Abstract
In cyanobacteria fatty acids destined for lipid synthesis can be synthesized de novo, but also exogenous free fatty acids from the culture medium can be directly incorporated into lipids. Activation of exogenous fatty acids is likely required prior to their utilization. To identify the enzymatic activity responsible for activation we cloned candidate genes from Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942 and identified the encoded proteins as acyl-acyl carrier protein synthetases (Aas). The enzymes catalyze the ATP-dependent esterification of fatty acids to the thiol of acyl carrier protein. The two protein sequences are only distantly related to known prokaryotic Aas proteins but they display strong similarity to sequences that can be found in almost all organisms that perform oxygenic photosynthesis. To investigate the biological role of Aas activity in cyanobacteria, aas knockout mutants were generated in the background of Synechocystis sp. PCC 6803 and S. elongatus PCC 7942. The mutant strains showed two phenotypes characterized by the inability to utilize exogenous fatty acids and by the secretion of endogenous fatty acids into the culture medium. The analyses of extracellular and intracellular fatty acid profiles of aas mutant strains as well as labeling experiments indicated that the detected free fatty acids are released from membrane lipids. The data suggest a considerable turnover of lipid molecules and a role for Aas activity in recycling the released fatty acids. In this model, lipid degradation represents a third supply of fatty acids for lipid synthesis in cyanobacteria.
Cyanobacteria present a diverse group of Gram-negative bacteria capable of oxygenic photosynthesis (Margulis, 1975). Their two photosystems, as well as other genetic and morphological similarities, identified them as putative predecessors of chloroplasts of eukaryotic plants (Wallace, 1982; Pakrasi, 1995). The structural similarities of cyanobacteria and chloroplasts are reflected in part by equivalence of biochemical pathways and their components. For instance, cyanobacterial fatty acid and glycerolipid compositions closely resemble those of the inner envelope and thylakoid membranes of chloroplasts (Roughan et al., 1980; Heinz and Roughan, 1983). In cyanobacteria, as well as in chloroplasts, fatty acids are synthesized by a type II fatty acid synthase (FAS) complex utilizing a freely dissociable acyl carrier protein (ACP; Froehlich et al., 1990). The products of FAS are released as acyl ACPs and may serve directly as substrates for acyltransferases, incorporating the fatty acids into membrane lipids (Frentzen et al., 1983). The substrate specificity of the acyltransferases establishes in cyanobacteria as well as in plastids the typical prokaryotic fatty acid pattern characterized by C16 fatty acids esterified to the sn-2 position. The correspondence of metabolic pathways between cyanobacteria and chloroplasts is reflected by the shared presence of closely related enzymes that catalyze key reactions. Besides the many similarities, however, there are also clear discrepancies that in part account for the fact that cyanobacteria are unicellular organisms, whereas chloroplasts are embedded in the metabolism of a eukaryotic cell. In terms of lipid metabolism, such differences become obvious if one considers the fact that the plastidial FAS also supplies the extraplastidic compartment with fatty acids (Browse et al., 1986). Fatty acid export from the chloroplast necessitates the release of synthesized acyl chains from ACP to allow transport across both envelope membranes. The release is achieved by the action of acyl-ACP thioesterases that hydrolyze the acyl-ACP thioester to liberate the fatty acid (Voelker et al., 1997). In cyanobacteria such export would obviously result in an unfavorable loss of fatty acids, and consequently homologous proteins to acyl-ACP thioesterases cannot be found here. Whereas cyanobacteria seem to be unable to release fatty acids enzymatically from their activated state, all cyanobacterial genomes available to date encode an activity most likely responsible for the activation of free fatty acids. The respective sequences are annotated as acyl-CoA synthetases. Conserved motifs in the amino acid sequence identify these proteins as members of the well-established superfamily of AMP-binding proteins. This protein family comprises several hundred amino acid sequences spreading across all organisms analyzed so far. The family members are annotated in the PROSITE database under entry number PS00455. Although these predicted fatty acid-activating enzymes of cyanobacteria are annotated as acyl-CoA synthetases due to their sequence similarity to proteins with such enzymatic activity, there is a much higher degree of similarity to certain AMP-binding proteins of plant origin with less-well-established function. These plant proteins are predicted to reside in chloroplasts and one member of this subgroup from Arabidopsis (Arabidopsis thaliana) designated as AAE15 was recently described as acyl-ACP synthetase. The conclusions were based on the comparison of enzymatic activity between plant extracts of wild-type and knockout mutant lines (Koo et al., 2005). Whereas the biological role of this activity remained largely elusive, it was shown that the capacity of plant extracts to elongate supplied medium fatty acids depended on AAE15 activity. Since the elongation of medium chain fatty acids in the plastid depends on the FAS requiring acyl ACPs, it was concluded that the fatty acids must have been activated by ACP. The elongated fatty acids ultimately appeared in membrane lipids. Together these findings suggested that AAE15 is an acyl-ACP synthetase.
Besides encoding a protein homologous to AAE15 from Arabidopsis, cyanobacteria are also able to utilize exogenous fatty acids like it was shown for isolated chloroplasts. It is well established that feeding different cyanobacteria with free fatty acids results in the incorporation of these fatty acids into membrane lipids. For this process the activation of the fatty acids is believed to be essential. This causal relationship was clearly shown at least for other unicellular organisms like Escherichia coli and yeast (Saccharomyces cerevisiae) where the deletion of acyl-CoA synthetase activity resulted in the inability to utilize exogenous fatty acids (Overath et al., 1969; Knoll et al., 1995). It is not easy to assess how regularly cyanobacterial cells are exposed to exogenous free fatty acids in nature but at least for marine strains this is most likely a rather artificial situation. Therefore, it can be speculated that the capacity to activate free fatty acids might be of different relevance in the lipid metabolism of cyanobacteria in vivo.
In this article, we investigated the fatty acid metabolism of cyanobacteria. We isolated candidate genes potentially encoding enzymes involved in fatty acid activation from the strains Synechocystis sp. PCC 6803 (hereafter Synechocystis) and Synechococcus elongatus PCC 7942 (hereafter Synechococcus) and performed heterologous expression in E. coli. The recombinant proteins were shown to possess acyl-ACP synthetase activity with broad substrate specificity. Knockout mutant strains deficient in acyl-ACP synthetase activity were characterized by secretion of endogenous free fatty acids into the culture medium. Combined with labeling experiments, the results suggest an essential role for acyl-ACP synthetase in fatty acid recycling in cyanobacteria.
RESULTS
Fatty Acid-Activating Enzymes from Cyanobacteria Display Acyl-ACP Synthetase Activity
To investigate the fatty acid metabolism in cyanobacteria, we sought to characterize the fatty acid activation process as the entry point for acyl chains into lipid metabolism. In a first step the enzymatic activity was studied in vitro. According to Cyanobase (www.kazusa.or.jp), Synechocystis encodes only a single candidate gene for fatty acid activation, annotated as long-chain fatty acid CoA ligase and designated as open reading frame slr1609. To obtain reliable results concerning fatty acid activation in cyanobacteria in general, we selected Synechococcus as a second model organism with considerable phylogenetic distance to Synechocystis. Based on sequence comparisons, Synechococcus also contained only a single candidate gene for fatty acid activation. The protein sequence is deposited in GenBank as YP_399935 and the coding sequence is located within the genome from base-pair positions 924079 to 926028. The Synechococcus protein is annotated as long-chain fatty acid CoA ligase and shares 50% identity and 64% similarity with slr1609 of Synechocystis.
To characterize the enzymatic function of the protein encoded by slr1609 from Synechocystis and its homolog from Synechococcus, both genes were cloned in frame with a C-terminal polyhistidine tag present in pET expression vectors, and the enzymes were expressed in the E. coli strain Rosetta(DE3)pLysS. Purification of the enzymes was achieved by ultracentrifugation of the cell lysate to yield a membrane fraction, which was solubilized in 2% Triton X-100 prior to metal affinity chromatography. Aliquots of the protein fractions collected after respective preparation steps were analyzed by SDS-PAGE (Supplemental Fig. S1). An expressed recombinant protein of approximately 66 kD was visible in the fractions collected from cells expressing acyl-acyl carrier protein synthetases (Aas) of either Synechocystis (AasPCC 6803) or Synechococcus (AasPCC 7942). Under conditions employed, the heterologous expression of AasPCC 7942 turned out to be significantly more robust compared to the AasPCC 6803. Consequently, it was easier to obtain sufficient amounts of purified AasPCC 7942 and, therefore, most enzymatic studies were conducted with this enzyme.
