Abstract
Levels of the enzymes that produce wound response mediators have to be controlled tightly in unwounded tissues. The Arabidopsis (Arabidopsis thaliana) fatty acid oxygenation up-regulated8 (fou8) mutant catalyzes high rates of α -linolenic acid oxygenation and has higher than wild-type levels of the α -linolenic acid-derived wound response mediator jasmonic acid (JA) in undamaged leaves. fou8 produces a null allele in the gene SAL1 (also known as FIERY1 or FRY1). Overexpression of the wild-type gene product had the opposite effect of the null allele, suggesting a regulatory role of SAL1 acting in JA synthesis. The biochemical phenotypes in fou8 were complemented when the yeast (Saccharomyces cerevisiae) sulfur metabolism 3′(2′), 5′-bisphosphate nucleotidase MET22 was targeted to chloroplasts in fou8. The data are consistent with a role of SAL1 in the chloroplast-localized dephosphorylation of 3′-phospho-5′-adenosine phosphosulfate to 5′-adenosine phosphosulfate or in a closely related reaction (e.g. 3′,5′-bisphosphate dephosphorylation). Furthermore, the fou8 phenotype was genetically suppressed in a triple mutant (fou8 apk1 apk2) affecting chloroplastic 3′-phospho-5′-adenosine phosphosulfate synthesis. These results show that a nucleotide component of the sulfur futile cycle regulates early steps of JA production and basal JA levels.
Wound-inducible defense and repair responses must be tightly regulated. In metazoans, wounding initiates rapid cell migration and tissue remodeling at the damage site (Gurtner et al., 2008; Stappenbeck and Miyoshi, 2009) whereas in damaged plant tissues there is no cell migration to the wound. A well-studied feature of the wound response in plants is efficient signal transfer to proximal and distal tissues. This leads to extensive transcriptome reprogramming and improves plant survival in the face of the most common form of wound infliction: attack by mobile herbivores (Reymond et al., 2004; Browse and Howe, 2008; Howe and Jander, 2008). Local and distal wound responses in plants are controlled to a large extent by the fatty acid-derived prohormone: jasmonic acid (JA; Farmer, 2007; Wasternack, 2007; Fonseca et al., 2009). Levels of JA in unstimulated tissues have to be very tightly regulated to avoid the misexpression of defense programs. Indeed, the levels of JA in healthy resting plants range from undetectable to less than 100 pmol per gram of fresh weight (Glauser et al., 2008), but JA accumulation is strongly induced by wounding (for review, see Wasternack, 2007; Schaller and Stintzi, 2008). The seat of JA precursor generation is the chloroplast where molecular O2 is first incorporated into triunsaturated fatty acids (chiefly α -linolenic acid [18:3]) by lipoxygenases (LOXs). This is followed by the formation of unstable allene oxide intermediates catalyzed by allene oxide synthase (AOS; Wasternack, 2007; Schaller and Stintzi, 2008). Interestingly, these two reactions remain coupled for several minutes in rapidly prepared plant extracts. Furthermore, they can be monitored in vitro with a radiochemical/thin-layer chromatography assay (Caldelari and Farmer, 1998), providing an easily accessible read out on the activity of these two chloroplastic enzymes acting early in JA synthesis. The protocol is sufficiently rapid to be employed for genetic screens aimed at isolating mutant plants in which there are deregulated activities of chloroplast enzymes of JA synthesis (Bonaventure et al., 2007a).
The first mutant to emerge from such a screen was fatty acid oxygenation up-regulated2 (fou2) that eliminates an inhibitory calcium-binding site in the voltage-gated cation channel TPC1 (Bonaventure et al., 2007a). Analysis of the transcriptome of fou2 revealed that it clustered closely with that of plants being attacked by the chewing insect Pieris rapae. The fou2 transcriptome also resembled that of plants under potassium (K) starvation (Bonaventure et al., 2007b), a phenomenon that was already known to recapitulate to a large extent the jasmonate-induced transcriptome (Armengaud et al., 2004). Consistent with this was the recent finding that there is increased K conductance through the mutated TPC1 channel in fou2 (Beyhl et al., 2009). Concerning oxylipin metabolism, fou2 displays elevated LOX2 transcript levels and increased AOS catalytic activity and both of these enzymes have confirmed roles in JA synthesis in wounded Arabidopsis (Arabidopsis thaliana) leaves (Bell et al., 1995; Park et al., 2002; Glauser et al., 2009). This fits well with the fact that the mutant has increased basal JA levels in resting leaves and is hypersensitive to wounding, producing 7 times more JA than wild-type plants (Bonaventure et al., 2007a). While the fou2 mutation affects both resting and wound-inducible JA levels it is possible that mutants affecting only resting or only wound-induced JA accumulation could be found.
Sulfur (S) is another macronutrient related to JA responses. Mutation of a major sulfate uptake facilitator, SULTR1;2, in Arabidopsis led to the strong accumulation of a group of jasmonate-related transcripts including those encoding two key enzymes in JA synthesis: LOX2 and AOS (Maruyama-Nakashita et al., 2003). Also induced to a high level in these plants was the transcript of VEGETATIVE STORAGE PROTEIN2 (VSP2), which has long been used as a marker for jasmonate responses (e.g. Berger et al., 1995; Reymond et al., 2004). Other evidence links both S reduction and oxidized S (SO42−) metabolism and JA responses. For example, treatment with exogenous jasmonate strongly up-regulates the expression of a number of genes involved in S reduction and in Cys, Met, and glutathione (GSH) synthesis (Jost et al., 2005; Sasaki-Sekimoto et al., 2005). A more recent study has greatly extended knowledge of how oxidized S metabolism may impinge on jasmonate responses in plants. Mugford et al. (2009) have shown that two Arabidopsis 5′-phosphosulfate kinases (APK1 and APK2) play roles in the synthesis of sulfated metabolites including glucosinolates and 12-sulfojasmonate. The double mutant apk1 apk2 has reduced levels of these compounds and of the JA precursor 12-oxophytodienoic acid. Since both kinases were shown by Mugford et al. (2009) to be chloroplast localized, the data underscore the potential of plastid S metabolism to influence jasmonate responses.
We conducted a new forward screen for mutants with deregulated fatty acid oxygenation rates and isolated fou8. Like fou2, fou8 has increased basal JA levels and increased LOX catalytic activity. However, unlike fou2, the wound-induced levels of JA in fou8 were similar to those in the wild type. The fou8 mutation was mapped to a gene implicated in S metabolism that has been isolated previously in other unrelated genetic screens and extensively studied under different names (SAL1, FIERY1, or FRY1; Quintero et al., 1996; Xiong et al., 2001, 2004; Gy et al., 2007; Kim and von Arnim, 2009; Wilson et al., 2009) but, to our knowledge, never in connection with JA synthesis. The gene product was identified as a phosphatase active in vitro on S-containing nucleotides, nucleotide bisphosphates, and on inositol phosphates (IPs; Quintero et al., 1996). Since then the literature has diverged with proposed SAL1 substrates being IPs (Xiong et al., 2004; Wilson et al., 2009), or S-free nucleotide bisphosphates (Gy et al., 2007; Kim and von Arnim, 2009). The possibility that SAL1 could use S-containing nucleotides such as phosphoadenosine phosphosulphate (PAPS) in vivo was considered unlikely (Kim and von Arnim, 2009). Recently, Wilson et al. (2009) have emphasized the need to identify the SAL1 substrate in vivo. Our analysis of fou8 using genetic, molecular, cellular, and biochemical approaches suggests that SAL1 acts in the chloroplast compartment to dephosphorylate PAPS or a close relative such as adenosine 3′,5′-bisphosphate (PAP). Mutation of the SAL1 gene led to deregulated LOX activity and JA accumulation in otherwise healthy leaves, an effect that could be reversed by a genetic strategy to remove the direct precursor of PAPS in chloroplasts.
