Abstract
A deformable registration method is proposed for registering a normal brain atlas with images of brain tumor patients. The registration is facilitated by first simulating the tumor mass effect in the normal atlas in order to create an atlas image that is as similar as possible to the patient’s image. An optimization framework is used to optimize the location of tumor seed as well as other parameters of the tumor growth model, based on the pattern of deformation around the tumor region. In particular, the optimization is implemented in a multiresolution and hierarchical scheme, and it is accelerated by using a principal component analysis (PCA)-based model of tumor growth and mass effect, trained on a computationally more expensive biomechanical model. Validation on simulated and real images shows that the proposed registration framework, referred to as ORBIT (optimization of tumor parameters and registration of brain images with tumors), outperforms other available registration methods particularly for the regions close to the tumor, and it has the potential to assist in constructing statistical atlases from tumor-diseased brain images.
Index Terms: Atlas registration, brain tumor, deformable registration, image attributes, tumor growth model
I. INTRODUCTION
Statistical atlases have been used in a variety of studies of normal brain development and aging, as well as of brain diseases [1]–[7], but they have rarely been used in studies of brain cancer. Building statistical models of brain tumor evolution could help gain insight into the brain tumor disease. For example, population-based statistical atlases can potentially indicate whether a multiparametric imaging, or proximity to certain fiber tracts, profile suggests higher likelihood of tumor progression in a particular direction. Moreover, augmenting these models with tumor size and location relative to brain structures, could further improve predictive accuracy. The construction of such statistical models requires the integration of a variety of patient data, such as conventional magnetic resonance imaging (MRI), perfusion, and diffusion-tensor imaging (DTI) of a large number of patients, into the same space and also requires the linking of all these data to outcome measures. For this purpose, a registration method is needed that can map all the imaging data to a common space (normal atlas). While the problem of coregistering brain images of healthy subjects has been addressed by many approaches in the literature, the normalization of tumor diseased images into a common template space, is still a very challenging problem that has motivated our work.
Besides their potential value in predicting tumor progression, anatomical and functional statistical atlases or models are also useful in neurosurgical treatment planning, since they can integrate diverse information about anatomical and functional variability, thereby helping design treatment plans that minimize the risk for significant functional impairment of the patient or facilitate safe dose escalation. Here again, a method that can register a statistical atlas (image without disease) to the patient-specific brain tumor image is required.
Most of the available registration methods in neuroimaging [8]–[29] are designed to register a normal atlas with generally normal neuroanatomies. Direct application of these methods to images of tumor patients can lead to poor registration around the tumor region, due to large deformations and lack of clear definition of anatomical detail in a patient’s images. Specifically, in the images with tumor, the fundamental assumption of topological equivalence between the atlas and the patient’s image, which is almost ubiquitous in deformable registration methods, is violated due to the anatomical changes caused by tissue death and tumor emergence. Moreover, the confounding effects of edema and tumor infiltration, which cause changes in the image intensities, render the task of finding correspondences very difficult. Finally, the large distortions caused by the mass effect of a growing tumor violate the usual assumption of smoothness of the deformation fields.
The framework proposed herein aims to facilitate the registration process by creating an atlas image that is as similar as possible to the patient’s image, in the sense that it contains a tumor and mass effect similar to those in the patient’s image. Specifically, it is based on the idea of first creating a topologically equivalent atlas image by replacing part of the healthy tissue with a tumor seed and then decoupling the total deformation (between atlas and patient’s image) into two components, i.e., the deformation caused by the tumor mass effect and the deformation due to the intersubject differences. The tumor-induced deformation can be calculated by a biomechanical model of soft tissue deformation [30], [31], whereas the inter-subject deformation can be calculated by a deformable registration method. In other words, the tumor modeling component aims to resolve both the geometric discrepancies from the physiologic process of tumor growth and the image differences from the tumor emergence. The registration component is then based on the assumption that 1) there is equivalent image content between the atlas with simulated tumor and the patient’s image, and 2) the deformation between the tumor-bearing images is smooth, similar to normal image registration.
Similar approaches of brain tumor image registration have already been investigated [32]–[36]. Some of these methods use a normal-to-normal brain matching method after shrinking the tumor in the patient’s images [32]. Some apply first an affine transformation between the normal atlas and the patient’s image and subsequently either use a rich tumor simulation model of mass-effect and invasion without further accounting for the intersubject differences [35], or use a simplified radial growth model [37] refined by a nonrigid deformation based on optical flow [33], [34]. Other methods just combine the Talairach transformation with the simple radial expansion model [36] in order to speed up the algorithm, or do not deal with tumor growth and mass effect at all, but manually place a mask on and around the tumor, in order to ignore those regions during the image matching process since they are regarded as unreliable [38]. These approaches are based on oversimplified assumptions, because 1) tumor growth cannot be simply modeled as a radial expansion process, 2) the tumor seed cannot be simply estimated by calculating the mass center of tumor or shrinking the tumor, and 3) the morphological variability across individuals can not be captured by an affine transformation. The study presented in this paper, addresses all of these three issues, but gives special emphasis to the last two, since the first issue has been addressed in previous work [30]–[32], [39].
An earlier approach of our group for registering brain tumor images to a normal atlas, which avoids the previous simplifications, has been presented in [40]. We proposed a maximum likelihood framework for estimating the tumor model parameters by collecting statistics obtained from mass effect simulations. Although this method reported promising results for small quasi spherical tumors, more experiments on different kind of tumors revealed some limitations, which motivated us to examine alternative methodologies. First, this method applied normal-to-normal registration, as part of both the model parameters estimation and the final warping of the tumor-bearing images, whereas the new approach uses a registration method developed for brain images under the presence of tumors (in corresponding or close locations to each other). Also, the method in [40] was a statistical approach; it did not apply optimization and therefore did not provide any measure of confidence on the final solution (optimality criterion). Additionally, the method in [40] retrieved the tumor model parameters based only on the tumor-induced deformation. Especially under the assumption of the brain being a homogenous material, that is, having the same material properties for white and gray matter, local information is lost. In the current method, additionally to the information from the tumor-induced deformation fields, we incorporate local information from the image content, thus increasing the sensitivity of the optimization procedure. Finally, perhaps most importantly, the statistically-based method of [40] required a very large number of simulations of tumor growth, in order for shape statistics to be gathered. This renders this approach quite limiting in many practical situations.
Built upon the idea of HAMMER registration algorithm developed for normal brain registration [20], in this paper we present a framework for optimization (of tumor model parameters) and registration of brain images with tumors, that we shall from here on refer to as ORBIT. The main advantages of the ORBIT algorithm are 1) the incorporation of a similarity criterion that uses two kinds of information, namely tissue properties and spatial location relative to tumor, and 2) the development of a deformation strategy that is robust to unreliable matches caused by the presence of tumor. Moreover, one of the novelties of the proposed framework is the estimation of the optimal tumor-related parameters (including the origin of the tumor and the amount of tissue death), via optimization of a criterion reflecting the elastic stretching energy and the image similarity. Robustness is achieved by applying the optimization in a multiresolution scheme. Also, efficiency is achieved by replacing the expensive nonlinear biomechanical model with a principal component analysis (PCA)-based model of tumor growth, as well as by refining the warping throughout the optimization only in the regions of low confidence.
The rest of the paper is organized as follows. In Section II, we describe the basic components of ORBIT, i.e., the PCA-based model for simulating tumor mass effect, the registration method for tumor-bearing images, and the criterion for optimizing tumor parameters. The implementation details of these methods in a multi-resolution framework are introduced in Section III. In Section IV, we show the efficacy of this framework on both synthetic and clinical cases, i.e., by providing the sensitivity of registration to the tumor parameter estimation. The paper concludes in Section V.
II. METHODS
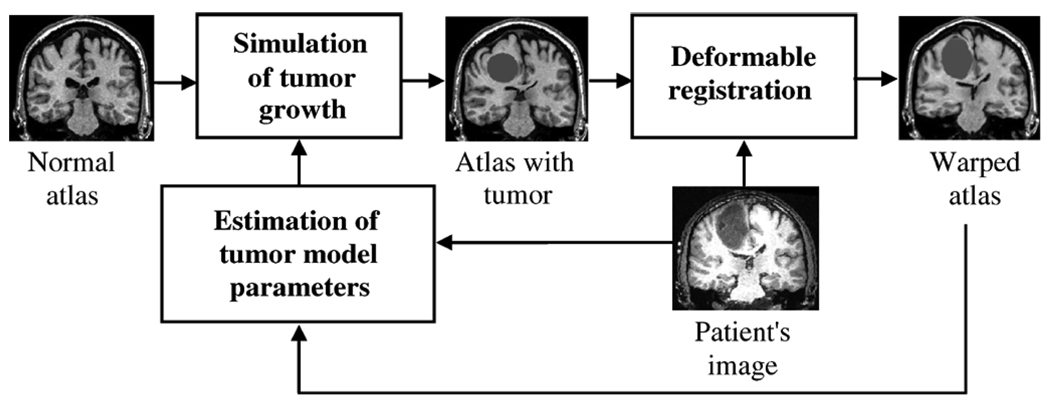
The basic components of ORBIT include: 1) a simulation model for tumor growth and mass effect, 2) a deformable registration method for tumor-bearing images, and 3)an optimization method for estimating the parameters of the tumor growth and mass effect model. Fig. 1 summarizes a closed-loop process for registering a normal brain atlas to a tumor-bearing image, using all three components. The illustrated mapping is useful for transferring information from the atlas to the patient’s space. The normalization of the patient’s image into a common (normal) atlas space can be performed by the reverse mapping.
Fig. 1.
Flowchart summarizing the basic steps for registration of a normal brain atlas with the image of a brain tumor patient.
A. Local PCA-Based Model for Simulating Tumor Mass Effect
The simulation of tumor mass effect is based on the application of a statistical model that describes the variation in deformability of the atlas brain due to different tumor model parameters. One way of modeling tumor growth and mass effect is via biomechanical simulators. Two examples of such simulators are based on 1) finite element models of nonlinear elasticity [30], [31] and 2) incremental linear elasticity models on regular grids [39]. However, incorporating such biomechanical models (especially the first one) into an iterative registration procedure could be computationally prohibitive. In this paper, we have circumvented this limitation by using the mass effect model of [30], [31] to train a PCA-based mass effect model and then using it in the registration framework. It is worth noting that the PCA-based tumor growth and mass effect simulation is extremely fast, since it can be achieved via linear combination of a relatively small number of principal components (deformations), thereby leaving the burden of simulation to offline training using the costly biomechanical model. As a note, any biomechanical simulator could be used for training purposes, subject always to the requirements of each application. As our experiments will show, this approach leads to very efficient approximation of the types of deformations caused by tumor growth, especially in view of the very approximating nature of any such modeling method.
