Abstract
Summary: Lindane, the γ-isomer of hexachlorocyclohexane (HCH), is a potent insecticide. Purified lindane or unpurified mixtures of this and α-, β-, and δ-isomers of HCH were widely used as commercial insecticides in the last half of the 20th century. Large dumps of unused HCH isomers now constitute a major hazard because of their long residence times in soil and high nontarget toxicities. The major pathway for the aerobic degradation of HCH isomers in soil is the Lin pathway, and variants of this pathway will degrade all four of the HCH isomers although only slowly. Sequence differences in the primary LinA and LinB enzymes in the pathway play a key role in determining their ability to degrade the different isomers. LinA is a dehydrochlorinase, but little is known of its biochemistry. LinB is a hydrolytic dechlorinase that has been heterologously expressed and crystallized, and there is some understanding of the sequence-structure-function relationships underlying its substrate specificity and kinetics, although there are also some significant anomalies. The kinetics of some LinB variants are reported to be slow even for their preferred isomers. It is important to develop a better understanding of the biochemistries of the LinA and LinB variants and to use that knowledge to build better variants, because field trials of some bioremediation strategies based on the Lin pathway have yielded promising results but would not yet achieve economic levels of remediation.
INTRODUCTION
The γ-isomer of hexachlorocyclohexane (HCH), formerly known as benzene hexachloride (BHC), is one of the now notorious organochlorine group of insecticides. About 600,000 tons were used throughout the world between the 1940s and the 1990s to control a wide range of agricultural, horticultural, and public health pests (80, 182, 183, 186, 187). Mounting concerns about its nontarget toxicity and persistence have since caused it to be deregistered in most countries (183, 189), but it is very stable in the environment (23, 192), and it is still being manufactured in India for local and export uses (187), so residue problems will continue for many decades.
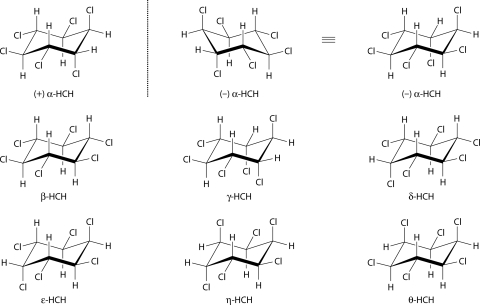
HCH is commercially manufactured by the photochemical chlorination of benzene in the presence of UV light. The technical-grade HCH produced in this way comprises mainly five variously stable isomers: α (60 to 70%), β (5 to 12%), γ (10 to 12%), δ (6 to 10%), and ɛ (3 to 4%) (183). These isomers differ in the spatial orientations of the chlorine atoms around the cyclohexane ring (Fig. 1). Although only the γ-isomer is insecticidal (163), technical HCH was used widely as a cheap but effective insecticide in developing countries. It is possible to extract and purify the γ-HCH isomer from technical HCH, and the purified γ-isomer, commonly known as lindane, was also commonly used as an insecticide, particularly in developed countries.
FIG. 1.
Axial versus equatorial arrangements of chlorine atoms in the five major isomers of HCH plus the less common η- and θ-isomers. Note that α-HCH also exists in two enantiomeric (+ and −) forms.
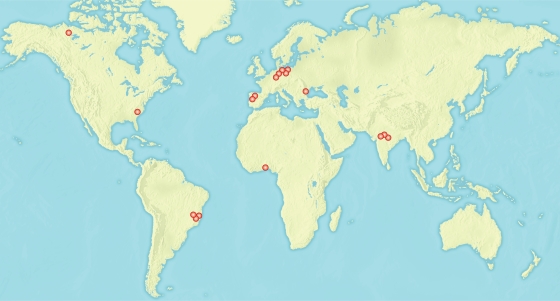
The production and use of HCH/lindane has led to two major residue problems. One involves the point source contamination of very high concentrations due to the open-air stockpiling of waste isomers during lindane production (183, 187); the production of 1 ton of lindane generates 8 to 12 tons of “HCH muck,” consisting primarily of the α-, β-, and δ-HCH isomers. The α-, β-, and δ-isomers are in many ways more problematic than lindane itself; α-HCH forms the major portion of technical HCH, and the β- and δ-isomers are significantly more stable than γ-HCH (181). Sites heavily contaminated with HCH have been reported from The Netherlands (180), Brazil (129), Germany (68, 159), Spain (29, 148), China (198), Greece (49), Canada (135), the United States (135), and India (139, 162) (Fig. 2). The 1998 Food and Agriculture Organization (FAO) inventory of obsolete, unwanted, and/or banned pesticides also found unused stockpiles of both technical HCH and lindane (2,785 tons of technical HCH, 304 tons of lindane, and 45 tons of unspecified HCH material) scattered in dump sites in Africa and the Near East (http://www.fao.org). Weber et al. (187) estimated that four to six million tons of various HCH materials have been dumped worldwide, which is similar in scale to the combined totals of dumped materials for all other persistent organic pollutants (POPs) defined by the Stockholm Convention (187).
FIG. 2.
Known locations of HCH dumps in excess of 50,000 tons in Brazil (38), Canada (135), Germany (159, 183), Spain (26, 29, 181), India (30, 139), and the United States (135). HCH-degrading sphingomonads have been recovered in six countries in Europe and Asia, although precise locations are often not reported (see text).
The second problem involves the diffuse contamination of the environment with lower HCH concentrations due either to dispersal from stockpiles or to the use of the insecticide. The majority of HCH waste has been discarded in the open or stored at various levels of containment near the production sites, although in some cases, the top layer of soil at such sites has been removed for use in filling material for the construction of roads and buildings (97, 145, 181). HCH residues at many of the sites have percolated into the soil and thence contaminated groundwater (145). Residues of HCH isomers have now been reported for many countries in samples of air (76, 79, 138), water (8, 71, 139), soil (29, 50, 71, 139, 144), food commodities (7, 17, 175, 184), milk (153, 197), fish (44, 87), and mammals (48) and even from human blood samples (15, 169) and adipose tissue (14, 73). Environmental residues from sites as remote from the major regions of production and use as the Arctic, Antarctic, and Pacific Ocean have been reported (63, 185). Despite its widespread deregistration, HCH thus continues to generate serious residue problems in a variety of circumstances around the world.
Reports on the anaerobic microbial degradation of HCH isomers started appearing in the 1960s, and subsequently, aerobic bacteria capable of degrading HCH isomers were also found. Genes involved in degradation (lin genes) have been isolated from aerobic bacteria and heterologously expressed, and the gene products have been characterized. We now have a reasonable understanding of the diversity, organization, and distribution of HCH catabolic genes in certain sphingomonads in particular (75), and there is growing interest from a variety of government, industry, and science agencies in using them to develop bioremediation technologies for the decontamination of HCH-contaminated sites (16, 19, 96, 144). This article outlines our knowledge of the microbial degradation of HCH and discusses the issues surrounding the potential uses of the system for the bioremediation of HCH-contaminated sites.
ANAEROBIC DEGRADATION
It was initially believed that HCH biodegradation is largely an anaerobic process, and variable levels of anaerobic degradation of α-, β-, γ-, and δ-HCH have indeed been observed. There are no reports as yet on the anaerobic degradation of the ɛ-isomer, but it is relatively unstable anyway. Isolates capable of degrading one or more of the other four HCH isomers under anaerobic conditions include Clostridium rectum (125), Clostridium sphenoides (58, 84), Clostridium butyricum, Clostridium pasteurianum (64), Citrobacter freundii (64), Desulfovibrio gigas, Desulfovibrio africanus, Desulfococcus multivorans (21), and a Dehalobacter sp. (178). The γ- and α-isomers are degraded more rapidly than δ- and β-HCH (34, 66, 85, 91, 141).
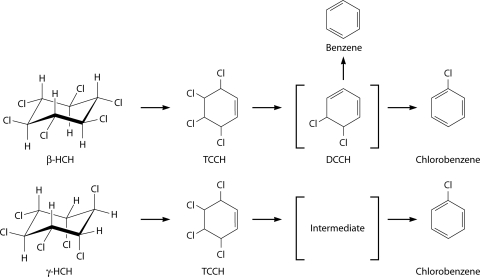
Several studies reported the production of chlorobenzene during the anaerobic degradation of γ-HCH (Fig. 3) (11, 21, 98, 126, 137). Ohisa et al. (126) proposed that the degradation proceeds through two dichloroeliminations, resulting in the formation of γ-3,4,5,6-tetrachloro-1-cyclohexene (γ-TCCH) and, subsequently, 5,6-dichlorocyclohexa-1,2-diene. Chlorobenzene is then produced by a dehydrochlorination reaction. Several reports confirmed the presence of γ-TCCH as an intermediate in the anaerobic degradation of γ-HCH (11, 58, 64, 126). The degradation of γ-HCH also results in the formation of small amounts of trichlorinated benzenes and benzene itself (64).
FIG. 3.
Consensus anaerobic degradation pathway of γ- and β-HCH. Note that two intermediates that have been proposed but not yet observed empirically are shown in square brackets. The structures of TCCH and DCCH are shown in the planar format because their stereochemistries have not been established.
The available data suggest that the anaerobic degradation of α-, β-, and δ-HCH also proceeds at least in part through successive dichloroeliminations and then dehydrochlorination to produce chlorobenzenes. Heritage and MacRae (58) originally proposed that the anaerobic degradation of α-HCH proceeds via δ-3,4,5,6-tetrachloro-1-cyclohexene (δ-TCCH), although the end product could not be identified. However, Middeldorp et al. (90) subsequently found that the anaerobic degradation of both α-HCH and δ-HCH produces chlorobenzene. Middeldorp et al. (98) also found that the anaerobic degradation of β-HCH (Fig. 3) results in the formation of dichlorocyclohexadiene via δ-TCCH by successive dichloroeliminations, some of which is further converted to chlorobenzene by dehydrochlorination. Interestingly, however, those authors also found that some of the dichlorocyclohexadiene was converted to benzene by an additional dichloroelimination reaction. Recent studies have confirmed that β-HCH can be converted to both benzene and chlorobenzene under anaerobic conditions (97, 178).
Quintero et al. (140) recently suggested that another anaerobic pathway could also generate chlorobenzene from all four of the major HCH isomers. They studied anaerobic liquid and soil slurry systems of undefined microbial contents and reported the production of pentachlorocyclohexane, followed by 1,2- and 1,3-dichlorobenzene and then chlorobenzene. For the α- and γ-isomers, those authors also observed the intermediate products tetrachlorocyclohexene and trichlorobenzene. How these findings relate to the consensus pathway from the other studies described above remains to be demonstrated, but it may well relate to the use of the slurry systems reported by Quintero et al. (141). One major factor restricting the microbial degradation of HCH isomers under anaerobic conditions is the strong adsorption of the isomers onto soil (146); degradation is much faster under liquid or slurry conditions (39, 141, 146). Significantly, the pentachlorocyclohexane and tetrachlorocyclohexene seen by Quintero et al. (141) in their laboratory slurry systems were recently seen in groundwater from HCH-contaminated sites but not from sediment samples from the same sites (145).
Most studies of anaerobic HCH degradation reported the accumulation of chlorobenzene and benzene (24, 98, 179), although Quintero et al. (140) detected only traces of these metabolites, and Jagnow et al. (64) reported the complete degradation of γ-HCH at least to chloride and chloride-free metabolites in mixed and pure cultures of Clostridium species. There have been other reports of the anaerobic mineralization of benzene (41, 147), but we are unaware of other reports of the anaerobic degradation of chlorobenzene. Both chlorobenzene and benzene, however, can be readily mineralized under aerobic conditions (36, 37, 45, 93; see below).
To the best of our knowledge, nothing has yet been reported for specific genes and enzymes involved in anaerobic HCH degradation.
AEROBIC DEGRADATION
Several studies have reported various levels of degradation of the four major HCH isomers under aerobic conditions (5, 107, 150-152) (Table 1). As with anaerobic degradation, essentially no work has yet been done on the biodegradation of the relatively unstable ɛ-HCH. Most of the HCH-degrading aerobes known to date are members of the family Sphingomonadaceae (74, 75); so far, 30 HCH-degrading sphingomonads from different parts of the globe have been reported (Table 1). It is not clear how soon after the first use of HCH these strains emerged; the first strain was reported in the late 1980s from Japan (155), but reports from other countries quickly followed. Most research has focused on 3 of the 30 strains, initially named Sphingomonas paucimobilis, UT26, B90A, and Sp+, isolated from HCH-contaminated soils in Japan (155), India (152), and France (27), respectively. A polyphasic approach including 16S rRNA gene analysis subsequently revealed that these strains are actually three distinct species of Sphingobium: Sphingobium japonicum UT26, Sphingobium indicum B90A, and Sphingobium francense Sp+, respectively (132). Hereafter, however, we simply call them UT26, B90A, and Sp+. The other 27 HCH-degrading sphingomonads were isolated from HCH-contaminated sites in Germany (20), Spain (105), China (83), Japan (62), and India (30). Although all these strains degrade HCH, there are some isomer-specific differences between strains in the early steps of the degradation pathway (20, 109), which are summarized in Fig. 4 and 5.
TABLE 1.
