Abstract
Bran from bread wheat (Triticum aestivum ‘Babbler’) grain is composed of many outer layers of dead maternal tissues that overlie living aleurone cells. The dead cell layers function as a barrier resistant to degradation, whereas the aleurone layer is involved in mobilizing organic substrates in the endosperm during germination. We microdissected three defined bran fractions, outer layers (epidermis and hypodermis), intermediate fraction (cross cells, tube cells, testa, and nucellar tissue), and inner layer (aleurone cells), and used proteomics to identify their individual protein complements. All proteins of the outer layers were enzymes, whose function is to provide direct protection against pathogens or improve tissue strength. The more complex proteome of the intermediate layers suggests a greater diversity of function, including the inhibition of enzymes secreted by pathogens. The inner layer contains proteins involved in metabolism, as would be expected from live aleurone cells, but this layer also includes defense enzymes and inhibitors as well as 7S globulin (specific to this layer). Using immunofluorescence microscopy, oxalate oxidase was localized predominantly to the outer layers, xylanase inhibitor protein I to the xylan-rich nucellar layer of the intermediate fraction and pathogenesis-related protein 4 mainly to the aleurone. Activities of the water-extractable enzymes oxalate oxidase, peroxidase, and polyphenol oxidase were highest in the outer layers, whereas chitinase activity was found only in assays of whole grains. We conclude that the differential protein complements of each bran layer in wheat provide distinct lines of defense in protecting the embryo and nutrient-rich endosperm.
Wheat grain (Triticum aestivum) is a major cereal crop and staple food in many parts of the world. The endosperm is the main nutritional component and is extracted in milling to produce base ingredients such as flour and semolina. Crop yield and quality may be compromised by both environmental and biological stresses. Wheat varieties are known to vary in their resistance to such stresses, probably due to individual differences in defense protein levels (Demeke and Morris, 2002; Bonnin et al., 2005; Yarullina et al., 2005). Cereal grain contains many defense proteins that have been categorized according to their mode of action and structural similarities. A major class of these is the pathogenesis-related (PR) proteins, which include PR-1, PR-2 (β -1,3-glucanases), PR-3 (chitinases), PR-4 (wheatwin1), and PR-5 (thaumatin-like proteins; Selitrennikoff, 2001; Desmond et al., 2006). Other known defense proteins are xylanase inhibitor proteins (XIPs) and α -amylase inhibitor proteins (Mundy et al., 1984; Payan et al., 2003). All of these defense proteins have both general and specific roles that contribute to plant survival, although little is known of their location within the various grain tissues, particularly the multiple layers that constitute bran.
Proteomic analysis of wheat grain has previously been applied to identify proteins in the germ and endosperm (Skylas et al., 2000; Wong et al., 2004; Mak et al., 2006), but analysis of bran and bran tissue fractions has not been reported. Collection of sufficiently pure bran tissue fractions has limited progress, mainly due to the strong bonds between the various bran tissue layers and endosperm in dry grain. Thus, a method to obtain bran layers free from contaminants, such as adjacent tissue and endosperm, is required to provide a sample suitable for proteomic analysis. Soaking whole grain in water causes the endosperm to soften, allowing it to be easily removed and washed from the bran; the bran becomes malleable enough to dissect. While this approach might not identify the proteome of dry grain fractions, it is the best available representation of the three distinct tissue fractions in grains, namely the outer layer (epidermis and hypodermis), intermediate layer (cross cells, tube cells, testa, and nucellar tissue), and inner layer (aleurone cells; Antoine et al., 2003, 2004). Using this method, water-soluble proteins that diffuse from the grain can be collected and identified.
In this study we aimed (1) to dissect bran into the three separate tissue fractions described above and to identify the protein complement of each fraction using proteomics, (2) to confirm the location of three major defense proteins identified (one from each microfraction) using immunolocalization, and (3) to identify water-soluble proteins and assay any defense-related proteins for enzymatic activity.
RESULTS
Light Microscopy of Bran Tissue Fractions
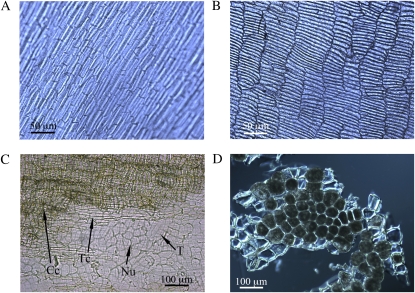
Microscopic examination of dissected tissue fractions showed that the cell types of each fraction were uniform and mostly free from cells of adjoining fractions. The distinctive cell patterns of the outer fraction (epidermis and hypodermis; Fig. 1A) and the intermediate fraction cross cells (Fig. 1B) confirmed the purity of each fraction. Four tissues (cross cells, tube cells, testa, and nucellar tissue) that make up the intermediate fraction were also distinguished (Fig. 1C). Finally, the inner fraction (aleurone) cells were free from endosperm and were also largely intact (Fig. 1D).
Figure 1.
Micrographs of the isolated bran fractions. A, Outer bran fraction (epidermis and hypodermis). B, Intermediate bran fraction (cross cells, tube cells, testa, and nucellar tissue). C, Detailed view of the individual layers in the intermediate fraction (Cc, cross cells; Nu, nucellar tissue; T, testa; Tc, tube cells). D, Aleurone cells. [See online article for color version of this figure.]
Protein Extraction from Bran Tissue Fractions
The outer bran layers and intermediate fraction contained significantly less protein than the inner fraction (aleurone): 0.4 mg protein g−1 was extracted from the outer layer (25% was water soluble), 3.6 mg protein g−1 was found in the intermediate fraction, and 156 mg protein g−1 was extracted from the inner layer.
Protein Identification from Two-Dimensional Electrophoresis Gels
The protein complement of the outer dead cell layers (outer layers and intermediate fraction) was much less diverse than that of the inner fraction (aleurone cells), as shown by the two-dimensional electrophoresis (2-DE) gels (Fig. 2; Supplemental Figs. S1–S6). Image analysis of triplicate 2-DE gels revealed 35 unique protein spots (30 identified using Mascot and 27 using The Global Proteome Machine [The GPM]) from the outer fraction (epidermis and hypodermis), 119 spots (106 identified using Mascot and 66 using The GPM) from the intermediate fraction (cross cells, tube cells, testa, and nucellar tissue), and 672 spots (606 identified using Mascot and 310 using The GPM) from the inner fraction extract. Although some proteins were identified in more than one fraction, there was minimal overlap of the most intense and abundant protein spots between fractions. This observation, together with microscopic examination of the microdissected tissue, confirms that there was negligible cross-contamination of proteins between the tissue types (Fig. 1).
Figure 2.
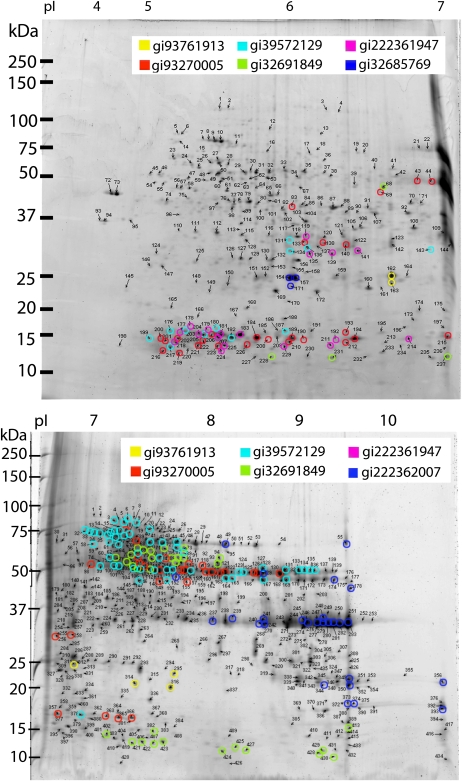
2-DE gels of the inner bran layer (aleurone). The highlighted spots show the different EST classes of 7S globulin, with the EST GenBank gi numbers shown in the legend. The unhighlighted gels are shown in Supplemental Figures S5 and S6. Top gel, pI 4 to 7; bottom gel, pI 6 to 11.
