Abstract
Mitochondria play an essential role in nitric oxide (NO) signal transduction in plants. Using the biotin-switch method in conjunction with nano-liquid chromatography and mass spectrometry, we identified 11 candidate proteins that were S-nitrosylated and/or glutathionylated in mitochondria of Arabidopsis (Arabidopsis thaliana) leaves. These included glycine decarboxylase complex (GDC), a key enzyme of the photorespiratory C2 cycle in C3 plants. GDC activity was inhibited by S-nitrosoglutathione due to S-nitrosylation/S-glutathionylation of several cysteine residues. Gas-exchange measurements demonstrated that the bacterial elicitor harpin, a strong inducer of reactive oxygen species and NO, inhibits GDC activity. Furthermore, an inhibitor of GDC, aminoacetonitrile, was able to mimic mitochondrial depolarization, hydrogen peroxide production, and cell death in response to stress or harpin treatment of cultured Arabidopsis cells. These findings indicate that the mitochondrial photorespiratory system is involved in the regulation of NO signal transduction in Arabidopsis.
Nitric oxide (NO) has emerged as a new chemical messenger in plant biology. It can interact with a variety of intracellular and extracellular targets, acting as either a cytotoxic or a cytoprotective agent. NO stimulates seed germination in different species, and a decrease in NO levels has been associated with fruit maturation and senescence of flowers (Beligni and Lamattina, 2001). NO production has been observed in response to several biotic and abiotic stimuli, such as pathogen infection, bacterial elicitors, high temperature, osmotic stress, and UV-B light (Durner et al., 1998; Barroso et al., 1999; Krause and Durner, 2004; Zeidler et al., 2004; Shapiro, 2005; Corpas et al., 2008; Kolbert et al., 2008; Zhao et al., 2009).
Despite the proven importance of NO, little is known about signaling pathways downstream from it. During both programmed cell death and defense responses, NO requires cGMP and cADP Rib as secondary messengers (Wendehenne et al., 2001). Furthermore, NO activates mitogen-activated protein kinases in different plant species during stress signaling (Nakagami et al., 2005). However, direct biological activity of NO arises from chemical reactions between proteins and NO itself (Foster and Stamler, 2004; Dahm et al., 2006). S-Nitrosylation is a labile posttranslational modification with a half-life of seconds to a few minutes and represents a very sensitive mechanism for regulating cellular processes (Hess et al., 2005). More than 100 candidate S-nitrosylated proteins were identified from extracts of Arabidopsis (Arabidopsis thaliana) cultured cells treated with the NO donor S-nitrosoglutathione (GSNO) and from Arabidopsis leaves treated with gaseous NO (Lindermayr et al., 2005). Using the same proteomic approach, changes were characterized in S-nitrosylated proteins in Arabidopsis leaves undergoing a hypersensitive response (Romero-Puertas et al., 2008).
In animals, mitochondria play a crucial role in S-nitrosylation-dependent NO signaling (Foster and Stamler, 2004). The mitochondrion is an essential organelle for normal cellular function, being an important site of ATP synthesis and an integrator for apoptotic signaling (Skulachev, 1999). Mitochondria interact with NO at several levels. One particularly well-characterized example is the inhibition of complex IV (cytochrome c oxidase) via binding of NO to its binuclear CuB/heme a3 site (Cleeter et al., 1994). There are several reasons why S-nitrosylation may be an important mitochondrial regulatory mechanism. For example, mitochondria contain sizeable pools of thiols and transition metals, all of which are known to modulate nitrosothiol (SNO) biochemistry (Foster and Stamler, 2004). In addition, mitochondria are highly membranous and accumulate lipophilic molecules such as NO. Interesting in this respect is the fact that the formation of the S-nitrosylating intermediate N2O3 is enhanced within membranes (Burwell et al., 2006).
The role of mitochondria in stress-related responses has been investigated in both animals and plants. Endogenous nitrosylation of the catalytic Cys site of a subset of mitochondrial caspases serves as an on/off switch regulating caspase activity during apoptosis (Mannick et al., 2001). Moreover, cytochrome c, which is modified by NO at its heme iron during apoptosis, is released from mitochondria into the cytoplasm, which plays a critical role in many forms of apoptosis by stimulating apoptosome formation and subsequent caspase activation (Schonhoff et al., 2003). We previously showed that a prime target of NO in plants is the mitochondrial apparatus, causing an inhibition of KCN-sensitive respiration and an activation of alternative respiration via alternative oxidase (AOX; Huang et al., 2002; Krause and Durner, 2004; Livaja et al., 2008).
The aim of this study was to identify possible targets for S-nitrosylation in mitochondria of Arabidopsis leaves in order to gain more insight into the regulatory function of NO at the protein level. Using a proteomic approach involving the highly specific biotin-switch method for detection and purification of S-nitrosylated proteins (Jaffrey and Snyder, 2001) in conjunction with liquid chromatography and tandem mass spectrometry (nanoLC/MS/MS), we could identify 11 mitochondrial proteins as targets for S-nitrosylation. Among these identified proteins, we focused our attention on the P-subunit of the Gly decarboxylase complex (GDC), which is an integral part of the photorespiratory system. Since the release of apoptotic factors from mitochondria may be a result of inhibition of respiration, transition of mitochondrial permeability, and formation of reactive oxygen species (ROS; Saviani et al., 2002; Taylor et al., 2004; Chen and Gibson, 2008), we investigated the molecular mechanism and the function of GDC-Cys modification in Arabidopsis.
RESULTS
Harpin-Induced Generation of NO in Mitochondria
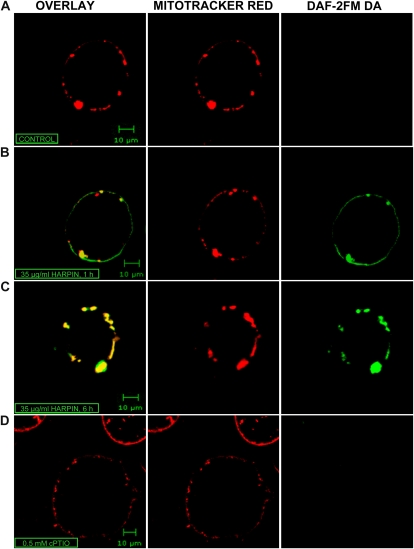
While a strict relationship between harpin-induced generation of NO in mitochondria was proven in different organisms and in different signaling events (Millar and Day, 1996; Cooper and Davies, 2000; Huang et al., 2002; Krause and Durner, 2004), direct evidence for NO production in plant leaf mitochondria is still missing. To elucidate NO localization in mitochondria, we treated Arabidopsis cultured cells with the bacterial elicitor harpin because we previously showed that this is able to induce NO production within a few hours (Krause and Durner, 2004). To analyze NO production in mitochondria, Arabidopsis cultured cells were loaded with the NO-specific fluorescent dye diaminofluorescein (DAF-2FM DA) and with the mitochondria-specific dye MitoTracker Red 580. Fluorescence was visualized by confocal laser microscopy (Fig. 1) as described (Foissner et al., 2000; Pedroso et al., 2000; Beligni et al., 2002; Yao et al., 2002). As a control, we applied carboxy-2-phenyl-4,4,5,5-tetramethylimidazolinone-3-oxide-1-oxyl (cPTIO), an NO scavenger that does not react with any ROS (Barchowsky et al., 1999). After a 1-h treatment with 35 μ g mL−1 harpin, an NO burst became visible in Arabidopsis mitochondria, with increasing intensity during the next hours (Fig. 1). The NO scavenger cPTIO was able to inhibit the elicited burst.
Figure 1.
Harpin-dependent NO production in mitochondria. Arabidopsis cell cultures were treated either with extracts of nontransformed E. coli (A) or with 35 μ g mL−1 recombinant partially purified harpin (B–D). Thirty minutes before microscopic analysis, cells were incubated with MitoTracker Red 580 as a mitochondria-specific dye (red fluorescence, center column) and DAF-2FM DA as an NO probe (green fluorescence, right column) and analyzed using a fluorescence confocal microscope (Zeiss LSM 510 NLO; 40 × water lens). For NO scavenging, cells were incubated with 0.5 mm cPTIO 20 min before application of the NO probe. Colocalization of both fluorescent signals appears yellow (left column). A, Cells treated with extracts of nontransformed E. coli. The images were taken 6 h after treatment. B and C, Cells were treated with recombinant harpin. The images were taken 1 h (B) and 6 h (C) after treatment. D, Cells were treated with recombinant harpin followed by incubation with 0.5 mm cPTIO and DAF-2FM DA. The images were taken 6 h after treatment. NO fluorescence was completely scavenged by the cPTIO preincubation.
Detection of S-Nitrosylated Proteins in Mitochondria
To identify S-nitrosylated proteins in mitochondria of Arabidopsis leaves, we used a modified biotin-switch method (Jaffrey and Snyder, 2001). The mitochondrial fraction from the leaves was prepared according to Keech et al. (2005). Since the mitochondrial proteome of Arabidopsis is well established (Kruft et al., 2001; Millar et al., 2001; Eubel et al., 2007), partially purified mitochondria were a useful experimental system to study posttranslational protein modification (Supplemental Fig. S1).