Aerobic HCH-degrading microorganisms
| Organism(s) | Reference(s) |
|---|---|
| Alcaligenes faecalis S-1 and A. faecalis S-2 | 52 |
| Arthrobacter citreus BI-100 | 34 |
| Bacillus circulans and Bacillus brevis | 51 |
| Bacillus sp. | 196 |
| Escherichia coli | 46 |
| Microbacterium sp. ITRC1 | 88 |
| Pandoraea sp. | 128 |
| Pseudomonas aeruginosa | 82 |
| Pseudomonas aeruginosa ITRC-5 | 70 |
| Pseudomonas putida | 12 |
| Pseudomonas sp. | 121 |
| Pseudomonas sp. | 177 |
| Rhodanobacter lindaniclasticus RP5557 | 120, 173 |
| Sphingobium chinhatense IP26 | 30 |
| Sphingobium indicum B90A | 152 |
| Sphingobium japonicum UT26 | 155 |
| Sphingobium francense Sp+ | 27 |
| Sphingobium quisquiliarum P25 | 9 |
| Sphingobium ummariense RL-3 | 160 |
| Sphingobium sp. BHC-A | 62 |
| Sphingobium sp. MI1205 | 83 |
| Sphingobium sp. SS04-1 | 193 |
| Sphingobium sp. SS04-2 | 193 |
| Sphingobium sp. SS04-3 | 193 |
| Sphingobium sp. SS04-4 | 193 |
| Sphingobium sp. SS04-5 | 193 |
| Sphingobium sp. UM2 | 31 |
| Sphingobium sp. HDU05 | 31 |
| Sphingobium sp. HDIP04 | 31 |
| Sphingobium sp. F2 | 31 |
| Sphingomonas sp. DS2 | 20 |
| Sphingomonas sp. DS2-2 | 20 |
| Sphingomonas sp. DS3-1 | 20 |
| Sphingomonas sp. UM1 | 31 |
| Sphingomonas sp. α1-2 | 105 |
| Sphingomonas sp. α4-2 | 105 |
| Sphingomonas sp. α4-5 | 105 |
| Sphingomonas sp. α16-10 | 105 |
| Sphingomonas sp. α16-12 | 105 |
| Sphingomonas sp. γ1-7 | 105 |
| Sphingomonas sp. γ12-7 | 105 |
| Sphingomonas sp. γ16-1 | 105 |
| Sphingomonas sp. γ16-9 | 105 |
| Streptomyces sp. M7 | 13 |
| Xanthomonas sp. ICH12 | 89 |
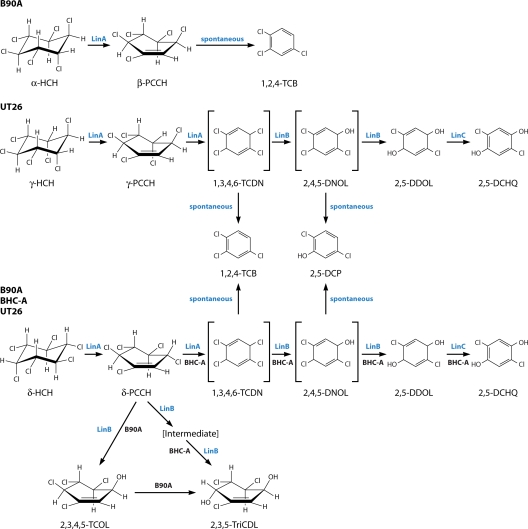
FIG. 4.
Upstream pathway for the aerobic degradation of α-, γ-, and δ-HCH initiated by two successive dehydrochlorination reactions. 1,3,4,6-TCDN and 2,4,5-DNOL are shown in square brackets because they have not been demonstrated empirically. A further intermediate in δ-HCH degradation that has not been demonstrated empirically in the BHC-A strain is also shown in square brackets. This intermediate may be 3,4,5,6-TCOL, as was demonstrated for B90A. Note that the LinA, LinB, and LinC enzymes believed to catalyze several of the reactions are indicated, although direct evidence for the role of these enzymes in the strains indicated is not always available (see text).
FIG. 5.
Upstream pathway for the aerobic degradation of β- and δ-HCH involving two successive hydrolytic dechlorination reactions. LinB has been directly implicated in these reactions in some although not all of the strains shown.
γ-HCH
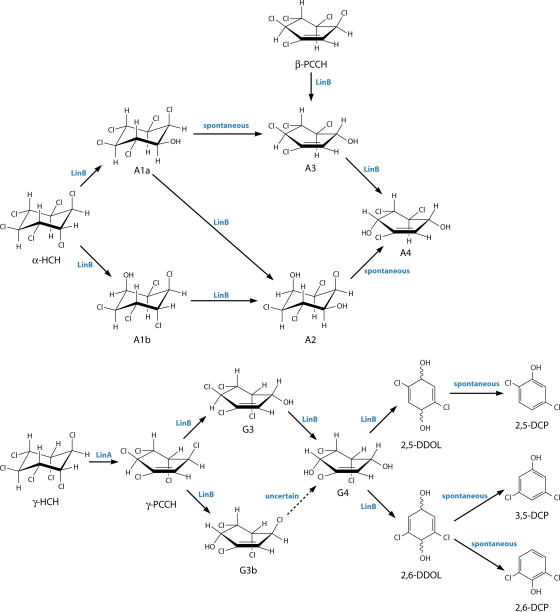
The aerobic degradation pathway of γ-HCH has been studied in some detail for UT26 (109, 115) (Fig. 4). It was suggested that two initial dehydrochlorination reactions produce the putative product 1,3,4,6-tetrachloro-1,4-cyclohexadiene (1,3,4,6-TCDN) via the observed intermediate γ-pentachlorocyclohexene (γ-PCCH) (61). Subsequently 2,5-dichloro-2,5-cyclohexadiene-1,4-diol (2,5-DDOL) is generated by two rounds of hydrolytic dechlorinations via a second putative metabolite, 2,4,5-trichloro-2,5-cyclohexadiene-1-ol (2,4,5-DNOL) (117). 2,5-DDOL is then converted by a dehydrogenation reaction to 2,5-dichlorohydroquinone (2,5-DCHQ) (118). The formation of 2,5-DCHQ completes what is known as the upstream degradation pathway.
It was suggested that the major upstream pathway reactions described above are enzymatically catalyzed, but two other, minor products, 1,2,4-trichlorobenzene (1,2,4-TCB) and 2,5-dichlorophenol (2,5-DCP), are produced, presumptively by spontaneous dehydrochlorinations of the two putative metabolites, 1,3,4,6-TCDN and 2,4,5-DNOL (106-108). Both 1,2,4-TCB and 2,5-DCP appear to be dead-end products in this strain.
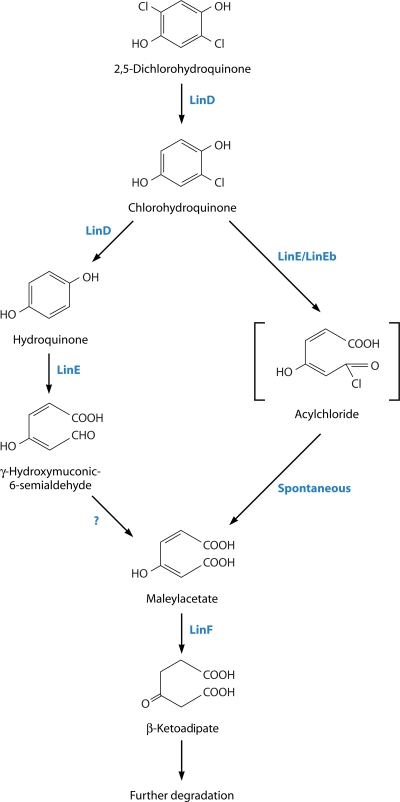
The first step in the subsequent, downstream degradation pathway is a reductive dechlorination of 2,5-DCHQ to chlorohydroquinone (CHQ) (102). The pathway then bifurcates, with the minor route being a further reductive dechlorination to produce hydroquinone (HQ), which is then ring cleaved to γ-hydroxymuconic semialdehyde (γ-HMSA). The major route involves the direct ring cleavage of CHQ to an acylchloride, which is further transformed to maleylacetate (MA) (100). MA is converted to β-ketoadipate (42) and then to succinyl coenzyme A (CoA) and acetyl-CoA, which are both metabolized in the citric acid cycle (109). These reactions are summarized in Fig. 6.
FIG. 6.
Downstream pathway for the aerobic degradation of the γ-HCH isomer in strain UT26 together with the Lin enzymes catalyzing various steps. The intermediate shown in the square brackets (acylchloride) is hypothetical, and the reaction shown by a question mark has not been demonstrated empirically. The enzyme catalyzing the conversion of γ-hydroxymuconic-6-semialdehyde to maleylacetate is not known.
Work on the aerobic degradation pathways of the α- and β-HCH isomers has so far focused on the initial steps in the upstream pathway (119, 143, 158). However, some differences from that observed for γ-HCH by UT26 described above are already emerging.
α-HCH
α-HCH degradation has been studied with the B90A strain (Fig. 4), and the first step is again dehydrochlorination (167, 168). α-HCH actually exists in two enantiomeric forms (Fig. 1), each of which is converted to its respective β-PCCH enantiomer. (+)-α-HCH is converted to β-(3S,4S,5R,6S)-1,3,4,5,6-PCCH, and (−)-α-HCH becomes β-(3R,4R,5S,6R)-1,3,4,5,6-PCCH (167).
Interestingly, these two β-PCCHs are then metabolized to 1,2,4-TCB (167). Although not established empirically, extrapolation from observations of γ-HCH (and δ-HCH [see below]) degradation suggests that it would also occur via TCDN and two further rounds of dehydrochlorination. TCDN would spontaneously be converted to 1,2,4-TCB, which again would be a dead-end product in B90A, as it was in UT26. Further empirical work on the route from β-PCCH to 1,2,4-TCB in B90A (which mineralizes α-HCH [R. Lal et al., unpublished data]) is needed because recent work with resting Escherichia coli cells heterologously expressing lin genes suggests the possibility of several routes (see LinB Haloalkane Dehalogenase below).
β-HCH
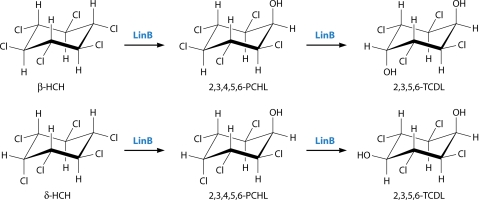
β-HCH is the most recalcitrant of all the major HCH isomers and does not undergo mineralization easily (6). Its relative stability is attributed to the fact that it is the only fully equatorially substituted HCH isomer (Fig. 1). This appears to be a barrier to the dehydrochlorination reactions, which are the first steps in the degradation of γ- and α-HCH described above and which apparently require axial chlorine atoms (see below). Instead, it seems that hydrolytic dechlorination reactions now become more feasible (Fig. 5). The transformation of β-HCH to 2,3,4,5,6-pentachlorocyclohexanol (PCHL) has been observed for all five strains tested. PCHL appears to be a terminal product in UT26 and Sp+ (119, 158), whereas in B90A, Sphingobium sp. MI1205, and Sphingobium sp. BHC-A (hereafter simply MI1205 and BHC-A), it is further converted to 2,3,5,6-tetrachlorocyclohexanediol (2,3,5,6-TCDL) (62, 142, 190). Thus, there is both a difference between strains in the metabolism of β-HCH and a difference within strains such as B90A and UT26 in the ways in which they metabolize β- versus γ- or α-HCH.
δ-HCH
Two upstream pathways have been proposed for the degradation of δ-HCH (Fig. 4 and 5). One, outlined by Wu et al. (191) for BHC-A at least, is similar to the dehydrochlorinase-initiated route by which γ-HCH is degraded in UT26. The other pathway, also seen in these strains plus some others, is essentially the same as the hydrolytic dechlorination-led route by which β-HCH is degraded as described above, together with the same strain-specific variations described above (142, 191). This duality of degradative pathways is most likely a consequence of δ-HCH possessing a single axial chlorine in the cyclohexane ring. The axial chlorine is evidently amenable to dehydrochlorination via the same mechanism as that of the axial chlorines in γ-HCH, whereas the equatorial chlorines are amenable to hydrolytic dechlorination via the same mechanism as that of the equatorial chlorines in β-HCH.
In the case of the hydrolytic dechlorination-led route, δ-HCH is converted to PCHL in UT26 and Sp+ but with no further metabolism of the PCHL evident in these strains (158). On the other hand, in B90A and BHC-A, PCHL is again generated but then further converted to TCDL (142, 158, 191). These strain differences are thus perfectly correlated with the pattern seen for β-HCH, implying a common enzymatic cause.
In the case of the dehydrochlorinase-initiated route, for both strains BHC-A and B90A, δ-HCH is initially converted to δ-PCCH (142, 191) and, as Wu et al. (191) suggested, for BHC-A at least, then on to the putative metabolite 1,3,4,6-TCDN in reactions analogous to those for γ-HCH. The metabolism of the putative 1,3,4,6-TCDN is then proposed to continue down a pathway that is precisely analogous to that suggested for γ-HCH by strain UT26 (191). This would involve two successive hydrolytic dechlorinations to produce first the presumptive 2,4,5-DNOL metabolite and then the empirically validated 2,5-DDOL. The dehydrogenase reaction to produce 2,5-DCHQ then follows. There are also, again, the minor, dead-end products 1,2,4-TCB and 2,5-DCP. Interestingly, however, BHC-A is also able to take δ-PCCH through two successive hydrolytic dechlorinations to generate 2,3,5-trichloro-5-cyclohexene-1,4-diol (2,3,5-TriCDL), probably via 2,3,4,5-tetrachloro-5-cyclohexene-1-ol (2,3,4,5-TCOL), as was also shown in the case of strain B90A (see above).