A list of the major proteins identified in this study is shown in Figure 3. Complete protein identification tables (Mascot and The GPM results) and representative 2-DE images with numbered spots are provided in Supplemental Tables S1 to S7 and Supplemental Figures S1 to S6. An individual false discovery rate (FDR) was only calculated for proteins identified using The GPM. This was because the proteins that were not identified in The GPM search, which employs peptide mass and sequence from tandem mass spectrometry (MS/MS), were identified by peptide mass fingerprinting (PMF) in the Mascot search (FDRs could not be calculated in the same way from PMF identifications and are thus not comparable). The average FDR for the 453 proteins identified using The GPM was 1.4%.
Figure 3.
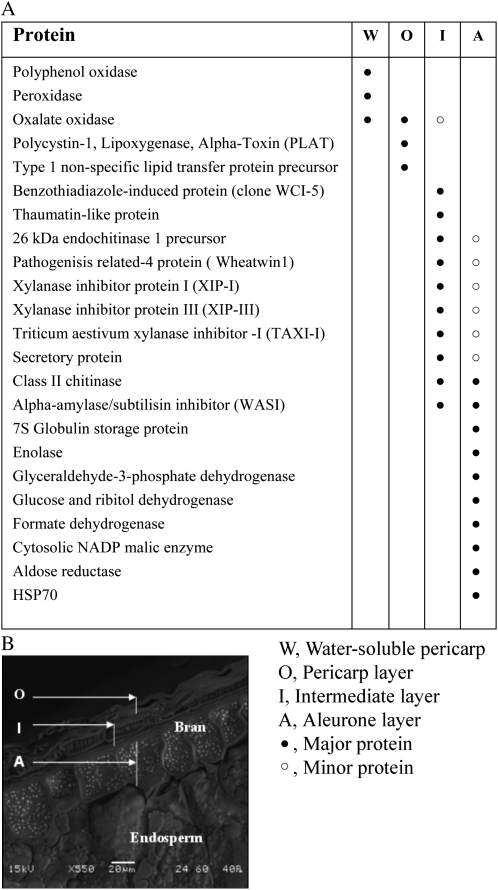
Summary of the major proteins identified in bran tissue fractions, supernatant from imbibed grain, and the outer tissue fraction. A, Table of major bran tissue and water-soluble proteins identified. B, Scanning electron micrograph of a cross-section of bran showing the bran tissue fractions.
Outer Fraction
The proteins identified in this fraction were mostly oxidative stress- and defense-related proteins such as oxalate oxidase (OXO), lipid transfer protein, and lipoxygenase (Supplemental Tables S1 and S2).
Intermediate Fraction
The intermediate fraction had a much more diverse set of defense proteins than the outer fraction. These proteins are oxidative stress- and defense-related proteins such as OXO, xylanase inhibitor I protein (XIP-I), chitinase and endochitinase, α-amylase/subtilisin inhibitor (WASI), wheatwin1, thaumatin-like protein, and benzothiadiazole clone of a wheat chemically induced protein (Supplemental Tables S3 and S4) . OXO was the only protein identified in this fraction that was also identified in the outer fraction and supernatant.
Inner Fraction (Aleurone Cells)
Mascot was used to search MS and MS/MS spectra against a cereal and wheat EST database, which identified 606 proteins. More than half (387 spots or 58%) were identified as globulin-like storage proteins from the cupin superfamily. The cumulative spot volumes of these 387 globulin-like storage protein spots represent 74.9% of the protein detected in the aleurone gels. Of the 387 spots, four were matched to a rice (Oryza sativa) globulin-like protein, one each to a putative rice globulin and a barley (Hordeum vulgare) embryo globulin, while 207 spots were matched to the wheat 7S globulin storage protein. The remaining 174 spots were matched to wheat ESTs, which were searched using National Center for Biotechnology Information (NCBI) BLAST. The BLAST search aligned 163 ESTs to 7S globulin storage protein from wheat and 11 to a rice cupin family-expressed protein.
Additionally, the MS/MS spectra that were identified as cupin superfamily proteins were searched against the wheat EST database using The GPM. This search identified 212 of the 387 cupin superfamily spots identified in the Mascot search. The majority of the 175 spots not identified in The GPM search were identified by Mascot as cupin superfamily proteins based on PMF data alone. A BLAST search of the 212 EST matches aligned 203 spots to a globulin 3 storage protein (homologous to the 7S globulin storage protein) and nine spots to a cupin family protein. Within the 212 spots identified in the EST database, there were seven distinct groups of ESTs. An unrooted phylogenetic tree was produced using the derived amino acid sequences from the ESTs of the seven groups, showing that they are distinct but related to each other (Supplemental Fig. S7). The 2-DE gels (pI 4–7 and 6–11) of the inner fraction show that the seven groups are uniquely distributed as highlighted in Figure 2. Proteins from the cupin superfamily are thus distributed into seven sequence-based groups with a total of 29 subgroups, according to significant differences in their molecular mass (Table I).
Table I. Subgroups of globulin proteins in inner aleurone fraction tissue separated on 2-DE gels.
| GenBank gi No. | Subgroup Average Mass | Subgroup Mass Range | Subgroup Average pI | Subgroup pI Range | No. of Spots | Percentage of Aleurone Proteins |
| kD | kD | |||||
| gi|39572129| | 13.8 | |||||
| 14.8 | 13–17 | 5.3 | 4.8–6.5 | 9 | 1.1 | |
| 30.3 | 29–32 | 5.8 | 5.7–5.9 | 3 | 0.3 | |
| 49.8 | 47–53 | 7.9 | 6.5–9.2 | 24 | 7.1 | |
| 59.0 | 58–60 | 7.1 | 6.9–7.4 | 3 | 0.5 | |
| 72.3 | 66–78 | 7.1 | 6.5–8.1 | 27 | 4.8 | |
| gi|93270005| | 14.5 | |||||
| 14.2 | 12–17 | 5.7 | 4.9–7.1 | 23 | 4.1 | |
| 31.0 | 28–34 | 6.1 | 5.8–6.4 | 5 | 0.8 | |
| 49.5 | 45–53 | 7.5 | 5.8–8.7 | 20 | 9.1 | |
| 57.3 | 55–59 | 7.5 | 7.1–7.7 | 3 | 0.3 | |
| 73.5 | 73–74 | 7.0 | 7.0–7.1 | 2 | 0.2 | |
| gi|32691849| | 2.2 | |||||
| 12.8 | 11–15 | 7.7 | 5.2–9.7 | 16 | 0.5 | |
| 49.5 | 48–52 | 7.0 | 6.5–7.4 | 4 | 0.5 | |
| 57.4 | 55–60 | 7.4 | 6.9–8.0 | 6 | 0.8 | |
| 69.2 | 67–72 | 7.4 | 7.0–8.0 | 9 | 0.5 | |
| gi|222362007| | 18.5 | |||||
| 18.7 | 17–21 | 9.9 | 9.3–10.8 | 7 | 0.9 | |
| 36.8 | 36–38 | 9.0 | 8.0–9.8 | 17 | 16.2 | |
| 47.0 | 44–50 | 8.8 | 7.5–9.6 | 4 | 1.1 | |
| 73.5 | 72–75 | 8.9 | 8.2–9.5 | 2 | 0.3 | |
| gi|222361947| | 3.5 | |||||
| 5.2 | 5.1–5.2 | 5.2 | 5.1–5.3 | 2 | 0.1 | |
| 14.4 | 13–15 | 5.5 | 5.0–6.7 | 12 | 2.6 | |
| 30.0 | 29–33 | 6.0 | 5.9–6.3 | 4 | 0.7 | |
| gi|93761913| | 22.8 | 20–25 | 6.9 | 6.4–7.5 | 6 | 1.0 |
| gi|32685769| | 24.5 | 23.5–25 | 5.8 | 5.8–5.8 | 3 | 1.4 |
The remaining 285 proteins identified in the Mascot search participate in a range of cell functions. They include carbohydrate metabolism (127 spots or 19%) and protein synthesis (23 spots or 3.4%), stress and defense (31 spots or 4.5%), and other/miscellaneous functions (38 spots or 5.7%; Supplemental Tables S5 and S6).
