Abstract
Summary: A trade-off between strategies maximizing growth and minimizing losses appears to be a fundamental property of evolving biological entities existing in environments with limited resources. In the special case of unicellular planktonic organisms, the theoretical framework describing the trade-offs between competition and defense specialists is known as the “killing the winner” hypothesis (KtW). KtW describes how the availability of resources and the actions of predators (e.g., heterotrophic flagellates) and parasites (e.g., viruses) determine the composition and biogeochemical impact of such organisms. We extend KtW conceptually by introducing size- or shape-selective grazing of protozoans on prokaryotes into an idealized food web composed of prokaryotes, lytic viruses infecting prokaryotes, and protozoans. This results in a hierarchy analogous to a Russian doll, where KtW principles are at work on a lower level due to selective viral infection and on an upper level due to size- or shape-selective grazing by protozoans. Additionally, we critically discuss predictions and limitations of KtW in light of the recent literature, with particular focus on typically neglected aspects of KtW. Many aspects of KtW have been corroborated by in situ and experimental studies of isolates and natural communities. However, a thorough test of KtW is still hampered by current methodological limitations. In particular, the quantification of nutrient uptake rates of the competing prokaryotic populations and virus population-specific adsorption and decay rates appears to be the most daunting challenge for the years to come.
INTRODUCTION
Organisms face the dilemma of how to allocate the limited resources available to them. In general terms, resources can be used to increase growth and reproduction or to counteract stress (e.g., predators, parasites, decay, unfavorable environmental conditions, or competitive ability). Thus, substantial investments into stress defense or avoidance strategies may result in a reduction in growth and reproduction and vice versa (39). This trade-off has been reported frequently in the literature for plants (27, 90) and for animals such as insects (22, 49), sponges (111), or birds (9, 97). It is less well know that such a trade-off between reproduction and stress defense holds even for viruses infecting prokaryotes (organisms lacking a nucleus, comprised of the domains Bacteria and Archaea; this grouping does not imply a phylogenetic relationship). De Paepe and Taddei (23) have shown that the mortality of a diverse array of viruses infecting Escherichia coli (viruses infecting prokaryotes are often referred to as phages) is positively correlated to their multiplication rate. In other words, viruses with a high multiplication rate degrade quickly, because under resource-limited conditions these viruses cannot invest higher levels of resources into the stability of their capsids but rather invest them into numbers of offspring. Those authors further demonstrated that the capsid thickness and the density of the packaged genome in the virus capsids account for the majority of the variation in mortality between the different viruses. Thus, De Paepe and Taddei (23) gave a mechanistic explanation for the trade-off between reproduction (multiplication rate) and stress defense (capsid thickness and density of packaged genome exerting pressure onto the capsid) in a very simple biological system. Nyström (67) went a step further and proposed that the trade-off between growth and stress (maintenance of cellular functions) in cells of Escherichia coli is facilitated by the intracellular availability of RNA polymerases (a limited resource) and the competition between different transcription factors. If correct, the hypothesis by Nyström (67) would extend the concept toward the molecular level. Given the many examples of a trade-off between strategies maximizing growth and minimizing losses, from the very small and simple to the very large and complex, one cannot avoid concluding that such a trade-off appears to be a fundamental property of evolving biological entities existing in environments with limited resources (10).
More than 20 years ago it was discovered that conventional culturing techniques grossly underestimate the abundance and number of different types (richness) of prokaryotes in natural environments (“great plate count anomaly”) (88). Soon thereafter it became evident that viruses are abundant and active members of aquatic food webs and constitute a major source of mortality for prokaryotes (7, 73). Since those early days, tremendous improvements in our understanding of the roles of both planktonic prokaryotes (4, 47) and viruses (91, 100) have been made. However, one can still claim that our understanding of the mechanisms regulating prokaryotic and viral abundance, community composition, and population dynamics in natural aquatic environments remains inadequate.
The concept of negative frequency-dependent selection states that the fitness of an organism (e.g., success in reproduction) decreases as its frequency (relative abundance) increases. If correct, this mechanism selects for rare types and thus maintains high diversity (see, e.g., references 28 and 99). Soon after the discovery of high viral abundance in the ocean, it was argued that viruses might be instrumental in maintaining high prokaryotic richness (29, 93), because the viral infection rate depends on, among other parameters, the abundance of host cells. Thus, as suggested by negative frequency-dependent selection, abundant prokaryotic types will be exposed to strong viral pressure. Eventually, this and other ideas were incorporated into what is now known as the “killing the winner” hypothesis (KtW) (92, 94), where “winner” refers not necessarily to the most abundant but to the most active prokaryotic population. KtW was mathematically formalized in the context of an idealized food web comprised of prokaryotes, viruses, and protozoans grazing nonselectively on prokaryotes and is based on Lotka-Volterra-type equations (8). The concept has some potential as a testable theoretical framework that can be challenged with observations and experiments, and it has indeed been used as such by a number of investigators. However, in many cases the comprehension of KtW appears to be based more on a heuristic and not always correct understanding than on an appreciation of the assumptions and limitations of the highly idealized mathematical versions of the hypothesis. For example, the concept of negative frequency-dependent selection is very appealing and can be understood almost intuitively; however, it is only one aspect of KtW, albeit the most prominent one. In order to further stimulate a fruitful dialogue between experimental or observational work and the development of the conceptual framework, our objectives are (i) to review the basic ideas and assumptions of KtW; (ii) to conceptually extend the framework by introducing size- or shape-selective grazing by protozoans, leading to a Russian doll-like hierarchy with KtW principles at work on different levels; (iii) to evaluate which aspects of present experimental and observational knowledge appear to be in agreement or disagreement with KtW; (iv) to point out fundamental limitations of KtW in its present form; and (v) to list challenges for the future that currently prevent a thorough test of the theory.
BASIC IDEAS AND ASSUMPTIONS OF KtW
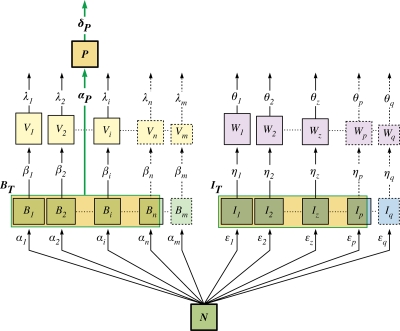
KtW in its general form is illustrated in Fig. 1. The assumption is that two populations (competition specialist and defense specialist) compete for a shared limiting resource (e.g., phosphate). This limiting resource either can exist in the free form that is directly available to the two competing populations or is being sequestered by the competitors and a predator or parasite in the form of biomass. Heuristically, one can see that if the selective loss of the competition strategist from predation or parasitism prevents this population from sequestering all of the resource, there will be more of the limiting resource available to the defense specialist. The optimal strategy in terms of dominating biomass thus depends on the environmental conditions. In an oligotrophic environment (low total available resource level) the competition specialist would be expected to dominate, whereas in a eutrophic environment (high total available resource level) the system would be dominated by the defense specialist. It is important to realize that the “winner” in KtW refers to the competition specialist, which may or may not correspond to the most abundant population. This simple three-population structure (Fig. 1) contains some of the basic elements that need to be incorporated into a general ecological framework. It links trophic interactions to biogeochemistry and food web dynamics to organism properties and strategies. Thus, KtW appears to be relevant for the present debate on a trait-based ecology (30, 54). Since it also contains the arms race perspective between the three populations, there is a clear evolutionary aspect implicit in this generalized structure. By allowing the two populations to coexist on one common limiting resource, KtW provides a simple solution to Hutchinson's paradox (40) without the need to invoke particular assumptions about spatial and/or temporal heterogeneities.
FIG. 1.
Schematic of the general structure of the “killing the winner” hypothesis.
KtW ON DIFFERENT LEVELS: A RUSSIAN DOLL-LIKE HIERARCHY
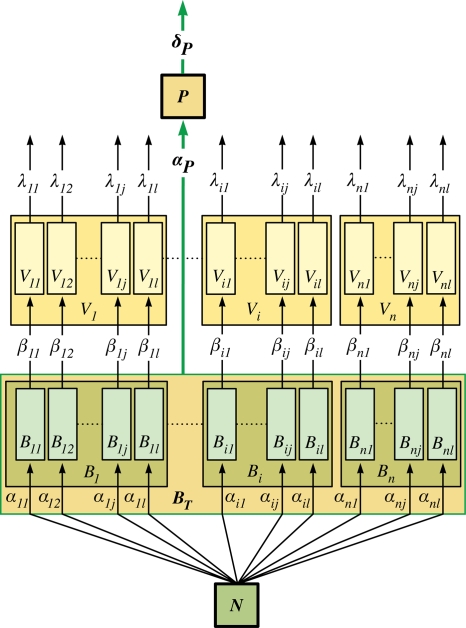
Not surprisingly, most attention to KtW has come from studies of the influence of lytic viruses on their host populations (see, e.g., references 11, 12, and 13). However, KtW in its general form (Fig. 1) is much more widely applicable. In the immediate trophic neighborhood of the pelagic food web, the prokaryote-phytoplankton-protozoan case has been explored both theoretically and in an idealized experimental system (70, 95). In more complex systems, the balance between edible and inedible populations as representatives of the competition and defense specialists, respectively, has been explored for the prokaryote-protozoan case (46, 71), as well as for the phytoplankton-mesozooplankton case (58). Thus, the principle appears to work, not only at different trophic levels in the food web but also for different levels of aggregation (e.g., species versus communities of edible and inedible types). Based on this observation, we conceptually expand the system analyzed by Thingstad (92) by introducing size- or shape-selective grazing due to protozoa, as illustrated in Fig. 2. While host-specific parasitism allows different prokaryotic types to coexist on one limiting resource, selective grazing by protozoa leads to the establishment of a group of inedible prokaryotic types (subject to resource availability). However, in natural environments selective grazing is likely to lead to a number of different groups of populations differing in the degree of their accessibility to the selective predator; thus, Fig. 2 illustrates the simplest case. The basic requirement is that the sum of the biomasses of all organisms in the system and the amount of free resource N must equal the amount of the total available resource NT (Table 1 gives a list of symbols used in the formulas and their meanings):
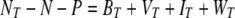 |
(1) |
Using the equations given by Thingstad (92) for the total biomass of edible prokaryotes (BT) and their viruses (VT) and following analogous arguments for the case of inedible prokaryotic types (IT) and their viruses (WT), we get
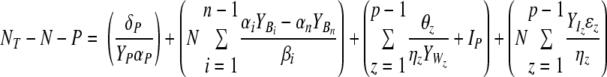 |
(2) |
The system in Fig. 2 is analogous to a Russian doll, where KtW principles are at work on an inner level (viruses) and an outer level (protozoan grazing). More intriguing, this hierarchy bears a resemblance to the self-similarity principle of fractal geometry (57), where the same structure is repeated at different levels of resolution within the system.
FIG. 2.
Schematic of the mechanisms underlying the “killing the winner” hypothesis, also incorporating size- or shape-selective grazing by protozoa. There are potentially m different edible and q different inedible prokaryotic types that are infected by as many different virus types. The populations Bn and Ip do not grow fast enough to sustain stable virus populations Vn and Wp (indicated by excluding part of the rectangles referring to the population sizes of Bn and Ip). Thus, population Bn is only subject to mortality due to grazing, and Ip is controlled solely by resource limitation. Populations not able to establish stably in the system are indicated by dashed lines. The meanings of the symbols are given in Table 1.
