Abstract
The WWOX gene, an archetypal fragile gene, encompasses a chromosomal fragile site at 16q23.2, and encodes the approximately 46-kDa Wwox protein, with WW domains that interact with a growing list of interesting proteins. If the function of a protein is defined by the company it keeps, then Wwox is involved in numerous important signal pathways for bone and germ-cell development, cellular and animal growth and death, transcriptional control and suppression of cancer development. Because alterations to genes at fragile sites are exquisitely sensitive to replication stress-induced DNA damage, there has been an ongoing scientific discussion questioning whether such gene expression alterations provide a selective advantage for clonal expansion of neoplastic cells, and a parallel discussion on why important genes would be present at sites that are susceptible to inactivation. We offer some answers through a description of known WWOX functions.
Keywords: breast cancer, chromosome fragile sites, mechanisms of tumor suppression, WW domain, Wwox interacting proteins
WWOX gene & gene product
The WWOX gene spans a genomic locus of more than 1 Mbp encompassing nine exons encoding an open reading frame of 1245 bp; the protein sequence includes two WW domains and a short-chain dehydrogenase/reductase (SDR) domain homologous to 17β-hydroxysterol reductase 3, which may be involved in sexsteroid metabolism. The gene spans the fragile site FRA16D and includes a genomic region involved in chromosome translocation in multiple myelomas and in hemi- and homozygous deletions (HDs) in cancers and cancer-derived cell lines; in addition, the WWOX promoter region is frequently hypermethylated in cancers (reviewed in [1–3]). Most cancer cell lines with FRA16D HDs also exhibit deletions in FRA3B and the FHIT gene, consistent with the finding that common fragile loci are highly susceptible to DNA damage and recombination. The mouse ortholog, Wwox, at murine chromosome 8E1, is also fragile and highly homologous to the human locus [4]. Wwox is expressed in most organs, but is expressed at highest levels in hormonally regulated, secretory epithelial cells such as those of breast, ovary, testes and prostate [1–3].
Wwox binds the proline-rich ligand PPxY and a number of proteins have been demonstrated to interact with its first WW domain; among these ligand-containing proteins are p73, Ap2α, Ap2γ, ErbB4, Jun and Runx2, which are described in more detail.
Characterization of mouse strains with targeted Wwox gene knockout has led to important clues to the roles of Wwox in tumorigenesis and metabolism (reviewed in [5]). At birth, homozygous Wwox-deficient (Wwox−/−) pups were indistinguishable from wild-type (WT) or heterozygous littermates; at 3 days, homozygous pups were smaller than littermates and all Wwox−/− mice died by 4 weeks after birth with severe metabolic defects [6–8]. Macroscopic and histological examination of the organs confirmed atrophy of organs, gonadal abnormalities and bone growth retardation in Wwox−/− mice [8]. A Wwox hypomorphic mouse has also been produced and demonstrated to have increased susceptibility to tumor induction [9]. In this mouse strain, it was difficult to demonstrate expression of Wwox protein, though some protein must be produced in multiple organs or these mice would not be normal. Testes from the homozygous males had high numbers of atrophic seminiferous tubules and reduced fertility compared with WT mice, and the hypomorphic allele led to a shorter lifespan.
Wwox expression in common human cancers
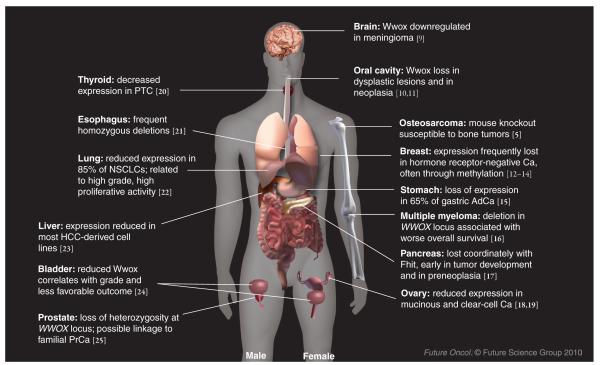
There are now approximately 100 reports concerning the correlation of the loss of Wwox expression with cancer development, including some reporting association of Wwox absence with poor prognosis and outcome in various cancer types (Figure 1 summarizes findings for cancers of many organs) [10–26]. Ectopically overexpressed Wwox has been reported to promote apoptosis, tumor suppression, suppression of anchorage-independent growth and colony formation in Matrigel™ (BD Biosciences, NJ, USA) [1–3]. Since many of these studies have been reviewed previously, we will highlight the most recent studies evaluating the role of Wwox in specific cancers.
Figure 1. Alteration of Wwox expression in common human cancers.
Summary of studies reporting loss or reduction of Wwox expression or alterations to WWOX alleles in cancers of many organs of both males and females.
AdCa: Adenocarcinoma; Ca: Carcinoma; HCC: Hepatocellular carcinoma; NSCLC: Non-small-cell lung cancer; PrCa: Prostrate cancer; PTC: Papillary thyroid carcinoma.
Very recently, Gourley et al. demonstrated that stable transfection of WWOX into human PEO1 ovarian cancer cells exhibiting WWOX HDs abolished tumorigenicity, but did not alter in vitro growth [27]. Rather, WWOX restoration or Wwox overexpression in ovarian cancer cells resulted in reduced attachment and migration on fibronectin, an extracellular matrix component linked to peritoneal metastasis. Conversely, siRNA-mediated knockdown of endogenous WWOX in ovarian cancer cells increased adhesion to fibronectin. There was not a WWOX-dependent difference in cell death in adherent cells but WWOX-transfected suspension cultures demonstrated enhanced apoptosis. WWOX expression also led to reduced membrane-associated integrin-α(3) protein, which mediates adhesion of ovarian cancer cells. The authors suggested a role for WWOX loss in dissemination of ovarian cancer, a function that may be amenable to therapeutic intervention [27].
In a high-throughput retroviral insertion site screen in mice, for mutations collaborating with p53 or p19 deficiency, Uren et al. identified 20 genes specifically mutated in p19-deficient, p53-deficient or WT mice, including candidate tumor suppressor genes [28]. Comparison with allele copy number data from human cancer cell lines revealed candidate tumor suppressors Wwox and Arfrp2 as retroviral insertion targets, suggesting that Wwox inactivation can cooperate with so called ‘classical’ tumor suppressor loss in tumor development.
Recent additional evidence appears to define Wwox as a central player in many physiological and pathological states, through connection of Wwox to the central Wnt–catenin signaling pathway. Bouteille et al. reported that Wwox physically interacts with the Dvl family signaling elements involved in the Wnt–catenin pathway, inhibiting the Wnt–catenin pathway transcriptional activity; the SDR domain was reportedly essential and sufficient for this inhibitory effect [29]. Wwox did not inhibit the Wnt–catenin pathway by disrupting the interaction of Dvl-2 with the β-catenin-degradation complex but by sequestering Dvl proteins in the cytoplasmic compartment, thereby inhibiting their function in β-catenin stabilization (illustrated in Figure 2). Aberrant activation of the Wnt–catenin pathway plays an important role in the development and progression of many human cancer types (reviewed in [30]); thus, negative regulation of this pathway by Wwox could have an inhibitory effect on both tumor initiation and progression towards an invasive phenotype.