Purified AasPCC 7942 and AasPCC 6803 were subjected to enzyme assays to evaluate their capacity to activate free fatty acids. Since the proteins are annotated as acyl-CoA synthetases but share also sequence similarity with the putative acyl-ACP synthetase AAE15 from Arabidopsis (Koo et al., 2005), CoA and ACP were both considered as possible acceptors of the acyl group. The results unequivocally showed acyl-ACP synthetase activity for both enzymes but no activity with CoA. The reaction was shown to be ATP dependent, which is consistent with the mechanism that had been described for proteins belonging to the superfamily of AMP-binding proteins (Babbitt et al., 1992). Figure 1A summarizes the results of activity assays with protein from Synechococcus but the results are representative for Aas from Synechocystis as well. Control measurements, either without protein (control I) or with protein denatured by boiling (control II) defined the background values of the assay. Under the conditions applied, the acylation rate measured for AasPCC 7942 was linear with time for at least 30 min and linear with protein concentration up to at least 1 μ g protein (data not shown).
Figure 1.
A, Putative fatty acid-activating enzyme of Synechococcus was expressed in E. coli, purified, and analyzed by in vitro assays conducted in presence of radiolabeled palmitic acid, ATP, and either CoA or ACP as an acyl acceptor. Control measurements were performed either without protein (control I) or with protein denatured by boiling (control II). B, Substrate specificity of Aas of Synechococcus. Purified enzyme was tested in acyl-ACP activity assays using six different fatty acids. Each assay was performed in triplicate. The error bars represent the sd.
To evaluate the substrate specificity of AasPCC 7942 the purified enzyme was tested by in vitro assays using different fatty acids as substrates. As indicated in Figure 1B, AasPCC 7942 displayed rather broad substrate specificity, accepting fatty acids with chain length between C12 and C18. To determine the kinetic properties of AasPCC 7942 the product formation was measured as a function of increasing concentration of radioactive oleic acid. The catalysis followed Michaelis-Menten-type kinetics with Km and kcat values of 10.49 (error 1.11) μm and 0.016 s−1, respectively (Supplemental Fig. S2).
Phylogenetic Analysis of Acyl-ACP Synthetases
Having characterized both cyanobacterial proteins unambiguously as Aas, we next analyzed their phylogenetic relationships. To this end, the amino acid sequences of both enzymes were used to identify orthologous proteins. To distinguish Aas from other members of the superfamily of AMP-binding proteins, it was essential to establish criteria defining the subfamily of acyl-ACP synthetases. The comparison identified the group of long-chain acyl-CoA synthetases (Lacs) to be most similar to the acyl-ACP synthetases. Both subfamilies differ from all other members of the AMP-binding protein family by the presence of a stretch of amino acids separating two highly conserved sequence motifs. Such a sequence insertion was first described for two Lacs of rat (Fujino and Yamamoto, 1992) and later discussed as a distinct general feature of Lacs of eukaryotic origin (Shockey et al., 2002). The insertion domain displays only weak sequence similarities between different members of the Lacs family but is characterized by both its position within the protein sequences and its highly conserved length of 40 to 45 amino acids. Strikingly, in case of the Aas candidate sequences, the position is conserved as well but the length of the insertion is extended to 68 to 74 amino acid residues. Besides the unusual length of the insertion, we also identified three specific residues or pair of residues that are absolutely conserved in the group of potential acyl-ACP synthetases and that are absent in any other member of 150 randomly chosen members of the AMP-binding family used for comparison. These conserved residues are indicated in the sequence comparison (Supplemental Fig. S3).
Applying these criteria, we identified 23 orthologous proteins and their relationships were assessed by phylogenetic analyses. The sequences were aligned with ClustalX (Thompson et al., 1997) and the resulting multiple alignment was used to estimate phylogenies with different software of the PHYLIP package (Felsenstein, 1989). Finally, a phylogenetic tree based on maximum-likelihood analysis was generated (Fig. 2) and displayed using TreeIllustrator (Trooskens et al., 2005). The corresponding information for the abbreviated sequence names including organism and database accession number is summarized in Table I. In the phylogenetic tree, two Lacs proteins of Arabidopsis (clade V) were included as reference for a related but also clearly distinguishable activity. Sequences with a higher degree of similarity to the identified Aas than these Lacs proteins were found in four distinct subfamilies. Clade I exclusively contains the Aas sequences of marine cyanobacteria. Clade II comprises the sequences of Synechocystis and Synechococcus and in addition, sequences from other fresh water and marine cyanobacteria. Clade III covers a wide range of organisms spanning from Volvox carteri to higher plants including also AAE15 (AtAAS1) from Arabidopsis, whereas clade IV contains only three sequences of the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum.
Figure 2.
Phylogenetic tree of acyl-ACP synthetases of photosynthetic organisms including two Lacs sequences of Arabidopsis as outgroup (clade V). The branch lengths of the tree are proportional to the calculated divergence. The 0.1 scale represents 10% sequence divergence, calculated as estimated numbers of replacement. The numbers indicate the confidence levels for various branches as determined by bootstrap analysis (Felsenstein, 1989). Abbreviations are given in Table I.
Table I. Acyl-ACP synthetases of photosynthetic organisms and related sequences.
Proteins are grouped by clade. If possible the proteins are identified by the GenBank accession number. In case of ongoing annotation by the U.S. Department of Energy Joint Genome Institute (JGI), the preliminary protein identification number is given or if assembled manually the coding region is denoted. For Arabidopsis also the locus identifiers defined by the Arabidopsis Genome Initiative (AGI) are given.
| Clade | Organism | Abbreviation | GenBank Accession No./JGI/AGI |
| I | Prochlorococcus marinus sp. pastoris str. CCMP1378 | Pro_CCMP1378 | ZP_00104400 |
| P. marinus sp. marinus str. CCMP1375 | Pro_SS120 | NP_874793 | |
| Synechococcus sp. CC9311 | Syn_CC9311 | YP_729826 | |
| P. marinus str. MIT 9313 | Pro_MIT9313 | NP_894048 | |
| Synechococcus sp. WH 8102 | Syn_WH8102 | NP_896762 | |
| II | Thermosynechococcus elongatus BP-1 | Therm_BP-1 | NP_682091 |
| Trichodesmium erythraeum IMS101 | Tri_IMS101 | YP_721558 | |
| Synechococcus elongatus PCC 7942 | Syn_PCC7942 | YP_399935 | |
| Synechococcus sp. PCC 7002 | Syn_PCC7002 | YP_001735222 | |
| Anabaena sp. PCC 7120 | Ana_PCC7120 | NP_487642 | |
| Synechocystis sp. PCC 6803 | Syn_PCC6803 | BAA17024 | |
| Crocosphaera watsonii WH 8501 | Cro_WH8501 | ZP_00514521 | |
| III | V. carteri | Vc_AAS | JGI Volca1 120723 |
| Physcomitrella patens ssp. patens | Pp_AAS1 | JGI Phypa1_1 169848 | |
| P. patens ssp. patens | Pp_AAS2 | JGI Phypa1_1 140960 | |
| Oryza sativa (japonica cultivar group) | Os_AAS | NP_001051878 | |
| Populus trichocarpa | Pt_AAS1 | JGI scaffold_88: 413600-428931a | |
| P. trichocarpa | Pt_AAS2 | JGI scaffold LG_I: 24797125-24808759a | |
| Arabidopsis | At_AAS1 | NP_193143At4g14070 | |
| Arabidopsis | At_AAS2 | AAM28629At3g23790 | |
| IV | Thalassiosira pseudonana CCMP 1335 | Tp_AAS1 | JGI chromosome 7: 511777-514965a |
| T. pseudonana CCMP 1335 | Tp_AAS2 | JGI chromosome 6: 1257611-1260333a | |
| Phaeodactylum tricornutum | Pht_AAS | JGI Phatr2 35735 | |
| V | Arabidopsis | At_LACS1 | NP_182246At2g47240 |
| Arabidopsis | At_LACS3 | NP_176622At1g64400 |
Assembled manually.