TABLE 1.
Explanation of symbols
| Symbol | Meaning |
|---|---|
| N | Concn of free limiting resource |
| NT | Total available limiting resource |
| BT and IT | Total prokaryotic biomass of edible and inedible types |
| Bi and Iz | Prokaryotic biomass of edible and inedible types |
| VT and WT | Total viral biomass infecting edible and inedible prokaryotic types |
| Vi and Wz | Biomass of viruses infecting edible and inedible prokaryotic types |
| mi and lz | Burst size of viruses infecting edible and inedible prokaryotic types |
| P | Protozoan biomass |
| YVi and YWz | Fraction of resource in prokaryotic host transferred to viruses |
| YBi and YIz | Fraction of resource transferred to prokaryotes |
| αi and ɛz | Prokaryotic affinity for limiting resource of edible and inedible types |
| βi and ηz | Viral adsorption constants of Vi and Wz |
| λi and θz | Viral decay rates of Vi and Wz |
| YP | Fraction of limiting resource transferred to protozoans |
| αP | Protozoan grazing rate |
| δP | Protozoan loss rate |
PREDICTIONS OF KtW
In this section we use key equations describing the biomasses of specific prokaryotic and viral populations in the case of nonselective (92) and size- or shape-selective protozoan grazing (Fig. 2) together with the equations for total prokaryotic and viral biomass given above to evaluate the evidence for and against specific predictions of KtW.
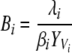 |
(3) |
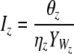 |
(4) |
The equations state that the prokaryotic abundance or biomass Bi (edible prokaryotes) or Iz (inedible prokaryotes) is directly proportional to the virus-specific decay rate λi or θz and indirectly proportional to the product of the fraction YVi or YWz of the limiting resource in Bi or Iz that is transferred to the virus population Vi or Wz and the virus-specific adsorption constant βi or ηz (Fig. 2).
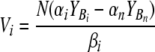 |
(5) |
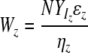 |
(6) |
In a system with nonselective protozoan grazing (92) and given a limiting resource N, the abundance Vi of a specific viral population is directly proportional to the difference in the products of the fraction YB of the limiting resource that is taken up by prokaryotic hosts and the prokaryotic affinity α for the limiting resource of the ith and nth populations and is indirectly proportional to the virus-specific adsorption constant βi (Fig. 2). Size- or shape-selective protozoan grazing creates a niche for inedible prokaryotic types. Given a limiting resource N, the biomass Wz of a specific viral population infecting inedible prokaryotic types is directly proportional to the fraction YIz of the limiting resource transferred into prokaryotes and the prokaryotic affinity ɛz and is indirectly proportional to the virus-specific adsorption constant ηz (Fig. 2).
These equations give rise to a number of predictions (Table 2). For example, an increase in the virus-specific decay rate λi increases the biomass Bi of a specific prokaryotic population (equation 3) and thus reduces the total number of prokaryotic types that can become established in the system (equations 1 and 2). Thus, an increase in the virus-specific decay rate λi causes a decrease in the evenness and richness of the prokaryotic assemblage, since less well adapted populations are becoming extinct due to a shortage of the available resource in the system (Fig. 2). However, the model as a whole is more complicated than the sum of its parts and gives rise to a number of less recognized predictions. In the case of nonselective protozoan grazing, the total viral biomass VT and the biomass of specific viral populations Vi depend on the difference in the adsorption coefficients α of the prokaryotic host populations (equations 1, 2, and 5). In other words, large differences between fast- and slow-growing prokaryotic host populations in a system result in a high total viral biomass VT. The biomasses Vi and Wz of viruses with high adsorption constants βi and ηz are small because the size of the prokaryotic host population declines correspondingly (equations 3 to 6). Also, the biomass of edible prokaryotes BT is controlled by grazing, whereas the biomass of grazing-resistant prokaryotes IT depends on the amount of free resource N (equations 1 and 2).
TABLE 2.
Specific predictions of the “killing the winner” hypothesis
| Equation(s) for indicated parameter(s) | Mechanism | Prediction |
|---|---|---|
| Bi and Iz | Increase in λi and θz | Bi and Iz increase, no. of different prokaryotic types decreases |
| Bi and Iz | Increase in βi and ηz | Bi and Iz decrease, no. of different prokaryotic types increases |
| Bi and Iz | Increase in YVi and YWz, increase in mi and li | Bi and Iz decrease, no. of different prokaryotic types increases |
| Bi and Iz | Change in N | No. of different prokaryotic types does not depend on N |
| BT | Increase in δP | Total prokaryotic abundance BT increases |
| BT | Increase in YP | Total prokaryotic abundance BT decreases |
| BT | Increase in αP | Total prokaryotic abundance BT decreases |
| IT and model | Increase in N | Increase in IT |
| Vi and VT, Wz and WT | Increase in N | Increase in Vi and Wz, increase in total viral abundance VT and WT |
| Vi and VT | Increase in αiYBi − αnYBn | Increase in Vi and VT |
| Vi and VT, Wz and WT | Increase in αiYBi and ɛzYIz | Increase in Vi and VT, Wz and WT |
| Vi and VT, Wz and WT | Increase in βi and ηz | Decrease in Vi and VT, Wz and WT |
| Model | Change in host community composition | Change in viral community composition |
| Model | High-productivity environment/high N | Predation regulates prokaryotic community composition |
SIZING UP THE EVIDENCE
Here we review the recent literature from 2000 onwards that is relevant to the KtW hypothesis to identify aspects of the theory that are in agreement or disagreement with experimental and observational work. Also, we will point out shortcomings of KtW that are revealed in the literature. In order to give a better overview, we distinguish experimental studies according to the nature of the manipulations and the type of organisms or environments studied (Table 3). Likewise, we have grouped observational studies according to the environments investigated (Table 4). For an overview of the general literature, not covered in this article, pertaining to all aspects of viral ecology, see reviews by Wommack and Colwell (110), Weinbauer (100), and Weinbauer and Rassoulzadegan (104).
TABLE 3.
Experimental studies with relevance to the “killing the winner” hypothesis, grouped according to the nature of the experimental manipulation and the organisms or environments studied
| Authors (reference) | Manipulations | Organisms and/or environments studied |
Main parameter(s) | |||
|---|---|---|---|---|---|---|
| Isolates | Marine | Freshwater | Estuary | |||
| Brockhurst et al. (19) | Viruses | × | Pseudomonas aeruginosa adaptive radiation | |||
| Holmfeldt et al. (38) | Viruses | × | Host strain susceptibility; phage host range | |||
| Lennon et al. (52) | Viruses | × | Cost of resistance in Synechococcus spp. | |||
| Middelboe et al. (61) | Viruses | × | Prokaryotic and viral abundance, colony morphology, phage susceptibility; BIOLOG profiles. lipopolysaccharide profiles of prokaryotes, partial 16S rRNA gene sequences, universally primed PCR profiles of prokaryotes | |||
| Middelboe et al. (63) | Viruses | × | Competition between Cellulophaga spp. and Photobacterium sp., prokaryotic and viral abundance, prokaryotic morphology | |||
| Middelboe et al. (60) | Viruses | × | competition between 4 isolates, batch and continuous cultures, clonal composition (resistant vs sensitive) | |||
| Bouvier and del Giorgio (16) | Viruses | × | FISH for distinct prokaryotic groups, prokaryotic production, growth rates of the community and distinct prokaryotic groups | |||
| Hewson and Fuhrman (34) | Viruses | × | Prokaryotic and viral abundance, community composition of Bacteria, richness of nifH gene (diazotrophs) | |||
| Middelboe and Lyck (62) | Viruses | × | Influence of viruses on prokaryotic net growth and respiration, prokaryotic and viral abundance, prokaryotic production | |||
| Schwalbach et al. (82) | Viruses | × | Prokaryotic abundance, community composition of Bacteria | |||
| Winter et al. (109) | Viruses | × | Prokaryotic abundance, community composition of Bacteria and Archaea | |||
| Auguet et al. (3) | Viruses | × | × | Influence of auto- and allochthonous viruses on prokaryotes. prokaryotic and viral abundance, community composition of Bacteria and viruses, prokaryotic production, viral production | ||
| Hewson et al. (35) | Viruses | × | Benthic study, community composition of microalgae and phytoplankton, prokaryotic abundance, photosynthesis and carbon fixation rates | |||
| Bonilla-Findji et al. (15) | Prokaryotes | × | × | Influence of nonindigenous prokaryotes on viruses, prokaryotic and viral abundance, prokaryotic production, prokaryotic respiration, dissolved organic carbon, community composition of Bacteria | ||
| Šimek et al. (85) | HNFa | × | Community composition of Bacteria, mortality due to viruses, FISH for distinct prokaryotic groups | |||
| Sime-Ngando and Pradeep Ram (87) | HNF | × | Concn of nutrients, prokaryotic and viral abundance, prokaryotic production, frequency of visibly infected cells, FISH for distinct prokaryotic groups | |||
| Bonilla-Findji et al. (14) | Viruses, HNF | × | Prokaryotic and viral abundance, prokaryotic production, inorganic nutrients, community composition of Bacteria and Archaea | |||
| Zhang et al. (113) | Viruses, HNF | × | Prokaryotic abundance and production, community composition of Bacteria and Archaea | |||
| Jacquet et al. (42) | Viruses, HNF | × | Prokaryotic and viral abundance, concn of nutrients, grazing rates, viral infection and burst size | |||
| Jardillier et al. (44) | Viruses, HNF | × | Prokaryotic and viral abundance, abundance of protists, community composition of Bacteria, FISH for distinct prokaryotic groups, prokaryotic production, viral infection of prokaryotes | |||
| Šimek et al. (86) | Viruses, HNF | × | Effects of predators on Flectobacillus sp. populations | |||
| Weinbauer et al. (103) | Viruses, HNF | × | Prokaryotic abundance and production, community composition of Bacteria and Archaea, FISH for distinct prokaryotic groups | |||
| Benmayor et al. (6) | Viruses, productivity, disturbance | × | Pseudomonas fluorescens adaptive radiation | |||
| Brockhurst et al. (18) | Viruses, temp | × | Outcome of competition between Pseudomonas fluorescens and Pseudomonas aeruginosa | |||
| Brockhurst et al. (20) | Viruses, spatial heterogeneity | × | Pseudomonas fluorescens adaptive radiation | |||
| Evans et al. (25) | Viruses, HNF, nitrate, phosphate | × | Chlorophyll a, concn of nutrients, abundance and composition of phytoplankton (Micromonas spp.), viral abundance, grazing and lysis rates | |||
| Hewson et al. (36) | Viruses, phosphate, ammonia | × | Benthic study, prokaryotic abundance, community composition of Bacteria, photosynthesis | |||
| Malits and Weinbauer (56) | Viruses, turbulence | × | Prokaryotic and viral abundance, prokaryotic production, viral production, prokaryotic cell vol, no. of organic aggregates, community composition of Bacteria and Archaea, concn of nitrate and phosphate | |||
| Weinbauer et al. (102) | Viruses, dissolved organic matter, dilution | × | Prokaryotic and viral abundance, prokaryotic production, abundance of Vibrio- and Rhodobacter-related populations using FISH | |||
| Bohannan and Lenski (11) | Glucose | × | Abundance of bacteria and viruses | |||
| Middelboe (59) | Different growth rates in chemostats | × | Host cell lysis and viral production, burst size, latent period, resistance | |||
| Riemann and Grossart (75) | Agarose beads | × | Abundance of bacteria and viruses | |||
| Jacquet et al. (43) | Nitrate, phosphate, glucose | × | Prokaryotic and viral abundance, prokaryotic cell size and morphology, distinction of prokaryotic and viral populations using flow cytometry | |||
| Larsen et al. (50) | Nitrate, phosphate | × | Abundance of prokaryotes, viruses, and algae; community composition of Bacteria and Eukarya; viral community composition; primary production; chlorophyll a; phytoplankton abundance | |||
| Øvreås et al. (68) | Nitrate, phosphate, glucose | × | Community composition of Bacteria and viruses, prokaryotic abundance and cell morphology, FISH for distinct prokaryotic groups | |||
| Riemann et al. (77) | Phosphate, silicate, nitrate, ammonia; induction of diatom blooms | × | Prokaryotic and viral abundance, HNF abundance, prokaryotic production, community composition of Bacteria, chlorophyll a, abundance of phytoplankton cells, ectoenzyme activity | |||
| Sandaa et al. (81) | Phosphate, glucose | × | Prokaryotic and viral abundance, chlorophyll a, community composition of Bacteria and viruses | |||
| Jardillier et al. (45) | HNF, zooplankton, nitrate, phosphate, ammonia | × | Concn of nitrate, phosphate, and ammonia; prokaryotic and viral abundance; community composition of Bacteria; prokaryotic production; abundance of protists and metazooplankton; grazing rates; FISH for distinct prokaryotic groups | |||
| Pradeep Ram and Sime-Ngando (72) | HNF, glucose, nitrate, phosphate | × | Prokaryotic and viral abundance, prokaryotic production and respiration, mortality due to viral lysis, FISH for distinct prokaryotic groups | |||
| Šimek et al. (84) and Weinbauer et al. (101) | HNF, availability of phosphorus and DOC | × | Prokaryotic and viral abundance, prokaryotic production, viral production and lysis rate, prokaryotic cell vol, FISH for distinct prokaryotic groups | |||
HNF, heterotrophic nanoflagellates.