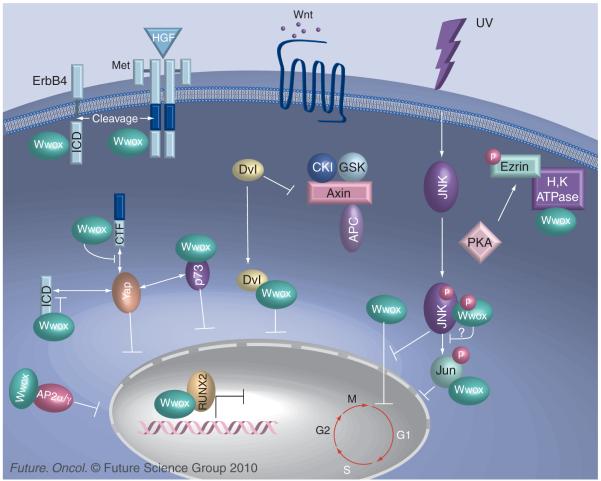
Figure 2. Wwox participates in multiple cancer-associated signal pathways through protein–protein interactions.
Wwox-partner protein interactions in signal pathways are affected by absence or reduction of Wwox expression. Wwox, via its first WW domain, binds PPxY domain-containing proteins and sequesters them in the cytoplasm, suppressing their transcriptional transactivation functions. Examples of these proteins are Ap2α/γ, p73, ICD of ErbB4 and juxtamembrane fragments of Met (CTF). Moreover, Wwox competes with other WW domain-containing proteins for binding to these interactor proteins; for example, Wwox outcompetes Yap for binding to the ErbB4 ICD and inhibits the Yap-induced ICD activity. In addition to sequestering the active ErbB4 and Met fragments, Wwox binds and stabilizes the full-length forms of these proteins. In osteoblasts and probably in some solid cancer cells, Wwox associates with chromatin-bound Runx2 and suppresses its transactivation function. Wwox also regulates the Wnt–β-catenin signaling pathway by preventing the nuclear import of the Dvl protein. In response to UV stress, the Wwox–Jun complex is significantly enhanced. This sequesters Jun in the cytoplasm, suppressing its transcriptional activity mediated through Jnk activation. According to work in Chang’s laboratory, Jnk1 may also bind to murine Wwox, blocking cell-cycle progression and inhibiting Wwox-mediated cell death [79]. Wwox physically interacts with ezrin after PKA-mediated phosphorylation of ezrin, facilitating proton pump H,K-ATPase recruitment to apical membrane during the parietal cell activation. It is likely that there is tissue specificity of individual pathways in signaling growth, stasis, cell or substrate interaction, metabolic activity, differentiation or cell death, with dependence on WW domain networks active in particular organs or contexts.
CKI: Casein kinase; CTF: C-terminal fragment; GSK: Glycogen synthase kinase; ICD: Intracellular domain; JNK: Jun N-terminal kinase; PKA: Protein kinase A.
Wwox WW domains & protein interactions
The Wwox N-terminal WW modules [31,32] mediate protein–protein interactions. WW domains are among the smallest modular domains known that mediate complexes associated with signaling pathways implicated in a variety of cellular processes, such as transcriptional regulation and protein stability [33]; they are composed of 35–40 amino acids that include two signature tryptophan residues, spaced approximately 20 amino acids apart, that are important in domain structure and function [33]. Based on ligand predilection, WW domains fall into two major and two minor groups. Major group I binds polypeptides with the minimal core consensus, PPxY, such as dystrophin, Nedd4 WW-3 and Yap [34], and major group II binds ligands with PPLP motif [35,36]. Group III WW domains select poly-P motifs flanked by arginine or lysine [37], whereas Group IV domains bind to short sequences with phosphoserine or phosphothreonine, followed by P, in a phosphorylation-dependent manner [38].
To ‘fish’ for Wwox partners in vitro, Hu et al. developed a biochemical approach to map WW domain peptide–protein interactions and determined that Wwox WW domains associated with PPxY-containing peptides [39]. Our in vivo validation studies confirmed these results and demonstrated that the first Wwox WW domain belongs to Group I WW [40], as previously reported [41]. Other laboratories have also reported and confirmed these results [9,42]. Although Wwox contains an SDR domain that is predicted to be involved in oxidation/reduction processes, Wwox signaling functions examined thus far are mainly determined by interaction of its WW domains with PPxY motifs in its partners.
Wwox–p73 association enhances apoptosis
The first Wwox partner to be identified was the p53 homolog, p73 [40]. A peptide derived from p73 (482PPPPY488) bound with high affinity to the first WW domain of Wwox, as predicted by Hu et al. [39], and co-immunoprecipitation results demonstrated strong complexes between Wwox and both p73α and β [40]. Under the same conditions where p73 interacts with Wwox, we were unable to recapitulate p53 binding to Wwox reported by another group [43], a discrepancy that is possibly related to different experimental conditions. Mutagenesis of Y487 of p73β abolished the Wwox–p73β interaction and p73 lacking a PPPY motif failed to bind Wwox. Furthermore, a mutation in Y33 in the first Wwox WW domain, but not Y61 in the second WW domain, abolished the interaction, indicating a specific association between the first WW domain and the PPPY motif in p73. Moreover, phosphorylation of Y33 by Src kinase enhanced Wwox–p73 interaction. Upon binding to Wwox, p73 is sequestered in the cytoplasm, whereas more p73 is translocated to the nucleus when Wwox is silenced by siRNA. In accordance with these findings, we observed a significant decrease in p73-transactivation ability upon Wwox co–expression, as well as a decrease in p21 protein level, due to decreased transcriptional activation of the gene encoding p21 by p73. This sequestration enhanced proapoptotic activity; Saos2 cells coexpressing Wwox and p73β exhibited an increased sub-G1 fraction, compared with Wwox or p73β alone, indicating that p73 binding to Wwox increases apoptotic activity independent of p73 transcriptional activity. While p73-dependent apoptosis seems to be primarily regulated by its ability to transcriptionally activate proapoptotic p53 target genes [44], some studies have suggested transactivation-independent apoptosis [45,46]. Therefore, it is possible that Wwox enhances p73 cytoplasmic apoptotic function. Another possibility is that Wwox can compete with other WW domain-containing proteins that bind and degrade p73 to potentiate or diminish p73 transcriptional and apoptotic activity [46]. Indeed, we have found that Wwox inhibits coactivation of p73 by Yap, while expression of Yap2 did not affect this suppression. When Wwox is in the nucleus together with p73, it still inhibits its association with Yap and thus prevents its coactivation, indicating that the effect of Wwox expression is superior to that of Yap. Recently, a caspase-cleaved p73 fragment was demonstrated to localize to the mitochondria and enhance TRAIL-induced apoptosis [45]. It is thus possible that following association with Wwox, p73 is cleaved in the cytoplasm and enhances transcription-independent apoptosis. Just as Wwox competes with Yap for p73, it may compete with Yap in the interaction with RUNX2 and determine its biological function.