It is, in fact, anomalous for δ-HCH to go through two dehydrochlorination reactions in this strain when it has only one axial Cl. This might be due to a peculiarity of the strain, or it might indicate that the proposed requirement for axial chlorine arrangements is not the full explanation for the isomer specificity of the dehydrochlorination reaction. An empirical demonstration of the putative 1,2,4,5-TCDN metabolite will be an important priority from here. This issue is considered again in the section on LinA below.
THE lin GENES
Genes necessary for the aerobic degradation of γ-HCH (called lin genes) were initially identified and characterized for UT26 (115) and were subsequently recovered from B90A (40, 72) as well. Very similar lin genes have also been identified for all the other HCH-degrading sphingomonads tested (20, 27, 62, 75, 190, 193). In UT26, where the system is best characterized (109) (Table 2), the pathway is comprised as follows: linA, encoding a dehydrochlorinase (61); linB, encoding a haloalkane dehalogenase (117); linC, encoding a dehydrogenase (118); linD, encoding a reductive dechlorinase (102); linE/linEb, encoding a ring cleavage oxygenase (42, 100); linF, encoding a maleylacetate reductase (42); linGH, encoding an acyl-CoA transferase (109); and linJ, encoding a thiolase (109), plus linR/linI, which are regulatory genes (101, 109).
TABLE 2.
HCH-degradative (lin) genes in HCH-degrading sphingomonadsa
| Gene | Amino acid residue(s) | Function | Strains (GenBank accession no.) |
|---|---|---|---|
| linA1/linA/linA2 | 154/156 | Dehydrochlorinase | UT26 (D90355), B90/B90A (AY331258, AY150580), Sp+ (AY903217, AY690622), DS2 (AJ871378), DS2-2 (AJ871379), γ1-7 (AJ871380), δ12-7 (AJ871381), γ16-1 (AJ871382), α1-2 (AJ871383), α4-2 (AJ871384), DS3-1 (AJ871385), BHC-A (DQ372106) |
| linB | 296 | Haloalkane dehalogenase | UT26 (D14594), B90A (AY331259), Sp+ (AY903216), BHC-A (DQ246619), MI1205 (AB278602), SS04-1 (AB304077), SS04-2 (AB304078), SS04-3 (AB304079), SS04-4 (AB304080), SS04-5 (AB304081) |
| linC | 250 | Dehydrogenase | UT26 (D14595), B90A (AY331258), Sp+ (DQ111068), BHC-A (DQ462464) |
| linD | 346 | Reductive dechlorinase | UT26 (D89733), B90A (AY334273), BHC-A (DQ480725) |
| linE | 321 | Ring cleavage oxygenase | UT26 (AB021867), B90A (AY334273), BHC-A (DQ399709) |
| linR | 303 | LysR family transcriptional regulator | UT26 (AB021863), B90A (AY334273), BHC-A (DQ399711) |
| linF | 352 | Maleylacetate reductase | UT26 (AB177985), BHC-A (DQ399710) |
| linEb | 319 | Ring cleavage oxygenase | UT26 (AB177985) |
| linG | 156 | Acyl-CoA transferase | |
| linH | 212 | Acyl-CoA transferase | |
| linI | 267 | IclR family transcriptional regulator | |
| linJ | 403 | Thiolase | |
| linK | 316 | Putative ABC transporter system, inner membrane protein | UT26 (AB267475) |
| linL | 282 | Putative ABC transporter, ATPase | UT26 (AB267475) |
| linM | 320 | Putative ABC transporter system, periplasmic protein | UT26 (AB267475) |
| linN | 202 | Putative ABC transporter system, lipoprotein | UT26 (AB267475) |
| linXb | 250 | Dehydrogenase | UT26 (D23722), B90/B90A (AY331258, AY150580), BHC-A (DQ486136) |
Data from Nagata et al. (115), Kumari et al. (72), Dogra et al. (40), Böltner et al. (20), Cérémonie et al. (27), Lal et al. (75), Ito et al. (62), Nagata et al. (109), Wu et al. (191), and Yamamoto et al. (193).
linX is apparently not required for the γ-HCH degradation pathway of S. japonicum UT26 (118).
Thus, linA to linC encode the enzymes responsible for the upper pathway, and linD to linJ encode those enzymes for the lower pathway. The organization of these genes in operons will be covered briefly below.
Evidence across a variety of strains indicates that the linA-encoded HCH dehydrochlorinase (LinA) (142, 167, 191) and the linB-encoded haloalkane dehalogenase (LinB) (62, 119, 142, 158, 190, 191) catalyze the dehydrochlorinase and hydrolytic dechlorinase reactions, respectively, in the upper pathway. There is unequivocal experimental evidence that LinA dehydrochlorinates γ-, α-, and δ-HCH, while LinB hydrolytically dechlorinates β- and δ-HCH in all strains examined. However, an interpretation of the roles of LinA and LinB in some of the subsequent reactions is not so straightforward because certain key metabolites have not been recovered experimentally for the dehydrochlorinase-led pathway for the γ-, α-, and δ-isomers, and there are strain differences with respect to the hydrolytic dechlorinase-led pathway for β- and δ-HCH.
LinA HCH DEHYDROCHLORINASE
Biochemistry
LinA is a homotetrameric protein with a 16.5-kDa molecular mass (111, 112), which appears to be located in the periplasm of sphingomonads (110), although a homologue in Rhodanobacter lindaniclasticus appears to be secreted extracellularly (120, 173). LinA is believed to be a unique type of dehydrogenase. It has no close relatives but shows low-level sequence similarity to a small group of proteins of diverse function and an α/β-crystatin-like structural fold (116, 176). Homology models based on these known structures suggest a catalytic mechanism similar to that of scytalone dehydratase, and the behaviors of mutants of key residues in LinA are consistent with this mechanism and the assignment of at least some parts of the substrate binding pocket (116). It is proposed that H73 and D25 form a catalytic dyad; H73 abstracts a proton from the HCH ring, and the resulting positively charged H73 residue is stabilized though its interaction with D25. Upon the abstraction of the proton and the formation of the double bond, the leaving-group chloride is expelled in a bimolecular elimination reaction.
Only qualitative work on the substrate range of LinA has been performed so far (111), and this suggests that its substrate range may be restricted to α-, γ-, and δ-HCH and their corresponding PCCH products. Since β-HCH, which is not hydrolyzed by LinA, is the only major isomer that lacks at least one adjacent trans-diaxial H/Cl pair (Fig. 1), it was proposed and subsequently confirmed that the dehydrochlorination of the other HCH isomers by LinA occurs stereoselectively at this hydrogen/chlorine pair (108, 111, 176). A similar reaction mechanism was proposed for the PCCHs, yielding TCDNs (111, 176, 190). Significantly, however, Wu et al. (191) reported that LinA catalyzes the dehydrochlorination of both δ-HCH and its product, δ-PCCH, even though δ-PCCH does not possess a trans-diaxial H/Cl pair. Additional work is clearly required to fully understand the molecular basis for the LinA reaction and its isomer specificities.
Variants of LinA
Most strains tested have one copy of the linA gene, but B90A and Pseudomonas aeruginosa ITRC-5 are known to have two, denoted linA1/linA2 and linAa/linAb (40, 72, 75, 161). There is also evidence from DNA hybridization studies that some other strains may also have two copies of the linA gene (31). The LinA1 and LinA2 enzymes from B90A are known to differ by about 10% in their amino acid sequences. The linA genes of UT26 and B90A were shown to be constitutively expressed (115, 168), although there is evidence for some inducible expression in Rhodanobacter lindaniclasticus (120, 173).
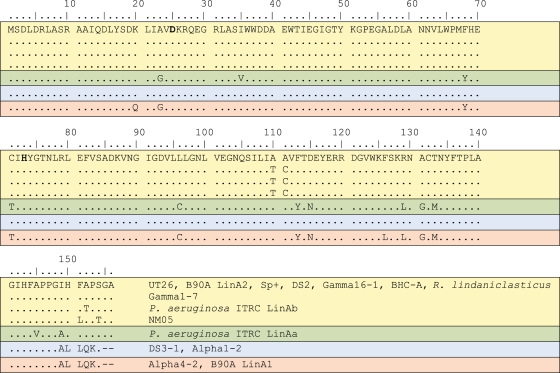
Figure 7 shows the differences among the sequences of LinA enzymes from known HCH-degrading bacteria, including sphingomonads (20, 27, 40, 61, 72, 109, 191), Pseudomonas aeruginosa ITRC-5 (161), Rhodanobacter lindaniclasticus (120, 173), and Xanthomonas sp. ICH12 (89). The sequences fall into four major groups.
FIG. 7.
Sequence differences among LinA variants from known HCH-degrading bacteria. Residues D25 and H73 that form the catalytic dyad are shown in boldface type. The four groups of sequences described in the text are separated by solid lines.
The first and largest group (10 sequences) includes the single LinA sequences from the well-characterized strains UT26 and BHC-A plus one (LinA2) of the two sequences from the well-characterized strain B90A. These sequences are in fact identical, although three other members of this group, found in P. aeruginosa LinAb, Sphingomonas sp. NM05, and Sphingomonas sp. γ1-7, differ by two to four residues.
Sequences in the second group show 12 other single-amino-acid differences. Four of these differences cluster around the catalytic dyad residues D25 and H73 (although D25 and H73 themselves are conserved), and there is another cluster of five changes around, and including, the R129 residue, which has also been implicated in substrate binding and the catalytic mechanism (see below). The only representative of this group is P. aeruginosa LinAa.
The third group differs from the first only by the insertion of a five-residue motif from the IS6100 insertion sequence (see below) very close to the C terminus of the protein. This third group includes sequences from two sphingomonads.
The fourth group includes most of the changes seen in the second group plus the C-terminal IS6100 insertion seen in the third group. It contains LinA1 of B90A and LinA of another sphingomonad strain, α4-2.
Data on functional differences between the four major LinA groups are as yet scant, but it is already clear that the first and fourth clades differ at least in enantiomer preferences. Suar et al. (167) were able to show that LinA1 from strain B90A preferentially dehydrochlorinates the (+)-α-HCH enantiomer, whereas LinA2 from this strain preferentially dehydrochlorinates the (−)-α-HCH enantiomer. Different chiral preferences in the degradation of α-HCH enantiomers have in fact been observed for different soil samples (24, 56, 57, 78, 131).
Recently, Mencía et al. (95) recovered a C132R mutant of LinA through an in vitro evolution experiment. This mutant has a sixfold increase in activity for γ-HCH. The mutation is located within the substrate binding pocket of the enzyme, as predicted by the modeling described previously by Nagata et al. (116), and is only three residues away from the R129 residue, which the site-directed mutagenesis described by Nagata et al. (116) showed was essential for HCH dehydrochlorinase activity. This mutant was found after only a single generation of in vitro evolution and suggests that the further evolution of LinA to enhance its activity and potential for bioremediation should be possible.
One of the priorities for future work on LinA will be kinetic analysis of heterologously produced versions of the different LinA variants against all the proposed substrates. This will be central to the resolution of its role in steps after the first dehydrochlorinase reaction in the metabolism of γ-, α-, and δ-HCH. Also highly desirable is a crystal structure of the LinA proteins, both to elucidate the sequence-structure-function relationships underpinning the kinetic differences described above and to allow a rational design approach for the development of new LinA variants with improved bioremediation potentials (see below).
LinB HALOALKANE DEHALOGENASE
Biochemistry
LinB is a monomeric 32-kDa protein (117), which, like LinA, is located in the periplasm of the sphingomonads tested (110). The major catalytic domain of LinB belongs to the large and well-characterized α/β-hydrolase fold superfamily of proteins, which characteristically carry out two-step hydrolytic reactions driven by a nucleophile (Asp) that is part of a catalytic triad and using an oxanion hole to stabilize the intermediate (69, 114, 117). LinB has been the subject of several crystallographic (92, 123, 166) and computational studies (32, 33, 69, 122, 130).
By analogy to the well-studied mechanism of another haloalkane dehalogenase (DhlA) from Xanthobacter autotrophicus (65), the catalytic mechanism of LinB is proposed to involve an interaction of the substrate with the catalytic triad (D108, H272, and E132, i.e., nucleophile-histidine-acid) and a group of halide-stabilizing residues (principally N38 and W109) (18, 32, 60, 130). It was suggested that nucleophilic attack by D108 at the carbon atom of the substrate results in C-Cl bond cleavage by an SN2 displacement mechanism, resulting in the formation of an acyl-enzyme intermediate. E132 functions to orient and increase the basicity of H272, which in turn activates a water molecule for the hydrolysis of the acyl-enzyme and regeneration of the active site. The NH groups of N38 and W109 stabilize the developing negative charge on the halide in the transition state of the SN2 displacement reaction.
The specificity of LinB for different HCH isomers has not been studied in any detail. However, data for purified, heterologously expressed LinB (69) reveal a very broad substrate preference for halogenated compounds up to a chain length of eight carbon atoms. The maximum catalytic efficiency (kcat/Km) was found to be 233 s−1 mM−1 for 1-iodohexane. LinB was also shown to catalyze the hydrolytic dechlorination of β- and δ-HCH although at considerably lower efficiencies (kcat/Km = 1 s−1 mM−1 for β-HCH) (119). As expected, heterologously expressed LinB does not catalyze the hydrolytic dechlorination of γ-HCH, although surprisingly, some low-level activity against the α-isomer was recently reported for LinB from B90A (143). Significantly, LinB from B90A and MI1205 is an order of magnitude more efficient than LinB from UT26 at catalyzing the dechlorination of β-HCH, and it also catalyzes the dechlorination of the product PCHL at the 4-position, resulting in the dihydroxylated product 2,3,5,6-TCDL (62, 158). The biochemical differences between LinB of B90A and LinB of UT26 provide an elegant explanation for the differences between these two strains in their performance of the hydrolytic dechlorination-led degradation of β-HCH.