RESULTS
Morphological and Genetic Characterization of the fou8 Mutant
To identify new mutants affected in the early steps of oxylipin biosynthesis we performed a genetic screen in resting Arabidopsis Columbia-0 (Col-0) leaves. Leaves from approximately 4,000 M2 ethyl methanesulfonate-mutagenized Arabidopsis plants were tested for their 18:3 oxygenation capacity using a previously described assay (Caldelari and Farmer, 1998; Bonaventure et al., 2007a). Fifteen plants were identified with an up-regulated 18:3 oxygenation rate and, after evaluation of the M3 generation, one showed a stable and heritable morphological and biochemical phenotype. We named this mutant fou8 keeping the nomenclature employed previously for similar mutants (Bonaventure et al., 2007a). The fou8 mutant presents a strong morphological phenotype with wrinkled leaves and shorter than wild-type petioles (Fig. 1A). The mutant also accumulates higher than wild-type levels of anthocyanins that are frequently associated with activity of the jasmonate pathway in leaves and stems. Flowering time is delayed in fou8 compared to the wild type (37 ± 2 versus 78 ± 2 d after germination in the wild type and fou8, respectively). The 18:3 oxygenation rate of juvenile plants (five-leaf stage) is similar in mutant and wild-type plants, but at adult stages the 18:3 oxygenation rate in extracts of fou8 leaves becomes higher than those in wild-type plants (Fig. 1B). An up-regulation of the JA pathway between the third and fourth week of development in wild-type plants was previously reported (Stenzel et al., 2003; Bonaventure et al., 2007a). In our experiment this increase is reflected in a higher 18:3 oxygenation rate between the 11- and 15-leaf stages in the wild type (Fig. 1B). This increase seems to occur earlier in fou8 (between five and seven leaves; Fig. 1B). Crushed fou8 leaves smelled faintly of crushed cress (Lepidium sativum) seedlings. It is possible that fou8 has altered levels of compounds similar to benzyl isothiocyanate but this was not investigated further.
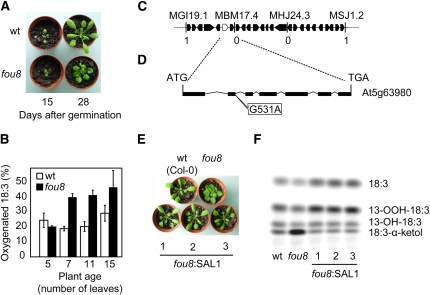
Figure 1.
Identification, mapping, and complementation of fou8. A, Rosette morphology of wild-type (wt) and fou8 plants. B, Oxygenation rate of radiolabeled 1-[14C]18:3 catalyzed by fou8 and wild-type leaf extracts. Leaves were harvested at different development stages and leaf juice (2 μ L) was incubated for 2 min with 1-[14C]18:3. Products were separated by thin-layer chromatography and the corresponding bands quantified using a phosphor imager. Total protein in the leaf extract was precipitated with TCA and quantified using bicinchoninic acid reagent. Reaction rate was measured as band intensity of oxygenation products normalized to total protein levels. C, Chromosome V segment indicating the markers used for fine mapping and the number of recombinant events for 1,600 chromosomes. Black boxes represent annotated genes and the white box indicates the SAL1 gene. D, Predicted structure of the SAL1 gene. Black boxes represent exons. The G to A transition in position 531 of the gene is responsible for the fou8 phenotype. E, Rosette morphology of the wild type, fou8, and three independent and representative fou8-SAL1 transgenic lines. F, Rate of 18:3 oxygenation catalyzed by extracts of the wild type, fou8, and three independent fou8-SAL1 transgenic lines.
Segregation analysis indicated that the fou8 phenotype is caused by a single, recessive mutation (P < 0.05; Pearson χ2; n = 109). The fou8 mutation was mapped to the bottom of chromosome V using microarray technology as described by Borevitz (2006; Supplemental Fig. S1A). Fine mapping narrowed the region to 27 candidate genes (Fig. 1C) and direct sequencing of these genes revealed a G to A transition in the splicing donor sequence of the second intron in SAL1 (At5g63980; Fig. 1D). This transition produces the skipping of the second exon during splicing (Supplemental Fig. S1B). To assure that the mutation found in the SAL1 gene was responsible for the fou8 phenotype, we complemented the mutant with a 4.5 kb genomic region including the SAL1 open reading frame under control of its native promoter. Ten independent transgenic lines were selected on hygromycin-containing growth medium and were then transferred to soil. In these 10 independent lines, the SAL1 gene successfully complemented the fou8 phenotype. All transformants showed wild-type-like petioles and normally developed leaves (Fig. 1E). Moreover, the 18:3 oxygenation rate was reduced to wild-type levels (Fig. 1F). The new splice variant in fou8 produces a change in the open reading frame creating a new stop codon in coding nucleotide position 249 and therefore suggesting that fou8 was a loss-of-function mutant. To test this, three independent T-DNA insertion lines in the SAL1 gene were identified (Supplemental Fig. S2A). The line sal1-1 (SALK_020882) contains a T-DNA insertion in the second exon. This line showed phenotypes similar to that of fou8 both in morphology (Supplemental Fig. S2B) and in increased 18:3 oxygenation rates compared to the wild type (Supplemental Fig. S2C). Additionally, a cross between fou8 and sal1-1 did not change these phenotypes (Supplemental Fig. S2, B and C), confirming that fou8 is a null allele of the SAL1 gene.
SAL1 Regulates LOX2 Protein Levels and JA Production in Resting, Undamaged Leaves
The higher than wild-type rates of 18:3 oxygenation catalyzed by fou8 leaf extracts might be explained if the amount of leaf LOXs was increased in fou8. Specific antibodies directed against LOX2 were generated to test this hypothesis. The LOX2 protein is known to function in JA synthesis in leaves (Bell et al., 1995; Glauser et al., 2009). Indeed, resting leaves of fou8 showed higher levels of LOX2 protein than the wild type (Fig. 2A). This result then prompted us to compare JA levels in wild-type and fou8 leaves. To quantify the in vivo levels of JA, we collected expanded leaf tissue from resting and wounded plants (90 min post damage infliction) from fou8 and wild-type samples and the JA content was analyzed by gas chromatography using a fully cognate heavy isotope-labeled (18O-JA) internal standard (Mueller et al., 2006). In agreement with results on 18:3 oxygenation rates, fou8 produced twice as much JA as wild-type plants in resting leaves (0.94 ± 0.165 versus 0.47 ± 0.027 nmol/g fresh weight, respectively; Fig. 2B). In contrast, the levels of JA in wild-type and mutant leaves 90 min after wounding were similar (6.87 ± 0.198 versus 7.5 ± 2.9 nmol/g fresh weight, respectively; Fig. 2B).