Wwox–Ap2 complexes in breast cancer
Another candidate peptide was derived from Ap2 (56PPPYFPPPY64), which bound with high affinity to the first WW domain of Wwox [39,47]. We demonstrated that Ap2α and γ interact with the first Wwox WW domain via their prolinerich motif PPPY. Like p73, Wwox sequesters the Ap2α/β transcription factors in the cytoplasm, suppressing transactivation ability. Ap2α/γ function modulation by Wwox may have clinical relevance. Ap2s comprise a family of highly homologous proteins (reviewed in [48]) that recognize and bind GC-rich DNA sequences of target genes, mediating both activating and repressing stimuli. Transcriptional activity of Ap2 factors is highly determined by interacting molecules such as SP1, p53 and Myc [49–51]. Our data suggest that non-DNA-binding factors such as Wwox may also contribute to regulation of Ap2α/γ transcriptional function.
Although clinical studies concerning Ap2γ in breast cancer are controversial, recognition of its importance in breast carcinogenesis came from studies demonstrating it to be an essential regulator of breast-cancer-related genes in vitro [52,53]; also, the chromosomal locus of the AP2γ gene is known to be amplified in breast cancer and elevated expression of the gene encoding Ap2γ is associated with poor prognosis in breast cancer [54,55]. Ap2γ is reportedly overexpressed in breast cancer and overexpression correlates with poor prognosis [55,56]. Thus, reduced Wwox expression could result in increased Ap2γ activity and increased tumorigenicity. In a study designed to examine the correlation between Wwox interactor sequestration in the cytoplasm and tamoxifen resistance, it was found that lost or reduced expression of Wwox and high-level expression of Ap2γ and Her2 were significantly correlated with tamoxifen resistance, and Wwox and Ap2γ were independent markers of tamoxifen resistance. While Wwox expression was better than progesterone receptor in prediction of resistance, in high-risk patients, nuclear Ap2γ expression was better than Her2, especially in low-risk patients [57]. Another study assessed the relation of the basal-like phenotype to expression scores for Fhit, Wwox, Ap2α and Ap2γ and observed a highly significant association of the basal-like phenotype with very low expression of Fhit and Wwox and high expression of Ap2γ. According to the authors, nuclear Ap2α/γ expression was also more frequent in basal-like tumors, perhaps partially because of Wwox reduction, which would release these factors from cytoplasm to act as transcriptional regulators in the nucleus [58].
Wwox regulates ErbB4 localization & stability in breast cancer cells
Wwox also interacts with the ErbB4 tyrosine receptor kinase through its PPxY motifs and sequesters it in the cytoplasm, suppressing transcriptional coactivation by its intracellular domain (ICD), mediated by Yap [59]. ErbB4 plays an important role in cellular differentiation and proliferation [60], suggesting involvement in the pathogenesis and progression of various types of cancer [60–62]. Moreover, the prognostic value of ErbB4 in breast cancer is unclear; some studies report correlation with good clinical outcomes, and others with poor ones [60]. In an attempt to assess the clinical significance of the Wwox–ErbB4 association, we found that membranous expression of ErbB4, together with Wwox expression, is associated with favorable survival when compared with expression of membranous ErbB4 in the absence of Wwox [63]. This may be explained by the fact that Wwox prevents translocation of ErbB4 ICD into the nucleus and stabilizes the full-length ErbB4 at the cell membrane. This favors signaling via the full-length ErbB4 as opposed to nuclear ErbB4, defining a subgroup of breast-cancer patients with a favorable outcome. Recent evidence has suggested that sequestration of ICD in the cytoplasm is an important effector of tamoxifen-induced apoptosis of breast tumor cells [64]. Authors demonstrated that by disrupting the growth-promoting ErbB4/estrogen receptor-α coactivator complex in the nucleus, ErbB4-ICD accumulates within mitochondria and can trigger apoptosis through the activity of an intrinsic cell-killing BCL-2 homology 3 domain. Thus, it is possible that Wwox plays a role in breastcancer response to tamoxifen by sequestering ErbB4 ICD in the cytoplasm and enhancing its entry to mitochondria to induce apoptosis.
Analysis of Wwox-ErbB4 association also revealed that Wwox can compete with other WW domain-containing proteins, Yap and Itch, for binding common target proteins, such as ErbB4 and p73, hence determining functional outcomes. In one study, it was demonstrated that whereas Yap coactivates ICD transactivation function [65], the presence of Wwox, by competing for interaction with ICD, suppresses this coactivation [59]. In another study, it was demonstrated that Itch ubiquitylates ErbB4 CYT-1 isoform and promotes its degradation [60]. Therefore, it is possible that the different WW domain-containing proteins regulate the expression, localization and function of common partners, depending on their affinity of interaction and expression profiles in different contexts.
In the same manner, Wwox appears to regulate the HGF/Met system [66]. It has been demonstrated that Wwox expression stabilized the full-length Met in MDA-MB231 cells and prevented nuclear accumulation of the Met C-terminal fragment (CTF), likely impairing constitutive Met transcriptional activity. It was suggested that this effect of Wwox on the HGF/Met signal pathway reduces MDA-MB231 cell migration, prompting the hypothesis that Wwox could be involved in tumor progression towards a metastatic phenotype (see Figure 2 for summary of Wwox signal pathways). Moreover, the authors suggested that endogenous Yaps maintained Met CTF-constitutive transactivating activity in MDA-MB231 cells, and that Met activity in MCF-7 cells was the reverse, because of elevated endogenous Wwox; exogenously expressed Yap1 and 2 increased Met CTF transactivating activity. This study gives another example of Wwox–Yap antagonistic effects; while Yaps maintain constitutively activated nuclear Met fragments that act as transcription factors, likely regulating genes modulating the motile phenotype, Wwox does the opposite.
Wwox associates with Jun following UV irradiation
Wwox was also defined as a partner of the transcription factor Jun, suppressing its transactivation ability. The two proteins physically interact, and overexpression of Mekk1 or UV radiation, which activate Jnk1, causing phosphorylation and activation of Jun [67], significantly enhances Wwox–Jun complex formation. Complex formation was abrogated by mutation of the first Wwox WW domain or the tyrosine in the Jun PPVY domain. Wwox sequesters Jun in the cytoplasm, suppressing the transcriptional activity mediated through Jnk activation. The Jun oncoprotein is extremely responsive to environmental signals, such as UV [68], while Wwox expression is reportedly reduced following UV radiation, perhaps owing to WWOX localization within a fragile site [69,70]. Given the role of Wwox as a tumor suppressor and potent regulator of Jun, its loss through UV exposure could be a novel mechanism for transformation and skin carcinogenesis.