The use of a statistical model for simulating the tumor mass effect involves the following steps. First, the parameters that are necessary for the simulation of tumor-induced deformation are defined. Then, the training step is presented. Finally, the interpolation step, on how to produce an estimator with continuous values of the model parameters, is given.
1) Definition of Tumor Model Parameters
The applied biomechanical model simulates the displacement of structures caused by the tumor mass effect and edema swelling, as well as tissue death in a somewhat simplistic way, since our goal here is merely to construct an atlas that looks similar to the patient’s images and to facilitate subsequent deformable registration. The tumor mass effect is simulated by replacing part of the brain tissue with a small tumor seed and applying a pressure normal to the seed’s boundary. In the current approach, the initial tumor seed is created by approximately warping the tumor from the patient’s to the atlas space and then applying multiple erosions. This provides an initial estimate of the tumor location, but since the initial warping of the patient’s image to the normal atlas is not very accurate, the exact seed location, defined by the center of mass of the tumor, is part of the tumor model parameters to be estimated. The amount of shrinkage determines the initial size of the seed, rt, and it is also one of the tumor model parameters.
2) Training
Consider the discrete displacement maps ui(x), i = 1,…,N, at the 3-D Cartesian coordinates x = (x(1), x(2),x(3)) due to the tumor mass effect, simulated by a biomechanical model with parameters θi = (xti,rti), i = 1,…,N in the atlas image. The displacement maps are first defined in a coordinate system x′, centered at each tumor center, in order to make the domain of all the maps the same and allow point-to-point comparison and collection of statistics. Moreover, the domain is restricted inside a region around each tumor center, where nonzero displacement due to mass effect is expected.
Under the assumption of a Gaussian distribution, each ui(x′) can be represented as
where μ the mean of the displacement at each voxel location, V the matrix containing the eigenvectors of the covariance matrix that correspond to the M largest eigenvalues, and the corresponding coefficients vector.
3) Interpolating
Provided that the statistical parameters μ and V have been determined from the training set,1 the displacement map û(x′) can be calculated for any new tumor parameters θ = (xt, rt) as û = µ + Vẑ, if ẑ is known. Since it is reasonable to assume that the coefficients change smoothly for small variations of θ, we approximate each coefficient ẑ(j) in the coefficients vector ẑ = [ẑ(1),…,ẑ(M)]T by interpolating between the corresponding coefficients of the training samples in θ-space, in order to produce an estimator with continuous values of the model parameters. For this purpose, two scattered interpolation methods were implemented and compared. The method with the smallest reconstruction error is chosen, as described in Appendix I. Finally, the tumor-induced deformation map is calculated by re-centering the displacements at the original coordinate system, û(x) = û(x′ + xt).
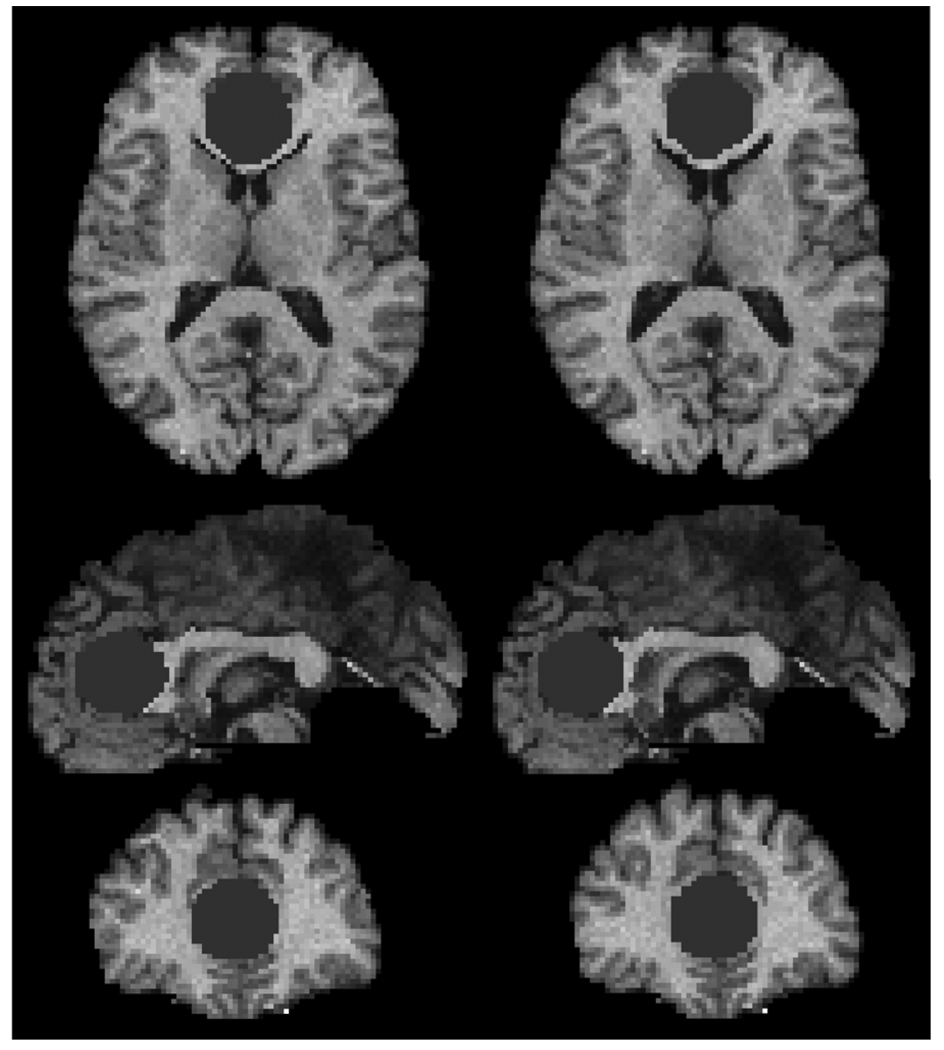
Fig. 2 shows an example of an image with simulated tumor using the biomechanical model and the proposed PCA-based model, respectively. The two simulation results are very similar and thereby indicate that, from the registration perspective, the substitution of the computationally expensive biomechanical model is justified.
Fig. 2.
Simulation of tumor growth using the biomechanical model (left) and the corresponding PCA-based model (right), respectively.
B. Deformable Registration Method
The elastic deformation field is calculated according to the hierarchical approximation of an energy function, which consists of the similarity matching criterion defined in the template space, a constraint on the inverse matching, and smoothness constraints on the displacement field, following the general framework of the HAMMER algorithm [20]. Critical parts of our formulation of the registration of tumor-bearing images are 1) the definition of the similarity criterion, 2) the deformation mechanism, and 3) the mechanism for improving the robustness of the method to slightly inaccurate estimates of the tumor simulation parameters.
1) Similarity Criterio
The similarity criterion is designed based on the similarity of attribute vectors, which are defined for each voxel in the image in order to capture the anatomical context (including healthy and malignant tissue) around it. Specifically, the attribute vector, a(x) = [a1(x) a2(x) a3(x) a4(x) a5(x)], reflects edge type (a1), tissue type (a2), and geometric moment invariants from all tissue types, respectively. a1 and a2 are scalars taking discrete labels, whereas a3 is a 1 × K vector comprising the geometric moment invariants of each tissue and is used to capture shape information, as described more analytically in [20]. In this application, only zero-order regular moments are used. The number of tissue types depends on the segmentation method applied to labeling brain tissue. Besides the attributes that capture brain structure information, the attribute vector captures also the geometric location relative to the brain tumor. Specifically, a4 is used to reflect the signed distance from the tumor boundary, and a5 to reflect the angular location with respect to the tumor center.
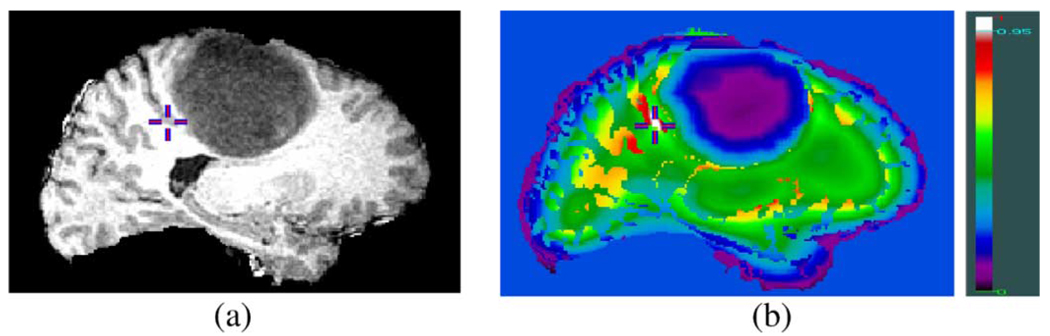
The elastic deformation field that spatially warps the template to the patient’s image is calculated by maximizing a similarity criterion reflecting the distance of attributes, as will be defined in the following. Fig. 3 demonstrates how such a similarity criterion can distinguish between different parts of a tumor-bearing brain image, which might otherwise be indistinguishable.
Fig. 3.
Demonstration of the distinctiveness of attribute vectors in brain tumor images. (a) Slice from a 3-D brain image with tumor. (b) Color-coded similarity of attribute vectors. The attribute vector based similarity between the gray matter voxel indicated by a cross in (a) and every other voxel in the 3-D data is shown in (b), with white reflecting high similarity (>0.95). The similarity is calculated from equation (1) without using edge type information (attribute a1), for simplicity of figure (b). The crosses correspond to the same location in both images.
The similarity of two voxels x and y is defined as the weighted summation of a similarity criterion matching the brain structures, SimB, and a similarity criterion matching the tumor geometry, SimT, as given below
| (1) |
where, as shown in the equation at bottom of page, w(x, y) is a weighting factor which decreases with the distance of x and y from each tumor respectively, and ci are positive constants. If at least one of the two images is normal (without tumor), w becomes zero and the similarity criterion matches only the brain structures, similar to HAMMER [20]. The use of spatially adapted weights ensures that the identification of corresponding points is driven mainly by one of the two matching criteria, whereas the spatially smooth decrease of w makes the total similarity Sim smooth. Regarding the intrinsic difference between the similarity criteria, we should note that the function values of both SimT and SunB are normalized in the range [0,1]. Moreover, the constants c1 and c2 determine the sensitivity (gradient) of SimT.
Notice that the incorporation of the similarity based on the tumor location SimT into the proposed similarity criterion serves the purpose of enforcing the warping of the tumor volumes, in order to facilitate the method for estimating the optimal parameters for the tumor growth model. As will be explained in the corresponding paragraph, the hypothesis is that the optimal tumor parameters minimize the discrepancy between the coregistered atlas and the patient’s images. Since the deformation pattern is most informative around the tumor, the similarity criterion is designed to put special emphasis on the registration of the tumor neighborhood.