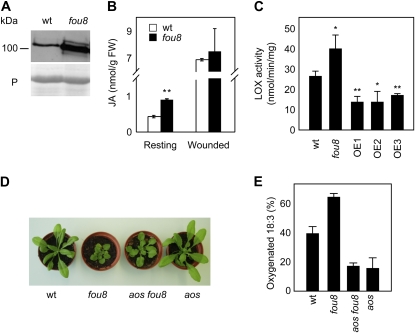
Figure 2.
SAL1 affects JA biosynthesis in resting leaves. A, LOX2 protein abundance in fou8 and wild-type (wt) samples. The bottom section represents the loading control: Rubisco large subunit stained with Ponceau red (P). B, JA levels in resting wild-type and fou8 leaves and in leaves 90 min post wounding. Values are means of three biological replicates ± se. **, P < 0.01 (t test). C, LOX activity in resting leaves of the wild type, fou8, and three SAL1 overexpression (OE) lines. Data are means of three biological replicates ± se. *, P < 0.05 (t test); **, P < 0.01 (t test). D, Rosette morphology of the aos fou8 double mutant. E, Oxidation rate of radiolabeled 18:3 in 4-week-old wild-type, fou8, aos, and aos fou8 double-mutant plants.
To test whether overexpression of the SAL1 gene inhibited the JA biosynthesis pathway we transformed wild-type plants with the wild-type SAL1 open reading frame under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The overexpression of SAL1 was confirmed by quantitative reverse transcription (qRT)-PCR (Supplemental Fig. S3A) and three independent lines were selected for further analysis. These three lines showed longer petioles and bigger rosettes than wild-type plants, features typical of plants in which JA synthesis or perception is impaired (e.g. Yan et al., 2007). We measured LOX catalytic activity in resting leaves of these transgenic lines using a spectrophotometric assay. The three overexpression lines produced significantly lower amounts of product than the wild type, indicating that LOX activity is reduced in the SAL1 overexpression lines (Fig. 2C) in contrast to the increased 18:3 oxygenation caused by loss of SAL1 function. To further explore the effects of SAL1 overexpression, we performed qRT-PCR measurements of LOX2 mRNA levels. In agreement with the LOX activity results, the three independent overexpression lines showed significantly lower levels of LOX2 mRNA (Supplemental Fig. S3B).
The Role of JA and Abscisic Acid in the fou8 Phenotype
The AOS gene encodes an enzyme dedicated to JA biosynthesis and disruption of this singleton gene completely blocks the JA biosynthesis and signaling pathways (Park et al., 2002). To test the dependence of the fou8 phenotype on an intact JA biosynthesis pathway, we introgressed the fou8 mutation into the aos background. The resulting double mutants partially suppressed the fou8 morphological phenotype. Although rosette size in the double mutant was still smaller than in the wild type, aos fou8 plants had wild-type-like petioles and did not accumulate higher levels of anthocyanins than the wild type (Fig. 2D). Importantly, all aos fou8 plants showed 18:3 oxygenation rates similar to aos and lower than those in wild-type and fou8 plants (Fig. 2E). These results indicate that the morphological phenotype of fou8 is partially dependent on JA production whereas the biochemical phenotype (18:3 oxygenation) is completely dependent on an intact JA biosynthesis pathway.
Since we reported previously that a mutation in the cation channel TPC1 increases the production of JA both in resting and wounded Arabidopsis leaves (Bonaventure et al., 2007a) we checked to what extent the biochemical mechanisms of JA synthesis activation in fou8 and fou2 are common. To do this, plants were crossed to produce fou8 fou2 double mutants. The double mutants showed a fou8-like phenotype during the first 3 weeks of development. After this period their leaves became epinastic, a characteristic of the fou2 mutant (Supplemental Fig. S4A). The SAL1 gene has been reported to be a repressor of abscisic acid (ABA) signaling (Xiong et al., 2001, 2004). To further investigate the role of ABA in the fou8 phenotype, we crossed fou8 and aba2-1 mutants. The aba2-1 mutant affects one of the last steps in ABA biosynthesis and produces between 20% and 25% of ABA wild-type levels (Leon-Kloosterziel et al., 1996). The mutated ABA2 gene in the fou8 background prevented the development of the double-mutant plants (Supplemental Fig. S4B).
The SAL1 Protein Localizes to Plastids
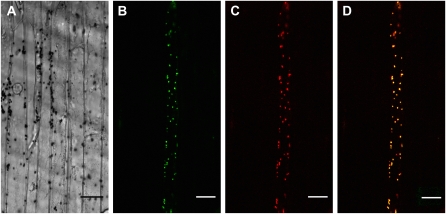
The SAL1 gene has a predicted chloroplast transit peptide (cTP) in the N terminus (Supplemental Fig. S5) and the SAL1 protein was previously detected by mass spectrometry in chloroplast stroma preparations (Peltier et al., 2006). However, in a recent publication, SAL1 protein was reported to localize to the nucleus and cytosol but not in plastids (Kim and von Arnim, 2009). Therefore, it was necessary to clarify SAL1's subcellular localization. In our hands, a SAL1:GFP fusion protein colocalized with pt-rk CD3-999 (Fig. 3), a red fluorescent plastid indicator (Nelson et al., 2007). Together with the data of Peltier et al. (2006), this confirmed that SAL1 localizes to chloroplasts.
Figure 3.
SAL1 is a chloroplast protein. A to D, Colocalization of SAL1:GFP and pt-rk CD3-999 (ssRbcTP:mCherry) fusion proteins in onion epidermal cells. Scale bar = 30 μ m. A, Differential interference contrast (DIC). B, SAL1:GFP green fluorescence. C, ssRbcTP:mCherry red fluorescence. D, Overlay of sections B and C.