Wwox associates with Runx2 & regulates osteoblast differentiation
Targeted ablation of the murine Wwox gene led to postnatal lethality, although by 3 weeks of age mice developed focal lesions along the diaphysis of their femurs resembling early osteosarcomas. Biochemical analysis of Wwox partners suggested that physical and functional association of Wwox with the master transcription factor specific for osteoblast differentiation, Runx2, might be responsible for development of osteosarcoma in Wwox-deficient mice [8]. This association suppresses Runx2 transactivation function. Interestingly, we observed impaired differentiation in osteoblasts isolated from Wwox-null mice, suggesting that osteosarcoma formation could be related to a differentiation defect in the osteoblast compartment. In fact, Wwox seems to be essential in regulating proliferation and maturation of osteoprogenitor cells during bone formation [8]. Runx2 levels increased in Wwox-deficient mice both in clavaria and femur bones. Since Wwox seems to have a central role in osteoblast differentiation, its loss might promote osteosarcoma formation. Of note, Runx2 is a target of other WW domain-containing proteins, including coactivators and ubiquitin ligases [71]. Therefore, in the absence of Wwox, the balance between the different WW domain adaptor proteins and Runx2 may determine the functional outcome of Runx2 expression. Interestingly, Wwox, which contains a nuclear localization signal [72] but is predominantly in the cytoplasm, interacts with Runx2 in the nucleus when the transcription factor is already bound to chromatin, and inhibits its transcriptional activity. Since Runx2 is upregulated in osteosarcoma [73], we speculate that Wwox loss may be partly responsible for this altered expression.
Other Wwox partners & functions
In addition to its role in transcriptional control and apoptosis induction, Wwox participates in other signaling functions. It has been reported that Wwox physically interacts with ezrin. The interaction was mediated through the first Wwox WW domain and the ezrin PPxY motif, and PKA-mediated phosphorylation of ezrin was essential and sufficient for the apical localization of Wwox protein. The disruption of this ezrin–Wwox interaction blocked remodeling of the apical membrane cytoskeleton associated with the translocation and insertion of H,K-ATPase into the apical membrane. Therefore, the authors speculated that the interaction between phosphoezrin and Wwox may mediate the apical membrane transformation from a resting to secreting state by facilitating proton pump H,K-ATPase recruitment to apical membrane during parietal cell activation [74]. Ezrin is the most ubiquitous ezrin/moesin/radixin (ERM) protein in epithelial cells and is thought to play a role in progression of several cancers. Ezrin is a member of the ERM family that acts as a linker between the plasma membrane and the actin cytoskeleton and generates propulsive forces driving cell migration. It has been reported that ezrin modulates remodeling of actin cytoskeleton and is implicated in tumor-cell migration and progression of certain tumors [75–77]. Thus, Wwox regulation of this protein may be a key event in preventing tumor progression.
Other Wwox-interacting partners, independent of the WW domain and PPxY motifs, have also been suggested. The murine Wwox (also called Wox1) protein reportedly interacts with p53 [78], Jnk1 [79], Tau [80] and Mdm2 ([43], reviewed in [72]).
Conclusion Wwox & fragility
Though we have not dealt with mechanisms of fragility (reviewed in [81]), location of the WWOX gene at one of the most active human chromosome fragile sites has had a major influence on the frequency of loss or reduction of Wwox expression in cancers (summarized in Figure 1). It seems highly unlikely that the frequent loss of Wwox expression does not contribute to a selective advantage for clonal expansion of cells within specific organs in some contexts, though the contexts have not been fully defined. Loss of Wwox expression is frequently correlated with hypermethylation of its regulatory regions in many cancers, rather than with allele deletion [1,3,82], a mechanism of silencing not known to be associated with susceptibility of fragile loci to replication stress.
The WWOX gene, like other fragile genes, has large introns, so that some replication stress-induced small deletions may fall entirely within introns, as has been observed for the FHIT locus, and may not contribute to clonal expansion, supporting arguments against a tumor suppressor role for fragile gene products. The WWOX locus has thus far not been examined in enough detail to precisely delineate deletion end points and will require further investigation to determine if such intron-only deletions occur.
The size of fragile loci, with large introns, could make the genes impervious to inactivation by some genetic alterations, such as exogenous DNA integration, or possibly to the use of fragile sites as targets for chromosomal evolution during speciation, as has been proposed [83]. It is also possible that fragile chromosome regions may serve as early warning systems for DNA damage [84–86]; when fragile sites are damaged, the cells must activate DNA damage response checkpoints, blocking further replication until errors are repaired. If some fragile site damage is not completely repaired in a few cells of some organs, no harm is done in the evolutionary sense, since such damage may not have consequences until well past reproductive age.
Bloom’s syndrome and Fanconi anemia are inherited syndromes associated with extreme susceptibility to cancer development, through mechanisms that have not been defined in detail. Both conditions are associated with chromosome instability. In very elegant studies of chromosome fragile sites, Chan et al. [87] and Naim and Roselli [88] have demonstrated that the Fanconi anemia proteins FANCD2 and FANCI specifically associate with common fragile site loci and have proposed that, after replication stress, sister chromatids are inter-linked at genetic loci with intrinsic replication difficulties, such as fragile sites; in Bloom’s syndrome cells, poor resolution of DNA linkages at fragile sites leads to increased numbers of anaphase bridges, micronuclei containing fragile-site DNA and deletions within fragile loci. Chan et al. proposed that cancer predisposition in Bloom’s syndrome patients may be due to ‘accumulated loss of tumor suppressor function of genes residing at fragile site loci’ [87].
Wwox as a tumor suppressor
Since Wwox protein expression is lost in cancers, rather than gained (gain is another possible consequence of fragility [89,90]), it was proposed, at its discovery, that it functions as a tumor suppressor [31]; its replacement in numerous Wwox-negative cancer-derived cell lines caused reduced cell growth in vitro and tumorigenicity in vivo [1–3].
In addition, analysis of Wwox-mutant mice demonstrated that Wwox functions as a bona fide tumor suppressor. Spontaneous osteosarcomas in juvenile Wwox−/− and lung papillary carcinomas in adult Wwox+/− mice were observed, and Wwox+/− mice developed significantly more ethyl nitrosourea-induced lung tumors and lymphomas and more N-nitrosomethylbenzylamine-induced forestomach tumors [6–8] in comparison with WT littermates. These tumors expressed Wwox protein, suggesting that haploinsufficiency of Wwox is cancer predisposing.