Generation of aas Knockout Mutants in Synechocystis and Synechococcus
To investigate the biological function of Aas we generated aas knockout mutant strains in the background of both Synechocystis and Synechococcus. In the knockout strains the aas was inactivated by replacing a substantial part of the respective coding region by a kanamycin resistance cassette. Since cyanobacteria contain multiple copies of the chromosome per cell (Mann and Carr, 1974), it was essential to establish the complete segregation of the wild-type allele. The complete replacement of wild-type alleles was confirmed by PCR (Supplemental Fig. S4). The first reaction used primers specific for the aas gene to show the absence of the wild-type gene in the aas mutant strains. The second reaction proved successful insertion and correct localization of the kanamycin cassette within the chromosome of the mutant strains. Together these data indicated the elimination of a functional aas gene in both strains.
Aas-Deficient Cells Were Unable to Utilize Exogenous Fatty Acids
Under laboratory conditions the aas mutant strains developed normally and showed growth rates comparable to wild type. To evaluate the effect of this mutation on cellular fatty acid metabolism we first tested the response of the mutant cells to exogenous fatty acids. Like other unicellular organisms many cyanobacteria are capable of utilizing fatty acids taken up from the surrounding medium. Preventing fatty acid activation in other unicellular organisms like yeast and E. coli by deletion of acyl-CoA synthetases largely disables the utilization of exogenous fatty acids (Overath et al., 1969; Knoll et al., 1995). To analyze if the aas gene plays a comparable role in this context in cyanobacteria, the mutant strains and the corresponding wild-type strains were grown in the presence of radiolabeled oleic acid. The fate of the fatty acids was examined by extracting total lipids from harvested cells and subsequent analysis of the extracts by thin-layer chromatography (TLC; Fig. 3). As expected, wild-type cells of both strains incorporated the radiolabeled fatty acids into various lipid classes, suggesting the presence of an enzymatic activity capable of fatty acid activation. In contrast, the aas knockout mutant strains were not able to channel exogenous oleic acid into either glycolipids or phospholipids; instead the radiolabeled fatty acid accumulated as free fatty acid within the mutant cells. In conclusion, the results are compatible with the assumption that the aas gene encodes the only enzyme for fatty acid activation in both cyanobacterial strains under investigation. To unequivocally prove this concept, a complementation experiment was performed. For this purpose, a wild-type allele of the aas gene from Synechococcus was transformed into the Aas-deficient mutant of Synechocystis. Feeding experiments with radiolabeled oleic acid demonstrated the reestablishment of the wild-type phenotype, confirming the strict dependence on Aas for fatty acid activation in this strain (Supplemental Fig. S5).
Figure 3.
Autoradiography of total lipid extracts of Synechocystis and Synechococcus wild-type cells (WT) and aas knockout mutant cells (ko) fed with radiolabeled oleic acid (18:1). Lipids were separated by TLC using acetone:toluene:water (91:30:8, v/v/v) as solvent system. The individual spots were identified by authentic standards as free fatty acids (FFAs), MGDG, digalactosyldiacylglycerol (DGDG), SQDG, and PG. Additional spots remained unidentified. In the extract of Synechocystis wild-type cells also monoglucosyldiacylglycerol being the precursor of MGDG is detectable as spot slightly above MGDG.
Aas-Deficient Cells Secreted Fatty Acids into the Medium
Besides the inability to activate exogenous fatty acids a visual phenotype was also observed in cultures of the aas mutant strains. In aerated cultures of wild-type cells, the inner surfaces of the glass flasks above the level of liquid media became overgrown with cells after a few days, due to spreading of cells by bursting air bubbles. In the mutant strains, the glass surfaces remained essentially free of cells. Further investigations revealed micromolar concentrations of free fatty acids in the culture medium of the aas mutant strains as responsible reason for this difference. For the analysis of the culture media the cells from dense cultures were removed by centrifugation and the used culture media was employed to prepare lipid extracts that were further analyzed by gas chromatography. In a representative experiment with three independent cultures each, the culture media of Aas-deficient Synechocystis and Synechococcus contained free fatty acid concentrations of 6.4 (sd 0.3) nmol mL−1 OD750−1 and 8.4 (sd 0.5) nmol mL−1 OD750−1, respectively, while in the culture media of wild-type strains only traces of fatty acids were detected (Fig. 4A). From these data it was concluded that the deletion of aas in both strains resulted in secretion of bulk amounts of free fatty acids into the culture medium. To investigate the obvious changes in fatty acid metabolism of the mutants in more detail, we compared the profiles of extracellular and intracellular fatty acids (Fig. 4). Samples were collected from cultures at OD750 of 2 to 3. To distinguish between free and esterified fatty acids cellular samples were split and subjected to two different protocols, resulting in either specific methylation of free fatty acids or in transmethylation of esterified fatty acids.
Figure 4.
Fatty acid profiles of cells and culture medium of wild-type and aas mutant strains. A, Free fatty acids in the culture media. B, Free fatty acids within the cells. C, Esterified fatty acids within the cells. Results are shown for Synechocystis wild type and aas knockout mutant (6803wt, 6803ko) as well as for the corresponding strains of Synechococcus (7942wt, 7942ko). For each strain three independent cultures were analyzed. The error bars represent the sd.
The profile of the esterified fatty acids showed only minor differences between wild-type and mutant strains (Fig. 4C). These data indicated that Aas activity had only negligible influence on the fatty acid composition of lipids of both cyanobacterial strains under the conditions employed. The concentration of free fatty acids within the cells, on the other hand, was significantly higher in both mutant strains compared to the respective wild-type strain (Fig. 4B). The concentrations of cellular free fatty acids were estimated to be 0.6 (sd 0.4) nmol mL−1 OD750−1 in wild-type cells and 2.5 (sd 0.4) nmol mL−1 OD750−1 in aas mutant cells in the case of Synechocystis. In Synechococcus, the corresponding values were 1.4 (sd 0.3) nmol mL−1 OD750−1 in wild-type cells and 3.1 (sd 0.8) nmol mL−1 OD750−1 in aas mutant cells. The composition of the pool of free cellular fatty acids proved to be almost identical between wild-type and mutant strains and reflected the corresponding composition of the esterified fatty acids. From this general statement two exceptions were found. In the aas mutant of Synechocystis the concentration of 18:0 was found to be significantly higher in the cellular pool of free fatty acids compared to wild type as well as compared to the pool of esterified fatty acids of the mutant. Second, in the aas mutant of Synechococcus the concentration of 16:0 in the cellular pool of free fatty acids was considerably increased compared to wild-type cells. Taken together, it could be concluded that the differences between wild-type and mutant strains in the pool of cellular free fatty acids were small but significant. Substantial differences were observed, on the other hand, when the culture media were analyzed (Fig. 4A). As mentioned above, free fatty acids were found in the media of the aas mutant strains only. In this pool all fatty acids found esterified in membrane lipids of the particular strain were detected. This finding provided important information regarding the origin of the secreted fatty acids. Double bonds in fatty acids are introduced in both strains by acyl-lipid-type desaturases, suggesting that these enzymes act only on fatty acids that are coupled by an ester bond to the glycerol backbone of membrane lipids (Sato et al., 1986). Therefore, the presence of the unsaturated fatty acids 18:1(9), 18:2, and 18:3 in the pool of free fatty acids in the culture medium strongly suggested that these fatty acids had been released from membrane lipids before being secreted.
