, 1936; Bommert and Werr, 2001). The upper chalazal cell originates the embryo proper and the lower micropylar cell forms the suspensor. Successive irregular cell divisions of the apical cell produce an undifferentiated globular cell structure (the proembryo), from which cell-specific lineages have yet to be determined. At the end of the proembryo stage, the protoderm specifies as a single layer of homogenously sized peripheral cells surrounding the embryo proper. Maize mutants impaired in the restriction of embryonic protoderm-specific genes have been shown to fail in establishing a normal radial organization and to enter the morphological phase of embryogenesis (Elster et al., 2000). Embryonic differentiation continues with the adaxial/abaxial regionalization that is marked by the expression of ZmDRN (Zimmermann and Werr, 2005), a potential ortholog of the Arabidopsis DORNRÖSCHEN (DRN), on the abaxial side, followed by lateral initiation of the shoot apical meristem (SAM) at the adaxial surface. Just above the suspensor, a second group of meristematic cells initiate the root apical meristem (RAM). The SAM and RAM define a secondary axis at an acute angle to the apical/basal axis of the zygote, and the two meristems are protected by two organs exclusively found in grass species, the coleoptile and the coleorhiza (Bommert and Werr, 2001). The SAM is organized into the L1 and the inner L2/L3 layers (Zimmermann and Werr, 2005) and initiates six leaf primordia before the onset of seed dormancy. Maize RAM organization is more complicated than in Arabidopsis: the quiescent center consists of 1,000 to 1,550 cells, compared with only four cells in Arabidopsis, and the root shows a more complex architecture, with a large number of ground tissue cell files (Feldman, 1994). The markedly different developmental embryonic programs of maize and Arabidopsis have recently been reviewed by Chandler et al. (2008), and the main stages of embryo development are reported in Figure 8 below.
Figure 8.
Overview of successive stages of maize embryogenesis and model for the ZmPIN1-mediated auxin transport during maize embryogenesis. Medial longitudinal sections of proembryo, transition, coleoptilar, and L1 and L2 stage maize embryos are shown, adapted from Bommert and Werr (2001). The localization of ZmPIN1 proteins in embryo plasma membrane is reported in red, and arrows indicate the polar auxin fluxes deduced by our results. After the first division of the zygote, a series of cell divisions in unpredictable planes gives rise, at the proembryo stage, to the small embryo proper and the larger suspensor. During the transition stage, adaxial/abaxial polarity is manifested by outgrowth of the scutellum at the abaxial side of the embryo. By the late transition stage, the SAM is evident as a group of cytoplasm-rich cells on the adaxial side. At the coleoptilar stage, the coleoptile becomes evident histologically above the SAM. Activity of the SAM is detected at leaf stage 1 (L1) by emergence of the first leaf primordium opposite the coleoptile. Note that the shoot-root axis is oriented in an oblique angle relative to the apical-basal polarity of the proembryo. The second leaf is initiated opposite the first leaf. The RAM is protected by the coleorhiza. clh, Coleorhiza; col, coleoptile; ep, embryo proper; I1, incipient primordium; P1 and P2, leaf primordia; scu, scutellum; su, suspensor.
Auxins are mobile plant signals, and their local asymmetric distribution in plant tissues contributes to apical-basal axis formation in embryo development, organogenesis, RAM maintenance, vascular tissue differentiation, lateral root initiation, and tropic growth (Benkova et al., 2003; Friml et al., 2003; Reinhardt et al., 2003; Blilou et al., 2005; De Smet and Jurgens, 2007). Auxins are passively taken up into cells but are also actively taken up by AUX/LAX permeases and by at least one reversible P-glycoprotein-ATP-binding cassette B (ABCB) transporter (Marchant et al., 1999; Terasaka et al., 2005). Auxins are exported by PINFORMED (PIN) plasma membrane efflux carriers (Petrasek et al., 2006) and a subset of ABCB transport proteins characterized by ABCB1 and ABCB19 (Geisler et al., 2005; Geisler and Murphy, 2006; Blakeslee et al., 2007). In particular, the asymmetrical cellular localization of PIN proteins determines the direction of cell-to-cell auxin flow in development (Benkova et al., 2003; Friml, 2003; De Smet and Jurgens, 2007).
During Arabidopsis embryogenesis, an apical-basal auxin gradient mediated by PIN1 and PIN7 triggers the specification of apical embryo structures and subsequent reorganization of the auxin gradient to specify the basal root pole (Friml et al., 2003; De Smet and Jurgens, 2007). Studies from wheat (Triticum aestivum) zygotic embryos indicate that heterogeneous auxin distribution plays an analogous role in monocot embryonic pattern formation (Fischer and Neuhaus, 1996; Fischer-Iglesias et al., 2001).
Several genes regulating the polar targeting of PIN proteins have been identified. In Arabidopsis, AtPIN1 basal localization is mediated by the GNOM ADP ribosylation factor/guanine nucleotide exchange factor, which functions in brefeldin A-sensitive endosomal vesicle formation (Steinmann et al., 1999; Geldner et al., 2004), while PINOID, a Ser/Thr protein kinase, controls the polarity of PIN localization by direct phosphorylation of specific PIN residues (Friml et al., 2004; Michniewicz et al., 2007).
In order to make up for the knowledge deficit on the role of PIN-mediated auxin transport during maize embryo and endosperm development (Chandler et al., 2008), we investigated the behavior of ZmPIN1 genes and auxin accumulation patterns during maize kernel differentiation in wild-type and defective endosperm-B18 (de*-B18) mutant plants (Torti et al., 1984, 1986). In a previous work, we characterized two ZmPIN1 genes, ZmPIN1a and ZmPIN1b, which were shown to play an important role in maize postembryonic development (Carraro et al., 2006); now, our results indicate that the ZmPIN1-mediated transport of auxin and auxin accumulation are related to cellular domain differentiation during seed formation, and these data are confirmed by treatments with an auxin transport inhibitor. On the basis of our findings, we formulate a model for the ZmPIN1-mediated transport of auxin and the related auxin fluxes during maize embryogenesis and endosperm development. Similarities and differences between this model and the one previously proposed for the Arabidopsis plant model system are discussed.
RESULTS
Maize contains at least three PIN1 orthologous genes, named ZmPIN1a (accession no. DQ836239), ZmPIN1b (accession no. DQ836240), and ZmPIN1c (accession no. EU570251; for the characterization of these genes, see Carraro et al. [2006] and Supplemental Figs. S1 and S2).
Analysis of the ZmPIN1 Transcript Levels during Maize Kernel Development
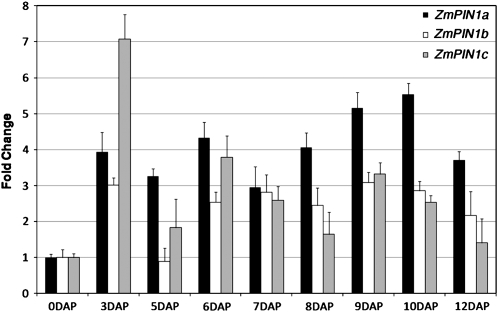
In this study, we focused on the analysis of the expression profile of maize members of the ZmPIN family during embryogenesis and endosperm development. To this end, we performed quantitative real-time reverse transcription (RT)-PCR using RNA extracted from kernels harvested from 3 to 12 DAP. Expression data in ovaries from unfertilized ear are used as reference. Our results showed that all ZmPIN1 genes were up-regulated after double fertilization and that their transcript levels fluctuated during kernel development, with different patterns for each gene (Fig. 1). In addition, ZmPIN1c exhibited a characteristic increase of its mRNA level (7-fold that of unfertilized ear) in 3-DAP kernels.
Figure 1.
ZmPIN1 quantitative expression analysis. Quantitative real-time PCR analysis reveals that ZmPIN1 genes show differential expression patterns during kernel development. All three genes are up-regulated after double fertilization of the maize embryo sac: ZmPIN1c shows an increase in expression levels (7-fold increase) in 3-DAP kernels, while ZmPIN1a expression increases during later stages. The average value from three replicates and two biological samples was estimated for the three ZmPIN1 mRNAs. This value was subsequently normalized by applying the Pfaffl (2001) method to the transcript level of the housekeeping genes 18S and GAPC2, in order to avoid differences in the RNA extraction and cDNA synthesis. Bar diagrams show standardized average levels of ZmPIN1 transcripts, expressed as fold change with respect to the mRNA levels measured in ovaries from unfertilized ears. sd bars are indicated.
ZmPIN1 Transcripts and Protein Localization during Endosperm Development
To investigate the role of ZmPIN1 genes in maize endosperm development, we analyzed the cellular localization patterns of their transcripts and proteins in kernels harvested at different developmental stages.
In situ hybridization experiments were initially performed using three different ZmPIN1 RNA probes designed on the 3 ′ untranslated regions of each ZmPIN1 transcript and specific for each gene. These three specific probes gave a weak and completely overlapping in situ hybridization signal (data not shown). Then we decided to design a fourth probe in a region common to the three ZmPIN1 RNAs and tested for cross-hybridization to all their transcripts. This common probe exhibited a stronger signal, and its localization profile was the same with each of the three specific probes (Fig. 2). A sense probe was used as a negative control for in situ hybridization experiments (Supplemental Fig. S3, A and B).
