Abstract
This paper investigates the performance of a new multivariate method for tensor-based morphometry (TBM). Statistics on Riemannian manifolds are developed that exploit the full information in deformation tensor fields. In TBM, multiple brain images are warped to a common neuroanatomical template via 3-D nonlinear registration; the resulting deformation fields are analyzed statistically to identify group differences in anatomy. Rather than study the Jacobian determinant (volume expansion factor) of these deformations, as is common, we retain the full deformation tensors and apply a manifold version of Hotelling’s T 2 test to them, in a Log-Euclidean domain. In 2-D and 3-D magnetic resonance imaging (MRI) data from 26 HIV/AIDS patients and 14 matched healthy subjects, we compared multivariate tensor analysis versus univariate tests of simpler tensor-derived indices: the Jacobian determinant, the trace, geodesic anisotropy, and eigenvalues of the deformation tensor, and the angle of rotation of its eigenvectors. We detected consistent, but more extensive patterns of structural abnormalities, with multivariate tests on the full tensor manifold. Their improved power was established by analyzing cumulative p-value plots using false discovery rate (FDR) methods, appropriately controlling for false positives. This increased detection sensitivity may empower drug trials and large-scale studies of disease that use tensor-based morphometry.
Index Terms: Brain, image analysis, Lie groups, magnetic resonance imaging (MRI), statistics
I. INTRODUCTION
Accurate measurement of differences in brain anatomy is key to understanding the effects of brain disorders, as well as changes associated with normal growth or variation within a population. Statistical methods that make a more complete use of the available data can greatly improve the analysis of temporal changes within a single subject, as well as intersubject variations. Given the subtle and distributed brain changes that occur with development and disease, improved statistical analyses are needed that detect group differences or time-dependent changes in brain structure with optimal power.
As computational anatomy studies expand into ever larger populations, voxel-based image analysis methods have become increasingly popular for detecting and analyzing group differences in magnetic resonance imaging (MRI) scans of brain structure [10]. These statistical mapping methods aim to identify brain regions where some imaging parameter differs between clinical groups, or covaries with age, gender, medication, or genetic variation [66], [95].
Some methods to map structural differences in the brain, such as voxel-based morphometry (VBM) [7] assess the distribution of specific tissue types, e.g., gray and white matter and cerebrospinal fluid (CSF). They can also be applied to generate voxelwise imaging statistics for relaxometric measures (such as parametric T1 or T2 images) or diffusion-weighted imaging parameters (such as fractional anisotropy or mean diffusivity [27], [55]). By contrast, deformation-based methods for analyzing brain structure compute information on regional volumetric differences between groups by warping brain images to a canonical template. The applied deformation is then used as an index of the volume differences between each subject and the template, and, subsequently, between groups.
Although these two approaches for structural image analysis are conceptually distinct, they can be combined, as in the “optimized VBM” [51], and RAVENS approaches (regional analysis of volumes examined in normalized space [38]). These hybrid approaches combine information on tissue distribution (based on image segmentation) and anatomical shape differences (from deformable image matching) in a unified setting. Other hybrid morphometric approaches have been developed, and some groups have combined statistical mapping methods with surface based models of the cortex [32], [43], [96], [100] or subcortical structures [37], [50], [92], [97] to detect changes in signals defined on anatomical surfaces, often adapting voxel-based methods to the parametric surface domain.
Among the many deformation-based approaches for analyzing anatomy, tensor-based morphometry (TBM) computes the spatial derivatives of the deformation fields that match a set of brain images to a common template. TBM may also be used in a longitudinal design where image warping algorithms are used to align sequentially acquired images and to recover information on brain growth or atrophy over time. TBM has yielded significant, new neuroscientific information on brain development and disease. It has been used to map the profile of volumetric differences throughout the brain in studies of Alzheimer’s disease [47], semantic dementia [64], [65], [85], HIV/AIDS [26], [86], Fragile X syndrome [62], neuropsychiatric disorders such as schizophrenia [48], twins [66], and normally developing children [94]. TBM has also been applied to large numbers of images collected over short intervals (e.g., two weeks) to measure the stability of different MRI acquisition protocols, thereby optimizing the ability to detect brain change [64]. Over intervals as short as one month, TBM approaches have identified the effects of medications, such as lithium, on human brain structure, suggesting the potential of the approach for gauging subtle changes in medication trials [65].
The broad application and increasing popularity of TBM methods makes it advantageous to optimize the statistical methods used in conjunction with it. The optimization of these methods is the focus of this paper. In tensor-based morphometry (TBM), a template T is matched to a study S using nonlinear registration, and the displacement vector u⃗(r⃗) is found such that T(r⃗ − u⃗) corresponds with S(r⃗). Here, r⃗ denotes the voxel location. To help estimate anatomical correspondences, features such as point, curve, and surface landmarks present in both datasets can be used, [58] or, more commonly, intensity-based cost functions are used based on normalized cross-correlation [34], mean squared intensity difference [8], [105], or divergence measures based on information theory [26], [33], [39], [87], [104].
The Jacobian matrix of the deformation field is defined (in 3-D) by
Its determinant, the Jacobian, is most commonly used to analyze the distortion necessary to deform the images into agreement. A value detJ(r⃗) > 1 implies that the neighborhood adjacent to r⃗ in the study was stretched to match the template (i.e., local volumetric expansion), while detJ(r⃗) < 1 is associated with local shrinkage. When many subjects’ images are aligned to the same standard template or atlas, maps of the Jacobians can be computed in the atlas coordinate system and group statistics can be computed at each voxel to identify localized group differences in anatomical shape or size. However, much of the information about the shape change is lost using this measure. As a toy example of this problem, let us consider an image voxel for which the eigenvalues of the Jacobian matrix are λ1,2,3 = {1,2,0.5}. In such a case, the value of the Jacobian determinant would be 1. Thus, though the eigenvalues clearly indicate directional shrinkage and growth, these changes would be left undetected, if only the Jacobian was analyzed.
In this work, we resolve this issue by making use of the deformation tensors for the analysis, which are defined as
| (1) |
The directional values of shape change are thus kept as variables in the analysis. More specifically, the eigenvalues of the deformation tensor are determined by the size of the deformation in the direction of the associated eigenvectors.
In particular, we apply Hotelling’s T2 test to obtain statistics for the deformation. However, the problem is complicated by the fact that for invertible transformations, the deformation tensors are constrained to the space of positive definite matrices. The latter form a cone in the space of matrices, which is itself a vector space. Thus, the deformation tensors do not form a vector space, and a manifold version of the statistics is needed. Several research groups have worked on the statistics of positive definite symmetric matrices.
The first person to investigate this problem was Fréchet [46], who defined the mean S̄ on a manifold as the point that leads to a minimum value for the sum of squared geodesic distances between S̄ and all the other points. S̄ yields the usual value for the mean in Euclidean space, but is not necessarily unique on other manifolds. Thus, the local value of the mean is generally used instead [59], [60].
To facilitate computations, Pennec et al. [72]–[74] and Fletcher and Joshi [44] independently proposed the use of the affine-invariant metric on the space of symmetric, positive-definite tensors. Pennec then proceeded to use the latter to define normal distributions on these matrices, as well as the Mahalanobis distance and χ2 law. Fletcher and Joshi [44], [45] used the metric to develop the notion of principal geodesic analysis, which extends principal component analysis to manifolds.
Several groups have used these techniques to assist with the computations involved in DTI smoothing [24], anisotropic filtering [31], segmentation [63], [103], fiber tracking [42], statistics on tensor fields [36], [77], [78], as well as to develop probabilistic matrices that describe fiber connectivity in the brain [20]. In a more recent development in DTI statistical analysis, Khurd et al. [61] used isometric mapping and manifold learning techniques (eigendecomposition of the geodesic distance matrix) to directly fit a manifold to the tensors, compute its dimensionality and distinguish groups using Hotelling’s T 2 statistics.
The relevance of techniques from Riemannian manifold theory to calculate means of Jacobian matrices was first suggested by Woods [106]. In [35], statistics on JJT were used to determine a weight factor for the regularizer in nonrigid registration. Furthermore, the Mahalanobis distance between the Cauchy-Green tensors was used as an elastic energy for the regularizer in a recent nonlinear registration technique [75]. However, to our knowledge this is the first time that the full deformation tensors have been used in the context of tensor-based morphometry. Once a metric is defined, the manifold-valued elements are projected into the tangent plane at the origin using the inverse of the exponential map, and the computations are done in this common space.
Recently, Arsigny et al. [2], [4], [5], considered a new family of metrics, the ’Log-Euclidean metrics.” These metrics make computations easier to perform, as they are chosen such that the transformed values form a vector space, and statistical parameters can then be computed easily using standard formulae for Euclidean spaces (see also e.g., [40] for a good review of the properties of exponential and log maps on manifolds). For two points S1 and S2 on the manifold, these metrics are of the form
where ‖.‖ denotes a norm. Following [4], in this work we will use
| (2) |
In this paper, we use a set of brain MRI scans from HIV/AIDS patients and matched healthy control subjects to illustrate our method. A preliminary version of this work can be found in [67]. We use a fluid registration algorithm [26] (see also [29]) to compute displacement fields and thus obtain deformation tensors. The fluid registration approach is guaranteed to generate only mappings with positive-definite deformation tensors. We refer to [11], [29], [44], [70] for background on these fluid models and their advantages; briefly, mappings regularized by linear elastic or Laplacian penalties are not guaranteed to be diffeomorphic (i.e., differentiable mappings with differentiable inverses). This potential problem with small-deformation models can be resolved by moving to fluid or other large-deformation models (E.g., LDDMM [14]) or geodesic shooting [56], [102]. Invertible mappings also have positive definite Jacobian matrices and deformation tensors, ensuring that the matrix logarithm operations are well defined in the following methods.
All our statistics are computed within the Log-Euclidean framework. A voxelwise Hotelling’s T 2 test is used as a measure of local anatomical differences between patients and controls. To assess the difference between our results and the ones found from the determinant of the Jacobian, these results are compared to the one-dimensional Student’s t-test on the determinants of the Jacobian matrices. The goal of the work was to find out whether multivariate statistics on the deformation tensors afforded additional power in detecting anatomical differences between patients and controls, and if so, which aspects of the tensor decomposition (rotation or magnitude of eigenvalues) were most influential in securing added sensitivity.