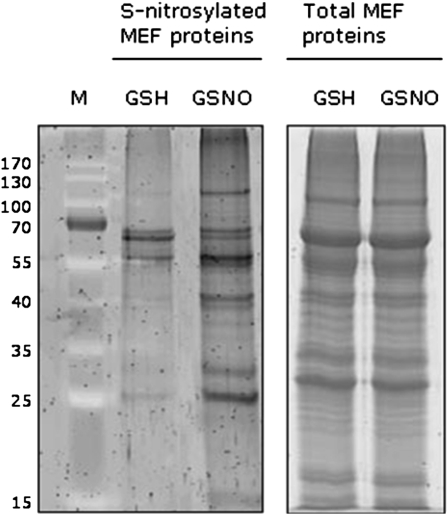
After treating mitochondrial proteins either with the trans-nitrosylating agent GSNO or with glutathione (GSH) as a negative control, S-nitrosylated proteins were biotinylated and separated by SDS-PAGE (Fig. 2; Supplemental Fig. S2). In order to identify S-nitrosylated proteins, GSNO- and GSH-derived eluates were subjected to nanoLC/MS/MS analysis. It was possible to identify 25 proteins with a significant score as judged by the Mascot search algorithm (score > 30). Five of these were chloroplast protein contaminations, while nine were not listed in the mitochondrial proteome (Millar et al., 2001) and did not harbor the mitochondrial targeting signal as identified by analysis using either TargetP 1.1 or iPSORT (Table I). Furthermore, mitochondrial fractions seemed to be contaminated by peroxisomes, as indicated by detection of catalase 3. Among 11 mitochondrial proteins in Table I, we could identify three subunits of the GDC, the key enzyme of the photorespiratory C2 cycle in plants, namely Gly dehydrogenase subunit P2 (15223217), Gly decarboxylase H1 (15226973), and Gly dehydrogenase subunit P1 (14596025).
Figure 2.
Detection of S-nitrosylated proteins in Arabidopsis mitochondria. Fifteen milligrams of protein of mitochondria-enriched fractions (MEF) was treated with 1 mm GSNO or GSH and labeled with biotin according to the biotin-switch method. Proteins were purified by affinity chromatography using neutravidin-agarose beads. A part of eluates was separated by 12% SDS-PAGE and visualized by SYPRO Ruby staining. Protein bands corresponding to predominant bands of the immunoblot analysis were identified by nanoLC/MS/MS. Additionally, to compare the amount of proteins in the different treatments, aliquots of mitochondria-enriched fractions were separated by SDS-PAGE (right gel). The relative masses of protein standards (M) are shown on the left (kD). The experiment was repeated three times with similar results.
Table I. S-Nitrosylated proteins from Arabidopsis mitochondria.
Mitochondria extracts treated with GSNO or GSH were subjected to the biotin-switch method and analyzed by nanoLC/MS/MS after tryptic digestion. The Mascot search engine was used to parse MS data to identify proteins from primary sequence databases. The best-matching peptide identifying the protein is given. If there were additional peptides found, the number of the peptides is given. The asterisks indicate proteins not listed in the mitochondrial proteome (Millar et al., 2001) but with mitochondrial targeting signal (TargetP). Proteome analysis was carried out by TopLab.
| Protein Identifier | Protein Name | Molecular Mass | Identified Peptides (Score) |
| D | |||
| 15223217 | Gly dehydrogenase subunit P2 | 18,000 | K.LTESPGLINSSPYEDGWMIK.V (87) +2 |
| 15221119 | Aminomethyltransferase | 44,759 | R.TGYTGEDGFEISVPDEHAVDLAK.A (80) +10 |
| 15235745 | Ser hydroxymethyltransferase | 57,535 | K.LIVAGASAYAR.L (70) +9 |
| 15226973 | Gly decarboxylase H1 | 18,050 | K.LTESPGLINSSPYEDGWMIK.V (87) +3 |
| 14916970 | ATP synthase subunit α | 55,296 | K.AVDSLVPIGR.G (44) +4 |
| 18394888 | Catalase 3 | 57,059 | R.LGPNYLQLPVNAPK.C (54) +4 |
| 30684419 | Lipoamide dehydrogenase 2 | 54,237 | K.HIIVATGSDVK.S (47) +7 |
| 15221044 | Lipoamide dehydrogenase 1 | 54,239 | K.HIIVATGSDVK.S (47) +4 |
| 79401911 | Unknown protein* | 78,638 | K.GSFSSVVSDK.S (42) +7 |
| 14596025 | Gly dehydrogenase subunit P1 | 113,852 | R.EYAAFPAPWLR.S (35) +11 |
| 21537215 | Unknown protein* | 32,979 | K.SNMSNCETSSEIQKPDYIHVR.A (28) +1 |
Mass Spectrometric Analyses of GSNO-Treated P Protein
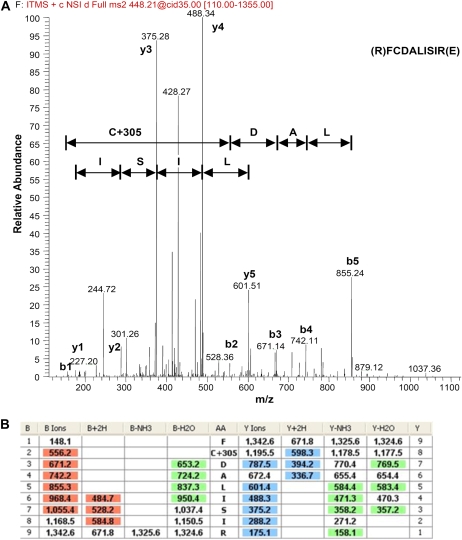
GSNO is able to S-nitrosylate or S-glutathiolate Cys residues, while both modifications can have different effects on protein conformation and activity. We investigated which kind of modification(s) resulted from the GSNO treatment of GDC, focusing our attention on the decarboxylating subunit, the P protein. In Arabidopsis, there are two genes encoding the P subunit, which share high homology. Since the production of recombinant P protein was not successful, we partially purified the P subunit from Arabidopsis mitochondria as described (Bourguignon et al., 1988) and tested its integrity by assaying it for the CO2-exchange reaction (Supplemental Fig. S3). S-Nitrosylation of this partially purified P protein was confirmed using the biotin-switch method (data not shown). In addition, we used mass spectrometric analyses to detect GSNO-mediated modifications of the Cys residues, since this method allows the detection of both S-nitrosylation and S-glutathionylation. Protein S-glutathionylation is promoted by nitrosative stress and transient S-nitrosylation, but it also occurs in unstressed cells (Dalle-Donne et al., 2009). After treating the P protein with GSNO and digesting it with trypsin, we analyzed the Cys-containing peptides for their modifications (Fig. 3; Supplemental Fig. S4). C402 and C463 were found to be S-glutathionylated after all treatments. In contrast, C98, C943, C777, and C1022 were S-glutathionylated only after treatment with 1 mm GSNO (Table II). No S-nitrosylated Cys residues were detected by mass spectrometric analysis, probably because of the transient nature of this modification at the P protein, as reported for several other proteins (Martinez-Ruiz and Lamas, 2007).
Figure 3.
Mass spectrometric analyses of partially purified GSNO-treated P protein. For mass spectrometric analyses, partially purified P protein was digested with trypsin at 37°C for 1 h, pH 6.5. Digested proteins were analyzed by nanoLC/MS/MS, with an automatic switch between MS, MS2, and MS3 acquisition. A, The S-glutathionylation was detected in the MS2 as a gain of 305 kD in mass. The spectra represent the MS3 fragmentation of the peptide FCDALISIR containing the glutathionylated C943. The spectrum is characterized by y-ion (y1–y5) and b-ion (b1–b5) series and shows that C943 was modified with GSH, as indicated by the presence of a b2-ion at m/z 556.2 (mass shift +305 D). B, Sequencing table showing the shift of C943. The table details peptide fragments (AA, amino acids; B, b-ions having the charge retained on the N-terminal fragment; Y, y-ions having the charge retained on the C-terminal fragment), b-ions and y-ions (single and double charged), and the neutral losses of ammonia and water for b-ions and y-ions.
Table II. Mass spectrometric analyses of the partially purified P protein.
P protein (20 μ g) was treated with 0.5 or 1 mm GSNO for 15 min. After removing the excess of GSNO, the protein was digested with trypsin and analyzed as described in “Materials and Methods.” Six peptides belonging to the P subunit (P1 gene) of the GDC showed a mass difference compared with the expected one of 305 D, which corresponds to the addition of one −GS group. No peptide showed a shift of 29 D (−NO) in the mass. The asterisks indicate peptides that showed a shift in mass after both the GSNO treatments.