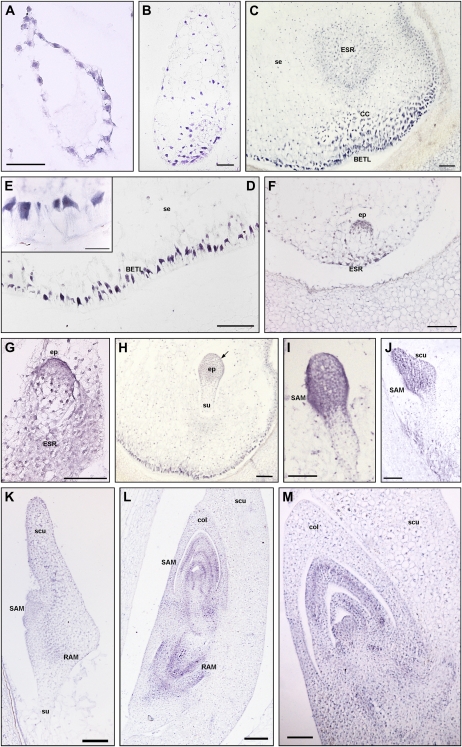
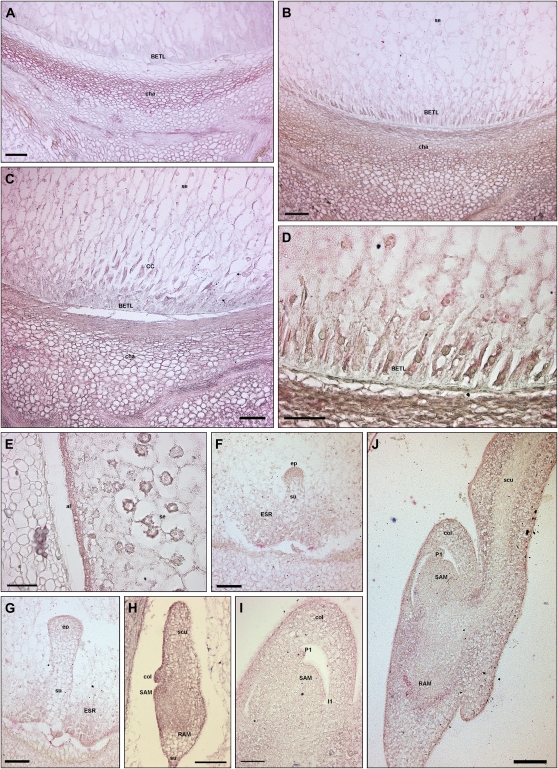
Figure 2.
In situ hybridization of ZmPIN1 mRNAs during maize kernel development. All the images represent longitudinal sections of B73 inbred kernels labeled with an antisense mRNA probe that hybridizes to all three ZmPIN1 transcripts. As soon as the double fertilization occurs, ZmPIN1 genes start to be expressed around the free dividing nuclei that form the syncytium (A), while at the end of cellularization phase, the endosperm shows a gradient of ZmPIN1 expression. The transcripts are most abundant in the basal part of the endosperm (B), a region that will differentiate in the BETL domain and in the cells of the ESR. At the end of differentiation, both these domains show a high expression of ZmPIN1 (C). In BETL cells, ZmPIN1 transcripts are very abundant and compartmentalized on the inner side of the cells (D and E); they also mark the above-located conductive cells (C). In the ESR domain, the ZmPIN1 hybridization signal is particularly strong during early stages of embryogenesis (C, F, and G). During embryogenesis, ZmPIN1 mRNAs are mainly expressed in the embryo proper at the proembryo stage (F and G). Later on, at the early transition stage, ZmPIN1 transcripts mark both the central group of cells in the embryo proper and the differentiating protoderm (H), while no ZmPIN1 expression is detectable in the suspensor (H). The arrow in H indicates the protoderm layer. During the late transition stage, ZmPIN1 mRNAs are mainly localized on the adaxial surface where SAM bulges out (I) and in the scutellum tip (J), whereas very weak hybridization signal is detectable on the scutellum abaxial side (J). A high expression of ZmPIN1 is detectable in the scutellum provascular tissues and in the RAM (K). During the morphogenetic phase, SAM produces five to six leaf primordia and ZmPIN1 genes are expressed in the corpus of the SAM, in the developing primordia, and in the vasculature of the differentiated leaves (L and M). The signal is still present in the RAM and coleorhiza (L). CC, Conductive cells; col, coleoptile; ep, embryo proper; scu, scutellum; se, starchy endosperm; su, suspensor. Bars = 200 μ m in L; 100 μ m in C, G, H to K, and M; 50 μ m in A, B, D, and F; and 25 μ m in E.
The localization profiles of ZmPIN1 proteins were assessed by immunolocalization assays (Fig. 3) using an anti-AtPIN1 monoclonal antibody (Boutte et al., 2006). This antibody was previously tested in maize and was shown to specifically recognize all the different ZmPIN1 polypeptides (Carraro et al., 2006; data not shown). For the negative controls of immunostaining experiments, we performed hybridization with the secondary antibody only (Supplemental Fig. S3, C and D).
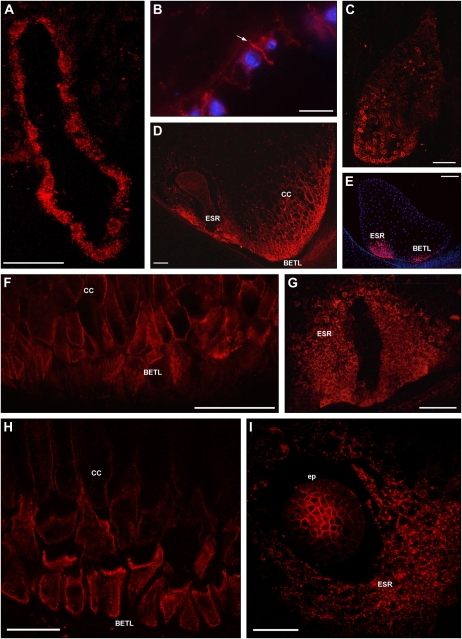
Figure 3.
ZmPIN1 protein localization during maize endosperm development. All images portray maize B73 kernel sections labeled with the anti-AtPIN1 antibody plus a secondary antibody carrying the Alexa568 fluorochrome. A, H, and I present laser confocal images, while B to G present epifluorescence images acquired with a Leica DC300F camera. Nuclei in B and E are blue due to DAPI staining. During the first events of endosperm development, ZmPIN1 proteins are detectable in the nuclear cytoplasmic domains of syncytial endosperm, where proteins are localized in endomembrane systems (A). ZmPIN1 proteins mark the formation of the first anticlinal cell plasma membrane at the beginning of the cellularization phase, while endomembranes of the syncytium are still labeled (B). The arrow in B indicates the newly formed anticlinal plasma membrane. At the end of the cellularization phase, ZmPIN1 proteins are detectable both in endomembranes and cell plasma membranes without any detectable polarity (C). A gradient of protein expression is evident: the proteins are more abundant in the basal-chalazal region of the endosperm (C), where BETL and ESR will differentiate. Both transfer layer and ESR cells show high ZmPIN1 levels (D and E), but these domains are characterized by two different ZmPIN1 localization patterns. In BETL and inner conductive cells, the proteins are localized in all the cell membranes, without any detectable polarization, suggesting a homeostasis of auxin levels between the cells of this endosperm domain (D, F, and H). In ESR, ZmPIN1s are detectable almost exclusively in the cell endomembranes (D, G, and I), suggesting a lack of auxin efflux mediated by PIN1 proteins from these cells. CC, Conductive cells; ep, embryo proper. Bars = 200 μ m in E; 100 μ m in A, C, D, F, and G; 75 μ m in I; 50 μ m in B; and 40 μ m in H.
Our results showed that ZmPIN1 transcripts and proteins accumulated in the endosperm after double fertilization occurred. The hybridization signal for the RNA antisense probe was detectable in the dividing free nuclei forming the syncytium, which surrounded a large central vacuole in the endosperm (Fig. 2A), and the proteins marked the nuclear cytoplasmic domains of the syncytial endosperm (Fig. 3A). At the beginning of the cellularization phase, ZmPIN1 proteins marked the anticlinal cell plasma membrane and endomembranes of the syncytium (Fig. 3B).
At the end of cellularization, a gradient of ZmPIN1 transcript (Fig. 2B) and protein (Fig. 3C) accumulation was evident toward the chalazal region of the endosperm, where the transfer cells and ESR cellular domain begin to differentiate. The proteins marked the endomembranes and cell boundaries without any detectable polarity (Fig. 3C).
In 6-DAP endosperms, the aleurone and endosperm initials differentiated into the basal endosperm transfer layer. At this developmental stage, the transfer cells showed a strong accumulation of ZmPIN1 transcripts, which was evident starting from the formation of the first cell wall ingrowths (Fig. 2D) up to the complete development of the above-located prismatic cells (Fig. 2C), also called conductive cells. Interestingly, the hybridization signal appeared strongly polarized in the differentiated transfer cells, because they polarized their nuclei and cytoplasms toward those cell membranes facing the central region of the endosperm (Fig. 2E). In the basal endosperm region, ZmPIN1 transcripts were also localized in the ESR cells (Fig. 2, C, F, and G). In the ESR, the hybridization signal with ZmPIN1 antisense probe was particularly strong from 6 to 12 DAP, when the early stages of embryogenesis occurred (Fig. 2, F and G).