2) Image Deformation Mechanism
The registration process starts by registering points with salient features (driving voxels), in order to reduce ambiguity in finding correspondence. The voxels that drive the deformation are selected hierarchically according to the distinctiveness of their attributes, i.e., points on the roots of sulci, crowns of gyri, and strong isolated edges. As the registration process proceeds, additional driving voxels are selected to increase local accuracy. Especially for the tumor area, the selection of driving voxels is not only based on the saliency of features, but also depends on the necessity of warping of the tumor volumes. When registration is used as part of the estimation process of the tumor model parameters, driving voxels are selected on the tumor boundaries in order to facilitate the warping of the tumor volumes. Upon parameters estimation and tumor growth simulation, the registration is performed by relaxing all forces that prioritize the matching of the tumor boundaries. The reason is that the final registration should not be affected by 1) not accurately determined tumor boundaries and 2) the residual variability in the tumor vicinity which is primarily due to fundamental differences in the growth process between a real and a simulated tumor.
The optimal correspondence of each driving voxel is determined by integrating the similarity of all voxels within a small spherical neighborhood around this driving voxel. If the similarity is high, then the respective driving voxel is displaced. This displacement is also interpolated in the neighborhood via a Gaussian kernel function. Upon interpolation of the displacement everywhere, a Laplacian-based smoothing is applied to ensure locally smooth displacement fields. The smoothing reduces with time, as the level of confidence in estimating the tumor model parameters increases.
3) Robustness
Robustness is achieved by requiring all the driving voxels (close and far from tumor) to deform together and nearly to a global affine transformation in each iterative registration step. Since the pattern of deformation around the tumor might deviate significantly from the pattern of deformation in the rest of the brain, e.g., some inaccurate estimate of the tumor location would result in a large translational component, all the driving voxels in the tumor neighborhood are required to deform at the same affine transformation as the driving voxels in the rest of the brain. The opposite requirement is not placed, and therefore the deformation in the healthy part of the brain does not depend on the selected tumor simulation parameters. This registration procedure is robust in registering the healthy part of the brain, even if the tumor simulation parameters are slightly inaccurately estimated.
C. Optimality Criterion for θ
The optimal set of tumor growth parameters, θ, is not known for a particular patient; it must be estimated from the patient’s image. The pattern of deformation around the tumor can be indicative of the accuracy in estimating the parameters of the tumor model, θ. If the estimation of θ is wrong and thus the tumor is incorrectly simulated in the atlas, unrealistic and severe deformations are expected around the tumor region, when trying to match the atlas with the patient’s image. Conversely, if tumor location and mass effect in the atlas are estimated in agreement with those in the patient, a relatively smooth deformation can be obtained. Additionally, due to the smoothness constraints applied during registration, the similarity of the two coregistered images is expected to be low around the tumor if the estimation of θ is inaccurate. Accordingly, we use the characteristics of the deformation field and the anatomical characteristics of the coregistered atlas and patient’s images around the tumor to define an optimality criterion, E, for estimating θ
Specifically, E is defined as the combination of three normalized measures: 1) the residual volume of overlap of the coregistered atlas and patient’s images (E1), 2) the distance of attribute vectors (E2), and 3) the Laplacian of the deformation field defined to reflect the smoothness property of the deformation field (E3), as mathematically given below
| (2) |
The constants ck are used to assign different weights on different measures, whereas hk(x) is used to assign different weights according to the voxel’s location x. hk(x) is selected to decrease with the distance from the tumor boundary for all three measures, and to increase on voxels lying on edges particularly for the image-related measures, i.e., E1 and E2. The constants ck are learned by evaluating the performance of each measure separately in the registration of patient images, i.e., how informative each measure is in estimating θ.ΩE is the volume over which E is calculated, and it is defined in the subject brain within a specific distance from tumor boundary, where the effects of misregistration are expected to be more prominent. The part of the image that has no tissue label due to low confidence in tissue segmentation, is excluded from ΩE.
III. IMPLEMENTATION DETAILS
The detailed procedure of the proposed registration framework is described next. First, the images are preprocessed as detailed in Section III-A. Then, the tumor center and initial seed size are optimized and the images are coregistered following a multiresolution strategy described in Section III-B. Afterwards the amount of tumor expansion is estimated, in order to allow the tumor growth simulation to finish before reaching the size of the delineated tumor in the patient’s image. This step is introduced to make the registration robust to inaccuracies in tumor segmentation in the patient’s image and is described in Section III-C. Since the deformation field displays almost negligible changes in the regions far away from the tumor during the iterative process of optimizing the tumor model parameters, the optimization is performed only in a subdomain in order to considerably speed up the implementation, as described with more details in Appendix I.
A. Preprocessing
The first task is to remove skull from the brain [41] and segment the atlas and individual MR images into four tissue types: white matter (WM), gray matter (GM), ventricular cerebrospinal fluid (CSF), and cortical CSF. For this purpose, the tumor is first delineated by an expert in the patient’s original image and then masked out. Herein, we would like to note that in this paper, we do not attempt to solve the problem of automated tumor segmentation, which has been actively treated in the literature. The images are segmented into WM, GM, and CSF using FAST (FMRIB’s Automated Segmentation Tool) [42]. After tissue segmentation, different labels are assigned to ventricular CSF and cortical CSF by using a modified version of HAMMER.
Subsequently, the patient’s scan is registered globally with the normal template without tumor (atlas A0) by applying an affine transformation [43]. For this purpose, the tumor region in the patient’s segmented image is first marked as GM, in order to achieve better affine registration. After global alignment, the tumor region in the patient’s segmented image is assigned a separate label, in order to provide appropriate information for the next step of elastic registration. If the uncertainty of segmentation is high due to confounding effects of edema and tumor infiltration, an additional label can be assigned for these regions that cannot be classified into any of the previous categories.
B. Workflow in a Multiresolution Strategy
The registration procedure that maps the normal atlas to the globally-aligned patient’s image, is performed in a coarse to fine resolution scheme in order to speed up the algorithm, reduce susceptibility to local minima in both registration and estimation, and achieve robustness. A detailed diagram is schematically shown in Fig. 4. In particular, each resolution level involves two steps. In the first step, the elastic deformation map is calculated from the patient’s scan S to the normal template (atlas) T, ϕ :ΩS → ΩT by applying the previously described registration method.
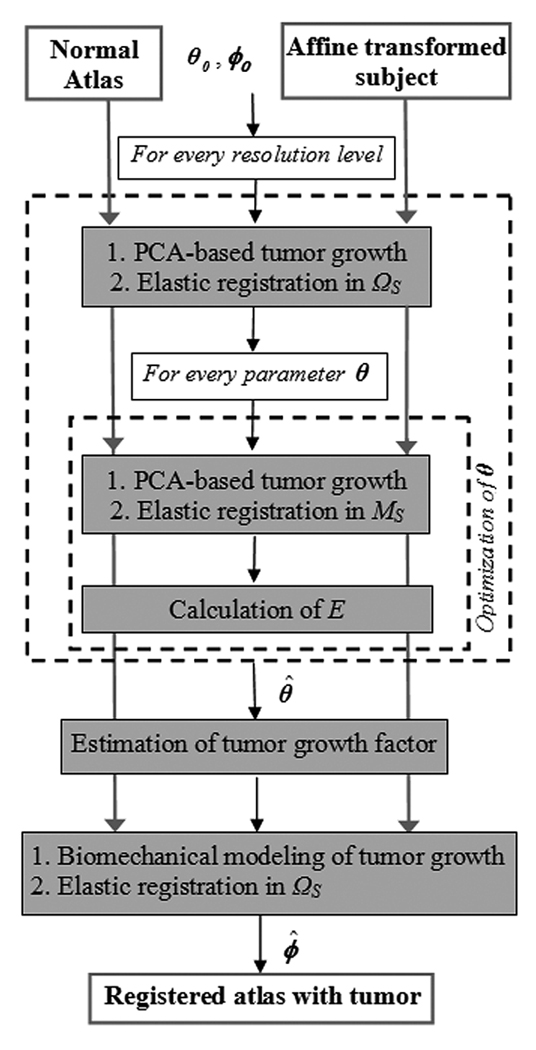
Fig. 4.
Schematic diagram of the multiresolution framework. In every resolution level, tumor growth is simulated in the normal atlas, producing an atlas with tumor which is subsequently registered to the subject. The optimization of θ is performed only in a region of focus MS ⊆ ΩS using the Downhill Simplex method with initial estimate obtained from the values calculated in the previous level. The procedure is initialized with θ0, an average of the tumor parameters used for training (see Appendix I for selection of training parameters), and ϕ0, the identity map for registration. Upon optimization of θ, the tumor growth model is estimated and the final registration is performed in ΩS.
Since the atlas does not include a tumor, the deformation is likely to be inaccurate in the regions close to the tumor. Therefore, in the second step, the deformation map is refined by it-eratively simulating the tumor growth in the normal atlas with different tumor model parameters θ and registering the tumor-bearing atlas with the patient’s image S, until a minimum error (as quantified by the optimality criterion E) is reached. In other words, this optimization step aims to estimate the tumor model parameters that reduce the discrepancies between the coregistered images as much as possible. The optimization is performed with the Downhill Simplex method. Since the deformation field ϕ remains unchanged in the regions far away from the tumor when parameters θ are changed slightly, the refinement of θ is performed only in a region of focus MS ⊆ ΩS in order to considerably reduce the computational cost. A modified deformation strategy is performed in order to avoid discontinuities of the deformation field on the boundary, ∂MS (see Appendix II for details). In every resolution level, the estimated tumor model parameters are used as an initial estimate for the next high-resolution level, and the optimized deformation field that coregisters the tumor-bearing atlas and the patient’s scan is upsampled and linearly interpolated to the next high resolution.
C. Estimation of the Tumor Growth Factor
After estimating θ in the fine resolution level, the final step is to estimate the tumor growth factor, defined as the ratio of the tumor volume in the patient’s image to the volume of the initial seed. During the previous steps of the optimization process, the final tumor volume has been assumed equal to the segmented tumor volume in the patient’s image. This is a good approximation, but the actual value might slightly deviate due to segmentation inaccuracies. Specifically, since part of the tissue inside the segmented tumor region might be tumor infiltration, which is assumed to cause negligible mass effect, the tumor mass effect simulation should terminate before the volume reaches the size in the segmented image. The estimation of the tumor growth factor (gf) is performed by simulating tumor growth using the estimated parameters, θ̂, for various stages of tumor expansion. The minimization of E from (2) (after registration) gives the optimum amount of expansion
In this registration stage, we are setting the weights for the features related to the tumor geometry (SimT) to zero, in (1), and let the deformation be guided only from the surrounding brain structures. We are also using the biomechanical model instead of the PCA-based model for simulating the mass effect. The reasons for using the biomechanical model are that 1) the number of simulations to be performed is small, 2) it provides the original solution whereas the PCA model is based on approximation, 3) the PCA model is trained in a lower resolution causing a reduction in accuracy due to the required upsampling, and 4) it allows tracking of all instances during tumor growth, whereas the local PCA-based model is trained only for final tumors exhibiting the same size as in the patient’s image.