TABLE 4.
Observational studies with relevance to the “killing the winner” hypothesis, grouped according to the environments studied
| Authors (reference) | Environment studied |
Main parameter(s) | |||
|---|---|---|---|---|---|
| Marine | Freshwater | Estuary | Other | ||
| Hewson and Fuhrman (33) | × | Prokaryotic and viral abundance, community composition of Bacteria, richness of nifH gene (diazotrophs), viral production, prokaryotic production | |||
| Hewson et al. (37) | × | Prokaryotic and viral Abundance, community composition of Bacteria and viruses | |||
| Larsen et al. (51) | × | Concn of nutrients, prokaryotic and viral abundance, abundance of phytoplankton, community composition of phytoplankton, community composition of viruses and Bacteria, particulate organic phosphorus, dissolved organic carbon, prokaryotic production, alkaline phosphatase activity | |||
| Parada et al. (69) | × | Prokaryotic and viral abundance, viral production, prokaryotic production, viral community composition in situ and newly produced viruses | |||
| Winter et al. (107) | × | Prokaryotic and viral abundance, viral infection, community composition of Bacteria and Archaea | |||
| Winter et al. (108) | × | Prokaryotic and viral abundance, prokaryotic production, community composition of Bacteria and Archaea | |||
| Filippini et al. (26) | × | Prokaryotic and viral abundance, prokaryotic production | |||
| Tijdens et al. (96) | × | Prokaryotic and viral abundance; abundance of phytoplankton and flagellates; community composition of Bacteria, cyanobacteria, and viruses | |||
| Yoshida et al. (112) | × | Microcystis aeruginosa, viruses infecting this species | |||
| Riemann et al. (76) | × | Nutrients, total organic carbon, chlorophyll a, concn of dissolved DNA, prokaryotic and viral abundance, prokaryotic production, viral production, community composition of viruses, alkaline phosphatase activity, prokaryotic DNA uptake rates | |||
| Wang and Chen (98) | × | Abundance of cyanobacteria and viruses, abundance of viruses infecting Synechococcus sp., richness of g20 genes (cyanomyoviruses) | |||
| Andersson and Banfield (1) | Mine biofilms | Community genomics | |||
| Sabet et al. (80) | Soda lake | Isolation of bacterial and viral strains, genetic analysis, probes targeting viral isolates | |||
The Issue of Resistance to Viral Infection
Studies focusing on the influence of viruses on prokaryotic isolates either alone or in competition experiments in the presence or absence of viruses could be considered the simplest type of experiment to conduct with respect to KtW. This type of experiment reveals aspects of the virus-host relationship that are difficult to attain in more complex setups. Many of the studies of isolates reviewed here demonstrate that the development of resistance to viral infection is rapid and that this mechanism can influence or completely change the clonal composition of the host population from vulnerable to resistant (6, 19, 20, 59, 60, 61, 75). However, in most cases the development of resistance was associated with a fitness penalty for the resistant population. This cost of resistance was investigated in detail by Lennon et al. (52) in the case of Synechococcus sp. isolates. The study found that when a cost of resistance was detected, it resulted in an ∼20% reduction in fitness compared to the ancestral strains. Furthermore, the cost of resistance was unaffected by the total number of viruses for which resistance occurred. However, under competitive conditions, the cost of resistance was dependent on the identity of the viruses. The essence of KtW is a trade-off between competition and defense specialists; thus, these results are in agreement with KtW (equations 1 and 2). Topically related work by Middelboe et al. (61) focused on the genetic, structural, and physiological differences of a Cellulophaga baltica strain that developed as a response to the presence of two viruses infecting this strain in chemostat cultures. The Cellulophaga baltica strain was initially sensitive to 24 tested viruses. During the incubations, the authors found a succession of strains, with the dominant strains eventually having acquired immunity to up to 22 of the tested viruses. At the end of the experiment, the cultures were dominated by strains resistant to either one or both of the viruses present during the incubations, with small populations of sensitive strains maintaining both viruses at abundances of above 106 ml−1. Loss of sensitivity to virus infection was associated with a reduction in the ability to use various carbon sources, thus demonstrating that the actions of viruses can lead to a functional diversification of the host population. Nevertheless, the degree of resistance was not correlated to the loss of metabolic potential. Thus, both studies (52, 61) found that the cost of resistance is not proportional to the degree of resistance. A fitness penalty depending on the specific virus type rather than on the total number of different viruses to which resistance developed implies the existence of different mechanisms of resistance. Many viruses infecting prokaryotes use as a docking site surface proteins that are also involved in the uptake of nutrients by the host cell (24, 66). A structural change in these surface proteins or in their abundance per host cell may result in resistance to viral infection and at the same time cause a reduction in the capabilities for uptake of nutrients (53). Lysogenic viruses may also confer resistance by initiating the production of repressor proteins that ensure the host's immunity to additional viral infection (21). Nevertheless, maintenance of such a repressor system is costly and incurs a fitness penalty for the cell. Recently, another highly specific mechanism of resistance to viral infection, based on clustered regularly interspaced short palindromic repeats (CRISPR), was described (5). The CRISPR system represents a DNA-encoded defense system based on genomic features that provides prokaryotes with immunity against viruses (and plasmids), likely employing RNA interference. While there presumably is a cost associated with running this defense system, the cost of adding a new virally derived CRISPR sequence to the library of the host cell seems minor. However, Andersson and Banfield (1) demonstrated that resistance to viral infection conferred by CRISPR sequences is rather short-lived due to recombination sufficiently shuffling the sequence motifs so that only the most recently acquired CRISPR sequences result in immunity. Based on the currently available knowledge, the CRISPR defense system appears to be difficult to incorporate into the KtW framework. The complexity of the issue of resistance to viral infection becomes even more apparent when considering a study by Holmfeldt et al. (38). Those authors investigated the susceptibility of 23 Bacteroidetes sp. strains to viral infection by 46 different viruses. Based on 16S rRNA gene and internal transcribed spacer DNA sequences, 21 of these bacterial isolates could be considered strains of Cellulophaga baltica (Flavobacteriaceae). The study found that all of the bacterial isolates showed unique virus susceptibilities and differed by up to 6 orders of magnitude in sensitivity to the same titer of virus. The tested viruses themselves showed variations in host ranges, infecting between 1 and 20 bacterial strains. This study reveals a major shortcoming of KtW as presented in Fig. 2: each host population is subject to mortality due to multiple co-occurring virus populations, not just one. The data suggest that a higher resolution is required also within the population boxes of Fig. 2 representing prokaryotic “species,” so that each prokaryotic population consists of different strains with profiles of different resistance to viruses. Thus, the hierarchical structure of KtW may be extended to the level of subpopulations representing different strains, as demonstrated in Fig. 3. However, if the control by lytic viruses is at the strain level, it is not immediately obvious how population size is controlled at the “species” level. Also, only 5 out of 46 different viruses tested by Holmfeldt et al. (38) were host specific, questioning the validity of the generally assumed specificity of the virus-host relationship and thus also making incorporation into a simple KtW structure difficult. The study also reveals a more general methodological problem. Many investigators studying the influence of viruses on natural communities rely on the use of marker genes such as the 16S rRNA gene to monitor the prokaryotic community. However, Holmfeldt et al. (38) suggest that these techniques may severely underestimate the complexity of virus-host relationships (Fig. 3) and in some cases may completely miss the effects of viral lysis on the host populations.
FIG. 3.
“Killing the winner” mechanisms on three levels. Experimental data suggest that different strains of the same “species” may have profiles of different resistance to viruses. Thus, it appears that the influence of viruses on prokaryotes is on the strain level (Bij; j = 1, 2, …, l) and not on the “species” level (Bi; i = 1, 2, …, n) commonly detected by PCR-based fingerprinting techniques targeting the 16S rRNA gene. This is illustrated here for the case of edible bacteria BT; however, similar arguments also hold for the case of inedible bacteria IT. The figure suggests that the abundance of the prokaryotic community BT is controlled by protozoan grazing and that the abundance at the strain level Bij is controlled by viral lysis. However, the controlling mechanism for population size at the “species” level Bi is not immediately obvious from this model. The meanings of the symbols are given in Table 1.