Proteins Identified in the Supernatant from Imbibed Grain and Isolated Outer Fraction
Water-soluble proteins in the supernatant from soaked grain and outer fraction were separated by one-dimensional (1-D) SDS-PAGE. Only the five major protein bands from the whole grain supernatant were selected for identification (Supplemental Fig. S8). The identified proteins were endochitinase, histone, OXO, peroxidase (POX), and polyphenol oxidase (PPO; Supplemental Table S7). The endochitinase and histone protein bands were not present in the outer fraction supernatant, suggesting that they have diffused from other parts of the grain such as the germ or inner bran layers.
Enzyme Activity Assays of the Water-Soluble Proteins Chitinase, OXO, POX, and PPO
OXO, POX, and PPO all showed enzymatic activity in the water-soluble protein extracts from whole grain and outer fraction, but no activity was detected in the endosperm extract. The activities of OXO and POX were much higher (28- and 7-fold, respectively) in the outer fraction supernatant compared with the whole grain supernatant, whereas PPO showed no significant difference in activity between these extracts.
Chitinase activity assays distinguished three different chitinases (exochitinase with chitobioside substrate, exochitinase [ β -N-acetylglucosaminidase], and endochitinase). Both endochitinase and exochitinase activities were detected in the whole grain water-soluble protein extracts and not in the outer fraction extracts (Table II). The exochitinase (β -N-acetylglucosaminidase) showed some activity in the water-soluble protein extract from the endosperm, but this was 6-fold less activity than in the whole grain water-soluble extract.
Table II. Enzyme activities of water-soluble protein extracts from whole grain, pericarp, and flour.
Values are presented as means ± se (n = 3) for OXO, POX, and PPO and means ± half-range (n = 2) for the chitinases.
| Enzyme | Whole Grain | Pericarp | Flour |
| nkat g−1 protein extract | |||
| OXO | 77 ± 56 | 2,219 ± 184 | 0 |
| POX | 1,998 ± 140 | 14,542 ± 106 | 0 |
| PPO | 188 ± 7 | 230 ± 100 | 0 |
| Exochitinase (chitobioside) | 6,909 ± 128 | 0 | 0 |
| Exochitinase (β -N-acetylglucosaminidase) | 4,286 ± 192 | 0 | 729 ± 103 |
Immunofluorescence Localization of OXO, XIP-I, and PR-4
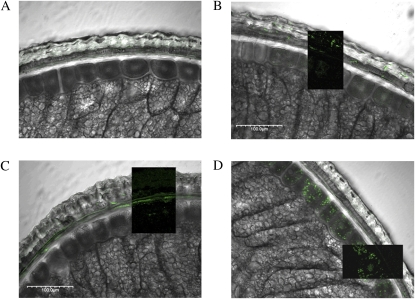
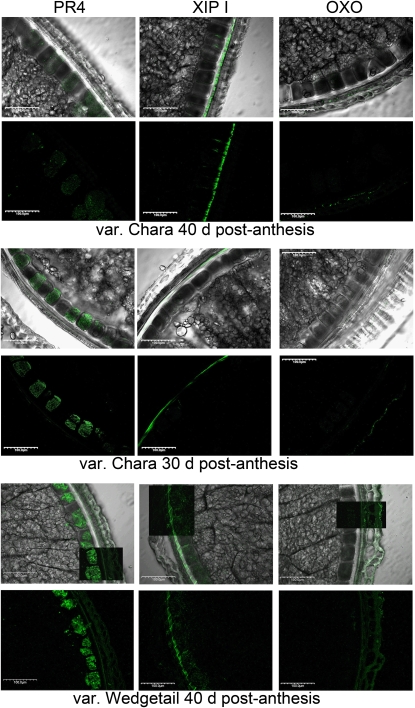
Affinity-purified antibodies to OXO, XIP-I, and PR-4 were used to confirm the localization of these proteins using cross-sections of wheat grain from cv Babbler (Fig. 4). OXO had a mosaic distribution throughout the outer layers (Fig. 4B). XIP-I was found predominantly in the nucellar tissue (Fig. 4C) immediately adjacent to the inner layer. PR-4 was found predominantly within the inner layer but was also distributed throughout the intermediate bran layers, including the testa (Fig. 4D). Immunolocalization of these proteins in two other wheat varieties, Chara and Wedgetail, confirmed that the localization was consistent in both genotypes, although the intensity of labeling and hence the relative levels of these proteins appears to vary considerably between varieties and developmental stages (Fig. 5).
Figure 4.
Fluorescence immunolocalization of defense proteins in bran cross-sections overlaid on differential interference contrast images of cross-sections (labeled). Dark inset overlays in images show fluorescence labeling without the differential interference contrast overlay. A, Control treated only with secondary antibody. B, OXO antibody. C, XIP-1 antibody. D, PR-4 antibody.
Figure 5.
Immunofluorescence localization of PR4, XIP-1, and OXO in wheat varieties Chara and Wedgetail. Images are arrayed in alternating rows of differential interference contrast overlaid with immunofluorescence image, followed by immunofluorescence-only image.
DISCUSSION
Proteomic analysis of bran tissue fractions from wheat grain revealed the location and distribution of many common plant defense-related proteins, which appear to be specific to certain tissue layers within the bran. The proteins identified within the outer layers (epidermis and hypodermis; Fig. 3A; Supplemental Tables S1 and S2) provide resistance to fungal and bacterial colonization and so fulfill a general defensive role rather than targeting specific biotic stresses. For example, OXO can degrade a fungus-derived toxin (oxalic acid) and produce hydrogen peroxide as an antifungal agent (Lane, 2002). The location of OXO was also confirmed by confocal immunomicroscopy, showing its distribution throughout these outer layers (Figs. 4B and 5). Other proteins identified in the outer fraction, and the water-soluble extract from this fraction, have similar general defense roles. These outer layer proteins include POX, which in the presence of hydrogen peroxide may act to strengthen cell walls through lignin cross-linking, thus inhibiting fungal penetration (as in the oxidative burst response in live plant tissue; Mellersh et al., 2002); PPO, which confers innate immunity against microorganisms in a number of plants (Niranjan Raj et al., 2006); polycystin-1 lipoxygenase α -toxin, which is expressed in acquired immune response against plant pathogens (Gorlach et al., 1996); and lipid transfer protein, which defends against fungi and bacteria (Blein et al., 2002; Breiteneder and Mills, 2005). Also, the activity of the enzymes OXO, POX, and PPO was readily detectable in the supernatant from imbibed outer tissue layers and whole grain, suggesting that their protective role is enhanced by their mobility and stability in the aqueous phase (Table II).