The SAL1 Protein Has a Nucleotide Phosphatase Activity in Vivo
SAL1 was first identified as the only Arabidopsis gene that complements the met22 mutant strain of yeast (Saccharomyces cereviseae), a strain that is impaired in S metabolism (Quintero et al., 1996). This suggests that SAL1 is an Arabidopsis ortholog of MET22. We aligned the sequence of MET22, SAL1, and four other Arabidopsis proteins annotated as 3′(2′), 5′-bisphosphate nucleotidases (Supplemental Fig. S5). The similarity between MET22 and all these proteins was between 51% and 53%. However, SAL1 was the only one of the Arabidopsis genes that shared with MET22 exactly the same set of amino acids that constitute the enzyme's active site. SAL1 was previously reported to have two different activities in vitro, inositol polyphosphate 1-phosphatase and 3′(2′), 5′-bisphosphate nucleotidase (Quintero et al., 1996). To investigate which of these putative SAL1 activities is responsible for the regulation of the JA pathway, we transformed the fou8 mutant with the yeast MET22 gene since the MET22 protein exclusively has a 3′(2′), 5′-bisphosphate nucleotidase activity and is involved in S metabolism in vivo (Murguia et al., 1995). To direct the yeast MET22 protein to the chloroplast, the cTP from the small subunit of Rubisco from pea (Pisum sativum) was fused to the N terminus of MET22. Five independent transgenic lines expressing the MET22 gene were recovered. All of these five lines showed larger-than-fou8 rosettes and wild-type-like petioles (Fig. 4A), a phenotype similar to that observed in the aos fou8 double mutant (Fig. 2D). Likewise, in all these lines the fatty acid oxygenation capacity of leaf extracts was reduced compared to fou8 levels (Fig. 4B), indicating that, in vivo, the 3′(2′), 5′-bisphosphate nucleotidase of SAL1 regulates basal 18:3 oxygenation and JA production in Arabidopsis.
Figure 4.
The MET22 gene from yeast partially complements the fou8 mutation. A, Rosette morphology of the wild type (wt), fou8, and five independent lines complemented with the MET22 gene. B, Production of radiolabeled 18:3-hydroperoxide produced after incubation with leaf juice from the wild type, fou8, and five independent fou8 lines complemented with the MET22 gene.
The fou8 Mutant Affects Chloroplastic PAPS Metabolism
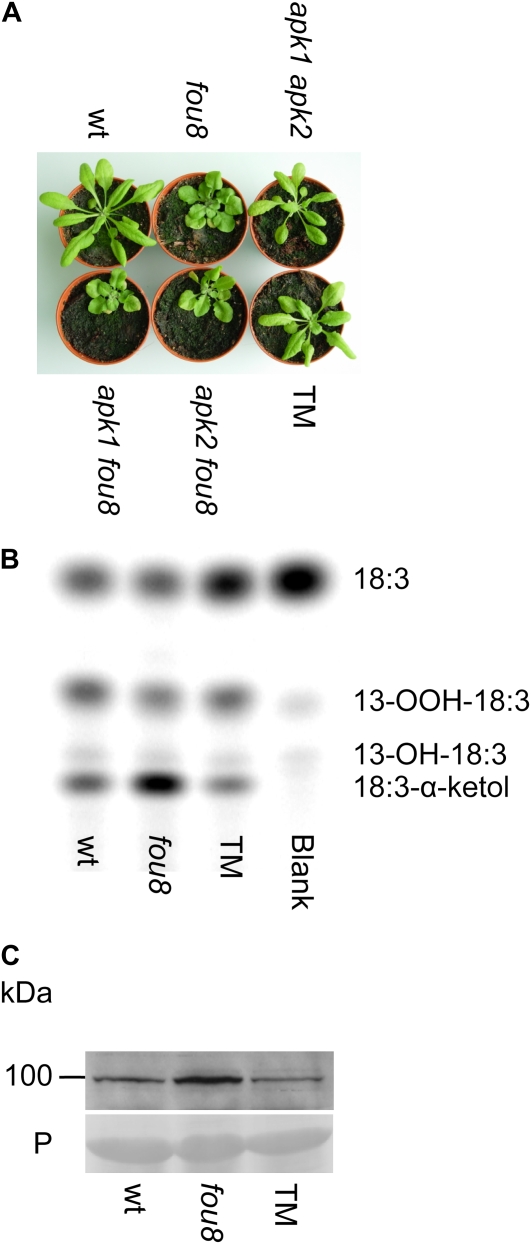
The 3′(2′), 5′-bisphosphate nucleotidase activity of SAL1 suggests a role in S metabolism by catalyzing the dephosphorylation of PAPS to 5′-adenosine phosphosulfate (APS). We cannot at present rule out a role in dephosphorylation of a molecule derived from PAPS, such as PAP. PAPS metabolism network is found in two cellular compartments, one in the cytosol and the other in chloroplasts (Pilon-Smith and Pilon, 2007). Since SAL1 protein is located in the chloroplast, this protein most probably affects the conversion of PAPS to APS in this compartment (Fig. 3, A–D). To test this hypothesis and to examine nucleotide pools from the wild type and fou8, we attempted to extract nucleotides from leaf extracts for analysis by liquid chromatography. However, all our efforts in this direction failed due to the complexity of the chromatograms, i.e. comigrating peaks. We then isolated chloroplasts from wild-type and fou8 leaves for nucleotide analysis. The liquid chromatography measurements (Supplemental Fig. S6A) showed that levels of several major nucleotides including AMP and ATP were similar in the two plants and that there was a possible accumulation of PAPS in fou8 chloroplasts but not in chloroplasts from the wild type. Significantly, the relative levels of APS were similar in both samples, suggesting that the main S-assimilation pathway is not affected in the fou8 mutant. If this were the case, levels of major S-containing reductants such as GSH should not be affected in fou8. To investigate this, total GSH and the percent of reduced GSH were quantified. No significant differences in either total or reduced GSH contents were observed between the wild type and fou8 (Supplemental Fig. S6B). These results indicate that the fou8 phenotype is not caused by a general response to S depletion but by a more specific regulatory pathway. APS and PAPS are interconnected through a so-called S futile cycle. To test the relevancy of this cycle in the fou8 phenotype, we crossed the fou8 mutant to two chloroplastic APS kinase mutants (apk1 and apk2; Mugford et al., 2009). As reported previously the single mutants apk1 and apk2 do not show any obvious morphological phenotype but the double mutant apk1 apk2 displays a slightly smaller rosette size than the wild type (Mugford et al., 2009). The double mutants apk1 fou8 and apk2 fou8 showed a phenotype similar to that of fou8 (Fig. 5A). Three independent triple mutants (apk1 apk2 fou8) were identified. All displayed a phenotype similar to that of the double mutant apk1 apk2 (Fig. 5A) and wild-type rates of 18:3 oxygenation and wild-type levels of LOX2 protein (Fig. 5, B and C). These crosses confirmed that both the morphological and biochemical phenotype of fou8 were due to its effect on chloroplastic PAPS metabolism.
Figure 5.
PAPS levels regulate the synthesis of JA. A, Rosette morphology of wild-type (wt), fou8, apk1 apk2, apk1fou8, apk2 fou8, and TM (apk1 apk2 fou8) plants. B, Oxygenation rate of radiolabeled 1-[14C]18:3 catalyzed by fou8, wild-type, and TM leaf extracts. Leaf juice (2 μ L) was incubated for 2 min with 1-[14C]18:3. Products were separated by thin-layer chromatography. C, LOX2 protein abundance in wild-type, fou8, and TM samples. The bottom section represents the loading control: Rubisco large subunit stained with Ponceau red (P).