The mechanism of tumor suppressor function of Wwox involves apoptosis and, according to a recent report, modulation of the interaction between tumor cells and the extracellular matrix [27]. Wwox appears to play an important role in tumor progression because it interacts with and modulates the function of different proteins involved in tumor migration, invasion and metastasis. These proteins include ezrin, Dvl and Met. It appears also that Wwox, indirectly, affects cellular interaction with fibronectin [27]. Data from several laboratories suggest that Wwox, via its WW domains, partners with PPxY-containing proteins and modulates their functions. PPxY-independent interactions have also been reported. Moreover, Wwox can regulate gene function by competing with other WW domain-containing proteins, such as coactivators and ubiquitin ligases, for binding with targets, thus affecting their transactivation and degradation rate. The nature of the various interacting partners with which Wwox can physically associate suggests that Wwox plays a central role in various signal transduction pathways. Therefore, when Wwox is lost, in precancerous or cancer cells, many of these signaling pathways could be altered, contributing to the multistep process of tumorigenesis.
Future perspective
Although the mechanisms of fragility have been investigated through complete sequencing of many fragile loci, investigation of time of replication during S phase, investigation of matrix attachment sites, chromosome map position relative to Giemsa light and dark bands, frequency of repetitive sequences and AT and GC content [81], we still do not fully understand what causes their extreme susceptibility to replication stress. It will be very important in the near future to understand thoroughly the chromatin configuration of the most fragile of these regions for new clues to their fragile nature. Also, it would be interesting to determine if genes at fragile sites are particular targets of hypermethylation, perhaps after being damaged during replicative stress.
For specific fragile genes it is important to continue to investigate in detail the relationship between the sensitivity of specific loci to replication stress and the types of genomic damage that occur in the associated genes, especially in noncancer-derived cell clones. An investigation of the biological consequences to ‘normal’ cell clones after surviving replication stress and carrying deleted WWOX or FHIT genes could be useful in characterization of their roles in subsequent clonal expansion.
Perhaps most importantly, a more complete characterization of functions of fragile gene products is necessary to understand the ramifications of loss of expression of these proteins in normal and preneoplastic cellular contexts for the health of the animal or human individual in an environment that inevitably allows exposure to endogenous or exogenous genotoxic agents. For understanding Wwox function this means increasing the focus on discovery of Wwox interacting proteins, definition of the WW domain interaction networks in specific cell types, description of the biochemical function of the SDR domain in normal and neoplastic contexts, and characterization of the consequences of Wwox loss on growth, death or differentiation of the given cell type. Detailed definition of Wwox functions, through characterization of its signaling partners and signaling pathways, may lead to identification of new targets for inter vention in tumor development or progression. Also, characterization of the extent of protection of fragile loci from genotoxic damage may be a useful surrogate marker for the effectiveness of antioxidants in cancer prevention.
Executive summary.
The WWOX/FRA16D locus is a very frequent target of replication stress, leading to its frequent inactivation in cancers.
More complete characterization of WWOX genome and epigenome alterations in precancers could contribute to understanding of the role of fragile sites in cancer development.
The WWOX promoter is frequently hypermethylated, leading to gene silencing.
Wwox protein interacts with a number of transcription factors through its WW domains, sequestering them in the cytoplasm and abrogating their transcriptional functions, suggesting pathways through which Wwox expression contributes to suppression of tumors.
There are numerous other WW-domain-containing proteins, suggesting hierarchies of competing interactions that determine the outcome of WW domain signal networks in regulating differentiation and other biological processes.
The networks must be defined for specific cell and organ types to fully understand the consequences of modulation of expression of individual WW-domain-containing proteins in specific cellular contexts.
Footnotes
Financial & competing interests disclosure The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Zaidoun Salah, The Lautenberg Center for Immunology, Institute for Medical Research Israel–Canada, The Hebrew University-Hadassah Medical School, Pharmacy Building, 5th floor, Room 543, POB 12272, Jerusalem 91120, Israel Tel.: +972 2675 8707 Fax: +972 2642 4653 zaidouns@ekmd.huji.ac.il.
Rami Aqeilan, The Lautenberg Center for Immunology, Institute for Medical Research Israel–Canada, The Hebrew University-Hadassah Medical School, Pharmacy Building, 5th floor, Room 543, POB 12272, Jerusalem 91120, Israel Tel.: +972 2675 8609 Fax: +972 2642 4653 aqeilan@cc.huji.ac.il.
Kay Huebner, Department of Molecular Virology, Immunology and Medical Genetics, The Ohio State University Comprehensive Cancer Center, Biomedical Research Tower Room 916, 460 W 12 Ave., Columbus, OH, USA Tel.: +1 614 292 4850 Fax: +1 614 688 8675 kay.huebner@osumc.edu.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Ramos D, Aldaz CM. Wwox, a chromosomal fragile site gene and its role in cancer. Adv. Exp. Med. Biol. 2006;587:149–159. doi: 10.1007/978-1-4020-5133-3_14. [DOI] [PubMed] [Google Scholar]
- 2.Lewandowska U, Zelazowski M, Seta K, Byczewska M, Pluciennik E, Bednarek AK. Wwox, the tumour suppressor gene affected in multiple cancers. J. Physiol. Pharmacol. 2009;60(Suppl 1):47–56. [PubMed] [Google Scholar]
- 3.Aqeilan RI, Croce CM. Wwox in biological control and tumorigenesis. J. Cell. Physiol. 2007;212(2):307–310. doi: 10.1002/jcp.21099. [DOI] [PubMed] [Google Scholar]
- 4.Krummel KA, Denison SR, Calhoun E, Phillips LA, Smith DI. The common fragile site FRA16D and its associated gene WWOX are highly conserved in the mouse at Fra8e1. Genes Chromosomes Cancer. 2002;34(2):154–167. doi: 10.1002/gcc.10047. [DOI] [PubMed] [Google Scholar]
- 5.Del Mare S, Salah Z, Aqeilan RI. Wwox: its genomics, partners, and functions. J. Cell. Biochem. 2009;108(4):737–745. doi: 10.1002/jcb.22298. [DOI] [PubMed] [Google Scholar]
- 6.Aqeilan RI, Trapasso F, Hussain S, et al. Targeted deletion of Wwox reveals a tumor suppressor function. Proc. Natl Acad. Sci. USA. 2007;104(10):3949–3954. doi: 10.1073/pnas.0609783104.▪▪ Confirmation of the tumor susceptibility of Wwox-knockout mice.
- 7.Aqeilan RI, Hagan JP, Aqeilan HA, Pichiorri F, Fong LY, Croce CM. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res. 2007;67(12):5606–5610. doi: 10.1158/0008-5472.CAN-07-1081.▪ Demonstration of the increased susceptibility of Wwox-deficient mice to carcinogen-induced tumors.
- 8.Aqeilan RI, Hassan MQ, De Bruin A, et al. The Wwox tumor suppressor is essential for postnatal survival and normal bone metabolism. J. Biol. Chem. 2008;283(31):21629–21639. doi: 10.1074/jbc.M800855200.▪▪ Describes the important role of Wwox in bone metabolism.
- 9.Ludes-Meyers JH, Kil H, Nunez MI, et al. Wwox hypomorphic mice display a higher incidence of B-cell lymphomas and develop testicular atrophy. Genes Chromosomes Cancer. 2007;46(12):1129–1136. doi: 10.1002/gcc.20497.▪▪ Report of tumor susceptibility of mice expressing very little Wwox.