The cell layers of the intermediate fraction (cross cells, tube cells, testa, and nucellar tissue) are the last line of defense against fungal hyphae, penetrating the metabolically active inner fraction (aleurone cells) and underlying endosperm. The protein complement of these intermediate fraction layers (Fig. 3A; Supplemental Tables S3 and S4) revealed an array of defense-related enzymes and inhibitor proteins that, together, form a challenging environment for invading microorganisms.
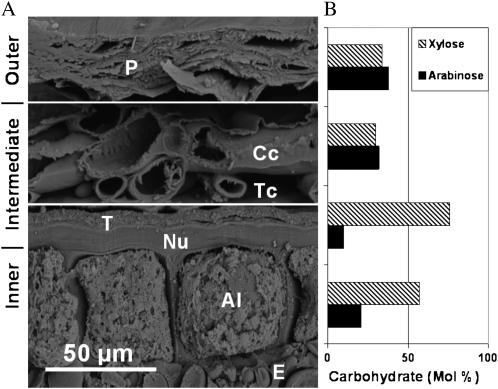
The two xylanase inhibitor-type proteins, XIP-I and TAXI (for T. aestivum xylanase inhibitor), were identified in the intermediate fraction. XIP-I inhibits only fungal xylanases, whereas TAXI inhibits both fungal and bacterial xylanases (Gebruers et al., 2002). XIPs were first characterized as being the most abundant inhibitor-type proteins in wheat (McLauchlin et al., 1999; Goesaert et al., 2004). Their location within the grain has not been identified explicitly and is usually assumed to be in the endosperm (Gebruers et al., 2001). Other reported proteomic analyses of wheat grain did not identify these proteins in the endosperm (Skylas et al., 2000) or embryo (germ; Mak et al., 2006), suggesting that they are likely to be exclusively located in the bran. Our proteomic and confocal immunomicroscopy results show that XIP-I is highly concentrated within the nucellar layer (Figs. 4C and 5). Based on this result, TAXI is also likely to be located within this layer and the inner layer (Fig. 3A). The carbohydrate composition of wheat bran (Fig. 6) clearly indicates that the nucellar layer has a higher ratio of Xyl to Ara than all the other bran tissues (Parker et al., 2005), which means that this tissue is highly susceptible to xylanase degradation. Thus, the location of XIPs in these high-Xyl tissues highlights their importance in protecting the inner layer and endosperm from biotic attack.
Figure 6.
Scanning electron micrograph of a cross-section of bran showing the bran tissue layers with corresponding Xyl and Ara contents. A, Outer (P, pericarp), intermediate (Cc, cross cells; Nu, nucellar tissue; T, testa [seed coat]; Tc, tube cells), and inner (Al, aleurone cells; E, endosperm) layers. B, Xyl and Ara (mol %) in corresponding bran tissue types (Parker et al., 2005).
The intermediate fraction also contains a series of PR proteins, which are induced by the defense-signaling elicitor molecules methyl jasmonate and ethylene in response to biotic and abiotic stress (Mauch and Staehelin, 1989; Yu and Muehlbauer, 2001; Desmond et al., 2006). Of these PR proteins, the PR-3 chitinases were well represented in the intermediate fractions, as both precursors and mature proteins. PR-3 chitinases are fungal growth inhibitor proteins, which hydrolyze the chitin of the fungal cell walls and so disrupt hyphal entry to the live inner fraction and endosperm (Selitrennikoff, 2001; Singh et al., 2007). However, the PR-2 class II chitinase (β -1,3-glucanase) identified here and in the inner layer has a different function (Mauch and Staehelin, 1989). Rather than primarily inhibiting fungal growth, this enzyme has a signaling role, releasing elicitor molecules from invading fungal cell walls. These elicitor molecules induce the expression of both PR-2 and PR-3 proteins, which are concentrated in the vacuole. The vacuole bursts when fungal hyphae penetrate the cell, releasing a high concentration of these chitinases (Mauch and Staehelin, 1989; Singh et al., 2007). The PR proteins wheatwin1 (PR-4) and thaumatin-like protein (PR-5) are also both inducible as part of a chemically elicited defense response (Desmond et al., 2006). PR-4 has a well-described structure (Caporale et al., 1999) and is induced in response to fungal attack (Caruso et al., 1999). Immunolocalization of PR-4 indicated its presence primarily within the testa of the intermediate fraction and the inner fraction (Figs. 4D and 5). The PR-5 protein is strongly antifungal in common monocots (Juge, 2006), killing hyphae by one or more mechanisms involving cell wall assembly or water relations (Selitrennikoff, 2001). Another defense protein identified in the intermediate fraction is the wheat chemically induced 5 (WCI-5) protein. It is induced by benzo-(1,2,3)-thiadiazole-7-carbothionic acid S-methyl ester to elicit systemic acquired resistance in plants (Schaffrath et al., 1997). The precise mode of action of this WCI-5 protein is unclear; however, it does confer some resistance to powdery mildew but not to all fungal pathogens (Gorlach et al., 1996; Yu and Muehlbauer, 2001). The spectrum of PR-related and other defense-related proteins identified in the intermediate bran layer of wheat seeds is certain to provide a hostile environment for invading hyphae. Identification of the inducible defense proteins in the dead cell layers suggests that they may be expressed in these cells during seed development or are expressed in the aleurone cells and diffuse to the intermediate fraction upon hydration.
The inner fraction is associated with a much greater diversity of protein function than in the other bran layers (Supplemental Tables S5 and S6). The majority of the proteins identified, corresponding to 150 of the 672 spots (22%), function in general metabolic activities such as protein synthesis, gene transcription, and associated energy metabolism. However, the major protein class in terms of spot number (387 spots or 58%) was identified as the 7S globulin storage protein (globulin 3 protein) of the cupin superfamily. Although previously reported in wheat grain (Robert et al., 1985), this protein was not identified as a component in the endosperm (Skylas et al., 2000) or germ (Mak et al., 2006) and has not been localized in wheat grain. It has been speculated that in addition to their storage protein function, 7S globulins may offer some protection against oxalic acid (Dunwell et al., 2000) produced by pathogenic fungi (Donaldson et al., 2001; Lane, 2002). It is also possible that some of our spots identified as 7S globulins may be the by-products of proteolytic processing to produce antimicrobial peptides, as has been found in other species (Marcus et al., 1999), and the large number of spots observed and the distinct molecular mass groups found suggest that this may be a function in wheat as well. Another explanation for the large number of spots may be the accumulation of breakdown products of 7S globulin proteins during grain softening. Proteolytic degradation of proteins in our sample by endogenous proteases during grain softening could not be prevented because the cells were not lysed during tissue collection and, therefore, protease inhibitors could not be effectively added. This grain softening inevitably triggers events associated with the very early stages of germination, which are known to involve the breakdown of specific storage proteins. However, the treatment used did not cause the aleurone to produce any detectable α -amylase, which indicates that initiation of these early germination processes was kept to a minimum under the treatment conditions used to isolate this fraction.