Influence of Environmental Conditions on Virus-Host Relationships Studied with Isolates
KtW predicts that under highly productive conditions, predation is the major regulatory mechanism for community composition, whereas in environments with low productivity, competition drives community composition. This prediction has been confirmed in a model system consisting of two strains of Escherichia coli with different vulnerabilities to the virus T2 (11). However, investigators also tested the influence of other environmental variables on virus-host relationships. Benmayor et al. (6) found that the presence of viruses increased diversity at environmental extremes of disturbance and productivity by imposing selection for virus-resistant types but decreased diversity in less stressful environments. Thus, viruses appear to have a mitigating effect on the influence of other environmental variables on the community composition of their hosts. The disturbance treatment by Benmayor et al. (6) consisted of repeated reinoculation of the culture into fresh culture medium. However, KtW is based on the assumption of steady-state conditions so that it refers to the climax situation at the end of a string of successional developments. Such a steady state may or may not be locally stable, and there may or may not be oscillations in the form of successive replacements of virus-host pairs (Fig. 4). The successional replacement of the most abundant hosts due to viral lysis is therefore a possible result of KtW but not a definition of the concept as sometimes presented (see, e.g., reference 17). In another study, Brockhurst et al. (20) tested the influence of spatial heterogeneity and the presence of parasites on the evolution of Pseudomonas fluorescens. Viruses increased total diversity in homogenous environments but had no effect in heterogenous environments. Heterogenous environments offer a number of different niches that can be occupied by the competing populations. KtW considers the case of competition for one limiting resource and is thus not relevant for heterogenous environments. However, for the case of homogenous environments, the results reported by Brockhurst et al. (20) are in agreement with KtW (equations 1 and 2). Brockhurst et al. (18) conducted competition experiments using communities consisting of Pseudomonas fluorescens and Pseudomonas aeruginosa and their viruses. The communities were incubated at two different environmental temperatures, which, without the presence of viruses, reversed the outcome of competition. The results show that coexistence was enhanced (evenness was increased) in the presence of viruses infecting the superior competitor and in the presence of viruses infecting both bacterial strains. In accordance with KtW, viruses reduced the abundance of the superior competitor, allowing the weaker competitor to increase its density (equations 1 and 2). However, coexistence of the two strains was not equally stable at the two different environmental temperatures.
FIG. 4.
Possible scenarios for temporal changes in host abundance as a consequence of viral lysis. In scenario 1, the abundance of the host population changes periodically, resembling predator-prey oscillations. Thus, viral lysis regulates numerically dominant populations (similar to negative frequency-dependent selection). In scenario 2, the host population is resistant to viral infection and thus can maintain high host abundance. This scenario is unlikely due to the transient nature of resistance and the associated costs of resistance. Scenario 3 depicts the case of a population that coevolves with its virus. The oscillations are due to a change in the population's sensitivity to the virus. In scenario 4, the sensitive host population is kept at a low abundance due to strong viral control.
Feedback Loops Generated by Release of Lysis Products
Viral lysis of host cells releases not only progeny virus particles but also a cocktail of sugars, proteins and peptides, amino acids, nucleic acids, etc., that could serve as a source of nutrients for the surviving community. A number of studies investigated whether these lysis products are taken up by the survivors and also whether this effect would be of any significance (35, 59, 62, 63, 76). Middelboe (59) studied the dynamics of a marine virus-host (Pseudoalteromonas sp.) system at different steady-state growth rates in chemostat experiments. The author found that cell lysis and virus production were, in agreement with KtW, positively correlated with the host growth rate (equations 5 and 6). The results also demonstrated that the burst size increased while the latent period decreased with increasing host growth rate and that the release of viral lysates stimulated growth of noninfected resistant cells. Thus, although resistance to viral infection may have an associated fitness penalty (52), resource enrichment due to the lysis of vulnerable cells may have a mitigating effect. Also, Hewson et al. (35) concluded that in oligotrophic sediments, viruses stimulated carbon fixation due to increased nutrient availability as a consequence of viral lysis of prokaryotes. Further studies of the role of viral lysates for prokaryotic growth demonstrated that this effect is of quantitative importance not only in model systems (63) but also for mixed marine prokaryotic communities (62, 76). In addition to liberating organic carbon bound in prokaryotic biomass, it has also been reported that 25% of the dissolved DNA pool in a phosphorus-limited estuary is derived from viral lysis and that the uptake of dissolved DNA accounts for 70% of the prokaryotic phosphorus demand in this environment (76). Based on the available evidence, the question of whether or not a feedback loop that routes resources from vulnerable (competition specialist) to resistant (defense specialist) populations exists can be answered in the affirmative, yet it is currently not included in KtW.
Availability of Resources
The effects of resource availability on mixed prokaryotic communities were investigated in studies of mesocosms inoculated with marine assemblages (43, 50, 68, 77, 81) or of a marine sediment-water mesocosm (36). These studies, although difficult to compare due to the distinct setups and inocula used, provide evidence relevant for specific aspects of KtW. Larsen et al. (50) followed the dynamics of phytoplankton, prokaryotes, and viruses in a seawater mesocosm enriched with nitrate and phosphate. Flagellates dominated the algal community, followed by the appearance of a number of different size classes of viruses. Similarly, Jacquet et al. (43) could link the development in the abundance of specific groups of autotrophic prokaryotes and eukaryotes to different groups of viruses in experiments with mesocosms enriched with nitrate, phosphate, and glucose. Thus, changes in the host community are followed by changes in the corresponding virus community, as predicted by KtW. Øvreås et al. (68) conducted mesocosm experiments to investigate the effects of the addition of inorganic nutrients (nitrate and phosphate) and of inorganic plus organic (glucose) nutrients on prokaryotic and viral communities. The authors found small changes in bacterial community composition upon addition of mineral nutrients alone; however, the combined addition of mineral nutrients and glucose resulted in major changes in the bacterial community composition. Also, in agreement with KtW, changes in the bacterial community caused similar changes in the virus community. Riemann et al. (77) studied the dynamics of prokaryotic activity, abundance, and bacterial community composition in mesocosms fertilized with inorganic nutrients to initiate a diatom bloom. Prokaryotic abundance abruptly decreased during the peak of the bloom concomitantly with the disappearance of three dominant phylotypes from the bacterial community. Increased flagellate and viral abundance during the diatom peak suggest that grazing and lysis could have caused this phylotype-specific mortality. Sandaa et al. (81) studied the effects of viral lysis and substrate limitation on the bacterial community in mesocosms amended with phosphate, glucose, and phosphate plus glucose. In agreement with KtW, these authors found that changes in the bacterial community composition were linked to changes in the viral community. Furthermore, data obtained by pulsed-field gel electrophoresis (PFGE) showed that the total number of virus populations was the same in all treatments; however, the composition of the viral community varied between the treatments. The authors concluded that the number of different virus-host pairs is controlled by viral lysis (top-down control, in agreement with KtW [equations 1 and 2]), whereas the identity of these virus-host pairs is controlled by substrate limitation (bottom-up control). Finally, Hewson et al. (36) investigated the effects of virus infection and nutrient enrichment on the microbial compartment of oligotrophic carbonate-based sediments. The addition of nutrients (phosphate and ammonia) resulted in elevated virus abundance and increased bacterial richness. These results are in principle in agreement with KtW (equations 1 and 2).
Manipulating Viral Abundance in Experimental Incubations
Studying mixed prokaryotic communities instead of isolates clearly bears a higher resemblance to studying natural systems. However, the results of such experiments are often more difficult to interpret and suffer from other biases, such as the adverse effects of sample handling and containment. Also, it is plausible to assume that not all prokaryotic communities coming from different environments will display similar reactions to experimental manipulations of viral abundance, so it is difficult to directly compare the results. Schwalbach et al. (82) and Winter et al. (109) incubated marine prokaryotic assemblages with significantly reduced or enhanced viral abundances to study the effects on prokaryotic community composition. Both studies found significant effects of sample manipulation and containment on the prokaryotic communities. Schwalbach et al. (82) reported modest but significant effects on bacterial community composition in virus-reduced incubations but no effects in virus-enriched cultures. The results of Winter et al. (109) showed that the effects of viruses were detected on the level of individual bacterial and archaeal phylotypes. In another study, Hewson and Fuhrman (34) investigated the influence of viral infection on bacterial communities at 15 stations in the North Atlantic, North Pacific, and Gulf of Mexico and in southern California. The authors performed batch culture incubations that were enriched in or depleted of ambient viruses and used diazotrophs as model organisms for rare prokaryotic types, which were hypothesized to benefit from viral activity. The effects of viruses were not consistent between sampling locations. Also, there was no significant difference in the relative abundances of common and rare bacterial types in the response to virally enriched or depleted incubations. In a study investigating the sediment-water interface, Hewson et al. (36) found that the addition of viruses to benthic flocculent layer samples increased bacterial richness compared with the addition of heat-killed viruses as a control. In what could be called a virus-transplant experiment, Auguet et al. (3) investigated the potential effect of freshwater viruses on the composition and activity of bacterial communities in an estuary (Marennes-Oléron Bay, France). Those authors found that confinement and incubation temperature were the two principle experimental factors influencing bacterial community composition. However, the addition of freshwater viruses had a significant effect on the bacterial communities and reduced net prokaryotic growth compared to the addition of inactivated viruses or ambient viruses. The study suggests that prokaryotic communities are adapted to ambient viruses, while the community needs to change in order to adjust to the presence of allochthonous viruses. In a similar study, Bonilla-Findji et al. (15) cross-transplanted freshwater and marine prokaryotic communities into batch cultures mimicking marine and freshwater environments. Those authors found that prokaryotic production was stimulated in the transplanted marine and freshwater communities, while bacterial richness decreased. Furthermore, mortality due to viruses increased in the transplanted marine community but decreased in the transplanted freshwater community, suggesting that freshwater viruses appear to be able to infect marine prokaryotes. All of the above-mentioned studies except for that of Hewson and Fuhrman (34) are in principle compatible with KtW. However, according to Holmfeldt et al. (38), current methodological limitations in the analysis of mixed prokaryotic communities may result in a severe underestimation of the effects of viral lysis on the host communities. Also, studies of specific virus-host systems (see, e.g., reference 18) suggest that a multitude of other environmental factors affect the sensitive balance between viruses and their hosts, and these might explain the varying results. One of those environmental factors rarely studied is turbulence. Malits and Weinbauer (56) found that turbulence stimulated prokaryotic production, likely by enhancing the formation of microaggregates and nutrient availability. However, the presence of viruses appeared to reduce the number of microaggregates. Furthermore, turbulence together with viruses increased prokaryotic cell length. Those authors also report that specific phylotypes appeared to be inhibited or stimulated by turbulence and/or viruses. Weinbauer et al. (102) studied the response of a prokaryotic community to the additions of dissolved organic carbon and viruses and to dilution, with particular focus on Vibrio- and Rhodobacter-related populations. Their results suggest that Vibrio- and Rhodobacter-related populations can be described as r and K strategists, respectively. K strategists are considered to be strong competitors and have a low reproductive output, whereas r strategists invest in large numbers of offspring but do not have strong competitive abilities (55). However, both populations were only weakly susceptible to viral infection despite a 2-fold-higher growth rate of Vibrio- compared to Rhodobacter-related populations. Finally, Bouvier and del Giorgio (16) incubated marine bacterioplankton in batch cultures that were greatly depleted of ambient viruses. The result was a dramatic increase in the relative abundance of bacterial groups that were undetectable in the in situ bacterioplankton community. Thus, host susceptibility is not necessarily proportional to host density, as is often assumed, and rare marine bacterioplankton groups may be more susceptible to virus-induced mortality because these groups may actually be the winners in the competition for nutrients (Fig. 4, scenario 4). This is in agreement with KtW, predicting that viruses with fast-growing hosts should be most abundant and that the viral biomass of specific populations Vi and Wz (equations 5 and 6) and the total viral biomass VT and WT (equations 1 and 2) are independent of host biomass.
