Abstract
A pseudo-testcross pedigree is widely used for mapping quantitative trait loci (QTL) in outcrossing species, but the model for analyzing pseudo-testcross data borrowed from the inbred backcross design can only detect those QTLs that are heterozygous only in one parent. In this study, an intercross model that incorporates the high heterozygosity and phase uncertainty of outcrossing species was used to reanalyze a published data set on QTL mapping in poplar trees. Several intercross QTLs that are heterozygous in both parents were detected, which are responsible not only for biomass traits, but also for their genetic correlations. This study provides a more complete identification of QTLs responsible for economically important biomass traits in poplars.
Keywords: QTL, pseudo-testcross, intercross marker, poplar
Introduction
Experimental crosses between different parents have served as a powerful tool for mapping quantitative trait loci (QTLs) that affect quantitatively inherited traits.1,2 Crossing two parents allows different but linked loci to be co-segregating, with which the segregation of an unknown QTL can be inferred from the segregation of observable markers. For many agricultural and experimental species, segregating crosses initiated with two contrasting inbred lines, such as the backcross, double haploid, RILs or F2, have been used for genetic mapping. A number of statistical models for QTL mapping have been originally established for these advanced crosses3–6 and have been instrumental for the mapping and identification of biologically meaningful QTLs.7,8
There is also a group of important species, like forest trees, whose biological properties prevent the generation of inbred lines and, therefore, of any advanced cross. However, because these species are highly heterozygous, their cross of one generation (F1) often displays substantial segregation and have many different types of segregation. Some loci may have four different alleles between the crossing parents, generating four genotype classes in the progeny. Many others may also follow the F2 pattern in a 1:2:1 ratio (called intercross loci) and the backcross pattern in a 1:1 ratio (called testcross loci).9 Using the testcross markers, i.e. those that are segregating in one parent but not in the other, Grattapaglia and Sederoff10 proposed a socalled pseudo-testcross strategy for linkage mapping in a controlled cross between two outbred parents. Although it only makes use of a portion of markers from the genome, this strategy provides a simple way for genetic mapping and has been widely utilized in practical mapping projects for outcrossing species.11,12
Current statistical mapping methods for the pseudo-testcross strategy are directly borrowed from available software developed for the backcross resulting from two inbred lines.13 However, although the markers used to estimate a putative QTL follow a testcross segregation type, the QTL may be segregating like an intercross gene given the parents’ outcrossing nature. The idea of mapping more heterozygous QTLs with less heterozygous markers was conceived by Haley et al.5 Stam14 developed a computer software package called JoinMap to map QTLs for outcrossing populations. More recently, Lin et al15 have proposed a likelihood model for estimating QTL locations and QTL effects in an outcrossed family by jointly considering possible QTL-marker linkage phases. For outcrossing parents, the linkage phase of alleles at the markers and a QTL bracketed by the markers is unknown. In this note, we use Lin et al’s intercross model to reanalyze a published data set for a poplar mapping project,16 in an attempt to provide a complete characterization of QTLs for biomass traits.
Materials and Methods
Mapping population
As one of the most important forest trees in biology and forestry, poplars have received an tremendous interest in genetic studies.17 We will use poplars as our study material to test the intercross model for QTL mapping. A mapping population used to map QTLs in hybrid poplars (Populus trichocarpa Torr. and A. Gray (T) × Populus deltoides Bartr. ex Marsh. (D)) was described by Yin et al12 and Wullschleger et al.16 It is an outbred backcross, TD1 × D2, which was produced by crossing a female interspecies hybrid derived from a T tree from Pacific Northwest and a D tree from Illinois with a different male D individual. Given the outcrossing nature of these two poplar species, this cross is expected to contain different types of heterozygous markers as listed in Lu et al.9 Using 171 genotypes from this cross, a number of segregating markers were generated from 92 microsatellite (simple sequence repeat, SSR) and 24 amplified fragment length polymorphism (AFLP) primer pairs. Of these markers, 92 SSR and 556 AFLP markers are segregating in maternal parent TD1 but not in paternal parent D2. A genetic linkage map was constructed from 544 of these testcross markers heterozygous in parent TD1. The map is composed of 19 linkage groups, equivalent to the Populus chromosome number. A complete description of genetic map construction was given in Yin et al.12
Poplar hybrids used for the mapping study were planted in the field using unrooted cuttings. A number of growth and biomass traits were measured for harvested poplar trees after one and two seasons of growth. These traits include the aboveground (leaf, branch, stem and cutting), belowground (fine and coarse root) and total biomass at two different years. The percentages of different organs over the total biomass were calculated (see for a detail16).