| Peptide Sequence | Peptide Identification Probability | Modifications Identified by Spectrum | Actual Peptide Mass | Peptide Start | Peptide Stop |
| % | |||||
| FCDALISIR | 95 | GSH (+305) | 1,341.61 | 942 | 950 |
| FCGFDHIDSLIDATVPK | 95 | GSH (+305) | 2,181.97 | 97 | 113 |
| TFCIPHGGGGPGMGPIGVK | 95 | GSH (+305) | 2,085.94 | 775 | 793 |
| DKATSNICTAQALLANMAAMYAVYHGPAGLK* | 95 | GSH (+305) | 3,498.65 | 395 | 425 |
| CSDAHAIADAASK* | 95 | GSH (+305) | 1,563.63 | 463 | 475 |
| LVCTLLPEEEQVAAAVSA | 95 | GSH (+305) | 2,147.02 | 1,020 | 1,037 |
Inhibition of Gly Decarboxylase Activity by GSNO
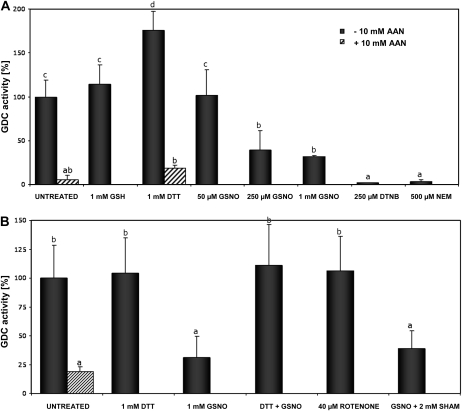
To elucidate the effects of S-nitrosylation/S-glutathionylation on GDC activity, we treated mitochondria with GSNO and monitored the GDC activity (Walker et al., 1982; Walker and Oliver, 1986). Following 14CO2 release from [1-14C]Gly (i.e. the activity of P and H subunits), we could observe a significant reduction of decarboxylation, which was dependent on GSNO concentration (Fig. 4A). After 20 min, the GDC activity of mitochondria incubated with 0.25 mm GSNO showed an inhibition of 60% as compared with the GDC activity of untreated organelles. Treatment with 1 mm GSNO further decreased the decarboxylation to about 30%, similar to the inhibition obtained with the GDC inhibitor aminoacetonitrile (AAN; 10 mm). The incubation of mitochondria with the reducing agents dithiothreitol (DTT) and GSH increased decarboxylation activity by 76% and 15% in vivo, respectively, demonstrating the redox-dependent regulation of the P protein and suggesting a change in the mitochondrial redox status during purification. To further characterize the redox dependency of GDC activity, we tested the effects of two Cys-modifying agents on the enzymatic activity: N-ethylmaleimide, a highly reactive agent that covalently and irreversibly alkylates free Cys thiol groups, and 5,5′-dithiobis-(2-nitrobenzoic acid), an oxidizing reagent. Both treatments resulted in a complete inhibition of P/H protein activity within 20 min (Fig. 4A).
Figure 4.
Modulation of Gly decarboxylase activity by thiol-modifying agents. A, Mitochondria-enriched fractions were preincubated with the indicated amounts of GSNO or SH-modifying agents for 20 min at room temperature in the dark, and enzyme activity was determined subsequently. The activity of the control (untreated) was set to 100%. B, Leaf slices were treated for 20 min with GSNO, rotenone, or SHAM at room temperature after equilibration in a MOPS buffer for 1 h at 30°C. For restoring GDC activity, 1 mm DTT was added to the inhibited enzymes and incubated for an additional 10 min. Treatment with the GDC inhibitor AAN (10 mm) served as a control. The activity of controls (untreated) was set to 100%. Presented data are means ± sd of three independent experiments. Different letters indicate values statistically different based on a one-way ANOVA followed by Tukey's honestly significant difference posthoc test (A, F = 22.46, df = 9, P < 0.01; B, F = 6.16, df = 4, P < 0.01).
Modulation of GDC activity by GSNO was confirmed using Arabidopsis leaf slices as described by Navarre and Wolpert (1995). Incubation with 1 mm GSNO for 20 min resulted in an inhibition of GDC activity by 75%, which could be restored by 1 mm DTT (Fig. 4B). Moreover, to test if modulation of GDC activity was due to a direct effect of GSNO on the photorespiratory system, Arabidopsis leaf slices were incubated with inhibitors of complex I of the mitochondrial respiration system (rotenone) and of AOX (salicylhydroxamic acid [SHAM]). In animal systems, complex I has been shown to be regulated by S-nitrosylation and to be involved in reversible ROS production (Borutaite and Brown, 2006; Burwell et al., 2006). In plants, it was shown that the NO-induced AOX suppressed the production of ROS and cell death induced by KCN-sensitive respiration inhibition (Huang et al., 2002). Therefore, the inhibition of GDC activity after GSNO treatment could have been an indirect effect of ROS induced by inhibition of complex I. It is well known that the activity of complex I of the mitochondrial electron transport chain is inhibited by nitrosothiols and peroxinitrite (Carreras et al., 2004; Dahm et al., 2006). This inhibition results in increased production of ROS, which can cause oxidative damage of GDC (Taylor et al., 2002). However, blocking of complex I in mitochondria via rotenone did not affect GDC activity within 30 min (Fig. 4B). Furthermore, neither increased ROS levels due to AOX inhibition nor reduced ROS levels due to catalase-dependent scavenging of ROS (data not shown) influenced the modulation of GDC activity.
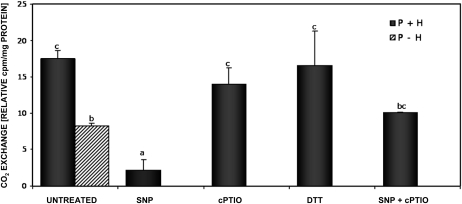
Since it was not possible to demonstrate GSNO-dependent S-nitrosylation of the P protein via MS, we tested the effect of sodium nitroprusside (SNP) on the activity of partially purified P protein. SNP is an NO donor that is not able to S-glutathionylate Cys residues. As shown by Fig. 5, 250 μm SNP was able to inhibit the CO2-exchange reaction, while the NO scavenger cPTIO (500 μm) almost completely prevented it. These data suggest that the activity of the P subunit of the GDC is inhibited by NO.
Figure 5.
CO2-exchange reaction modulated by NO. The activity of P protein was determined by measuring the amounts of [14C]bicarbonate fixed to the carboxyl group carbon atom of Gly in the presence of a saturating amount of H protein. Proteins were treated with 1 mm DTT, 250 μm SNP, and/or 500 μm cPTIO for 10 min, and the activity was measured 30 min after the addition of bicarbonate. As a source of H protein, 100 μ g of proteins of the light fraction from size-exclusion chromatography was used. P protein derived by chromatography, 30 μ g; black bars, H + P proteins; striped bar, P protein alone. Each value represents the mean of at least three technical replicates. Different letters indicate values statistically different based on a one-way ANOVA followed by Tukey's honestly significant difference posthoc test (F = 12.8, df = 5, P < 0.01).
Inhibition of Gly Decarboxylase Activity by Harpin
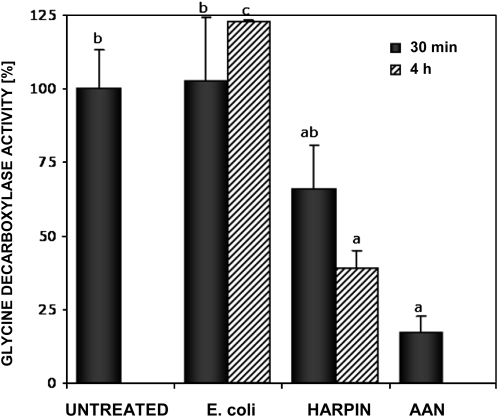
Since protein S-nitrosylation is emerging as a specific and fundamental mechanism in NO signal transduction (e.g. during plant-pathogen interactions; Romero-Puertas et al., 2004), we tested the involvement of GDC inhibition in a stress-related response. Harpin is a proteinaceous bacterial elicitor of Erwinia, Pseudomonas, and Xanthomonas strains that can induce an oxidative burst and programmed cell death in various host plants. Leaf slices were incubated at room temperature with 35 μ g mL−1 recombinant harpin protein or with an extract of untransformed Escherichia coli DH5 α as a control. GDC activity was recorded over a 4-h period. An inhibition of Gly decarboxylation in harpin-treated leaves was detected after 30 min and increased after 4 h to 61% in comparison with untreated and E. coli-treated samples (Fig. 6).
Figure 6.
Modulation of Gly decarboxylase activity by harpin. Leaf slices were treated with 35 μ g mL−1 recombinant harpin or with an extract of nontransformed E. coli at room temperature after equilibration in MOPS buffer for 1 h at 30°C. Treatment with the GDC inhibitor AAN was used as a control. The Gly decarboxylase activity in untreated control was set to 100%. Data shown are means ± sd of three independent experiments. Different letters indicate values statistically different based on a one-way ANOVA followed by Tukey's honestly significant difference posthoc test (F = 7.8, df = 5, P < 0.01).
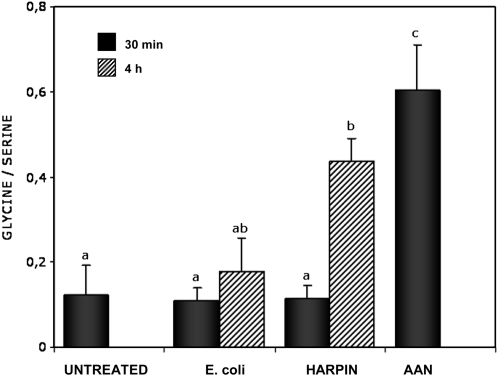
The Gly-Ser interconversion, catalyzed by GDC in conjunction with the Ser hydroxymethyltransferase, is an integral part of the photorespiratory metabolic pathway (Douce et al., 2001; Bauwe and Kolukisaoglu, 2003). Therefore, we investigated the role of GDC during plant defense responses in vivo by monitoring the amino acid content in Arabidopsis leaves (Blackwell et al., 1990; Abe et al., 1997; Heineke et al., 2001). The concentration of harpin used was able to cause cell death after 24 h when infiltrated into Arabidopsis leaves, but not after 30 min (in vitro experiments; data not shown). The inhibition of GDC activity observed after harpin treatment led to a distinctly elevated leaf Gly-Ser ratio after 4 h relative to the control treatment with E. coli extract (Fig. 7). As a second control, leaves were infiltrated with the GDC inhibitor AAN, which also resulted in a fast increase in Gly-Ser within 30 min (Fig. 7).
Figure 7.
Determination of Gly-Ser ratio to monitor GDC inhibition. The inhibition of the activity of the GDC was monitored following the increase in Gly content and decrease in Ser content in Arabidopsis leaves. Amino acid content analysis was performed using 100 mg of fresh tissue and a HPLC system. Each value represents the ratio between Gly and Ser (μ mol g−1 fresh weight). Leaves were treated for the indicated times with extracts of nontransformed E. coli or recombinant harpin (35 μ g mL−1). The GDC inhibitor AAN was used as a control. Experiments were repeated three times with similar results. Different letters indicate values statistically different based on a one-way ANOVA followed by Tukey's honestly significant difference posthoc test (F = 8.5, df = 5, P < 0.01).