- 10.Aarhus M, Bruland O, Bredholt G, et al. Microarray analysis reveals down-regulation of the tumour suppressor gene WWOX and up-regulation of the oncogene TYMS in intracranial sporadic meningiomas. J. Neurooncol. 2008;88(3):251–259. doi: 10.1007/s11060-008-9569-6. [DOI] [PubMed] [Google Scholar]
- 11.Pimenta FJ, Gomes DA, Perdigao PF, et al. Characterization of the tumor suppressor gene WWOX in primary human oral squamous cell carcinomas. Int. J. Cancer. 2006;118(5):1154–1158. doi: 10.1002/ijc.21446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pimenta FJ, Cordeiro GT, Pimenta LG, et al. Molecular alterations in the tumor suppressor gene WWOX in oral leukoplakias. Oral Oncol. 2008;44(8):753–758. doi: 10.1016/j.oraloncology.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guler G, Uner A, Guler N, et al. The fragile genes FHIT and WWOX are inactivated coordinately in invasive breast carcinoma. Cancer. 2004;100(8):1605–1614. doi: 10.1002/cncr.20137. [DOI] [PubMed] [Google Scholar]
- 14.Nunez MI, Ludes-Meyers J, Abba MC, et al. Frequent loss of Wwox expression in breast cancer: correlation with estrogen receptor status. Breast Cancer Res. Treat. 2005;89(2):99–105. doi: 10.1007/s10549-004-1474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guler G, Uner A, Guler N, et al. Concordant loss of fragile gene expression early in breast cancer development. Pathol. Int. 2005;55(8):471–478. doi: 10.1111/j.1440-1827.2005.01855.x. [DOI] [PubMed] [Google Scholar]
- 16.Aqeilan RI, Kuroki T, Pekarsky Y, et al. Loss of Wwox expression in gastric carcinoma. Clin. Cancer Res. 2004;10(9):3053–3058. doi: 10.1158/1078-0432.ccr-03-0594. [DOI] [PubMed] [Google Scholar]
- 17.Jenner MW, Leone PE, Walker BA, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110(9):3291–3300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- 18.Nakayama S, Semba S, Maeda N, Aqeilan RI, Huebner K, Yokozaki H. Role of the WWOX gene, encompassing fragile region FRA16D, in suppression of pancreatic carcinoma cells. Cancer Sci. 2008;99(7):1370–1376. doi: 10.1111/j.1349-7006.2008.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gourley C, Paige AJ, Taylor KJ, et al. WWOX mRNA expression profile in epithelial ovarian cancer supports the role of WWOX variant 1 as a tumour suppressor, although the role of variant 4 remains unclear. Int. J. Oncol. 2005;26(6):1681–1689. doi: 10.3892/ijo.26.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nunez MI, Rosen DG, Ludes-Meyers JH, et al. Wwox protein expression varies among ovarian carcinoma histotypes and correlates with less favorable outcome. BMC Cancer. 2005;5:64. doi: 10.1186/1471-2407-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dias EP, Pimenta FJ, Sarquis MS, et al. Association between decreased Wwox protein expression and thyroid cancer development. Thyroid. 2007;17(11):1055–1059. doi: 10.1089/thy.2007.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nancarrow DJ, Handoko HY, Smithers BM, et al. Genome-wide copy number analysis in esophageal adenocarcinoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2008;68(11):4163–4172. doi: 10.1158/0008-5472.CAN-07-6710. [DOI] [PubMed] [Google Scholar]
- 23.Donati V, Fontanini G, Dell’omodarme M, et al. Wwox expression in different histologic types and subtypes of non-small cell lung cancer. Clin. Cancer Res. 2007;13(3):884–891. doi: 10.1158/1078-0432.CCR-06-2016. [DOI] [PubMed] [Google Scholar]
- 24.Park SW, Ludes-Meyers J, Zimonjic DB, Durkin ME, Popescu NC, Aldaz CM. Frequent downregulation and loss of WWOX gene expression in human hepatocellular carcinoma. Br. J. Cancer. 2004;91(4):753–759. doi: 10.1038/sj.bjc.6602023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramos D, Abba M, Lopez-Guerrero JA, et al. Low levels of Wwox protein immunoexpression correlate with tumour grade and a less favourable outcome in patients with urinary bladder tumours. Histopathology. 2008;52(7):831–839. doi: 10.1111/j.1365-2559.2008.03033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange EM, Beebe-Dimmer JL, Ray AM, et al. Genome-wide linkage scan for prostate cancer susceptibility from the University of Michigan prostate cancer genetics project: suggestive evidence for linkage at 16q23. Prostate. 2009;69(4):385–391. doi: 10.1002/pros.20891.▪ Suggests that the WWOX locus could be involved in familial prostate cancer.
- 27.Gourley C, Paige AJ, Taylor KJ, et al. WWOX gene expression abolishes ovarian cancer tumorigenicity in vivo and decreases attachment to fibronectin via integrin α3. Cancer Res. 2009;69(11):4835–4842. doi: 10.1158/0008-5472.CAN-08-2974.▪ Presents evidence that Wwox loss could have an important role in the spread of ovarian cancers.
- 28.Uren AG, Kool J, Matentzoglu K, et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133(4):727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouteille N, Driouch K, Hage PE, et al. Inhibition of the Wnt/β-catenin pathway by the Wwox tumor suppressor protein. Oncogene. 2009;28(28):2569–2580. doi: 10.1038/onc.2009.120.▪▪ Demonstrates a role for Wwox in a very important ‘classical’ tumor suppressor pathway.
- 30.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 31.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. Wwox, a novel WW domain-containing protein mapping to human chromosome 16q23.3–24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60(8):2140–2145.▪▪ The original report of WWOX cloning and characterization.
- 32.Ried K, Finnis M, Hobson L, et al. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 2000;9(11):1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 33.Sudol M, Recinos CC, Abraczinskas J, Humbert J, Farooq A. WW or WOW: the WW domains in a union of bliss. IUBMB Life. 2005;57(12):773–778. doi: 10.1080/15216540500389039. [DOI] [PubMed] [Google Scholar]
- 34.Rentschler S, Linn H, Deininger K, Bedford MT, Espanel X, Sudol M. The WW domain of dystrophin requires EF-hands region to interact with β-dystroglycan. Biol. Chem. 1999;380(4):431–442. doi: 10.1515/BC.1999.057. [DOI] [PubMed] [Google Scholar]
- 35.Bedford MT, Chan DC, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16(9):2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ermekova KS, Zambrano N, Linn H, et al. The WW domain of neural protein Fe65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J. Biol. Chem. 1997;272(52):32869–32877. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- 37.Bedford MT, Sarbassova D, Xu J, Leder P, Yaffe MB. A novel pro-Arg motif recognized by WW domains. J. Biol. Chem. 2000;275(14):10359–10369. doi: 10.1074/jbc.275.14.10359. [DOI] [PubMed] [Google Scholar]
- 38.Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283(5406):1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 39.Hu H, Columbus J, Zhang Y, et al. A map of WW domain family interactions. Proteomics. 2004;4(3):643–655. doi: 10.1002/pmic.200300632.▪▪ Important report of use of bioinformatics and in vitro testing to predict WW domain.