In addition to the presence of these fatty acids in the culture medium itself, the composition of this extracellular pool is also noteworthy. The relative amounts of the different fatty acids revealed remarkable differences when compared to the cellular pools of either free fatty acids or esterified fatty acids. This fact is especially obvious when comparing the ratio of 16:0 to 16:1 in the different pools. Compared to the relative concentration of 16:1 in the pool of cellular free fatty acids its concentration in the culture media was significantly elevated in both mutant strains.
Beside the spectrum of fatty acids found in the esterified pool, in both mutant strains an additional fatty acid was detected in substantial amounts in extracts of the culture medium. In the pool of free fatty acids in the culture media this compound accounted for about 9.9% in the case of Synechocystis and about 14.8% for Synechococcus. The compound was identified to be 3-hydroxymyristic acid by authentic standard (Fig. 5A) and its identity was confirmed via gas chromatography-mass spectrometry analysis (Supplemental Fig. S6). 3-Hydroxymyristic acid is known to be a component of lipid A, a hydrophobic domain of lipopolysaccharide (LPS) in the outer membrane of Gram-negative bacteria. The detailed composition of LPS in the strains employed in our study is not known; nevertheless LPS represents the most likely source for 3-hydroxymyristic acid found in the medium. To examine whether 3-hydroxymyristic acid is indeed a compound of LPS of the strains under investigation we isolated LPS from wild-type cells of Synechocystis and subjected the isolated lipid fraction to acidic methanolysis. The resulting methyl esters were analyzed by gas chromatography, and one of the signals observed was clearly identified as 3-hydroxymyristic acid by coelution with an authentic standard (Fig. 5B). These findings suggested that 3-hydroxymyristic acid found in the culture medium was either released from mature LPS or secreted directly upon its own biosynthesis.
Figure 5.
Analysis by gas chromatography of methyl esters obtained from fatty acids secreted into the culture media by aas mutant cells of Synechocystis (A) and from the LPS fraction of Synechocystis (B). The presence of 3-hydroxymyristic acid (3-OH-14:0) in both extracts was confirmed by comigration with authentic standard (dotted line). Other fatty acids were identified by standards: 16:0 (a), 16:1 (b), internal standard 17:0 (c), 18:0 (d), 18:1 (e), 3-OH-14:0 (f), 18:2 (g), and 18:3 (h; A). Other peaks observed in the LPS fraction derived either from the extraction reagent or were not further investigated (B).
Complex Lipids Provide Fatty Acids for Aas-Dependent Lipid Synthesis
As described above, the finding of unsaturated fatty acids in the pool of free fatty acids in the aas mutant strains suggested their release from complex lipids. To further investigate the origin of the elevated concentrations of free fatty acids in the aas mutant strains we performed two different labeling studies. In the first experiment aas mutant cells were grown in the presence of 14C-acetate to achieve labeling of de novo synthesized fatty acids. Upon adding 14C-acetate to the culture medium, the label rapidly appeared in complex lipids like monoglucosyldiacylglycerol and monogalactosyldiacylglycerol (MGDG) with a lag phase of less than 10 min (Fig. 6A). At this early stage labeled free fatty acids were not yet detectable. Instead, labeled free fatty acids were found only after establishment of the label in complex lipids for at least 20 min. The chronology of the distribution of the label is compatible with the model describing free fatty acids originating from complex lipids. As control wild-type cells were incubated with labeled acetate (Fig. 6B). These cells accumulated label in all lipid classes as well but in contrast to aas mutant cells the pool of labeled free fatty acids was rather small and only transiently detectable.
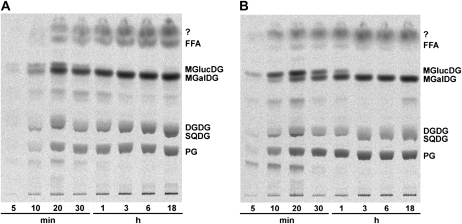
Figure 6.
Free fatty acids in cyanobacteria were released from complex lipids. aas mutant cells (A) and wild type (B) of Synechocystis were incubated with radiolabeled acetate to achieve labeling of fatty acids. The labeled fatty acids were subsequently incorporated into complex lipids. Following the addition of acetate, aliquots of the culture were removed at the times indicated. Lipid extracts of the cells were separated by TLC as described before. In aas mutant cells the label was clearly detectable in complex lipids after 10 min of incubation but did not yet accumulate in the pool of free fatty acids. In free fatty acids the label showed up with some delay, suggesting a release of fatty acids from lipid molecules. In wild-type cells label was incorporated into complex lipids but was detectable in only minor amounts in the pool of free fatty acids. The individual spots were identified as FFAs, monoglucosyldiacylglycerol (MGlucDG), MGDG (MGalDG), digalactosyldiacylglycerol (DGDG), SQDG, and PG. Analysis of the spot close to the solvent front (?) by two-dimensional TLC revealed a mixture of at least four different components that were not further investigated.
In a second set of experiments, designed to unequivocally prove the origin of the free fatty acids from complex lipids, the cells were fed with different purified 14C-labeled lipids. It was shown earlier that complex lipids like phosphatidylglycerol (PG) can be taken up efficiently by cyanobacteria (Hagio et al., 2000). For the experiment the lipids sulfoquinovosyldiacylglycerol (SQDG) and PG-containing 14C-labeled palmitic acid and 14C-labeled oleic acid were generated as described in “Material and Methods.” Cultures of wild-type and aas mutant cells were incubated individually with the purified lipid classes for 20 h before the cells were collected and the lipid extracts were analyzed by TLC. In wild-type as well as in aas mutant cells, the label did not remain trapped in the supplemented lipid but was detected in additional spots, indicating lipid remodeling (Fig. 7). No matter which one of both lipid class was provided or which strain was used, in all cases labeled free fatty acids as well as the corresponding lyso lipid were detected, suggesting partial degradation of SQDG and PG. Especially aas mutant cells accumulated huge amounts of free fatty acids. Wild-type cells on the other hand showed smaller amounts of free fatty acids but were instead able to channel labeled fatty acids to MGDG (Fig. 7, A and B). Supplementation of aas mutant cells with PG resulted in addition in the accumulation of an unidentified substance of reduced polarity (Fig. 7B). The results obtained clearly indicated an Aas-dependent exchange of fatty acids between different lipid classes. Wild-type cells achieve this exchange most likely by a couple of consecutive steps defined as the release of fatty acids from complex lipids, the establishment of a pool of free fatty acids, the reactivation of these fatty acids to provide substrate for acyltransferases, and finally lipid synthesis to produce again complex lipid molecules.
Figure 7.
Purified lipids labeled with 14C-oleic acid and 14C-palmitic acid were added individually to cultures of Synechocystis wild-type cells (WT) as well as to the corresponding aas mutant strain (ko). The cells were harvested after 20 h and the lipid extracts were analyzed by TLC as described before. In both strains the supplemented lipids were partially degraded to their corresponding lyso lipids and to free fatty acids but only the wild-type cells were able to utilize the released fatty acids to synthesize new lipid molecules. Upon supplementation with either SQDG (A) or PG (B), wild-type cells incorporated the released fatty acids into MGDG. The individual spots were identified by authentic standards as FFAs, MGDG, SQDG, PG, and lyso PG. The abbreviation of the supplemented lipid is highlighted in gray. Additional spots present in both strains could not be identified by available lipid standards.