The usual measure of anisotropy for tensor-valued data is the fractional anisotropy (used widely in diffusion tensor imaging) but this measure is not valid for positive definite symmetric manifolds, as it relies on a Euclidean measure for the distance. Thus, in [13], the authors proposed a new measure which depends instead solely on distances computed on the manifold. They define the geodesic anisotropy as the shortest geodesic distance between the tensor and the closest isotropic tensor within the affine-invariant metric framework. In the specific case where two tensors commute, affine invariant and Log-Euclidean metrics give identical distances between them. The geodesic anisotropy GA defined above obeys this condition, as isotropic tensors are proportional to the identity matrix I. When affine-invariant and Log-Euclidean metrics are based on the same norm, the definition for GA is identical for both frameworks, and gives
| (3) |
with
Before proceeding, we note that some groups have already made significant advances by analyzing deformations at a voxel level using other scalar, vector or tensor measures than the deformation magnitude or the Jacobian determinant. In [91] and [93], we computed a set of 3-D deformation vectors aligning parameterized brain surfaces from individual subjects to a group average surface. These vectors were treated as observations from a Hotelling’s T2 distributed (or χ2 distributed) random field, with known mean and anisotropic covariance estimated from the data. Abnormal anatomic deviations were then assessed using 3-D Gaussian distributions on the deformation vectors at each anatomical point (see also [41] for additional modeling of the covariance of deformation vector fields). Related multivariate statistical work by Schormann et al. [79] modeled the spatial uncertainty of anatomical landmarks in 2-D histologic images using Rayleigh-Bessel distributions. Cao and Worsley [21], [107] extended this work by developing analytical formulae for the expected values of statistical maxima in these multivariate random fields extending algebraic results on the Euler characteristics of the excursion sets of Gaussian random fields. Thirion et al. [89], [90] also studied the statistical dissymmetry in a set of deformation mappings. They analyzed asymmetry vector fields using the Hotelling’s T2 field as their null distribution to assess the significance of departures from the “normal” degree and direction of anatomic asymmetry. In [31], Chung et al. formulated a unified framework for deformation morphometry using several novel descriptors of localized deformation that were also treated as observations from t or T2 distributed random fields. Specifically, they modeled brain growth over time using deformations, whose vorticity and expansion rates were analyzed at each voxel using the general linear model. For analyzing surface deformations, the Lipschitz constants [69], conformal factors [103], and other differential parameters of grids representing deformations have also been assessed. Rey et al. [76] experimented with different vector and tensor measures for assessing brain deformation due to multiple sclerosis and mechanical mass-effects due to tumor growth. They combined the norm of the deformation with its local divergence, among other descriptors, to create maps that emphasized both shift and local expansions associated with tumor growth.
Finally, we note that not all multivariate analyses of deformations operate at the voxel level. The entire deformation vector field can be treated as a single multivariate vector and analyzed using thin-plate splines and Riemannian shape spaces [16], principal components analysis (PCA) [12], or canonical variates analysis [6] either for the purpose of dimensionality reduction or for characterizing the principal modes of variation. In Grenander and Miller’s formulation of pattern theory [54], anatomical template deformations are regarded as arising via the stochastic differential equation Lu = n, where n is (vector-valued) noise, u is the displacement field, and L is an infinite-dimensional self-adjoint differential operator regularizing the deformation (which therefore has a countable basis and real eigenvalues). Using spectral methods, the deformations can then be expressed as multivariate vectors whose components are coefficients of the eigenfunctions of the governing operator. This method has been used in computational anatomy for multivariate analysis of deformations representing hippocampal or cortical shape using spherical harmonics or Oboukhov expansions [57], [88], [91], elliptical Fourier descriptors [81], or eigenfunctions of the Laplace-Beltrami operator on the cortex [31]. We also note that spectral modeling of anatomic variation is implicit in many registration methods, which often estimate deformation fields in a hierarchical multiscale fashion, using splines, wavelets, or eigenfunctions of differential operators to parameterize the deformation.
The unique contribution of this paper, relative to prior work, is to examine how multivariate analysis of the deformation tensor affects the results in tensor-based morphometry. We include theoretical arguments regarding the distributions of deformation tensors and show in empirical tests how they compare with simpler statistics based on eigenvalues, or Jacobian determinants, while still controlling false positives at the expected rate. This leads naturally to the use of the Log-Euclidean framework developed in [4], which has not yet been applied in the context of TBM.
This paper is organized as follows. In the following section, we briefly describe our statistical method. We then proceed to illustrate the use of our statistical analysis in 2-D on data from the corpus callosum of patients and healthy controls. We compare the performance of different univariate and multivariate measures derived from the deformation tensors using the false discovery rate (FDR). Finally, we use our tensor measures to analyze the 3-D pattern of anatomical deficits in the brains of HIV/AIDS patients, relative to healthy controls.
II. STATISTICAL ANALYSIS
In order to apply Hotelling’s T 2 test, we need to compute the mean and covariance matrices of the tensor-valued data. Thus, here we provide a brief summary of the method to find those quantities. In Rn, the mean S̄ of a set of n-dimensional vectors Si, i = 1,…, m is the point that minimizes the summed squared distance d to all the Si. For data on a manifold A, d becomes the geodesic distance, so S̄ is given by [46]
| (4) |
In the Log-Euclidean framework, computations are simplified by transforming the space of symmetric, positive-definite matrices into a vector space on the tangent plane at the origin, and then moving it back to the manifold using the exponential map once the mean is taken. Thus, the formula for the mean is easily shown to be [4]
| (5) |
Arsigny et al. demonstrate that the covariance is
where dP is the probability measure, Ω is the space of possible outcomes and wεΩ. For discrete data, this becomes
| (6) |
Thus, we obtain the Mahalanobis distance M
| (7) |
To avoid assuming a normal distribution for the observations at each voxel, we performed a voxelwise permutation test [71], for which we randomly reassigned the labels of patients and controls, and compared the p-values so generated to those of the data. This assembles a nonparametric reference distribution—in fact a different distribution for each voxel—against which the likelihood of group differences can be calibrated. All our statistics were computed using 5000 permutations (this is easily sufficient to define a null distribution for nonparametric estimation of p-values at the voxel level).
A. FDR
Overall significance, correcting for the multiple spatial comparisons implicit in computing an image of statistics, was assessed by positive false discovery rate (pFDR) methods [83] for strong control over the likelihood of false rejections of the null hypotheses with multiple comparisons. The pFDR method directly measures the pFDR under a given primary threshold, and is more powerful than the popular sequential p-value FDR method (in [15] and [49]), as it estimates the probability that the null hypothesis is true from the empirical distribution of observed p-values [68]. The pFDR measure is theoretically more suitable for representing the rate at which discoveries are false than the FDR measure when the primary rejection region is relatively small [83].
Briefly, pFDR is the false discovery rate conditioned on the event that positive findings, rejecting the null hypothesis, have occurred, and is given by
| (8) |
where πo = Pr(H = 0) is the probability that the null hypothesis is true, and γ is the rejection threshold for the individual hypothesis, which was set to 0.01 in our experiments. We refer readers to [82] and [83] for the details of the estimation procedures to obtain pFDR for statistical maps. By conventions, a statistical map with pFDR < 0.05, i.e. the false discovery rate is less than 5%, was considered to be significant. We note that the voxel-level t and T 2 statistics, used here for comparing methods, are not assumed to have parametric distributions, as the cumulative (rank-order) distribution of p-values in FDR methods is independent of the distributions of the statistics themselves.
III. APPLICATIONS
A. Description of Data
Twenty-six HIV/AIDS patients (age: 47.2 ± 9.8 years; 25 M/1 F; CD4+ T-cell count: 299.5 ± 175.7 per µl; log10 viral load: 2.57 ± 1.28 RNA copies per ml of blood plasma) and 14 HIV-seronegative controls (age: 37.6 ± 12.2 years; 8M/6F) underwent 3-D T1-weighted MRI scanning; subjects and scans were the same as those analyzed in the cortical thickness study by Thompson et al. [97], where more detailed neuropsychiatric data from the subjects is presented. All patients met Center for Disease Control criteria for AIDS, stage C and/or 3 (Center for Disease Control and Prevention, 1992), and none had HIV-as-sociated dementia. Health care providers in Allegheny County, PA, served as a sentinel network for recruitment. All AIDS patients were eligible to participate, but those with a history of recent traumatic brain injury, CNS opportunistic infections, lymphoma, or stroke were excluded. All patients underwent a detailed neurobehavioral assessment within the four weeks before their MRI scan, involving a neurological examination, psychosocial interview, and neuropsychological testing, and were designated as having no, mild, or moderate (coded as 0, 1, and 2, respectively) neuropsychological impairment based on a factor analysis of a broad inventory of motor and cognitive tests performed by a neuropsychologist [97].
All subjects received 3-D spoiled gradient recovery (SPGR) anatomical brain MRI scans (256 × 256 × 124 matrix, TR = 25 ms, TE = 5 ms; 24-cm field of view; 1.5-mm slices, zero gap; flip angle = 40°) as part of a comprehensive neurobehavioral evaluation. The MRI brain scan of each subject was coregistered with scaling (nine-parameter transformation) to the ICBM53 average brain template, and extracerebral tissues (e.g., scalp, meninges, brainstem, and cerebellum).
The corpus callosum of each subject was hand-traced according to previously published criteria [98], using the interactive segmentation software.
B. Registration Algorithm
The images were nonlinearly registered using a fluid registration algorithm that was recently implemented [26]. In this approach, the template is treated as a viscous fluid, for which the velocity of the fluid particles satisfies a Navier-Stokes equation. A force term derived from the joint distribution of intensities between the images is used to drive the registration. This model allows for the stress of the transformation to relax over time, thereby permitting large deformations.
In practice, solving the velocity PDE is very time consuming. Thus, we use a filter based on the Green’s function of the governing operator of the fluid equation, with sliding boundary conditions [28] as was proposed by Bro-Nielsen and Gramkow [17], [18], [53] to increase the speed of the registration.
As a cost function, we chose to maximize a modified version of the Jensen-Rényi divergence (JRD). A more detailed description of our registration method can be found in [25], [26]. To save computation time and memory requirements, the source and the target images were filtered with a Hann-windowed sinc kernel, and isotropically downsampled by a factor of 2. As in other TBM studies [38], [85], we preferred registration to a typical control image rather than a multisubject average intensity atlas as it had sharper features. The resulting deformation field was trilinearly interpolated at each iteration to drive the source image towards the target at the original resolution to obtain the warped image.
C. Corpus Callosum Maps
In the case of the determinant, it is straightforward to understand the meaning of the statistic, as it is directly related to volume change. The interpretation for statistics on the deformation tensors is not so straightforward. Here we decompose the deformation tensor into various components in order to achieve a better understanding of the statistics.
We first examine the 2-D case, as it is easier to visualize. To do so, we generated p-maps (significance maps for group differences) based on various statistics derived from the deformation tensors on the corpus callosum. The corpus callosum traces were turned into binary maps, which were fluidly registered. This conversion to binary data was made to allow exact matching of the boundaries, and the resulting interior profile of deformation did not depend on the intensity-based fidelity term or on the Lagrange multiplier used in the registration cost function. In the next section, we develop a more formal comparison of the different statistics using the FDR, which adjusts the observed findings for multiple comparisons that are implicit in evaluating an entire image of statistics.