Combined Effects of Viral Lysis and Protistan Grazing
There are two experimental studies, both conducted in freshwater environments, that manipulated grazing pressure as the sole experimental factor (85, 87). Both studies found that prokaryotic and viral activity increased in the presence of grazers and suggest that this stimulation is due to grazer-mediated resource enrichment. Similar results were also obtained in a study manipulating grazer abundance and the concentration of nutrients (72). Thus, grazing appears to increase the amount of resources available to the surviving populations in a feedback loop similar to viral lysis. Such a feedback loop is currently not included in KtW. However, in agreement with KtW, Sime-Ngando and Pradeep Ram (87) and Šimek et al. (85) also noted the appearance of grazing-resistant forms and suggested that the biomass of edible prokaryotes BT is regulated by grazing (Fig. 2 and equations 1 and 2). Above we have introduced size- or shape-selective grazing to enhance the applicability of KtW (Fig. 2). As Fig. 2 makes clear, the presence of both viruses and grazers should result in the highest prokaryotic richness, subject to the availability of resource N. However, the experimental evidence is more differentiated than that. Jardillier et al. (44) investigated the influence of viruses and protistan grazing on freshwater prokaryotic communities in batch culture experiments. In agreement with KtW, those authors concluded that prokaryotic abundance was controlled mainly by protistan grazing, whereas viruses were mostly responsible for changes in the composition of the prokaryotic community (equations 1 to 4). Jacquet et al. (42) investigated the temporal variation of prokaryotic mortality due to viruses and grazers in a lake. Those authors found that mortality due to viruses increased and that due to grazers decreased during the summer compared to spring. Also, Evans et al. (25) found that mortality of Micromonas spp. due to grazing was similar to or exceeded mortality due to viral lysis. Šimek et al. (86) subjected bacterioplankton to additions of flagellates or viruses plus flagellates to identify the influence of these mortality factors on Flectobacillus sp. populations. Filamentous prokaryotic forms appeared in both treatments but were twice as abundant, long, and active in incubations with both predators than in the flagellate treatment alone. In a companion paper, Weinbauer et al. (103) found that the vulnerability to the two sources of mortality (viruses and flagellates) was distinctly different between prokaryotic groups (see also reference 38). However, in contradiction to KtW (Fig. 2), the results showed that prokaryotic richness was highest in the virus treatment and lowest when both predators were present and that flagellates reduced prokaryotic richness but increased viral production. In total, there are two marine studies investigating the effects of grazers and viruses on the prokaryotic communities in an offshore (14) and a coastal (113) marine environment. Bonilla-Findji et al. (14) used size fractionation to conduct batch culture experiments with virus, flagellate, and virus-plus-flagellate treatments. Those authors found that the addition of viruses or viruses plus flagellates increased prokaryotic abundance and richness but decreased prokaryotic production. However, the results were not consistent in the flagellate treatments compared to the control. Based on flow cytometric data, the relative abundance of large prokaryotic cells with a high nucleic acid content was lower in the virus and virus-plus-flagellate treatments than in the control or the flagellate treatment. The data suggest that viruses and viruses combined with flagellates sustain prokaryotic richness (equations 1 to 4) and control prokaryotic production by regulating the abundance of highly active members of the community (equations 5 and 6), in accordance with KtW. Similarly, Zhang et al. (113) found that the effects of viruses and flagellates combined were consistent and closely resembled natural conditions and that viral lysis and protistan grazing acted additively to reduce prokaryotic production and sustain prokaryotic richness. Overall, there is some variability in the literature as to the effects of protistan grazing on prokaryotic communities, particularly in combination with viral lysis. However, as shown above for isolates (11), the availability of resources is another important factor modifying the complex interactions between grazers, prokaryotes, and their viruses. In the case of mixed prokaryotic communities, Šimek et al. (84) and Weinbauer et al. (101) studied the effects of resource enrichment and grazing on viral production and bacterioplankton communities. In contradiction to KtW, predation affected bacterial community composition in resource-limited prokaryotic communities but was of minor importance under resource-replete conditions. However, in agreement with KtW, the effect of virus-induced mortality was higher in resource-replete treatments, where prokaryotic and viral abundance, prokaryotic and viral production, and virus-induced lysis rates increased (equations 5 and 6). Also, grazing stimulated viral abundance, production, and virus-induced mortality, as found in other studies (72, 85, 87). Jardillier et al. (45) studied the influence of nutrients, grazing, and viral lysis on the prokaryotic community composition in an oligomesotrophic lake and in a eutrophic lake. In agreement with KtW, those authors found that the prokaryotic community in the oligomesotrophic lake was affected mainly by the availability of nutrients and that in the eutrophic lake by the concentration of nutrients and mortality due to grazing and viral lysis (equations 1 to 4). In summary, the availability of resources appears to modify the effects of grazing on prokaryotic communities.
Observational Studies
Although observational studies may not always provide the hard evidence obtainable through experimental manipulation, they are nevertheless important because investigators can gain information about how the complex relationships between prokaryotes and their environment play out in nature. In accordance with KtW, a number of studies in marine and freshwater environments found that virus and host communities of either specific populations or entire assemblages are linked (26, 37, 33, 51, 69, 96, 98, 107, 108, 112), and there is evidence suggesting similarly coupled prokaryotic and viral communities in a soda lake (80). The details vary, however. More specifically, Winter et al. (108) studied the relationship between prokaryotic richness and the abundance of prokaryotes and viruses, prokaryotic production, and other physicochemical parameters in the North Sea. The results showed that bacterial richness decreased with increasing viral abundance and prokaryotic production. The authors conclude that high prokaryotic production was sustained by a relatively small number of highly active bacterial populations that also maintained high viral abundance. These results are in principle in agreement with KtW, because viruses with fast growing hosts are predicted to be the most abundant ones (equations 5 and 6). In another study, Hewson et al. (37) investigated the relationship between viral and bacterial communities in oligotrophic waters of the West Florida shelf. Consistent with KtW, the study found that biomass and community richness were independent (equations 1 to 4) and that temporal patterns in the bacterial community were closely related to changes in the viral community. In a comparative study, Hewson and Fuhrman (33) investigated the covariation of viral parameters (abundance and production) with bacterial community composition and richness in the water column and sediments of the North Pacific, Amazon River plume, North Atlantic, Gulf of Mexico, Southern California Bight, and Coral Sea. The results suggest that there appears to be no universally applicable relationship between viruses and their host community, probably due to the different effects of other environmental parameters. However, the community composition and richness of diazotrophs, as a model of rare but opportunistic organisms, were significantly negatively correlated to viral parameters across all sites. The authors concluded that viral lysis may have positive effects on bacterial richness in oligotrophic oceans, by regulating the abundance of dominant competitors. However, KtW predicts that predation/parasitism should be more important under eutrophic conditions, whereas under oligotrophic conditions competition is predicted to be the most important factor in shaping prokaryotic community composition (equations 1 and 2). The study also suggests that rare taxa may be more susceptible to viral attack due to opportunistic lifestyles (see also reference 16). Winter et al. (107) related bacterial and archaeal community composition and richness in the tropical Atlantic Ocean to the geographic distance between the sampling stations, the frequency of infected cells, and physicochemical parameters to identify factors influencing the prokaryotic community composition. Those authors found no detectable effect of geographic distance or differences between water masses on bacterial and archaeal community composition. Bacterial communities changed with depth, whereas changes in the archaeal community were related to temperature and the frequency of infected cells. Thus, the results suggest that KtW mechanisms are also relevant for archaeal virus-host relationships under natural conditions. Parada et al. (69) demonstrated that newly produced viruses represent only a fraction of the ambient viral community and that the in situ viral community was fairly stable over time. The results show that viral infection and lysis are dynamic processes changing on time scales of hours to days. In accordance with KtW, the results demonstrate the fine temporal scale at which changes in the host community are mirrored by changes in the virus community. Larsen et al. (51) studied the succession and community composition of algae, prokaryotes, and viruses in coastal waters around the time of the spring phytoplankton bloom. In agreement with KtW, those authors' results show that viruses were closely linked to the developments in the microbial community. Also, Wang and Chen (98) investigated the population dynamics of cyanovirus communities, and Yoshida et al. (112) studied the dynamics of viruses infecting the toxic bloom-forming cyanobacterium Microcystis aeruginosa. Both studies found that changes in the virus communities were related to changes in the host communities, supporting KtW. The link between viruses and hosts was further corroborated for entire freshwater assemblages by Filippini et al. (26) and Tijdens et al. (96). Filippini et al. (26) showed that changes in prokaryotic abundance were synchronized with changes in viral abundance. Tijdens et al. (96) found that viral abundance did not correlate with prokaryotic and cyanobacterial abundance during the entire study period. However, as suggested by KtW, viral community composition did correlate with bacterial and cyanobacterial community composition during short periods of strong fluctuations.
FUNDAMENTAL LIMITATIONS OF KtW
KtW remains a model, and as such it is based on highly idealized relationships and assumptions that may not always be correct. Among its shortcomings is that the model relies on the assumption of steady-state conditions. Only very few environments actually remain in such a deadlocked situation for long. However, the problem is really one of temporal resolution. A good example is the study by Parada et al. (69), which found that although the ambient viral community is relatively stable over time, only a small number of viral types are actually produced at any given time. Thus, small and rapid changes in the activity of specific viral populations give rise to an overall stable viral community. Another obvious problem in the assumptions underlying KtW is that in most environments prokaryotic hosts are vulnerable to more than one co-occurring virus population. To complicate matters even more, the vulnerability of distinct host populations to viral infection varies widely (38) (Fig. 4). KtW also assumes that virus-host relationships are specific, i.e., that one virus infects only one host. The evidence, although based solely on isolates, suggests that this may not be correct (38) and that viruses with a wide host range may be more abundant than currently acknowledged. Another aspect of KtW that might be perceived as a limitation is that the model considers the case of competition for one limiting resource as stated in Hutchinson's paradox (40). Different resources limit the activity of the different prokaryotic populations in a community/assemblage. As a whole, a prokaryotic community/assemblage usually depends on the availability of a small subset of these resources (see, e.g., reference 68). The fact that KtW considers only one limiting resource therefore does not appear to be a major limitation. However, both viral lysis and protistan grazing create feedback loops by liberating resources that would otherwise be locked up in biomass. These freed-up resources further appear to stimulate growth of the surviving populations (defense specialist) and represent a conceptual problem for the trade-off between resource and competition specialists that is at the heart of KtW.