IV. RESULTS
The registration accuracy is assessed by both synthetic and real brain tumor images. In particular, the synthetic brain tumor images are produced by first simulating anatomical variability by the method in [45], thereby creating synthetic brains from a template brain with exactly known displacement fields. Then, tumor growth is simulated in the synthetic brains, and the final images are treated as individual brain tumor images. Since the same mass effect model [30], [31] is utilized for tumor simulation, as used for the training of our PCA model, this set of synthesized images provides primarily a means of assessing the accuracy of our method. We assessed the sensitivity of both the proposed optimality criterion and the registration method as a function of the applied θ, as described, respectively, in Sections IV-A and IV-B. The total registration error, by using this synthetic data, is also reported. It is expected that, for the brain images with large tumors, the use of our proposed framework should outperform normal brain registration algorithms. Thus, here we select the synthetic cases with small tumor, to show that even for such cases, the registration can benefit from the simulation of tumor growth in the atlas. In particular, the final tumor volume in the synthetic images is 8.1 cc, with an initial radius of seed 5 mm.
Moreover, ORBIT is applied to register real brain images, with results presented in Section IV-C. All real images are registered with a normal brain image serving as a template, with image size 256 × 256 × 198 voxels and voxel size 1×1×1 mm3. It should be noted that the same set of parameters is used for all the results shown in this paper, and two levels of resolution are used in the multiresolution framework, corresponding to the original image resolution (high resolution) and a subsampled version by a factor of two (midresolution).
A. Sensitivity of the Optimality Criterion E
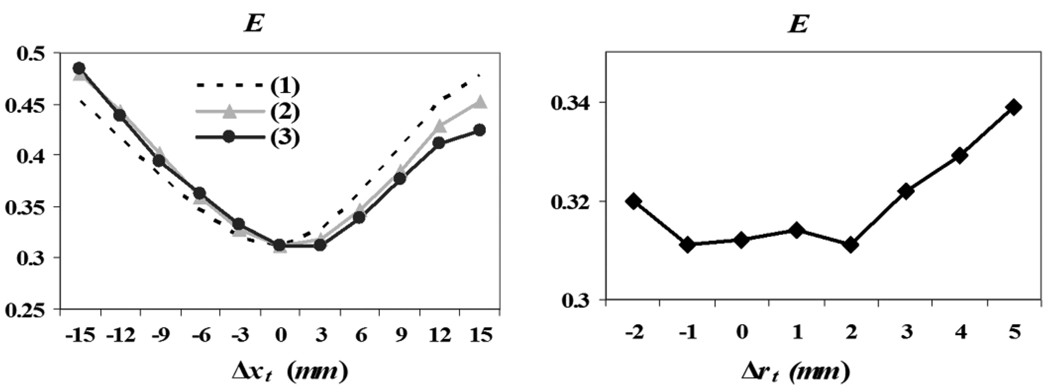
In order to test the potential of optimizing the tumor growth parameters using the proposed optimality criterion, we have plotted E as a function of the error in estimating θ, using one of the synthetic images. The proposed criterion has a global minimum at the correct tumor center, and it is also minimized around the correct size of the initial tumor seed. Therefore, this experiment demonstrates that the optimality criterion E can be used for estimating the parameters of the tumor growth model. E seems to vary across rt (Fig. 5, right) less smoothly than across xt (Fig. 5, left). This ambiguity is likely due to the very small increment of tumor size (1 mm), used for evaluating E.
Fig. 5.
Sensitivity of E as a function of θ (tumor seed location on the left, with the different lines corresponding to each of the 3-D Cartesian coordinates, and initial seed size on the right). E is optimal for the correct value of θ, which indicates that it is a good measure for estimating θ.
B. Sensitivity of Registration as a Function of the Applied θ
The purpose here is to investigate the robustness of the proposed registration algorithm to the estimated tumor parameters θ. The root-mean square (rms) and the maximum (max) registration errors of the estimated deformations from synthetic images to the atlas are calculated separately over the tumor neighborhood Ω and the rest of the brain Ω̅as detailed next.
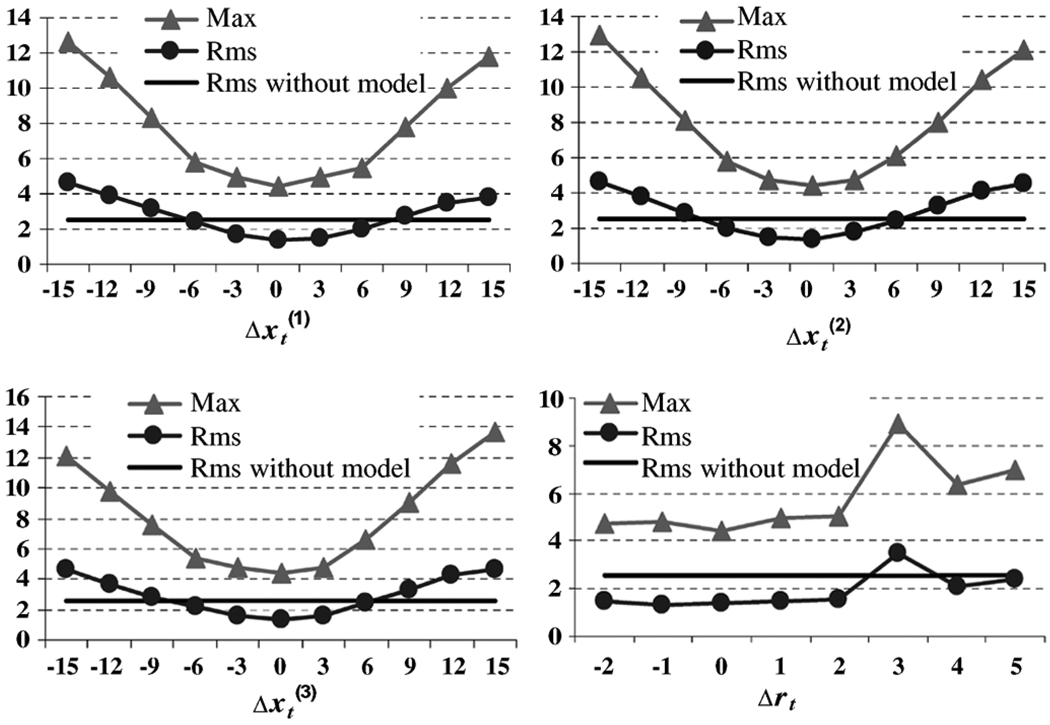
In the tumor neighborhood Ω, the rms registration error without the application of the tumor growth model is 2.5 mm, and the max registration error is 14.6 mm, for the same synthetic brain image as in Section IV-A. If the tumor growth model is applied, the registration accuracy varies with the model parameters used, as shown in Fig. 6. If the error of estimating tumor center is less than 7 mm, the rms registration error decreases to a value of 1.4 mm with the application of the tumor growth model prior to registration. The rms registration error is less sensitive to the initial tumor radius. On the other hand, the max registration error decreases significantly from 14.6 to 4.4 mm, when the tumor growth model is applied in the atlas. As a conclusion, if the tumor-mass effect model is applied prior to registration, the max registration error decreases significantly, and so does the rms registration error in the tumor vicinity for a range of the model parameters around the true values.
In the rest of the brain Ω̅, the deformation field is almost the same, whether the tumor mass effect model is used or not (rms = 0.8 mm, max = 5.1 mm, for the same synthetic brain image as above). Also, if the tumor mass effect model is used, the registration far from the tumor is not sensitive to the tumor parameters applied, as indicated by averaging the registration error map over the nine different synthetic brain images. The registration error maps, calculated by using optimal and suboptimal tumor parameters (tumor center misplaced by 5 mm), showed that the registration accuracy is affected only in the regions very close to the tumor seed. This observation prompted our decision to optimize the deformation only in a region around the tumor MS during the process of estimating the tumor model parameters (as described in Section III-B).
Fig. 6.
Sensitivity of ORBIT with respect to the estimation of tumor parameters θ. Both rms and max registration errors in the tumor neighborhood Ω are shown in millimeters. In particular, the max registration error without use of the biomechanical model is 14.6 mm.
Finally, after assessing the sensitivity of the two basic steps in ORBIT (θ-estimation and registration), we applied those steps to the same nine synthetic images in order to assess the finally achieved registration accuracy. The average rms registration error is 1.9 ± 0.2 mm for the tumor vicinity, and 0.8 mm for the rest of the brain. These results show that the rms registration error of ORBIT on synthetic images is at subvoxel accuracy for the healthy brain part and at the order of the diagonal voxel distance for the region close to the tumor.
C. Registration Assessment of ORBIT on Real Brain Tumor Patient Scans
Ten T1-weighted tumor-bearing brain images were selected for registration with the normal template which include tumors of different types, grades, and sizes. For comparison, these ten images were registered using ORBIT, the Image Registration Toolkit (ITK) [18], [46], [47], and HAMMER, respectively. The global alignment of each tumor-bearing brain image with the normal atlas is completed by using affine transformation with twelve degrees of freedom.
The ITK-based nonrigid image registration was executed with the following options: three resolution levels, 64 bins, 20 iterations, and four steps of length 5. The similarity measure was the normalized mutual information. The parameter Lamda, used to control the smoothness of deformation, was chosen such that the deformation field seemed to preserve topology (i.e., Lamda = 0.03).
For the other two methods, i.e., ORBIT and HAMMER, these ten brain images were preprocessed, as described in Section III-A. The tumor region is used in HAMMER as a mask to avoid the mismatch in the tumor vicinity due to the missing tumor in the normal template. It is worth noting that the use of such a mask has increased significantly the registration accuracy of HAMMER in the tumor vicinity, as compared to previous results [40], where the automatic segmentation of the brain (including the tumor) into normal tissue classes was directly used. The purpose here is to see if ORBIT can further increase the registration accuracy. Notice that the selection of the best possible options in ITK-based registration and the use of a tumor mask in HAMMER are expected to provide the best possible results for these two methods, thus removing potential biases in favor of our current method.
The execution time for the 256 × 256 × 198 images was | 17 h for ITK-based registration in a PC with Intel Pentium 4 and CPU 1.7 GHz. HAMMER algorithm took | 1.5 h in a Linux cluster with Dual Intel Xeon 2.80 GHz, running on a single CPU. The overall computational cost for the ORBIT pipeline on the same Linux cluster on a single CPU was |14 h, with most computations used for estimation of the tumor model parameters. We would like to acknowledge here that this algorithm, in its current state, is not intended for real-time usage, but rather as an offline preplanning tool. The performance of these three methods is evaluated quantitatively and visually as detailed next.