The measures of deformation we will study below are the log10 of the determinant, the principal log10 (eigenvalue), that is the largest eigenvalue in log space, log10 (trace), the geodesic anisotropy and the rotation angle of the principal eigenvector to the horizontal axis. A Student’s t-test was performed on these scalar statistics. Similarly, we computed the Hotelling’s T2 p-values for two multivariate statistics, namely the eigenvalues of the deformation tensors, and their logarithm.
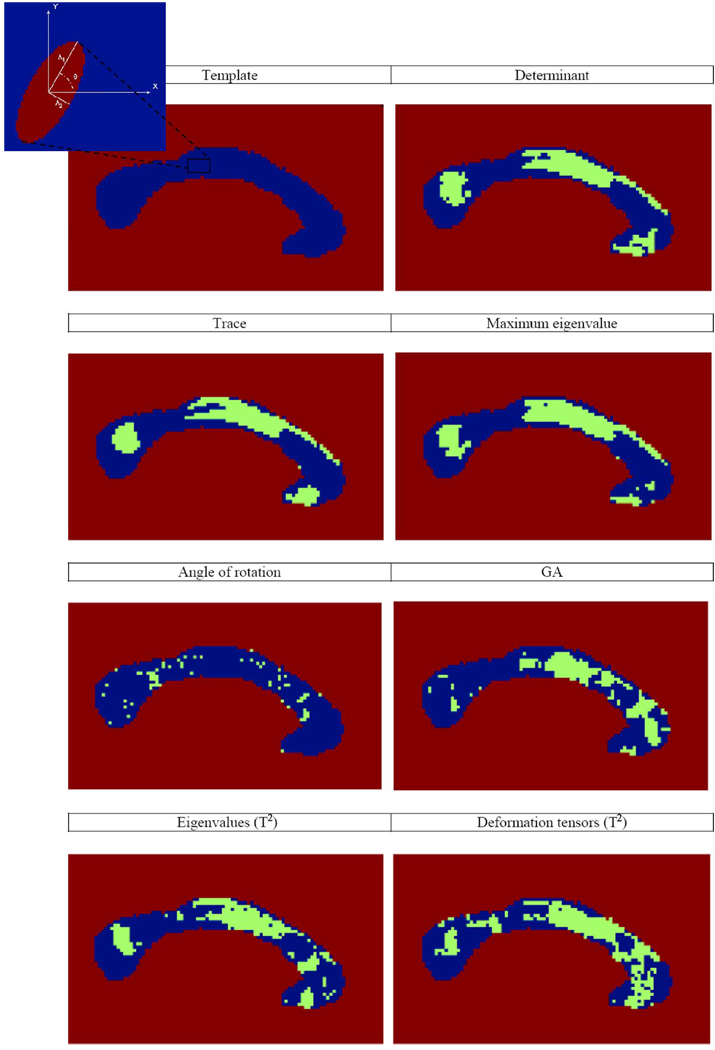
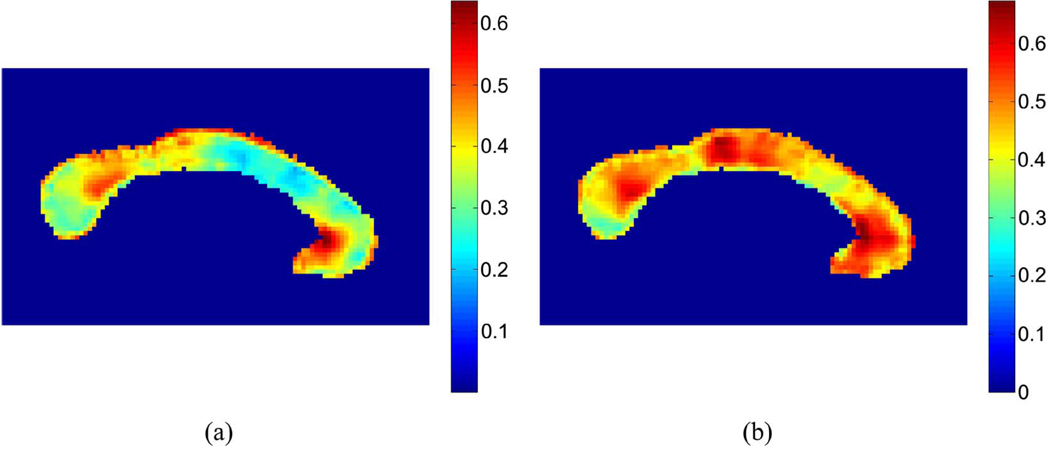
Fig. 2 shows the p-values from the Student’s t-test for all of the distributions. The TBM maps of the trace, determinant and maximum eigenvalues show atrophy mainly in the region of the splenium and the body. These correspond reasonably well with one of our prior studies which mapped the corpus callosum thickness (see [98]), where these two regions were significantly affected by the disease. In that study, the genu was also atrophied. Here, this result can only be seen via the multivariate statistics. Statistically significant differences in FA in the genu were also found in DTI analyses of the corpus callosum in HIV [99]. The total corpus callosum area is on average 11.5% lower in HIV/AIDS, and significant atrophy of the posterior body is detected in HIV patients. The trace, determinant, and principal eigenvalues give similar results, while the rotation angle is noisy and not very powerful for detecting regions of significant shape differences. This result may be due in part to the uncertainty in the estimation of the principal eigenvector when the tensors are oblate (i.e., two eigenvalues are similar). In this situation, the choice of rotation angle becomes arbitrary, which is important to consider if the statistics are defined on the deformation tensor, as the estimate of the rotation in the decomposition of the tensor may become quite noisy. Even so, the noise level is relatively stable even in regions where the average value of tanh(GA) is large, as shown in Fig. 1.
Fig. 2.
Green regions indicate p-values for which p < 0.05. From left to right and top to bottom. The template corpus callosum. The corner figure is an illustration of a deformation tensor at one voxel. The axes of the ellipse point in the directions of the eigenvectors, and their lengths represent the size of the associated eigenvalues. Scalar t-tests for group differences (AIDS versus controls) for various alternative measures of shape differences: the determinant (λ1 λ2) of the deformation tensor (pFDR = 0.018), trace (λ 1 + λ2) (pFDR = 0.028), maximum eigenvalue (λ1) (pFDR = 0.019), angle of rotation (θ) of the eigenvectors (pFDR = 0.44), and the geodesic anisotropy (pFDR = 0.034). Also shown are maps constructed from the multivariate Hotellings T2 tests: on the vector composed of the two eigenvalues (λ1, λ2) (pFDR = 0.016), and on the log of the deformation tensors (pFDR = 0.010). The shape alterations in the genu (the most anterior part) of the corpus callosum are not detected unless a multivariate test is used. Even areas with more subtle fiber degeneration are identified with the multivariate statistics on the full tensor.
Fig. 1.
Left: Average of tanh(GA) for the controls. Right: Average of tanh(GA) for the patients. (a) Controls. (b) Patients.
It is interesting to note that regions of significance for the geodesic anisotropy are similar to those where the deformation tensors perform better than the Jacobian determinant. This makes sense, as the two features tap different channels of information regarding the deformation, each of them is a scalar summary emphasizing different aspects of the deformation tensor. The GA describes the directionality of the shape dissimilarities. Thus, a high GA value indicates that the maximum eigenvalue (λ1) of the deformation tensor is large compared to the minimum one (λ2 in 2-D). However, as demonstrated by the toy model in Section I, the total volume difference λ1λ2 may still be small. Similarly, a high determinant says nothing about the anisotropy of the change.
Even though the log clearly performs better in most regions, we notice regions where the determinant outperforms the full tensor for this test. This can occur for several possible reasons. One such situation is when the eigenvalues are not very noisy, but the eigenvectors are, as the former are associated with the determinant, while the deformation tensors take into account both data. Any noise in the rotational component of the deformations tends to reduce the power of the deformation tensor relative to the determinant (as the determinant ignores rotations). A second situation is when the viscosity constants of the Navier operator are set such that the penalty on the Laplacian is much less than the penalty on the grad-div term (a volume conservation term). This can lead to greater degrees of rotation in the flows (as would be typically found in an incompressible fluid). The rotation term can become very noisy, especially when the Jacobian determinant term is heavily regularized (close to a constant everywhere).
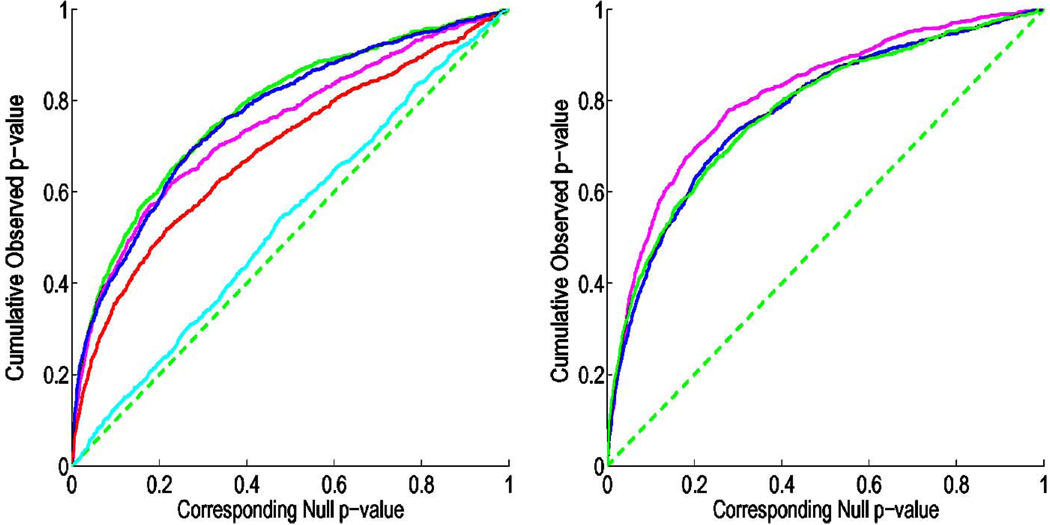
In Fig. 3, the cumulative distribution function of the p-values observed for the contrast of patients versus controls is plotted against the corresponding p-value that would be expected, under the null hypothesis of no group difference, for the different scalar and multivariate statistics. For null distributions, the cumulative distribution of p-values is expected to fall approximately along the x = y line (represented by the dotted line); large deviations from that curve are associated with significant signal, and greater effect sizes represented by larger deviations (the theory of false discovery rates gives formulae for thresholds that control false positives at a known rate). We note that the deviation of the statistics from the null distribution generally increases with the number of parameters included in the multivariate statistics, with statistics on the full tensor typically outperforming scalar summaries of the deformation based on the eigenvalues, trace or determinant.
Fig. 3.