Both the BETL and ESR cellular domains exhibited a high level of ZmPIN1 proteins (Fig. 3, D and E). In the transfer cells, the proteins were localized in all the cell plasma membrane, without any evident polarization (Fig. 3, D, F, and H). In the BETL cellular domain, the accumulation of ZmPIN1 proteins was detectable in both cells characterized by the extensive cell wall ingrowths facing the maternal tissue of the kernels and in the conductive cells (Fig. 3, D, F, and H). In the ESR domain, ZmPIN1 proteins showed a peculiar localization exclusively in the cytoplasm of the cells surrounding the embryo suspensor (Fig. 3, D, G, and I).
In both BETL and ESR domains, the ZmPIN1 transcript and protein levels decreased during the maturation phase of endosperm development and were not detectable in the other cellular domains, such as the aleurone and starchy endosperm (Figs. 2, C and H, and 3, D and E).
Overall, our results indicated that ZmPIN1 transcript and protein localization profiles overlap and that ZmPIN1 genes are expressed in the maize endosperm from the first developmental events; however, unlike other tissues (Carraro et al., 2006), the ZmPIN1 proteins were never polarly localized in plasma membrane of endosperm cells.
ZmPIN1 Transcript and Protein Localization during Embryo Development
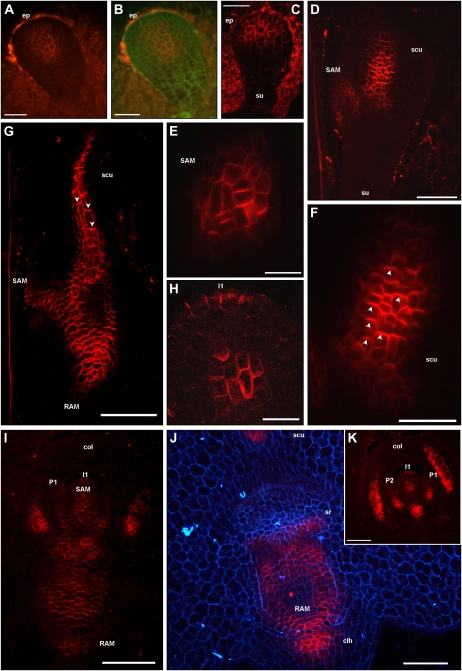
The localization of ZmPIN1 transcripts and corresponding proteins was also analyzed in the maize embryo during embryogenesis. Our results showed that during early embryogenesis, the ZmPIN1 genes were mainly expressed in the embryo proper of the proembryo (Fig. 2, F and G). ZmPIN1 proteins started to accumulate when the boundary between the embryo proper and the suspensor became evident. A group of cells placed in the middle of the proembryo showed ZmPIN1 localization in all cell plasma membranes without any detectable polarity (Figs. 3I and 4, A and B). At this stage, the embryo proper was made up of an undifferentiated globular cell mass, but slightly later it began to differentiate a distinct outer cell layer: the embryonic protoderm, where ZmPIN1 transcripts gave a strong hybridization signal (Fig. 2H). ZmPIN1 proteins accumulated in the upper cells of the proembryo and were also detectable and polarized only in the anticlinal membranes of differentiating embryo protoderm, indicating an auxin flux directed upward and converging toward the embryo tip (Fig. 4C).
Figure 4.
ZmPIN1 protein localization during maize embryogenesis. All images portray maize B73 embryo sections labeled with the anti-AtPIN1 antibody plus a secondary antibody carrying the Alexa568 fluorochrome. G to I and K present laser confocal images, while A to F and J present epifluorescence images acquired with a Leica DC300F camera. Green and blue color in B and J represents tissue autofluorescence collected with GFP and DAPI filters, respectively. E and F represent bigger magnification of D, while H reports a detail of K. During the proembryo stage, ZmPIN1 proteins mark a group of cells placed in the middle of the proembryo, without any evident polarity, while no signal is detectable in the suspensor (A and B). In the early transition phase, ZmPIN1 proteins are expressed in the upper cells of the proembryo and detectable and polarized only in the anticlinal membranes of differentiating embryo protoderm, indicating an auxin flux directed upward and converging toward the embryo tip (C). Later on, at the transition stage, ZmPIN1 proteins mark the initials of the SAM and inner tissues of the developing scutellum (D). The SAM initials are characterized by a central group of cells presenting apolar ZmPIN1 protein localization (E), while along the scutellum, the proteins mark the apical cell membranes, indicating the presence of an acropetal auxin flux directed toward the tip of the scutellum (F). Arrowheads in F indicate the polarization of ZmPIN1 proteins in apical cell membranes. At the late coleoptilar stage, an inversion of ZmPIN1 polarization is evident in the scutellum: after scutellum vasculature differentiation, the ZmPIN1 proteins are localized in the basal cell membranes, suggesting an auxin flux directed downward (G). Arrowheads in G indicate the polarization of ZmPIN1 proteins in basal cell plasma membranes. ZmPIN1 protein relocation at the level of the scutellum corresponds to the establishment of a continuous basally polarized localization of the ZmPIN1 proteins at the level of embryo axis (G). Therefore, two basipetal auxin fluxes can be detected at this stage: one from the scutellum tip to the SAM and RAM, and another from the SAM toward the root pole (G and I). Maize SAM initiates up to six true leaf primordia prior to seed dormancy, and during these processes, ZmPIN1s localize in all the cell membranes of the central group of cells in the SAM, while in the L1 outer layer ZmPIN1s localize only at the level of the incipient primordia (H, I, and K). ZmPIN1 proteins are basally localized in primordia provasculature, suggesting auxin fluxes directed from the primordia to the inner tissues of the SAM and then to the root pole (I and K). ZmPIN1 protein polarization also suggests basipetal auxin transport from the SAM to the RAM (I and J), and the proteins are also detectable in the coleorhiza (I and J) and at the sites of seminal root initiation (J). Moreover, when coleoptile is fully developed, a longitudinal section shows that ZmPIN1 proteins mark the apical cell membranes, suggesting auxin fluxes directed toward the coleoptile tip (K). clh, Coleorhiza; col, coleoptile; ep, embryo proper; I1, incipient primordium; P1 and P2, leaf primordia; scu, scutellum; sr, seminal root; su, suspensor. Bars = 100 μ m in D, G, I, and J; 75 μ m in K; 50 μ m in A to C, E, and F; and 20 μ m in H.
During the transition stage, when the anlage of the SAM started at the adaxial surface of the embryo and the abaxial portion of the embryo formed the scutellum, ZmPIN1 transcripts were more abundant at the adaxial surface and at the top of developing scutellum (Fig. 2, I and J). ZmPIN1 proteins marked the initials of the SAM and the inner tissues of the developing scutellum (Fig. 4D). In the SAM initials, the protein showed apolar localization (Fig. 4E), while along the scutellum, the proteins marked the apical cell membranes, indicating the presence of acropetal auxin fluxes directed toward the tip of the scutellum (Fig. 4F).
By the coleoptilar stage, ZmPIN1 transcripts marked the scutellum vasculature and the incipient root apical meristem (Fig. 2K). At this stage, the ZmPIN1 proteins were localized in the cell membranes of the coleoptile that was evident above the SAM. The proteins were also present in the vasculature of the scutellum, in the inner tissues of the SAM, and in the initials of the RAM (data not shown). At the late coleoptilar stage, a relocation of ZmPIN1 proteins was evident in the scutellum; indeed, during the differentiation of the scutellum vascular tissue, the PIN1 proteins were localized in the apical cell membrane, while after the vasculature was differentiated, the PIN1 proteins localized in the basal cell membrane (Fig. 4G). This observation suggests an auxin flux directed downward. Furthermore, ZmPIN1 protein relocation at the level of the scutellum corresponded to the establishment of a continuous polarized expression of the PIN1 proteins also at the level of the embryo: the ZmPIN1 proteins mainly marked the basal membranes of the cells of the SAM and the RAM, indicating an auxin flux toward the root pole (Fig. 4G).
During the morphogenetic phase of embryogenesis, from L1 to L6, the SAM initiated six leaves and ZmPIN1 transcripts were detectable in the corpus of the SAM in the developing primordia and vasculature of the differentiated leaves (Fig. 2, L and M). The hybridization signal was also detected in the RAM in the coleorhiza and primordia of the seminal roots (Fig. 2L; data not shown). At the L1 stage, the longitudinal sections of the embryo indicated that the ZmPIN1 proteins had accumulated in the two vascular strands of the scutellum in the SAM and root (Fig. 4I). In the SAM, the proteins were localized in the corpus and L1 layer at the site where a new leaf primordium was differentiating (Fig. 4, H and I). In the root, the signal was diffuse and mainly polarized in the basal membrane of cells. Both seminal root primordia and coleorhiza were marked (Fig. 4, I and J). At this stage, beneath the corpus of the SAM, ZmPIN1 localized in the basal cell membrane of all cell types accumulating these proteins. This observation suggests that the following auxin fluxes can occur: from the tip of the scutellum to the SAM and root; from the forming primordia toward the inner tissues of the SAM; from the SAM to the root toward the RAM; and from the RAM to the coleorhiza toward the suspensor (Fig. 4, I and J).
During the L1 to L5 stages, ZmPIN1 protein localization was quite modular, because the SAM was characterized by a central group of cells carrying the protein in all the cell membranes and the signal was localized in the outer L1 layer only at the level of the incipient primordia (Fig. 4K). At the level of the fully developed coleoptile, longitudinal sections showed that ZmPIN1 proteins marked the apical cell membranes, suggesting auxin fluxes directed toward the coleoptile tip (Fig. 4K). ZmPIN1 protein localization and polarization in the scutellum and root were conserved during the later stages of embryogenesis, thus indicating an auxin flux direct toward the suspensor (data not shown). After the L1 stage of embryo development, ZmPIN1 protein accumulation was observed in files of cells of the suspensor, indicating an auxin flow toward the endosperm (data not shown).