Mitochondrial and Cellular Responses to GDC Inhibition
To confirm the role of GDC in the harpin-induced plant defense response, Arabidopsis plants and cultured cells were treated with the GDC inhibitor AAN, which has no side effects on dark respiration and CO2 fixation (Usuda and Edwards, 1980; Creach and Stewart, 1982) and is able to mimic the toxin victorin-induced mitochondrial oxidative burst in oat (Avena sativa) cells (Yao et al., 2002).
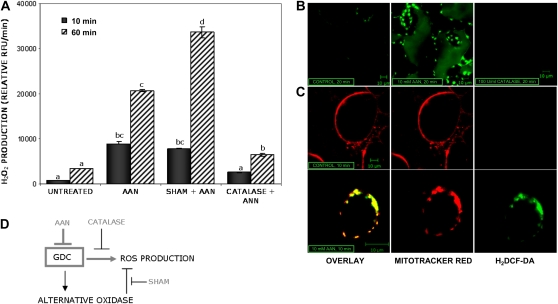
Stress-induced ROS generation has been reported as an essential signal for activation of resistance in several plants (Desikan et al., 1998; Delledonne et al., 2001; Krause and Durner, 2004). Energy dissipation mechanisms such as photorespiration or the water-water cycle (Asada, 2006) prevent excessive photoreduction of oxygen, which could potentially generate an excess of ROS, leading to photooxidation (Kozaki and Takeba, 1996; Osmond et al., 1997). Thus, inhibition of GDC should result in enhanced ROS. Photometric determination of hydrogen peroxide (H2O2) was performed using the ROS-specific fluorescent dye dichlorofluorescein (H2DCF-DA; Allan and Fluhr, 1997; Yao et al., 2002). ROS production in Arabidopsis cultured cells was monitored over time with a fluorescence plate reader (Fig. 8A), whereas the localization of ROS in the cells was analyzed with a confocal microscope (Fig. 8, B and C). The fluorescent signal of ROS increased already 10 min after addition of AAN to 1-week-old cultured cells, whereas untreated cells showed almost no fluorescence even after 60 min. In the presence of Gly, ROS accumulation increased dramatically (Supplemental Fig. S5), which could possibly result from glyoxylate-induced ROS production in peroxisomes. Catalase completely scavenged the H2O2 produced in the assay, demonstrating the specificity of the fluorescent dye. In contrast, pretreatment with the AOX inhibitor SHAM increased H2O2 accumulation after 60 min in Arabidopsis cells treated with AAN. H2O2 production after inhibition of the Gly decarboxylase was further confirmed in Arabidopsis leaves using H2DCF-DA. The site of ROS production was identified with double staining using MitoTracker Red 580 (Yao et al., 2002; Krause and Durner, 2004). The intracellular localization of H2O2 production in the AAN-treated samples matched that of the mitochondria-staining dye (Fig. 8C).
Figure 8.
AAN-dependent ROS production in cultured cells and leaves of Arabidopsis. A, AAN, an inhibitor of the GDC, was used to study the effect of GDC inhibition in cultured cells of Arabidopsis. The ROS-dependent fluorescence of the H2DCF-DA dye was monitored with a Tecan GENios fluorescence plate reader. Catalase was used to scavenge H2O2 production in the assay, and its relative fluorescence unit (RFU) value, corresponding to the dye background, was subtracted from other samples. SHAM, 2 mm; catalase, 100 units mL−1; AAN, 10 mm. Data shown are means ± sd of three independent experiments. Different letters indicate values statistically different based on a one-way ANOVA followed by Tukey's honestly significant difference posthoc test (F = 166.2, df = 7, P < 0.01). B, Arabidopsis leaf slices were incubated with 10 μm ROS-sensitive dye (H2DCF-DA). After 20 min, the leaf slices were treated with 10 mm AAN for 20 min (middle) or buffer (control; left). Produced H2O2 was scavenged by preincubation with 100 units mL−1 catalase (right). Confocal microscopy images were obtained using Zeiss LSM 510 NLO with a 40 × water lens. C, Colocalization of mitochondria and ROS production in Arabidopsis. Cells were incubated with 0.5 μm MitoTracker Red 580 as a specific mitochondrial marker (red fluorescence, center column) and subsequently with H2DCF-DA (10 μm; green fluorescence, right column). After 20 min, cells were treated either with 10 mm AAN or buffer for 10 min. Confocal microscopy images were obtained using Zeiss LSM 510 NLO with a 40 × water lens. The experiment was repeated three times with similar results. D, Model proposed to explain the relation between GDC and AOX in modulating ROS production.
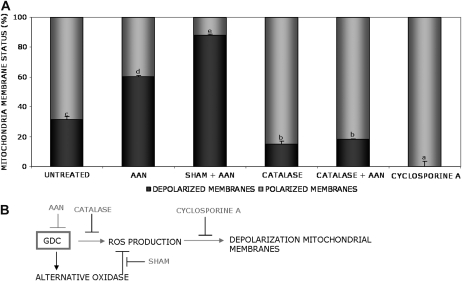
Plant mitochondrial responses to stress signals are similar to those of animal mitochondria and include a collapse of the transmembrane potential (Δ Ψm), release of cytochrome c, and decrease in ATP production (Saviani et al., 2002; Tiwari et al., 2002; Krause and Durner, 2004). Normally, the mitochondrial H2O2 burst appears to precede the breakdown in the Δ Ψm, probably as a result of perturbation of the mitochondrial electron transport chain (Pellinen et al., 1999; Yao et al., 2002). JC-1 was used as a dye to probe the mitochondrial membrane potential in AAN-treated cultured cells of Arabidopsis. At 30 min after treatment with AAN, approximately 50% of cells showed a decrease in Δ Ψm compared with 30% of the control cells (Fig. 9A). The depolarization observed in the control was most likely due to a stress condition of cells in the plate reader and was completely suppressed by pretreatment with cyclosporine A, which is an inhibitor of the mitochondrial transition pore (Saviani et al., 2002). In addition, catalase was able to reduce the Δ Ψm in untreated or AAN-treated cells, confirming that the oxidative burst is essential for the breakdown of the mitochondrial membrane potential (Fig. 9A). Furthermore, blocking of the ROS-scavenging ability of AOX (via SHAM) resulted in increased loss of red fluorescence, with approximately 90% of mitochondrial membranes depolarized (Fig. 9).
Figure 9.
AAN-induced depolarization of mitochondrial membranes. AAN was used to study the effect of GDC inhibition in Arabidopsis. A, One-week-old cultured Arabidopsis cells were treated for 30 min with different inhibitors on the 96-well plate in the grow chamber. Afterward, JC-1 dye (5 μ g mL−1), to probe the mitochondrial membrane potential, and 10 mm AAN were added and the measurement was started. The mitochondrial depolarization was indicated by a decrease in the red-green fluorescence intensity ratio. The data represent the status of the mitochondrial membrane 30 min after AAN treatment. Black bars indicate the percentage of depolarization. SHAM, 2 mm; catalase, 100 units mL−1; cyclosporine A, 50 μm. Cyclosporine A was used as a control for complete polarization (100%). The experiment was repeated three times with similar results. Different letters indicate values statistically different based on a one-way ANOVA followed by Tukey's honestly significant difference posthoc test (F = 532.6, df = 7, P < 0.01). B, Model proposed to explain the relation between GDC and AOX in modulating depolarization mitochondrial membranes.
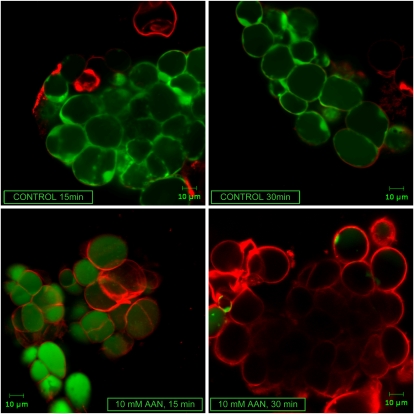
From experiments with toxins of plant pathogens or mutants of mitochondrial proteins, it seems that loss of mitochondrial function and concomitant mitochondrial ROS formation and signaling are necessary for subsequent cell death (Robson and Vanlerberghe, 2002; Vanlerberghe et al., 2002; Yao et al., 2002; Krause and Durner, 2004). To monitor AAN-induced cell death, we measured the loss of plasma membrane integrity by double staining of Arabidopsis suspension cells with Evans blue and fluorescein diacetate (Huang et al., 2002). Both fluorescent signals were analyzed by confocal microscopy. Untreated cells showed clear green fluorescence, while 15 to 30 min after treatment with AAN, a decrease in fluorescein signal and an increase in red fluorescence could be observed (Fig. 10).
Figure 10.
Visualization of AAN-induced cell death in Arabidopsis cell cultures. Confocal microscopy images were obtained using Zeiss LSM 510 NLO with a 40 × water lens. Cell death, measured as loss of plasma membrane integrity, was detected by double staining with Evans blue (red fluorescence, dead cells) and fluorescein diacetate (green fluorescence, living cells). After incubation with the dyes for 5 min, 10 mm AAN was added to the cell cultures and the fluorescence was visualized. The experiment was repeated three times with similar results.