This figure shows the cumulative distribution of p-values versus the corresponding cumulative p-value that would be expected from a null distribution for a) the various scalar statistics: trace (magenta), geodesic anisotropy (red), angle (cyan), maximum eigenvalue (blue), and the determinant (green). b) the determinant (green), the eigenvalues (blue) and the log of the deformation tensor (magenta). In FDR methods, any cumulative distribution plot that rises steeply is a sign of a significant signal being detected, with curves that rise faster denoting higher effect sizes. This steep rise of the cumulative plot relative to p-values that would be expected by chance can be used to compare the detection sensitivity of different statistics derived from the same data.
D. Whole Brain Statistics
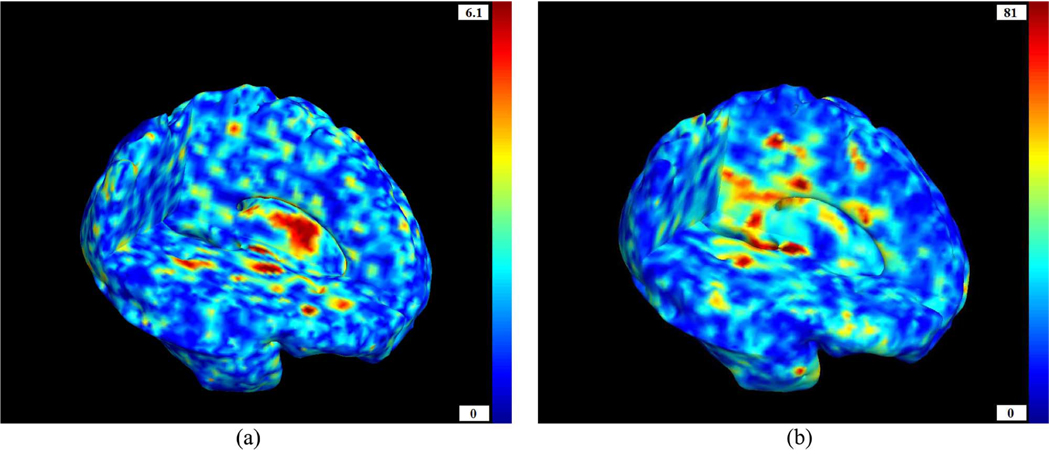
The geodesic anisotropy GA was found at each voxel using; tanh(GA) and is displayed in Fig. 4. The hyperbolic tangent of the geodesic anisotropy was used rather than GA itself, as it takes values in the interval [0,1], while those of GA span the interval [0,∞) (see [13]). We notice widespread directionality in the deformation tensors, which strengthens the idea that valuable information may be found from the anisotropic components of the changes.
Fig. 4.
Left: Voxelwise geodesic anisotropy calculated from 3. Right: Its hyperbolic tangent. Most of the brain shows anisotropy, meaning that volume elements in the average anatomy of the HIV/AIDS brain are not just isotropically contracted versions of their homologs in the normal brain; there are some preferred directions to the contraction of tissue. This directional preference is a type of information that is discarded when the Jacobian determinants are examined in TBM, but can be exploited by multivariate tests on the full tensor, providing additional power to detect disease effects. (a) GA. (b) tanh(GA).
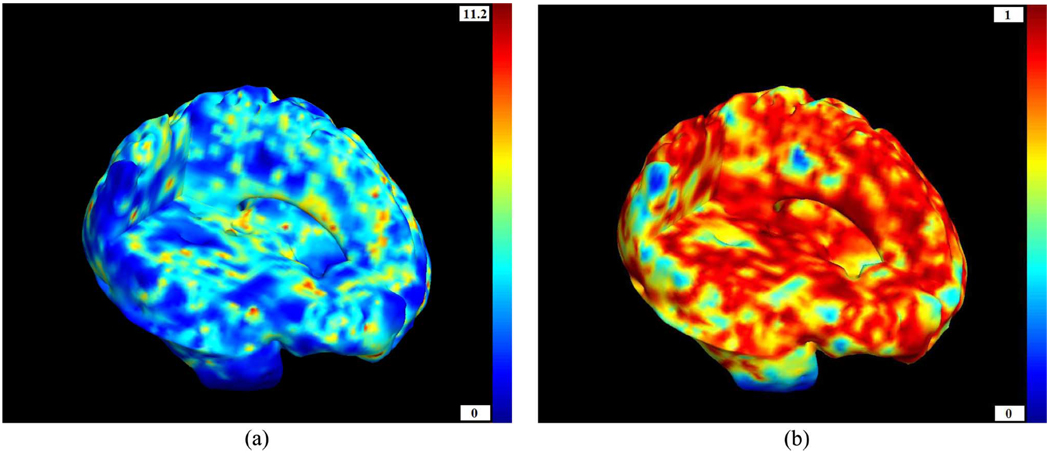
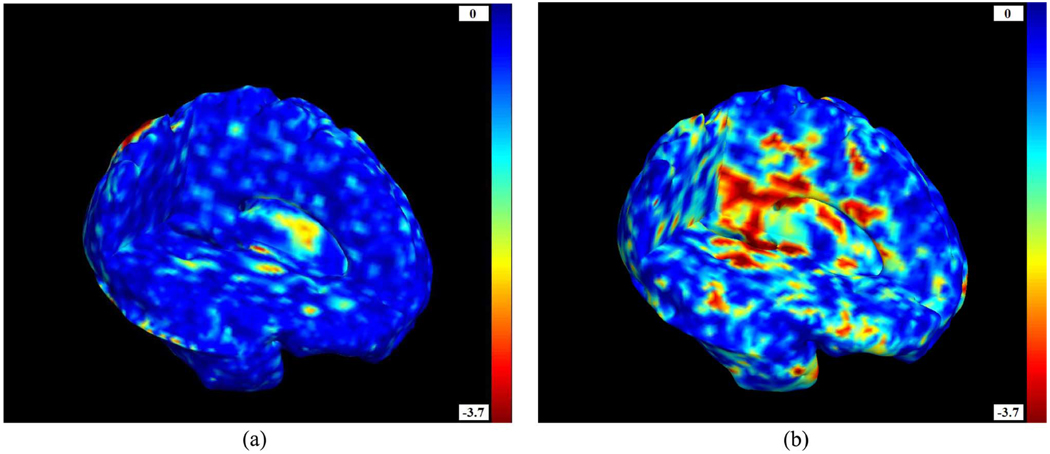
Fig. 6(a) shows the p-values from the Student’s t-test [Fig. 5(a)] applied to the log10(J) distribution. T 2 values and their corresponding p-values are shown in Fig. 5(b) and Fig. 6(b), respectively. The p-values are considerably lower in the case of the Hotelling’s T 2 test. The T 2 test is thus more sensitive, and can detect differences that are not detected with the conventional method, even when operating on the same deformation fields. Group differences in brain structure between HIV/AIDS patients and healthy subjects are visible throughout the brain, with the greatest effect sizes in the corpus callosum and basal ganglia. That result essentially replicates the distribution of atrophy observed in our prior TBM studies of HIV/AIDS where conventional statistics were used [25], [26], and is consistent with results found in other volumetric studies (see [101] for a review of neuroimaging studies of HIV and AIDS patients). The white matter exhibits widespread atrophy, as shown by many prior studies of the neuropathology of HIV/AIDS (see for instance [1], [84]), (some of which imply that the virus invades brain tissue by traveling radially through the white matter from the ventricles, which are enriched with the virus). Atrophic effects associated with HIV/AIDS are not detected in the vicinity of the cortex, perhaps because the registration method is intensity-based and may not perform so well in that area.
Fig. 6.
Left: Voxelwise p- values given by the Student’s t-test for the logarithm of the Jacobian determinant log10(J) (pFDR = 0.29). Right: Voxelwise p-values computed from the Hotelling’s T2 test on the deformation tensors. P-values are shown on a log10 scale; —3 denotes a voxel level p-value of 0.001. A brain with three cutplanes is shown, with the brain facing to the right. As expected from the literature on the neuropathology of HIV/AIDS, greatest atrophic effects are found in the subcortical regions, which border on ventricular regions enriched in the virus. The multivariate methods show comparable patterns of atrophy to the standard method, but with much greater sensitivity (i.e., better signal to noise). (a) p(t). (b) p(T2).
Fig. 5.
Left: Voxelwise t-values for log10 J. Right: Voxelwise T2 for log(S). (a) t. (b) T2.
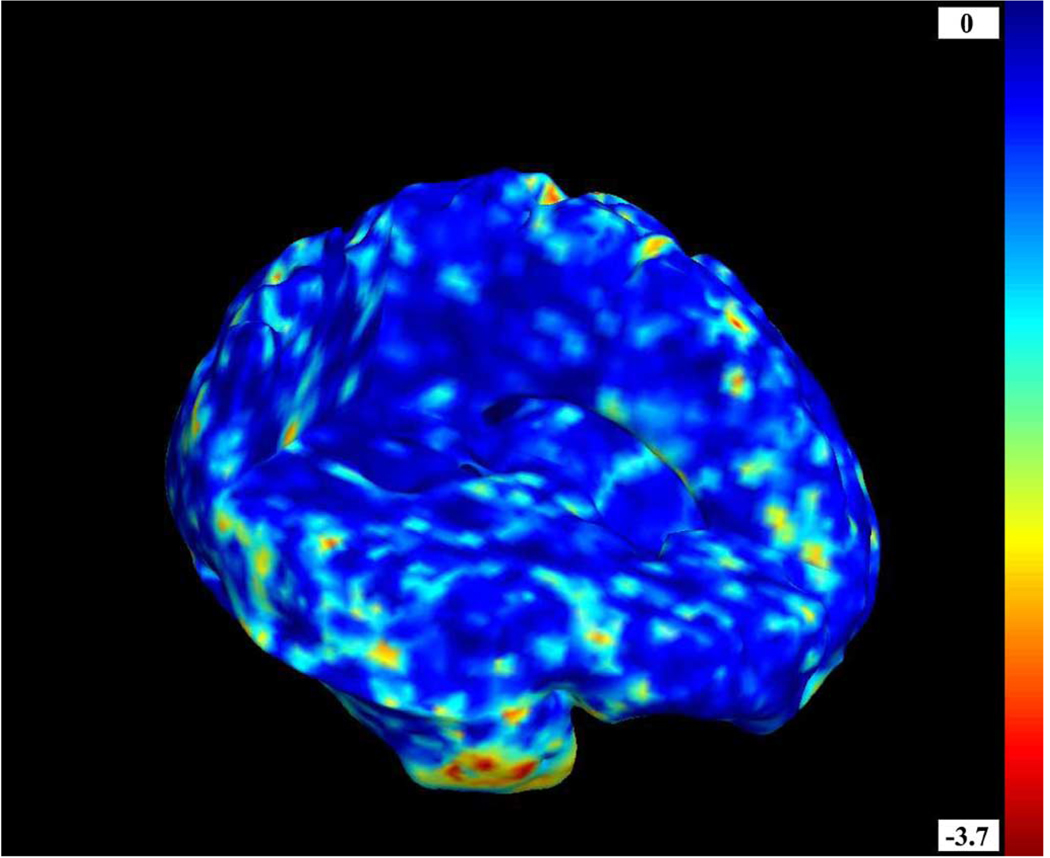
As the p-values were so low, we also performed experiments to make sure no effects were detected in truly null groupings of subjects where no disease-related difference was present. We distributed the data into two groups of randomly selected patients and controls (6 controls and 13 patients per group), and created a statistical map of evidence for group differences. The results of this test are shown in Fig. 7, which shows that the multiple statistical tests are controlled for Type I errors (false positives) at the appropriate expected rate.