1) Quantitative Assessment Based on Landmarks
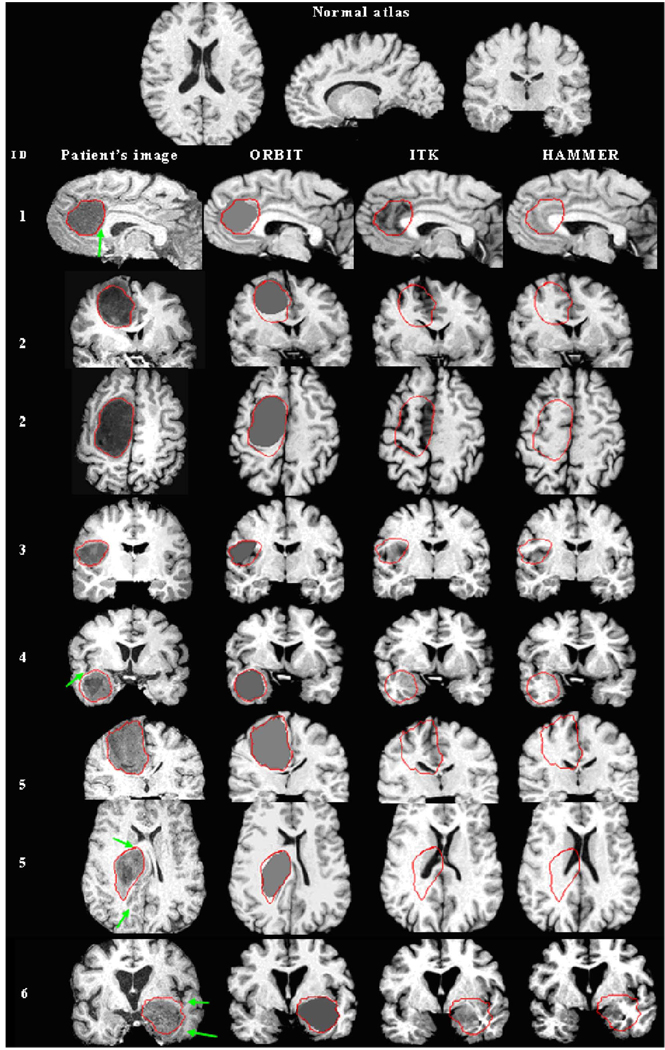
In order to quantitatively assess the registration accuracy, landmark points were manually placed in the patient’s images by a neuroradiologist with expertise on brain tumors, in anatomical regions displaced by the tumor and also in anatomical regions that were not displaced by the tumor. Similarly, the corresponding landmarks are manually identified in the atlas. This set of landmarks shall be referred to as the first set of landmarks. In order to ensure the consistency in the identification of landmarks, the reverse procedure was followed a few weeks later. The same expert first looked at the selected landmarks locations in the atlas, and then identified the corresponding points in the patient’s images. This set of landmarks is labeled as the second set of landmarks. The minimum (min), mean, maximum (max), and standard deviation (stdev) of the landmarks distance (between mapped and actual landmarks in the atlas) for the regions displaced by the tumor are shown in Table I. For each patient’s image, the first row in the table indicates the intrarater variability in placing the landmarks; the other three rows show the statistics of landmarks error for each of the first and second set of landmarks, shown at each cell, left and right, respectively. The landmark-based results show that on average ORBIT has higher registration accuracy than ITK and HAMMER in the tumor vicinity, especially in the case of large tumors. For patient 5, the mean error decreases |40%. The only cases where the mean error is slightly larger are patient 3 and patient 6. Regarding patient 3, it should be noted that the tumor exhibits a very small size, and the use of a tumor mass effect model is not expected to improve registration. Regarding patient 6, we would like to note that, despite the reported landmarks errors, the registration using ORBIT, as visually assessed later in Fig. 7, seems better than with the other two methods. Moreover, in order to determine the significance of the assessment based on landmark errors, we performed an independent t-test between each two methods, ORBIT and ITK or ORBIT and HAMMER under the null hypothesis that there is “no difference” between the landmark errors of the two methods
where NL is the number of landmarks and the indices 1 and 2 correspond to the first and second method, respectively. With a significance level α = 0.09, the null hypothesis could be rejected only for patient 5. This indicates that the landmarks errors are not statistically different for most patients, including the ones ORBIT performs slightly worse, but they are statistically different for the case ORBIT performs better than the other two methods (patient 5).
TABLE I.
Statistics of landmarks error (in millimeters) using orbit, hammer, and ITK-based registration, respectively, in areas displaced by tumor
| min | mean | max | stdev | ||
|---|---|---|---|---|---|
| Variability | 1.4 | 7.6 | 16.9 | 4.3 | |
| ORBIT | 1.3/2.4 | 10.2/8.6 | 21.2/27.7 | 6.4/7.4 | |
| ITK | 0.6/2.1 | 11.4/10.3 | 27.9/32.8 | 8.7/8.0 | |
| HAMMER | 2.1/2.8 | 11.1/10.1 | 21.7/27.2 | 6.3/6.3 | |
| Variability | 1.0 | 6.9 | 17.7 | 5.5 | |
| ORBIT | 2.0/0.4 | 9.4/10.3 | 15.4/18.6 | 4.1/4.9 | |
| ITK | 3.7/2.8 | 9.9/10.1 | 16.9/17.8 | 4.5/4.9 | |
| HAMMER | 1.9/0.8 | 10.4/11.3 | 20.0/20.4 | 4.9/6.3 | |
| Variability | 1.0 | 4.8 | 10.0 | 2.9 | |
| ORBIT | 1.2/1.6 | 5.9/7.4 | 10.7/15.2 | 2.9/4.3 | |
| ITK | 2.0/1.1 | 6.6/7.3 | 13.1/18.1 | 3.6/5.1 | |
| HAMMER | 1.7/2.1 | 5.8/6.8 | 11.4/16.1 | 3.1/4.4 | |
| Variability | 1.4 | 7.1 | 16.9 | 5.5 | |
| ORBIT | 0.8/2.7 | 5.7/8.7 | 18.2/18.3 | 4.1/5.4 | |
| ITK | 1.0/4.3 | 6.3/9.8 | 15.9/16.1 | 4.3/4.0 | |
| HAMMER | 1.6/3.3 | 7.2/10.1 | 17.4/18.7 | 4.8/5.0 | |
| Variability | 1 | 4.4 | 16.6 | 4.5 | |
| ORBIT | 1.9/2.4 | 11.5/11.0 | 27.3/16.8 | 7.0/5.1 | |
| ITK | 9.3/8.6 | 15.8/14.9 | 24.0/21.5 | 5.2/4.4 | |
| HAMMER | 10.0/5.4 | 16.9/15.9 | 23.0/25.0 | 5.0/6.2 | |
| Variability | 1 | 3.8 | 10.6 | 3.2 | |
| ORBIT | 1.4/2.9 | 9.0/8.4 | 20.1/17.9 | 5.3/4.5 | |
| ITK | 3.6/3.6 | 8.5/8.0 | 13.8/18.8 | 3.6/4.0 | |
| HAMMER | 3.7/3.3 | 8.7/8.1 | 13.3/12.8 | 3.3/12.8 | |
| Variability | 0 | 6.1 | 13.2 | 3.9 | |
| ORBIT | 13.8/8.0 | 25.2/24.2 | 41.8/41.8 | 10.4/12.5 | |
| ITK | 17.0/10.0 | 30.1/29.0 | 54.9/54.9 | 12.7/15.3 | |
| HAMMER | 12.3/9.0 | 25.0/25.4 | 43.8/43.8 | 10.3/12.1 | |
| Variability | 0 | 3.3 | 7.1 | 2.1 | |
| ORBIT | 1.8/2.2 | 9.0/10.1 | 23.0/23.4 | 6.4/7.4 | |
| ITK | 2.8/2.2 | 9.9/10.9 | 27.1/27.2 | 8.4/8.1 | |
| HAMMER | 3.7/2.7 | 9.9/10.8 | 27.1/27.3 | 7.7/8.4 | |
| Variability | 0 | 1.7 | 3.7 | 1.2 | |
| ORBIT | 3.2/2.8 | 8.2/8.4 | 14.1/14.4 | 4.2/4.3 | |
| ITK | 2.9/3.8 | 10.7/10.6 | 23.4/23.9 | 6.9/7.1 | |
| HAMMER | 3.3/2.9 | 10.6/10.8 | 21.7/21.7 | 6.8/7.1 | |
| Variability | 1 | 4 | 16.4 | 4.7 | |
| ORBIT | 4.0/3.7 | 12.2/9.6 | 36.8/17.4 | 9.9/4.9 | |
| ITK | 6.4/4.4 | 13.7/11.5 | 33.2/22.2 | 9.1/4.9 | |
| HAMMER | 5.6/4.8 | 13.0/10.6 | 37.4/22.6 | 9.9/5.6 | |
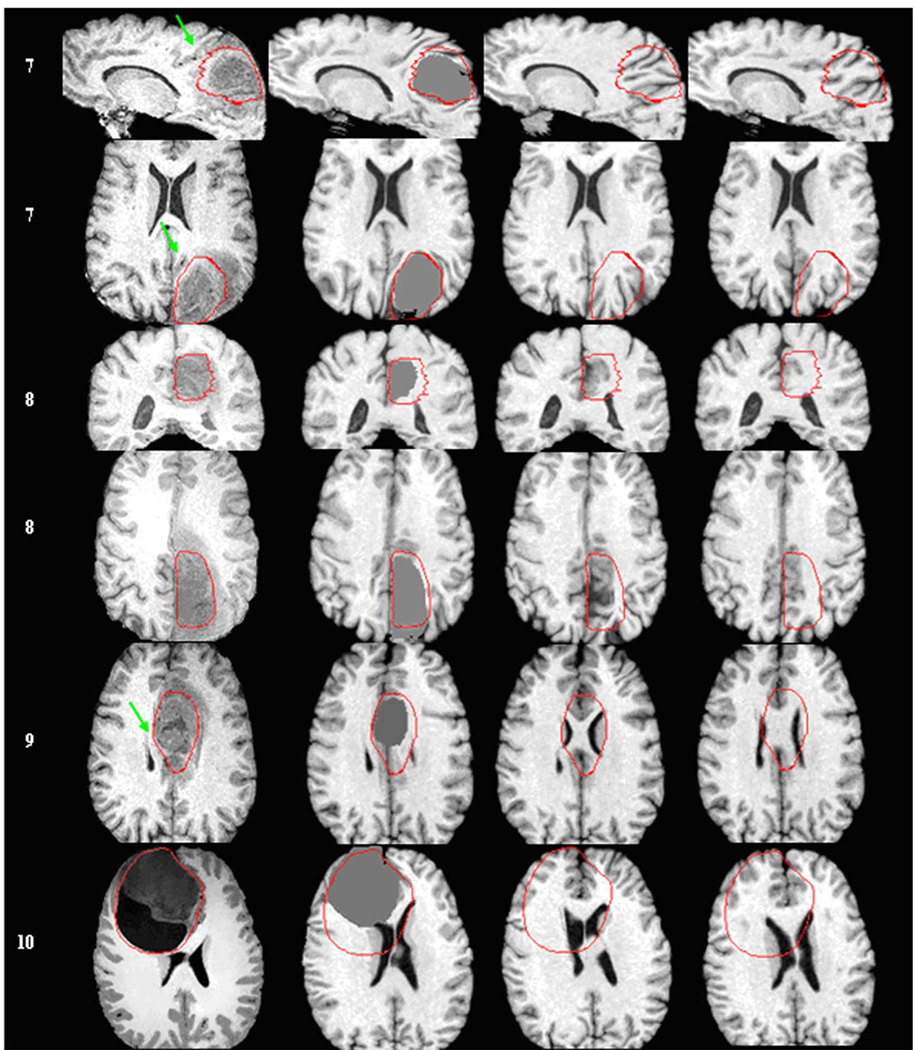
Fig. 7.