DISCUSSION
Programmed cell death as observed in the form of a hypersensitive response in several plant-pathogen interactions exhibits similarities to programmed cell death as seen in animal apoptosis, including chromatin condensation and fragmentation (Mur et al., 2008) and caspase-like proteolytic activity (Lam and del Pozo, 2000). Furthermore, it has been shown that NO and ROS play major regulatory and antimicrobial roles in plant defense responses (Delledonne et al., 2001). In animal cells, mitochondria are major sites of the formation of redox compounds and major targets of NO/ROS-induced damage (Skulachev, 1996). By contrast, large quantities of ROS and NO are produced in plastids and peroxisomes of plants, especially in photosynthetic cells (del Rio et al., 2003). Nonetheless, plant mitochondria also are responsible for the production of ROS and are a site of oxidative damage (Taylor et al., 2002). In this paper, we not only showed that NO and ROS accumulate in Arabidopsis mitochondria during stress responses but also that NO is capable of modulating an essential mitochondrial function, photorespiratory metabolism, as an integral part of this response.
Inhibition of GDC by Harpin and NO
GDC, together with Ser hydroxymethyltransferase, is responsible for the conversion of Gly to Ser during photorespiration. During this metabolic process, CO2 and NH3 are produced while ATP and reducing equivalents are consumed, which makes photorespiration an energetically wasteful process (Wingler et al., 2000). It has been described that oat GDC is involved in the response to the toxin victorin produced by the fungus Cochliobolus victoriae (Navarre and Wolpert, 1995; Yao et al., 2002). The P subunit of the GDC was found to bind victorin in vivo, but only in susceptible genotypes. The H subunit bound victorin in vivo in both susceptible and resistant genotypes (Navarre and Wolpert, 1995). In addition, application of the GDC inhibitor AAN resulted in apoptosis-like cell death and disease symptoms (Yao et al. 2002). In this study, we observed a clear modulation of the GDC activity by redox agents in both intact mitochondria and plant leaves following 14CO2 release from [1-14C]Gly (i.e. the H-dependent P protein reaction). The decarboxylation activity of GDC was significantly inhibited by GSNO as well as after blocking of the Cys residues with Cys-modifying chemicals. By contrast, strongly reducing agents such as DTT promoted GDC activity (Fig. 4A). The requirement of a reduced environment for both CO2 exchange and decarboxylase activity of GDC in vitro has been described previously (Hiraga and Kikuchi, 1980; Walker and Oliver, 1986). Inactivation of GDC after treatment of mitochondria with N-ethylmaleimide or 5,5′-dithiobis-(2-nitrobenzoic acid) showed that Cys residues are important for the enzymatic activity.
In animal systems, the toxicity of NO has been associated with inhibition of cellular respiration (Clementi et al., 1998). Interaction of NO with cytochrome c oxidase (complex I) has dramatic biological consequences, such as the increase in the mitochondrial ROS level and the induction of apoptosis (Brunori et al., 1999). Therefore, the reason for NO toxicity is not a shutdown of respiration per se but an overreduction of the ubiquinone pool. This inhibitory effect of NO on cytochrome c oxidase has also been described for soybean (Glycine max) and mung bean (Vigna radiata) mitochondria (Millar and Day, 1996; Yamasaki et al., 2001). Consequently, the modulation of GDC activity observed after GSNO treatment could be a secondary effect due to inhibition of the mitochondrial electron transport chain and subsequent accumulation of ROS. Using inhibitors of complex I (rotenone) and AOX (SHAM), we showed that a direct action of NO on the GDC is responsible for its decreased activity (Fig. 4B).
Posttranslational Modification of GDC by NO
MS analysis of partially purified P protein (Table II) suggested the possibility that the modulation of the GDC was due to an S-glutathionylation and not to an S-nitrosylation of specific Cys residues. Although the biotin-switch method is specific for detection of SNO-modified Cys, we tested the activity of the purified P protein after treatment with the nonglutathionylating agent SNP and analyzed if NO is able to modify GDC activity (Fig. 5). The inhibition of GDC activity observed after SNP treatment and the ability of cPTIO to scavenge this effect proved that NO is able to modulate GDC activity per se, confirming the strict connection between the two posttranslational modifications. In general, the role of S-nitrosothiols as primers for S-glutathionylation should be considered, with special emphasis given to GSNO as a reagent that can induce not only S-nitrosylation but also S-glutathionylation of proteins (Martinez-Ruiz and Lamas, 2007). In several proteins, “competition” between S-glutathionylation and S-nitrosylation has been compared, which showed that there is considerable variation among different proteins regarding their tendencies to undergo either modification.
The modulation of a key enzyme of the photorespiratory system via S-nitrosylation suggests a direct role of this metabolic pathway in the regulation of the redox state of the cell. In addition, GDC was found in root and etiolated tissue of different plants, in C4 plants, and in a wide range of organisms from bacteria to humans. Taken together, the data suggest a nonphotorespiratory role of the GDC complex (Navarre and Wolpert, 1995). For this reason, we investigated the state of GDC during a plant stress response. Using the Arabidopsis-harpin interaction as a model system (Xie and Chen, 2000; Krause and Durner, 2004; Livaja et al., 2008), we demonstrated that GDC activity is inhibited in Arabidopsis leaves after harpin treatment (Figs. 6 and 7). Thus, harpin inhibits not only the cytochrome c-dependent respiration (Xie and Chen, 2000; Livaja et al., 2008) but also the photorespiratory system in Arabidopsis. This reduces the cellular capacity of ATP synthesis and increases the production of ROS, which leads to an overreduction and overenergization of the entire cell. Recent studies revealed the importance of energy dissipation in mitochondria. Oxidative stress by excess light is involved in the regulation of respiratory gene expression and the modulation of respiratory properties, especially up-regulation of AOX (Yoshida et al., 2006, 2007, 2008).
Potential Involvement of GDC in Plant Defense Responses
The possible role of GDC in pathogen responses has been investigated upon victorin treatment of oat. In this system, application of the GDC-specific inhibitor AAN resulted in a highly localized accumulation of H2O2 within mitochondria followed by disease symptoms similar to those induced by victorin and during apoptosis-like cell death (Yao et al., 2002). Here, we demonstrated that AAN is able to induce an oxidative burst in cultured cells and in leaves of Arabidopsis (Fig. 8). Whereas catalase was able to scavenge AAN-induced ROS, inhibition of AOX (by SHAM) further increased the ROS signal 1 h after incubation (Fig. 8). The use of a mitochondria-specific dye allowed us to localize the ROS burst in Arabidopsis mitochondria, although peroxisomal ROS production cannot by excluded, since peroxisomes are localized next to mitochondria. The direct relationship between inhibition of GDC activity and ROS levels is difficult to explain. The oxidation of Gly by GDC provides NADH for the mitochondrial electron transport chain. Inhibition of GDC would lead to undersupply of NADH to the respiratory chain. This shortage of the electron donor pool might cause the production of partially reduced oxygen species. However, there might be other, still unknown connections between the inhibition of GDC on the one hand and ROS production as a result of perturbation of the energy metabolism in mitochondria on the other hand.
In animal cells, the increase of mitochondrial ROS, together with an elevation of cytosolic Ca2+, contributes to the opening of the mitochondrial permeability transition pore (PTP). The PTP depolarizes mitochondria, leading to mitochondrial swelling and subsequent release of cytochrome c from the intermembrane space (Goldstein et al., 2000). It has been demonstrated that the mitochondrial PTP participates in NO-induced cell death in plants (Saviani et al., 2002). In concurrence, our data show a 50% loss of membrane potential in Arabidopsis cells 30 min after treatment with AAN, whereas this loss is limited by the action of the alternative respiratory pathway (Fig. 9). Moreover, the scavenging effect of catalase proves that ROS are directly involved in activating depolarization of mitochondrial membranes in plants. Among the proteins released from mitochondria after depolarization of the mitochondrial membrane are cytochrome c and apoptosis-inducing factor. The latter protein moves directly to the nucleus, where it causes chromatin condensation and nuclear fragmentation. Cytochrome c activates a caspase signaling cascade that selectively cleaves vital substrates in the cell, including the nuclease responsible for DNA fragmentation (Green and Reed, 1998). In agreement with this scheme of actions, inhibition of Arabidopsis GDC by AAN resulted in cell death (Fig. 10).
In conclusion, we showed that GSNO is able to modulate the photorespiratory pathway by S-nitrosylation/S-gluthationylation of critical Cys residues of the P subunit of the GDC in Arabidopsis leaves. Moreover, we observed that this inhibition is part of the stress-related response of Arabidopsis to the bacterial elicitor harpin and that GDC inhibition alone is able to activate a redox response that triggers mitochondria perturbation and cell death. Taken together, these data reinforce the model of cross talk between NO/ROS and mitochondria in the activation of stress-related responses in plants.
MATERIALS AND METHODS
Harpin Purification
The bacterial elicitor harpin was purified by Escherichia coli DH5 α transformed with a pBluescript SK+ vector carrying the full-length Pseudomonas syringae 61 hrpZ open reading frame as described by Krause and Durner (2004). Proteins of nontransformed Escherichia coli were purified as the one carrying the harpin-encoding vector and used as a control.
Growth Conditions of Cell Culture and Plant
Arabidopsis (Arabidopsis thaliana) plants (ecotype Columbia-0) were cultivated as described by Lindermayr et al. (2005). Five- to 6-week-old plants were used for experiments. Cultured cells of Arabidopsis, originating from the ecotype Columbia-0, were grown as described by Krause and Durner (2004). All experiments were performed using cells in the logarithmic growth phase, 5 to 6 d after subculturing.