Fig. 7.
P-values are shown on a log10 scale from a permutation distribution assembled by comparing two groups of subjects who do not differ in diagnosis. Two subsets of 13 randomly selected patients and six randomly selected controls were compared, i.e., the sample was split in half and assigned to two groups randomly, while ensuring each group had equal numbers of patients and controls. As expected, no significant morphometric difference is detected. In any null random field, on average a proportion t of the voxels will have p-values lower than any fixed threshold t in the range zero to one. This accounts for the appearance of low p- values in some brain regions (e.g., the right temporal pole, shown here in red and yellow colors).
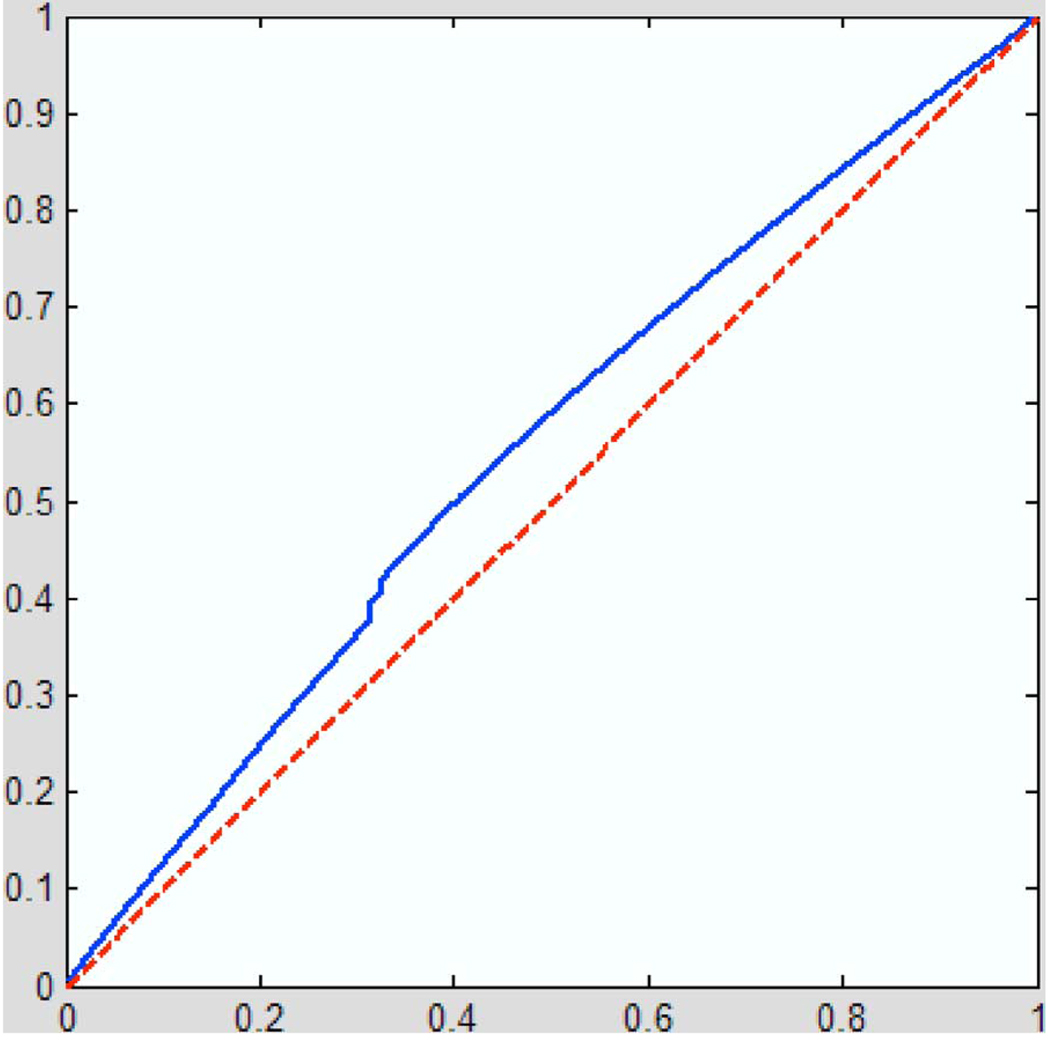
A serious concern in evaluating the added detection sensitivity of a proposed statistic is to ensure that it has the appropriate null distribution when no signal is present. In an independent experiment based on a large cohort of normal subjects, we also examined at the cumulative distribution function of the p-values for the Hotelling’s T 2 statistic, for a group of 100 controls divided into two gender- and age-matched subsets of 50 subjects each. The results are shown in Fig. 8. As expected for a null distribution (i.e., a statistical contrast that has been deliberately designed so that no effect is present), the results fall close to the diagonal. Strictly speaking many repeated large and independent samples would be required to prove that this p-value distribution were uniform on the interval [0,1], but this large control population suggest that the multivariate statistics are able to control for false positives at the expected rate.
Fig. 8.
Cumulative distribution function of the p-values versus the p-values for 100 controls divided into two equal size groups matched for age and gender (solid line). The dashed line shows the expected value of the null distribution.
IV. CONCLUSION
Here we used a combination of tensor-based morphometry, metrics on Riemannian manifolds, and multivariate statistics to detect systematic differences in anatomy between HIV/AIDS patients and healthy controls. The anatomical profile of group differences is in line with studies using traditional volumetric methods, as the HIV virus is known to cause widespread neuronal loss and corresponding atrophy of gray and white matter, especially in subcortical regions. The multivariate method presented here shows this finding with a much greater power than conventional TBM. In TBM, the Jacobian of the deformation (or its logarithm) is commonly examined, and multiple regression is applied to the scalar data from all subjects at each voxel, to identify regions of volumetric excess or deficit in one group versus another. This is clinically relevant especially given the large efforts, for example in drug trials of neurodegenerative disease, to detect subtle disease effects on the brain using minimal numbers of subjects (or the shortest possible follow-up interval in a longitudinal imaging study). As such it is important to optimize not only the nonlinear registration methods that gauge alterations in brain structure, but also the statistical methods for analyzing the resulting tensor-valued data.
In neuroscientific studies using TBM, any added statistical power could be helpful in detecting anatomical differences, for instance in groups of individuals with neurodegenerative diseases, or in designs where the power of treatment to counteract degeneration is evaluated, such as a drug trial. Even so, a major caveat is necessary regarding the interpretation of this data. Strictly speaking, we do not have ground truth regarding the extent and degree of atrophy or neurodegeneration in HIV/AIDS. So, although an approach that finds greater disease effect sizes is likely to be more accurate than one that fails to detect disease, it would be better to compare these models in a predictive design where ground truth regarding the dependent measure is known (i.e., morphometry predicting cognitive scores or future atrophic change). We are collecting this data at present, and any increase in power for a predictive model may allow a stronger statement regarding the relative power of multivariate versus scalar models in TBM.
Whether or not the multivariate statistics presented here offer greater value to a clinician or neuroscientist in a particular TBM study depends to some extent on the problem being studied. In some studies, it is of primary interest to know whether the volume of a particular structure is systematically reduced in disease, or whether the regional volume of gray matter, for example, is changing. In that context, the scalar Jacobian determinant would be a sufficient statistic for testing the hypothesis of regional alteration in volume. Even so, in longitudinal studies of growth for example, it is often clear from the deformations tracking growth processes that the expansion is far greater in a specific direction, and this is both interpretable physically and relevant to understanding the factors that drive growth in the brain, as well as beneficial for optimizing the statistics to detect growth. In cases where the full multivariate tensor statistics outperform simpler scalar summaries, efforts may still be required in interpreting and understanding the sources of directional and rotational differences, which may be less intuitive but more powerful as indices of morphometry in development or disease.
For the measures based on eigenvalues or Jacobian determinants, there is a clear physical interpretation to the detected differences in terms of directionally preferential effects or changes over time. For the geodesic measures that combine rotational and scale differences, there is no immediately clear interpretation of the effects unless the statistics of the rotations and eigenvalues are also presented, as these decompositions allow the sources of variation to be examined independently. In the case of the corpus callosum, we have noted regions where the rotational and scaling components of the tensor show statistical differences in partially overlapping regions of the structure, while the statistics on the full tensor pick up differences in both of these areas. This is clearly a case where the best SNR is provided by the more general statistic but interpretation of results is eased by examination of statistics on the factorized components, which can readily be presented as well. The geodesic measures that include rotational components may be hypothesized a priori as more relevant than the scalar Jacobian when measuring brain growth over time for example. In that application, it is known that there is a significant arching of the brain as a whole during embryonic development and regionally after birth (see e.g. [92]), and so a study may deliberately seek to find differences in relative rotations between anatomical structures. The same issue arises in studying developmental brain asymmetry, in which the orientation of structures in one hemisphere versus the other changes progressively between childhood and adolescence, and this asymmetry (which is a rotational as well as volume asymmetry) is of neurodevelopmental interest as a morphometric index of maturation and lateralization [80]. Even in these cases, where geodesic and other rotationally-sensitive measures may be of a priori interest, it would still be beneficial to show statistics on components of the full tensor to identify the partially independent effects of rotation and scale during the growth process.
The current work can be expanded in several ways. First, in a multivariate setting, deformation fields could be analyzed using differential operators or tensors other than the deformation tensor or deformation gradient; Ashburner [9] proposed examining the Hencky strain tensors, instead of the logarithm of the scalar Jacobian. The current form of the deformation tensors is, in some respects, a natural extension of TBM. There are many commonly used alternative definitions of deformation tensors in the continuum mechanics literature, and another possibility would be to use instead. We also could have chosen JT J, but taking the square root allows for a more natural comparison with the usual TBM statistics (the determinant of the scalar Jacobian determinant) as det(S) = det(J). Using the deformation tensor as defined in this work removes the local component of the rotation, while retaining information on the local directional size differences.