Two other biotic defense proteins that also appear as “major proteins” in the inner fraction (Fig. 3A) are the class II chitinase, as described earlier, and a WASI. α -Amylase inhibitors were discovered in the 1970s (Deponte et al., 1976) and fall into several classes with a range of inhibitory activities against mammalian, invertebrate, and, less commonly, endogenous plant α -amylases (Juge and Svensson, 2006). WASI, the wheat-specific form of this enzyme, has also been found in the endosperm in a previous study (Skylas et al., 2000). The family of α -amylase/subtilisin inhibitors has now been studied extensively and shown to inhibit a range of exogenous α -amylases but not all starch-degrading enzymes (Juge et al., 2004). Furthermore, α -amylase/subtilisin inhibitors disarm proteases from the subtilisin family, such as the subtilisin-like Fusarium proteinase (Pekkarinen and Jones, 2003).
CONCLUSION
A combination of bran layer microdissection, proteomics, and immunomicroscopy has localized many proteins to specific wheat bran layers and placed them into a functional framework. The wheat bran proteome is predominantly a sophisticated defense structure that has evolved to fortify the bran layers and to protect the embryo and nutrient-rich endosperm.
MATERIALS AND METHODS
Bran Tissue Microdissection
Wheat (Triticum aestivum ‘Babbler’) grains (four lots of 0.5 g) were placed on separate weigh boats, wetted with Milli-Q water, and left on the bench for 5 min. They were then stored at −20°C for 30 min, followed by thawing and removal of the outer layers using forceps with the aid of a dissecting microscope. Processing 0.5 g of grains at a time was done to minimize migration and loss of water-soluble proteins. The collected tissue (195 mg of total tissue from 2 g of grain) was placed into a 15-mL plastic tube and stored at −20°C.
To obtain the intermediate and inner fractions, grains were soaked in Milli-Q water for 41 h at an average temperature of 10.2°C (range, 2°C to 17.5°C over the period). This treatment caused sufficient softening of the endosperm to allow it to be scraped off and washed from the bran with a dissecting microscope. The bran was then separated into individual components by microdissection (Fig. 1). Total tissue collected from the intermediate layer was 42 mg, and the inner fraction was 45 mg.
Protein Extraction of Bran Tissue Fractions
Proteins were extracted from the three bran tissue fractions according to Wang et al. (2003). Samples were kept on ice when in solution during protein extraction. The tissues were placed into 2-mL screw-cap plastic tubes and washed twice with 1 mL of cold acetone, vortexed for 30 s, and centrifuged at 18,000g for 3 min each time. The tissue was left to dry inside the plastic tubes at room temperature. Once dry, the tissue was ground down to a fine powder with a mortar and pestle with the aid of a small amount of acid-washed sand. The powder was returned to the 2-mL plastic tubes and washed three times with cold 10% TCA in acetone (1 mL). For each wash, the sample was vortexed for 30 s and then centrifuged at 14,000g for 3 min. The sample was then washed twice with cold 10% TCA in water (1 mL) and finally washed twice with cold 80% acetone (1 mL). Again, for each step, the sample was vortexed for 30 s and centrifuged at 14,000g for 3 min. The tissue was left to dry at room temperature overnight. To the dry tissue was added 800 μ L of Tris-buffered phenol, pH 8.0, followed by 800 μ L of SDS buffer (30% Suc, 2% SDS, 0.1 m Tris-HCl at pH 8.0, and 5% 2-mercaptoethanol). The sample was then vortexed for 30 s and centrifuged at 14,000g for 3 min. The top phenol layer was removed and placed into a fresh 2-mL plastic tube. Five volumes of 0.1 m ammonium acetate in cold methanol was added to the phenol extracts and stored for 1 h at −20°C to allow for protein precipitation. The protein precipitate was centrifuged at 14,000g for 5 min, and the supernatant was removed. The protein pellet was washed twice with cold methanol and 0.1 m ammonium acetate and then twice with cold 80% acetone. Each time, the pellet was vortexed for 30 s and centrifuged for 5 min at 14,000g. After the final wash, the pellet was left to air dry to evaporate any acetone. Protein pellets were solubilized in 500 μ L of rehydration buffer (5 m urea, 2 m thiourea, 65 mm dithiothreitol, 2% [w/v] CHAPS, 2% [w/v] sulfobetaine 3-10, 1% [w/v] carrier ampholytes [Amersham Biosciences, GE], 40 mm Tris, 0.002% [w/v] bromphenol blue dye, and Milli-Q water) for isoelectric focusing (IEF).
Water-Soluble Protein Extraction from Whole Grain and Isolated Outer Fraction for 1-D SDS-PAGE
Whole grain (10 g) and outer layers tissue (100 mg) were placed into separate 50-mL and 1.5-mL plastic tubes in 15 mL and 1 mL of Milli-Q water, respectively. The tubes were placed on a rotating wheel for approximately 3 h at 4°C. Supernatant was collected into fresh 15-mL and 1.5-mL plastic tubes, respectively, and centrifuged for 20 min at 5,500g. Supernatant was collected into 50-mL plastic tubes and freeze dried overnight. Protein extracts were resuspended in 500 μ L and 200 μ L of Milli-Q water, respectively. The protein extracts were then separated by 1-D SDS-PAGE.
Water-Soluble Protein Extraction from Whole Grain, Isolated Outer Fraction, and Endosperm for Enzyme Activity Assays
Whole grain (10 g) was passed through a laboratory mill grain crusher (RM 001, custom made). Endosperm (5 g) was collected after passing through a 180- μ m sieve (Endecotts) and placed into a 50-mL plastic tube. The above protein extraction was repeated for endosperm, whole grain, and outer fraction up to the centrifugation step. The supernatant was collected in plastic tubes and stored at −20°C until needed for assaying.
Protein Estimation
Protein concentration was estimated using the Bradford method (Bio-Rad) with bovine serum albumin (Sigma-Aldrich) as the standard (Bradford, 1976).
2-DE
Prior to IEF, the samples were reduced with 5 mm tributylphosphine and alkylated in 10 mm acrylamide, followed by incubation for 1 h at room temperature. After incubation, the samples were centrifuged at 20,000g for 10 min at 4°C. Subsamples from each tissue (200 μ g of protein from aleurone cells, 25 μ g of protein from the intermediate layer, and 13 μ g of protein from the outer layer) were separated by IEF in triplicate using immobilized pH gradient (IPG) strips: 17-cm IPG strips pH 4 to 7 (Bio-Rad) and 18-cm IPG strips pH 6 to 11 (GE Healthcare). In the acidic pH range, samples were loaded (300 μ L) onto the IPG strips using passive hydration; for the alkaline pH range, the samples were cup loaded (120 μ L). IEF was performed with a step-wise protocol to 128 and 115 kV h−1, respectively. After IEF, the strips were stored at −80°C. The IPG strips were removed from the −80°C freezer, thawed at room temperature, and equilibrated in 6 m urea, 2% SDS, 0.375 m Tris-HCl, pH 8.8, 20% glycerol, 5 mm tributylphosphine, and 2.5% acrylamide for 15 min. This was repeated twice, and after each time the equilibration solution was poured off and replaced with fresh solution. IPG strips were embedded on top of 8% to 18% gradient polyacrylamide gels (17 cm × 17 cm) using hot agarose (0.5% agarose, 0.001% bromophenol blue, 192 mm Gly, 0.1% SDS, and 24.8 mm Tris base, pH 8.3). The gels were electrophoresed in Protean II multicell tanks (Bio-Rad) using a power box (Bio-Rad Power Pac 3000) set at 5 mA per gel for 30 min and then at 40 mA per gel for approximately 4.5 h or until the dye front had run off the gel. After electrophoresis, the gels were removed from their casts and placed into fixing solution (30% methanol and 7.5% acetic acid) for at least 1 h to prepare for Lava Purple (Fluorotechnics) staining. The solution was poured off and replaced with 200 mm sodium carbonate for 1 h to alkalinate the gels. Sodium carbonate solution was replaced with water, adding approximately 10 × the gel volume. Lava Purple stain was added to the water to make a final dilution of 1 in 200. The gels were covered with foil and left to stain overnight on a rocker. The stain was poured off and destained twice with 1% acetic acid. The gels were immediately scanned using a Typhoon variable mode imager (Amersham Biosciences). The gel image was scanned in fluorescence mode, 610 BP Deep Purple emission filter, green (532) laser, and with 100- μ m pixel resolution.