Partial Purification of Mitochondria
Crude, well-coupled mitochondria were isolated and purified by differential centrifugation as described by Keech et al. (2005), with a nonreducing medium replacing the extraction buffer to avoid interference with the biotinylation process. All procedures were carried out at 4°C in detergent-free vessels. Arabidopsis leaves (15 g, from 5- to 6-week-old plants) were ground with 20 mL of grinding medium (0.3 m Suc, 60 mm TES, 10 mm EDTA, 25 mm tetrasodium pyrophosphate, 1% [w/v] polyvinylpyrrolidone-40, 0.5% [w/v] defatted bovine serum albumin, 10 mm potassium phosphate [pH 8.0], 1 mm phenylmethylsulfonyl fluoride, 1 μm leupeptin, and 1 μm pepstatin) using a pestle and a small amount of quartz (fine granular, washed and calcined, guaranteed reagent grade; Merck). The extract was filtered through a 20- μ m nylon mesh and centrifuged at 2,500g for 5 min to remove most of the intact chloroplasts and thylakoid membranes. The supernatant was transferred to a new tube and centrifuged at 15,000g for 15 min. The pellet obtained was suspended in wash medium (0.3 m Suc, 10 mm TES, 10 mm potassium phosphate [pH 7.5], and Complete Protease Inhibitor Cocktail [Roche Applied Science]) and centrifuged at 15,000g for 15 min. The resulting supernatant was discarded, and the pellet containing the crude mitochondria was resuspended in 0.5 mL of wash medium or a specific assay medium. Cytochrome c oxidase activity, succinate-dependent respiration rate, and total protein concentration were estimated as described (Bradford, 1976; Neuburger et al., 1985; Livaja et al., 2008). Mitochondria were used immediately after preparation.
Detection of S-Nitrosylated Proteins
To detect S-nitrosylated proteins, we adopted the biotin-switch method, a three-step procedure that converts S-nitrosylated Cys residues into biotinylated Cys residues (Jaffrey and Snyder, 2001). Mitochondria (around 15 mg total wet weight) were incubated with the S-nitrosylating agent GSNO (1 mm; Alexis) or with GSH (1 mm; Sigma-Aldrich) in the dark at room temperature for 20 min with frequent vortexing. Mitochondrial proteins were precipitated with 10 volumes of cold acetone for 20 min at −20°C to remove the excess of GSNO/GSH. The samples were centrifuged at 10,000g for 10 min, and pellets were resuspended at a final concentration of 1 mg mL−1 and assayed with the biotin-switch method. The remaining proteins were analyzed by nanoLC/MS/MS essentially as described (Lindermayr et al., 2005).
Purification of P Protein from Mitochondria Extract
P protein purification from mitochondria extract was essentially as described by Bourguignon et al. (1988). Arabidopsis mitochondria (about 70 mg of protein) were diluted in dilution buffer (5 mm MOPS, 5 mm Tris, 1 mm β -mercaptoethanol, 1 mm EGTA, 20 μm pyridoxal phosphate, 1 mm Ser, 4 μm leupeptin, and 0.1% Triton X-100, pH 7.0) to 40 mg mL−1. Total release of the matrix protein was achieved by three cycles of freeze thaw (liquid N2 for 2 min followed by 30°C until thawed). The suspension of broken mitochondria was centrifuged at 100,000g for 2 h to remove all the mitochondrial membranes. The supernatant-containing soluble protein was concentrated with a Centricon Plus-20 Pl 10 kD (Millipore) by centrifugation at 3,600g for 30 min in a swilling rotor (Rotanta 460R; Hettich Zentrifugen). P protein was purified as described (Bourguignon et al., 1988). After an initial size-exclusion chromatography step with a HiPrep 16/60 Sephacryl S-300 HR (GE Healthcare) column connected to an ÄKTAexplorer FPLC device, an anion-exchange step was applied (HiTRAP DEAE FF; GE Healthcare). Final fractions were combined, concentrated with Amicon Ultra-4 10 kD (Millipore), and stored at −80°C (on addition of 20% glycerol).
Identification of S-Nitrosylated Proteins: Matrix-Assisted Laser-Desorption Ionization Time of Flight MS and NanoLC/MS/MS
Proteins were dissolved in 50 mL of 0.1 m NH4HCO3 containing 10% acetonitrile and digested with 3 mg of trypsin at 37°C overnight. Protein spots were visualized by colloidal Coomassie Brilliant Blue staining, excised from SDS gels, and completely destained by washing in 25 mm NH4HCO3. Digested peptides were extracted by vortexing for 3 h with 100 mL of 5% formic acid. All tryptic peptide samples were dried and redissolved in 50 mL of 0.1% trifluoroacetic acid and 5% acetonitrile. Peptides were separated by reverse-phase chromatography using an UltiMate Capillary/Nano liquid chromatography system (LC Packings) and eluted and fractionated on a self-packed analytical column (75 μ m × 120 mm) packed with YMC-Gel ODS-A (3 mm C18; YMC) with a gradient of 5% to 55% acetonitrile at a flow rate of 150 nL over 40 min. Eluted peptides were continuously delivered to a Q-Tof Ultima mass spectrometer (Waters/Micromass) by electrospray and analyzed by MS/MS employing data-dependent analysis (three most abundant ions in each cycle; 0.3 s MS, mass-to-charge ratio [m/z] 400–2,000, and maximum 4.8 s MS/MS, m/z 50–3,000; continuum mode, 60 s dynamic exclusion). The MS/MS raw data were processed and converted into Micromass pkl format using MassLynx 4.0 ProteinLynx. Resulting pkl files were compared with those of theoretical trypsin digestions and searched against predicted masses derived from the National Center for Biotechnology Information genomic database using ProFound software (Genomic Solutions). The Mascot search engine was used to parse MS data to identify proteins from primary sequence databases. Proteome analysis was carried out by TopLab.
Posttranslational Modifications of GDC: Nano-HPLC-MS2/3 and Data Analysis
For mass spectrometric analyses, partially purified P protein was digested by trypsin at 37°C for 1 h in 100 mm Tris-HCl, pH 6.5. The reaction was done in the dark to avoid light-dependent decomposition of the modifications. The used trypsin:protein ratio was 1:20. Protein digests were analyzed by online nanoLC/MS/MS. The samples were separated on an in-house-made 10-cm reverse-phase capillary emitter column (i.d. 75 μ m, 5 μ m ProntoSIL 120–5-C18 ace-EPS) using 120-min linear gradients from 0% to 40% acetonitrile/0.1% formic acid with a Flux Rheos 2200 nanoflow system (Flux Instruments) at a flow rate of 270 nL min−1. The LC setup was connected to an LTQ-Orbitrap classic (Thermo Fisher) equipped with a nanoelectrospray ion source (Proxeon Biosystems). The mass spectrometer was operated in the data-dependent mode to automatically switch between MS, MS2, and MS3 acquisition. Survey full-scan MS spectra (m/z 350–1,800) were acquired in the Orbitrap with resolution R = 7,500 at m/z 400. The six most intense ions were then sequentially fragmented in the linear ion trap using collisionally induced dissociation at normalized collision energy of 35. In the case of a resulting neutral loss of 9.7 m/z or 14.5 m/z in the MS2 spectra in the three most abundant peaks, indicating the loss of NO, these fragments were selected for further MS3 fragmentation. Former target ions selected for MS2 were dynamically excluded for 30 s. Total cycle time was approximately 3 s. The general mass spectrometric conditions were as follows: spray voltage, 1.4 kV; no sheath and auxiliary gas flow; ion transfer tube temperature, 200°C. Ion selection thresholds were as follows: 500 counts for MS2 and 500 counts for MS3. An activation q = 0.25 and activation time of 30 ms were applied in both MS2 and MS3 acquisitions.
Peptides and proteins were identified via automated database searching (Bioworks 3.3.1; SP1) of all MS2 and MS3 against an in-house-curated database (36,361 protein sequences). Spectra were normally searched with a mass tolerance of 1.5 atomic mass units for the parent mass and 1 atomic mass unit for the MS2 and MS3 fragment masses, with semitryptic specificity allowing two miscleavages. All modifications were set to be variable: oxidation of Met, nitrosylation and glutathionylation of Cys. Proteome analysis was carried out in cooperation with the Helmholtz Zentrum Munich core facility.
Gly Decarboxylase Activity
GDC activity in Arabidopsis leaf slices and intact mitochondria was determined following the Gly decarboxylase activity in vitro, by means of the P/H subunit activity (Somerville and Ogren, 1981). Leaf slices were prepared as described (Navarre and Wolpert, 1995; 2- × 7-mm slices). GDC activity was assayed with 8 mm [14C]Gly (8.75 μ Ci, labeled at the carboxyl group) to 300 μ L of assay buffer (4 mm sodium phosphate, pH 7.2, 0.3 m sorbitol, 20 mm MOPS, pH 7.2, 8 mm KCl, 4 mm MgCl2, and 0.1% bovine serum albumin) containing 100 μ g of mitochondrial protein or 10 leaf slices. Reaction mixtures were placed in 2-mL Eppendorf tubes suspended over 1.4 mL of Oxysolve C400 scintillator fluid (Zinsser Analytic) in 20-mL scintillation vials. Reactions were initiated by adding [14C]Gly and terminated by injecting 100 μ L of 6 m acetic acid into the reaction mixture. The vials were left overnight to permit trapping of 14CO2, the reaction tubes were removed, and the trapped 14C radioactivity was determined by scintillation counting (Beckman LS6000SC).
Gas-Exchange Measurements
The photorespiration rate in vivo was monitored following the response of the photosynthesis (A) to the intracellular CO2 mole fraction (Ci). Rates of photosynthesis and dark respiration in leaves were determined under the growth conditions using a portable infrared gas analyzer system (LI-6400; LI-COR Biosciences). One leaf was placed into the leaf chamber of the LI-6400 and exposed to 75, 250, or 400 μ mol m−2 s−1 light at a flow rate of 300 μ mol s−1. Leaf temperature was maintained at 25°C during measurement. To calculate the CO2 compensation point, a cycle with six CO2 reference concentrations was used: 400, 300, 200, 100, 50, and 400 pm.