We note that by examining the deformation tensor in this work, we are examining only the symmetric positive-definite part of the Jacobian matrix, and three remaining degrees of freedom (a rotational term) are still discarded and not used. The Log-Euclidean framework can be extended to analyze the full Jacobian matrices, performing computations on that space (see [5] for extensions of the Log-Euclidean framework to general matrix spaces). Going from six to nine parameters may therefore further increase the power of the multivariate statistics [106]. For sample sizes such as the one in this study, we decided to use statistics based on 6 deformation parameters to lessen the likelihood that artifacts from individual brains might appear in the results due to the large number of fitted parameters. There is also a need to study the potential interactions between the chosen deformation prior (regularizer) and the resulting null distributions of rotations arising from using the prior as a regularizer. We have found that priors that tend to conserve volume more strongly (e.g., some fluid models or priors regularizing the logarithm of the Jacobian determinant) may also result in large local rotations when image regions need to be displaced considerably to optimize the deformation energy. Future larger samples will allow us to estimate empirically the benefit of nine versus six parameters in the multivariate analysis and their interaction with the chosen regularizer [19].
The optimal tensor to use for detecting group differences in TBM may depend on the directional properties of the underlying disease or developmental process being analyzed, as well as the differential operator used to regularize the deformation. The standard regularization operators used in nonlinear image registration tend to penalize Laplacians and spatial gradients in the divergences of the deformation fields. Depending on the relative influence of these terms (which is controlled in the governing Navier-Stokes equations by viscosity coefficients), a redistribution of disease-related signal may occur between the rotational versus scaling components of the deformation tensor, and the level of noise in each of these tensor components may be differentially penalized by the partial differential operators controlling the deformation. These regularization effects are complex and will be the topic of further study. Finally, analytic formulae for the null distribution of the volume of the excursion sets of T 2-distributed random fields were recently computed by Cao and Worsley [22], [23], and these may also be applied in a Riemannian space setting to optimize signal detection. Before a complete random field theory of tensor-valued signals can be developed, further work is required to extend the concept of roughness and smoothness to tensor-valued data, as these tensors arise in the statistical flattening and omnibus significance testing for signal detection in scalar-valued random fields. Studies are also underway to incorporate the Log-Euclidean framework in the cost functions (or statistical priors) that regularize the nonlinear registrations themselves [75]. This suggests that these methods may be advantageous for both the registration and statistical analysis of tensor-valued data, which are in many respects complementary problems.
Acknowledgments
This work was supported by the National Institute for Biomedical Imaging and Bioengineering. The work of J. T. Becker was supported by the National Institute on Aging (under Grant AG016570 and Grant AG021431). J. T. Becker was also the recipient of a Research Scientist Development Award—Level II (MH01077). The work of P. M. Thompson was supported by the National Center for Research Resources under Grant EB01651, RR019771.
Contributor Information
Natasha Lepore, Laboratory of Neuro Imaging, Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, CA 90095 USA.
Caroline Brun, Laboratory of Neuro Imaging, Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, CA 90095 USA.
Yi-Yu Chou, Laboratory of Neuro Imaging, Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, CA 90095 USA.
Ming-Chang Chiang, Laboratory of Neuro Imaging, Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, CA 90095 USA.
Rebecca A. Dutton, Laboratory of Neuro Imaging, Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, CA 90095 USA
Kiralee M. Hayashi, Laboratory of Neuro Imaging, Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, CA 90095 USA
Eileen Luders, Laboratory of Neuro Imaging, Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, CA 90095 USA.
Oscar L. Lopez, Department of Psychiatry, University of Pittsburgh Medical Center, Pittsburgh, PA 15213 USA
Howard J. Aizenstein, Department of Neurology, University of Pittsburgh, Pittsburgh, PA 15213 USA
Arthur W. Toga, Laboratory of Neuro Imaging, Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, CA 90095 USA
James T. Becker, Department of Psychiatry, University of Pittsburgh Medical Center, Pittsburgh, PA 15213 USA
Paul M. Thompson, Laboratory of Neuro Imaging, Department of Neurology, David Geffen School of Medicine, University of California, Los Angeles, CA 90095 USA
REFERENCES
- 1.Archibald SL, Masliah E, Fennema-Notestine C, Marcotte TD, Ellis RJ, McCutchan A, Heaton RK, Grant I, Mallory M, Miller A, Jernigan TL. Correlation of in-vivo neuroimaging abnormalities with postmortem Human Immunodeficiency Virus encephalitis and dendritic loss. Arch. Neurol. 2004;vol. 61:369–376. doi: 10.1001/archneur.61.3.369. [DOI] [PubMed] [Google Scholar]
- 2.Arsigny V, Fillard P, Pennec X, Ayache N. Fast and simple computations on tensors With log-Euclidean metrics INRIA, Research Report 5584. 2005 [Google Scholar]
- 3.Arsigny V, Commowick O, Pennec X, Ayache N. A fast and log-Euclidean polyaffine framework for locally affine registration INRIA, Res. Rep. 5865. 2006 [Google Scholar]
- 4.Arsigny V, Fillard P, Pennec X, Ayache N. Log-Euclidean metrics for fast and simple calculus on diffusion tensors. Magn. Reson. Med. 2006;vol. 56:411–421. doi: 10.1002/mrm.20965. [DOI] [PubMed] [Google Scholar]
- 5.Arsigny V. Ph.D. dissertation. Paris, France: Ecole Polytechnique; 2006. Processing data in lie groups: An algebraic approach. Application to non-linear registration and diffusion tensor MRI. [Google Scholar]
- 6.Ashburner J, Friston K. High-dimensional image registration using symmetric priors. NeuroImage. 1999;vol. 9:619–628. doi: 10.1006/nimg.1999.0437. [DOI] [PubMed] [Google Scholar]
- 7.Ashburner J, Friston K. Voxel-based morphometry—The methods. NeuroImage. 2000;vol. 11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 8.Ashburner J, Andersson JLR, Friston KJ. Image registration using a symmetric prior in three dimensions. Human Brain Mapp. 2000;vol. 9:212–225. doi: 10.1002/(SICI)1097-0193(200004)9:4<212::AID-HBM3>3.0.CO;2-#. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashburner J. Ph.D. dissertation. London, U.K.: Univ. College London; 2000. Computational neuroanatomy. [Google Scholar]
- 10.Ashburner J, Csernansky J, Davatzikos C, Fox NC, Frisoni G, Thompson PM. Computer-assisted imaging to assess brain structure in healthy and diseased brains. Lancet Neurol. 2003;vol. 2:79–88. doi: 10.1016/s1474-4422(03)00304-1. [DOI] [PubMed] [Google Scholar]
- 11.Avants B, Gee JC. Geodesic estimation for large deformation anatomical shape averaging and interpolation. NeuroImage. 2004;vol. 23 no. Suppl 1:139–150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Barillot C, Gee JC, Le Briquer L, Le Goualher G. Fusion Intraet Inter- Individus en Imagerie Médicale Appliquée á la modélisation Anatomique du Cerveau Humain. Traitement du signal. 1994;vol. 11:513–523. [Google Scholar]
- 13.Batchelor P, Moakher M, Atkinson D, Calamante F, Connelly A. A rigorous framework for diffusion tensor calculus. Magn. Reson. Med. 2005;vol. 53:221–225. doi: 10.1002/mrm.20334. [DOI] [PubMed] [Google Scholar]
- 14.Beg MF, Miller MI, Trouvé A, Younes L. Computing large deformation metric mappings via geodesic flow on diffeomorphisms. Int. J. Computer Vision. 2005;vol. 61:139–157. [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc.: Series B (Statistical Methodology) 1995;vol. 57:289–300. [Google Scholar]
- 16.Bookstein FL. Morphometric Tools for Landmark Data: Geometry and Biology. Cambridge, U.K.: Cambridge Univ. Press; 1991. [Google Scholar]
- 17.Bro-Nielsen M, Gramkow C. Fast fluid registration of medical images. Proc. 4th Int. Conf. Visualization Biomed. Comput.; Sep. 22–25, 1996; Hamburg, Germany. pp. 267–276. [Google Scholar]
- 18.Bro-Nielsen M. Ph.D. dissertation. Copenhagen, Denmark: Danish Tech. Univ.; 1996. Medical image registration and surgery simulation. [Google Scholar]
- 19.Brun C, Lepore N, Pennec X, Chou Y-Y, Lopez OL, Aizenstein HJ, Becker JT, Toga AW, Thompson PM. Comparison of standard and Riemannian elasticity for tensor- based morphometry in HIV/AIDS. presented at the IEEE Computer Soc. Workshop Math. Methods Biomed. Image Anal. (MMBIA); Oct. 1, 2007; Rio de Janeiro, Brazil. [Google Scholar]
- 20.Cachier P, Mangin J-F. Registration of cortical connectivity matrices. presented at the Int. Conf. Math. Methods Biomed. Image Anal. (MMBIA); Jun. 17–18, 2006; New York. [Google Scholar]
- 21.Cao J. Ph.D. dissertation. Montreal, QC, Canada: Dept. Math. Stat., McGill Univ.; 1997. The excursion set of random fields with applications to human brain mapping. [Google Scholar]
- 22.Cao J, Worsley KJ. The detection of local shape changes via the geometry of the Hotelling’s T2 fields. Ann. Stat. 1999;vol. 27:925–942. [Google Scholar]
- 23.Cao J, Worsley K. Application of random fields in human brain mapping. In: Moore M, editor. Spatial Statistics: Methodological Aspects and Applications. vol. 159. New York: Springer; 2001. pp. 169–182. [Google Scholar]
- 24.Castano-Moraga CA, Lenglet C, Deriche R, Ruiz-Alzola J. A fast rigorous anisotropic smoothing method for DT-MRI. Proc. Int. Symp. Biomed. Imag. (ISBI); Apr. 6–9, 2006; Arlington, VA. pp. 93–96. [Google Scholar]