Taken together, our results indicate that, similar to what is observed in the endosperm, ZmPIN1 transcripts and proteins also colocalize in developing embryos. However, unlike endosperm, the ZmPIN1 proteins are polarly localized in the embryo cell plasma membrane from the first developmental stages, and several ZmPIN1-mediated auxin fluxes can be inferred.
Indole-3-Acetic Acid Localization in Maize Kernels
In order to correlate the direction of auxin active transport, mediated by ZmPIN1 proteins, with the actual amount of auxin present in maize kernels during development, an anti-indole-3-acetic acid (IAA) monoclonal antibody was used to analyze IAA distribution (Fig. 5; Supplemental Fig. S3, E–I). Several negative controls were performed to test the antibody specificity: no signal was visualized when the primary (Supplemental Fig. S3, E and F) or the secondary (Supplemental Fig. S3, G and H) antibody was omitted in the immunolocalization experiments. In kernels not prefixed with N-ethyl-N′ -(3-dimethylaminopropyl)carbodiimide hydrochloride (EDAC), the immune signal was not detectable and sections were as stained as the negative controls (Supplemental Fig. S3I).
Figure 5.
IAA localization during maize kernel development. An anti-IAA monoclonal antibody was employed to determine the auxin distribution and accumulation in developing maize endosperm and embryos. IAA accumulation is detectable in the BETL domain at 8 DAP (B) and 12 DAP (C and D), in conductive cells at 12 DAP (C), and in the chalazal maternal tissues from 6 to 16 DAP (A–C), while only a weak signal is visible in the starchy endosperm (B, C, and E). Free IAA accumulation is detectable in ESR cells (F and G) and in the aleurone layer (E). Embryos at the globular stage are characterized by high auxin accumulation at the top of the embryo proper, while a very low signal is detectable in the suspensor (F). At the early and late transition stages, the auxin accumulation is evident in the protodermal layer and inner cells of the embryo proper, whereas suspensor shows the IAA minimum (G). The coleoptilar stage embryos are characterized by gradients of IAA distribution: the signal is high in the tip of the scutellum and in all the protoderm cells at the adaxial side, while it decreases toward the suspensor (H). Auxin maxima are detectable at the tip of the developing coleoptile, in the SAM, and in embryo root pole (H). During the SAM morphogenetic phase, auxin accumulation is evident in the tip of leaf primordia (I and J), in the corpus of the SAM (I and J), and in the RAM (J). al, Aleurone; CC, conductive cells; cha, chalaza; col, coleoptile; ep, embryo proper; I1, incipient primordium; P1, leaf primordium; pro, protoderm; scu, scutellum; se, starchy endosperm; su, suspensor. Bars = 100 μ m in A to C, F to H, and J; and 50 μ m in D, E, and I.
Analysis of 8-DAP endosperm cross-sections and longitudinal sections showed that the hybridization signal was evident in three cellular domains: the transfer layer, ESR, and aleurone (Fig. 5, B and F; data not shown). At 12 DAP, just before the endosperm started to accumulate starch, a strong signal persisted in the transfer cells (Fig. 5, C and D), ESR (Fig. 5G), and aleurone (Fig. 5E). At this developmental stage, only a faint signal was visible in the starchy endosperm cellular domain (Fig. 5E). In addition, we observed that, in kernels harvested from 6 to 12 DAP, the IAA signal was high in the maternal region corresponding to chalaza (Fig. 5, A–C).
During the globular stages of embryogenesis, the distribution of IAA in the embryos was evident at the top of the embryo proper, while a very weak signal was detectable in the suspensor (Fig. 5F). At the early transition stage, auxin maximum was detectable in the differentiating protoderm (Fig. 5G). At the late transition stage, the auxin accumulation was still detectable in the protodermal layer and inner cells of the embryo proper, whereas the suspensor showed an IAA minimum (data not shown). At the coleoptilar stage, a gradient of IAA distribution characterized the embryo: the signal was high in the tip of the scutellum and decreased toward the suspensor. In addition, auxin accumulation was detectable in all the protoderm cells at the adaxial side, and auxin maxima were also detectable at the tip of the developing coleoptile and SAM (Fig. 5H). IAA accumulation was also evident in the embryonic root (Fig. 5H). During the morphogenetic phase of embryo development, the auxin accumulation was evident in the tip of the leaf primordia, corpus of the SAM, RAM, and embryo epidermal layer (Fig. 5, I and J). These results showed that in the endosperm, three cellular domains (aleurone, BETL, and ESR) accumulate free auxin, which also has a maximum in the maternal chalazal region of the kernel. In the embryo, an IAA maximum always correlates with the differentiation of a tissue and organ definition.
ZmPIN1 Expression, Auxin Accumulation, and BETL Differentiation in de*-B18 Mutant Kernels
The roles of PIN1-mediated polar auxin fluxes and accumulation during maize endosperm development were analyzed in the maize de*-B18 mutant recently mapped on maize chromosome 10 (Pasini et al., 2008). This mutant was identified as a dek mutant (mutants showing defective kernels with small or incompletely developed endosperms and embryos) showing reduction of dry matter accumulation in the endosperm during all kernel developmental stages (Torti et al., 1984, 1986). Auxin measurement in the mutant endosperm tissues revealed levels of free and bound IAA at least 15 times lower than in the wild-type kernels. Taking into account that exogenous auxin applications compensate the growth capacity of the mutant kernels (Torti et al., 1986), it has been suggested that the de*-B18 mutation is involved in IAA metabolism. During kernel development, quantitative expression analysis by real-time PCR revealed only a weak delay in ZmPIN1 gene expression in mutant kernels compared with wild-type kernals (data not shown).
In situ hybridization assays showed that in kernels with a severe phenotype, ZmPIN1 transcripts were not detectable in BETL cells (Fig. 6, A and D). This lack of ZmPIN1 efflux carrier expression was associated with an altered differentiation of the transfer cells that are round shaped, not polarized, and with the nucleus in a central position (Fig. 6, D and E). Abnormal transfer layer differentiation in de*-B18 mutant kernels was confirmed by the lack of BETL1 gene expression in the transfer layer and above-located conductive cells (Fig. 6, B and E). No additional cytological defects were detected in other endosperm domains and in the developing embryo (data not shown).
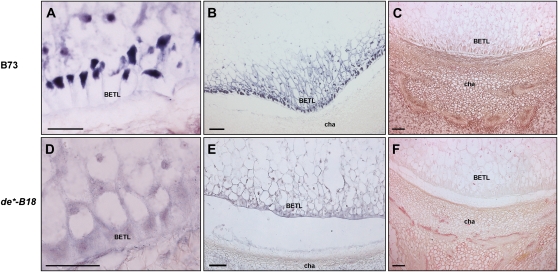
Figure 6.
IAA accumulation and ZmPIN1 and BETL1 transcript localization in de*-B18 endosperm transfer layer cells. Longitudinal sections of kernels at 12 DAP of B73 (A–C) and de*-B18 (D–F) were labeled with an antisense mRNA probe that hybridizes to all three ZmPIN1 transcripts (A and D) and with an antisense BETL1 probe (B and E; Hueros et al., 1995). Anti-IAA monoclonal antibody was used to determine the auxin distribution and accumulation in developing BETL cells (C and F). Unlike in B73 wild-type kernels (A), de*-B18 maize mutant kernels lack ZmPIN1 expression at the level of BETL cells (D) and show an altered differentiation pattern of these cells, which are round shaped and not polarized (D). Compared with the wild type (B), also the BETL1 gene is not expressed in mutant transfer and conductive cells (E). These alterations are accompanied by a reduction of auxin accumulation in BETL cells (C and F), while IAA accumulates as in the wild type in the chalazal maternal region. cha, Chalaza; se, starchy endosperm. Bars = 100 μ m in B, C, E, and F; and 50 μ m in A and D.
Immunolocalization experiments with the anti-IAA antibody showed a reduced accumulation of auxin in BETL cells (Fig. 6, C and F), while IAA accumulated as in the wild type in ESR, aleurone, and chalazal maternal regions (Fig. 6, C and F; data not shown).
Polar Auxin Transport Inhibition Affects Maize Kernel Development, PIN1 Expression and Localization, and Auxin Distribution
To determine whether auxin transport is determinant for embryogenesis and endosperm development, maize plants were treated with N-1-naphthylphthalamic acid (NPA), at two different concentrations, via daily watering for 2 weeks starting from the tasseling stage as described previously (Wu and McSteen, 2007). Embryo morphology, endosperm differentiation, ZmPIN1 transcript and protein localization, and auxin accumulation patterns were analyzed in kernels harvested from 6 to 17 DAP. Both 80 and 120 μm NPA had similar strong effects on ZmPIN1 expression, auxin accumulation, and endosperm and embryo development (Fig. 7).