Statistical analysis
All the biomass partitioning traits were analyzed by a testcross model and Lin et al’s15 intercross model incorporating the segregation pattern of heterozygous QTLs in both parents TD1 and D2. Model selection criteria (AIC and BIC) were calculated to determine the optimal model that explains the mapping data. As a tutorial-type article on QTL mapping published in this journal, we will provide a procedure of deriving the intercross model and its computational algorithm in the Appendix, aimed to help interested readers understand the methodological development of QTL mapping. One of the most important and difficult issues in QTL mapping is to determine the critical threshold for claiming the significance of a QTL. Because of the unknown distribution of the likelihood ratio test statistic, it is difficult to derive an analytical solution of the threshold. Permutation tests that do not rely on the distribution of a test statistic18 will be used to determine the threshold.
Results
Both the testcross and intercross models were employed to detect the QTL that affect various biomass traits in poplar hybrids at the first two years of growth. The critical thresholds for declaring the existence of a QTL were determined from permutation tests. A number of QTLs were detected by these two models, but only those detected genomewide are reported in this report, in order to increase the standard of QTL detection.
Table 1 tabulates the results about the chromosomal positions and genetic effects of QTLs obtained from the intercross and testcross model. In year 1, QTLs were mostly detected to locate on linkage group 4, with one on linkage groups 1, 3 and 6, respectively. Two QTLs on linkage group 4 and one on linkage group 1 were detected by both models. The AIC and BIC values calculated under the two models consistently support the testcross model (Table 2). Thus, all the QTLs detected in year 1 are segregating due to maternal parent TD1. Leaf biomass, above-ground biomass, and total biomass were affected by the same QTL linked with marker S_29 on linkage group 4, suggesting the pleiotropic control of this QTL. Linkage group 4 was also found to harbor a QTL (near marker P_204_C) responsible for cutting biomass percentage. Above-ground biomass percentage includes two QTLs, one linked with marker S15_8 on linkage group 1 and the other linked with marker S8_29 on linkage group 4. Fine-root biomass percentage is controlled by a QTL linked with T4_7 on linkage group 3. All the QTLs detected each explain about 9%–15% of the phenotypic variation for biomass traits.
Table 1.
QTL detection and the parental origin of favorable QTL alleles for biomass partitioning at the first two years in the field for a hybrid poplar family
| Trait |
Intercross model |
Testcross model |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Linkage group | Marker position | p̂ | â | d̂ | LR | R | Linkage group | Marker position | â | LR | R | |
| Year 1 | ||||||||||||
| Leaf biomass | 4 | S8_29 | 0.84 | 0.73 | 0.15 | 22.88 | 0.14 | 4 | S8_29 | 0.91 | 21.87 | 0.14 |
| Cutting % | – | 4 | P_204_C | 0.05 | 21.54 | 0.08 | ||||||
| Above-ground biomass | – | 4 | S8_29 | 0.85 | 19.03 | 0.11 | ||||||
| Above-ground % | 1 | S15_8 | 1.00 | 0.05 | −0.02 | 26.11 | 0.15 | 1 | S15_8 | 0.05 | 24.88 | 0.15 |
| 4 | S8_29 | 1.00 | 0.06 | −0.01 | 22.44 | 0.11 | 4 | S8_29 | 0.06 | 21.23 | 0.11 | |
| Fine-root % | – | 3 | T4_7 | 0.41 | 15.91 | 0.10 | ||||||
| Stem % | 6 | P_204_C2 | 0.71 | 0.15 | −0.02 | 29.51 | 0.09 | – | ||||
| Total biomass | – | 4 | S8_29 | 0.75 | 15.61 | 0.10 | ||||||
| Year 2 | ||||||||||||
| Branch biomass | 13 | T4_10 | 0.82 | 0.92 | 0.23 | 35.03 | 0.14 | – | ||||
| Branch % | – | 8 | O_268_B | −0.04 | 17.19 | 0.24 | ||||||
| Cutting biomass | – | 12 | T11_4 | 0.39 | 15.96 | 0.13 | ||||||
| Leaf biomass | 13 | T4_10 | 0.81 | 0.79 | 0.18 | 31.03 | 0.14 | – | – | |||
| Above-ground biomass | 13 | T4_10 | 0.82 | 0.83 | 0.19 | 32.84 | 0.13 | – | – | – | ||
| Coarse-root biomass | – | 12 | S12_10 | 0.51 | 16.98 | 0.14 | ||||||
| Total biomass | 13 | T4_10 | 0.81 | 0.80 | 0.19 | 31.47 | 0.13 | – | – | – | ||
Notes: p̂, diplotype probability of maternal parent TD1; â, additive effect; d̂, dominant effect; LR, the log-likelihood ratio; R, the percentage of the phenotypic variation explained by a QTL.