CHALLENGES TO BE OVERCOME FOR A THOROUGH TEST OF KtW
Over time, KtW has provided numerous testable hypotheses. Nevertheless, many aspects of KtW still remain untested or are not fully explored due to current methodological limitations. Essentially all of the studies investigating the influence of viral lysis and/or protistan grazing on prokaryotic community composition reviewed here used either (i) PCR-based fingerprinting techniques mainly targeting the 16S rRNA gene or the internal transcribed spacer region between rRNA genes or (ii) fluorescent in situ hybridization (FISH) targeting various groups of prokaryotes. However, particularly the study by Holmfeldt et al. (38) demonstrates that these techniques do not provide the resolution necessary to capture the changes occurring at the strain level in prokaryotic communities. It becomes even harder to obtain adequate data for viral communities, because viruses lack a universally conserved gene that could be used for fingerprinting (32, 79). Here, data on the composition of the virus community were generally based on PFGE. PFGE separates the members of the viral community based on their genome size, and thus the data have no genetic aspect, in the sense of nucleic acid sequence information (89). Recently, Winget and Wommack (106) introduced randomly amplified polymorphic DNA PCR (RAPD-PCR) analysis to determine viral community composition by circumventing the need for a universally conserved gene. Although RAPD-PCR represents a significant advance in our capabilities to study viral communities, we do not expect that it is able to provide enough resolution to capture the relevant changes in the virus community. Thus, presently our methods allow for a rough overview of the processes regulating prokaryotic community composition by viruses and/or protistan grazing, but we are largely ignorant of the fine-scale picture. Currently, the most detailed data on prokaryotic and viral community composition are provided by metagenomics, that is, the genomic analysis of a population or community of microorganisms (31) and viruses (see, e.g., reference 2). However, metagenomic techniques still carry a large price tag, and the huge data sets obtained can be difficult to interpret (41). We expect that as the costs of metagenomic analysis of prokaryotic and viral communities drop and the bioinformatic methods to tame and analyze the flood of data mature, it will be possible to employ these large-scale sequencing techniques to study temporal and spatial developments of prokaryotic and viral communities in experiments and the natural environment. Even so, it will be necessary for more traditional microbial ecologists to start speaking “metagenomics” (48) and for the scientists familiar with these relatively novel techniques to look beyond nucleic acid sequences and consider ecological concepts in their work (74). First steps into this direction have recently been carried out by Andersson and Banfield (1), who matched viruses to their prokaryotic hosts by identifying the viral CRISPR sequences in a metagenomic data set, and by Rodriguez-Valera et al. (78), who suggested that the differences between strains of the same bacterial species detected by metagenomics appear to be caused mainly by the actions of lytic viruses.
The second and probably most daunting challenge for the future lies in obtaining rate measurements for individual prokaryotic communities (especially affinity for the limiting resource) and viral communities (adsorption and decay rates) necessary to constrain the formalized version of KtW. This has been done before for simple model systems employing chemostats (see, e.g., reference 59) but presently seems not to be feasible for complex mixed communities and even more so at the level of individual strains. It is difficult to foresee if and when suitable techniques will be available to achieve this ambitious goal; however, we think that in the coming years a fruitful approach would be to analyze the pool of mRNAs of entire communities (metatranscriptomics [83]) or even of proteins (metaproteomics [see, e.g., reference 105]) in combination with metagenomics. Such an approach should yield a detailed picture of prokaryotic and viral community composition as well as which genes are transcribed and translated. Nevertheless, particularly proteomics presently requires large amounts of proteins that are not easily obtainable from prokaryotic and viral communities, and the amount of data generated is very large, demanding extensive bioinformatics skills and capabilities.
CONCLUDING REMARKS
Every model needs to strike a delicate balance between its complexity and its understandability. Thus, the trade-off for being as realistic as possible is a decrease in the model's usefulness, because overly complex models, although they may resemble natural systems more closely, will be harder to understand. KtW in its present form is a relatively simple model, and we think that the simplicity and intuitive understanding of some of its aspects led to the widespread dispersal of the idea. Despite all its problems, KtW in its simplicity appears to capture many aspects of the relationship between viruses, protozoan grazers, and prokaryotes, so that the basic principle of KtW (Fig. 1) is relatively well established. However, attempts to assemble this principle into a complete model for all the hierarchical levels that is consistent with observations still have a long way to go. Thus, we would like to caution the scientific community not to apply predictions based on KtW too rigorously and not to fall victim to the thought that the model is set in stone. Evidence will and always should be at the heart of research, yet KtW has proven to be very effective and useful in developing testable hypothesis. It is precisely the discrepancies between data and model that should be used to further develop KtW (64, 65) in order to better understand the underlying mechanisms.
Acknowledgments
C.W. thanks all his colleagues, past and present, for their strong support over the years. Special thanks go to his supervisors, including Gerhard J. Herndl and Curtis A. Suttle, for affording C.W. the freedom to pursue his own ideas. T.B. thanks all his colleagues for their encouragement since the start of his career. He is particularly grateful to M. Troussellier for his continuous support and to P. A. del Giorgio for his mentorship and inspiration in microbial ecology. We thank two anonymous reviewers for improving the article with their comments.
This work was supported by a grant from the French National Research Agency (no. ANR07 BIODIV 015) to M.G.W., T.F.T., and T.B.
Biography
 Christian Winter studied ecology and microbiology at the University of Vienna (Austria) and subsequently received a doctorate from the University of Groningen (The Netherlands) in 2004. He started to become interested in viral ecology during work on his doctoral thesis conducted at the Netherlands Institute for Sea Research, The Netherlands (1999 to 2003). Viral and microbial ecology continued to be at the center of his scientific interests during his stays as a postdoc at the Laboratoire d'Océanographie de Villefranche, France (2005 to 2006), and at the University of British Columbia, Canada (2007 to 2008). Since 2009, Christian Winter has been a University Assistant in the Department of Marine Biology at the University of Vienna.
Christian Winter studied ecology and microbiology at the University of Vienna (Austria) and subsequently received a doctorate from the University of Groningen (The Netherlands) in 2004. He started to become interested in viral ecology during work on his doctoral thesis conducted at the Netherlands Institute for Sea Research, The Netherlands (1999 to 2003). Viral and microbial ecology continued to be at the center of his scientific interests during his stays as a postdoc at the Laboratoire d'Océanographie de Villefranche, France (2005 to 2006), and at the University of British Columbia, Canada (2007 to 2008). Since 2009, Christian Winter has been a University Assistant in the Department of Marine Biology at the University of Vienna.
 Thierry Bouvier, a microbial ecologist, graduated in 1998 from the University of Montpellier, France, and from the Free University of Brussels, Belgium, after completing his Ph.D. thesis on the microbial dynamics in the Danube River estuary. He spent 2 years at the University of Maryland (1999 to 2000) and at the Université du Québec à Montréal, Canada (2001), as an assistant professor in Paul del Giorgio's lab. He is currently a scientist at the University of Montpellier-CNRS, France. He uses observational and experimental approaches to understand the underlying causal mechanisms governing the distribution of bacterial phylogenetic and physiological diversity. His research in environmental virology aims to refine the ecological concepts of virus-induced mortality in shaping of prokaryotic diversity.
Thierry Bouvier, a microbial ecologist, graduated in 1998 from the University of Montpellier, France, and from the Free University of Brussels, Belgium, after completing his Ph.D. thesis on the microbial dynamics in the Danube River estuary. He spent 2 years at the University of Maryland (1999 to 2000) and at the Université du Québec à Montréal, Canada (2001), as an assistant professor in Paul del Giorgio's lab. He is currently a scientist at the University of Montpellier-CNRS, France. He uses observational and experimental approaches to understand the underlying causal mechanisms governing the distribution of bacterial phylogenetic and physiological diversity. His research in environmental virology aims to refine the ecological concepts of virus-induced mortality in shaping of prokaryotic diversity.
 Markus G. Weinbauer studied zoology and marine biology at the University of Vienna (Austria), where he obtained his Dr. Rer. Nat. degree in 1994. He was a postdoc at the University of Texas at Austin Marine Science Institute (UTMSI) from 1994 to 1996. From 1996 to 1998 he worked as a postdoc at the National Research Center for Biotechnology, AG Microbial Ecology (Germany). At the same institute, he was a European Union visiting scientist (TMR grant) from 1998 to 2000. From 2000 to 2001, he was a postdoc at the Netherlands Institute for Sea Research (NIOZ). Since 2002, he has been a scientist at the Laboratoire d'Océanographie de Villefranche (France), where he is currently head of the Department of Microbial Ecology and Biogeochemistry. He has been working on viral community ecology since 1991. The fascination of the sea, the diversity of microorganisms, and the major role of microorganisms in driving marine ecological and biogeochemical processes are the motivations of his research.
Markus G. Weinbauer studied zoology and marine biology at the University of Vienna (Austria), where he obtained his Dr. Rer. Nat. degree in 1994. He was a postdoc at the University of Texas at Austin Marine Science Institute (UTMSI) from 1994 to 1996. From 1996 to 1998 he worked as a postdoc at the National Research Center for Biotechnology, AG Microbial Ecology (Germany). At the same institute, he was a European Union visiting scientist (TMR grant) from 1998 to 2000. From 2000 to 2001, he was a postdoc at the Netherlands Institute for Sea Research (NIOZ). Since 2002, he has been a scientist at the Laboratoire d'Océanographie de Villefranche (France), where he is currently head of the Department of Microbial Ecology and Biogeochemistry. He has been working on viral community ecology since 1991. The fascination of the sea, the diversity of microorganisms, and the major role of microorganisms in driving marine ecological and biogeochemical processes are the motivations of his research.
 T. Frede Thingstad obtained a degree in biophysics from the University of Oslo in 1973. Since then he has worked at the University of Bergen, Norway, in different positions in the microbiology/biology department, where he is now a professor. Ideas leading up to the discussions in this article can be traced back to his experimental and theoretical work on phytoplankton-bacterial coexistence in the mid-1980s and its later extension in the 1990s to lytic viruses and their hosts.
T. Frede Thingstad obtained a degree in biophysics from the University of Oslo in 1973. Since then he has worked at the University of Bergen, Norway, in different positions in the microbiology/biology department, where he is now a professor. Ideas leading up to the discussions in this article can be traced back to his experimental and theoretical work on phytoplankton-bacterial coexistence in the mid-1980s and its later extension in the 1990s to lytic viruses and their hosts.
REFERENCES
- 1.Andersson, A. F., and J. F. Banfield. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047-1050. [DOI] [PubMed] [Google Scholar]
- 2.Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auguet, J. C., H. Montanié, H. J. Hartmann, P. Lebaron, E. O. Casamayor, P. Catala, and D. Delmas. 2008. Potential effect of freshwater virus on the structure and activity of bacterial communities in the Marennes-Oléron Bay (France). Microb. Ecol. 57:295-306. [DOI] [PubMed] [Google Scholar]
- 4.Azam, F., and F. Malfatti. 2007. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5:782-791. [DOI] [PubMed] [Google Scholar]
- 5.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. [DOI] [PubMed] [Google Scholar]
- 6.Benmayor, R., A. Buckling, M. B. Bonsall, M. A. Brockhurst, and D. J. Hodgson. 2008. The interactive effects of parasites, disturbance, and productivity on experimental adaptive radiations. Evolution 62:467-477. [DOI] [PubMed] [Google Scholar]
- 7.Bergh, Ø., K. Y. Børsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 8.Berryman, A. A. 1992. The origins and evolution of predator-prey theory. Ecology 73:1530-1535. [Google Scholar]
- 9.Bertrand, S., C. Alonso-Alvarez, G. Devevey, B. Faivre, J. Prost, and G. Sorci. 2006. Carotenoids modulate the trade-off between egg production and resistance to oxidative stress in zebra finches. Oecologia 147:576-584. [DOI] [PubMed] [Google Scholar]
- 10.Bohannan, B. J. M., B. Kerr, C. M. Jessup, J. B. Hughes, and G. Sandvik. 2002. Trade-offs and coexistence in microbial microcosms. Antonie Van Leeuwenhoek 81:107-115. [DOI] [PubMed] [Google Scholar]
- 11.Bohannan, B. J. M., and R. E. Lenski. 2000. The relative importance of competition and predation varies with productivity in a model community. Am. Nat. 156:329-340. [DOI] [PubMed] [Google Scholar]
- 12.Bohannan, B. J. M., and R. E. Lenski. 1999. Effect of prey heterogeneity on the response of a model food chain to resource enrichment. Am. Nat. 153:73-82. [DOI] [PubMed] [Google Scholar]
- 13.Bohannan, B. J. M., and R. E. Lenski. 1997. Effect of resource enrichment on a chemostat community of bacteria and bacteriophage. Ecology 78:2303-2315. [Google Scholar]
- 14.Bonilla-Findji, O., G. J. Herndl, J.-P. Gattuso, and M. G. Weinbauer. 2009. Viral and flagellate control of prokaryotic production and community structure in offshore Mediterranean waters. Appl. Environ. Microbiol. 75:4801-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonilla-Findji, O., E. Rochelle-Newall, M. G. Weinbauer, M.-D. Pizay, M.-E. Kerros, and J.-P. Gattuso. 2009. Effect of seawater-freshwater cross-transplantations on viral dynamics and bacterial diversity and production. Aquat. Microb. Ecol. 54:1-11. [Google Scholar]
- 16.Bouvier, T., and P. A. del Giorgio. 2007. Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ. Microbiol. 9:287-297. [DOI] [PubMed] [Google Scholar]
- 17.Breitbart, M., M. Middelboe, and F. Rohwer. 2008. Marine viruses: community dynamics, diversity and impact on microbial processes, p. 443-479. In D. L. Kirchman (ed.), Microbial ecology of the oceans, 2nd ed. Wiley-Blackwell, Hoboken, NJ.