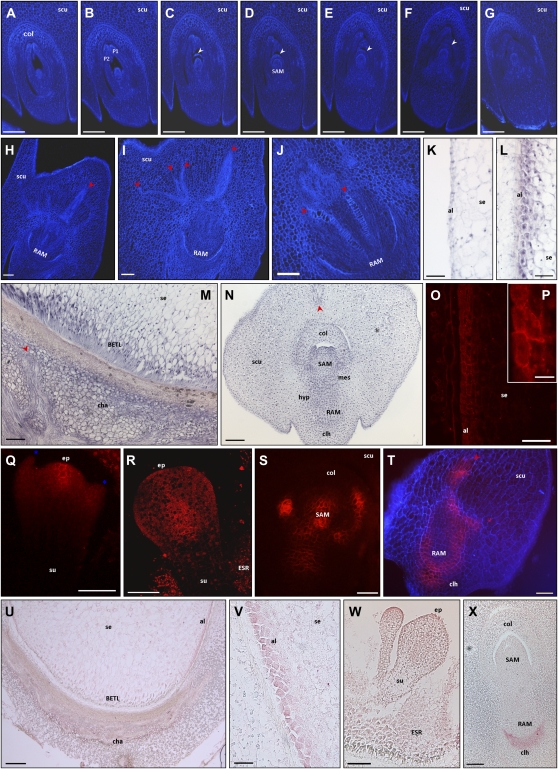
Figure 7.
Effects of NPA treatment on embryo and endosperm differentiation, ZmPIN1 expression, and auxin accumulation. The figure represents Calcofluor staining (A–J), ZmPIN1 in situ hybridization (K–N), ZmPIN1 immunolocalization (O–T), and IAA localization (U–X) on longitudinal sections of kernels at different developmental stages collected from plants watered for 2 weeks with 80 or 120 μm NPA. Q and R present laser confocal images, while A to J, O, P, S, and T present epifluorescence images acquired with a Leica DC300F camera. Blue color in T represents tissue autofluorescence collected with a DAPI filter. P reports a detail of O. The serial sections through a 17-DAP embryo collected from a plant watered for 19 d with 80 μm NPA (A–G) show that NPA induces the formation of a newly semicircular ridge of cells above the meristem, while the two leaf primordia only partially enclose the SAM. At the same stage, NPA treatments result in abnormal scutellum growth with complete loss of symmetry (H) and altered differentiation of vascular bundles in both scutellum (H and I) and embryonic root (J). Growth of misspecified apical structures is observed also in a 9-DAP embryo treated for 15 d with 120 μm NPA (Q), while the most severe phenotype, with two embryos in the same kernel, was observed in 11-DAP seeds treated for 15 d with 80 μm NPA (W). NPA treatments induce ZmPIN1 gene and protein ectopic expression. In 15-DAP kernels treated for 18 d with 80 μm NPA, ZmPIN1 transcripts (L) and proteins (O and P) mark the pluristratified aleurone, while no hybridization signal is detectable in the mock control treated with DMSO only for 18 d (K). ZmPIN1 protein plasma membrane insertion is observable in some aleurone cells (O and P). ZmPIN1 ectopic expression is detectable in the maternal tissues corresponding to the chalaza (M). At the proembryo stage (9-DAP embryo treated for 15 d with 120 μm NPA), ZmPIN1 protein localization is restricted to the apical part of the embryo proper (Q), while at the early transition stage (11-DAP embryo treated for 15 d with 120 μm NPA), the proteins were not inserted in the plasma membranes of the outer cell layer at the protoderm differentiation stage (R). See Figure 4, A to C, for ZmPIN1 protein localization in untreated embryos at the proembryo and early transition stages. During the embryo morphogenetic phase (15-DAP embryo treated for 19 d with 120 μm NPA), ZmPIN1 transcripts are detectable in the whole embryo (N) and in particular in embryo mesocotyl and hypocotyl (N), regions in which no hybridization signal is detectable in untreated kernels (Fig. 2M). In 15-DAP embryo treated for 18 d with 80 μm NPA, ZmPIN1 proteins mark all the cell plasma membranes without any evident polarity (S), and an altered localization of ZmPIN1 proteins is also detectable in the scutellum provascular tissues and in the abnormal embryonic root (T). See Figure 2, I and J, for ZmPIN1 protein localization in untreated embryos. Inhibition of polar auxin transport causes alteration of auxin gradients inside the kernel and embryo. In 13-DAP kernels treated for 15 d with 120 μm NPA, a very high IAA accumulation is visible in the altered aleurone (U and V), while a uniform IAA distribution is evident in the transfer cells toward the central endosperm (U). NPA treatments result in loss of the auxin gradient inside the embryo and in a uniform IAA distribution in both the embryo and the scutellum, with a higher IAA level in the coleorhiza of 15-DAP embryos treated for 19 d with 120 μm NPA (X). See Figure 5 for auxin accumulation patterns in untreated endosperms and embryos. White arrowheads indicate the semicircular structure that surrounds the NPA-treated SAM, while red arrowheads point to the abnormal, enlarged, and irregular vascular tissues. Blue asterisks mark two abnormal structures flanking the embryo proper. al, Aleurone; cha, chalaza; clh, coleorhiza; col, coleoptile; hyp, hypocotyl; mes, mesocotyl; P1 and P2, first and second leaf primordia; scu, scutellum; se, starchy endosperm; su, suspensor. Bars = 200 μ m in A to G and U; 100 μ m in H to J, M to O, and W; 75 μ m in Q and R; 50 μ m in K, L, S, T, V, and X; and 20 μ m in P.
The embryo defects observed ranged from cup-shaped embryos with misspecified apical structures (Fig. 7Q) to development of two embryos in the same kernel (Fig. 7W). Serial sections of 15- and 17-DAP NPA-treated kernels revealed defects in scutellum symmetry and morphology (Fig. 7, H and I; Supplemental Fig. S4) and the formation of a semicircular ridge of cells above the meristem (Fig. 7, A–G). At the same stages of development, NPA-treated embryonic root showed abnormal differentiation of the vascular tissues in the central cylinder (Fig. 7J). Vasculature differentiation was also defective in the enlarged and very irregularly shaped scutellum (Fig. 7, H and I; Supplemental Fig. S4).
Compared with untreated sample (Fig. 7K), in the endosperm NPA inhibited the correct development of the aleurone, leading to the formation of a pluristratified epidermic layer (Fig. 7, L, O, P, and V), while no evident morphological alterations were detectable in BETL and ESR domains (Fig. 7, M, R, U, and W).
In situ hybridization and immunolocalization experiments were performed on NPA-treated kernels to assess the effects of auxin transport inhibition on ZmPIN1 expression. The ectopic expression of ZmPIN1 transcripts was observed in the pluristratified aleurone (Fig. 7, K and L) and in the chalaza (Fig. 7M). Accordingly, immunolocalization showed ZmPIN1 protein expression in the aleurone cells (Fig. 7, O and P). In the aleurone plasma membrane, protein insertion was observed just in some cells (Fig. 7, O and P), while the strong autofluorescence signal of the chalaza region of the seeds did not allow a clear localization of ZmPIN protein (data not shown).
During early stages of embryogenesis, ZmPIN1 transcripts were localized to the embryo proper, as described previously in nontreated kernels (Fig. 2, G–I). Anti-PIN1 antibody mainly marked the apical region of the embryo proper (Fig. 7Q), but ZmPIN1 proteins were not inserted in the plasma membrane of the outer cell layer at the protoderm differentiation stage (Fig. 7R).
During the embryo morphogenetic phase, ZmPIN1 transcripts were expressed throughout the embryo (Fig. 7N), and in particular in embryo mesocotyl and hypocotyl (Fig. 7N), which did not show any hybridization signal in nontreated kernels (Fig. 2M; data not shown). At this stage, ZmPIN1 proteins marked all the cell plasma membrane without any evident polarity (Fig. 7S). An altered expression of ZmPIN1 proteins was also detectable in the scutellum provascular tissues and in the abnormal embryonic root (Fig. 7T).
To verify whether polar auxin transport inhibition exerted its effect on IAA distribution during kernel development, immunolabeling experiments with the anti-IAA antibody on NPA-treated kernel sections were performed. From 11 to 15 DAP, alterations in IAA distribution were observed in the endosperm. A higher IAA accumulation was visible in the altered aleurone (Fig. 7, U and V), while a uniform IAA distribution was evident in the transfer cells toward the central endosperm (Fig. 7U).
During the morphogenetic phase, the auxin gradient evident inside the embryo, with a higher accumulation detectable in the embryo root (Fig. 5, H and J), was not observed in NPA-treated kernels (Fig. 7X). Indeed, a uniform auxin distribution characterized both the embryo and the scutellum, with a higher IAA level in the coleorhiza (Fig. 7X) and in the scutellum epidermis (data not shown).
DISCUSSION
In this study, we analyzed the cellular localization patterns of three members of the maize PIN1 gene family during embryogenesis and endosperm development in order to better understand the role of maize PIN1-like genes in the developmental programs of these organs. Three ZmPIN1 genes, named ZmPIN1a, ZmPIN1b, and ZmPIN1c, have been identified in maize so far. Sequences and phylogenetic analysis showed that these three genes are very likely members of a family that contains the maize orthologs of the Arabidopsis PIN1 gene. Quantitative expression analysis revealed that the three ZmPIN1 genes were up-regulated after double fertilization of the maize embryo sac and that their mRNA levels fluctuated during kernel development, exhibiting different expression profiles. However, the three ZmPIN1 transcripts showed an overlapping cellular localization profile in different stages of kernel development. These results may suggest a certain degree of functional redundancy among the members of the ZmPIN1 family, as was shown by mutant analysis for Arabidopsis PIN genes (Vieten et al., 2005). Nevertheless, the identification and analysis of mutants specifically affecting the activity of each member of the ZmPIN1 family is required to fully elucidate whether and to what extent the three ZmPIN1 genes are functionally redundant.