DISCUSSION
We performed a genetic screen to identify mutants affected in the regulation of the early steps in oxylipin biosynthesis. A new mutant (fou8) was identified on the basis of the ability of leaf extracts to catalyze higher than wild-type levels of 18:3 oxygenation rates in vitro. The mutant displayed an increase in LOX activity and LOX2 protein levels in resting tissue with respect to the wild type. LOX2 is a major LOX in JA synthesis in leaves (Bell et al., 1995; Schaller and Stintzi, 2008) and contributes to the production of high JA levels in tissues proximal to wounds (Bell et al., 1995; Glauser et al., 2009) and to the synthesis of arabidopsides (esterified cyclopentenone jasmonates; Glauser et al., 2009). Prior to isolating fou8 another mutant, fou2, was isolated with the same strategy (Bonaventure et al., 2007a). In contrast to fou8, fou2 shows a higher than wild-type production of JA starting between the third and fourth week of development either in resting conditions or after wounding. The creation of a fou2 fou8 double mutant with fou2 revealed that the effects of the two mutations were independent and additive.
We mapped the fou8 mutation to the SAL1 gene and showed that it is a loss-of-function mutant. The SAL1 gene has previously been reported to play a role as negative regulator of ABA stress signaling (Xiong et al., 2001; Wilson et al., 2009) and light-mediated repressor of cell elongation (Kim and von Arnim, 2009). To investigate to what extent ABA in fou8 could be responsible for its phenotype, we crossed fou8 with the aba2.1 mutant (Leon-Kloosterziel et al., 1996). The double mutant invariably died after 2 weeks (Supplemental Fig. S4B), strongly suggesting that wild-type levels of ABA are necessary for fou8 mutant development by playing a protective role in these plants. Microarray data from GENEVESTIGATOR (Zimmermann et al., 2004) show a slight increase of the SAL1 transcript in plants germinated in a medium with 0.5 μm ABA and in plants treated with 10 μm methyl jasmonate. This might suggest a function of this gene in a JA-ABA cross-regulation pathway.
Most of the features of fou8 (shorter than wild-type petioles, wrinkled leaves, and anthocyanin accumulation) have previously been associated with an enhanced activity in the jasmonate pathway (e.g. Bonaventure et al., 2007a; Yan et al., 2007; Zhang and Turner, 2008). Petiole length, accumulation of anthocyanins, and flowering time are rescued in the double mutant with aos, suggesting that, to a large extent, the fou8 phenotype is due to higher accumulation of JA in resting leaves (Fig. 2D). Consistent with our results, Wilson et al. (2009) found that the mutation alx8, another mutant allele of SAL1, produces strong changes in gene expression that are correlated with drought, heat, cold, and wound transcriptomes. Three of the most highly up-regulated transcripts in alx8 are VSP1, VSP2, and MYC2 (Wilson et al., 2009). These are well-known markers of JA responses (Benedetti et al., 1995; Lorenzo et al., 2004). Additionally, transcripts encoding the JA biosynthesis enzyme OPR3 were overexpressed in alx8 (supplemental table 1 in Wilson et al., 2009). To test whether the increase in LOX activity and LOX2 protein level that we observed in fou8 was reflected in higher levels of JA production, we measured the JA content in resting and wounded tissue. In agreement with the LOX2 activity results, and consistent with the microarray data of Wilson et al. (2009), the levels of JA in fou8 are higher than in the wild type in resting tissue. However, the mutant and wild-type genotypes produce similar levels of JA after wounding. To our knowledge, fou8 is the first mutant reported to have effects on the basal levels of JA in resting leaves without having a marked effect on JA levels in wounded tissues. Arabidopsis plants overexpressing SAL1 displayed lower than wild-type levels of 18:3 oxygenation, a phenotype that was opposite to that of the fou8 mutant. Importantly, these loss- and gain-of-function data show that SAL1 regulates steps in the synthesis of JA in vivo in resting tissue. The observation ruled out the possibility that the increased basal JA biosynthesis observed in fou8 is simply due to the toxicity of the substrate for SAL1 and support a bona fide role for SAL1 in LOX2 regulation.
SAL1 has been the subject of extensive research (Quintero et al., 1996; Xiong et al., 2001, 2004; Gy et al., 2007; Kim and von Arnim, 2009; Wilson et al., 2009) and the SAL1 protein has at least two putative activities in vivo, 3′(2′), 5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase (Quintero et al., 1996). All our data are consistent with SAL1 performing the former activity in vivo since the yeast S metabolism gene MET22 partially rescues the morphological phenotype of fou8 and restores the 18:3 oxygenation rate of fou8 to that observed in the wild type (Fig. 4). The SAL1 protein thus has 3′(2′), 5′-bisphosphate nucleotidase activity in vivo. In our experiments the complementation of fou8 with the MET22 cDNA was partial. One possibility is that the chloroplast offers a different environment to the enzyme than does the yeast cytosol. We also considered the possibility that the wild-type SAL1 gene produces multiple transcript variants with and without cTPs. However, our data on the localization of the SAL1 protein coupled to GFP only provided evidence for a chloroplastic version of the protein. Although the active site of the SAL1 enzyme is unknown, residues in this protein could be aligned with the residues that compose the active site of MET22 and the alignment showed exact positional matches between these potentially critical residues in SAL1. The potential importance of such amino acids in SAL1 was revealed by the fry1-1 and fry1-2 mutants, in which a single substitution in one of the amino acids corresponding to the putative active site yielded loss-of-function mutations (Xiong et al., 2001). In vitro, SAL1 efficiently hydrolyzes nucleotide-related compounds (PAPS or PAP) and can also hydrolyze IPs with lower efficiency (Quintero et al., 1996). Xiong et al. (2001) have reported an accumulation of IP3 in a loss-of-function mutant of SAL1. This observation could be explained as an indirect effect of higher JA accumulation in these mutants (Mosblech et al., 2008).
Where does the SAL1 gene fit into S metabolism in plants? S-assimilation pathways have two main functions: the donation of oxidized S (SO42−) to acceptor molecules and S reduction. Chloroplasts are the sites of S reduction and the synthesis of Cys and subsequently GSH through a pathway that uses APS as a key intermediate (Kopriva, 2006). In oxidized S metabolism, PAPS is known to act as a SO42− donor in the cytosol (Klein and Papenbrock, 2004). Cytosolic reactions can produce PAPS (Rotte and Leustek, 2000; Mugford et al., 2009) although chloroplasts also contain the APS kinase enzymes capable of generating PAPS from APS (Mugford et al., 2009). PAPS can be hydrolyzed back in a futile cycle to APS by a 5′-bisphosphate nucleotidase (Kopriva, 2006). All our data indicate that SAL1 dephosphoryates PAPS (or a PAPS derivative) and that this enzyme functions in plastids.