Registration of a normal atlas image to ten patients’ images using ORBIT, ITK, and HAMMER. First row illustrates the normal atlas. Every other row shows from left to right: the patient ID, a section of the skull stripped T1-weighted patient’s image (axial, sagittal, or coronal), and the corresponding section of the atlas warped with ORBIT, ITK, and HAMMER correspondingly. For some patients, more than one sections are shown. The tumor segmentation, as manually performed by the expert, is illustrated on all images with a red line. Arrows point to structures that are displaced correctly only by ORBIT. (a) Patients 1 –6. Registration of a normal atlas image to ten patients’ images using ORBIT, ITK, and HAMMER. First row illustrates the normal atlas. Every other row shows from left to right: the patient ID, a section of the skull stripped T1-weighted patient’s image (axial, sagittal, or coronal), and the corresponding section of the atlas warped with ORBIT, ITK, and HAMMER correspondingly. For some patients, more than one sections are shown. The tumor segmentation, as manually performed by the expert, is illustrated on all images with a red line. Arrows point to structures that are displaced correctly only by ORBIT. (b) Patients 7–10.
Additionally, we assessed the registration accuracy with a rater-independent measure, such as the surface distance of the ventricles (VN–dist) between the coregistered images. We calculated VN – dist as the mean Euclidean distance of the ventricular boundaries in both directions, from the patient’s image to the warped atlas image and reversely. Table II shows VN – dist calculated only in the tumor vicinity, in order to emphasize differences between the three methods. It can be seen that ORBIT, in comparison to HAMMER, exhibits the smallest distance for all patients and also it performs better than ITK for all patients except patient 7, for whom ITK shows a marginally smaller error. It should be noted that the tumor in patient 7 is far from the ventricles and there is no actual deformation of the ventricles due to tumor mass effect.
TABLE II.
Deformable registration error of the ventricles surface (VN-dist, measured in millimeters) in the tumor vicinity, using orbit, hammer, and ITK-based registration, respectively
| patient | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| ORBIT | 1.895 | 1.266 | 0.462 | 1.597 | 1.190 | 1.645 | 1.100 | 1.519 | 1.561 | 3.407 |
| ITK | 2.465 | 1.903 | 0.484 | 6.358 | 2.186 | 2.940 | 1.004 | 1.577 | 2.208 | 6.213 |
| HAMMER | 2.207 | 1.418 | 0.663 | 3.360 | 1.650 | 2.687 | 1.597 | 1.707 | 2.095 | 4.424 |
In the following, we evaluated the registration accuracy in areas not displaced by tumor by calculating the error of the remaining landmarks. The landmarks error statistics of ORBIT, HAMMER, and ITK-based registration, showed that the three registration methods are comparable (a t-test between ORBIT-ITK and ORBIT-HAMMER showed p-values larger than 0.4 for all patients). The errors, averaged across the two sets of landmarks and across all patients for ORBIT (min = 2.415, mean = 7.635, max = 15.62, stdev = 4.58), for ITK (min = 2.32, mean = 8.015, max = 18.32, stdev = 5.46), and for HAMMER (min = 2.345, mean = 7.335, max = 16.205, stdev = 4.62), respectively, showed that ORBIT exhibits the smallest average maximum error and the smallest average standard deviation.
2) Visual Assessment
Besides providing the quantitative errors by landmarks, the registration accuracy of ORBIT can be also visually assessed. Fig. 7 illustrates the registration result of those ten patients’ images with the normal template (without tumor) using the three registration methods, respectively. It is notable that ORBIT outperforms ITK and HAMMER for all patients’ images. High similarity between the warped tumor-bearing template with ORBIT and the patient’s image can be observed for all cases, except for patient 10. Patient 10 has a tumor consisting of a solid and a cystic portion, both together exhibiting a size of |200 cc. The deformation caused on the ventricles is disanalogously larger than on the rest of the structures, and also it resembles more to rigid motion. It is possible that the current tumor mass effect model is not adequate for cystic tumors and that the presence of a cyst requires the application of a more accurate biophysical model compensating also for the incompressibility effects of the cystic fluids.
Moreover, the effect of the tumor segmentation refinement step in the ORBIT algorithm, is visible in many of the warped images with ORBIT. These warped images have tumors exhibiting smaller size than the tumors in the patient’s image. They indicate that the part of the tumor causing brain tissue displacement during tumor growth is smaller than the one manually segmented. Also, the presence of tumorous tissue outside this bulk region indicates the existence of tumor that infiltrates the brain without displacing structures.
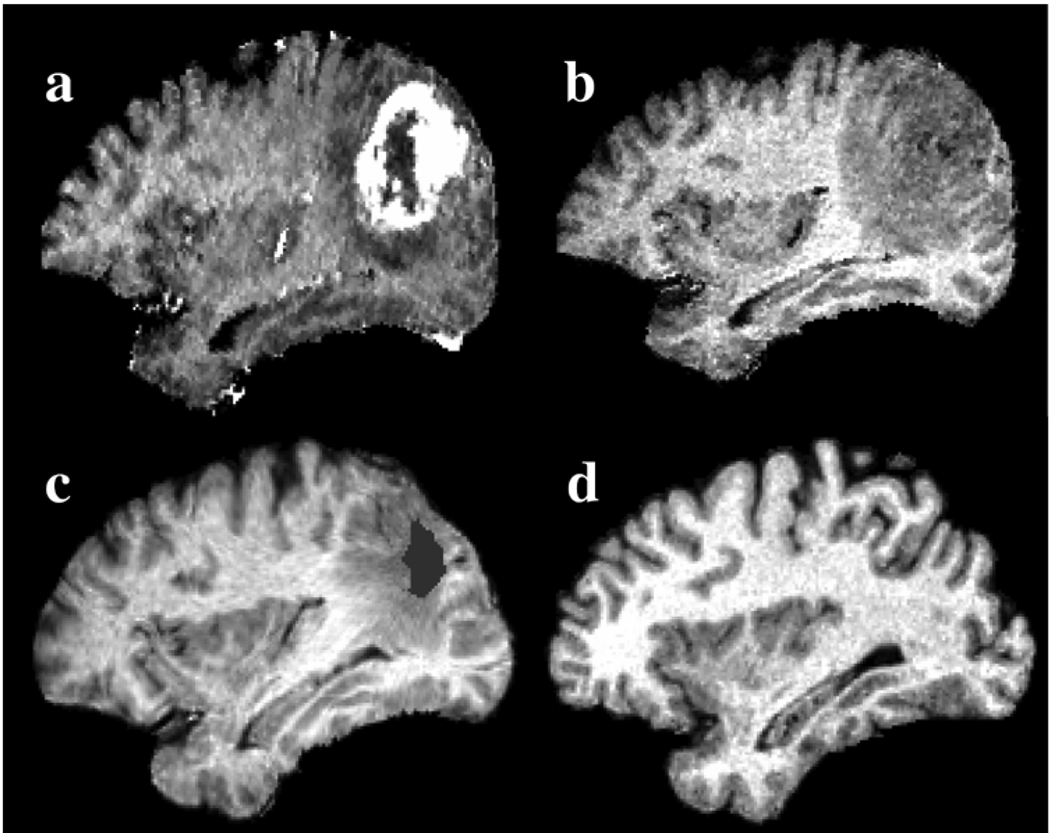
Finally, an example of inverse mapping is shown in Fig. 8 for patient 7. In this case, the tumor consists of an enhancing and a nonenhancing region, as illustrated by the T1-weighted image with gadolinium. The nonenhancing region indicates the presence of edema or tumor infiltration. The patient image is warped to the normal atlas space by reversing the deformation field produced by ORBIT, û o φ̂ through the concatenation of the two components. This warping causes relaxation of the mass effect and correction of the intersubject differences facilitating the detection of the two tumorous regions: 1) the initial seed (as estimated by ORBIT) showing the tissue that is replaced by tumor, and 2) the surrounding region that is infiltrated by tumor or edema.
Fig. 8.
Registration of patient 7 into the normal atlas space using ORBIT. The top row shows the T1-weighted patient’s image (a) with and (b) without gadolinium rigidly registered to the atlas in (d). The image in (b) after de-formable registration to the normal atlas, which causes relaxation of the mass effect and correction of the intersubject differences, is shown in (c). The initial tumor seed, shown with gray color in (c), represents tissue death and indicates the location of initial tumor appearance, as defined in the atlas. The surrounding peri-tumor edema or infiltration, as mapped in the normal atlas, is also visible.
V. DISCUSSION AND FUTURE WORK
A framework for deformable registration of brain tumor images (ORBIT) has been presented, which incorporates an elastic feature-based registration method, along with a tumor mass-effect model. Moreover, an optimality criterion has been proposed for optimization of the parameters pertinent to the tumor of a specific patient, such as tumor location in the atlas space, amount of tissue death, and amount of expansion (growth factor). After optimization is achieved, the proposed registration method shows good performance for both tumor and other brain regions, as validated by both simulated and real tumor cases.
ORBIT has been compared with two other registration methods, HAMMER and Image Registration Toolkit (ITK), using the best possible options for these two methods. The efficacy of the three registration methods is not easy to compare in brain regions far from the tumor due to the ambiguity in defining correspondence between different brains. The landmarks error seems to be quite large for all registration methods and also indicates that the methods have a similar performance in brain regions not displaced by the tumor. However, this is not conclusive since the intrarater variability in placing the landmarks was also relatively large.