Interestingly, while most HCH-degrading strains have only a single linB gene, Sphingobium sp. MI1205 and P. aeruginosa ITRC-5 were reported to contain an IS6100-flanked duplication of a linB variant similar to the linB sequences found in strains B90A and BHC-A (62, 161). This cluster is also identical to the one found for plasmid pLB1, isolated from soil, suggesting a horizontal transfer of lin genes (103). The linB gene is known to be constitutively expressed in UT26 and B90A at least (115, 168).
Variants of LinB
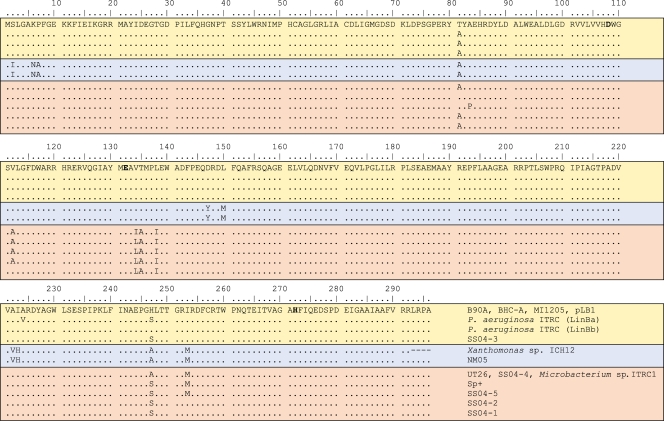
Figure 8 summarizes the amino acid differences among sequenced LinB proteins from known HCH-degrading sphingomonads (20, 27, 40, 62, 109, 117, 190, 191) as well as the HCH-degrading Microbacterium sp. ITRC1 (88), Xanthomonas sp. strain ICH12 (89), and Pseudomonas aeruginosa ITRC-5 (161). Figure 8 also contains a sequence from plasmid pLB1, also obtained from HCH-contaminated soils (103). These sequences fall into three major groups. One group, comprising seven sequences, includes sequences from the well-characterized strains B90A, BHC-A, and MI1205 plus one from another sphingomonad, one from a plasmid (pLB1) isolated directly from soil, and the two genes from P. aeruginosa strain ITRC-5. A second group (containing just two sequences from the sphingomonad NM05 and Xanthomonas sp. ICH12) is quite closely related to the first but contains seven unique substitutions and two others shared with the third group. The third group, including sequences from the well-characterized strains UT26 and Sp+ plus four from other sphingomonads and Microbacterium sp. ITRC1, is characterized by a cluster of three unique changes near E132 of the catalytic triad plus a small number of changes elsewhere in the sequence.
FIG. 8.
Sequence differences among LinB variants from known HCH-degrading bacteria. Residues D108, E132, and H272 that form the catalytic triad are shown in boldface type. The three groups of sequences described in the text are separated by solid lines. Note that the C terminus of the Xanthomonas sp. ICH12 LinB enzyme has not been determined.
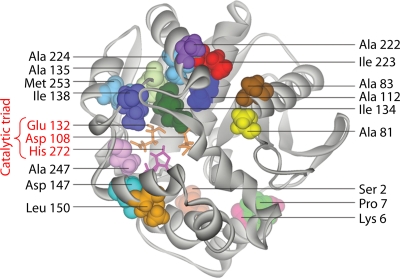
The sequence differences between the first and third groups described above are particularly interesting because the well-characterized members of the first group (B90A, BHC-A, and MI1205) have an ability to degrade PCHL, which the well-characterized members of the third group (UT26 and Sp+) lack (see above). Site-directed mutagenesis and structural modeling suggest that seven amino acid differences between B90A/BHC-A/MI1205 and UT26/Sp+ explain this difference, with V134I and H247A (B90A/BHC-A/MI1205 amino acid first) of particular importance (62) (Fig. 9). Specifically, V134 and H247 are essential for the correct orientation of PCHL for SN2 attack. The other five differences, T81, V112, T135, L138, and I253, also lie in the active site (T135 and L138 close to V134 and the catalytic triad E132) and contribute to the activity difference in a more subtle fashion.
FIG. 9.
Variant positions in the structure of LinB. The active-site residues D108, E132, and H272 are marked with red text. The coordinates used were those reported under Protein Data Bank accession number 1MJ5 (166).
Unresolved Substrate Specificity Issues
Very recently, Raina et al. (143) showed that resting cells of E. coli heterologously expressing the linB gene from B90A can act on α-HCH and on β- and γ-PCCH, which are the products of LinA action on α- and γ-HCH (see above). Two stereoisomers of 3,4,5,6-tetrachloro-2-cyclohexene-1-ol and 2,5,6-trichloro-2-cyclohexene-1,4-diol were identified as being products of these reactions (Fig. 10). These metabolites had not been reported for HCH-degrading strains (see above), and α-HCH had not previously been reported as a substrate for any LinB variant. Moreover, the kinetics of these reactions were not reported by Raina et al. (143). Nevertheless, given that the two key metabolites in the pathway of LinA/dehydrochlorinase-led degradation of the γ-, α-, and δ-isomers of HCH proposed by Wu et al. (190) have not been observed either, some aspects of this pathway currently remain open to question. Detailed comparative kinetics of purified, heterologously expressed LinA and LinB enzymes from key strains with several metabolites will be crucial to the full elucidation of the pathway.
FIG. 10.
Reactions of α-HCH and β- and γ-PCCH catalyzed by heterologously expressed LinB from B90A observed by Raina et al. (143). Product nomenclature was described previously by Raina et al. (143).
LinC DEHYDROGENASE
Less is currently known about the third upstream pathway enzyme, LinC, or any of the downstream pathway proteins.
LinC is a 2,5-DDOL dehydrogenase in the short-chain alcohol dehydrogenase family (118, 133). A general catalytic mechanism was proposed for these enzymes (67), involving the participation of a conserved Ser-Tyr-Lys catalytic triad and an NAD+ cofactor. Specifically, the tyrosine hydroxyl is stabilized in a deprotonated state by the amino group of the lysine. The deprotonated tyrosine then participates in proton abstraction from the hydroxyl group of the substrate, which is further activated through a hydrogen-bonding interaction with the serine. Finally, hydride transfer from the substrate to NAD+ forms NADPH and the reduced product, i.e., the conversion of 2,5-DDOL to 2,5-DCHQ.
linC genes have been recovered from several HCH-degrading sphingomonads (20, 27, 40, 118, 190), as a single copy in both UT26 and B90A (40, 118) but in two copies in Sp+ (75). Variants of LinC have also been amplified directly from HCH-contaminated soils (47). Amino acid identities among the available sphingomonad LinC sequences are 98 to 100%. Like linA and linB, linC was found to be constitutively expressed in strains UT26 and B90A (115, 168). More divergent (97 to 98% amino acid identities) linC genes have also been recovered from HCH-degrading Pseudomonas aeruginosa ITRC-5 (161) and Microbacterium sp. ITRC1 (88).
Nagata et al. (118) also discovered a linX gene in the vicinity of the linA gene of UT26 that encodes a protein with 33% amino acid identity to LinC and also some limited 2,5-DDOL dehydrogenase activity (Table 2). linX genes have now been found in most of the HCH-degrading sphingomonads, generally in multiple copies (20, 40). Three copies are found in the vicinity of the linA1/linA2 genes in B90A; linX1 and linX3 are identical, and linX2 is 67% different at the amino acid level (40). Nagata et al. (118) showed that LinX is not necessary for the degradation of γ-HCH in vivo, but its real physiological role is not clear.
THE DOWNSTREAM PATHWAY
Most of the HCH-degrading sphingomonads tested, with the notable exception of Sphingomonas sp. γ12-7, contain the genes linD and linE, which encode the first two (reductive dechlorinase and ring cleavage oxygenase, respectively) steps in the downstream pathway, as well as the regulatory gene linR, which encodes the LysR-type transcriptional regulator LinR (20, 27, 40, 115, 191). These three genes as well as linF (encoding maleylacetate reductase) are also present in the HCH-degrading Pseudomonas aeruginosa ITRC-5 (161). In strain UT26, the linD and linE genes are located near each other and may form an operon (100), and their transcription is induced by LinR in the presence of their respective substrates 2,5-DCHQ (LinD), CHQ (LinD), and HQ (LinE) (101) (Fig. 6). Intriguingly, Suar et al. (168) reported that the linD and linE genes can also be induced with α- and γ- but not β- and δ-HCH in strain B90A; as noted above, this strain will produce the 2,5-DCHQ, CHQ, and HQ substrates for LinD and LinE from γ- and δ-HCH. Further work is needed to understand how these patterns of gene induction relate to metabolite flow through the pathways and the operon arrangements of the lin genes.
Strain UT26 also has a second ring cleavage oxygenase, LinEb, which is 53% identical to LinE and also contributes to the degradation of CHQ (42). The linEb gene is located close to the linF gene, and an open reading frame located between the two encodes another LysR-type regulator, which is implicated in their regulation (42).
The linGH and linJ genes, which encode the acyl-CoA transferase and thiolase required for the conversion of β-ketoadipate to succinyl-CoA and acetyl-CoA, were recently identified in UT26, as was the linI gene, which encodes an IclR-type transcriptional regulator that may regulate their expression (109).
The linK, linL, linM, and linN genes recently identified in UT26 encode a permease, ATPase, periplasmic protein, and lipoprotein, respectively, which together form a putative ABC-type transporter system (43). This system is required for the utilization of γ-HCH, probably by conferring tolerance to toxic dead-end metabolites such as 2,5-DCP (43). Homologues showing high levels of similarity to the linKLMN genes have been found only in sphingomonads, suggesting that they might be important for the high metabolic activity of sphingomonads toward a range of xenobiotic compounds.
EVOLUTIONARY ISSUES AND SCOPE FOR IMPROVEMENT
There are some striking similarities in the mechanisms by which at least the aerobic HCH-degradative pathways have evolved and those that have been found for other xenobiotics (174). Notable here are the roles of horizontal gene transfer across species and insertion sequence-mediated transposition within species in the rapid assembly (<50 years) of a novel catabolic pathway (75). However, there are also some very distinctive aspects to the evolution of the lin system, and these suggest that there is substantial scope for further improvement in various key aspects of the pathway.
One line of evidence indicating that the lin system has been disseminated across sphingomonads and other species by horizontal gene transfer is the strong similarity in the sequences of the characterized lin genes from different species. Most notable here is the finding of closely similar linA, linB, and linC genes in the Gram-positive Microbacterium sp. ITRC1 (88), which is phylogenetically well removed from the sphingomonads and other Gram-negative organisms that account for all the other reported lin gene occurrences. The inferred LinB sequence of ITRC1 was identical to that of UT26, and its inferred LinC differed from the UT26 counterpart by only 2 of 141 residues. Similarly, although the organizations of the genes are quite variable among species (see below), this variation bears no relationship to the phylogeny of the organisms. For example, linA, linB, and linDER from Pseudomonas aeruginosa ITRC-5 show microsyntenous arrangements that are strikingly similar to those of their counterparts in UT26 and B90A (161).
Interestingly, codon usage and GC content are very different in linA than they are in other lin genes or, indeed, most other sphingomonad genes (40, 115). This suggests that linA, at least, may have been recruited from a different source organism than other lin genes, although the others could have sphingomonad origins.
A second line of evidence for the role of horizontal gene transfer events in the spread of the lin genes across species is the location of many of them in plasmids. Several other genes encoding xenobiotic degradation have been found on plasmids in sphingomonads (10, 124), and lin genes have been recovered from plasmids in UT26 (113), Sp+ (27), and B90A (86). At least some of the lin gene occurrences outside the sphingomonads are also associated with plasmids (173). Evidence for the conjugative nature of several of these plasmids (103, 113, 173) supports their possible role in the spread of lin genes among species.
However, not all lin genes are located on plasmids. In UT26, for example, they are dispersed across at least three replicons: linA to linC are on chromosome I, linF is on chromosome II, and linDER is on plasmid pCHQ1 (113). Some of the spread of lin genes across species, and in particular within species, is instead attributed to their association with IS6100 insertion sequences. Most of the lin genes found to date are in fact associated with IS6100, both within (20, 27, 31, 40, 62, 103, 109) and outside (161) (Table 3) the Sphingomonadaceae. IS6100 sequences have also been associated with the spread of genes for degrading antibiotics (170, 172) and other pesticides (59, 81). The data for the lin genes implicate a role of IS6100 sequences in the dispersion of individual lin genes both among different hosts and among replicons within species.
TABLE 3.