A Model for ZmPIN1-Mediated Auxin Transport during Embryogenesis
The analysis of ZmPIN1 expression patterns and protein localization and its comparison with auxin accumulation during axis formation and early development in maize embryos indicate that ZmPIN1 proteins play an important role in mediating the auxin transport during maize embryogenesis. The sequence of events we observed during embryogenesis (Fig. 8) suggests a model in which ZmPIN1 polarization is associated with cell and tissue differentiation. According to this model, the auxin fluxes mediated by ZmPIN1s would be created to trigger patterning and differentiation during maize embryogenesis.
This model is supported by a number of observations arising from our study. First, in the embryo proper, ZmPIN1 proteins marked all cell boundaries of the undifferentiated proembryo core, without any polarity, and auxin was mainly localized in the apical part of the embryo. As soon as polar localization of ZmPIN1 was observed in the apical anticlinal membranes of the embryo proper outer cell layer, differentiation of the protoderm started. It has been reported that the protoderm is the first evidence of differentiation in maize embryo (Elster et al., 2000), and we observed that this differentiation is correlated with high auxin accumulation.
Second, we observed that ZmPIN1s exhibit protein polarization also during scutellum differentiation. At the early transition stage, ZmPIN1 proteins marked the provascular cells of the differentiating scutellum, suggesting an auxin flux toward the tip of the single maize cotyledon. After differentiation of the scutellum was induced and auxin accumulated at its tip, an inversion of ZmPIN1 polarity was observed and an auxin flux toward the root pole was established in the scutellum.
Third, the inversion of ZmPIN1 polarity marked the shift from a radial to a bilateral symmetry, and the established auxin flux allowed the differentiation of the SAM and RAM in the embryonic axis. A gradient of auxin distribution was observed from the scutellum to the suspensor. When scutellum differentiation was triggered, ZmPIN1 proteins were detectable at the place where SAM initiation started, without any polarity. After SAM anlagen, ZmPIN1 protein localization indicated auxin transport from the SAM to the developing RAM, which showed an IAA maximum.
Fourth, during the morphogenetic phase, the embryonic SAM was characterized by a central group of cells showing apolar ZmPIN1 localization in all the plasma membranes, while the signal in the external L1 layer was detectable only at the level of the primordium initiation site. Auxin accumulation at this site seems to be fundamental for organ initiation, while a subsequent ZmPIN1 polarization in basal plasma membrane is necessary for auxin canalization from the primordium and its connection with the inner vascular tissues. These findings are also in agreement with our previous observations on vegetative SAM (Carraro et al., 2006).
In previous studies, a model was proposed of auxin distribution and transport during embryogenesis in wheat (Fischer-Iglesias et al., 2001). The authors concluded that diffusion but not active or low-active transport of auxin occurred in radially symmetrical embryos and associated the shift from radial to bilateral symmetry of the embryos with a change in auxin distribution. They also hypothesized the existence of a bidirectional polar transport of auxin toward the scutellum and the SAM from the root pole of the embryo (Fischer-Iglesias et al., 2001). These results are in agreement with our observations on the shift from radial to bilateral symmetry, which is associated with an inversion of PIN1 polarization and auxin flux mediated by these proteins along the embryo. In our results, ZmPIN1 protein polarization in the embryonic axis suggests a flux of auxin toward the root pole, and a gradient of auxin localization toward the RAM was observed. It may be that other transporters are involved in mediating auxin transport from the root pole to the SAM, as was observed in wheat.
The comparison of our results with findings from studies on the model plant Arabidopsis indicates that the PIN1-mediated auxin transport and accumulation during embryogenesis share conserved features also between monocotyledons and dicotyledons. Arabidopsis embryogenesis is characterized by the apical localization of PIN7 in the basal cells of the zygote, thus allowing auxin accumulation in the apical cell for embryo apical pole specification. At the globular stage, when the embryo begins to produce auxin, the localization of PIN7 reversed, shifting to the basal cell of the suspensor. Concomitantly, the PIN1 basal localization was established in the provascular cells, establishing a downward transport of auxin toward the region of the developing root pole. Subsequently, PIN4 expression at the basal pole of the embryo supported the action of both PIN7 and PIN1, triggering root pole specification (Friml et al., 2003). Therefore, in both Arabidopsis and maize, auxins accumulate very early in the apical embryo-forming cells, while no accumulation is detectable in the suspensor cells. Furthermore, maize embryo proper and Arabidopsis globular stage embryos are characterized by PIN1 localization without polarization.
On the other hand, we also observed that some differences exist in PIN1 localization between Arabidopsis and maize. In Arabidopsis heart stage embryos, PIN1 localizes in apical membranes of epidermal cells and in basal membranes of internal tissues, mediating IAA accumulation in the cotyledon tip and the subsequent auxin canalization at the level of cotyledon provascular tissues (Tanaka et al., 2006). We reported that the double localization also characterizes the differentiation of maize scutellum, the monocot single cotyledon, but this occurs in two different moments. At the transition stage, ZmPIN1s first mark the apical cell membrane of developing scutellum, driving IAA accumulation in the tip. Then a switch in ZmPIN1 localization in the basal plasma membrane drives auxin removal and vascular tissue differentiation. Auxin fluxes from cotyledon to the embryo inner tissues drive auxin accumulation in the basal root pole in both maize and Arabidopsis, allowing the RAM establishment. However, the more complex architecture of the maize embryo is accompanied by a more elaborate ZmPIN1 localization pattern. The investigations on proteins involved in polar localization of ZmPIN1 in maize embryo could be facilitated by the recent characterization of the Barren inflorescence2 gene (McSteen et al., 2007), which encodes a maize co-ortholog of the PINOID Ser/Thr kinase, an essential protein for correct polar PIN targeting in Arabidopsis (Friml et al., 2004).
ZmPIN1 Patterning during Endosperm Development
The early developmental events that precede the accumulation of storage materials are essential for endosperm growth in cereals. In previous studies on maize endosperm, the timing of developmental events and regulatory roles of auxin were examined, showing that IAA concentration abruptly increased from 9 to 11 DAP. This increase in auxin levels, and the resulting decrease in the cytokinin-auxin ratio, induces cellular differentiation events, endoreduplication, and expression of particular zein storage proteins (Lur and Setter, 1993). Accordingly, we found that a high accumulation of auxin is observed in BETL, aleurone, and ESR domains in correspondence to the differentiation phase of endosperm development. On the contrary, the auxin level decreased in the starchy endosperm. A high accumulation of auxin was also observed in the ESR in seeds of Arabidopsis (Avsian-Kretchmer et al., 2002). Our results show that the ZmPIN1-mediated transport of auxin is also involved in cellular differentiation in the early events of endosperm development. Specifically, the auxin distribution mediated by ZmPIN1 is defined during the cellularization phase, in particular during first anticlinal membrane formation, and is related to the differentiation of the transfer layer. In the highly specialized transfer cells, the presence of the apolar localization of ZmPIN1 proteins and the high auxin content support the hypothesis that a constant auxin flux may be required to maintain a high level of polarization in these “modified aleurone cells.” In accordance with this hypothesis, it has been shown that Arabidopsis PINs, in addition to the polarized transport of auxin, may also mediate apolar auxin efflux from cells. Indeed, apolar membrane orientations were observed for all PINs with the exception of PIN2/PIN1 in the globular embryos (Friml et al., 2003) and in the SAM (Heisler et al., 2005), PIN3 in the columella cells (Friml et al., 2002b), PIN4 in the quiescent center-surrounding cells (Friml et al., 2002a), and PIN7 in cotyledons, hypocotyl epidermis, and the columella (Blakeslee et al., 2007). In columella cells of Arabidopsis, the apolar PIN3 localization is associated with a maximum of auxin (Sabatini et al., 1999).
Although our results indicate that the ZmPIN1-mediated transport of auxin is related to cellular differentiation during endosperm development, it is also likely that additional factors and mechanisms contribute to the establishment of auxin accumulation. The maize de*-B18 mutant shows reduced levels of free and bound IAA in the endosperm that lead to a reduction in dry matter accumulation (Torti et al., 1986). Anti-IAA immunolocalization experiments showed a reduced accumulation of auxin in de*-18 endosperm, which mainly concerns the transfer cell layer. It might be hypothesized that the down-regulation of ZmPIN1 expression and the lower auxin content in BETL cells result in the disruption of transfer cell polarity and activity. Defective transfer cells are less active in nutrient import into the developing endosperm, explaining the reduced dry matter accumulation in mutant kernels.
A further outcome from this study regards the peculiar subcellular localization of ZmPIN1 proteins in the cytoplasm of the ESR cell domain. This observation raises questions about why the ZmPIN1 proteins in the ESR cellular domain are not targeted to the cell membrane, as usually occurs in other cell types, and whether ZmPIN1 accumulation in the internal cellular compartments is related to particular aspects of their function. Better characterization of the ESR domain at cytological, physiological, and molecular levels is needed in order to elucidate its role in supporting the embryo growth. These investigations could also be useful for understanding the peculiar ZmPIN1 protein localization and IAA accumulation in the ESR cellular domain. At the same time, ZmPIN1 plasma membrane localization studies could confirm the presence of sequence-specific signals mediating cell membrane targeting.
NPA Treatment Confirms the Strong Correlation between ZmPIN1-Mediated Auxin Fluxes, Auxin Accumulation, and Kernel Differentiation
In order to investigate whether ZmPIN1-mediated polar auxin transport has a role in endosperm and embryo patterning, we modified IAA distribution by blocking its transport with the auxin transport inhibitor NPA. Although NPA effects have been reported for multiple targets besides PIN1 (ABCB1 [Noh et al., 2001], ABCB19 [Noh et al., 2001; Nagashima et al., 2008], and Aminopeptidase M1 [APM1; Peer et al., 2009]), our results allowed us to assess the relationship between auxin gradients and correct seed development.