- 40.Aqeilan RI, Pekarsky Y, Herrero JJ, et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc. Natl Acad. Sci. USA. 2004;101(13):4401–4406. doi: 10.1073/pnas.0400805101.▪▪ First report of a Wwox WW domain interacting protein.
- 41.Ludes-Meyers JH, Bednarek AK, Popescu NC, Bedford M, Aldaz CM. WWOX, the common chromosomal fragile site, FRA16D, cancer gene. Cytogenet. Genome Res. 2003;100(1–4):101–110. doi: 10.1159/000072844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludes-Meyers JH, Kil H, Bednarek AK, Drake J, Bedford MT, Aldaz CM. Wwox binds the specific proline-rich ligand PPxY: identification of candidate interacting proteins. Oncogene. 2004;23(29):5049–5055. doi: 10.1038/sj.onc.1207680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang NS, Doherty J, Ensign A, Schultz L, Hsu LJ, Hong Q. Wox1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylated p53. J. Biol. Chem. 2005;280(52):43100–43108. doi: 10.1074/jbc.M505590200. [DOI] [PubMed] [Google Scholar]
- 44.Melino G, De Laurenzi V, Vousden KH. p73: friend or foe in tumorigenesis. Nat. Rev. Cancer. 2002;2(8):605–615. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- 45.Sayan AE, Sayan BS, Gogvadze V, et al. p73 and caspase-cleaved p73 fragments localize to mitochondria and augment Trail-induced apoptosis. Oncogene. 2008;27(31):4363–4372. doi: 10.1038/onc.2008.64. [DOI] [PubMed] [Google Scholar]
- 46.Pietsch EC, Sykes SM, McMahon SB, Murphy ME. The p53 family and programmed cell death. Oncogene. 2008;27(50):6507–6521. doi: 10.1038/onc.2008.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aqeilan RI, Palamarchuk A, Weigel RJ, Herrero JJ, Pekarsky Y, Croce CM. Physical and functional interactions between the Wwox tumor suppressor protein and the Ap-2γ transcription factor. Cancer Res. 2004;64(22):8256–8261. doi: 10.1158/0008-5472.CAN-04-2055.▪ First report of interaction of Wwox with a transcription factor important in breast cancer.
- 48.Pellikainen JM, Kosma VM. Activator protein-2 in carcinogenesis with a special reference to breast cancer – a mini review. Int. J. Cancer. 2007;120(10):2061–2067. doi: 10.1002/ijc.22648. [DOI] [PubMed] [Google Scholar]
- 49.Batsche E, Muchardt C, Behrens J, Hurst HC, Cremisi C. Rb and c-Myc activate expression of the E-cadherin gene in epithelial cells through interaction with transcription factor Ap-2. Mol. Cell. Biol. 1998;18(7):3647–3658. doi: 10.1128/mcb.18.7.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McPherson LA, Loktev AV, Weigel RJ. Tumor suppressor activity of Ap2α mediated through a direct interaction with p53. J. Biol. Chem. 2002;277(47):45028–45033. doi: 10.1074/jbc.M208924200. [DOI] [PubMed] [Google Scholar]
- 51.Pena P, Reutens AT, Albanese C, et al. Activator protein-2 mediates transcriptional activation of the Cyp11a1 gene by interaction with Sp1 rather than binding to DNA. Mol. Endocrinol. 1999;13(8):1402–1416. doi: 10.1210/mend.13.8.0335. [DOI] [PubMed] [Google Scholar]
- 52.Bosher JM, Totty NF, Hsuan JJ, Williams T, Hurst HC. A family of Ap-2 proteins regulates c-ErbB-2 expression in mammary carcinoma. Oncogene. 1996;13(8):1701–1707. [PubMed] [Google Scholar]
- 53.DeConinck EC, McPherson LA, Weigel RJ. Transcriptional regulation of estrogen receptor in breast carcinomas. Mol. Cell. Biol. 1995;15(4):2191–2196. doi: 10.1128/mcb.15.4.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanner Mm, Tirkkonen M, Kallioniemi A, et al. Increased copy number at 20q13 in breast cancer: defining the critical region and exclusion of candidate genes. Cancer Res. 1994;54(16):4257–4260. [PubMed] [Google Scholar]
- 55.Zhao C, Yasui K, Lee CJ, et al. Elevated expression levels of NCOA3, TOP1, and TFAP2C in breast tumors as predictors of poor prognosis. Cancer. 2003;98(1):18–23. doi: 10.1002/cncr.11482. [DOI] [PubMed] [Google Scholar]
- 56.Tanner MM, Tirkkonen M, Kallioniemi A, et al. Expression of AP-2 transcription factors in human breast cancer correlates with the regulation of multiple growth factor signalling pathways. Cancer Res. 1998;58(23):5466–5472. [PubMed] [Google Scholar]
- 57.Guler G, Iliopoulos D, Guler N, Himmetoglu C, Hayran M, Huebner K. Wwox and Ap2γ expression levels predict tamoxifen response. Clin. Cancer Res. 2007;13(20):6115–6121. doi: 10.1158/1078-0432.CCR-07-1282. [DOI] [PubMed] [Google Scholar]
- 58.Guler G, Huebner K, Himmetoglu C, et al. Fragile histidine triad protein, WW domain-containing oxidoreductase protein Wwox, and activator protein 2γ expression levels correlate with basal phenotype in breast cancer. Cancer. 2009;115(4):899–908. doi: 10.1002/cncr.24103.▪▪ Reports significant correlation between absence of Fhit and Wwox fragile gene products and the triple-negative subclass of breast cancer.
- 59.Aqeilan RI, Donati V, Palamarchuk A, et al. WW domain-containing proteins, Wwox and Yap, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;65(15):6764–6772. doi: 10.1158/0008-5472.CAN-05-1150.▪▪ Describes the beginning of a signal network for Wwox, through its WW domains, with relevance to breast cancer.