Gly-Ser Content
For amino acid determination, 100 mg of leaf material was ground in liquid nitrogen and extracted in 1 mL of 80% ethanol for 30 min. After centrifugation for 10 min at 24,000g, the supernatant was vacuum dried and the dried extract was dissolved in 1 mL of 0.3 m sodium phosphate (pH 6.8) and 0.4% tetrahydroflurane. Samples were incubated for 30 min and then diluted 1:50 with the same phosphate buffer. Individual amino acids were separated by HPLC and quantified as described (Hagemann et al., 2005).
Cytofluorometric Analysis of NO and ROS Formation
For the detection of NO or ROS in mitochondria, untreated or harpin-treated (35 μ g mL−1) cultured cells of Arabidopsis were stained in the dark with 0.5 μm MitoTracker Red 580 (Invitrogen) as mitochondria-specific marker (Yao et al., 2002; Krause and Durner, 2004). After incubation for 20 to 30 min in the presence or absence of the NO scavenger cPTIO (0.5 mm; Alexis), 2 μm NO-specific dye DAF-2FM DA (Molecular Probe) or 10 μm ROS-sensitive dye H2DCF-DA (Invitrogen) was added to the cells and incubated for 10 min under continuous shaking. Fluorescence development was observed and photographed with a Zeiss confocal microscope (LSM 510 NLO). The mitochondria-dependent red fluorescence was excited using a 543-nm laser and detected through a long-pass filter LP 565 to 615 nm. The NO- or ROS-dependent green fluorescence was excited with a 488-nm argon laser and visualized through a band-pass filter BP 500 to 550 nm. Images were analyzed with LSM Image Browser (Zeiss).
Determination of Mitochondrial Δ Ψm
Changes in mitochondrial Δ Ψm in response to different elicitors were monitored using the mitochondrial potential sensor JC-1 (Invitrogen; Yao et al., 2002). Cultured cells of Arabidopsis were stained with the 5 μ g mL−1 JC-1 dye and incubated for 30 min in the dark with continuous shaking. Stained cells were diluted with PS medium, placed on a black 96-well microplate (Nunc), and treated with the elicitors. Changes in fluorescence intensities were measured immediately after treatment, every 5 min over 100 min, using a Tecan GENios reader (excitation, 488 nm; emission 1, 535 nm; emission 2, 595 nm).
Cell Death Assay
Cell death, measured as loss of plasma membrane integrity, was detected by double staining of harvested cultured cells of Arabidopsis with Evans blue (Sigma-Aldrich) to stain dead cells and fluorescein diacetate to stain live cells. The excitation and emission wavelengths of the two dyes were different and not interfering (Evans blue: excitation, 543 nm and LP filter 650 nm; fluorescein diacetate: excitation, 488 nm and BP filter 505–530 nm). Harvested suspension cells were stained with 0.005% Evans blue for 5 min. Stained cells were then incubated for a further 5 min with 0.01% fluorescein diacetate and treated with different inhibitors or water. Confocal microscopy images were obtained using a Zeiss LSM 510 NLO, and images were analyzed with LSM Image Browser (Zeiss).
Statistical Analysis
All experiments had more than two levels of the independent variable. For this reason, we carried out a one-way ANOVA, followed by Tukey's honestly significant difference posthoc test, in which sets of two means at a time were compared. Letters indicate statistically different values.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT4G33010 and AT2G26080.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Mitochondria fraction purification degree.
Supplemental Figure S2. Controls for the biotin-switch method.
Supplemental Figure S3. Activity assay for purified P protein.
Supplemental Figure S4. Mass spectrometric analyses of partially purified P protein.
Supplemental Figure S5. AAN-induced ROS production in Arabidopsis cell cultures.
Acknowledgments
We thank Jahnke Kathrin for excellent technical assistance and Benjamin Zeitler for helpful discussions and instructions. We are thankful to Corina Vlot for carefully editing the manuscript and for scientific discussions.
References
- Abe J, Takahashi M, Ishida M, Lee JD, Berk BC. (1997) c-Src is required for oxidative stress-mediated activation of big mitogen-activated protein kinase 1 (BMK1). J Biol Chem 272: 20389–20394 [DOI] [PubMed] [Google Scholar]
- Allan AC, Fluhr R. (1997) Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9: 1559–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. (2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchowsky A, Klei LR, Dudek EJ, Swartz HM, James PE. (1999) Stimulation of reactive oxygen, but not reactive nitrogen species, in vascular endothelial cells exposed to low levels of arsenite. Free Radic Biol Med 27: 1405–1412 [DOI] [PubMed] [Google Scholar]
- Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma JM, Lupianez JA, del Rio LA. (1999) Localization of nitric-oxide synthase in plant peroxisomes. J Biol Chem 274: 36729–36733 [DOI] [PubMed] [Google Scholar]
- Bauwe H, Kolukisaoglu U. (2003) Genetic manipulation of glycine decarboxylation. J Exp Bot 54: 1523–1535 [DOI] [PubMed] [Google Scholar]
- Beligni MV, Fath A, Bethke PC, Lamattina L, Jones RL. (2002) Nitric oxide acts as an antioxidant and delays programmed cell death in barley aleurone layers. Plant Physiol 129: 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Lamattina L. (2001) Nitric oxide: a non-traditional regulator of plant growth. Trends Plant Sci 6: 508–509 [DOI] [PubMed] [Google Scholar]
- Blackwell RD, Murray AJ, Lea PJ. (1990) Photorespiratory mutants of the mitochondrial conversion of glycine to serine. Plant Physiol 94: 1316–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borutaite V, Brown GC. (2006) S-Nitrosothiol inhibition of mitochondrial complex I causes a reversible increase in mitochondrial hydrogen peroxide production. Biochim Biophys Acta 1757: 562–566 [DOI] [PubMed] [Google Scholar]
- Bourguignon J, Neuburger M, Douce R. (1988) Resolution and characterization of the glycine-cleavage reaction in pea leaf mitochondria: properties of the forward reaction catalysed by glycine decarboxylase and serine hydroxymethyltransferase. Biochem J 255: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 77: 248–254 [DOI] [PubMed] [Google Scholar]
- Brunori M, Giuffre A, Sarti P, Stubauer G, Wilson MT. (1999) Nitric oxide and cellular respiration. Cell Mol Life Sci 56: 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. (2006) Direct evidence for S-nitrosation of mitochondrial complex I. Biochem J 394: 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras MC, Franco MC, Peralta JG, Poderoso JJ. (2004) Nitric oxide, complex I, and the modulation of mitochondrial reactive species in biology and disease. Mol Aspects Med 25: 125–139 [DOI] [PubMed] [Google Scholar]
- Chen Y, Gibson SB. (2008) Is mitochondrial generation of reactive oxygen species a trigger for autophagy?. Autophagy 4: 246–248 [DOI] [PubMed] [Google Scholar]
- Cleeter MW, Cooper JM, Darley-Usmar VM, Moncada S, Schapira AH. (1994) Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide: implications for neurodegenerative diseases. FEBS Lett 345: 50–54 [DOI] [PubMed] [Google Scholar]
- Clementi E, Brown GC, Feelisch M, Moncada S. (1998) Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci USA 95: 7631–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CE, Davies NA. (2000) Effects of nitric oxide and peroxynitrite on the cytochrome oxidase K(m) for oxygen: implications for mitochondrial pathology. Biochim Biophys Acta 1459: 390–396 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Chaki M, Fernandez-Ocana A, Valderrama R, Palma JM, Carreras A, Begara-Morales JC, Airaki M, del Rio LA, Barroso JB. (2008) Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol 49: 1711–1722 [DOI] [PubMed] [Google Scholar]
- Creach E, Stewart CR. (1982) Effects of aminoacetonitrile on net photosynthesis, ribulose-1,5-bisphosphate levels, and glycolate pathway intermediates. Plant Physiol 70: 1444–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahm CC, Moore K, Murphy MP. (2006) Persistent S-nitrosation of complex I and other mitochondrial membrane proteins by S-nitrosothiols but not nitric oxide or peroxynitrite: implications for the interaction of nitric oxide with mitochondria. J Biol Chem 281: 10056–10065 [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. (2009) Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci 34: 85–96 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C. (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio LA, Corpas FJ, Sandalio LM, Palma JM, Barroso JB. (2003) Plant peroxisomes, reactive oxygen metabolism and nitric oxide. IUBMB Life 55: 71–81 [DOI] [PubMed] [Google Scholar]
- Desikan R, Reynolds A, Hancock JT, Neill SJ. (1998) Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochem J 330: 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rebeille F. (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6: 167–176 [DOI] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF. (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Lee CP, Kuo J, Meyer EH, Taylor NL, Millar AH. (2007) Free-flow electrophoresis for purification of plant mitochondria by surface charge. Plant J 52: 583–594 [DOI] [PubMed] [Google Scholar]
- Foissner I, Wendehenne D, Langebartels C, Durner J. (2000) In vivo imaging of an elicitor-induced nitric oxide burst in tobacco. Plant J 23: 817–824 [DOI] [PubMed] [Google Scholar]
- Foster MW, Stamler JS. (2004) New insights into protein S-nitrosylation: mitochondria as a model system. J Biol Chem 279: 25891–25897 [DOI] [PubMed] [Google Scholar]
- Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. (2000) The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant. Nat Cell Biol 2: 156–162 [DOI] [PubMed] [Google Scholar]
- Green DR, Reed JC. (1998) Mitochondria and apoptosis. Science 281: 1309–1312 [DOI] [PubMed] [Google Scholar]