Table 2.
Model selection based on both AIC and BIC criteria.
| Year | Trait | Linkage group |
Intercross model |
Testcross model |
||||
|---|---|---|---|---|---|---|---|---|
| LR | AIC | BIC | LR | AIC | BIC | |||
| 1 | Above-ground percentage | 1 | 26.11 | 7.17 | 12.76 | 24.88 | 5.21 | 8.54 |
| 4 | 22.44 | 7.30 | 13.18 | 21.23 | 5.35 | 8.98 | ||
| 1 | Leaf biomass | 4 | 22.88 | 7.28 | 13.13 | 21.87 | 5.32 | 8.90 |
In year 2, different QTLs were detected to affect biomass traits (Table 1). The testcross model detected a QTL near marker O_268_B on linkage group 8 for branch biomass percentage, a QTL near marker T11_4 on linkage group 12 for cutting biomass and a QTL near marker S12_10 on linkage group 12 for coarse-root biomass. Each of these QTLs explains about 13%–24% of the phenotypic variation for these traits. Because markers T11_4 and S12_10 are located on the same region of a chromosome, the linkage of different QTLs may be contribute to the correlation between cutting biomass and coarse-root biomass.
It is interesting to see that the intercross model identifies significant QTLs which could not be detected by the testcross model (Table 1). As compared to year 1, an increasing number of traits is controlled by intercross QTLs. In year 1, only one intercross QTL for stem biomass was mapped to marker P_204_C2 on linkage group 6, whereas an intercross QTL near marker T4_10 on linkage group 13 was observed for stem biomass, leaf biomass percentage, aboveground biomass, and total biomass in year 2. These intercross QTLs operate in an additive (d/a ~ 0) manner in year 1 and in a partially dominant (d/a = 0.20–0.30) manner in year 2, where a and d are the additive and dominant effects of a QTL, respectively. It is also interesting to find that QTL alleles in a coupling phase e of marker alleles derived from the maternal parent TD1 contribute to favorable effects on increasing values of these biomass traits. The intercross QTLs detected in year 2 account for about 13%–14% of the phenotypic variation.
Discussion
Although statistical models for QTL mapping have been well developed since the publication of Lander and Botstein’s3 seminal paper, the model development of QTL mapping in outbred populations, a group of species of great environmental and economical importance, has received limited attention. Stam14 and Lin et al15 proposed models and algorithms for QTL mapping of outcrossing traits, although the research from these two groups has a different focus. The latter incorporates the uncertainty on linkage phase, typical of outbred parents, into the mapping model, allowing a general formulation of mapping models. In this note, we used Lin et al’s model to map intercross QTLs that are segregating in both parents for a full-sib family of poplars. It is also our hope that, by providing a detailed procedure for model derivation (Appendix), interested readers of this journal can better understand general statistical principles behind QTL mapping, ultimately helping their result interpretations.
Although there is a set of QTLs segregating only in hybrid poplar TD1 (see also),16 the intercross model also finds those QTLs that are heterozygous in both maternal parent TD1 and paternal parent D2. Interestingly, different QTLs were detected between years 1 and 2. It is possible that the genetic control of quantitative traits is subjected to developmental changes. In a similar mapping experiment for poplar hybrids, Wu et al19 also detected different QTLs that affect growth traits between the first two years in the field. In the establishment year, trees grow from unrooted cuttings, which needs to allocate more energy to adapt themselves to the environment of the field. But after trees develop a rooting system, the pattern of their growth will be alternated to better use resources. It is likely that these processes in years 1 and 2 are controlled by QTLs located at different chromosomal positions and with different segregating patterns.