Connections between S assimilation and jasmonate responses have been observed previously. JA application strongly activates the expression of several genes of the S-reduction pathway in Arabidopsis (Jost et al., 2005; Sasaki-Sekimoto et al., 2005). Likewise, plants undergoing S-starvation responses show enhanced expression of JA-related genes (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003). More recently, a double mutant in the two chloroplastic kinases apk1 apk2 was found to produce lower levels of glucosinolates and lower levels of the JA precursor 12-oxophytodienoic acid as well as the JA metabolite 12-sulfojasmonate (Mugford et al., 2009). Based on the identification of APK1 and APK2 as chloroplastic APS kinases (Mugford et al., 2009) we constructed fou8 apk1 apk2 triple mutants. All three independent triple mutants we isolated displayed a phenotype similar to that of the double mutant apk1 apk2, largely abolishing the short petiole phenotype that is characteristic of fou8 (Fig. 5A). Moreover, the higher 18:3 oxygenation rates and higher levels of LOX2 protein displayed by fou8 were reduced to wild-type levels in the triple mutant (Fig. 5, B and C). These results pinpointed chloroplastic PAPS or another closely related molecule produced directly or indirectly by APK1 and APK2 as regulators of the fatty acid oxidation capacity of the leaf and of basal JA levels. Liquid chromatography of chloroplast extracts from fou8 indicate that the molecule likely to influence JA production is not APS since APS levels were similar to those in chloroplasts from the wild type. PAPS, a molecule involved in the synthesis of a broad range of sulfated compounds via reactions mediated by cytosolic sulfotransferases (Varin et al., 1997; Bick and Leustek, 1998), is the most likely nucleotide influencing JA levels. While our data point to plastids as the site of SAL1 action it is also conceivable that PAPS exerts its effects on JA levels by acting outside plastids. It was suggested that PAPS might be exported to the cytosol (Mugford et al., 2009).
The possible broad effects of the fou8 mutant on S metabolism required further investigation. The fact that the levels of total and reduced GSH were similar in fou8 and wild-type plants (Fig. S6B) indicated that fou8 might not be under strong S starvation. Morphologically, the fou8 and fou2 mutants share similarity (Supplemental Fig. S4A) and the fou2 fou8 double mutant displayed combined (i.e. additive) characteristics of each individual mutant, creating a strongly dwarfed but viable plant. Gathering evidence suggests that fou2 affects subcellular K levels (Bonaventure et al., 2007b; Beyhl et al., 2009). It will be interesting to explore a potential effect on K homeostasis in fou8.
In summary, we show that the activity of a LOX in JA synthesis is regulated by a component of the S futile cycle to maintain the resting levels of a potent wound response regulator below a threshold that, if crossed in a healthy and undamaged leaf, would result in growth impairment. While there is a great deal of knowledge on LOX catalytic mechanisms (Schneider et al., 2007) less is known of the regulation of the levels of these important and widespread catalysts.
MATERIALS AND METHODS
Genetic Screen
Approximately 4,000 M2 ethyl methanesulfonate-mutagenized Col-0 Arabidopsis (Arabidopsis thaliana) plants (Lehle seeds) were screened for LOX activity using a previously described assay (Caldelari and Farmer, 1998; Bonaventure et al., 2007a). Plants were grown in long-day conditions (16 h light, 8 h dark) at 22°C under white light (100 μ E m−2 s−1). Leaf juice from one leaf was harvested 4 weeks after sowing and incubated immediately with 1-[14C]18:3. The products were separated by thin-layer chromatography in silica gel (20 × 20 cm; 200 μ m thickness; Merck) and radioactive bands were detected and quantified using a Storm 820 phosphor imager and ImageQuant software (Amersham Biosciences), respectively. M3 progeny from putative M2 mutants were retested to identify mutants with heritable changes in the level of leaf LOX activity. Total leaf protein was quantified using bicinchoninic acid assay (Pierce) after precipitation with 8% (v/v) trichloroacetic acid.
Genetic Mapping
To obtain a mapping population, the fou8 mutant was outcrossed to wild-type Landsberg erecta. Total genomic DNA was extracted from a single leaf of 4-week-old F2 plants. DNA from 50 wild-type and 50 mutant plants was pooled and each pool was hybridized to ATH1 Arabidopsis genechips (Affymetrix) as described (Borevitz, 2006). After positioning the mutation in a region of 1.2 Mb at the bottom of chromosome V, fine mapping was performed using 782 F2 fou8 homozygous plants. To narrow the region containing the fou8 mutation, new single sequence length polymorphism and cleaved-amplified polymorphic sequences markers were designed based in Col-0 and Landsberg erecta polymorphism (Supplemental Table S1). The new markers were named according to their location in bacterial artificial chromosomes.
JA Measurements
Resting expanded leaves from 4-week-old plants were harvested and frozen in liquid nitrogen for subsequent measurements. For wounding experiments the apical third of the leaf was crushed with forceps, harvested after 90 min, and frozen in liquid nitrogen. JA measurements were carried out using an oxygen-18 internal JA standard as described (Mueller et al., 2006). This result was confirmed with an independent technology as described (Glauser et al., 2008; data not shown).
Protein Immunoblotting
Rabbit antibodies were raised against two LOX2 peptides (KNREEVGEFTKFERI and SLITWEIVEREVKG) immobilized to keyhole limpet hemocyanin (Eurogenetec). The antibody was used for protein immunoblots at 1/1,000 dilution. Inmunoblotting was performed using standard protocols.
Cloning and Transformation
All constructs were cloned using Gateway technology (Invitrogen). To complement the fou8 mutation, a genomic fragment (4.5 kb) including the SAL1 gene and its native promoter was amplified by PCR (LA Taq, Takara Bio Inc.) and cloned into the binary vector pMDC100 (Curtis and Grossniklaus, 2003). To obtain wild-type lines constitutively expressing SAL1, the coding sequence was amplified by PCR (LA Taq, Takara Bio Inc.) and cloned in the binary vector pMDC32 (Curtis and Grossniklaus, 2003) under the control of the CaMV 35S promoter. For fou8 complementation using MET22 (the yeast [Saccharomyces cerevisiae] ortholog of SAL1), the MET22 coding sequence was amplified by PCR from yeast clone YSC3867 (Open Biosystems). To direct the MET22 protein into the chloroplast, the gene was fused with the cTP of the small subunit of Rubisco from pea (Pisum sativum). This cTP sequence was amplified from plasmid prSS2-1 kindly provided by Dr. John E. Froehlich, using Phusion high-fidelity polymerase (Finnzymes Oy). The MET22 and cTP amplicons included new restriction targets for KpnI (Pharmacia) in their 5′and 3′end, respectively. Fragments were digested with KpnI and ligated using T4 DNA ligase (Promega) and the final product was cloned in the binary vector pMDC32. Plants were transformed using the floral-dip method and selected in Murashige and Skoog medium containing the appropriate antibiotic. Primers used for plasmid constructions are listed in Supplemental Tables S2 and S3.