On the other hand, the assessment through quantitative and visual criteria in the tumor vicinity, which is the main region of interest for this study, showed that ORBIT outperforms the other two methods. It is notable that the landmark-based quantitative evaluation did not show a significant improvement achieved by ORBIT, relative to the other two methods, except for one patient. We attribute this largely to the fact that the neuroradiologist tended not to place landmarks extremely close to the tumor, since it was difficult to reliably define the anatomy in that region. Therefore, the landmarks tend to reflect performance not too close to the tumor, i.e., in regions where we expect all three methods to be relatively comparable. Visual evaluation (e.g., Fig. 7) shows more marked improvement achieved by ORBIT, which is also in agreement with our expectation, since ORBIT uses a tumor growth and mass effect model to simulate tumor before registration. Moreover, the registration error, calculated on the ventricular boundary in the tumor vicinity, shows that ORBIT outperforms the other two methods. It should be noted that we selected the ventricles for validation, because they are structures with distinct boundaries providing accurate segmentation.
A linear PCA model of tumor growth is used in order to interpolate between precalculated solutions. Besides the reduction of the computational cost, the replacement of the complex biomechanical model insures that the template with simulated tumor will be produced for any seed location inside the brain parenchyma and for any seed size. On the contrary, the biomechanical model does not converge in some cases, e.g., if the tumors are large and very close to the skull or the ventricular boundary, since remeshing of the 3-D finite element grid does not necessarily guarantee that the large elements distortions will be eliminated. In these cases, the failure to converge could mislead the optimization procedure of the model parameters to wrong optimum. Our experiments showed that the PCA-based approximation did not significantly affect registration accuracy; hence it was adopted, due to the significant computational savings in comparison to using a biomechanical model.
It is worth noting here, that the training step in the proposed framework is not very costly computationally, since it is performed only in the selected template. On the contrary, the training in method in [40] is computationally very expensive because it requires the performance of tumor growth simulations for a large number of subjects, in order to capture the statistical variation of tumor mass effect for each set of model parameters. In both methods, the tumor model parameters used for training are chosen to cover the range expected for the subject to be registered, i.e., selected locally. Therefore, the training step has to be repeated for each new tumor-bearing image to be registered. In the future, the PCA models used by each method could be trained once for every possible set of tumor growth parameters, thus eliminating completely the training step.
One of the contributions of this work is the procedure for estimation of the optimal set of parameters of the tumor growth and mass effect model. Our results are generally promising, in that the tumor model parameters can be estimated accurately. However, more experimentation along this direction is necessary in future work, especially with respect to sensitivity of this parameter estimation process for different growth models. It is notable (Fig. 5) that tumor position seems to be relatively easier to estimate, as opposed to initial tumor seed size. This was indeed our expectation prior to developing this estimation procedure, since wrong tumor location estimates will generate atlases that are very different from the patient’s images close to the tumor, whereas small variations in tumor seed size cause much more subtle differences.
One of the motivations of the current work is the coregistration of patient images in a common atlas space, as illustrated in the example shown in Fig. 8. This inverse registration procedure causes relaxation of the tumor growth mass effect and removal of the intersubject differences. Such mappings can facilitate the correlation of treatment parameters with therapy outcome. For example, the correlation of tumor recurrence with radiation dose profiles (inevitably defined in the common atlas space) can be studied for patients with tumors emerging from similar anatomical locations. Also, tumor model parameters, e.g., size of initial seed indicating tissue death or amount of infiltration, can be quantified and compared across patients, since they are measured in a common space.
Since a great deal of effort has been devoted by the computational biology community to the development of mathematical and simulation tools that help predict how cancer evolves, in the future we plan to efficiently exploit this knowledge to improve the component related to tumor modeling. As an example, more advanced biophysical models of tumor infiltration and edema spread [35], [48], [49] could be integrated to the applied biomechanical model of tumor mass effect to provide better initializations for the registration component. It also should be noted, that the biomechanical model in [30] and [31] can also simulate the swelling caused by possible peri-tumor edema. Currently, we do not simulate edema swelling, first because the expansion caused by edema, if present, is not large and can therefore be captured by the deformable registration method, and second, in order to reduce the computational cost by limiting the number of modeling parameters. Moreover, edema can have very diverse size and shape causing the parameterization to be very difficult.
A current limitation of our approach is that it is based on the prior tissue segmentation, which poses considerable difficulties in practice, especially in the region around the tumor that often displays edema and infiltration. The proposed framework is mostly suitable for tumors with distinct tumor boundaries, which are not very challenging from segmentation perspective. However, it is important to note that the current implementation is robust to inaccuracies in tumor segmentation because the simulated tumor is not forced to expand until it reaches the manually segmented tumor in the patient’s image. The amount of tumor expansion is rather determined by optimizing the defined optimality criterion. One future extension of the proposed registration method could be the transition from hard tissue segmentation into a fuzzy or probabilistic segmentation framework, which is more appropriate for the inherently diffuse and infiltrative brain tumors, since in those cases the tumorous area can only be characterized through probabilistic tissue abnormality maps. On-going work in our laboratory investigates pattern classification methods that use multiacquisition imaging profiles, including T1, T1-GAD-enhanced, FLAIR, DTI, and aims to achieve a more accurate tissue classification, thereby assisting in the registration process.
ACKNOWLEDGMENT
The authors would like to thank Dr. Z. Xue at the Section of Biomedical Image Analysis for providing the synthetic data and Dr. E. R. Melhem in the Department of Radiology, University of Pennsylvania, for providing the patient’s data.
This work was supported by the National Institutes of Health under Grant NS042645. Asterisk indicates corresponding author.
Appendix I
Implementation Details on the PCA-Based Model of Tumor Mass Effect
In the following, we present some implementation details of the PCA model of tumor growth, which provides the tumor-induced deformation field for generating the tumor-bearing atlas image through the placement of initial seed in the atlas. The criterion of selecting sample deformations for training the PCA model of tumor growth and the technique for interpolating between the coefficients vectors are both explained next.
1) Selection of Training Samples
Tumor mass effect simulations are conducted in the atlas space for building a set of training samples with tumor parameters, θi = (xti,rti), i = 1,…,N. Tumor center in the atlas space can be roughly estimated as the center of mass of the tumor of the globally aligned and approximately warped patient’s image. Thus, the locations of tumor samples can be selected close to this center, with a sampling rate that is higher in areas close to the cortical or ventricular boundary. Notice that the sampling rate should be spatially adjusted, since the tumor deformations close to the boundary show larger variation due to different boundary conditions, compared to the ones developed in uniform regions, with deformation pattern similar to radial expansion. Moreover, samples that lie on the different sides of the ventricle are not jointly used, since their deformation patterns are very different.
As to the size of the initial seed, there is no immediate way of acquiring an initial estimate. Therefore, two or three values are chosen for the training, between the lower and upper limits of the initial seed size. In particular, the upper limit is smaller than the final tumor in the patient’s image, whereas the lower limit is larger than the grid discretization of the biomechanical model.
2) Selection of Interpolation Method
The coefficient vector used to reconstruct the tumor-induced deformation field for new model parameters can be interpolated from the coefficient vectors of the training samples. Two scattered interpolation methods, based on either inverse distance weighting or local fitting, were tested. The inverse distance weighting interpolation method was finally chosen and given below, since it provided slightly smaller reconstruction errors
where
M is the number of retained principal components and g is the bivariate Gaussian distance function prescribing that the influence of the sample θi fades away with the distance from the fitting point θ. K is a kernel used to control the number of neighboring samples contributing for interpolation, and h1 and h2 corresponding constants
Appendix II
Optimization in a Subdomain
In order to speed up the execution, the optimization of θ is performed in a subdomain of the subject space, which is larger than the tumor neighborhood ΩE used to evaluate the optimality criterion, ΩE ⊂ MS. The optimization method Downhill Simplex [44] is applied. A modified deformation strategy is performed in order to avoid discontinuities of the deformation field on the boundary ∂MS. First of all, driving voxels are selected only inside MS. Since the identification of corresponding voxels is based on maximization of the similarity of the attributes in a sphere around each driving voxel, in order to avoid partial volume effects, MS is expanded to include also parts of the spherical subvolume that might fall outside the initial subdomain.
Then, when deforming the subvolume of a template driving voxel x, the deformation is propagated on every neighboring voxel y according to a Gaussian kernel that should leave the boundary voxels of the kernel unchanged. In order to preserve the continuity of the displacement field around ∂MS, the displacement field is scaled according to the distance of y to ∂MS, in addition to the Gaussian scaling based on the distance ∥x – y∥.
After deforming the subvolume, Laplacian smoothness constraints are applied to refine the displacement field. The smoothing is applied only in the subdomain MS, in order to retain the estimated deformations outside MS. Additional to the minimization of the Laplacian cost term which leads to smooth first derivatives, a constraint on the second derivatives is applied inside MS as well as close to the boundary ∂MS, thus leading to well behaved deformation fields.
Footnotes
Details about the model parameters selection for creating sample deformations for training the local PCA-based model of tumor growth, can be found in Appendix I.
Contributor Information
Evangelia I. Zacharaki, Email: eva.zacharaki@uphs.upenn.edu, Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania, 3600 Market Street, Philadelphia, PA 19104 USA.
Dinggang Shen, Email: dinggang.shen@uphs.upenn.edu, Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania, Philadelphia, PA 19104 USA.
Seung-Koo Lee, Email: lee.seung-koo@uphs.upenn.edu, Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania, Philadelphia, PA 19104 USA.
Christos Davatzikos, Email: christos.davatzikos@uphs.upenn.edu, Section of Biomedical Image Analysis, Department of Radiology, University of Pennsylvania, Philadelphia, PA 19104 USA.