- 18.Brockhurst, M. A., A. Fenton, B. Roulston, and P. B. Rainey. 2006. The impact of phages on interspecific competition in experimental populations of bacteria. BMC Ecol. 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brockhurst, M. A., A. Buckling, and P. B. Rainey. 2005. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen Pseudomonas aeruginosa. Proc. R. Soc. Lond. B 272:1385-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brockhurst, M. A., P. B. Rainey, and A. Buckling. 2004. The effect of spatial heterogeneity and parasites on the evolution of host diversity. Proc. R. Soc. Lond. B 271:107-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell, A. 1994. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 48:193-222. [DOI] [PubMed] [Google Scholar]
- 22.Cotter, S. C., L. E. B. Kruuk, and K. Wilson. 2004. Costs of resistance: genetic correlations and potential trade-offs in an insect immune system. J. Evol. Biol. 17:421-429. [DOI] [PubMed] [Google Scholar]
- 23.De Paepe, M., and F. Taddei. 2006. Viruses' life history: towards a mechanistic basis of a trade-off between survival and reproduction among phages. PLoS Biol. 4:e193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupont, K., T. Janzen, F. K. Vogensen, J. Josephsen, and B. Stuer-Lauridsen. 2004. Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl. Environ. Microbiol. 70:5825-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans, C., S. D. Archer, S. Jacquet, and W. H. Wilson. 2003. Direct estimates of the contribution of viral lysis and microzooplankton grazing to the decline of a Micromonas spp. population. Aquat. Microb. Ecol. 30:207-219. [Google Scholar]
- 26.Filippini, M., N. Buesing, and M. O. Gessner. 2008. Temporal dynamics of freshwater bacterio- and virioplankton along a littoral-pelagic gradient. Freshw. Biol. 53:1114-1125. [Google Scholar]
- 27.Fine, P. V. A., Z. J. Miller, I. Mesones, S. Irazuzta, H. M. Appel, M. H. H. Stevens, I. Sääksjärvi, J. C. Schultz, and P. D. Coley. 2006. The growth-defense trade-off and habitat specialization by plants in Amazonian forests. Ecology 87:S150-S162. [DOI] [PubMed] [Google Scholar]
- 28.Fitzpatrick, M. J., E. Feder, L. Rowe, and M. B. Sokolowski. 2007. Maintaining a behaviour polymorphism by frequency-dependent selection on a single gene. Nature 447:210-212. [DOI] [PubMed] [Google Scholar]
- 29.Fuhrman, J. A., and C. A. Suttle. 1993. Viruses in marine planktonic systems. Oceanography 6:51-63. [Google Scholar]
- 30.Green, J. L., B. J. M. Bohannan, and R. J. Whitaker. 2008. Microbial biogeography: from taxonomy to traits. Science 320:1039-1043. [DOI] [PubMed] [Google Scholar]
- 31.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendrix, R. W. 1999. Evolution: the long evolutionary reach of viruses. Curr. Biol. 9:R914-R917. [DOI] [PubMed] [Google Scholar]
- 33.Hewson, I., and J. A. Fuhrman. 2007. Covariation of viral parameters with bacterial assemblage richness and diversity in the water column and sediments. Deep Sea Res. I 54:811-830. [Google Scholar]
- 34.Hewson, I., and J. A. Fuhrman. 2006. Viral impacts upon marine bacterioplankton assemblage structure. J. Mar. Biol. Assoc. UK 86:577-589. [Google Scholar]
- 35.Hewson, I., J. M. O'Neil, C. A. Heil, G. Bratbak, and W. C. Dennison. 2001. Effects of concentrated viral communities on photosynthesis and community composition of co-occuring benthic microalgae and phytoplankton. Aquat. Microb. Ecol. 25:1-10. [Google Scholar]
- 36.Hewson, I., G. A. Vargo, and J. A. Fuhrman. 2003. Bacterial diversity in shallow oligotrophic marine benthos and overlying waters: effects of virus infection, containment, and nutrient enrichment. Microb. Ecol. 46:322-336. [DOI] [PubMed] [Google Scholar]
- 37.Hewson, I., D. M. Winget, K. E. Williamson, J. A. Fuhrman, and K. E. Wommack. 2006. Viral and bacterial assemblage covariance in oligotrophic waters of the West Florida Shelf (Gulf of Mexico). J. Mar. Biol. Assoc. UK 86:591-603. [Google Scholar]
- 38.Holmfeldt, K., M. Middelboe, O. Nybroe, and L. Riemann. 2007. Large variabilities in host strain susceptibility and phage host range govern interactions between lytic marine phages and their Flavobacterium hosts. Appl. Environ. Microbiol. 73:6730-6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holt, R. D., J. Grover, and D. Tilman. 1994. Simple rules for interspecific dominance in systems with exploitative and apparent competition. Am. Nat. 144:741-771. [Google Scholar]
- 40.Hutchinson, G. E. 1961. The paradox of the plankton. Am. Natur. 95:137-145. [Google Scholar]
- 41.Hugenholtz, P., and G. W. Tyson. 2008. Microbiology: metagenomics. Nature 455:481-482. [DOI] [PubMed] [Google Scholar]
- 42.Jacquet, S., I. Domaizon, S. Personnic, A. S. Pradeep Ram, M. Heldal, S. Duhamel, and T. Sime-Ngando. 2005. Estimates of protozoan- and viral-mediated mortality of bacterioplankton in Lake Bourget (France). Freshw. Biol. 50:627-645. [Google Scholar]
- 43.Jacquet, S., H. Havskum, T. F. Thingstad, and D. Vaulot. 2002. Effects of inorganic and organic nutrient addition on a coastal microbial community (Isefjord, Denmark). Mar. Ecol. Prog. Ser. 228:3-14. [Google Scholar]
- 44.Jardillier, L., Y. Bettarel, M. Richardot, C. Bardot, C. Amblard, T. Sime-Ngando, and D. Debroas. 2005. Effects of viruses and predators on prokaryotic community composition. Microb. Ecol. 50:557-569. [DOI] [PubMed] [Google Scholar]
- 45.Jardillier, L., D. Boucher, S. Personnic, S. Jacquet, A. Thénot, D. Sargos, C. Amblard, and D. Debroas. 2005. Relative importance of nutrients and mortality factors on prokaryotic community composition in two lakes of different trophic status: microcosm experiments. FEMS Microb. Ecol. 53:429-443. [DOI] [PubMed] [Google Scholar]
- 46.Jürgens, K., and C. Matz. 2002. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek 81:413-434. [DOI] [PubMed] [Google Scholar]
- 47.Karl, D. M. 2007. Microbial oceanography: paradigms, processes and promise. Nat. Rev. Microbiol. 5:759-769. [DOI] [PubMed] [Google Scholar]
- 48.Kirchman, D. L. 2009. Mysteries of metagenomics revealed. LNO Bull. 18:2-6. [Google Scholar]
- 49.Kraaijeveld, A. R., J. Ferrari, and H. C. J. Godfray. 2002. Costs of resistance in insect-parasite and insect-parasitoid interactions. Parasitology 125:S71-S82. [DOI] [PubMed] [Google Scholar]
- 50.Larsen, A., T. Castberg, R. A. Sandaa, C. P. D. Brussaard, J. Egge, M. Heldal, A. Paulino, R. Thyrhaug, E. J. van Hannen, and G. Bratbak. 2001. Population dynamics and diversity of phytoplankton, bacteria and viruses in a seawater enclosure. Mar. Ecol. Prog. Ser. 221:47-57. [Google Scholar]
- 51.Larsen, A., G. A. Fonnes Flaten, R.-A. Sandaa, T. Castberg, R. Thyrhaug, S. R. Erga, S. Jacquet, and G. Bratbak. 2004. Spring phytoplankton bloom dynamics in Norwegian coastal waters: microbial community succession and diversity. Limnol. Oceanogr. 49:180-190. [Google Scholar]
- 52.Lennon, J. T., S. A. M. Khatana, M. F. Marston, and J. B. H. Martiny. 2007. Is there a cost of virus resistance in marine cyanobacteria? ISME J. 1:300-312. [DOI] [PubMed] [Google Scholar]
- 53.Lenski, R. E. 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli. I. Variation in competitive fitness among mutants resistant to virus T4. Evolution 42:425-432. [DOI] [PubMed] [Google Scholar]
- 54.Litchman, E., and C. A. Klausmeier. 2008. Trait-based community ecology of phytoplankton. Annu. Rev. Ecol. Evol. Syst. 39:615-639. [Google Scholar]
- 55.MacArthur, R. H., and E. O. Wilson. 1967. The theory of island biogeography, p. 203. Princeton University Press, Princeton, NJ.
- 56.Malits, A., and M. G. Weinbauer. 2009. Effect of turbulence and viruses on prokaryotic cell size, production and diversity. Aquat. Microb. Ecol. 54:243-254. [Google Scholar]
- 57.Mandelbrot, B. B. 1983. The fractal geometry of nature, p. 468. W. H. Freeman and Co., New York, NY.
