Abstract
We examine the ability of current state-of-the-art methods in protein structure prediction to discriminate topologically distant folds encoded by highly similar (>90% sequence identity) designed proteins in blind protein structure prediction experiments. We detail the corresponding prognosis for the protein fold recognition field and highlight the features of the methodologies that successfully deciphered this folding riddle.
Introduction and context
Natural proteins with over 35% sequence similarity tend to fold into similar conformations [1], yet several evolutionarily related natural protein pairs with up to 40% similarity have been observed to produce substantially different topologies [2,3]. Two sequences bearing the same length and only three nonidentical residues were posted as sequential targets in the recent 8th Community-Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction (CASP8). Targets T0498 and T0499 therefore posed a riddle for the international protein fold prediction community to determine whether the conformations of these 95% identical sequences maintain the same topological folds or adopt different ones.
The proteins were artificially produced in the group of John Orban and Philip Bryan [4] as a study of the tolerance of sequence identity to maintain the 3-α and α/β folds of streptococcal protein G domain A (GA) and domain B (GB), respectively. The two 16% identity domains of protein G were brought together in sequence space first by adding terminal tails to GA to make it equal in length to GB and then by progressively mutating sites of nonidentity. The key in this approach was linking each fold to its natural function: human serum albumin binding for the 3-α GA fold and IgG binding for the α/β GB fold. This linkage of fold to function allowed the application of powerful biologic selection methods to determine clusters of sites in each protein, which could be substituted with the corresponding amino acid in the other protein. Iteratively combining mutations identified by the selection methods resulted in two 88% identical proteins [4]. More recently, two 95% identical sequences possessing the same fold, GA95 and GB95, were designed and provided as the two CASP8 targets discussed here [5]. The designed protein pairs maintain the fold and specific binding function of the proteins from which they were derived, with immeasurable structural or functional character of the domain represented in the alternate protein [4].
A prerequisite of recognizing a fold is prior observation of the fold. Structural genomics consortia contribute thousands of new protein structures each year, yet previously unobserved folds are seldom found [6]. This pattern seems to indicate that the majority of folds that can be detected by current laboratory techniques have already been observed. The completeness of the structural fold space has been addressed using a subset of 1,489 proteins covering the protein data bank [7] at the level of 35% sequence identity; all but two folds can be resolved using templates found within the same set [8]. Thus, template-based modeling appears to be feasible given the best template(s) within the set. The search for the best template for a given query protein is known as ‘fold recognition’.
Major recent advances
The best performing freely available fold recognition web server methods are maintained by Yang Zhang [9] within the local meta-threading server (LOMETS) fold recognition pipeline of I-TASSER (iterative threading assembly refinement algorithm), the best performing protein structure prediction server in the past two CASP experiments. As an isolated meta-threading server, LOMETS uses local implementation to avoid the destructive aspects of internet dynamic regulation corrupting so many meta-servers [10]. The nine methods of LOMETS are representative of the fold recognition field (normally targeted toward naturally occurring proteins) and can be summarized as various combinations of the following: comparing target to known structure sequence profiles, secondary structure preferences, environmental fitness, pairwise contact probabilities, structure profiles, simulated mutations, single-body or residue-specific knowledge-based potentials, and profile hidden Markov models (HMMs) [10].
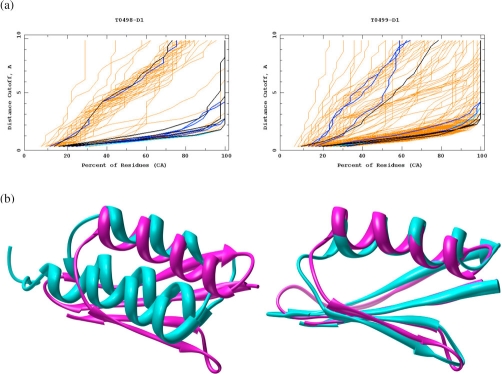
Most web server groups predicted both T0498 and T0499 to adopt the α/β fold of protein GB (Figure 1a). For example, our own predictions for T0498 did not significantly resemble the target structure [37.2 global distance test total score (GDT-TS); Figure 1b, left], yet all five of our predictions for T0499 were within the top 10 total predictions (88.4 GDT-TS; Figure 1b, right). The models for T0499 exemplify progress in another major challenge in protein structure prediction: refinement of model quality from the best template [11].