Association of lin genes with IS6100 in HCH-degrading sphingomonads
| Organism | Associationa |
Reference(s) | ||||
|---|---|---|---|---|---|---|
| linA/IS6100 | linB/IS6100 | linC/IS6100 | linDER/IS6100 | linF/IS6100 | ||
| S. japonicum UT26 | +/+ | +/− | +/+ | +/+ | +/+ | 109 |
| S. indicum B90A | +/+ | +/+ | +/+ | +/+ | +/ND | 40, 75 |
| S. francense Sp+ | +/+ | +/+ | +/ND | +/+ | +/ND | 27, 75 |
| S. ummariense RL-3 | +/+ | +/+ | +/+ | +/+ | ND/ND | 31, 160 |
| S. chinhatense IP26 | +/+ | +/+ | +/+ | +/+ | ND/ND | 30, 31 |
| Sphingomonas sp. DS2 | +/− | +/+ | +/− | +/ND | ND/ND | 20 |
| Sphingomonas sp. DS2-2 | +/− | +/+ | +/− | +/ND | ND/ND | 20 |
| Sphingomonas sp. DS3-1 | +/+ | +/+ | +/+ | +/ND | ND/ND | 20 |
| Sphingomonas sp. α1-2 | +/+ | +/+ | +/+ | +/ND | ND/ND | 20 |
| Sphingomonas sp. α4-2 | +/+ | +/+ | +/+ | +/ND | ND/ND | 20 |
| Sphingomonas sp. γ1-7 | +/− | +/+ | +/+ | +/ND | ND/ND | 20 |
| Sphingomonas sp. γ12-7 | +/− | +/+ | +/− | −/ND | ND/ND | 20 |
| Sphingomonas sp. γ16-1 | +/− | +/+ | +/− | +/ND | ND/ND | 20 |
| Sphingobium sp. MI1205 | ND/ND | +/+ | ND/ND | ND/ND | ND/ND | 62 |
| Sphingobium sp. BHC-A | +/ND | +/ND | +/ND | +/ND | +/ND | 191 |
| Sphingomonas sp. UM1 | +/+ | +/+ | +/+ | +/+ | ND/ND | 31 |
| Sphingobium sp. UM2 | +/+ | +/+ | +/+ | +/+ | ND/ND | 31 |
| Sphingobium sp. HDU05 | +/+ | +/+ | +/+ | +/+ | ND/ND | 31 |
| Sphingobium sp. HDIP04 | +/+ | +/+ | +/+ | +/+ | ND/ND | 31 |
| Sphingobium sp. F2 | +/+ | +/+ | +/+ | +/+ | ND/ND | 31 |
| Sphingobium sp. SS04-1 | +/ND | +/+ | +/ND | +/ND | ND/ND | 193 |
| Sphingobium sp. SS04-2 | +/ND | +/+ | +/ND | +/ND | ND/ND | 193 |
| Sphingobium sp. SS04-3 | +/ND | +/+ | +/ND | +/ND | ND/ND | 193 |
| Sphingobium sp. SS04-4 | +/ND | +/+ | +/ND | +/ND | ND/ND | 193 |
| Sphingobium sp. SS04-5 | +/ND | +/+ | +/ND | +/ND | ND/ND | 193 |
ND, not determined.
The latter movements within species in turn lead to some of the changes in lin gene copy numbers and the coalescence of certain lin genes into operons. For example, the duplication of linA genes and the deletion of linDER genes in B90A and the deletion of one copy of linA in Sp+ have all been attributed to IS6100-mediated recombination events (40). Similarly, the colocalization of linX, linX2, and linA in strains B90A, Sp+, DS3-1, α1-2, and α4-2 has been attributed to recombination events mediated by IS6100 (20, 40, 75).
While the prominent roles of insertion sequences (including IS6100) and conjugative plasmids in assembling and dispersing novel catabolic pathways are common features of many microbial xenobiotic detoxification systems, there are also some other, unusual aspects of the HCH/Lin system that are informative for both fundamental biochemical and applied bioremediation perspectives.
One unusual feature of aerobic HCH degradation is that essentially only one pathway and one genetic system have as yet been found. Multiple systems have been reported for several other pesticides and toxins. For example, at least three and often more have been described for organophosphates, carbamates, glyphosate, triazines, 2,4,6-trinitrotoluene, and even other organochlorines like endosulfan and trichloroethylene (3, 28, 81, 154, 156, 157, 164, 188; C. J. Hartley, S. J. Dorrian, L. J. Briggs, M. R. Williams, R. J. Russell, and J. G. Oakeshott, international patent application PCT/Au2007/000640). This unusual feature of the lin system may reflect the relative recalcitrance of HCH to degradation, as is also suggested by the length of the lin pathway. All the examples described above involve catabolic pathways that are shorter than the lin pathway.
Another unusual feature of the lin system is that it has been found mainly in a single genus. Seventeen of just over 20 well-characterized lin isolates are sphingomonads, and most of the others are also from relatively closely related Gram-negative organisms. To our knowledge, there is just a single report of the lin system in a Gram-positive organism (see above). In contrast, some of the other systems described above are widely distributed across diverse Gram-positive and -negative genera (59, 156), and a couple that are more restricted, like the esd/ese system for endosulfan degradation, tend to be restricted largely to the Gram-positive Actinomycetes, which are well known for the versatility of their secondary metabolisms (188). It was suggested that the concentration of HCH metabolic capabilities in the sphingomonads may be because much of the downstream pathway and perhaps also some of the progenitors for the missing upstream steps preexisted for other reasons in certain sphingomonads (40, 115).
The highly variable organization of lin genes across species is also a distinctive feature of this system. Catabolic genes for many other xenobiotics are generally organized into operons and coordinately regulated (53, 55, 99, 156, 157, 195). Individual lin genes vary in copy number and genetic localization, and while there is some condensation into clusters and operons, the constitutions of these clusters and operons again vary across strains (27, 86, 113). Interestingly, dispersed distributions of catabolic genes have been found for a few other xenobiotics among the sphingomonads, for example, for the genes involved in the degradation of pentachlorophenol in Sphingobium chlorophenolicum ATCC 39723 (25), dibenzo-p-dioxin by Sphingomonas sp. RW1 (2), and protocatechuate by Sphingomonas paucimobilis SYK-6 (94). The scattered and variable arrangement of lin genes may thus partly reflect some unusual feature of sphingomonad genetics. However, given both the refractory nature of HCH and its isomeric complexity, we suggest that it also could reflect a degradative system still evolving toward an optimal state(s).
Part of our suspicion that the lin system is still evolving quite rapidly comes from a consideration of its enzymatic efficiency. The first detoxification step in other xenobiotic catabolic systems, for example, the Oph/OpdA organophosphate-degrading enzyme, approach diffusion-limited kinetics (194), and some of those for glyphosate, carbamates, and atrazine also have specificity constants in excess of 104 s−1 M−1 (28, 154). However, the specificity constants of LinB for its preferred HCH isomer are nearly 3 orders of magnitude lower, and the specificity constant of LinA has not been estimated at this time. The isomeric complexity of the HCHs and their early-step metabolites also present a challenge to the reaction biology of LinA and LinB, and we are already seeing divergent copies of both enzymes emerge that have distinct isomer preferences, albeit still very modest enzyme efficiencies (see above).
HCH BIOREMEDIATION
Bioremediation technologies potentially have four types of applications involving the cleanup of contaminated soils, liquid wastes, stockpiles, and commodities (149, 171). All four are relevant to HCH, even though it is no longer in widespread use as an insecticide. Its persistence and the processes used in its production mean that contaminated soils and stockpiles are a particular priority, with contamination of liquid wastes (e.g., agricultural runoff) and commodities also still being problematic because of diffusion from heavily contaminated soils and stockpiles. Essentially all the work on HCH bioremediation to date involves contaminated soils.
The two major approaches taken to soil bioremediation to this point have been biostimulation and bioaugmentation. Biostimulation involves the addition of oxygen and/or inorganic nutrients to stimulate the growth of resident bacteria that have some capacity to break down the contaminant in question. Bioaugmentation involves the introduction of additional bacteria that have the capacity, acquired naturally or by genetic modification in the laboratory, to break down the contaminant. The two approaches are not mutually exclusive. Both have been applied to HCH-contaminated sites, with various levels of success.
Biostimulation
The first attempt at full-scale in situ HCH bioremediation was carried out at an industrial site contaminated with low levels (<1 mg kg−1) of HCH muck in The Netherlands (77, 180). The biostimulation of an anaerobic zone with an electron donor degraded all HCH isomers to benzene or chlorobenzene, and this plume was then extracted to a wastewater treatment facility where the benzene and chlorobenzene were mineralized aerobically. Costs of over a million euros and time frames of years were nevertheless justified because of the high value of the land once remediated.
Phillips et al. (135) used proprietary biostimulation agents known as Daramend products derived from natural plant fibers to treat 1,100 tons of soil contaminated with high HCH concentrations (∼5,000 mg kg−1) in the United States. Based on developmental work in laboratory microcosms (134), two Daramend treatment approaches (cycled anaerobic/aerobic treatment and a strictly aerobic treatment) using two different Daramend products (D6390 and D6386, respectively) were used. Some tillage to enhance aerobic degradation was also used, and reductions averaging just under 50% were achieved over a year of treatment.
Rubinos et al. (148) reported the use of another, less elaborate biostimulation-plus-tillage trial of a soil with high HCH concentrations (>5,000 mg kg−1) in Spain. The process, which they called land farming, involved the mixing of nitrogen and phosphate nutrients and lime (to correct otherwise acidic pH) plus regular irrigation and tillage over an 11-month period. Reductions in α- and γ-HCH concentrations of over 80% were achieved, although no decrease in the β-HCH concentration was observed. Interestingly, the major metabolites were PCCH and TCCH, suggesting a mix of aerobic and anaerobic degradation.
Most recently, an ex situ biostimulation process has also shown promise for HCH bioremediation (31). In this experiment, soil with indigenous microflora from a dump site for HCH muck was mixed with pristine garden soil, and aeration, moisture, and nutrients were provided intermittently. Reductions in total HCH concentrations (starting at 75 mg kg−1, summed over the α-, β-, and δ-isomers) were found to be ∼3% and 30% at the end of 24 and 240 days, respectively. A comparison of genetic markers over time showed changes in microbial community structure and stimulation of the indigenous sphingomonad population during the experiment. More elaborate studies are now needed to clarify the role of culturable and unculturable microbes during the course of biostimulation-mediated degradation of HCH.
Thus, the consensus from the four biostimulation trials described above is that significant reductions in HCH concentrations can be achieved, even with relatively inexpensive protocols. However, time frames of many months are required, and for the more heavily contaminated sites, only partial reductions were achieved, even for the less stable isomers, hence the interest in bioaugmentation.
Bioaugmentation
The first report of a bioaugmentation trial with HCH, by Bidlan et al. (16), demonstrated the removal of all four major HCH isomers from spiked soils in the laboratory after the addition of an HCH-degrading microbial consortium. Starting concentrations were very low (0.25 mg kg−1), but no detectable HCH remained 5 days after the addition of an HCH-degrading microbial consortium. Mertens et al. (96) then demonstrated the removal of around half of the γ-HCH added (50 mg liter−1 added every few days) to either liquid or soil microcosms by the inoculation of a single HCH-degrading isolate, Sphingomonas sp. γ1-7, encapsulated in open-ended silicone tubes.
Most recently, Raina et al. (144) demonstrated the removal of most of a mix of HCH isomers by the inoculation of strain B90A (at 3 × 1010 cells kg−1 soil, immobilized on corncob powder) both into pits of transplanted contaminated soil (60 to 70 mg kg−1 HCH in 100 kg soil) and in situ at an HCH-contaminated agricultural site (∼4 mg kg−1 HCH in 3 by 3 m2 up to a depth of 20 cm soil). The viability of the added cells was about 20% after 8 days, after which more cells were inoculated. Degradation was uneven across isomers, with over 80% of the α- and γ-HCH removed but only about half of the β- and δ-HCH removed. Nevertheless, this study represents an important proof of concept in achieving a significant cleanup of all the major HCH isomers in the field by bioaugmentation under conditions approaching economic viability.
One refinement of the bioaugmentation process is to use degradative strains that are good colonizers of the rhizosphere and to inoculate them into the root zone of compatible crops in contaminated soil, a process known as rhizoremediation. The potential benefit of the method is the development and maintenance of high levels of degradative bacteria in the soil without the need for high-titer inoculations initially or ongoing supplementation. Böltner et al. (19) proposed the potential of this method for the treatment of various organic chemicals. The rhizoremediation of laboratory soils spiked with 500 mg kg−1 γ-HCH by the incubation of pregerminated corn seeds covered with two HCH-degrading sphingomonads (strains GOF-203 and Ans-PL0) resulted in about a 30% reduction in γ-HCH levels after 25 days (19). Field studies are now needed to demonstrate the full potential of this approach.
Free-Enzyme Bioremediation
While we could find no reports of HCH bioremediation in stockpiles, liquid wastes, or commodities, there is considerable work on the cleanup of other pollutants from such environments. Some of the most promising advances come not from the use of live microbes but from the use of free enzymes (22, 171). The concept of enzymatic bioremediation centers on the use of enzymes that can catalyze the substantive detoxification of the pollutant(s) in question in a single step with reasonable kinetics (>104 s−1 M−1) and without the need for expensive cofactors. These enzymes are heterologously expressed in commercial microbial expression vectors, minimally purified, and formulated in various ways to address the particular stability requirements pertaining to the environments of their use.
Although genetic manipulation (GM) technology is used in the production of free-enzyme bioremediants, no live organisms remain in the formulated products, which therefore avoids any potentially contentious issues around the release of live genetically modified organisms (GMOs). Moreover, free- enzyme bioremediation does not require microbial growth for activity. Hence, it can be effective in short time frames (less than a day, depending on the application) and in circumstances where microbial growth might be either problematic technically (e.g., very concentrated point source contamination) or unacceptable (e.g., potable water supplies). The technology does depend on a liquid, or at least very moist, environment to allow the enzyme access to the substrate. Nevertheless, enzymatic bioremediants are now commercially available for the cleanup of certain pesticides in large-scale liquid wastes (1, 15, 28, 154) and show promise for the treatment of various stockpiles, soils, and, in some cases, external surfaces of commodities (22, 35). The good results for the enzymatic treatment of stockpiles are of particular interest with respect to very concentrated HCH deposits like dump sites, where microbial growth is severely limited by the availability of nutrients.