Chloroplast Extraction
Chloroplasts were extracted following Heinemeyer et al. (2004) with some modifications. Leaves of 4-week-old plants were homogenized in 200 mL of extraction buffer (20 mm Tricine/KOH, 10 mm EDTA, 1 mm MnCl2, 450 mm sorbitol, 10 mm NaHCO3, 0.05% bovine serum albumin, 1 mm phenylmethylsulfonyl fluoride, pH 8.4) with a Waring blender and then filtered through two layers of Miracloth. Subsequently, chloroplasts were pelleted by centrifugation for 5 min at 1,500g (4°C) and the pellet was carefully resuspended in 1 mL of resuspension buffer (20 mm Tricine/KOH, 2.5 mm EDTA, 5 mm MgCl2, 300 mm sorbitol). Two purification steps were carried out by centrifugation in a bilayer Percoll gradient (40% [v/v]–85% [v/v]) for 10 min at 4,000g (4°C). Finally chloroplasts were resuspended in 1 volume of storage buffer containing 30% (v/w) ethylene glycerol, 20 mm Tris pH 7.8, 200 mm Suc, and 125 mm KCL with a pH adjusted to 6.9.
Extraction and Detection of Nucleotides
Nucleotides were extracted according to Napolitano and Shain (2005). Fifty microliters of chloroplast solution were mixed with 50 μ L of 7% (v/v) perchloric acid and incubated for 10 min at 4°C. Samples were then centrifuged at 4,000g for 5 min. The supernatant was recovered and the pH adjusted to approximately 7 with 4 m KOH and 1 m K2HPO4. The mixture was vortexed, centrifuged for 5 min (14,000g at 4°C), and 100 μ L transferred to a new tube for liquid chromatography. HPLC was performed using a LaChrom elite HPLC system (Merck-Hitachi). HPLC was controlled with the EZChrom elite program version 3.1.7 (Merck-Hitachi). Samples (80 μ L) were separated with a Nucleodur column (C18, 5 μ m, 4.6 × 250 mm; Macherey-Nagel) with isocratic gradient of 5% methanol in 83.3 mm triethylammonium adjusted to pH 6 with phosphoric acid, at a flow rate of 1 mL/min. Nucleotides were detected with a UV detector at a wavelength of 254 nm.
qRT-PCR
qRT-PCR was conducted following the recommendations of Udvardi et al. (2008). Total RNA was extracted using the RNeasy plant mini kit (Qiagen) and its quality checked on an agarose gel. Two micrograms of total RNA were reverse transcribed using Superscript II (Invitrogen) and oligo dT (20) according to the manufacturer's instructions. qRT-PCR was performed in a 25 μ L reaction with the FullVelocity SYBR Green master mix (Stratagene). Gene-specific primers where designed to have a melting temperature of approximately 60°C and to target amplicons between 200 and 250 bp (Supplemental Table S4). Five different reference genes (Supplemental Table S4) were tested and analyzed using LingRegPCR (Ramakers et al., 2003) and geNorm (Vandesompele et al., 2002) to determine the most stably expressed gene for normalization purposes. qPCR reactions were carried out on a Mx3000P spectrofuorometric thermal cycler (Stratagene).
Transient Transformation of Epidermal Onion Cells
The full-length SAL1 cDNA was amplified by PCR using a Phusion Taq (NEB). Due to low yield in the first amplification a second PCR was performed using the first amplicon as template. The product was cloned into the binary vector pMDC83 (Curtis and Grossniklaus, 2003) using Gateway technology (Invitrogen) under control of CaMV 35S promoter. The pt-rk CD3-999 construct (that includes the target signal from tobacco [Nicotiana tabacum] Rubisco small subunit cDNA fused with mCherry; Nelson et al., 2007) was purchased from the Arabidopsis Biological Resource Center. Plasmids were precipitated onto gold beads and transformed into onion (Allium cepa) epidermis as described (Mueller et al., 1997). Microscopy was performed with a Leica microsystems DM5000B microscope equipped with a Leica DFC340F camera.
Spectrophotometric Assay for LOX Activity
One-hundred milligrams of fresh leaf tissue from wild-type and fou8 plants were extracted in 100 μ L of reaction buffer (MOPS-KOH pH 7, 0.01% Tween). The reaction was carried out at room temperature in a quartz cuvette (1.0 cm light path) by mixing 890 μ L of reaction buffer with 100 μ L of 2.5 mm 18:3 (Cayman Chemical). This solution was used to set the reference absorption at 234 nm. Ten microliters of leaf juice were then added, quickly mixed by pipetting, and the reaction followed for up to 2 min at 22°C. A molar extinction coefficient of 28,000 L per mole per cm was used to convert absorbance readings to moles of product (Smith and Lands, 1972).
Generation of the fou8 apk1 apk2 Triple Mutant
Single mutants fou8 and apk1 (SALK_053427C) were crossed to obtain the F1. This population was crossed to apk2 (SALK_025296C) and the triple heterozygote selected by PCR and self pollinated. All possible combinations of single, double, and triple mutants were selected using primers listed in Supplemental Table S5.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers BT005993 (SAL1), X72847 (MET22), NM_116594 (TPC1), NM_127039 (APK1), NM_120157 (APK2), NM_123629 (AOS), and NM_104113 (ABA2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Positional cloning of the fou8 gene.
Supplemental Figure S2. fou8 is a loss-of-function mutant.
Supplemental Figure S3. SAL1 overexpression lines.
Supplemental Figure S4. fou8 fou2 and fou8 aba2.1 double mutants.
Supplemental Figure S5. The SAL1 and MET22 proteins share the same sequence in the active site.
Supplemental Figure S6. The fou8 mutation specifically affects chloroplastic PAPS metabolism.
Supplemental Table S1. Primers and enzymes used for the newly designed markers.
Supplemental Table S2. Primers used for cloning SAL1.
Supplemental Table S3. Primers used for cTP-MET22 fusion and cloning.
Supplemental Table S4. Primers used for qRT-PCR.
Supplemental Table S5. Primers used to identify mutant plants.
Acknowledgments
We thank J. Froehlich (Michigan State University) for cTP cDNA; the staff of the Lausanne DNA array facility for support; G. Glauser (University of Geneva) for confirmatory JA analyses; Emanuel Schmid (University of Lausanne) for chloroplast extractions; Hatem Rouached, Yves Poirier, Esther Dohmann, and Ivan Acosta (University of Lausanne) for helpful comments; and two reviewers for constructive suggestions.