- 58.McCauley, E., and F. Briand. 1979. Zooplankton grazing and phytoplankton species richness. Limnol. Oceanogr. 24:243-252. [Google Scholar]
- 59.Middelboe, M. 2000. Bacterial growth rate and marine virus-host dynamics. Microb. Ecol. 40:114-124. [DOI] [PubMed] [Google Scholar]
- 60.Middelboe, M., Å. Hagström, N. Blackburn, B. Sinn, U. Fischer, N. H. Borch, J. Pinhassi, K. Simu, and M. G. Lorenz. 2001. Effects of bacteriophages on the population dynamics of four strains of pelagic marine bacteria. Microb. Ecol. 42:395-406. [DOI] [PubMed] [Google Scholar]
- 61.Middelboe, M., K. Holmfeldt, L. Riemann, O. Nybroe, and J. Haaber. 2009. Bacteriophages drive strain diversification for phage resistance and physiological properties. Environ. Microb. 11:1971-1982. [DOI] [PubMed] [Google Scholar]
- 62.Middelboe, M., and P. G. Lyck. 2002. Regeneration of dissolved organic matter by viral lysis in marine microbial communities. Aquat. Microb. Ecol. 27:187-194. [Google Scholar]
- 63.Middelboe, M., L. Riemann, G. F. Steward, V. Hansen, and O. Nybroe. 2003. Virus-induced transfer of organic carbon between marine bacteria in a model community. Aquat. Microb. Ecol. 33:1-10. [Google Scholar]
- 64.Miki, T., T. Nakasawa, T. Yokokawa, and T. Nagata. 2008. Functional consequences of viral impacts on bacterial communities: a food-web model analysis. Freshw. Biol. 53:1142-1153. [Google Scholar]
- 65.Miki, T., and N. Yamamura. 2005. Intraguild predation reduces bacterial species richness and loosens the viral loop in aquatic systems: ‘kill the killer of the winner’ hypothesis. Aquat. Microb. Ecol. 40:1-12. [Google Scholar]
- 66.Mizoguchi, K., M. Morita, C. R. Fischer, M. Yoichi, Y. Tanji, and H. Unno. 2003. Coevolution of bacteriophage PP01 and Escherichia coli O157:H7 in continous culture. Appl. Environ. Microbiol. 69:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nyström, T. 2004. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition. Mol. Microbiol. 54:855-862. [DOI] [PubMed] [Google Scholar]
- 68.Øvreås, L., D. Bourne, R.-A. Sandaa, E. O. Casamayor, S. Benlloch, V. Goddard, G. Smerdon, M. Heldal, and T. F. Thingstad. 2003. Response of bacterial and viral communities to nutrient manipulations in seawater mesocosms. Aquat. Microb. Ecol. 31:109-121. [Google Scholar]
- 69.Parada, V., A.-C. Baudoux, E. Sintes, M. G. Weinbauer, and G. J. Herndl. 2008. Dynamics and diversity of newly produced virioplankton in the North Sea. ISME J. 2:924-936. [DOI] [PubMed] [Google Scholar]
- 70.Pengerud, B., E. F. Skjoldal, and T. F. Thingstad. 1987. The reciprocal interaction between degradation of glucose and ecosystem structure—studies in mixed chemostat cultures of marine-bacteria, algae, and bacterivorous nanoflagellates. Mar. Ecol. Prog. Ser. 35:111-117. [Google Scholar]
- 71.Pernthaler, J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3:537-546. [DOI] [PubMed] [Google Scholar]
- 72.Pradeep Ram, A. S., and T. Sime-Ngando. 2008. Functional responses of prokaryotes and viruses to grazer effects and nutrient additions in freshwater microcosms. ISME J. 2:498-509. [DOI] [PubMed] [Google Scholar]
- 73.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60-62. [Google Scholar]
- 74.Prosser, J. I., B. J. M. Bohannan, T. P. Curtis, R. J. Ellis, M. K. Firestone, R. P. Freckleton, J. L. Green, L. E. Green, K. Killham, J. J. Lennon, A. M. Osborn, M. Solan, C. J. van der Gast, and J. P. W. Young. 2007. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5:384-392. [DOI] [PubMed] [Google Scholar]
- 75.Riemann, L., and H.-P. Grossart. 2008. Elevated lytic phage production as a consequence of particle colonization by a marine Flavobacterium (Cellulophaga sp.). Microb. Ecol. 56:505-512. [DOI] [PubMed] [Google Scholar]
- 76.Riemann, L., K. Holmfeldt, and J. Titelman. 2009. Importance of viral lysis and dissolved DNA for bacterioplankton activity in a P-limited estuary, northern Baltic Sea. Microb. Ecol. 57:286-294. [DOI] [PubMed] [Google Scholar]
- 77.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rodriguez-Valera, F., A.-B. Martin-Cuadrado, B. Rodriguez-Brito, L. Pašić, T. F. Thingstad, F. Rohwer, and A. Mira. 2009. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7:828-836. [DOI] [PubMed] [Google Scholar]
- 79.Rohwer, F., and R. Edwards. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sabet, S., W. Chu, and S. C. Jiang. 2006. Isolation and genetic analysis of haloalkaliphilic bacteriophages in a North American soda lake. Microb. Ecol. 51:543-554. [DOI] [PubMed] [Google Scholar]
- 81.Sandaa, R.-A., L. Gómez-Consarnau, J. Pinhassi, L. Riemann, A. Malits, M. G. Weinbauer, J. M. Gasol, and T. F. Thingstad. 2009. Viral control of bacterial biodiversity—evidence from a nutrient enriched marine mesocosm experiment. Environ. Microbiol. 11:2585-2597. [DOI] [PubMed] [Google Scholar]
- 82.Schwalbach, M. S., I. Hewson, and J. A. Fuhrman. 2004. Viral effects on bacterial community composition in marine plankton microcosms. Aquat. Microb. Ecol. 34:117-127. [Google Scholar]
- 83.Shi, Y., G. W. Tyson, and E. F. DeLong. 2009. Metatranscriptomics reveals unique microbial small RNAs in the ocean's water column. Nature 459:266-269. [DOI] [PubMed] [Google Scholar]
- 84.Šimek, C., K. Hornák, M. Masín, U. Christaki, J. Nedoma, M. G. Weinbauer, and J. R. Dolan. 2003. Comparing the effects of resource enrichment and grazing on a bacterioplankton community of a meso-eutrophic reservoir. Aquat. Microb. Ecol. 31:123-135. [Google Scholar]
- 85.Šimek, K., J. Pernthaler, M. G. Weinbauer, K. Hornák, J. R. Dolan, J. Nedoma, M. Masín, and R. Amann. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Šimek, K., M. G. Weinbauer, K. Hornák, J. Jezbera, J. Nedoma, and J. R. Dolan. 2007. Grazer and virus-induced mortality of bacterioplankton accelerates development of Flectobacillus populations in a freshwater community. Environ. Microbiol. 9:789-800. [DOI] [PubMed] [Google Scholar]
- 87.Sime-Ngando, T., and A. S. Pradeep Ram. 2005. Grazer effects on prokaryotes and viruses in a freshwater microcosm experiment. Aquat. Microb. Ecol. 41:115-124. [Google Scholar]
- 88.Staley, J. T., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 89.Steward, G. F., J. L. Montiel, and F. Azam. 2000. Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol. Oceanogr. 45:1697-1706. [Google Scholar]
- 90.Strauss, S. Y., J. A. Rudgers, J. A. Lau, and R. E. Irwin. 2002. Direct and ecological costs of resistance to herbivory. Trends Ecol. Evol. 17:278-285. [Google Scholar]
- 91.Suttle, C. A. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801-812. [DOI] [PubMed] [Google Scholar]
- 92.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 93.Thingstad, T. F., M. Heldal, G. Bratbak, and I. Dundas. 1993. Are viruses important partners in pelagic food webs? Trends Ecol. Evol. 8:209-213. [DOI] [PubMed] [Google Scholar]
- 94.Thingstad, T. F., and R. Lignell. 1997. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13:19-27. [Google Scholar]
- 95.Thingstad, T. F., and B. Pengerud. 1985. Fate and effect of allochthonous organic material in aquatic microbial ecosystems. An analysis based on chemostat theory. Mar. Ecol. Prog. Ser. 21:47-62. [Google Scholar]
- 96.Tijdens, M., H. L. Hoogveld, M. P. Kamst-van Agterveld, S. H. H. Simis, A.-C. Baudoux, H. J. Laanbroek, and H. J. Gons. 2008. Population dynamics and diversity of viruses, bacteria and phytoplankton in a shallow eutrophic lake. Microb. Ecol. 56:29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tschirren, B., and H. Richner. 2006. Parasites shape the optimal investment in immunity. Proc. R. Soc. Lond. B 273:1773-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang, K., and F. Chen. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat. Microb. Ecol. 34:105-116. [Google Scholar]
- 99.Weeks, A. R., and A. A. Hoffmann. 2008. Frequency-dependent selection maintains clonal diversity in an asexual organism. Proc. Natl. Acad. Sci. U. S. A. 105:17872-17877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 101.Weinbauer, M. G., U. Christaki, J. Nedoma, and K. Šimek. 2003. Comparing the effects of resource enrichment and grazing on viral production in a meso-eutrophic reservoir. Aquat. Microb. Ecol. 31:137-144. [Google Scholar]
- 102.Weinbauer, M. G., R. Christen, and M. G. Höfle. 2006. The response of Vibrio- and Rhodobacter-related populations of the NW Mediterranean Sea to additions of dissolved organic matter, phages, or dilution. Microb. Ecol. 51:336-344. [DOI] [PubMed] [Google Scholar]
- 103.Weinbauer, M. G., K. Hornák, J. Jezbera, J. Nedoma, J. R. Dolan, and K. Šimek. 2007. Synergistic and antagonistic effects of viral lysis and protistan grazing on bacterial biomass, production and diversity. Environ. Microbiol. 9:777-788. [DOI] [PubMed] [Google Scholar]
- 104.Weinbauer, M. G., and F. Rassoulzadegan. 2004. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 6:1-11. [DOI] [PubMed] [Google Scholar]
- 105.Wilmes, P., and P. L. Bond. 2009. Microbial community proteomics: elucidating the catalysts and metabolic mechanisms that drive the Earth's biogeochemical cycles. Curr. Opin. Microbiol. 12:310-317. [DOI] [PubMed] [Google Scholar]
- 106.Winget, D. M., and K. E. Wommack. 2008. Randomly amplified polymorphic DNA PCR as a tool for assessment of marine viral richness. Appl. Environ. Microbiol. 74:2612-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Winter, C., M. M. Moeseneder, G. J. Herndl, and M. G. Weinbauer. 2008. Relationship of geographic distance, depth, temperature, and viruses with prokaryotic communities in the eastern tropical Atlantic Ocean. Microb. Ecol. 56:383-389. [DOI] [PubMed] [Google Scholar]
- 108.Winter, C., A. Smit, G. J. Herndl, and M. G. Weinbauer. 2005. Linking bacterial richness with viral abundance and prokaryotic activity. Limnol. Oceanogr. 50:968-977. [Google Scholar]
- 109.Winter, C., A. Smit, G. J. Herndl, and M. G. Weinbauer. 2004. Impact of virioplankton on archaeal and bacterial community richness as assessed in seawater batch cultures. Appl. Environ. Microbiol. 70:804-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wulff, J. L. 2005. Trade-offs in resistance to competitors and predators, and their effects on the diversity of tropical marine sponges. J. Anim. Ecol. 74:313-321. [Google Scholar]
- 112.Yoshida, M. T., A. Kashima, Y. Takashima, N. Hosoda, K. Nagasaki, and S. Hiroishi. 2008. Ecological dynamics of the toxic bloom-forming cyanobacterium Microcystis aeruginosa and its cyanophages in freshwater. Appl. Environ. Microbiol. 74:3269-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang, R., M. G. Weinbauer, and P.-Y. Qian. 2007. Viruses and flagellates sustain apparent richness and reduce biomass accumulation of bacterioplankton in coastal marine waters. Environ. Microbiol. 9:3008-3018. [DOI] [PubMed] [Google Scholar]