Results from this study help to provide the explanations about genetic mechanisms for trait correlations. Strong correlations among leaf biomass, above-ground biomass (also its percentage), and total biomass16 in year 1 may be due to a pleiotropic QTL detected on linkage group 4. It turns out that these correlations in year 2 are mediated by a different QTL on linkage group 13. On the other hand, a strong correlation between leaf biomass and cutting biomass percentage in year 1 may be due to the linkage of different QTLs located on a similar region of a chromosome (linkage group 4). The linkage of different QTLs on linkage group 12 may be responsible for the association between cutting biomass and coarse-root biomass in year 2. A detailed understanding of the genetic mechanisms for trait correlations and the developmental change of such correlations deserves further investigation.
Acknowledgments
We thank two anonymous reviewers for their constructive comments on this manuscript. RW’s group was supported by Joint NSF/NIH grant DMS/NIGMS-0540745 and the Changjiang Scholarship Award at Beijing Forestry University. Support of TY, SDW, and GAT was provided by U.S. Department of Energy (DOE), Office of Science, Biological and Environmental Research (BER). Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the DOE under contract DE-AC05-00OR22725.
Appendix.
Pseudo-testcross design
In what follows, we give a tutorial-type procedure for deriving the statistical model for mapping intercross QTLs in a pseudo-testcross pedigree for outcrossing species. Consider an F1 family for QTL mapping derived from two outbred parents P1 and P2. Suppose there is an intercross QTL of alleles Q and q that is inferred by two flanking testcross markers each with alleles 1 and 0. Thus, the genotypes of the two parents should be 10/Qq/10 (for P1 heterozygous at the markers) and 00/Qq/00 (for P2 homozygous at the markers), respectively. It is possible that the P1 genotype has two different diplotypes Φ1 = 1Q1|0q0 and Φ2 = 1q1|0Q0, assuming that the linkage phase between the two markers is known. Let p and (1 – p) be the probabilities for the F1 to carry diplotype Φ1 and Φ2, respectively. We use r, r1 and r2 to denote the recombination fractions between the two markers, between the first marker and QTL, and between the QTL and second marker, respectively. Under each diplotype, the frequencies of eight genotypes (comprising of two markers and one QTL) in the outcross between the P1 and P2 can be derived by assuming crossover independence and expressed in matrix form as
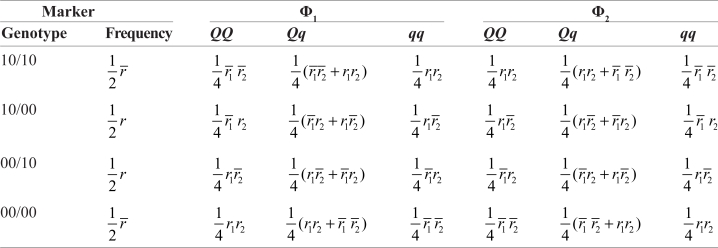 |
(1) |
where r̄ = 1 – r,r̄1 = 1 – r1, r̄2 = 1 – r2, and p̄ = 1 – p. Considering two possible diplo-types, the expected genotype frequencies in the crosss should be a mixture of the genotype frequencies weighted by the diplotype probabilities, expressed as
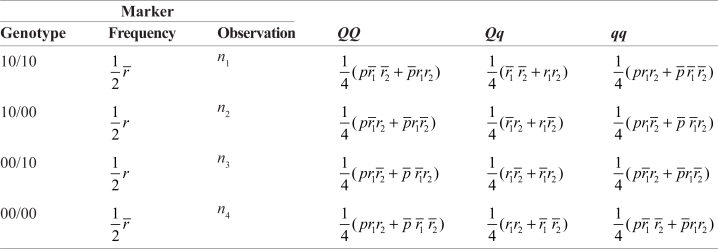 |
(2) |
where n1, …, n4 that sums to n are the observations of each marker genotype in the outcross. According to Bayes’ theorem, the conditional probability (denoted by ωj|i) of QTL genotype j (j = 2 for QQ, 1 for Qq and 0 for qq) given the marker genotype of individual i in the cross can be calculated using the probabilities in matrix (2). Obviously, such conditional probabilities for an outcross are different from those for a backcross, as expressed in matrix (1), that has a fixed parental diplotype.
Likelihood and estimation
In the outcross produced by the P1 and P2, n individuals are genotyped for a set of testcross markers (M) and phenotyped for a quantitative traits (y) that is controlled by an outcross QTL. The likelihood of observed phenotypes and markers can be expressed, in terms of three possible QTL genotypes in the outcross, as
| (3) |
where the mixture proportion, ωj|i, i.e. the conditional probability of a QTL genotype given the marker genotype, can be derived from matrix (2), and fj(y) is a normal distribution of the phenotypic trait, with mean μj and variance σ2.