- Hagemann M, Vinnemeier J, Oberpichler I, Boldt R, Bauwe H. (2005) The glycine decarboxylase complex is not essential for the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Biol (Stuttg) 7: 15–22 [DOI] [PubMed] [Google Scholar]
- Heineke D, Bykova N, Gardestrom P, Bauwe H. (2001) Metabolic response of potato plants to an antisense reduction of the P-protein of glycine decarboxylase. Planta 212: 880–887 [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. (2005) Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6: 150–166 [DOI] [PubMed] [Google Scholar]
- Hiraga K, Kikuchi G. (1980) The mitochondrial glycine cleavage system: functional association of glycine decarboxylase and aminomethyl carrier protein. J Biol Chem 255: 11671–11676 [PubMed] [Google Scholar]
- Huang X, von Rad U, Durner J. (2002) Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215: 914–923 [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001: PL1. [DOI] [PubMed] [Google Scholar]
- Keech O, Dizengremel P, Gardeström P. (2005) Preparation of leaf mitochondria from Arabidopsis thaliana. Physiol Plant 124: 403–409 [Google Scholar]
- Kolbert Z, Bartha B, Erdei L. (2008) Osmotic stress- and indole-3-butyric acid-induced NO generation are partially distinct processes in root growth and development in Pisum sativum. Physiol Plant 133: 406–416 [DOI] [PubMed] [Google Scholar]
- Kozaki A, Takeba G. (1996) Photorespiration protects C3 plants from photooxidation. Nature 384: 557–560 [Google Scholar]
- Krause M, Durner J. (2004) Harpin inactivates mitochondria in Arabidopsis suspension cells. Mol Plant Microbe Interact 17: 131–139 [DOI] [PubMed] [Google Scholar]
- Kruft V, Eubel H, Jansch L, Werhahn W, Braun HP. (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol 127: 1694–1710 [PMC free article] [PubMed] [Google Scholar]
- Lam E, del Pozo O. (2000) Caspase-like protease involvement in the control of plant cell death. Plant Mol Biol 44: 417–428 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J. (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livaja M, Palmieri MC, von Rad U, Durner J. (2008) The effect of the bacterial effector protein harpin on transcriptional profile and mitochondrial proteins of Arabidopsis thaliana. J Proteomics 71: 148–159 [DOI] [PubMed] [Google Scholar]
- Mannick JB, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B. (2001) S-Nitrosylation of mitochondrial caspases. J Cell Biol 154: 1111–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Ruiz A, Lamas S. (2007) Signalling by NO-induced protein S-nitrosylation and S-glutathionylation: convergences and divergences. Cardiovasc Res 75: 220–228 [DOI] [PubMed] [Google Scholar]
- Millar AH, Day DA. (1996) Nitric oxide inhibits the cytochrome oxidase but not the alternative oxidase of plant mitochondria. FEBS Lett 398: 155–158 [DOI] [PubMed] [Google Scholar]
- Millar AH, Sweetlove LJ, Giege P, Leaver CJ. (2001) Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol 127: 1711–1727 [PMC free article] [PubMed] [Google Scholar]
- Mur LA, Kenton P, Lloyd AJ, Ougham H, Prats E. (2008) The hypersensitive response: the centenary is upon us but how much do we know?. J Exp Bot 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H. (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10: 339–346 [DOI] [PubMed] [Google Scholar]
- Navarre DA, Wolpert TJ. (1995) Inhibition of the glycine decarboxylase multienzyme complex by the host-selective toxin victorin. Plant Cell 7: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuburger M, Day DA, Douce R. (1985) Transport of NAD in Percoll-purified potato tuber mitochondria: inhibition of NAD influx and efflux by N-4-azido-2-nitrophenyl-4-aminobutyryl-3′-NAD. Plant Physiol 78: 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond B, Badger M, Maxwell K, Björkman O, Leegood R. (1997) Too many photons: photorespiration, photoinhibition and photooxidation. Trends Plant Sci 2: 119–121 [Google Scholar]
- Pedroso MC, Magalhaes JR, Durzan D. (2000) A nitric oxide burst precedes apoptosis in angiosperm and gymnosperm callus cells and foliar tissues. J Exp Bot 51: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Pellinen R, Palva T, Kangasjarvi J. (1999) Short communication: subcellular localization of ozone-induced hydrogen peroxide production in birch (Betula pendula) leaf cells. Plant J 20: 349–356 [DOI] [PubMed] [Google Scholar]
- Robson CA, Vanlerberghe GC. (2002) Transgenic plant cells lacking mitochondrial alternative oxidase have increased susceptibility to mitochondria-dependent and -independent pathways of programmed cell death. Plant Physiol 129: 1908–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Puertas MC, Campostrini N, Matte A, Righetti PG, Perazzolli M, Zolla L, Roepstorff P, Delledonne M. (2008) Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 8: 1459–1469 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Perazzolli M, Zago ED, Delledonne M. (2004) Nitric oxide signalling functions in plant-pathogen interactions. Cell Microbiol 6: 795–803 [DOI] [PubMed] [Google Scholar]
- Saviani EE, Orsi CH, Oliveira JF, Pinto-Maglio CA, Salgado I. (2002) Participation of the mitochondrial permeability transition pore in nitric oxide-induced plant cell death. FEBS Lett 510: 136–140 [DOI] [PubMed] [Google Scholar]
- Schonhoff CM, Gaston B, Mannick JB. (2003) Nitrosylation of cytochrome c during apoptosis. J Biol Chem 278: 18265–18270 [DOI] [PubMed] [Google Scholar]
- Shapiro AD. (2005) Nitric oxide signaling in plants. Vitam Horm 72: 339–398 [DOI] [PubMed] [Google Scholar]
- Skulachev VP. (1996) Why are mitochondria involved in apoptosis? Permeability transition pores and apoptosis as selective mechanisms to eliminate superoxide-producing mitochondria and cell. FEBS Lett 397: 7–10 [DOI] [PubMed] [Google Scholar]
- Skulachev VP. (1999) Mitochondrial physiology and pathology: concepts of programmed death of organelles, cells and organisms. Mol Aspects Med 20: 139–184 [DOI] [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. (1981) Photorespiration-deficient mutants of Arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol 67: 666–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NL, Day DA, Millar AH. (2002) Environmental stress causes oxidative damage to plant mitochondria leading to inhibition of glycine decarboxylase. J Biol Chem 277: 42663–42668 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Day DA, Millar AH. (2004) Targets of stress-induced oxidative damage in plant mitochondria and their impact on cell carbon/nitrogen metabolism. J Exp Bot 55: 1–10 [DOI] [PubMed] [Google Scholar]
- Tiwari BS, Belenghi B, Levine A. (2002) Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol 128: 1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usuda H, Edwards GE. (1980) Localization of glycerate kinase and some enzymes for sucrose synthesis in C(3) and C(4) plants. Plant Physiol 65: 1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Robson CA, Yip JY. (2002) Induction of mitochondrial alternative oxidase in response to a cell signal pathway down-regulating the cytochrome pathway prevents programmed cell death. Plant Physiol 129: 1829–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GH, Sarojini G, Oliver DJ. (1982) Identification of a glycine transporter from pea leaf mitochondria. Biochem Biophys Res Commun 107: 856–861 [DOI] [PubMed] [Google Scholar]
- Walker JL, Oliver DJ. (1986) Glycine decarboxylase multienzyme complex: purification and partial characterization from pea leaf mitochondria. J Biol Chem 261: 2214–2221 [PubMed] [Google Scholar]
- Wendehenne D, Pugin A, Klessig DF, Durner J. (2001) Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci 6: 177–183 [DOI] [PubMed] [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC. (2000) Photorespiration: metabolic pathways and their role in stress protection. Philos Trans R Soc Lond B Biol Sci 355: 1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Chen Z. (2000) Harpin-induced hypersensitive cell death is associated with altered mitochondrial functions in tobacco cells. Mol Plant Microbe Interact 13: 183–190 [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Shimoji H, Ohshiro Y, Sakihama Y. (2001) Inhibitory effects of nitric oxide on oxidative phosphorylation in plant mitochondria. Nitric Oxide 5: 261–270 [DOI] [PubMed] [Google Scholar]
- Yao N, Tada Y, Sakamoto M, Nakayashiki H, Park P, Tosa Y, Mayama S. (2002) Mitochondrial oxidative burst involved in apoptotic response in oats. Plant J 30: 567–579 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Terashima I, Noguchi K. (2006) Distinct roles of the cytochrome pathway and alternative oxidase in leaf photosynthesis. Plant Cell Physiol 47: 22–31 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Terashima I, Noguchi K. (2007) Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol 48: 606–614 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Watanabe C, Kato Y, Sakamoto W, Noguchi K. (2008) Influence of chloroplastic photo-oxidative stress on mitochondrial alternative oxidase capacity and respiratory properties: a case study with Arabidopsis yellow variegated 2. Plant Cell Physiol 49: 592–603 [DOI] [PubMed] [Google Scholar]
- Zeidler D, Zahringer U, Gerber I, Dubery I, Hartung T, Bors W, Hutzler P, Durner J. (2004) Innate immunity in Arabidopsis thaliana: lipopolysaccharides activate nitric oxide synthase (NOS) and induce defense genes. Proc Natl Acad Sci USA 101: 15811–15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MG, Chen L, Zhang LL, Zhang WH. (2009) Nitric reductase-dependent nitric oxide production is involved in cold acclimation and freezing tolerance in Arabidopsis. Plant Physiol 151: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]