- 25.Chiang MC, Dutton RA, Hayashi KM, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. Fluid Registration of medical images using Jensen-Rényi divergence reveals 3-D profile of brain atrophy in HIV/AIDS. Proc. Int. Symp. Biomed. Imag. (ISBI); Apr. 6–9, 2006; Arlington, VA. pp. 1172–1175. [Google Scholar]
- 26.Chiang MC, Dutton RA, Hayashi KM, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. 3-D pattern of brain atrophy in HIV/AIDS visualized using tensor-based morphometry. NeuroImage. 2007;vol. 34:44–60. doi: 10.1016/j.neuroimage.2006.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang MC, Leow AD, Dutton RA, Barysheva M, Rose S, McMahon KL, de Zubicaray GI, Toga AW, Thompson PM. Fluid registration of diffusion tensor imaging using information theory. IEEE Trans. Med. Imag. doi: 10.1109/TMI.2007.907326. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen GE. D.Sc. dissertation. St. Louis, MO: Electr. Eng. Dept., Washington Univ.; 1994. Deformable shape models for anatomy. [Google Scholar]
- 29.Christensen GE, Rabbitt RD, Miller MI. Deformable templates using large deformation kinematics. IEEE Trans. Image Process. 1996 Oct.vol. 5(no. 10):1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- 30.Christensen GE, Sonka M. Information processing in medical imaging. presented at the Inf. Process. Med. Imag. (IPMI); Jul. 11–15, 2005; Glenwood Springs, CO. [Google Scholar]
- 31.Chung MK, Lazar M, Alexander AL, Lu Y, Davidson R. Probabilistic connectivity measure in diffusion tensor imaging via anisotropic kernel smoothing. Wisconsin: Dept. Stat., Univ.; 2003. [Google Scholar]
- 32.Chung MK, Robbins SM, Dalton RJ, Davidson AL, Alexander ACE. Cortical thickness analysis in autism with heat kernel smoothing. NeuroImage. 2005;vol. 25:1256–1265. doi: 10.1016/j.neuroimage.2004.12.052. [DOI] [PubMed] [Google Scholar]
- 33.Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G. Automated multimodality medical image using information theory. Proc. Inf. Process. Med. Imag. (IPMI); Jun. 26–30, 1995; Ile de Berder, France. pp. 263–274. [Google Scholar]
- 34.Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3-D intersubject registration of MR volumetric data in standardized Talairach space. J. Computer Assisted Tomogr. 1994;vol. 18:192–205. [PubMed] [Google Scholar]
- 35.Commowick O, Stefanescu R, Fillard P, Arsigny V, Ayache N, Pennec X, Malandain G. Incorporating statistical measures of anatomical variability in atlas-to-subject registration for conformal brain radiotherapy. Proc. 8th Int. Conf. Med. Image Computing Computer-Assisted Intervention (MICCAI); Oct. 26–30, 2005; Palm Springs, CA. pp. 927–934. [DOI] [PubMed] [Google Scholar]
- 36.Corouge I, Fletcher PT, Joshi S, Gilmore JH, Gerig G. Fiber tract-oriented statistics for quantitative diffusion tensor MRI analysis. Proc. Int. Symp. Biomed. Imag. (ISBI); Apr. 6–9, 2006; Arlington, VA. pp. 93–96. [DOI] [PubMed] [Google Scholar]
- 37.Csernansky JG, Joshi S, Wang L, Haller JW, Gado M, Miller JP, Grenander U, Miller MI. Hippocampal morphometry in schizophrenia by high dimensional brain mapping. NeuroImage. 1998;vol. 95:11406–11411. doi: 10.1073/pnas.95.19.11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davatzikos C, Genc A, Xu D, Resnick S. Voxel-based morphometry using the RAVENS maps: Methods and validation using simulated longitudinal atrophy. NeuroImage. 2001;vol. 14:1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- 39.D’Agostino E, Maes F, Vandermeulen D, Suetens P. A viscous fluid model for multimodal nonrigid registration using mutual information. Med. Image Anal. 2003;vol. 7:565–575. doi: 10.1016/s1361-8415(03)00039-2. [DOI] [PubMed] [Google Scholar]
- 40.DoCarmo M. Riemannian Geometry. Cambridge, MA: Birkhauser; 1992. [Google Scholar]
- 41.Fillard P, Arsigny V, Pennec X, Thompson PM, Ayache N. Extrapolation of sparse tensor fields: Application to the modeling of brain variability. Proc. Int. Symp. Biomed. Imag. (ISBI); Apr. 6–9, 2006; Arlington, VA. pp. 786–789. [DOI] [PubMed] [Google Scholar]
- 42.Fillard P, Arsigny V, Pennec X, Ayache N. Clinical DTI estimation, smoothing and fiber tracking with the log-Euclidean metric. Proc. Int. Symp. Biomed. Imag. (ISBI); Apr. 6–9, 2006; Arlington, VA. pp. 786–789. [Google Scholar]
- 43.Fischl B, Liu A, Dale AM. Automated manifold surgery: Constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans. Med. Imag. 2001 Jan.vol. 20(no. 1):70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- 44.Fletcher PT, Joshi S. Principal geodesic analysis on symmetric spaces: Statistics of diffusion tensors. Proc. Eur. Conf. Computer Vis. (ECCV); Prague, Czech Republic. May 11–14, 2004; pp. 87–98. [Google Scholar]
- 45.Fletcher PT. Ph.D. dissertation. North Carolina, Chapel Hill: Dept. Comput. Sci., Univ.; 2004. Statistical variability in nonlinear spaces: Application to shape analysis and DT-MRI. [Google Scholar]
- 46.Fréchet M. Les éléments aléatoires de nature quelconque dans un espace distancié. Ann. Inst. H. Poincaré. 1948;vol. 10:215–310. [Google Scholar]
- 47.Fox NC, Cousens S, Schill R, Harvey RJ, Rossor MN. Using serial registered brain magnetic resonance imaging to measure progression Alzheimer disease: Power calculations and estimates of sample size to detect treatment effects. Arch. Neurol. 2000;vol. 57:339–344. doi: 10.1001/archneur.57.3.339. [DOI] [PubMed] [Google Scholar]
- 48.Gaser C, Nenadic I, Buchsbaum BR, Hazlett EA, Buchsbaum MS. Ventricular enlargement in schizophrenia related to volume reduction of the Thalamus, Striatum, and Superior Temporal Cortex. Am. Psychiatry. 2004;vol. 161:154–156. doi: 10.1176/appi.ajp.161.1.154. [DOI] [PubMed] [Google Scholar]
- 49.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. NeuroImage. 2002;vol. 15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 50.Gerig G, Gouttard S, Corouge I. Analysis of brain white matter via fiber tract modeling. Proc. IEEE Eng. Med. Biol. (IMBS); Sep. 1–5, 2004; San Francisco, CA. pp. 4421–4424. [DOI] [PubMed] [Google Scholar]
- 51.Good CD, Johnsrude IS, Ashburner J, Henson RNA, Friston KJ, Frckowiak RSJ. A voxel-based morphometric study of aging in 465 normal adult human brains. NeuroImage. 2001;vol. 14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- 52.Gramkow C, Bro-Nielsen M. Comparison of three filters in the solution of the Navier-Stokes equation in registration. Proc. Scandinavian Conf.Image Anal. SCIA; Jun. 9–11, 1997; Lappeenranta, Finland. pp. 795–802. [Google Scholar]
- 53.Gramkow C. M.S. thesis. Copenhagen, Denmark: Danish Tech. Univ.; 1996. Registration of 2-D and 3-D medical images. [Google Scholar]
- 54.Grenander U, Miller MI. Computational anatomy: An emerging discipline. Q. Appl. Math. 1998;vol. 56:617–694. [Google Scholar]
- 55.Hendry J, DeVito T, Gelman N, Densmore M, Rajakumar N, Pavlosky W, Williamson P, Thompson PM, Drost D, Nicolson R. White matter abnormalities in autism detected through transverse relaxation time imaging. NeuroImage. 2005;vol. 29:1049–1057. doi: 10.1016/j.neuroimage.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 56.Holm DD, Ratnanather A, Trouvé A, Younes L. Soliton dynamics in computational anatomy. NeuroImage. 2004;vol. 23:170–178. doi: 10.1016/j.neuroimage.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 57.Joshi SC. Ph.D. dissertation. St. Lois, MO: Univ. Washington; 1998. Large deformation diffeomorphisms and Gaussian random fields for statistical characterization of brain sub-manifolds. [Google Scholar]
- 58.Joshi AA, Shattuck DW, Thompson PM, Leahy RM. Simultaneous surface and volumetric brain registration using harmonic mappings. IEEE Trans. Med. Imag. doi: 10.1109/tmi.2007.901432. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karcher H. Riemannian center of mass and mollifier smoothing. Commun. Pure Appl. Math. 1977;vol. 30:509–541. [Google Scholar]
- 60.Kendall WS. Probability, convexity and harmonic maps with small image-I: Uniqueness and fine existence. Proc. Lond. Math. Soc. 1990;vol. 3:371–406. [Google Scholar]
- 61.Khurd P, Verma R, Davatzikos On characterizing and analyzing diffusion tensor images by learning the underlying manifold structure. Proc. IEEE Workshop Math. Methods Biomed. Imag. Anal. (MMBIA); Jun. 17–18, 2006; New York. pp. 61–68. [Google Scholar]
- 62.Lee AD, Leow AD, Lu A, Reiss AL, Hall S, Toga AW, Thompson PM. 3-D pattern of brain abnormalities in fragile X syndrome visualized using tensor-based morphometry. NeuroImage. 2006 doi: 10.1016/j.neuroimage.2006.09.043. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lenglet C, Rousson M, Deriche R. A statistical framework for DTI segmentation. Proc. Int. Symp. Biomed. Imag. (ISBI); Apr. 6–9, 2006; Arlington, VA. pp. 794–797. [Google Scholar]
- 64.Leow AD, Geng A, Becker J, Davis SW, Toga AW, Thompson PM. Inverse consistent mapping in 3-D deformable image registration: Its construction and statistical properties. presented at the Inf. Process. Med. Imag. (IPMI); Jul. 10–15, 2005; Glenwood Springs, CO. [DOI] [PubMed] [Google Scholar]
- 65.Leow AD, Soares JC, Hayashi KM, Klunder AD, Lee AD, Bearden CE, Monkul ES, Nicoletti MA, Cerchiari AP, Trakhen-broit M, Brambilla P, Sassi RB, Mallinger AG, Toga AW, Thompson PM. Asymmetrical effects of lithium on brain structure mapped in healthy individuals. J. Neurosci. accepted for publication. [Google Scholar]
- 66.Lepore N, Chou Y-Y, Brun C, Mani M, de Zubricaray GI, McMahon K, Wright M, Martin N, Toga AW, Thompson PM. Genetic influences on brain architecture and fiber architecture mapped using diffusion tensor imaging and tensor-based morphometry. 12th Annu. Meeting Organization Human Brain Mapp.; Jun. 11–15, 2006; Florence, Italy. [Google Scholar]