Proteomic Analysis
The scanned images were uploaded into the image-analysis software Sample Spots version 2.0 (Nonlinear Dynamics) to overlay the triplicate gel images and to determine true protein spots. These processed images were transferred to Progenesis PG240 version 2006 (Nonlinear Dynamics) software to annotate spots and to calculate spot volumes for each of the triplicate gels. The gels were all counterstained with colloidal Coomassie Brilliant Blue G-250 stain (17% ammonium sulfate, 3% phosphoric acid, 0.1% Coomassie Brilliant Blue G-250, 34% methanol, and Milli-Q water to make up 1 L total; Neuhoff et al., 1988) and left on a rocker overnight. Background destaining was done using 1% acetic acid. The Coomassie Brilliant Blue-stained images of all the gels were scanned on an automated spot cutter (Bio-Rad, EXQuest), and only spots that could be clearly seen with Coomassie Brilliant Blue stain were manually selected for excision and identification.
Gel plugs were manually destained three times in 120 μ L of wash solution (50% [v/v] acetonitrile and 25 mm ammonium bicarbonate). For each wash, the gel plugs were placed onto an orbital shaker and incubated at 37°C for 10 min, replacing wash solution each time. The gel plugs were vacuum dried using a Savant Speed Vac Plus SC210A.
To each dry gel plug were added 8- μ L portions of 15 ng μ L−1 sequencing grade trypsin (Promega) in 25 mm ammonium bicarbonate, pH 7.8, followed by incubation for 1 h at 4°C to allow the trypsin to be absorbed. Excess trypsin was removed, and the gel plugs were sealed and incubated overnight at 37°C. Peptides were extracted in 10 μ L of extraction solution (0.1% trifluoroacetic acid [TFA]) with the aid of a water bath sonicator (Transsonic 700/H; Elma) for 20 min.
Zip Tips were activated with 10 μ L of 70% acetonitrile and 0.1% TFA by pipetting 10 μ L up and down three times. The tips were washed with 0.1% TFA in the same manner. The peptide extraction solution was taken up into the Zip Tip, drawing up and down 8 μ L ten times to concentrate the peptides onto the column. The Zip Tip was further washed three times with 10 μ L of 0.1% TFA. Four microliters of extraction solution (4 mg mL−1 matrix, α -cyano-4-hydroxycinnamic acid, 70% acetonitrile, and 0.1% TFA) was drawn up into the Zip Tip. The extraction solution in the tip was drawn up and down at least five times, forming a drop at the end of the tip to elute the peptides from the column. Finally, 2 μ L of this solution was spotted onto a designated circle marked on the ABI plate. A standard (prep mix with matrix) was also spotted following each sample on the ABI plate to externally calibrate using near-point calibration with four peptide standards (bradykinin, angiotensin I, neurotensin, and adrenocorticotropic hormone fragment).
Samples were dried and analyzed using an Applied Biosystems 4700 MALDI MS/MS apparatus with time of flight (TOF)/TOF optics in reflector mode for positive ion detection. A Nd.YAG laser with wavelength and repetition rate of 355 nm and 200 Hz, respectively, was used. All MS spectra resulted from accumulation of 4,000 laser shots (20 subspectra were accumulated with 200 shots per subspectra). Laser intensity varied between 3,000 and 4,000. Data were collected over a mass range of 750 to 3,500 D. Mass spectral data were analyzed using Mascot (Matrix Science). Peak detection criteria for mass lists were as follows: for MS, mass range 500 to 4,000 D, maximum 30 peaks per 200 D, minimum signal-to-noise ratio of 20, minimum area 200, maximum peak/spot 200; for MS/MS, mass range 60 D to precursor −15, maximum 20 peaks per 200 D, minimum signal-to-noise ratio of 18, minimum area 300, maximum peak/spot 60. This converts the mass lists into Mascot- and The GPM-compatible text files. Mascot and X-Tandem were used to search cereal entries in the NCBI nonredundant databases and a translated wheat EST database (PlantGDB database Triticum aestivum EST assembly, February 2006). Identification of proteins with high scores and low e-values (less than −1.0) with good peptide matches and coverage was tabulated with corresponding spot number and location on the 2-DE gel. Some spots that did not reveal significant protein identification were further analyzed using electrospray ionization MS/MS.
Digested peptides were separated by nano-liquid chromatography (LC) using a CapLC system (Agilent 1100 Series; Agilent Technologies). Sample (39 μ L) was injected onto a peptide trap (Michrome peptide Captrap) for preconcentration and desalted with 0.1% formic acid at 10 μ L min−1. The peptide trap was then switched into line with the analytical column containing C18 RP silica (SGE ProteCol C18, 300A, 3 μ m, 150 μ m × 10 cm). Peptides were eluted from the column using a linear solvent gradient with steps from water:CH3CN (95:5 + 0.1% formic acid) to water:CH3CN (20:80 + 0.1% formic acid) at 600 nL min−1 over 45 min. The LC eluent was subjected to positive ion nanoflow electrospray analysis on an Applied Biosystems QSTAR XL mass spectrometer. The QSTAR was operated in an information-dependent acquisition mode in which a TOF/MS survey scan was acquired (mass-to-charge ratio 400–2,000, 1.0 s), with the four largest multiply charged ions (counts > 25) in the survey scan sequentially subjected to MS/MS analysis. MS/MS spectra were accumulated for 1 s (mass-to-charge ratio 50–2000). The LC-MS/MS data were used to search cereal entries in the NCBI nonredundant protein database using Mascot. High scores in the database searches indicated a likely match, confirmed or qualified by inspection of the spectra and search results.
The GPM was used as a secondary search engine using the PlantGDB for T. aestivum to both confirm the Mascot identity and to calculate a FDR for all MS/MS spectra. FDR was calculated using the mixture model according to Choi and Nesvizhskii (2007). Search results and analyses from both Mascot and The GPM are provided as Supplemental Tables S1 to S7 in Excel format. Protein identifications have only been listed when their Mascot scores are greater than 19. Of the 748 proteins identified in the supplemental tables, 98% (732) of the identifications have a Mascot score greater than 50 and/or greater than five matching peptides and/or greater than 25% peptide coverage.
OXO and POX Activity Assay
To measure OXO activity, 50 μ L of substrate solution (20 mm ammonium oxalate) was mixed with 50 μ L of o-phenylenediamine dihydrochloride (OPD; 0.08 g) in 10 mL of substrate buffer (0.1 m citric acid and 0.2 m disodium phosphate buffer, pH 5.0). Substrate solution for measuring POX activity was 100 μ L of OPD (0.04 g) in 10 mL of substrate buffer with 5 μ L of 35% peroxide. A portion (50 μ L) of each protein extract was placed into wells of a 96-well flat-bottom microtiter plate (Greiner Bio-One) in triplicate, together with 50 μ L of sodium succinate buffer. Finally, 100 μ L of the ammonium oxalate with OPD (OXO assay) and 100 μ L of the OPD with peroxide (POX assay) were added separately to each protein extract. Absorbance was read at 492 nm using a plate reader (Thermo Multiskan EX) at time intervals of 0, 30, 60, and 120 min.