Since neither LinA nor LinB requires cofactors and each removal of a chlorine atom from the cyclohexane ring reduces toxicity by severalfold, it is well worth considering their potential as starting resources for the development of an enzymatic bioremediation technology for HCH.
FUTURE PROSPECTS
Although scientific interest in HCH degradation was initially motivated by environmental and health concerns due to HCH residues, its aerobic degradation is now proving an excellent model for investigating fundamental issues in microbial and molecular evolution. Because HCH is such a chemically refractory molecule, the evolution of an effective detoxification and utilization pathway has required the acquisition of qualitatively new functions for certain individual enzymes plus the assembly of these and other enzymes into a coordinately regulated pathway. The isomer complexity of HCH adds a further degree of difficulty, with major differences between isomers in the reactions by which their breakdown can be catalyzed, at least under aerobic conditions. These have apparently necessitated the recruitment of alternative enzymes for different isomers in certain upstream steps in the aerobic pathway. Work to date indicates that several microbes, often but not always sphingomonads, are assembling the requisite capabilities, although the phenotype remains variable and probably not optimized in either its genetics or its biochemical efficacy.
Although evidence for the anaerobic degradation of HCH was reported nearly 2 decades earlier than evidence for its aerobic degradation, work on the latter has advanced far further since. Even for the latter, however, there remain several crucial gaps in our knowledge.
One fundamental gap concerns the reactions in the upper pathway, where the most biochemically problematic detoxification steps occur. Two sets of observations indicate that certain key elements of the widely accepted scheme for this pathway may be incorrect. One disconcerting observation has been that the presumptive dehydrochlorination of the putative δ-HCH metabolite δ-PCCH by LinA is inexplicable in terms of the bimolecular elimination reaction characteristic of enzymes from the family in which LinA sits (142). The second recent surprise has been the finding that heterologously expressed LinB (from B90A at least) has a wider substrate specificity across HCH and PCCH isomers in vitro (albeit with unknown kinetics) than the accepted pathway assumed, and moreover, it produces metabolites from some of these substrates that do not sit in the accepted pathway either (143). There is now considerable doubt as to the validity of certain steps in the pathway that have hitherto been inferred rather than empirically demonstrated.
The resolution of these uncertainties about the upper metabolic pathway clearly requires more detailed metabolite analyses of HCH degradation in HCH-degrading bacteria. Another important goal will be to better understand the biochemistry of the LinA and LinB enzymes. Particular priorities in the latter are detailed kinetic analyses of heterologously expressed LinA and LinB with a range of resolved isomers of putative substrates within the pathway. LinA has recently been crystallized (127), so its structure should soon be published. This would also be an invaluable aid to understand its mechanism and the structure-function relationships underpinning that mechanism.
There are also other important aspects of the Lin pathway that we currently do not understand. One of these involves the mysterious LinX enzymes, which are distantly related to LinC and show some LinC function in vitro but are apparently not essential for γ-HCH degradation, at least in vivo. One possibility is that LinX catalyzes the transformation of γ-hydroxymuconic-6-semialdehyde to maleylacetate in the downstream pathway. This, like the upstream step catalyzed by LinC, requires a dehydrogenation step. This step is not essential for HCH degradation in vivo because a bifurcation in the downstream pathway two steps prior to the production of γ-hydroxymuconic-6-semialdehyde provides another option for the catabolic process.
Another major gap in our current knowledge of the system concerns the extent and biological significance of genetic variation both in the organization of the various lin genes and in the coding sequences of the key upstream linA and linB genes. Although some condensation of lin genes into operons has been reported, the complements and structures of the operons reported are still highly variable, and some lin genes remain apparently unlinked to the operons, either on plasmids or elsewhere in the various genomes. Such variation is unusual and suggests that the evolution of lin operons is still a work in progress. Several studies have found transposable elements, in particular IS6100 elements, in the vicinity of the lin genes. This is not unusual for recently assembled catabolic operons, but it is certainly consistent with a dynamic, ongoing process of the organization of lin genes into arrangements suited to coordinate control. Time course studies of the genetics of HCH degradation in highly contaminated sites could prove very informative in this context.
Evidence to date indicates high levels of polymorphism in the amino acid sequences of both the LinA and LinB enzymes, with convincing if indirect evidence that at least some of it affects function. The LinB proteins of UT26 and B90A in particular seem to differ qualitatively in their HCH isomer preferences. Similarly profound differences appear to exist between the α-HCH enantiomer preferences of LinA1 and LinA2. However, there could well be many other qualitative and quantitative differences in the activities of other LinA and LinB variants. Further work is needed to elucidate the extent of the variation, the molecular basis of isomer-specific differences in the functions of some of the variants, and the role of these differences in generating the metabolic diversity needed to deal with the plethora of isomers involved in the first four steps of the upstream pathway.
The anaerobic pathway for HCH degradation remains poorly understood. Few metabolites have been demonstrated empirically, differences in the pathway between isomers remain unresolved possibilities, and essentially nothing is known about the enzymology or genetics of the process. Upstream dichloroelimination reactions are implicated, with hydrolytic dechlorination likely further downstream. However, enzymes to catalyze these reactions could come from a variety of protein families (165). Much basic biochemical work is needed, and genome sequencing could well prove rewarding for some of the anaerobes involved.
Although the Lin pathway has now been recovered from many different sphingomonads and a few other bacteria isolated from HCH-contaminated sites, there is still a great deal that we do not know about the function of the pathway in this ecosystem. We cannot even yet conclude that the sphingomonads are the major hosts for the pathway, since many recent studies have selected primarily for sphingomonads (by a facile antibiotic screen) and then secondarily tested them for their HCH-degradative abilities. More importantly, we do not know to what extent the evolution of HCH-degrading organisms at contaminated sites is accelerating the rate at which the HCH load is being reduced. This must be an urgent priority for further work. Equally, however, unless the rate has been massively accelerated, the size of the stockpiles and the stability of the compounds are such that many environments heavily contaminated with HCH will remain with us for decades, if not centuries, hence the priority to develop effective bioremediation technologies.
Both the biostimulation and bioaugmentation approaches taken thus far for the remediation of HCH-contaminated soils have met with some success, for certain isomers at least. Both technologies are still in early developmental stages, and substantial further improvements should still be quite readily achievable. While some cleanup situations involving land of potentially high value could withstand high costs, there are many more situations, particularly in the developing world, where costs will need to be low if treatment is to be a realistic option. Further work on application protocols and, for bioaugmentation at least, on strain development is clearly needed. The sphingomonads are clearly the group of first choice for further work on strain development. Their niche in the rhizosphere also predisposes them to many bioremediation applications, and strains with improved growth and viability properties under conditions of use should be quite readily selectable. Many sphingomonads are now quite tractable to isolation, laboratory culture, and selection. Importantly, the remarkable diversity of secondary metabolism seen across the sphingomonads is now attracting the interest of the biotechnology industry for a range of different applications (4), so the infrastructure of knowledge and resources for their genetic analysis and manipulation and commercial fermentation is also rapidly expanding.
The Lin pathway is clearly the most accessible (and possibly the only) biochemical system of the sphingomonads, or other bacteria, for aerobic HCH detoxification. The substrate and isomer coverage and kinetics of LinA and LinB will be critical properties to consider both for the development of improved strains to be used in soil bioremediation and for the development of enzymatic bioremediation strategies for stockpile and liquid contamination.
The enzymes should be amenable to various modern research technologies for obtaining improved variants either by the isolation of new variants from uncultured environmental samples/bacteria or by the improvement of existing variants by techniques of in vitro evolution. Early work on both approaches has yielded promising new variants of LinA and/or LinB (95). The availability of high-throughput screening assays, for some of the reactions at least (90, 104, 136), plus the crystal structures of the enzymes will help these efforts enormously.
We believe that there are good prospects for developing economically viable HCH bioremediation technologies based on the sphingomonad/Lin systems: soil bioremediation through various biostimulation/bioaugmentation approaches and stockpile and, as necessary, liquid remediation through direct enzymatic approaches. Significant work on enzyme characterization, particularly for LinA, and strain and enzyme improvement is still needed. However, the promise of the system evident thus far and the potential of modern microbial and enzyme research technologies to make radical improvements give us confidence that the development of successful technologies for a most pernicious pollutant is quite achievable.
Acknowledgments
We thank Robyn Russell, Simran Jit, and Kiran Bala for invaluable discussions.
Part of the work was supported by grants under the Indo-Australian Biotechnology Fund and Indo-Swiss Collaboration in Biotechnology from the Department of Education Science and Technology (DEST), Australia; the Swiss Agency for Development and Cooperation (SDC), Switzerland; and the Department of Biotechnology (DBT), India. P.S. and K.K. acknowledge CSIR-UGC, Government of India, for providing their research fellowships.
Biography
 Rup Lal is a Professor in Molecular Biology at the Department of Zoology, University of Delhi. Born in a village, Kanoh, in Himachal Pradesh, Rup Lal did his Ph.D. at the University of Delhi (1980). He is the awardee of an Alexander von Humboldt Fellowship, a DBT Overseas Fellowship, and an ASM Indo-US Professorship and is a visiting scientist at the University of Cambridge. Currently, he is the Head, Department of Zoology, and Dean, Faculty of Science. His primary research interests include microbial diversity at pesticide-polluted sites, genetics and biochemistry of hexachlorocyclohexane (HCH) degradation, and development of HCH bioremediation technology. Rup Lal is also interested in the evolution of lin genes and the genetic manipulation of the rifamycin producer Amycolatopsis mediterranei. His group is sequencing genomes of Amycolatopsis mediterranei and Sphingobium indicum. He has published over 100 papers and is the Editor-in-Chief of the Indian Journal of Microbiology and Fellow of the National Academy of Agricultural Sciences and the Association of Microbiologists of India.
Rup Lal is a Professor in Molecular Biology at the Department of Zoology, University of Delhi. Born in a village, Kanoh, in Himachal Pradesh, Rup Lal did his Ph.D. at the University of Delhi (1980). He is the awardee of an Alexander von Humboldt Fellowship, a DBT Overseas Fellowship, and an ASM Indo-US Professorship and is a visiting scientist at the University of Cambridge. Currently, he is the Head, Department of Zoology, and Dean, Faculty of Science. His primary research interests include microbial diversity at pesticide-polluted sites, genetics and biochemistry of hexachlorocyclohexane (HCH) degradation, and development of HCH bioremediation technology. Rup Lal is also interested in the evolution of lin genes and the genetic manipulation of the rifamycin producer Amycolatopsis mediterranei. His group is sequencing genomes of Amycolatopsis mediterranei and Sphingobium indicum. He has published over 100 papers and is the Editor-in-Chief of the Indian Journal of Microbiology and Fellow of the National Academy of Agricultural Sciences and the Association of Microbiologists of India.
 Gunjan Pandey completed his master's degree in biochemistry from G. B. Pant University of Agriculture and Technology, Pantnagar, India, in 1999 and his doctorate from the Institute of Microbial Technology (IMTECH), Chandigarh, India, in 2004. His doctoral research involved both basic and applied studies of the biodegradation of nitroaromatic compounds, a major class of environmental pollutants. In 2004, Gunjan joined CSIRO Entomology in Canberra, Australia, as a postdoctoral fellow and worked to develop commercial enzyme products for the detoxification of several pesticide chemistries. He was subsequently appointed as an indefinite research scientist at CSIRO Entomology. He is now leading a research team that is broadly interested in biodegradation, biotransformation, and biocatalysis, with special emphasis on discovering and developing commercially viable enzymes for pesticide detoxification.
Gunjan Pandey completed his master's degree in biochemistry from G. B. Pant University of Agriculture and Technology, Pantnagar, India, in 1999 and his doctorate from the Institute of Microbial Technology (IMTECH), Chandigarh, India, in 2004. His doctoral research involved both basic and applied studies of the biodegradation of nitroaromatic compounds, a major class of environmental pollutants. In 2004, Gunjan joined CSIRO Entomology in Canberra, Australia, as a postdoctoral fellow and worked to develop commercial enzyme products for the detoxification of several pesticide chemistries. He was subsequently appointed as an indefinite research scientist at CSIRO Entomology. He is now leading a research team that is broadly interested in biodegradation, biotransformation, and biocatalysis, with special emphasis on discovering and developing commercially viable enzymes for pesticide detoxification.
 Pooja Sharma is a graduate student with Professor Rup Lal. She submitted her doctoral thesis to the University of Delhi, India, in September 2009 on the enzymatic bioremediation of hexachlorocyclohexane (HCH) by the action of a microbial enzyme, HCH dehydrochlorinase (LinA). She completed her master's degree in zoology with a specialization in cell biology in 2004 at the University of Delhi. She has a keen interest in microbes that degrade pesticides and is currently involved in deploying bacteria or their enzymes to restore environmental health.
Pooja Sharma is a graduate student with Professor Rup Lal. She submitted her doctoral thesis to the University of Delhi, India, in September 2009 on the enzymatic bioremediation of hexachlorocyclohexane (HCH) by the action of a microbial enzyme, HCH dehydrochlorinase (LinA). She completed her master's degree in zoology with a specialization in cell biology in 2004 at the University of Delhi. She has a keen interest in microbes that degrade pesticides and is currently involved in deploying bacteria or their enzymes to restore environmental health.