DISCUSSION
Fatty acid activation is a central step in fatty acid metabolism. In this study, we aimed to address the mechanism and the biological role of this enzymatic activity in cyanobacteria. The genomes of sequenced cyanobacterial strains encode only one protein predicted to be responsible for fatty acid activation. In vitro assays performed with purified Aas protein of Synechococcus revealed acyl-ACP synthetase activity with broad substrate specificity. The enzyme converted medium and long-chain fatty acids with different degrees of unsaturation with similar rates to acyl-ACP thioesters. This result is in line with the metabolic context in cyanobacteria, employing acyltransferases as immediate consumers of activated fatty acids with a strong preference for acyl-ACP over acyl-CoA (Weier et al., 2005). Orthologous proteins were found exclusively in almost all available genomes of organisms performing oxygenic photosynthesis. Whereas cyanobacterial genomes contain one of these aas genes, eukaryotic organisms encode one—or more often two—of these proteins. The sole exceptions to this rule were found in the cyanobacteria Gloeobacter violaceus PCC 7421 and in the microalgae of the class Prasinophytes containing the genera Ostreococcus and Micromonas, where no homologous sequences could be identified. G. violaceus PCC 7421 is a member of an early branching lineage within the radiation of cyanobacteria (Honda et al., 1999) and is characterized by the lack of thylakoids (Rippka et al., 1974). To evaluate the question if the appearance of thylakoids and Aas activity is somehow connected to the genomic sequence information of additional ancient cyanobacterial strains would be important. The genome of Ostreococcus tauri, on the other hand, was described as strikingly small and compact but this compactness was assigned to rare repeated sequences and the reduction of intergenic spaces rather than to a fundamental loss of genes (Keeling, 2007). Nevertheless, the absence of homologous Aas sequences in all four genomes of Prasinophytes suggests a loss of at least this type of sequence.
Fatty Acid Secretion as Consequence of Disabled Fatty Acid Recycling
A biological role of the sequences under investigation was clearly revealed by the analysis of the aas mutant strains. In both strains, the deletion of acyl-ACP synthetase resulted in increased levels of intracellular free fatty acids. In addition, free fatty acids were also detected in the culture media of the mutant strains (Fig. 4C). The extracellular and intracellular free fatty acid profiles suggested processes of lipid remodeling or lipid degradation as origin of these fatty acids. This was indicated by the fact that also unsaturated fatty acids were detected, which are synthesized in cyanobacteria exclusively by desaturases of acyl lipid type. In addition, it was concluded from the fatty acid profile of Synechocystis that the fatty acids are released from both sn positions of the glycerol backbone since sn-2-specific 16:0 as well as C18 fatty acids were detected that are predominantly esterified to the sn-1 position (Wada and Murata, 1990). Strong support for the concept describing lipids as the source of the accumulated free fatty acids was finally obtained by labeling studies. Labeling with acetate suggested that de novo synthesized fatty acids did not directly contribute to the accumulation of free fatty acids. This finding is in agreement with the hypothesis that acyl-ACP as product of fatty acid synthesis can most likely not be cleaved in the cyanobacterial strains under investigation. This view is supported by the fact that no acyl-ACP thioesterase can be identified by sequence comparison within the annotated genomes. Direct evidence for the release of fatty acids from complex lipids, on the other hand, was obtained by feeding the cells with labeled complex lipids. The internalized lipid molecules were partially degraded, and free fatty acids were liberated. In strains with functional Aas, the released fatty acids served as substrate for the synthesis of complex lipids again. The results are compatible with a more general hypothesis, proposing a comprehensive fatty acid recycling process in wild-type cells. In the course of this procedure, fatty acids are released from membrane lipids to appear transiently in the pool of free fatty acids, before being reactivated to become substrates for lipid synthesis. In the mutant cells, on the other hand, fatty acids, which have been released from lipids could not reenter lipid metabolism. Instead, these fatty acids accumulated inside the cell and were finally secreted into the culture medium. The differences between the intracellular and the extracellular fatty acid profiles might be explained by a preference of the export process for unsaturated fatty acids. On the other hand, we cannot exclude the possibility that preferentially sn-1-specific enzymes might be able to release fatty acids directly into the culture medium.
The reactivation of free fatty acids in wild-type cells seems to take place at the plasma membrane as it was shown by proteomic studies that Aas is localized at the plasma membrane (Pisareva et al., 2007) but is absent in thylakoid membranes (Srivastava et al., 2005). This localization is compatible with the involvement of Aas in the utilization of exogenous fatty acids but also with its ability to prevent secretion of free fatty acid. Plasma membrane localization might also be beneficial for the recycling of components of the bacterial outer membrane. In addition to the typical C16 and C18 fatty acids of membrane lipids the free fatty acid pool of the culture medium of aas mutant cells contained also R-3-hydroxymyristic acid (Fig. 5). This fatty acid is known to be an important component of lipid A in many Gram-negative bacteria and was detected recently also in marine cyanobacteria (Snyder et al., 2009). Lipid A represents the lipophilic element of the LPS in the bacterial outer membrane. The detection of this substance in the culture medium of aas mutant strains suggests a similarly defective recycling process for R-3-hydroxymyristic acid as proposed for the fatty acids derived from membrane lipids. In analogy to the origin of C16 and C18 fatty acids from glycerolipids, it can be speculated that also R-3-hydroxymyristic acid might be released from the intact lipid A molecule. In wild-type cells, rapid reactivation by Aas followed by incorporation into lipid A seems to prevent the accumulation of free R-3-hydroxymyristic acid. In aas mutant cells on the other hand, this return into lipid synthesis is obviously inhibited.
The proposed model of fatty acid recycling could also explain the significantly increased concentration of 18:0 in all fatty acid pools of the mutant cells (Fig. 4). In cells with active Aas 18:0 was barely detectable at all. In wild-type cells the pool of fatty acids available for lipid synthesis is presented most likely by a mixture of both de novo synthesized saturated fatty acids and partially unsaturated fatty acids deriving from the recycling process. In contrast, the lipid synthesis in aas mutant cells depends completely on de novo synthesized, and hence saturated fatty acids. The accordingly increased demand for desaturation capacity might delay the conversion of saturated to unsaturated fatty acids, resulting eventually in higher levels of 18:0 bound to lipids. Under such conditions, the activities responsible for the release of fatty acids from lipid molecules appear to access their substrate sometimes even faster than the Δ 9-desaturase, resulting in the detection of substantial amounts of 18:0 also in the pools of free fatty acids.
The data reported here for cyanobacterial strains with disabled fatty acid activation are surprisingly similar to those found for mutants of yeast with a comparable defect (Scharnewski et al., 2008). In yeast, the inactivation of two out of five existing acyl-CoA synthetases resulted in very similar phenotypes as described here for the deletion of aas in cyanobacteria. In both cases, exogenous fatty acids were no longer used as substrates for lipid synthesis, and in addition the mutant cells were characterized by secretion of free fatty acids into the culture medium. In cyanobacterial cells as well as in yeast cells, membrane lipids were identified as source of the released fatty acids, indicating permanent and significant lipid turnover. In both cases, wild-type cells were able to reintroduce released free fatty acids back into the lipid metabolism. We conclude that the recycling of endogenous fatty acids is probably one of the most important tasks of fatty acid activation in general. In addition, the striking similarities between distantly related organisms might indicate a well-conserved strategy of lipid turnover behind a deceiving front of apparent lipid stability. Currently, we can only speculate about the reasons for the establishment of such a rather energy-costly procedure. One possible explanation could be a permanent adaptation of the lipid composition or the molecular lipid species to changes of the environmental conditions. This might be economically unviable under artificially constant parameters present in the laboratory but might be beneficial to cope with challenging conditions in the natural environment.
In conclusion, we have shown that the activation of free fatty acids by Aas in cyanobacteria is essential for the incorporation of exogenously supplied free fatty acids into cellular lipid metabolism. The more important role of Aas seems to be, nevertheless, the activation of endogenous free fatty acids permanently released from membrane lipids. The resulting recycling of fatty acids can be observed in distantly related species like yeast as well, suggesting a well-conserved mechanism in cellular lipid metabolism in general.
MATERIALS AND METHODS
Strains and Growth Conditions
Liquid cultures of the Glc-tolerant Synechocystis sp. PCC 6803 and Synechococcus elongatus PCC 7942 were grown photoautotrophically in BG 11 media (Rippka et al., 1979) at 30°C. The cultures were grown under constant illumination at a photosynthetic photon flux density of approximately 38 μ mol photons m−2 s−1 and with aeration by sterile air. The growth was monitored by measurement of OD750. For growth on solid media BG 11 was supplemented with 20 mm HEPES-NaOH, pH 7.5, 0.3% (w/v) sodium thiosulfate pentahydrate, and 1.5% (w/v) agar. Mutants of Synechocystis and Synechococcus were grown in the presence of antibiotic for selection (15 μ g mL−1 kanamycin or 10 μ g mL−1 chloramphenicol, respectively).