OPD Standard Curve
Excess horseradish POX (Sigma) in sodium succinate buffer was added to the OPD solution (0.04 g of OPD in 10 mL of substrate buffer [0.1 m citric acid and 0.2 m disodium phosphate buffer, pH 5.0] and 5 μ L of 35% peroxide). A 50% dilution series was prepared upon reaction completion. A portion (200 μ L) of each dilution was added in duplicate to wells of a flat-bottom 96-well microtiter plate. Absorbance at 492 nm on a 96-well plate reader was used to calculate the micromoles of OPD in each well.
PPO Assay
The protocol for the PPO activity assay was modified from Espín et al. (1998). Substrate solutions hydroquinone monomethyl ether (25 mg) in 50 mL of water and 3-methylbenzothiazolin-2-one hydrazone (20 mg) in 2 mL of 70% ethanol were prepared separately. The two solutions were added together prior to the assay. Fifty microliters of each sample, together with 20 μ L of 1 m sodium acetate, pH 5, and 430 μ L of Milli-Q water, were placed into a 1-mL cuvette with a 1-cm light path. Finally, 500 μ L of substrate solution was added to each sample in the cuvette (Sarstedt). Absorbance was read at 492 nm at time points 0, 30, 60, and 120 min. The reaction rate in nkat g−1 protein was calculated using the hydroquinone monomethyl ether extinction coefficient (Espín et al., 1998).
Chitinase Assay
The chitinase assays for exochitinase (chitobioside substrate), exochitinase (β -N-acetylglucosaminidase), and endochitinase activity were performed in accordance with the manufacturer's instructions (Sigma; chitinase assay kit, colorimetric, product code CS0980). The assay was modified slightly such that the blank contained the sample in stop solution instead of the substrate in stop solution.
Generation of Antibodies and Affinity Purification
Polyclonal antibodies were generated in New Zealand White rabbits immunized with peptides to XIP-I (H-CNQNLGWEGSWDHKWTA-NH2 and H-AGGKTGQCSLIKYYA-OH), PR-4 (H-FTKIDTNGIGYQQGHC-NH2 and H-ATYHYYRCRDNN-OH), and OXO (H-AKAGNTSTPNGSAVTC-NH2 and H-TDPDPLQCSKFAAGF-OH) covalently linked to diphtheria toxoid using a thioether linkage. The polyclonal antibodies were affinity purified by conjugating free peptides to an activated beaded agarose SulfoLink Coupling Gel (Pierce) according to the manufacturer's instructions.
Sectioning and Microscopy
Wheat grains were soaked in phosphate-buffered saline (PBS) for 0.5 to 2 h at room temperature before both ends of the grain were removed with a razor blade. Grains were then fixed in 2% to 4% paraformaldehyde in PBS for at least 2 d at 4°C. Fixed grains were sectioned on a Leica VT 1000S Vibratome (speed 7, frequency 4) taking 60- to 100- μ m sections directly into PBS. All processing was performed at room temperature. Grain sections were blocked with 10% fetal bovine serum (FBS) in PBS for 30 min followed by primary antibody diluted in 10% FBS for 1 h. Sections were washed five times in PBS (5 min per wash) before secondary antibody (anti-rabbit AlexaFluor 488; Molecular Probes) diluted in 10% FBS was applied for 1 h in darkness. Sections were again washed five times in PBS before mounting the sections with Gel/Mount (Biomeda) and sealing the coverslips with nail polish. Sections were stored in the dark at 4°C overnight before visualizing on an Olympus IX 70 confocal microscope using Fluoview FV300 software.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Outer fraction protein separation. pI range 4 to 7.
Supplemental Figure S2. Outer fraction protein separation. pI range 6 to 11.
Supplemental Figure S3. Intermediate fraction protein separation. pI range 4 to 7.
Supplemental Figure S4. Intermediate fraction protein separation. pI range 6 to 11.
Supplemental Figure S5. Inner fraction (aleurone) protein separation. pI range 4 to 7.
Supplemental Figure S6. Inner fraction (aleurone) protein separation. pI range 6 to 11.
Supplemental Figure S7. Unrooted tree comparison of 7S globulin-like sequences from sequences identified in Figure 2 and shown in Table I.
Supplemental Figure S8. SDS-PAGE of water-soluble extracts of whole grain and pericarp.
Supplemental Table S1. Outer fraction protein identification. pI range 4 to 7.
Supplemental Table S2. Outer fraction protein identification. pI range 6 to 11.
Supplemental Table S3. Intermediate fraction protein identification. pI range 4 to 7.
Supplemental Table S4. Intermediate fraction protein identification. pI range 6 to 11.
Supplemental Table S5. Inner fraction (aleurone) protein identification. pI range 4 to 7.
Supplemental Table S6. Inner fraction (aleurone) protein identification. pI range 6 to 11.
Supplemental Table S7. Water-soluble fraction protein identification. SDS-PAGE.
Acknowledgments
We thank Debra Birch from Macquarie University for technical assistance and advice with the confocal microscopy and the technical staff at the Australian Proteome Analysis Facility for sharing their expertise in all aspects of the proteomic analysis.
References
- Antoine C, Peyron S, Lullien-Pellerin V, Abecassis J, Rouau X. (2004) Wheat bran tissue fractionation using biochemical markers. J Cereal Sci 39: 387–393 [Google Scholar]
- Antoine C, Peyron S, Mabille F, Lapierre C, Bouchet B, Abecassis J, Rouau X. (2003) Individual contribution of grain outer layers and their cell wall structure to the mechanical properties of wheat bran. J Agric Food Chem 51: 2026–2033 [DOI] [PubMed] [Google Scholar]
- Blein JP, Coutos-Thévenot P, Marion D, Ponchet M. (2002) From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci 7: 293–296 [DOI] [PubMed] [Google Scholar]
- Bonnin E, Daviet S, Gebruers K, Delcour JA, Goldson A, Juge N, Saulnier L. (2005) Variation in the levels of the different xylanase inhibitors in grain and flour of 20 French wheat cultivars. J Cereal Sci 41: 375–379 [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Breiteneder H, Mills ENC. (2005) Plant food allergens: structural and functional aspects of allergenicity. Biotechnol Adv 23: 395–399 [DOI] [PubMed] [Google Scholar]
- Caporale C, Caruso C, Facchiano A, Nobile M, Leonardi L, Bertini L, Colonna G, Buonocore V. (1999) Probing the modelled structure of Wheatwin1 by controlled proteolysis and sequence analysis of unfractionated digestion mixtures. Proteins 36: 192–204 [PubMed] [Google Scholar]
- Caruso C, Chilosi G, Caporale C, Leonardi L, Bertini L, Magro P, Buonocore V. (1999) Induction of pathogenesis-related proteins in germinating wheat seeds infected with Fusarium culmorum. Plant Sci 140: 87–97 [Google Scholar]
- Choi H, Nesvizhskii AI. (2007) False discovery rates and related statistical concepts in mass spectrometry-based proteomics. J Proteome Res 7: 47–50 [DOI] [PubMed] [Google Scholar]
- Demeke T, Morris C. (2002) Molecular characterization of wheat polyphenol oxidase (PPO). Theor Appl Genet 104: 813–818 [DOI] [PubMed] [Google Scholar]