- 67.Lepore N, Brun C, Chou Y-Y, Chiang M-C, Dutton RA, Hayachi KM, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. Multivariate statistics of the Jacobian matrices in tensor-based morphometry and their application to HIV/AIDS. Proc. Int. Conf. Med. Image Computing Computer Assisted Intervention (MICCAI); Oct. 1–6, 2006; Copenhagen, Denmark. pp. 191–198. [DOI] [PubMed] [Google Scholar]
- 68.Manly KF, Nettleton D, Hwang JT. Genomics, prior probability and statistical tests of multiple hypotheses. Genome Res. 2004;vol. 14:997–1001. doi: 10.1101/gr.2156804. [DOI] [PubMed] [Google Scholar]
- 69.Memoli F, Thompson PM, Sapiro G. Brain surface warping via minimizing Lipschitz extensions. Proc. Workshop Math. Foundations Computational Anatomy (MFCA); Oct. 1, 2006; Copenhagen, Denmark. pp. 58–67. [Google Scholar]
- 70.Miller MI. Computational Anatomy: Shape, Growth and Atrophy Comparison via Diffeomorphisms. NeuroImage. 2004;vol. 23:19–33. doi: 10.1016/j.neuroimage.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 71.Nichols TE, Holmes AP. Non parametric permutation tests for functional neuroimaging: A primer with examples. Human Brain Mapp. 2001;vol. 15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pennec X, Fillard P, Ayache N. A riemannian framework for tensor computing. Int. J. Computer Vis. vol. 66:41–66. [Google Scholar]
- 73.Pennec X. Probabilities and statistics on Riemannian manifolds: Basic tools for geometric measurements. Proc. IEEE-EURASIP Workshop Non-Linear Signal Image Process. (NSIP); Jun. 20–23, 1999; Antalya, Turkey. pp. 194–198. [Google Scholar]
- 74.Pennec X. Probabilities and statistics on Riemannian manifolds: A geometric approach INRIA. Res. Rep. 5255. 2004 [Google Scholar]
- 75.Pennec X, Stefanescu R, Arsigny V, Fillard P, Ayache N. Riemannian elasticity: A statistical regularization framework for non-linear registration. Proc. 8th Int. Conf. Med. Image Comput. Computer-Assisted Intervention (MICCAI); Oct. 26–30, 2005; Palm Springs, CA. pp. 943–950. [DOI] [PubMed] [Google Scholar]
- 76.Rey D, Subsol G, Delingette H, Ayache N. Automatic detection and Segmentation of Evolving Processes in 3-D Medical Images: Application to Multiple Sclerosis. Proc. Inf. Process. Med. Imag. (IPMI); Jun. 28–Jul. 2 1999; Visegrad, Hungary. pp. 154–167. [DOI] [PubMed] [Google Scholar]
- 77.Schwartzman A, Dougherty RF, Taylor JE. Cross-subject comparison of principal diffusion direction maps. Magn. Reson. Med. 2005;vol. 52:1423–1431. doi: 10.1002/mrm.20503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwartzman A. Ph.D. dissertation. Stanford, CA: Dept. Stat., Stanford Univ.; 2006. Random ellipsoids and false discovery rates: Statistics for diffusion tensor imaging data. [Google Scholar]
- 79.Schormann T, Dabringhaus A. Statistics of nonlinear spatial distortions in histological images. In: Moore M, editor. Spatial Statistics: Methodological Aspects and Applications, Lecture notes in Statistics. vol. 159. New York: Springer; 2001. pp. 247–262. [Google Scholar]
- 80.Sowell ER, Thompson PM, Rex DE, Kornsand DS, Jernigan TL, Toga AW. Mapping Sulcal pattern asymmetry and local cortical surface gray matter distribution in vivo: Maturation in Perisylvian cortices. Cerebral Cortex. 2002;vol. 12:17–26. doi: 10.1093/cercor/12.1.17. [DOI] [PubMed] [Google Scholar]
- 81.Staib L, Duncan J. Deformable Fourier models for surface finding in 3-D images. Visualization Biomed. Comput.; Oct. 13–16, 1992; Chapel Hill, NC. pp. 90–104. [Google Scholar]
- 82.Storey JD, Tibshirani R. Estimating False Discovery Rates Under Dependence, With Applications to DNA Microarrays. Stanford, CA: Dept. Stat., Stanford Univ.; 2001. Tech. Rep. 2001–28. [Google Scholar]
- 83.Storey JD. A direct approach to false discovery rates. J. R. Stat. Soc.: Series B (Statistical Methodology) 2002;vol. 64:479–498. [Google Scholar]
- 84.Stout JC, Ellis RJ, Jernigan TL, Archibald SL, Abramson I, Wolfson T, McCutchan A, Wallace MR, Atkinson JH, Grant I. HIV Neurobehavioral Reseach Center group, Progressive cerebral volume loss in human immunodeficiency virus infection. Arch. Neurol. 1998;vol. 55:161–168. doi: 10.1001/archneur.55.2.161. [DOI] [PubMed] [Google Scholar]
- 85.Studholme C, Cardenas V, Schuff N, Rosen H, Miller B, Weiner M. Detecting spatially consistent structural differences in Alzheimer’s and fronto-temporal dementia using deformation morphometry. Proc. Int. Conf. Med. Image Computing Computer Assisted Intervention (MICCAI); Oct. 14–17, 2001; Utrecht, The Netherlands. pp. 41–48. [Google Scholar]
- 86.Studholme C, Cardenas V, Lampiris H, Meyerhoff D, Weiner M. Evidence for ongoing brain tissue loss in HIV+ subjects on retroviral therapy. presented at the 12th Ann. Meeting Organization Human Brain Mapp.; Jun. 11–15, 2006; Florence, Italy. [Google Scholar]
- 87.Studholme C, Drapaca C, Iordanova B, Cardenas V. Deformation-based mapping of volume change from serial brain MRI in the presence of local tissue contrast change. IEEE Trans. Med. Imag. 2006 May;vol. 25(no. 5):626–639. doi: 10.1109/TMI.2006.872745. [DOI] [PubMed] [Google Scholar]
- 88.Styner M, Gerig G, Lieberman J, Jones D, Weinberger D. Statistical shape analysis of neuroanatomical structures based on medial models. Med. Image Anal. 2003;vol. 7:207–220. doi: 10.1016/s1361-8415(02)00110-x. [DOI] [PubMed] [Google Scholar]
- 89.Thirion J-P. Fast non-rigid matching of 3-D medical images INRIA, Res. Rep. 2547. 1995 [Google Scholar]
- 90.Thirion J-P, Prima S, Subsol G, Roberts N. Statistical analysis of normal and abnormal dissymmetry in volumetric medical images. IEEE Workshop Biomed. Image Anal. (WBIA); Jun. 26–27, 1998; Santa Barbara, CA. pp. 74–83. [DOI] [PubMed] [Google Scholar]
- 91.Thompson PM, Schwartz C, Toga AW. High resolution random mesh algorithms for creating a probabilistic 3-D surface atlas of the human brain. NeuroImage. 1996;vol. 3:19–34. doi: 10.1006/nimg.1996.0003. [DOI] [PubMed] [Google Scholar]
- 92.Thompson PM, Toga AW. A surface-based technique for warping 3-dimensional images of the brain. IEEE Trans. Med. Imag. 1996 Aug.vol. 15(no. 4):1–16. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- 93.Thompson PM, Toga AW. Detection, visualization and animation of abnormal anatomic structure with a deformable brain atlas based on random vector field transformation. Med. Image Anal. 1997;vol. 1:271–294. doi: 10.1016/s1361-8415(97)85002-5. [DOI] [PubMed] [Google Scholar]
- 94.Thompson PM, Giedd JN, Woods RP, MacDonald D, Evans AC, Toga AW. Growth patterns in the developing brain detected by using continuum-mechanical tensor maps. Nature. 2000;vol. 404:190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- 95.Thompson PM, Cannon TD, Narr KL, van Erp T, Khaledy M, Poutanen V-P, Huttunen M, Lonnqvist J, Standertskjold-Nordenstam C-G, Kaprio J, Dail R, Zoumalan CI, Toga AW. Genetic influences on brain structure. Nature Neurosci. vol. 4:1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- 96.Thompson PM, Hayashi KM, Sowell ER, Gogtay N, Giedd JN, Rapoport JL, De Zubicaray GI, Janke AL, Rose SE, Semple J, Doddrell DM, Wang YL, Van Erp TGM, Cannon TD, Toga AW. Mapping cortical change in Alzheimer’s disease, brain development, and schizophrenia. NeuroImage. 2004;vol. 23 no. Suppl 1:S2–S18. doi: 10.1016/j.neuroimage.2004.07.071. [DOI] [PubMed] [Google Scholar]
- 97.Thompson P, Dutton R, Hayashi K, Toga A, Lopez O, Aizenstein H, Becker J. Thinning of the cerebral cortex visualized in HIV/AIDS reflects CD4+T-lymphocyte decline. Proc. Nat. Acad. Sci. USA. 2005;vol. 102:15647–15652. doi: 10.1073/pnas.0502548102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thompson PM, Dutton RA, Hayashi KM, Lu A, Lee SE, Lee JY, Lopez OL, Aizenstein HJ, Toga AW, Becker JT. 3-D mapping of ventricular and corpus callosum abnormalities in HIV/AIDS. NeuroImage. 2006;vol. 31:12–23. doi: 10.1016/j.neuroimage.2005.11.043. [DOI] [PubMed] [Google Scholar]
- 99.Thurnher MM, Castillo M, Stadler A, Rieger A, Schmid B, Sundgren PC. Diffusion tensor MR imaging of the brain in human immunodeficiency virus-positive patients. Am J. Neuroradiol. 2005;vol. 26:2275–2281. [PMC free article] [PubMed] [Google Scholar]
- 100.Tosun D, Reiss AL, Lee AD, Dutton RA, Hayashi KM, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Thompson PM. Use of 3-D cortical morphometry for mapping increased cortical gyrification and complexity in Williams Syndrome. Proc. Int. Symp. Biomed. Imag. (ISBI); Apr. 6–9, 2006; Arlington, VA. pp. 1172–1175. [Google Scholar]
- 101.Tucker KA, Robertson KR, Lin W, Smith JK, An H, Chen Y, Aylward SR, Hall CD. Neuroimaging in human immunodeficiency virus infection. J. Neuroimmunol. 2004;vol. 157:153–162. doi: 10.1016/j.jneuroim.2004.08.036. [DOI] [PubMed] [Google Scholar]
- 102.Vaillant M, Miller MI, Younes L, Trouvé A. Statistics on diffeomorphisms via tangent space representations. NeuroImage. 2004;vol. 23:161–169. doi: 10.1016/j.neuroimage.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 103.Wang J, Cohen MF. An iterative optimization approach for unified image segmentation and matting. Proc. IEEE Int. Conf. Computer Vision (ICCV); Oct. 17–20, 2005; Bejing, China. pp. 936–943. [Google Scholar]
- 104.Wells WM, Viola P, Atsumi H, Nakajima S, Kikinis R. Multimodal volume registration by maximization of mutual information. Med. Image Anal. 1996;vol. 31:35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- 105.Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration. J. Comput. Assist. Tomogr. 1998;vol. 22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- 106.Woods RP. Characterizing volume and surface deformation in an atlas framework: Theory, applications and implementation. NeuroImage. 2003;vol. 18:769–788. doi: 10.1016/s1053-8119(03)00019-3. [DOI] [PubMed] [Google Scholar]
- 107.Worsley KJ, Cao J, Paus T, Petrides M, Evans AC. Application of random field theory to functional connectivity. Human Brain Mapp. 1998;vol. 6:364–367. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<364::AID-HBM6>3.0.CO;2-T. [DOI] [PMC free article] [PubMed] [Google Scholar]