DNA Isolation and Generation of aas Knockout Mutants in Cyanobacteria
Genomic DNA from Synechocystis and Synechococcus was isolated according to a protocol described before (Porter, 1988). The aas mutants in the background of Synechocystis and Synechococcus were created by replacing part of the coding region by a cassette encoding a kanamycin resistance gene via homologous recombination. The constructs were prepared as follows: The open reading frame of slr1609 was amplified from genomic DNA of Synechocystis with the specific primer pair GGAATTCATATGGACAGTGGCCATGGCGCT, 6803_KOfor and AGAATTCTCGAGAAACATTTCGTCAATTAAATGTTG, 6803_KOrev and was subsequently cloned into the SmaI site of the vector pUC19. A 230-bp fragment was excised from the gene by digestion with NaeI and SmaI and replaced by a kanamycin resistance cassette released from the vector pKRP11 (Reece and Phillips, 1995) by SmaI. An analogous strategy was applied to prepare the construct for disruption of the homologous gene from Synechococcus. Genomic DNA was used as a template to amplify the open reading frame including fragments of 957 bp upstream and of 645 bp downstream the coding region using primers ACACGCATGCTTAAATGACTTCTTGTGGAAAG, 7942_KOfor and AGAGATCTAGAGACGGCACCTCAACTCCTAGGT, 7942_KOrev. The obtained PCR product was cloned into pGEM-T (Promega), a fragment of 1,704 bp was removed by digestion with EcoRV and SmaI, and the kanamycin resistance cassette was inserted into the vector.
For the construction of a plasmid required to achieve genetic complementation of the aas mutant strain of Synechocystis a variant of the knockout construct described above was used as a starting point. This construct contained a chloramphenicol resistance cassette instead of the described kanamycin cassette. The chloramphenicol cassette was released from the vector pKRP10 (Reece and Phillips, 1995). Fragments of Synechocystis DNA, flanking the chloramphenicol cassette, served as borders for homologous recombination. In one of the flanking fragments 25 bps distant from the chloramphenicol cassette 3 bps, were changed (QuikChange II site-directed mutagenesis kit, Stratagene) to introduce an EagI restriction site (GATGAACTACACCAGCGGCCGCCATTTCAAGGG, 6803_QCfor and CCCTTGAAATGGCGGCCGCTGGTGTAGTTCATC, 6803_QCrev). A 3,920 bps fragment, including the open reading frame of aas of Synechococcus plus promoter and terminator, was amplified by PCR with forward and reverse primers (ACAGCGGCCGCGATCGCGTCTCGAATCG, 7942_for, and ACAGCGGCCGCGCAGCACGATTTCAACTTGC, 7942_rev), adding NotI restriction sites to both ends. The PCR fragment was cloned first into pGEM-T vector and then transferred into the newly created EagI restriction site of the complementation construct. The resulting vector was used to transform Synechocystis aas knockout cells. Cyanobacterial cells were transformed as described before (Porter, 1988). For initial selection of transformants the DNA/cells mixture was spread on solid BG 11 medium. After 24 h the appropriate antibiotic (0.45 mg kanamycin or 0.2 mg chloramphenicol) was added into three evenly distributed wells punched into the agar. Homozygous mutants were obtained by successive streaking on BG 11 plates containing antibiotic.
Both the correct integration of the knockout cassette and the complete segregation of the wild-type alleles were confirmed by PCR. For Synechocystis the complete segregation was evaluated by 6803_con1 (CTCTACATCCTAGAAGACAGC) combined with 6803_con2 (GAATCCAATTCCCGTACTTGGTGC) and the correct insertion of the kanamycin resistance cassette was confirmed by Kan_con3 (GATTCAGTCGTCACTCATGGTG) combined with 6803_con4 (CACAGCCGGGGCACACCGACAATG). For Synechococcus the complete segregation was evaluated by the combination of 7942_con1 (CCGTAATCAGCGTGTAGATGATGG) and 7942_con2 (GATCGAACCGCTGTCCTCTAAGACG). The correct insertion of the kanamycin resistance cassette was confirmed by Kan_con3 as given above combined with 7942_con4 (GTGTTCCGCGACAACGTTGCGACG). The successful integration of the construct used for complementing the aas mutant of Synechocystis was confirmed by testing for the presence of aas of Synechococcus using the primer combination 7942_con5 (CCGTAATCAGCGTGTAGATGATGG) and 7942_con6 (GATCGAACCGCTGTCCTCTAAGACG).
Cloning of Cyanobacterial aas Genes for Expression in Escherichia coli
The open reading frames of the candidate genes were amplified from genomic DNA of Synechocystis and Synechococcus using specific primer pairs: GGAATTCATATGGACAGTGGCCATGGCGCT, 6803_ORFfor, combined with AGAATTCTCGAGAAACATTTCGTCAATTAAATGTTG, 6803_ORFrev, and ACACCATGGCTGGAACCGCCCTCGCGCAAC, 7942_ORFfor, combined with ACAGCGGCCGCACTCGCCGATTCAAACATCCCGT, 7942_ORFrev, respectively. Primers were designed to convert the existing GTG start codons into ATG and to remove the stop codons of both genes. The fragments obtained were cloned into either pET24c or pET24d vectors, resulting in pETaas6803 and pETaas7942, respectively. Both genes were cloned in frame with the polyhistidine-tag sequence of the vector. The primers were constructed according to data provided by CyanoBase (http://www.kazusa.or.jp/cyano/cyano.html) and the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov). The integrity of all cloned fragments was confirmed by sequencing.
Enzyme Overproduction and Purification
Two hundred milliliter cultures of Rosetta (DE3)pLysS cells (Novagen) carrying a pETaas construct were grown at 37°C with shaking to an OD600 0.8. The protein expression was induced after 2 h incubation at 16°C by the addition of 1 mm isopropylthio- β -galactoside and the cultures were further incubated for 66 h at 16°C. The cultures were chilled on ice and the cells were harvested by centrifugation at 3,200g. For protein purification some modifications were introduced to the method described before (Shanklin, 2000). The cellular pellet was resuspended in 5 mL of extraction buffer (50 mm Tris-HCl, pH 8.0) and cells were disrupted by sonification. The cellular extract was clarified by centrifugation at 16,000g for 25 min at 4°C. The supernatant was collected and MgCl2 was added to a final concentration of 20 mm. The membrane fraction was collected by ultracentrifugation at 100,000g for 1 h at 4°C. The supernatant was discarded and the membrane pellet was resuspended in 300 μ L of extraction buffer. An equal volume of the buffer containing detergent (50 mm Tris-HCl, 20 mm MgCl2, 4% [v/v] Triton X-100, pH 8.0) was added to solubilize the membranes. To improve the removal of membrane-bound proteins the fraction was incubated on ice with agitation for 30 min before clarification by centrifugation at 100,000g for 30 min at 4°C. During centrifugation BD TALON resin (BD Biosciences) was equilibrated with the column buffer (50 mm Tris-HCl, 20 mm MgCl2, 2% [v/v] Triton X-100, pH 8.0). The clarified and solubilized membrane fraction was applied to the resin and gently agitated at 4°C for 1 h on a platform shaker to allow the polyhistidine-tagged protein to bind the resin. The resin was transferred to a 5 mL gravity-flow column and washed sequentially with three column volumes of the column buffer followed by three column volumes of the column buffer containing 20 mm imidazole to remove nonspecifically bound proteins. The target protein was eluted with the column buffer containing 100 mm EDTA. A total of 130 to 150 μ L fractions containing the polyhistidine-tagged protein were collected and dialyzed overnight against 400 mL of the column buffer at 4°C. Protein fractions were analyzed by SDS-PAGE carried out as described by Laemmli (1970) and gels were stained with Coomassie Brilliant Blue (Serva).