Following NPA treatment, ectopic up-regulation of ZmPIN1 expression was observed in the embryo proper, in the whole embryo during the morphogenetic phase, and in aleurone cells in the endosperm. Similar ectopic up-regulation was observed for AtPIN1, AtPIN2, and AtPIN4 in Arabidopsis roots after NPA treatments (Vieten et al., 2005).
Blocking auxin transport during the early phases of embryogenesis produced extreme phenotypes, such as the differentiation of two embryos connected at the level of the suspensor or the formation of misspecified apical structures (Fig. 7). Similar strong embryonic defects have been recently observed in apm1 loss-of-function Arabidopsis mutants, while pin1pin7 mutants do not exhibit these alterations (Peer et al., 2009), suggesting that the existence of multiple NPA targets should be taken into consideration also in maize.
NPA treatments also caused the perturbation of the proper auxin gradient in the scutellum, which accumulates auxin in epidermis and failed to develop symmetrically, showing abnormal vasculature proliferation. Indeed, as previously demonstrated in Arabidopsis, NPA applications correlated the spatial patterns of auxin distribution with cotyledon specification and differentiation (Friml et al., 2003).
In contrast to untreated seeds, NPA treatments caused the differentiation of up to four layers of aleurone cells in the endosperm. In each layer, the ectopic expression of ZmPIN1 genes and proteins was detected, along with auxin accumulation. Based on the observation that auxin accumulation in the aleurone layer does not involve ZmPIN1 proteins, the identification of the auxin transporters expressed in the aleurone is a prerequisite to elucidate the molecular mechanism creating this phenotype and to clarify the role of auxin in aleurone epidermal cell fate specification.
MATERIALS AND METHODS
Plant Material
Maize (Zea mays) kernels from the B73 inbred line were used for immunolocalization and in situ hybridization experiments. The maize mutant line de*-B18, characterized by Torti et al. (1986), was kindly supplied by Prof. Adriano Marocco (Università Cattolica Piacenza). Plants were self-pollinated during field trials, and kernels were harvested and analyzed at different DAP.
Genomic DNA, RNA Extraction, and Quantitative Real-Time RT-PCR
Genomic DNA was extracted from maize tissues according to the Nucleon Phytopure extraction kit (Amersham). Total RNA was extracted from maize kernels according to the RNeasy Plant Mini Kit (Qiagen) and subjected to on-column DNase treatment. cDNA synthesis was performed with the SuperScript III reverse transcriptase kit (Invitrogen), according to the manufacturer's instructions. One microgram of total RNA were used as a template together with 1 μ L of oligo(dT)12–18 (0.5 μ g μ L−1). ZmPIN1a (forward, 5′-GCAGGGCAAGGCGAACAAGTACGGCCAG-3 ′ ; reverse, 5′-GGCGACGGCCATGCGGACCTCCTTGAC-3 ′), ZmPIN1b (forward, 5′-CCGACCCGTGGGTGATGGCAATGGCA-3 ′ ; reverse, 5′-CTCAAGCCATCAAACTCCGGAGGTGAGC-3 ′), and ZmPIN1c (forward, 5′-TGACCGTCTCATCCCCATGGAGTCGAGG-3 ′ ; reverse, 5′-ATCTCTCCTGTCATAACTCAGAATGG-3 ′) specific primer combinations were used for quantitative expression analysis of the three transcripts. Both maize 18S (forward, 5′-GGAGCCATCCCTCCGTAGTTAGCTTCTT-3 ′ ; reverse, 5′-CCTGTCGGCCAAGGCTATATACTCGTTG-3 ′) and GAPC2 (forward, 5′-AATGGCAAGCTCACTGGC-3 ′ ; reverse, 5′-CTGTCACCGGTGAAGTCG-3 ′) transcripts were used as endogenous references for normalization of the mRNA target. Amplification efficiencies were determined for all primer combinations preparing a standard curve.
The real-time RT-PCR was performed using an ABI 7500 Real-Time PCR System (Applied Biosystems) and the POWER SYBR GREEN PCR Master Mix (Applied Biosystems), following the manufacturer's guidelines. Real-time conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. For each PCR, we observed product melting curves by heating from 60°C to 95°C at 0.2°C s−1. For all transcripts, this procedure allowed the identification of a single product, which we confirmed by analysis on 2% agarose gels. Two different biological samples, obtained by different RNA preparations from separate kernel pools, were processed; three replicates were carried out for each primer combination and for each biological sample. The relative quantification of ZmPIN1 genes was performed with the method of Pfaffl (2001), entering previously determined amplification efficiencies in the qBASE software (Hellemans et al., 2007).
In Situ Hybridization
In situ hybridization experiments were conducted as described (Varotto et al., 2003). Maize kernels were fixed in 4% paraformaldehyde (Sigma) in 0.1 m phosphate buffer (pH 7.2) for 16 h at 4°C and embedded in Paraplast Plus (Sigma-Aldrich). Sections (7–10 μ m) were cut using a microtome (RM2135; Leica) and collected on xylane-coated slides. Slides were deparaffinized and treated with 10 μ g mL−1 proteinase K. In vitro transcription of the digoxigenin-UTP (Roche)-labeled RNA sense and antisense probes was obtained using T7 and Sp6 polymerases. The probe used to detect all three ZmPIN1 transcripts concomitantly corresponds to a 500-bp fragment of the ZmPIN1a cDNA and overlaps the sequence of the peptide recognized by the anti-AtPIN1 antibody (Carraro et al., 2006); this probe has a nucleotide sequence identity of 89% with ZmPIN1b and 85% with ZmPIN1c and was proven to cross-hybridize to the three mRNAs. The three probes specific for each ZmPIN1 mRNA are designed on the 3 ′ untranslated region of each ZmPIN1 transcript, and the sequences are available upon request. The hybridization was performed in a 50% formamide buffer at 48°C overnight. Digoxigenin detection and signal visualization were done using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche), following the manufacturer's instructions. Slides were air dried and mounted with DPX mounting medium (Fluka Biochemika).
Immunolocalization
Immunostaining was performed on wax-embedded (9:1 PEG400 distearate:1-hexadecanol 99%; Sigma) kernels and embryos. Material was previously fixed in 4% paraformaldehyde in 1 × phosphate-buffered saline (PBS) for 16 h at 4°C. Embedded kernels were cut with a Leica RM2135 microtome and stained overnight with the anti-AtPIN1 antibody (Boutte et al., 2006) at a 1:200 dilution in 1 × PBS and 1% bovine serum albumin. Subsequently, the sections were washed twice for 10 min in 1 × PBS and stained for 1 h with the secondary antibody (anti-rabbit IgG conjugated with Alexa568; Molecular Probes) diluted 1:400 in 1 × PBS. Slides were mounted with Vectashield with 4 ′,6-diamino-phenylindole (DAPI) Mounting Medium (Vector Laboratories). For the negative controls of the immunostaining experiments, we performed incubation with the secondary antibody only (Supplemental Fig. S3, C and D). Five to 10 samples for each developmental stage were analyzed. Contradictory localization patterns were never observed in wild-type maize kernels.
IAA Localization
A monoclonal anti-IAA antibody (Biofords PMD09346/0096) was used for IAA immunolocalization. This antibody was tested to specifically recognize free IAA (Weiler et al., 1981). Kernels were prefixed in 3% (w/v) EDAC (Sigma) and 0.1 m PBS, postfixed in 4% paraformaldehyde in the same buffer, and embedded in Paraplast as described above. Some kernels were not prefixed with EDAC to test the specificity of the antibody reaction. Sections (7–10 μ m) were cut using a microtome, collected on polysine slides, and processed as reported previously (Avsian-Kretchmer et al., 2002). The anti-IAA primary antibody was used at a concentration of 0.05 mg mL−1. The secondary antibody, an anti-mouse IgG-AP (Santa Cruz Biotechnology), was used at a 1:250 dilution. Signal visualization was done using SigmaFast FastRed (Sigma), which contains an inhibitor of endogenous alkaline phosphatase activity, following the manufacturer's instructions. For each series of slides derived from one kernel, two negative controls (omission of primary and secondary antibody, respectively) were used, and after 15 to 20 min, the color reaction was stopped in water. Slides were mounted with CC/Mount (Sigma).
Polar Auxin Transport Inhibition and Histological Analysis
B73 maize plants were grown in the greenhouse with supplemental lighting during winter/spring. From the tasseling stage, plants were watered for 15 to 20 d with NPA (ChemService) previously dissolved in dimethyl sulfoxide (DMSO; Sigma) and diluted to the appropriate concentration with tap water. All plants were watered on all days with 150 mL of solution as described (Wu and McSteen, 2007). We tested two different concentrations: 80 and 120 μm NPA. Ten to 15 plants were used for each concentration, and kernels from treated ears were collected at different DAP. Control plants were watered with tap water containing an equal amount of DMSO. Kernels were collected from 6 to 15 DAP, fixed, and embedded as described previously for in situ hybridization or ZmPIN1 or IAA localization.
For histological observations, slides were stained in 0.01% Calcofluor (Fluorescent Brightener; Sigma) and mounted with DPX mounting medium (Fluka Biochemika). About 50 treated kernels were sectioned and observed with the microscope.