REFERENCES
- 1.Miller M, Banerjee A, Christensen G, Joshi S, Khaneja N, Grenander U, Matejic L. Statistical methods in computational anatomy. Statist. Meth. Med. Res. 1997;vol. 6:267–299. doi: 10.1177/096228029700600305. [DOI] [PubMed] [Google Scholar]
- 2.Thompson PM, Mega MS, Woods RP, Zoumalan CI, Lindshield CJ, Blanton RE, Moussai J, Holmes CJ, Cummings JL, Toga AW. Cortical change in alzheimer’s disease detected with a disease-specific population-based brain atlas. Cerebral Cortex (Cary) 2001 Jan.vol. 11(no. 1):1–16. doi: 10.1093/cercor/11.1.1. 2001. [DOI] [PubMed] [Google Scholar]
- 3.Thompson PM, Giedd JN, Woods RP, MacDonald D, Evans AC, Toga AW. Growth patterns in the developing brain detected by using continuum-mechanical tensor maps. Nature. 2000 Mar.vol. 6774:190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- 4.Mazziotta J. A four-dimensional probabilistic atlas of the human brain. J. Amer. Med. Inform. Assoc. 2001;vol. 8:401–430. doi: 10.1136/jamia.2001.0080401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashburner J, Csernansky JG, Davatzikos C, Fox NC, Frisoni GB, Thompson PM. Computer-assisted imaging to assess brain structure in healthy and diseased brains. Lancet. 2003;vol. 2:79–88. doi: 10.1016/s1474-4422(03)00304-1. [DOI] [PubMed] [Google Scholar]
- 6.Chung MK, Worsley KJ, Paus T, Cherif C, Collins DL, Giedd JN, Rapoport JL, Evanst AC. A unified statistical approach to deformation-based morphometry. NeuroImage. 2001 Sep.vol. 14(no. 3):595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- 7.Roland PE, Graufelds CJ, Wahlin J, et al. Human brain atlas: For high-resolution functional and anatomical mapping. Human Brain Mapp. 1994;vol. 1:173–184. doi: 10.1002/hbm.460010303. [DOI] [PubMed] [Google Scholar]
- 8.Ashburner J, Friston K. Nonlinear spatial normalization using basis functions. Hum. Brain Mapp. 1999;vol. 7(no. 4):254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashburner J, Friston KJ. Voxel-based morphometry: The methods. NeuroImage. 2000 Jun.vol. 11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 10.Christensen GE, Johnson HJ. Consistent image registration. IEEE Trans. Med. Imag. 2001 Jul.vol. 20(no. 7):568–582. doi: 10.1109/42.932742. [DOI] [PubMed] [Google Scholar]
- 11.Chiu E, Vaisey J, Atkins MS. Wavelet based space-frequency compression of ultrasound images. IEEE Trans. Inf. Technol. Biomed. 2001 Dec.vol. 5(no. 4):300–310. doi: 10.1109/4233.966105. [DOI] [PubMed] [Google Scholar]
- 12.Chui HL, Rangarajan A. A new point matching algorithm for non-rigid registration. Comput. Vision Image Understand. 2003;vol. 89:114–141. [Google Scholar]
- 13.Davatzikos C. Spatial transformation and registration of brain images using elastically deformable models. Comput. Vis. Image Understand. 1997 May;vol. 66:207–222. doi: 10.1006/cviu.1997.0605. [DOI] [PubMed] [Google Scholar]
- 14.Ferrant M, Warfield S, Guttman C, Mulkern R, Jolesz F, Kikinis R. 3-D Image matching using a finite element based elastic deformation model. Proc. MICCAI’98. 1999:202–209. [Google Scholar]
- 15.Gee JC. On matching brain volumes. Pattern Recognit. 1999 Jan.vol. 32(no. 1):99–111. [Google Scholar]
- 16.Johnson HJ, Christensen GE. Consistent landmark and intensity-based image registration. IEEE Trans. Med. Imag. 2002 May;vol. 21(no. 5):450–461. doi: 10.1109/TMI.2002.1009381. [DOI] [PubMed] [Google Scholar]
- 17.Pizer S, Fritsch DS, Yushkevich PA, Johnson VE, Chaney EL. Segmentation, registration and measurement of shape variation via image object shape. IEEE Trans. Med. Imag. 1999 Oct.vol. 18(no. 10):851–865. doi: 10.1109/42.811263. [DOI] [PubMed] [Google Scholar]
- 18.Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: Application to breast MR images. IEEE Trans. Med. Imag. 1999 Aug.vol. 18(no. 8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz-Alzola J, Westin CF, Warfield SK, Alberola C, Maier S, Kikinis R. Nonrigid registration of 3-D tensor medical data. Med. Image Anal. 2002;vol. 6(no. 2):143–161. doi: 10.1016/s1361-8415(02)00055-5. [DOI] [PubMed] [Google Scholar]
- 20.Shen D, Davatzikos C. HAMMER: Hierarchical attribute matching mechanism for elastic registration. IEEE Trans. Med. Imag. 2002 Nov.vol. 21(no. 11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 21.Shen D, Davatzikos C. Measuring temporal morphological changes robustly in brain MR images via 4-dimensional template warping. NeuroImage. 2004 Apr.vol. 21:1508–1517. doi: 10.1016/j.neuroimage.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Studholme C, Cardenas V, Blumenfeld K, Schuff N, Rosen HJ, Miller B, Weiner M. Deformation tensor morphometry of sematic dementia with quantitive validation. NeuroImage. 2004;vol. 21(no. 4):1387–1398. doi: 10.1016/j.neuroimage.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Thompson P, Toga AW. A surface-based technique for warping three-dimensional images of the brain. IEEE Trans, Med. Imag. 1996 Aug.vol. 15(no. 4):402–417. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- 24.Vemuri BC, Ye J, Chen Y, Leonard CM. Image registration via level-set motion: Applications to atlas-based segmentation. Med. Image Anal. 2003;vol. 7:1–20. doi: 10.1016/s1361-8415(02)00063-4. [DOI] [PubMed] [Google Scholar]
- 25.Pluim JP, Maintz JB, Viergever MA. Mutual-information-based registration of medical images: A survey. IEEE Trans. Med. Imag. 2003 Aug.vol. 22(no. 8):986–1004. doi: 10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 26.D'Agostino FME, Vandermeulen D, Suetens P. An information theoretic approach for non-rigid image registration using voxel class probabilities. Med. Image Anal. vol. 10:413–431. doi: 10.1016/j.media.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Knops JBAMZF, Viergever MA, Pluim JPW. Normalized mutual information based registration using k-means clustering and shading correction. Med. Image Anal. vol. 10:432–439. doi: 10.1016/j.media.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Lorenzen MPP, Davis B, Gerig G, Bullitt E, Joshi S. Multimodal image set registration and atlas formation. Med. Image Anal. vol. 10:440–451. doi: 10.1016/j.media.2005.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Periaswamy HFS. Medical image registration with partial data. Med. Image Anal. 2003;vol. 10:452–464. doi: 10.1016/j.media.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Mohamed A, Davatzikos C. Finite element modeling of brain tumor mass-effect from 3-D medical images. Proc. MICCAI. 2005:400–408. doi: 10.1007/11566465_50. [DOI] [PubMed] [Google Scholar]
- 31.Mohamed A. Ph.D. dissertation. Baltimore, MD: Dept. Comput. Sci. Baltimore, Johns Hopkins Univ; 2005. Combining statistical and biomechanical models for estimation of anatomical deformations. [Google Scholar]
- 32.Kyriacou SK, Davatzikos C, Zinreich SJ, Bryan RN. Nonlinear elastic registration of brain images with tumor pathology using a biomechanical model. IEEE Trans. Med. Imag. 1999 Jul.vol. 18(no. 7):580–592. doi: 10.1109/42.790458. [DOI] [PubMed] [Google Scholar]
- 33.Dawant BM, Hartmann SL, Gadamsetty S. Brain atlas deformation in the presence of large space-occupying tumours. Proc. MICCAI’99. 1999;vol. 1679:589–596. doi: 10.1002/igs.10029. [DOI] [PubMed] [Google Scholar]
- 34.Cuadra MB, Pollo C, Bardera A, Cuisenaire O, Villemure J-G, Thiran J-P. Atlas-based segmentation of pathological MR brain images using a model of lesion growth. IEEE Trans. Med. Imag. 2004 Oct.vol. 23(no. 10):1301–1314. doi: 10.1109/TMI.2004.834618. [DOI] [PubMed] [Google Scholar]
- 35.Clatz O, Sermesant M, Bondiau P-Y, Delingette H, Warfield SK, Malandain G, Ayache N. Realistic simulation of the 3-D growth of brain tumors in MR images coupling diffusion with mass effect. IEEE Trans. Med. Imag. 2005 Oct.vol. 24(no. 10):1334–1346. doi: 10.1109/TMI.2005.857217. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowinski WL, Belov D. Toward atlas-assisted automatic interpretation of MRI morphological brain scans in the presence of tumor. Acad. Radiol. 2005;vol. 12:1049–1057. doi: 10.1016/j.acra.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 37.Ganser KA, Dickhausa H, Metznerb R, Wirtzb CR. A deformable digital brain atlas system according to talairach and tournoux. Med. Image Anal. 2004 Mar.vol. 8:3–22. doi: 10.1016/j.media.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Stefanescu R, Commowick O, Malandain G, Bondiau P-Y, Ayache N, Pennec X. Non-rigid atlas to subject registration with pathologies for conformal brain radiotherapy; Proc. 7th Int. Conf. Med. Image Comput. Computer-Assist. Intervention—MICCAI 2004; 2004. pp. 704–711. [Google Scholar]
- 39.Hogea CS, Abraham F, Biros G, Davatzikos C. A framework for soft tissue simulations with applications to modeling brain tumor mass-effect in 3-D images. 3rd Canadian Conf. Comput. Robot Vision (CRV’06); Quebec City, QC, Canada. 2006. pp. 24–33. [Google Scholar]
- 40.Mohamed A, Zacharaki EI, Shen D, Davatzikos C. Deformable registration of brain tumor images via a statistical model of tumor-induced deformation. Med. Image Anal. 2006;vol. 10:752–763. doi: 10.1016/j.media.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Smith SM. Fast robust automated brain extraction. Human Brain Mapp. 2002;vol. 17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden markov random field model and the expectation maximization algorithm. IEEE Trans. Med. Imag. 2001 Jan.vol. 20(no. 1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 43.Jenkinson M, Bannister PR, Brady JM, Smith SM. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002 Oct.vol. 17(no. 2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 44.Press W, Teukolsky S, Vetterling W, Flannery B. Numerical Recipes in C. Cambridge, U.K.: Cambridge Univ. Press; 1992. [Google Scholar]
- 45.Xue Z, Shen D, Karacali B, Davatzikos C. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2005. New York: Springer; 2005. Statistical representation and simulation of high-dimensional deformations: Application to synthesizing brain deformations; pp. 500–508. [DOI] [PubMed] [Google Scholar]
- 46.Schnabel JA, Rueckert D, Quist M, Blackall JM, Castellano-Smith AD, Hartkens T, Penney GP, Hall WA, Liu H, Truwit CL, Gerritsen FA, Hill DLG, Hawkes DJ. A generic framework for non-rigid registration based on non-uniform multi-level free-form deformations. In: Niessen WJ, Viergever MA, editors. Proceedings of the 4th International Conference on Medical Image Computing and Computer-Assisted Intervention; New York: Springer Verlag; 2001. pp. 573–581. Lecture Notes in Computer Science. [Google Scholar]
- 47.Rueckert D. Image registration toolkit [Online] Available: http://wwwhomes.doc.ic.ac.uk/~dr/software.
- 48.Nagashima T, Tamaki N, Takada M, Tada Y. Formation and resolution of brain edema associated with brain tumors. A comprehensive theoretical model and clinical analysis. Acta Neurochir. Suppl. (Wien) 1994;vol. 60:165–167. doi: 10.1007/978-3-7091-9334-1_44. [DOI] [PubMed] [Google Scholar]
- 49.Nagashima T, Horwitz B, Rapoport SI. A mathematical model for vasogenic brain edema. Adv. Neurol. 1990;vol. 52:317–326. [PubMed] [Google Scholar]