Acyl-ACP Synthetase and Acyl-CoA Synthetase in Vitro Assays
The acyl-ACP synthetase activity was measured according to a modified method described previously (Rock and Cronan, 1981). The assay was conducted in 1.5 mL microcentrifuge tubes in a volume of 40 μ L. The assay mixture contained 2.5 mm Tris-HCl, pH 8.0, 2 mm dithiothreitol, 10 mm MgCl2, 5 mm ATP, 10 mm LiCl, 2% (v/v) Triton X-100, 15 μm ACP, 30 μm 14C-fatty acid (specific activity 53.7–60 mCi mmol−1), and defined amount (0.1–1 μ g) of protein sample.
The acyl-CoA synthetase activity assay was performed in the same way as the acyl-ACP synthetase activity assay. The composition of the assay mixture followed a slightly modified protocol described previously (Joyard and Stumpf, 1981). In detail, the assay mixture was composed of 100 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 5 mm ATP, 2.5 mm dithiothreitol, 0.5 mm CoA, 2% (v/v) Triton X-100, 30 μm 14C-fatty acid (specific activity 53.7–60 mCi mmol−1), and protein sample in a final volume of 40 μ L.
Phylogenetic Analysis
The amino acid sequences of Aas candidate proteins were aligned together with Lacs sequences serving as outgroup using ClustalX (Thompson et al., 1997). The resulting alignments were used to investigate phylogenetic relationships by employing the following software of the PHYLIP program package (Felsenstein, 1989): PROTDIST in combination with FITCH as a neighbor-joining method, and PROML as maximum-likelihood method. Parameters used were: the Jones-Taylor-Thornton model, global rearrangements, and randomized input order of sequences with three jumbles. The bootstrap values were calculated with SEQBOOT and CONDENSE 100 replicates. Finally, the tree was displayed using TreeIllustrator (Trooskens et al., 2005).
Lipid Analytical Methods
For analysis of fatty acid profiles in cells and media 8 mL aliquots of cultures at OD750 of about 2 to 3 were collected. Cells were harvested by centrifugation and washed twice with 1 mL 0.1 m NaHCO3. Intracellular and extracellular lipid extractions were performed according the protocol established previously (Bligh and Dyer, 1959). To recover free fatty acids quantitatively the extraction mixture was acidified by addition of 10 μ L 0.1 m hydrochloric acid in case of the cellular pellet and 100 μ L 0.1 m hydrochloric acid in case of the culture medium. Prior to extraction, heptadecanoic acid (17:0) as internal standard for free fatty acids (15 μ g) and triheptadecanoylglycerol as internal standard for esterified fatty acids (20 μ g) were added.
Free fatty acids from intracellular and extracellular lipid extracts were methylated according to a modified protocol described earlier (Stumpe et al., 2001). In short, the lipid extract (40 μ L) was transferred to a new glass tube and dried under a stream of nitrogen. Methanol (400 μ L) was added together with 10 μ L of 1-ethyl-3-3-dimethylaminopropylcarbodiimide (0.1 mg μ L−1 in methanol) and incubated for 2 h at 22°C. The reaction was stopped by adding 200 μ L of 0.1 m Tris-HCl, pH 7.5. The methyl esters of free fatty acids were extracted with 1 mL of hexane followed by centrifugation at 1,000g for 2 min. The upper hexane phase was transferred to a 1.5 mL microcentrifuge tube, dried, and resuspended in acetonitrile (12 μ L) and analyzed by gas chromatography.
Esterified fatty acids from intracellular lipid extracts were transmethylated according to a modified protocol described earlier (Christie, 1982). Lipid extract (40 μ L) was transferred to a 2-mL microcentrifuge tube and dried under a stream of nitrogen. A total of 333 μ L methanol:toluene (1:1, v/v) and 167 μ L 0.5 m sodium methoxide (CH3NaO) were added and the mixture was incubated at 22°C. After 20 min, the reaction was stopped by adding 500 μ L of 1 m NaCl and 50 μ L of 32% hydrochloric acid. The methyl esters were extracted with 1 mL of hexane followed by centrifugation at 2,300g for 2 min. The upper hexane phase was transferred to a new 1.5 mL microcentrifuge tube, dried, and resuspended in acetonitrile (15 μ L). The fatty acid methyl esters were analyzed by gas chromatography using an Agilent 6890 series gas chromatograph equipped with a capillary DB-23 column (Agilent).
Extraction and Analysis of LPS
LPS was extracted from the cells of Synechocystis with the LPS extraction kit (Intron Biotechnology, Molecular Solutions Europe) according to the manufacturer's protocol and subjected to acidic methanolysis that was performed as described previously (Miquel and Browse, 1992). The resultant methyl esters were analyzed by gas chromatography. The analysis of the hydroxy fatty acids was carried out using Agilent 5973 network mass selective detector connected to Agilent 6890 gas chromatograph. Electron energy of 70 eV, an ion source temperature of 230°C, and a temperature of 260°C for the transfer line was used.
Fatty Acid Uptake Assay
Cyanobacterial cells were collected from 10 mL cultures at OD750 of about 3.6 by centrifugation, resuspended in 2 mL of fresh BG 11 medium, and transferred to 2 mL microcentrifuge tubes. Radiolabeled fatty acid [1-14C]18:1 (Amersham Biosciences; specific activity 56 mCi mmol−1) was individually added in amounts corresponding to 0.25 μ Ci and the tubes were placed on a platform shaker under light. After 1 h incubation 0.5 mL of each culture were transferred to a new microcentrifuge tube and the cells were collected by centrifugation at 3,000g. The remaining 1.5 mL were further incubated for additional 15 h before another aliquot of 0.5 mL were recovered from each culture and treated as described above. A total of 0.4 mL of each supernatant was used to determine the radioactivity present in the culture medium by liquid scintillation counting. The residual supernatant was discarded. Cell pellets were washed twice with 0.1 m NaHCO3 before being subjected to total lipid extraction as described above. Different lipid classes were separated by TLC using acetone/toluene/water (91/30/8, v/v/v) as solvent and were visualized by fluorography.
Synthesis of 14C-Labeled Glycerolipids
Radiolabeled lipids were generated by incubating 20 mL Synechocystis wild-type cells at OD750 5 with 3 μ Ci of [1-14C]palmitic acid and 3 μ Ci [1-14C]oleic acid (Amersham Biosciences). After 20 h incubation on the platform shaker under light at 30°C, cells were harvested, washed with 0.1 m NaHCO3, and subjected to lipid extraction. Lipid extract was fractionated into neutral lipids, glycolipids, and phospholipids using Strata SI-1 silica column (Phenomenex). The individual lipid classes were purified twice by TLC and finally dissolved in 2.5% (v/v) Triton X-100.
Labeling Experiments
A total of 1.5 μ Ci of radiolabeled [1-14C]acetate (Amersham Biosciences) was added to 8 mL culture of Synechocystis aas knockout strain at OD750 13. Culture was grown for 24 h under light, at 30°C, aerated by shaking on a platform shaker. Aliquots of 0.5 mL were collected at defined time points. Cells were harvested by centrifugation, washed with 0.1 m NaHCO3, and subjected to total lipid extraction followed by TLC.
Radiolabeled lipids generated as described above were added in volumes of 20 to 40 μ L corresponding to 0.1 μ Ci to 20 mL culture of wild type and aas knockout strain of Synechocystis at OD750 0.9. Cultures were grown for 16 h under light, at 30°C, aerated by shaking on a platform shaker. Cells were harvested by centrifugation and lipids were extracted and analyzed as described above.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers YP_399935 (amino acid sequence of Aas of S. elongatus PCC 7942) and BAA17024 (amino acid sequence of Aas of Synechocystis sp. PCC 6803).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Heterologous expression of cyanobacterial aas genes in E. coli.
Supplemental Figure S2. Michaelis-Menten plot for Aas of Synechococcus assayed with oleic acid.
Supplemental Figure S3. Alignment of the amino acid sequences of AasPCC 7942 and AasPCC 6803.
Supplemental Figure S4. Confirmation of the successful disruption of aas genes in Synechocystis and Synechococcus by PCR.