- Deponte R, Parlamenti T, Petrucci V, Silano V, Tomasi M. (1976) Albumin α -amylase inhibitor families from wheat flour. Cereal Chem 53: 805–820 [Google Scholar]
- Desmond OJ, Edgar CI, Manners JM, Maclean DJ, Schenk PM, Kazan K. (2006) Methyl jasmonate induced gene expression in wheat delays symptom development by the crown rot pathogen Fusarium pseudograminearum. Physiol Mol Plant Pathol 67: 171–179 [Google Scholar]
- Donaldson PA, Anderson T, Lane BG, Davidson AL, Simmonds DH. (2001) Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotina sclerotiorum. Physiol Mol Plant Pathol 59: 297–307 [Google Scholar]
- Dunwell JM, Khuri S, Gane PJ. (2000) Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol Mol Biol Rev 64: 153–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espín JC, Tudela J, García-Cánovas F. (1998) 4-Hydroxyanisole: the most suitable monophenolic substrate for determining spectrophotometrically the monophenolase activity of polyphenol oxidase from fruits and vegetables. Anal Biochem 259: 118–126 [DOI] [PubMed] [Google Scholar]
- Gebruers K, Brijs K, Courtin CM, Goesaert H, Proost P, Van Damme J, Delcour JA. (2002) Affinity chromatography with immobilised endoxylanases separates TAXI- and XIP-type endoxylanase inhibitors from wheat (Triticum aestivum L.). J Cereal Sci 36: 367–375 [Google Scholar]
- Gebruers K, Debyser W, Goesaert H, Proost P, Van Damme J, Delcour JA. (2001) Triticum aestivum L. endoxylanase inhibitor (TAXI) consists of two inhibitors, TAXI I and TAXI II, with different specificities. Biochem J 353: 239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goesaert H, Elliott G, Kroon PA, Gebruers K, Courtin CM, Robben J, Delcour JA, Juge N. (2004) Occurrence of proteinaceous endoxylanase inhibitors in cereals. Biochim Biophys Acta 1696: 193–202 [DOI] [PubMed] [Google Scholar]
- Gorlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, Oostendorp M, Staub T, Ward E, Kessmann H, et al. (1996) Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8: 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge N. (2006) Plant protein inhibitors of cell wall degrading enzymes. Trends Plant Sci 11: 359–367 [DOI] [PubMed] [Google Scholar]
- Juge N, Payan F, Williamson G. (2004) XIP-I, a xylanase inhibitor protein from wheat: a novel protein function. Biochim Biophys Acta 1696: 203–211 [DOI] [PubMed] [Google Scholar]
- Juge N, Svensson B. (2006) Proteinaceous inhibitors of carbohydrate-active enzymes in cereals: implication in agriculture, cereal processing and nutrition. J Sci Food Agric 86: 1573–1586 [Google Scholar]
- Lane BG. (2002) Oxalate, germins, and higher-plant pathogens. IUBMB Life 53: 67–75 [DOI] [PubMed] [Google Scholar]
- Mak Y, Skylas DJ, Willows R, Connolly A, Cordwell SJ, Wrigley CW, Sharp PJ, Copeland L. (2006) A proteomic approach to the identification and characterisation of protein composition in wheat germ. Funct Integr Genomics 6: 322–337 [DOI] [PubMed] [Google Scholar]
- Marcus JP, Green JL, Goulter KC, Manners JM. (1999) A family of antimicrobial peptides is produced by processing of a 7S globulin protein in Macadamia integrifolia kernels. Plant J 19: 699–710 [DOI] [PubMed] [Google Scholar]
- Mauch F, Staehelin LA. (1989) Functional implications of the subcellular localization of ethylene-induced chitinase and β -1,3-glucanase in bean leaves. Plant Cell 1: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlin WR, Garcia-Conesa MT, Williamson G, Roza M, Ravestein P, Maat J. (1999) A novel class of protein from wheat which inhibits xylanases. Biochem J 338: 441–446 [PMC free article] [PubMed] [Google Scholar]
- Mellersh DG, Foulds IV, Higgins VJ, Heath MC. (2002) H2O2 plays different roles in determining penetration failure in three diverse plant-fungal interactions. Plant J 29: 257–268 [DOI] [PubMed] [Google Scholar]
- Mundy J, Hejgaard J, Svendsen I. (1984) Characterization of a bifunctional wheat inhibitor of endogenous [alpha]-amylase and subtilisin. FEBS Lett 167: 210–214 [Google Scholar]
- Neuhoff V, Arold N, Taube D, Ehrhardt W. (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9: 255–262 [DOI] [PubMed] [Google Scholar]
- Niranjan Raj SN, Sarosh BR, Shetty HS. (2006) Induction and accumulation of polyphenol oxidase activities as implicated in development of resistance against pearl millet downy mildew disease. Funct Plant Biol 33: 563–571 [DOI] [PubMed] [Google Scholar]
- Parker ML, Ng A, Waldron KW. (2005) The phenolic acid and polysaccharide composition of cell walls of bran layers of mature wheat (Triticum aestivum L. cv. Avalon) grains. J Sci Food Agric 85: 2539–2547 [Google Scholar]
- Payan F, Flatman R, Porciero S, Williamson G, Juge N, Roussel A. (2003) Structural analysis of xylanase inhibitor protein I (XIP-I), a proteinaceous xylanase inhibitor from wheat (Triticum aestivum, var. Soisson). Biochem J 372: 399–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekkarinen AI, Jones BL. (2003) Purification and identification of barley (Hordeum vulgare L.) proteins that inhibit the alkaline serine proteinases of Fusarium culmorum. J Agric Food Chem 51: 1710–1717 [DOI] [PubMed] [Google Scholar]
- Robert LS, Adeli K, Altosaar I. (1985) Homology among 3S and 7S globulins from cereals and pea. Plant Physiol 78: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffrath U, Freydl E, Dudler R. (1997) Evidence for different signaling pathways activated by inducers of acquired resistance in wheat. Mol Plant Microbe Interact 10: 779–783 [Google Scholar]
- Selitrennikoff CP. (2001) Antifungal proteins. Appl Environ Microbiol 67: 2883–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Kirubakaran SI, Sakthivel N. (2007) Heterologous expression of new antifungal chitinase from wheat. Protein Expr Purif 56: 100–109 [DOI] [PubMed] [Google Scholar]
- Skylas DJ, Mackintosh JA, Cordwell SJ, Basseal DJ, Walsh BJ, Harry J, Blumenthal C, Copeland L, Wrigley CW, Rathmell W. (2000) Proteome approach to the characterisation of protein composition in the developing and mature wheat-grain endosperm. J Cereal Sci 32: 169–188 [Google Scholar]
- Wang W, Scali M, Vignani R, Spadafora A, Sensi E, Mazzuca S, Cresti M. (2003) Protein extraction for two-dimensional electrophoresis from olive leaf, a plant tissue containing high levels of interfering compounds. Electrophoresis 24: 2369–2375 [DOI] [PubMed] [Google Scholar]
- Wong JH, Cai N, Balmer Y, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB. (2004) Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry 65: 1629–1640 [DOI] [PubMed] [Google Scholar]
- Yarullina LG, Troshina NB, Maksimov IV, Khairullin RM. (2005) The effect of pathogens and phytohormones on the rate of oxidation of phenols by oxalate oxidase in wheat seedlings. Biol Bull 32: 143–146 [PubMed] [Google Scholar]
- Yu GY, Muehlbauer GJ. (2001) Benzothiadiazole-induced gene expression in wheat spikes does not provide resistance to Fusarium head blight. Physiol Mol Plant Pathol 59: 129–136 [Google Scholar]