Microscopy
Immunostained slides were observed with a Leica TCS SP2 laser confocal microscope. Images were collected frame by frame with the acusto-optical tunable filter using helium/neon laser line 543/594 nm. Images were coded red for Alexa568 and green/blue for autofluorescence. Histological analysis, in situ hybridization, and anti-IAA immunolocalization images were taken with a Leica DM4000B digital microscope equipped with a Leica DC300F camera and Leica Image Manager 50 software.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ836239 (ZmPIN1a), DQ836240 (ZmPIN1b), and EU570251 (ZmPIN1c).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structural comparison between AtPIN1 and ZmPIN1 genes and proteins.
Supplemental Figure S2. PIN1 phylogenetic analysis.
Supplemental Figure S3. In situ hybridization and immunolocalization negative controls.
Supplemental Figure S4. Effects of NPA treatments on scutellum development and symmetry.
Supplemental Materials and Methods S1. Isolation of the ZmPIN1c gene and phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank V. Rossi, N. Carraro, and J. Traas for discussion during early stages of this project, A. Garside for checking the English, and T. Pengo for plant care and supporting experiments. The anti-AtPIN1 antibody was kindly provided by J. Traas.
References
- Avsian-Kretchmer O, Cheng JC, Chen L, Moctezuma E, Sung ZR. (2002) Indole acetic acid distribution coincides with vascular differentiation pattern during Arabidopsis leaf ontogeny. Plant Physiol 130: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balandin M, Royo J, Gomez E, Muniz LM, Molina A, Hueros G. (2005) A protective role for the embryo surrounding region of the maize endosperm, as evidenced by the characterisation of ZmESR-6, a defensin gene specifically expressed in this region. Plant Mol Biol 58: 269–282 [DOI] [PubMed] [Google Scholar]
- Barrero C, Royo J, Grijota-Martinez C, Faye C, Paul W, Sanz S, Steinbiss HH, Hueros G. (2009) The promoter of ZmMRP-1, a maize transfer cell-specific transcriptional activator, is induced at solute exchange surfaces and responds to transport demands. Planta 229: 235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW. (2001) Cell fate specification in the cereal endosperm. Semin Cell Dev Biol 12: 387–394 [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J. (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al. (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19: 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Bommert P, Werr W. (2001) Gene expression patterns in the maize caryopsis: clues to decisions in embryo and endosperm development. Gene 271: 131–142 [DOI] [PubMed] [Google Scholar]
- Bonello JF, Opsahl-Ferstad HG, Perez P, Dumas C, Rogowsky PM. (2000) Esr genes show different levels of expression in the same region of maize endosperm. Gene 246: 219–227 [DOI] [PubMed] [Google Scholar]
- Boutte Y, Crosnier MT, Carraro N, Traas J, Satiat-Jeunemaitre B. (2006) The plasma membrane recycling pathway and cell polarity in plants: studies on PIN proteins. J Cell Sci 119: 1255–1265 [DOI] [PubMed] [Google Scholar]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S. (2006) ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142: 254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J, Nardmann J, Werr W. (2008) Plant development revolves around axes. Trends Plant Sci 13: 78–84 [DOI] [PubMed] [Google Scholar]
- De Smet I, Jurgens G. (2007) Patterning the axis in plants: auxin in control. Curr Opin Genet Dev 17: 337–343 [DOI] [PubMed] [Google Scholar]
- Elster R, Bommert P, Sheridan WF, Werr W. (2000) Analysis of four embryo-specific mutants in Zea mays reveals that incomplete radial organization of the proembryo interferes with subsequent development. Dev Genes Evol 210: 300–310 [DOI] [PubMed] [Google Scholar]
- Feldman L. (1994) The maize root. Freeling M, Walbot V, , The Maize Handbook. Springer Laboratory, New York, pp 29–37 [Google Scholar]
- Fischer C, Neuhaus G. (1996) Influence of auxin on the establishment of bilateral symmetry in monocots. Plant J 9: 659–669 [Google Scholar]
- Fischer-Iglesias C, Sundberg B, Neuhaus G, Jones AM. (2001) Auxin distribution and transport during embryonic pattern formation in wheat. Plant J 26: 115–129 [DOI] [PubMed] [Google Scholar]
- Friml J. (2003) Auxin transport: shaping the plant. Curr Opin Plant Biol 6: 7–12 [DOI] [PubMed] [Google Scholar]
- Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jurgens G, et al. (2002a) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673 [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jurgens G. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Friml J, Wisniewska J, Benkova E, Mendgen K, Palme K. (2002b) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865 [DOI] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, et al. (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44: 179–194 [DOI] [PubMed] [Google Scholar]
- Geisler M, Murphy AS. (2006) The ABC of auxin transport: the role of P-glycoproteins in plant development. FEBS Lett 580: 1094–1102 [DOI] [PubMed] [Google Scholar]
- Geldner N, Richter S, Vieten A, Marquardt S, Torres-Ruiz RA, Mayer U, Jurgens G. (2004) Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131: 389–400 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueros G, Varotto S, Salamini F, Thompson RD. (1995) Molecular characterization of BET1, a gene expressed in the endosperm transfer cells of maize. Plant Cell 7: 747–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lur HS, Setter TL. (1993) Role of auxin in maize endosperm development (timing of nuclear DNA endoreduplication, zein expression, and cytokinin). Plant Physiol 103: 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. (1999) AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J 18: 2066–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Malcomber S, Skirpan A, Lunde C, Wu X, Kellogg E, Hake S. (2007) barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol 144: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, et al. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056 [DOI] [PubMed] [Google Scholar]
- Muniz LM, Royo J, Gomez E, Barrero C, Bergareche D, Hueros G. (2006) The maize transfer cell-specific type-A response regulator ZmTCRR-1 appears to be involved in intercellular signalling. Plant J 48: 17–27 [DOI] [PubMed] [Google Scholar]
- Nagashima A, Uehara Y, Sakai T. (2008) The ABC subfamily B auxin transporter AtABCB19 is involved in the inhibitory effects of N-1-naphthyphthalamic acid on the phototropic and gravitropic responses of Arabidopsis hypocotyls. Plant Cell Physiol 49: 1250–1255 [DOI] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP. (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA. (2001) Endosperm development: cellularization and cell fate specification. Annu Rev Plant Physiol Plant Mol Biol 52: 233–267 [DOI] [PubMed] [Google Scholar]
- Opsahl-Ferstad HG, Le Deunff E, Dumas C, Rogowsky PM. (1997) ZmEsr, a novel endosperm-specific gene expressed in a restricted region around the maize embryo. Plant J 12: 235–246 [DOI] [PubMed] [Google Scholar]
- Pasini L, Stile M, Puja E, Valsecchi R, Francia P, Carletti G, Salamini F, Marocco A. (2008) The integration of mutant loci affecting maize endosperm development in a dense genetic map using an AFLP-based procedure. Mol Breed 22: 527–541 [Google Scholar]
- Peer WA, Hosein FN, Bandyopadhyay A, Makam SN, Otegui MS, Lee GJ, Blakeslee JJ, Cheng Y, Titapiwatanakun B, Yakubov B, et al. (2009) Mutation of the membrane-associated M1 protease APM1 results in distinct embryonic and seedling developmental defects in Arabidopsis. Plant Cell 21: 1693–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al. (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph LF. (1936) Developmental morphology of the caryopsis in maize. J Agric Res 53: 881–916 [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, et al. (1999) An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Slocombe S, Maitz M, Hueros G, Becker HA, Yan G, Mueller M, Varotto S, Santandrea G, Thompson RD. (1999) Genetic control of endosperm development. Russo VEA, Cove DJ, Edgar LG, Jaenisch R, Salamini F, , Development Genetics, Epigenetics and Environmental Regulation. Springer-Verlag, New York, pp 185–197 [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Galweiler L, Palme K, Jurgens G. (1999) Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286: 316–318 [DOI] [PubMed] [Google Scholar]
- Tanaka H, Dhonukshe P, Brewer PB, Friml J. (2006) Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cell Mol Life Sci 63: 2738–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaka K, Blakeslee JJ, Titapiwatanakun B, Peer WA, Bandyopadhyay A, Makam SN, Lee OR, Richards EL, Murphy AS, Sato F, et al. (2005) PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17: 2922–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RD, Hueros G, Becker H, Maitz M. (2001) Development and functions of seed transfer cells. Plant Sci 160: 775–783 [DOI] [PubMed] [Google Scholar]
- Torti G, Lombardi L, Manzocchi LA, Salamini F. (1984) Indole-3-acetic acid content in viable defective endosperm mutants of maize. Maydica 29: 335–343 [Google Scholar]
- Torti G, Manzocchi L, Salamini F. (1986) Free and bound indole-acetic acid is low in the endosperm of the maize mutant defective endosperm-B18. Theor Appl Genet 72: 602–605 [DOI] [PubMed] [Google Scholar]
- Varotto S, Locatelli S, Canova S, Pipal A, Motto M, Rossi V. (2003) Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol 133: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benkova E, Benjamins R, Beeckman T, Luschnig C, Friml J. (2005) Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531 [DOI] [PubMed] [Google Scholar]
- Weiler EW, Jourdan PS, Conrad W. (1981) Levels of indole-3-acetic acid in intact and decapitated coleoptiles as determined by a specific and highly sensitive solid-phase enzyme immunoassay. Planta 153: 561–571 [DOI] [PubMed] [Google Scholar]
- Wu X, McSteen P. (2007) The role of auxin transport during inflorescence development in maize (Zea mays, Poaceae). Am J Bot 94: 1745–1755 [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Werr W. (2005) Pattern formation in the monocot embryo as revealed by NAM and CUC3 orthologues from Zea mays L. Plant Mol Biol 58: 669–685 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.