Figure 1. Difficulties in fold recognition for the redesigned streptococcal protein domains GA95 versus GB95.
(a) Only four out of over 150 contributing team groups recognized the difference in fold caused by three nonidentical residues in the 56 residue proteins: HHpred (cyan), Feig (black), FOLDpro (blue), and Coma (others are in orange). The results are shown in global distance test (GDT) plot format, in which the alpha carbon atoms of the predicted model and experimental structure are spatially aligned within distance cutoffs of 0.5 Å, 1 Å, and 1.5 Å up to 10 Å, such that lower lines denote higher accuracy. A common trend of these four groups was to predict the alternate fold as a lower confidence model. Most groups correctly identified the GB95 T0499 fold, yet most models were no better than random for GA95 T0498. The ability for four automated servers to disentangle this riddle provides a positive outlook for the fold recognition field. (b) Predictions made by our group (purple) for T0498 and T0499 compared with the experimental structures for GA95 and GB95 (cyan), respectively. While our predictions were among the very best for T0499/GB95 (the only group with all five submissions in the top 10), the incorrect fold assignment led to highly inaccurate predictions for T0498/GA95. As with so many other protein structure prediction groups, we failed to predict that profoundly similar sequences would produce different folds. CA, alpha carbon; GA95, the artificial protein with GA fold and 95% sequence identity to GB95; GB95, the artificial protein with GB fold and 95% sequence similarity to GA95.
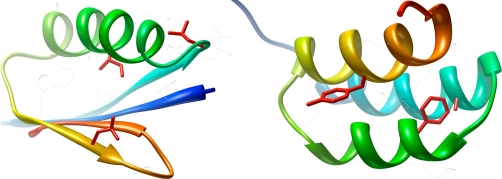
The side chain interactions visible in the experimental structures of GA95 and GB95, as well as the simulated mutant models depicted in Figure 2, demonstrate interactions within a relatively stable, folded state, which are not necessarily illustrative of those interactions occurring during the folding process. Even when the structures are known, it is difficult to ascertain exactly what makes the two proteins follow different fold trajectories. Yet fold recognition methods do not simulate folding. Rather, they rely on calculated interactions within simulated mutants of these folded structures to test the accuracy of fit for a possible template; thus, even with a perfect energy function, mistakes in fold recognition could occur.
Figure 2. Putative causes of CASP8 fold recognition failure and success for redesigned streptococcal protein GA95 versus GB95.
A sequence-to-structure cross of GA95 and GB95 is presented to demonstrate determinants of fold recognition from side chain packing of the nonidentical residues (red). The lack of profound steric clashes created by applying the side chain identities from T0498 to the structure of GB95 (left) misleads predictors to identify an incorrect fold topology. Conversely, the clash that occurs between F30 and A20 when applying the side chain identities from T0499 to the structure of GA95 (right) illustrates an incorrect fold for predictors. GA95, the artificial protein with GA fold and 95% sequence identity to GB95; GB95, the artificial protein with GB fold and 95% sequence similarity to GA95.
In this case, a multitude of experimentally derived structures for GA and GB and detectable sequence similarity within this group reasonably limit the fold search to these topologies. Crossing fold assignments for GA95 and GB95 enables interrogation of side chain packing for the three nonidentical residues (Figure 2). The clash occurring between F30 and A20 when the nonidentities from T0499 are applied to the structure of GA95 (Figure 2, right) implicates an incorrect fold to predictors. Conversely, minimal steric clashes emerge when the T0498 sequence is applied to the structure of GB95 (Figure 2, left). This absence of incriminating evidence for the T0498 GB95 sequence fold pair could mislead predictors to select this fold topology.
Out of over 150 contributing teams, four groups recognized the difference in fold caused by three nonidentical residues in the 56 amino acid proteins: HHpred, FOLDpro, Feig, and Coma. The accurate predictions of these groups demonstrate sensitivity to subtle changes affecting folding not previously demonstrated in a bona fide blind prediction scenario.