The likelihood (3) contains the unknown parameters that define the the marker-QTL diplotype probability of the F1 (p), QTL position (r1, r2), QTL effects (μ2, μ1, μ0) and variance (σ2). These parameters are estimated by implementing the standard EM algorithm. The estimation process is described below.
The EM algorithm is implemented to estimate the unknown parameters (μj, σ2, r1, r2, p). In the E step, calculate the posterior probability of an individual i to carry a particular QTL genotype j by
In the M step, estimate the unknown parameters based on the calculated posterior probabilities, expressed as
where
An iteration is repeated between steps E and M until the estimates of the parameters converge. The values at the convergence are the maximum likelihood estimates of the parameters.
Hypothesis testing
The intercross model allows for the estimates of the additive (a) and dominant (d) genetic effects for a heterozygous QTL. The significance of a QTL is tested by formulating hypotheses
H0: a = d = 0,
H1: At least one of the equalities above does not hold.
Under each hypothesis, the plugging-in likelihood values are calculated, from which a log-likelihood ratio test statistics is estimated. The critical threshold for declaring the existence of a QTL can be determined on the basis of permutation tests.18 The additive and dominance effects of a QTL can be further tested.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors report no conflicts of interest.
References
- 1.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sinauer Associates; Sunderland, MA: 1998. [Google Scholar]
- 2.Wu RL, Ma CX, Casella G. Statistical Genetics of Quantitative Traits: Linkage, Maps, and QTL. Springer-Verlag; New York: 2007. [Google Scholar]
- 3.Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–99. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–68. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haley CS, Knott SA, Elsen JM. Mapping quantitative trait loci between outbred lines using least squares. Genetics. 1994;136:1195–207. doi: 10.1093/genetics/136.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu S. Estimating polygenic effects using markers of the entire genome. Genetics. 2003;163:789–801. doi: 10.1093/genetics/163.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frary A, Nesbitt TC, Frary A, Grandillo S, et al. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–8. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 8.Li CB, Zhou AL, Sang T. Rice domestication by reducing shattering. Science. 2006;311:1936–9. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 9.Lu Q, Cui YH, Wu RL. A multilocus likelihood approach to joint modeling of linkage, parental diplotype and gene order in a full-sib family. BMC Genetics. 2004;5:20. doi: 10.1186/1471-2156-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grattapaglia D, Sederoff RR. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics. 1994;137:1121–37. doi: 10.1093/genetics/137.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cervera MT, Storme V, Ivens B, Gusmao J, et al. Dense genetic linkage maps of three Populus species (Populus deltoides, P. nigra and P. trichocarpa) based on AFLP and microsatellite markers. Genetics. 2001;158:787–809. doi: 10.1093/genetics/158.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin TM, DiFazio SP, Gunter LE, Riemenschneider D, Tuskan GA. Large-scale heterospecific segregation distortion in Populus revealed by a dense genetic map. Theor Appl Genet. 2004;109:451–63. doi: 10.1007/s00122-004-1653-5. [DOI] [PubMed] [Google Scholar]
- 13.Lincoln S, Daly M, Lander ES.MAPMAKER/EXP. and MAPMAKER/QTL (Version 1.1b)Whitehead Institute Technical Report, 1992.
- 14.Stam P. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 1993;3:739–44. [Google Scholar]
- 15.Lin M, Lou XY, Chang M, Wu RL. A general statistical framework for mapping quantitative trait loci in non-model systems: Issue for characterizing linkage phases. Genetics. 2003;165:901–13. doi: 10.1093/genetics/165.2.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wullschleger SD, Yin TM, DiFazio SP, et al. Phenotypic variation in growth and biomass distribution for two advanced-generation pedigrees of hybrid poplar (Populus spp.) Can J For Res. 2005;35:1779–89. [Google Scholar]
- 17.Wullschleger SD, Jansson S, Taylor G. Genomics and forest biology: Populus emerges as the perennial favorite. Plant Cell. 2002;14:2651–5. doi: 10.1105/tpc.141120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–71. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu R, Bradshaw HD, Stettler RF. Developmental quantitative genetics of growth inPopulus. Theor Appl Genet. 1998;97:1110–9. [Google Scholar]