References
- Armengaud P, Breitling R, Amtmann A. (2004) The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol 136: 2556–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E, Creelman RA, Mullet JE. (1995) A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA 92: 8675–8679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti CE, Xie D, Turner JG. (1995) Coi1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol 109: 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Bell E, Sadka A, Mullet JE. (1995) Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol 27: 933–942 [DOI] [PubMed] [Google Scholar]
- Beyhl D, Hortensteiner S, Martinoia E, Farmer EE, Fromm J, Marten I, Hedrich R. (2009) The fou2 mutation in the major vacuolar cation channel TPC1 confers tolerance to inhibitory luminal calcium. Plant J 58: 715–723 [DOI] [PubMed] [Google Scholar]
- Bick JA, Leustek T. (1998) Plant sulfur metabolism-the reduction of sulfate to sulfite. Curr Opin Plant Biol 1: 240–244 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Gfeller A, Proebsting WM, Hortensteiner S, Chetelat A, Martinoia E, Farmer EE. (2007a) A gain-of-function allele of TPC1 activates oxylipin biogenesis after leaf wounding in Arabidopsis. Plant J 49: 889–898 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Gfeller A, Rodriguez VM, Armand F, Farmer EE. (2007b) The fou2 gain-of-function allele and the wild-type allele of Two Pore Channel 1 contribute to different extents or by different mechanisms to defense gene expression in Arabidopsis. Plant Cell Physiol 48: 1775–1789 [DOI] [PubMed] [Google Scholar]
- Borevitz J. (2006) Genotyping and mapping with high-density oligonucleotide arrays. Methods Mol Biol 323: 137–145 [DOI] [PubMed] [Google Scholar]
- Browse J, Howe GA. (2008) New weapons and a rapid response against insect attack. Plant Physiol 146: 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldelari D, Farmer EE. (1998) A rapid assay for the coupled cell free generation of oxylipins. Phytochemistry 47: 599–604 [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE. (2007) Jasmonate perception machines. Nature 448: 659–660 [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R. (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5: 344–350 [DOI] [PubMed] [Google Scholar]
- Glauser G, Dubugnon L, Mousavi SA, Rudaz S, Wolfender JL, Farmer EE. (2009) Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J Biol Chem 284: 34506–34513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauser G, Grata E, Dubugnon L, Rudaz S, Farmer EE, Wolfender JL. (2008) Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J Biol Chem 283: 16400–16407 [DOI] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, Longaker MT. (2008) Wound repair and regeneration. Nature 453: 314–321 [DOI] [PubMed] [Google Scholar]
- Gy I, Gasciolli V, Lauressergues D, Morel JB, Gombert J, Proux F, Proux C, Vaucheret H, Mallory AC. (2007) Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19: 3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer J, Eubel H, Wehmhoner D, Jansch L, Braun HP. (2004) Proteomic approach to characterize the supramolecular organization of photosystems in higher plants. Phytochemistry 65: 1683–1692 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K. (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33: 651–663 [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Jost R, Altschmied L, Bloem E, Bogs J, Gershenzon J, Hahnel U, Hansch R, Hartmann T, Kopriva S, Kruse C, et al. (2005) Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynth Res 86: 491–508 [DOI] [PubMed] [Google Scholar]
- Kim BH, von Arnim AG. (2009) FIERY1 regulates light-mediated repression of cell elongation and flowering time via its 3′(2′),5′-bisphosphate nucleotidase activity. Plant J 58: 208–219 [DOI] [PubMed] [Google Scholar]
- Klein M, Papenbrock J. (2004) The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J Exp Bot 55: 1809–1820 [DOI] [PubMed] [Google Scholar]
- Kopriva S. (2006) Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot (Lond) 97: 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M. (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655–661 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R. (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H. (2003) Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol 132: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosblech A, König S, Stenzel I, Grzeganek P, Feussner I, Herilmann I. (2008) Phosphoinositide and inositolpolyphosphate signalling in defense responses of Arabidopsis thaliana challenged by mechanical wounding. Mol Plant 1: 249–261 [DOI] [PubMed] [Google Scholar]
- Mueller LA, Hinz U, Uzé M, Sautter C, Zryd JP. (1997) Biochemical complementation of the betalain biosynthetic pathway in Portulaca grandiflora by a fungal 3,4-dihydroxyphenylalanine dioxygenase. Planta 203: 260–263 [Google Scholar]
- Mueller MJ, Mene-Saffrane L, Grun C, Karg K, Farmer EE. (2006) Oxylipin analysis methods. Plant J 45: 472–489 [DOI] [PubMed] [Google Scholar]
- Mugford SG, Yoshimoto N, Reichelt M, Wirtz M, Hill L, Mugford ST, Nakazato Y, Noji M, Takahashi H, Kramell R, et al. (2009) Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21: 910–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murguia JR, Belles JM, Serrano R. (1995) A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267: 232–234 [DOI] [PubMed] [Google Scholar]
- Napolitano MJ, Shain DH. (2005) Quantitating adenylate nucleotides in diverse organisms. J Biochem Biophys Methods 63: 69–77 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R. (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Cai Y, Sun Q, Zabrouskov V, Giacomelli L, Rudella A, Ytterberg AJ, Rutschow H, van Wijk KJ. (2006) The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol Cell Proteomics 5: 114–133 [DOI] [PubMed] [Google Scholar]
- Pilon-Smith EAH, Pilon M. (2007) Sulfur metabolism in plastids. Wise RA, Hoober JK, , The Structure and Function of Plastids. Springer, Dordrecht, The Netherlands, pp 387–402 [Google Scholar]
- Quintero FJ, Garciadeblas B, Rodriguez-Navarro A. (1996) The SAL1 gene of Arabidopsis, encoding an enzyme with 3′(2′),5′-bisphosphate nucleotidase and inositol polyphosphate 1-phosphatase activities, increases salt tolerance in yeast. Plant Cell 8: 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RMP, Krishnamurthy V, Dicke M, Farmer EE. (2004) A conserved transcript pattern in response to a especialist and a generalist herbivore. Plant Cell 16: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotte C, Leustek T. (2000) Differential subcellular localization and expression of ATP sulfurylase and 5′-adenylylsulfate reductase during ontogenesis of Arabidopsis leaves indicates that cytosolic and plastid forms of ATP sulfurylase may have specialized functions. Plant Physiol 124: 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Taki N, Obayashi T, Aono M, Matsumoto F, Sakurai N, Suzuki H, Hirai MY, Noji M, Saito K, et al. (2005) Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J 44: 653–668 [DOI] [PubMed] [Google Scholar]
- Schaller A, Stintzi A. (2008) Direct defenses in plants and their induction by wounding and insect herbivores. Schaller A, , Induced Plant Resistance to Herbivory. Springer, Dordrecht, The Netherlands, pp 349–366 [Google Scholar]
- Schneider C, Pratt DA, Porter NA, Brash AR. (2007) Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem Biol 14: 473–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WL, Lands WE. (1972) Oxygenation of unsaturated fatty acids by soybean lipoxygenase. J Biol Chem 247: 1038–1047 [PubMed] [Google Scholar]
- Stappenbeck TS, Miyoshi H. (2009) The role of stromal stem cells in tissue regeneration and wound repair. Science 324: 1666–1669 [DOI] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Miersch O, Kurz T, Maucher H, Weichert H, Ziegler J, Feussner I, Wasternack C. (2003) Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Mol Biol 51: 895–911 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Czechowski T, Scheible WR. (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varin L, Marsolais F, Richard M, Rouleau M. (1997) Sulfation and sulfotransferases 6: biochemistry and molecular biology of plant sulfotransferases. FASEB J 11: 517–525 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PB, Estavillo GM, Field KJ, Pornsiriwong W, Carroll AJ, Howell KA, Woo NS, Lake JA, Smith SM, Harvey Millar A, et al. (2009) The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J 58: 299–317 [DOI] [PubMed] [Google Scholar]
- Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK. (2001) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15: 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Huang R, Zhu JK. (2004) A single amino acid substitution in the Arabidopsis FIERY1/HOS2 protein confers cold signaling specificity and lithium tolerance. Plant J 40: 536–545 [DOI] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chetelat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. (2007) A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Turner JG. (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS One 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]