 Kirti Kumari completed her master's degree in zoology (specialization in cell biology) from the University of Delhi, India, in 2005 and her Ph.D. under the supervision of Professor Rup Lal from the University of Delhi, India, in November 2009. Her doctoral research involved studying the kinetics and diversity of the enzyme haloalkane dehalogenase (LinB) in the HCH degradation pathway. She is currently working on the enzymatic bioremediation of HCH-contaminated soils. Her research interests include deciphering bioremediation options for the decontamination of pesticide residues in the environment.
Kirti Kumari completed her master's degree in zoology (specialization in cell biology) from the University of Delhi, India, in 2005 and her Ph.D. under the supervision of Professor Rup Lal from the University of Delhi, India, in November 2009. Her doctoral research involved studying the kinetics and diversity of the enzyme haloalkane dehalogenase (LinB) in the HCH degradation pathway. She is currently working on the enzymatic bioremediation of HCH-contaminated soils. Her research interests include deciphering bioremediation options for the decontamination of pesticide residues in the environment.
 Shweta Malhotra was awarded her master's degree in zoology (specialization in cell biology) and subsequently her Ph.D. degree from the University of Delhi, India. She worked as a postdoctoral fellow at the University of Louisville, Louisville, Kentucky. Currently, she is working as a postdoctoral fellow in the field of metagenomics at the Dong-A University, Busan, South Korea. Her research interests include metagenomics, mobile genetic elements (plasmids and transposons), the biodegradation of organic compounds, and actinomycete biology.
Shweta Malhotra was awarded her master's degree in zoology (specialization in cell biology) and subsequently her Ph.D. degree from the University of Delhi, India. She worked as a postdoctoral fellow at the University of Louisville, Louisville, Kentucky. Currently, she is working as a postdoctoral fellow in the field of metagenomics at the Dong-A University, Busan, South Korea. Her research interests include metagenomics, mobile genetic elements (plasmids and transposons), the biodegradation of organic compounds, and actinomycete biology.
 Rinku Pandey was born in New Delhi, India, in 1977. She attended the University of Delhi, where she obtained a master's degree in zoology specializing in cell and molecular biology in 2000. She joined the group of Professor Rup Lal at the University of Delhi in 2001, where she worked on the molecular analysis of the lin genes involved in hexachlorocyclohexane (HCH) degradation and the phylogenetic analysis and reclassification of the three major HCH-degrading bacteria and in 2005 earned a Ph.D. in molecular biology. She joined CSIRO Entomology as a postdoctoral fellow in 2006 to work in the field of microbially enhanced oil recovery and the enzymatic bioremediation of cyclic organochlorine residues.
Rinku Pandey was born in New Delhi, India, in 1977. She attended the University of Delhi, where she obtained a master's degree in zoology specializing in cell and molecular biology in 2000. She joined the group of Professor Rup Lal at the University of Delhi in 2001, where she worked on the molecular analysis of the lin genes involved in hexachlorocyclohexane (HCH) degradation and the phylogenetic analysis and reclassification of the three major HCH-degrading bacteria and in 2005 earned a Ph.D. in molecular biology. She joined CSIRO Entomology as a postdoctoral fellow in 2006 to work in the field of microbially enhanced oil recovery and the enzymatic bioremediation of cyclic organochlorine residues.
 Vishakha Raina completed her master's degree in zoology with a specialization in cell and molecular biology (1997 to 1999) at the University of Delhi. Later, she fulfilled her Ph.D. in molecular biology under Professor Rup Lal at the University of Delhi (1999 to 2004), where she worked on the physiology and genetics of a hexachlorocyclohexane (HCH)-degrading microbe, Sphingobium indicum B90A, and its applications for the bioremediation of HCH-contaminated soils. During this time she visited EPFL, Lausanne, Switzerland (July to December 2001), as a fellow. Later, she joined the group of Dr. Hans-Peter E. Kohler as a postdoctoral fellow at EAWAG (Swiss Federal Institute of Aquatic Science and Technology), Duebendorf, Switzerland (2005 to 2007), where she worked extensively on the biochemistry of HCH isomers. She elucidated the metabolic degradation pathways of all HCH isomers in Sphingobium indicum B90A. She has been working on pesticide degradation for 10 years. Currently, she is an Assistant Professor at KIIT University, Orissa, India.
Vishakha Raina completed her master's degree in zoology with a specialization in cell and molecular biology (1997 to 1999) at the University of Delhi. Later, she fulfilled her Ph.D. in molecular biology under Professor Rup Lal at the University of Delhi (1999 to 2004), where she worked on the physiology and genetics of a hexachlorocyclohexane (HCH)-degrading microbe, Sphingobium indicum B90A, and its applications for the bioremediation of HCH-contaminated soils. During this time she visited EPFL, Lausanne, Switzerland (July to December 2001), as a fellow. Later, she joined the group of Dr. Hans-Peter E. Kohler as a postdoctoral fellow at EAWAG (Swiss Federal Institute of Aquatic Science and Technology), Duebendorf, Switzerland (2005 to 2007), where she worked extensively on the biochemistry of HCH isomers. She elucidated the metabolic degradation pathways of all HCH isomers in Sphingobium indicum B90A. She has been working on pesticide degradation for 10 years. Currently, she is an Assistant Professor at KIIT University, Orissa, India.
 Hans-Peter E. Kohler is a Senior Scientist at the Swiss Federal Institute of Aquatic Science and Technology (EAWAG). He earned an M.S. in biochemistry at the Swiss Federal Institute of Technology (ETHZ), Zürich, Switzerland (1980), and a Ph.D. in microbiology at ETHZ (1986). Since 1998, he has headed the research group Environmental Biochemistry of the Department of Environmental Microbiology at EAWAG. In 1998, he was a visiting research professor at the Department of Microbiology and the Center for Global and Regional Environmental Research (CGRER) of the University of Iowa. His primary research interests include microbial degradation and environmental fate of recalcitrant organic pollutants, biochemistry of mono- and dioxygenases, and microbial transformation of chiral compounds. In particular, elucidation of microbial degradation pathways, isolation and characterization of competent microbes, and identification and characterization of crucial enzymes are his central research themes. The research of Dr. Kohler also aims to develop bioremediation strategies for the decontamination of polluted sites and to promote biotechnological processes for the sustainable production of organic compounds, e.g., β-peptides.
Hans-Peter E. Kohler is a Senior Scientist at the Swiss Federal Institute of Aquatic Science and Technology (EAWAG). He earned an M.S. in biochemistry at the Swiss Federal Institute of Technology (ETHZ), Zürich, Switzerland (1980), and a Ph.D. in microbiology at ETHZ (1986). Since 1998, he has headed the research group Environmental Biochemistry of the Department of Environmental Microbiology at EAWAG. In 1998, he was a visiting research professor at the Department of Microbiology and the Center for Global and Regional Environmental Research (CGRER) of the University of Iowa. His primary research interests include microbial degradation and environmental fate of recalcitrant organic pollutants, biochemistry of mono- and dioxygenases, and microbial transformation of chiral compounds. In particular, elucidation of microbial degradation pathways, isolation and characterization of competent microbes, and identification and characterization of crucial enzymes are his central research themes. The research of Dr. Kohler also aims to develop bioremediation strategies for the decontamination of polluted sites and to promote biotechnological processes for the sustainable production of organic compounds, e.g., β-peptides.
 Christof Holliger was born in Zürich, Switzerland. He studied at the Swiss Federal Institute of Technology in Zürich (ETHZ) and received his diploma in biology in 1984. He obtained his Ph.D. from the Agricultural University, Wageningen, The Netherlands, in 1992, with a thesis on reductive dechlorination by anaerobic bacteria. He was appointed group leader at the Swiss Institute of Aquatic Research (EAWAG) in 1992 and Assistant Professor in 1998 at the Swiss Federal Institute of Technology in Lausanne (EPFL), Switzerland. In 2004, he was promoted to Associate Professor at the same institution. His research interests are the molecular biology, genetics, biochemistry, physiology, as well as microbial ecology of bacteria degrading chlorinated compounds. In addition, the laboratory that he heads investigates and develops processes treating wastewaters, solid wastes, and waste gas with a special emphasis on the microbial consortia involved.
Christof Holliger was born in Zürich, Switzerland. He studied at the Swiss Federal Institute of Technology in Zürich (ETHZ) and received his diploma in biology in 1984. He obtained his Ph.D. from the Agricultural University, Wageningen, The Netherlands, in 1992, with a thesis on reductive dechlorination by anaerobic bacteria. He was appointed group leader at the Swiss Institute of Aquatic Research (EAWAG) in 1992 and Assistant Professor in 1998 at the Swiss Federal Institute of Technology in Lausanne (EPFL), Switzerland. In 2004, he was promoted to Associate Professor at the same institution. His research interests are the molecular biology, genetics, biochemistry, physiology, as well as microbial ecology of bacteria degrading chlorinated compounds. In addition, the laboratory that he heads investigates and develops processes treating wastewaters, solid wastes, and waste gas with a special emphasis on the microbial consortia involved.
 Colin Jackson was born in Hamilton, New Zealand, and received his B.Sc. (Hons) in 2002 from the University of Otago. In 2006 he completed his Ph.D. under the supervision of Professor David Ollis at the Australian National University. He subsequently completed a short postdoctoral fellowship under Dr. John G. Oakeshott at the Commonwealth Scientific and Industrial Research Organization (CSIRO, Australia) before being appointed as an indefinite research scientist at CSIRO. He is currently on leave to take up a Marie Curie Research Fellowship at the Institut de Biologie Structurale in Grenoble, France. He is broadly interested in the structural and chemical determinants of enzyme catalysis and enzyme engineering.
Colin Jackson was born in Hamilton, New Zealand, and received his B.Sc. (Hons) in 2002 from the University of Otago. In 2006 he completed his Ph.D. under the supervision of Professor David Ollis at the Australian National University. He subsequently completed a short postdoctoral fellowship under Dr. John G. Oakeshott at the Commonwealth Scientific and Industrial Research Organization (CSIRO, Australia) before being appointed as an indefinite research scientist at CSIRO. He is currently on leave to take up a Marie Curie Research Fellowship at the Institut de Biologie Structurale in Grenoble, France. He is broadly interested in the structural and chemical determinants of enzyme catalysis and enzyme engineering.
 John G. Oakeshott is the Chief Scientist at the Division of Entomology of Australia's Commonwealth Scientific and Industrial Research Organization (CSIRO). His expertise lies in enzyme biochemistry, molecular biology, and genomics, and he has focused much of his work over the last 20 years on pesticide-degrading enzymes of both prokaryotes and eukaryotes. He has a long-term fundamental interest in molecular evolution and in particular the means by which qualitatively new biochemical functions arise. However, he also has strong applied interests and has led projects to develop free-enzyme bioremediation technologies for removing pesticide residues from contaminated liquids and wettable surfaces. John has published over 170 papers, including over 130 in ISI-listed journals (over 3,500 citations), plus nearly 20 patents/patent applications. John served for many years on Australia's peak gene technology regulatory committees and is a member of the Australian Academy of Technological Sciences and Engineering.
John G. Oakeshott is the Chief Scientist at the Division of Entomology of Australia's Commonwealth Scientific and Industrial Research Organization (CSIRO). His expertise lies in enzyme biochemistry, molecular biology, and genomics, and he has focused much of his work over the last 20 years on pesticide-degrading enzymes of both prokaryotes and eukaryotes. He has a long-term fundamental interest in molecular evolution and in particular the means by which qualitatively new biochemical functions arise. However, he also has strong applied interests and has led projects to develop free-enzyme bioremediation technologies for removing pesticide residues from contaminated liquids and wettable surfaces. John has published over 170 papers, including over 130 in ISI-listed journals (over 3,500 citations), plus nearly 20 patents/patent applications. John served for many years on Australia's peak gene technology regulatory committees and is a member of the Australian Academy of Technological Sciences and Engineering.