HHpred
The Söding group uses HMM emission sequences to evaluate target template matches. The emission sequence of HMMs includes position-specific insertion and deletion probabilities along with the sequence distributions found in multiple sequence alignment profiles. HHsearch specifically includes secondary structures via a substitution matrix derived from comparing measurements on the template to target predictions and to the confidence thereof. To interrogate alignments, the HHsearch method maximizes the coemission log-odds probability for the pair of HMMs derived for a given protein pair. HHsearch directs the structural similarity search hierarchically by searching databases of alignments organized by fold family rather than lists of disconnected sequences [12]. The CSI-BLAST (context-specific iterative basic local alignment search tool) sequence similarity search method recently published by the group was likely used to build the profile input to the HMMs for each alignment [13].
FOLDpro
The Cheng group uses a supervised classification approach previously used for fold classification, invoking support vector machines to combine global profile-profile alignment, secondary structure, solvent accessibility, contact map, and strand hydrogen bond pairing [14].
Feig
The Feig group used a very typical set of methods, including fold recognition functions overlapping those in LOMETS (including HHsearch/HHpred), standard model construction, and a modified cluster calculation using a standard discriminatory potential function [15]. Other promising work by this group in the refinement category includes the use of an implicit continuum dielectric solvent based on generalized Born theory to drive lattice-based course grain searches, Monte Carlo molecular dynamics, and restrained normal mode sampling [16].
Coma
The Venclovas group invokes a profile comparison method for detection of distant evolutionary relationships across profile databases, adding a modified two-level SEG (segment sequences by local complexity) algorithm to filter noninformative profile regions, variable gap penalties, and adaptive parameterization. The underlying sequence similarity search is driven by their PSI-BLAST-ISS (position-specific iterative BLAST intermediate sequence search), which evaluates and refines output profile alignments [17]. The manual submissions by this group displayed the overall best performance in CASP8.
Future directions
A handful of the automated algorithms were able to recognize the fold switch caused by the three nonidentical residues of GA95 and GB95 (Figure 2). However, the experimentally unobserved 60% of naturally occurring proteins [6] and the prospect of designing new folds heralded by Top7 [18] demand more methods sensitive enough to detect subtle triggers in fold switching and predict previously unobserved topologies.
Developments in the protein fold prediction field can often be limited to incremental engineering optimizations. In this fold recognition problem, the proper application of support vector machines and HMM methods enabled success for two groups. Also, two groups created their own improvements on PSI-BLAST [19]: CSI-BLAST [13] and PSI-BLAST-ISS [17], which both enhance quality and relevance of a search by interrogating low-quality regions in the alignment by context and together comprise the first significant improvements on the enormously popular algorithm in a decade. The novel algorithmic adjustments in fold recognition used in CASP8 demonstrate significant progress amounting to new tools for the field.
Future developments are anticipated to include the steady stream of mathematical enhancements observed since the inception of the protein structure prediction field but also include new conceptual paradigms such as functional signatures [20] and the use of template-free modeling [21] to drive the difficult fold recognition problems.
Acknowledgments
The authors thank the Samudrala CompBio group for thoughtful comments and advice. RS was supported in this work by a National Science Foundation (NSF) CAREER award and GEMSEC, an NSF-MRSEC at the University of Washington (DMR 0520567). JH was supported by the National Institute of Dental and Craniofacial Research (NIDCR) Ruth L Kirschstein National Research Service Award (NRSA): NIH F30DE017522.
Abbreviations
- BLAST
basic local alignment search tool
- CASP8
8th Community-Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction
- CSI
context-specific iterative
- GA
streptococcal protein G domain A
- GA95
the artificial protein with GA fold and 95% sequence identity to GB95
- GB
streptococcal protein G domain B
- GB95
the artificial protein with GB fold and 95% sequence similarity to GA95
- GDT-TS
global distance test total score
- HMM
hidden Markov model
- ISS
intermediate sequence search
- I-TASSER
iterative threading assembly refinement algorithm
- LOMETS
local meta-threading server
- PSI
position-specific iterative
- SEG
segment sequences by local complexity
Competing interests
The authors declare that they have no competing interests.