- 60.Sundvall M, Iljin K, Kilpinen S, Sara H, Kallioniemi OP, Elenius K. Role of ErbB4 in breast cancer. J. Mammary Gland Biol. Neoplasia. 2008;13(2):259–268. doi: 10.1007/s10911-008-9079-3. [DOI] [PubMed] [Google Scholar]
- 61.Gullick WJ. C-Erbb-4/Her4: friend or foe? J. Pathol. 2003;200(3):279–281. doi: 10.1002/path.1335. [DOI] [PubMed] [Google Scholar]
- 62.Junttila TT, Sundvall M, Maatta JA, Elenius K. ErbB4 and its isoforms: selective regulation of growth factor responses by naturally occurring receptor variants. Trends Cardiovasc. Med. 2000;10(7):304–310. doi: 10.1016/s1050-1738(01)00065-2. [DOI] [PubMed] [Google Scholar]
- 63.Aqeilan RI, Donati V, Gaudio E, et al. Association of Wwox with ErbB4 in breast cancer. Cancer Res. 2007;67(19):9330–9336. doi: 10.1158/0008-5472.CAN-07-2147. [DOI] [PubMed] [Google Scholar]
- 64.Naresh A, Thor AD, Edgerton SM, Torkko KC, Kumar R, Jones FE. The HER4/4ICD estrogen receptor coactivator and BH3-only protein is an effector of tamoxifen-induced apoptosis. Cancer Res. 2008;68(15):6387–6395. doi: 10.1158/0008-5472.CAN-08-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein Yap associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003;278(35):33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 66.Matteucci E, Bendinelli P, Desiderio MA. Nuclear localization of active HGF receptor Met in aggressive MDA-MB231 breast carcinoma cells. Carcinogenesis. 2009;30(6):937–945. doi: 10.1093/carcin/bgp080. [DOI] [PubMed] [Google Scholar]
- 67.Gaudio E, Palamarchuk A, Palumbo T, et al. Physical association with Wwox suppresses c-Jun transcriptional activity. Cancer Res. 2006;66(24):11585–11589. doi: 10.1158/0008-5472.CAN-06-3376. [DOI] [PubMed] [Google Scholar]
- 68.Devary Y, Gottlieb RA, Lau LF, Karin M. Rapid and preferential activation of the c-Jun gene during the mammalian UV response. Mol. Cell. Biol. 1991;11(5):2804–2811. doi: 10.1128/mcb.11.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ishii H, Mimori K, Inageta T, et al. Components of DNA damage checkpoint pathway regulate UV exposure-dependent alterations of gene expression of FHIT and WWOX at chromosome fragile sites. Mol. Cancer Res. 2005;3(3):130–138. doi: 10.1158/1541-7786.MCR-04-0209. [DOI] [PubMed] [Google Scholar]
- 70.Thavathiru E, Ludes-Meyers JH, MacLeod MC, Aldaz CM. Expression of common chromosomal fragile site genes, WWOX/FRA16D and FHIT/FRA3B is downregulated by exposure to environmental carcinogens, UV, and BPDE but not by IR. Mol. Carcinog. 2005;44(3):174–182. doi: 10.1002/mc.20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lian JB, Stein GS, Javed A, et al. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev. Endocr. Metab. Disord. 2006;7(1–2):1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 72.Chang NS, Hsu LJ, Lin YS, Lai FJ, Sheu HM. WW domain-containing oxidoreductase: a candidate tumor suppressor. Trends Mol. Med. 2007;13(1):12–22. doi: 10.1016/j.molmed.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 73.Papachristou DJ, Papavassiliou AG. Osteosarcoma and chondrosarcoma: new signaling pathways as targets for novel therapeutic interventions. Int. J. Biochem. Cell Biol. 2007;39(5):857–862. doi: 10.1016/j.biocel.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 74.Jin C, Ge L, Ding X, et al. PKA-mediated protein phosphorylation regulates ezrin–Wwox interaction. Biochem. Biophys. Res. Commun. 2006;341(3):784–791. doi: 10.1016/j.bbrc.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 75.Fais S. A role for ezrin in a neglected metastatic tumor function. Trends Mol. Med. 2004;10(6):249–250. doi: 10.1016/j.molmed.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 76.Khanna C, Wan X, Bose S, et al. The membrane–cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat. Med. 2004;10(2):182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 77.Bruce B, Khanna G, Ren L, et al. Expression of the cytoskeleton linker protein ezrin in human cancers. Clin. Exp. Metastasis. 2007;24(2):69–78. doi: 10.1007/s10585-006-9050-x. [DOI] [PubMed] [Google Scholar]
- 78.Chang NS, Pratt N, Heath J, et al. Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity. J. Biol. Chem. 2001;276(5):3361–3370. doi: 10.1074/jbc.M007140200. [DOI] [PubMed] [Google Scholar]
- 79.Chang NS, Doherty J, Ensign A. Jnk1 physically interacts with WW domain-containing oxidoreductase (Wox1) and inhibits Wox1-mediated apoptosis. J. Biol. Chem. 2003;278(11):9195–9202. doi: 10.1074/jbc.M208373200. [DOI] [PubMed] [Google Scholar]
- 80.Sze CI, Su M, Pugazhenthi S, et al. Down-regulation of WW domain-containing oxidoreductase induces Tau phosphorylation in vitro. A potential role in Alzheimer’s disease. J. Biol. Chem. 2004;279(29):30498–30506. doi: 10.1074/jbc.M401399200. [DOI] [PubMed] [Google Scholar]
- 81.Arlt MF, Durkin SG, Ragland RL, Glover TW. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst.) 2006;5(9–10):1126–1135. doi: 10.1016/j.dnarep.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 82.Kuroki T, Yendamuri S, Trapasso F, et al. The tumor suppressor gene WWOX at FRA16D is involved in pancreatic carcinogenesis. Clin. Cancer Res. 2004;10(7):2459–2465. doi: 10.1158/1078-0432.ccr-03-0096. [DOI] [PubMed] [Google Scholar]
- 83.Ruiz-Herrera A, Castresana J, Robinson TJ. Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol. 2006;7(12):R115. doi: 10.1186/gb-2006-7-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Debatisse M, El Achkar E, Dutrillaux B. Common fragile sites nested at the interfaces of early and late-replicating chromosome bands: cis acting components of the G2/m checkpoint? Cell Cycle. 2006;5(6):578–581. doi: 10.4161/cc.5.6.2574. [DOI] [PubMed] [Google Scholar]
- 85.Pichiorri F, Palumbo T, Suh SS, et al. Fhit tumor suppressor: guardian of the preneoplastic genome. Future Oncol. 2008;4(6):815–824. doi: 10.2217/14796694.4.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okumura H, Ishii H, Pichiorri F, Croce CM, Mori M, Huebner K. Fragile gene product, Fhit, in oxidative and replicative stress responses. Cancer Sci. 2009;100(7):1145–1150. doi: 10.1111/j.1349-7006.2009.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 2009;11(6):753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 88.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 2009;11(6):761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 89.Debatisse M, Coquelle A, Toledo F, Buttin G. Gene amplification mechanisms: the role of fragile sites. Recent Results Cancer Res. 1998;154:216–226. doi: 10.1007/978-3-642-46870-4_13. [DOI] [PubMed] [Google Scholar]
- 90.Ciullo M, Debily MA, Rozier L, et al. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum. Mol. Genet. 2002;11(23):2887–2894. doi: 10.1093/hmg/11.23.2887. [DOI] [PubMed] [Google Scholar]