Supplemental Figure S5. The aas mutant strain of Synechocystis can be complemented by the aas gene from Synechococcus.
Supplemental Figure S6. Mass spectra of 3-hydroxymyristic acid.
Supplementary Material
Acknowledgments
We are grateful to Kay Marin for providing the strain Synechocystis sp. PCC6803. We thank Jay Shockey, Ingo Heilmann, and Ivo Feussner for critical comments on the manuscript and for valuable discussions. We also thank Conny Goebel for her help with the identification of 3-hydroxymyristic acid.
References
- Babbitt PC, Kenyon GL, Martin BM, Charest H, Slyvestre M, Scholten JD, Chang KH, Liang PH, Dunaway-Mariano D. (1992) Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry 31: 5594–5604 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR. (1986) Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem J 235: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie WW. (1982) A simple procedure for rapid transmethylation of glycerolipids and cholesteryl esters. J Lipid Res 23: 1072–1075 [PubMed] [Google Scholar]
- Felsenstein J. (1989) PHYLIP—Phylogeny Interference Package (Version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Frentzen M, Heinz E, McKeon TA, Stumpf PK. (1983) Specificities and selectivities of glycerol-3-phosphate acyltransferase and monoacylglycerol-3-phosphate acyltransferase from pea and spinach chloroplasts. Eur J Biochem 129: 629–636 [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Poorman R, Reardon E, Barnum SR, Jaworski JG. (1990) Purification and characterization of acyl carrier protein from two cyanobacteria species. Eur J Biochem 193: 817–825 [DOI] [PubMed] [Google Scholar]
- Fujino T, Yamamoto T. (1992) Cloning and functional expression of a novel long-chain acyl-CoA synthetase expressed in brain. J Biochem 111: 197–203 [DOI] [PubMed] [Google Scholar]
- Hagio M, Gombos Z, Varkonyi Z, Masamoto K, Sato N, Tsuzuki M, Wada H. (2000) Direct evidence for requirement of phosphatidylglycerol in photosystem II of photosynthesis. Plant Physiol 124: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz E, Roughan PG. (1983) Similarities and differences in lipid metabolism of chloroplasts isolated from 18:3 and 16:3 plants. Plant Physiol 72: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda D, Yokota A, Sugiyama J. (1999) Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. J Mol Evol 48: 723–739 [DOI] [PubMed] [Google Scholar]
- Joyard J, Stumpf PK. (1981) Synthesis of long-chain acyl-CoA in chloroplast envelope membranes. Plant Physiol 67: 250–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. (2007) Ostreococcus tauri: seeing through the genes to the genome. Trends Genet 23: 151–154 [DOI] [PubMed] [Google Scholar]
- Knoll LJ, Johnson DR, Gordon JI. (1995) Complementation of Saccharomyces cerevisiae strains containing fatty acid activation gene (FAA) deletions with a mammalian acyl-CoA synthetase. J Biol Chem 270: 10861–10867 [DOI] [PubMed] [Google Scholar]
- Koo AJ, Fulda M, Browse J, Ohlrogge JB. (2005) Identification of a plastid acyl-acyl carrier protein synthetase in Arabidopsis and its role in the activation and elongation of exogenous fatty acids. Plant J 44: 620–632 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–684 [DOI] [PubMed] [Google Scholar]
- Mann N, Carr NG. (1974) Control of macromolecular composition and cell division in the blue-green algae Anacystis nidulans. J Gen Microbiol 83: 399–405 [DOI] [PubMed] [Google Scholar]
- Margulis L. (1975) Symbiotic theory of the origin of eukaryotic organelles; criteria for proof. Symp Soc Exp Biol 1975: 21–38 [PubMed] [Google Scholar]
- Miquel M, Browse J. (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis: biochemical and genetic characterization of a plant oleoyl-phosphatidylcholine desaturase. J Biol Chem 267: 1502–1509 [PubMed] [Google Scholar]
- Overath P, Pauli G, Schairer HU. (1969) Fatty acid degradation in Escherichia coli: an inducible acyl-CoA synthetase, the mapping of old mutants, and the isolation of regulatory mutants. Eur J Biochem 7: 559–574 [PubMed] [Google Scholar]
- Pakrasi HB. (1995) Genetic analysis of the form and function of photosystem I and photosystem II. Annu Rev Genet 29: 755–776 [DOI] [PubMed] [Google Scholar]
- Pisareva T, Shumskaya M, Maddalo G, Ilag L, Norling B. (2007) Proteomics of Synechocystis sp. PCC 6803: identification of novel integral plasma membrane proteins. FEBS J 274: 791–804 [DOI] [PubMed] [Google Scholar]
- Porter RD. (1988) DNA transformation. Methods Enzymol 167: 703–712 [DOI] [PubMed] [Google Scholar]
- Reece KS, Phillips GJ. (1995) New plasmids carrying antibiotic-resistance cassettes. Gene 165: 141–142 [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111: 1–61 [Google Scholar]
- Rippka R, Waterbury JB, Cohen-Bazire G. (1974) A cyanobacterium which lacks thylakoids. Arch Microbiol 100: 419–436 [Google Scholar]
- Rock CO, Cronan JE., Jr (1981) Acyl-acyl carrier protein synthetase from Escherichia coli. Methods Enzymol 71: 163–168 [DOI] [PubMed] [Google Scholar]
- Roughan PG, Holland R, Slack CR. (1980) The role of chloroplasts and microsomal fractions in polar-lipid synthesis from [1-14C]acetate by cell-free preparations from spinach (Spinacia oleracea) leaves. Biochem J 188: 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Seyama Y, Murata N. (1986) Lipid-linked desaturation of palmitic acid in monogalactosyl diacylglycerol in the blue-green alga (cyanobacterium) Anabaena variablis studies in vivo. Plant Cell Physiol 27: 819–835 [Google Scholar]
- Scharnewski M, Pongdontri P, Mora G, Hoppert M, Fulda M. (2008) Mutants of Saccharomyces cerevisiae deficient in acyl-CoA synthetases secrete fatty acids due to interrupted fatty acid recycling. FEBS J 275: 2765–2778 [DOI] [PubMed] [Google Scholar]
- Shanklin J. (2000) Overexpression and purification of the Escherichia coli inner membrane enzyme acyl-acyl carrier protein synthase in an active form. Protein Expr Purif 18: 355–360 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Fulda MS, Browse JA. (2002) Arabidopsis contains nine long-chain acyl-coenzyme A synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol 129: 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder DS, Brahamsha B, Azadi P, Palenik B. (2009) Structure of compositionally simple lipopolysaccharide from marine synechococcus. J Bacteriol 191: 5499–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Pisareva T, Norling B. (2005) Proteomic studies of the thylakoid membrane of Synechocystis sp. PCC 6803. Proteomics 5: 4905–4916 [DOI] [PubMed] [Google Scholar]
- Stumpe M, Kandzia R, Gobel C, Rosahl S, Feussner I. (2001) A pathogen-inducible divinyl ether synthase (CYP74D) from elicitor-treated potato suspension cells. FEBS Lett 507: 371–376 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trooskens G, De Beule D, Decouttere F, Van Criekinge W. (2005) Phylogenetic trees: visualizing, customizing and detecting incongruence. Bioinformatics 21: 3801–3802 [DOI] [PubMed] [Google Scholar]
- Voelker TA, Jones A, Cranmer AM, Davies HM, Knutzon DS. (1997) Broad-range and binary-range acyl-acyl-carrier protein thioesterases suggest an alternative mechanism for medium-chain production in seeds. Plant Physiol 114: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Murata N. (1990) Temperature-induced changes in the fatty acid composition of the cyanobacterium, Synechocystis PCC6803. Plant Physiol 92: 1062–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. (1982) Structure and evolution of organelle genomes. Microbiol Rev 46: 208–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weier D, Muller C, Gaspers C, Frentzen M. (2005) Characterisation of acyltransferases from Synechocystis sp. PCC6803. Biochem Biophys Res Commun 334: 1127–1134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.