The electronic version of this article is the complete one and can be found at: http://www.F1000.com/Reports/Biology/content/1/69
References
- 1.Gan HH, Perlow RA, Roy S, Ko J, Wu M, Huang J, Yan S, Nicoletta A, Vafai J, Sun D, Wang L, Noah JE, Pasquali S, Schlick T. Analysis of protein sequence/structure similarity relationships. Biophys J. 2002;83:2781–91. doi: 10.1016/S0006-3495(02)75287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grishin N. Fold change in evolution of protein structures. J Struct Biol. 2001;134:167–85. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Rob Russell 12 Oct 2001
- 3.Roessler CG, Hall BM, Anderson WJ, Ingram WM, Roberts SA, Montfort WR, Cordes MH. Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds. Proc Natl Acad Sci U S A. 2008;105:2343–8. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander PA, He Y, Chen Y, Orban J, Bryan PN. The design and characterization of two proteins with 88% sequence identity but different structure and function. Proc Natl Acad Sci U S A. 2007;104:11963–8. doi: 10.1073/pnas.0700922104. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8.1 ExceptionalEvaluated by Barry Stoddard 24 Jul 2007, Nick Grishin 26 Jul 2007, Tobin Sosnick 08 Jan 2008
- 5.He Y, Chen Y, Alexander P, Bryan PN, Orban J. NMR structures of two designed proteins with high sequence identity but different fold and function. Proc Natl Acad Sci U S A. 2008;105:14412–7. doi: 10.1073/pnas.0805857105. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.6 Must ReadEvaluated by H Jane Dyson 23 Oct 2008, Andras Fiser 02 Jan 2009, Nick Grishin 27 Jan 2009
- 6.Chandonia JM, Brenner SE. The impact of structural genomics: expectations and outcomes. Science. 2006;311:347–51. doi: 10.1126/science.1121018. [DOI] [PubMed] [Google Scholar]
- 7.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc Natl Acad Sci U S A. 2005;102:1029–34. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Mark Nelson 07 Apr 2005
- 9.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69(8):108–17. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 10.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–82. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu T, Horst JA, Samudrala R. A novel method for predicting and using distance constraints of high accuracy for refining protein structure prediction. Proteins. 2009;77:220–34. doi: 10.1002/prot.22434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–60. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Nick Grishin 21 Jan 2005
- 13.Biegert A, Söding J. Sequence context-specific profiles for homology searching. Proc Natl Acad Sci U S A. 2009;106:3770–5. doi: 10.1073/pnas.0810767106. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 9.0 ExceptionalEvaluated by Kevin Karplus 09 Mar 2009
- 14.Cheng J, Baldi P. A machine learning information retrieval approach to protein fold recognition. Bioinformatics. 2006;22:1456–63. doi: 10.1093/bioinformatics/btl102. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Ram Samudrala 15 Sep 2006
- 15.Stumpff-Kane A, Feig M. A correlation-based method for the enhancement of scoring functions on funnel-shaped energy landscapes. Proteins. 2006;63:155–64. doi: 10.1002/prot.20853. [DOI] [PubMed] [Google Scholar]
- 16.Stumpff-Kane AW, Maksimiak K, Lee MS, Feig M. Sampling of near-native protein conformations during protein structure refinement using a coarse-grained model, normal modes, and molecular dynamics simulations. Proteins. 2008;70:1345–56. doi: 10.1002/prot.21674. [DOI] [PubMed] [Google Scholar]
- 17.Margelevičius M, Venclovas Č. PSI-BLAST-ISS: an intermediate sequence search tool for estimation of the position-specific alignment reliability. BMC Bioinformatics. 2005;6:185. doi: 10.1186/1471-2105-6-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–8. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]; F1000 Factor 11.3 ExceptionalEvaluated by Kristina Downing 28 Nov 2003, Gideon Schreiber 05 Dec 2003, Rob Russell 09 Dec 2003, Wolfgang Guba 16 Dec 2003, Nick Grishin 18 Dec 2003, Padmanabhan Balaram 19 Dec 2003, Robert Kelley 05 Jan 2004, Kevin Plaxco 06 Jan 2004, Sachdev Sidhu 03 Feb 2004
- 19.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Horst J, Cheng G, Nickle D, Samudrala R. Protein meta-functional signatures from combining sequence, structure, evolution and amino acid property information. PLoS Comput Biol. 2008;4:e1000181. doi: 10.1371/journal.pcbi.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–71. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]; F1000 Factor 9.0 ExceptionalEvaluated by Gideon Schreiber 29 Sep 2005