REFERENCES
- 1.Alcalde, M., M. Ferrer, F. J. Plou, and A. Ballesteros. 2006. Environmental biocatalysis: from remediation with enzymes to novel green processes. Trends Biotechnol. 24:281-287. [DOI] [PubMed] [Google Scholar]
- 2.Armengaud, J., B. Happe, and K. N. Timmis. 1998. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J. Bacteriol. 180:3954-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arp, D. J., C. M. Yeager, and M. R. Hyman. 2001. Molecular and cellular fundamentals of aerobic cometabolism of trichloroethylene. Biodegradation 12:81-103. [DOI] [PubMed] [Google Scholar]
- 4.Aso, Y., Y. Miyamoto, K. M. Harada, K. Momma, S. Kawai, W. Hashimoto, B. Mikami, and K. Murata. 2006. Engineered membrane superchannel improves bioremediation potential of dioxin-degrading bacteria. Nat. Biotechnol. 24:188-189. [DOI] [PubMed] [Google Scholar]
- 5.Bachmann, A., W. de Bruin, J. C. Jumelet, H. H. Rijnaarts, and A. J. Zehnder. 1988. Aerobic biomineralization of alpha-hexachlorocyclohexane in contaminated soil. Appl. Environ. Microbiol. 54:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachmann, A., P. Walet, P. Wijnen, W. de Bruin, J. L. Huntjens, W. Roelofsen, and A. J. Zehnder. 1988. Biodegradation of alpha- and beta-hexachlorocyclohexane in a soil slurry under different redox conditions. Appl. Environ. Microbiol. 54:143-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bajpai, A., P. Shukla, B. S. Dixit, and R. Banerji. 2007. Concentrations of organochlorine insecticides in edible oils from different regions of India. Chemosphere 67:1403-1407. [DOI] [PubMed] [Google Scholar]
- 8.Bakore, N., P. J. John, and P. Bhatnagar. 2004. Organochlorine pesticide residues in wheat and drinking water samples from Jaipur, Rajasthan, India. Environ. Monit. Assess. 98:381-389. [DOI] [PubMed] [Google Scholar]
- 9.Bala, K., P. Sharma, and R. Lal. 2009. Sphingobium quisquiliarum sp. nov., a hexachlorocyclohexane (HCH) degrading bacterium isolated from HCH contaminated soil. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] doi: 10.1099/ijs.0.010868-0. [DOI] [PubMed]
- 10.Basta, T., A. Keck, J. Klein, and A. Stolz. 2004. Detection and characterization of conjugative degradative plasmids in xenobiotic-degrading Sphingomonas strains. J. Bacteriol. 186:3862-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beland, F. A., S. O. Farwell, A. E. Robocker, and R. D. Geer. 1976. Electrochemical reduction and anaerobic degradation of lindane. J. Agric. Food Chem. 24:753-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benezet, H. J., and F. Matsumura. 1973. Isomerization of γ-BHC to α-BHC in the environment. Nature 243:480-481. [Google Scholar]
- 13.Benimeli, C. S., G. R. Castro, A. P. Chaile, and M. J. Amoroso. 2006. Lindane removal induction by Streptomyces sp. M7. J. Basic Microbiol. 46:348-357. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Michael, E., F. Grauer, C. Raphael, Z. Sahm, and E. D. Richter. 1999. Organochlorine insecticide and PCB residues in fat tissue of autopsied trauma victims in Israel 1984 to 1986. J. Environ. Pathol. Toxicol. Oncol. 18:297-303. [PubMed] [Google Scholar]
- 15.Bhatnagar, V. K., R. Kashyap, S. S. Zaidi, P. K. Kulkarni, and H. N. Saiyed. 2004. Levels of DDT, HCH, and HCB residues in human blood in Ahmedabad, India. Bull. Environ. Contam. Toxicol. 72:261-265. [DOI] [PubMed] [Google Scholar]
- 16.Bidlan, R., M. Afsar, and H. K. Manonmani. 2004. Bioremediation of HCH-contaminated soil: elimination of inhibitory effects of the insecticide on radish and green gram seed germination. Chemosphere 56:803-811. [DOI] [PubMed] [Google Scholar]
- 17.Blasco, C., C. M. Lino, Y. Pico, A. Pena, G. Font, and M. I. Silveira. 2004. Determination of organochlorine pesticide residues in honey from the central zone of Portugal and the Valencian community of Spain. J. Chromatogr. A 1049:155-160. [PubMed] [Google Scholar]
- 18.Bohac, M., Y. Nagata, Z. Prokop, M. Prokop, M. Monincova, M. Tsuda, J. Koca, and J. Damborsky. 2002. Halide-stabilizing residues of haloalkane dehalogenases studied by quantum mechanic calculations and site-directed mutagenesis. Biochemistry 41:14272-14280. [DOI] [PubMed] [Google Scholar]
- 19.Böltner, D., P. Godoy, J. Muρoz-Rojas, E. Duque, S. Moreno-Morillas, L. Sánchez, and J. L. Ramos. 2007. Rhizoremediation of lindane by root-colonizing Sphingomonas. Microb. Biotechnol. 1:87-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Böltner, D., S. Moreno-Morillas, and J. L. Ramos. 2005. 16S rDNA phylogeny and distribution of lin genes in novel hexachlorocyclohexane-degrading Sphingomonas strains. Environ. Microbiol. 7:1329-1338. [DOI] [PubMed] [Google Scholar]
- 21.Boyle, A. W., M. M. Haggblom, and L. Y. Young. 1999. Dehalogenation of lindane (γ-hexachlorocyclohexane) by anaerobic bacteria from marine sediments and by sulfate-reducing bacteria. FEMS Microbiol. Ecol. 29:379-387. [Google Scholar]
- 22.Briseno-Roa, L., J. Hill, S. Notman, D. Sellers, A. P. Smith, C. M. Timperley, J. Wetherell, N. H. Williams, G. R. Williams, A. R. Fersht, and A. D. Griffiths. 2006. Analogues with fluorescent leaving groups for screening and selection of enzymes that efficiently hydrolyze organophosphorus nerve agents. J. Med. Chem. 49:246-255. [DOI] [PubMed] [Google Scholar]
- 23.Brown, A. W. A. 1978. Ecology of pesticides. John Wiley & Sons, New York, NY.
- 24.Buser, H. R., and M. D. Muller. 1995. Isomer and enantioselective degradation of hexachlorocyclohexane isomers in sewage sludge under anaerobic conditions. Environ. Sci. Technol. 29:664-672. [DOI] [PubMed] [Google Scholar]
- 25.Cai, M., and L. Xun. 2002. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J. Bacteriol. 184:4672-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvelo-Pereira, R., M. Camps-Arbestain, B. R. Garrido, F. Macias, and C. Monterroso. 2006. Behaviour of α-, β-, γ- and δ- hexachlorocylcohexane in the soil-plant system of a contaminated site. Environ. Pollut. 144:210-217. [DOI] [PubMed] [Google Scholar]
- 27.Cérémonie, H., H. Boubakri, P. Mavingui, P. Simonet, and T. M. Vogel. 2006. Plasmid-encoded γ-hexachlorocyclohexane degradation genes and insertion sequences in Sphingobium francense (ex-Sphingomonas paucimobilis Sp+). FEMS Microbiol. Lett. 257:243-252. [DOI] [PubMed] [Google Scholar]
- 28.Cheesman, M. J., I. Horne, K. M. Weir, G. Pandey, M. R. WilIiams, C. Scott, R. J. Russell, and J. G. Oakeshott. 2007. Carbamate pesticides and their biological degradation: prospects for enzymatic bioremediation. Am. Chem. Soc. Symp. Ser. 966:288-305. [Google Scholar]
- 29.Concha-Graña, E., M. I. Turnes-Carou, S. Muniategui-Lorenzo, P. López-Mahia, D. Prada-Rodriguez, and E. Fernández-Fernández. 2006. Evaluation of HCH isomers and metabolites in soils, leachates, river water and sediments of a highly contaminated area. Chemosphere 64:588-595. [DOI] [PubMed] [Google Scholar]
- 30.Dadhwal, M., S. Jit, H. Kumari, and R. Lal. 2009. Sphingobium chinhatense sp. nov., a hexachlorocyclohexane (HCH)-degrading bacterium isolated from an HCH dumpsite. Int. J. Syst. Evol. Microbiol. 59:3140-3144. [DOI] [PubMed] [Google Scholar]
- 31.Dadhwal, M., A. Singh, O. Prakash, S. K. Gupta, K. Kumari, P. Sharma, S. Jit, M. Verma, C. Holliger, and R. Lal. 2009. Proposal of biostimulation for hexachlorocyclohexane (HCH)-decontamination and characterization of culturable bacterial community from high-dose point HCH-contaminated soils. J. Appl. Microbiol. 106:381-392. [DOI] [PubMed] [Google Scholar]
- 32.Damborsky, J., and J. Koca. 1999. Analysis of the reaction mechanism and substrate specificity of haloalkane dehalogenases by sequential and structural comparisons. Protein Eng. 12:989-998. [DOI] [PubMed] [Google Scholar]
- 33.Damborsky, J., E. Rorije, A. Jesenska, Y. Nagata, G. Klopman, and W. J. Peijnenburg. 2001. Structure-specificity relationships for haloalkane dehalogenases. Environ. Toxicol. Chem. 20:2681-2689. [PubMed] [Google Scholar]
- 34.Datta, J., A. K. Maiti, D. P. Modak, P. K. Chakrabartty, P. Bhattacharyya, and P. K. Ray. 2000. Metabolism of γ-hexachlorocyclohexane by Arthrobacter citreus strain BI-100: identification of metabolites. J. Gen. Appl. Microbiol. 46:59-67. [DOI] [PubMed] [Google Scholar]
- 35.Dawson, R. M., S. Pantelidis, H. R. Rose, and S. E. Kotsonis. 2008. Degradation of nerve agents by an organophosphate-degrading agent (OpdA). J. Hazard. Mater. 157:308-314. [DOI] [PubMed] [Google Scholar]
- 36.Deeb, R. A., and L. Alvarez-Cohen. 2000. Aerobic biotransformation of gasoline aromatics in multicomponent mixtures. Bioremed. J. 4:171-179. [Google Scholar]
- 37.Deeb, R. A., J. C. Spain, and L. Alvarez-Cohen. 1999. Mineralization of benzene, toluene, ethylbenzene, m-xylene, and p-xylene by two Rhodococcus species, p. 553. Abstr. 99th Gen. Meet. Am. Soc. Microbiol., Chicago, IL, 30 May to 3 June 1999.
- 38.de Oliveira, R. M., L. H. P. Bastos, A. E. de Oliveira e Dias, S. A. da Silva, and J. C. Moreira. 2003. Residual concentration of hexachlorocyclohexane in a contaminated site in Cidade dos Meninos, Duque de Caxias, Rio de Janeiro, Brazil, after calcium oxide treatment. Cad. Saúde Pública 19:447-453. [DOI] [PubMed] [Google Scholar]
- 39.Doelman, P., L. Haanstra, E. de Ruiter, and J. Slange. 1985. Rate of microbial degradation of high concentration of α-hexachlorocyclohexane in soil under aerobic and anaerobic conditions. Chemosphere 14:565-570. [Google Scholar]
- 40.Dogra, C., V. Raina, R. Pal, M. Suar, S. Lal, K. H. Gartemann, C. Holliger, J. R. van der Meer, and R. Lal. 2004. Organization of lin genes and IS6100 among different strains of hexachlorocyclohexane-degrading Sphingomonas paucimobilis: evidence for horizontal gene transfer. J. Bacteriol. 186:2225-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edwards, E. A., and D. Grbic-Galic. 1992. Complete mineralization of benzene by aquifer microorganisms under strictly anaerobic conditions. Appl. Environ. Microbiol. 58:2663-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Endo, R., M. Kamakura, K. Miyauchi, M. Fukuda, Y. Ohtsubo, M. Tsuda, and Y. Nagata. 2005. Identification and characterization of genes involved in the downstream degradation pathway of γ-hexachlorocyclohexane in Sphingomonas paucimobilis UT26. J. Bacteriol. 187:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Endo, R., Y. Ohtsubo, M. Tsuda, and Y. Nagata. 2007. Identification and characterization of genes encoding a putative ABC-type transporter essential for utilization of γ-hexachlorocyclohexane in Sphingobium japonicum UT26. J. Bacteriol. 189:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erkmen, B., and D. Kolankaya. 2006. Determination of organochlorine pesticide residues in water, sediments and fish samples from the Meric Delta Turkey. Int. J. Environ. Anal. Chem. 86:161-169. [Google Scholar]
- 45.Fairlee, J. R., B. L. Burback, and J. J. Perry. 1997. Biodegradation of groundwater pollutants by a combined culture of Mycobacterium vaccae and a Rhodococcus sp. Can. J. Microbiol. 43:841-846. [DOI] [PubMed] [Google Scholar]
- 46.Francis, A. J., R. J. Spanggord, and G. I. Ouchi. 1975. Degradation of lindane by Escherichia coli. Appl. Microbiol. 29:567-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuchu Genki, Y. O., M. Ito, R. Miyazaki, A. Ono, Y. Nagata, and M. Tsuda. 2008. Insertion sequence-based cassette PCR: cultivation-independent isolation of γ-hexachlorocyclohexane-degrading genes from soil DNA. Appl. Microbiol. Biotechnol. 79:627-632. [DOI] [PubMed] [Google Scholar]
- 48.Gangwar, M. P., B. D. Lakhchaura, and S. C. Sud. 1979. In vitro studies on the effect of certain pesticides on rumen microorganisms of the Buffalo. Indian Vet. J. 56:1001-1006. [PubMed] [Google Scholar]
- 49.Golfinopoulos, S. K., A. D. Nikolaou, M. N. Kostopoulou, N. K. Xilourgidis, M. C. Vagi, and D. T. Lekkas. 2003. Organochlorine pesticides in the surface waters of Northern Greece. Chemosphere 50:507-516. [DOI] [PubMed] [Google Scholar]
- 50.Gong, Z. M., F. L. Xu, R. Dawson, J. Cao, W. X. Liu, B. G. Li, W. R. Shen, W. J. Zhang, B. P. Qin, R. Sun, and S. Tao. 2004. Residues of hexachlorocyclohexane isomers and their distribution characteristics in soils in the Tianjin area, China. Arch. Environ. Contam. Toxicol. 46:432-437. [DOI] [PubMed] [Google Scholar]
- 51.Gupta, A., C. P. Kaushik, and A. Kaushik. 2000. Degradation of hexachlorocyclohexane (HCH; α, β, γ and δ) by Bacillus circulans and Bacillus brevis isolated from soil contaminated with HCH. Soil Biol. Biochem. 32:1803-1805. [Google Scholar]
- 52.Gupta, A., C. P. Kaushik, and A. Kaushik. 2001. Degradation of hexachlorocyclohexane isomers by two strains of Alcaligenes faecalis isolated from a contaminated site. Bull. Environ. Contam. Toxicol. 66:794-800. [DOI] [PubMed] [Google Scholar]
- 53.Harayama, S., and M. Rekik. 1990. The meta cleavage operon of TOL degradative plasmid pWW0 comprises 13 genes. Mol. Gen. Genet. 221:113-120. [DOI] [PubMed] [Google Scholar]
- 54.Reference deleted.
- 55.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 56.Hegeman, W. J., and R. W. Laane. 2002. Enantiomeric enrichment of chiral pesticides in environment. Rev. Environ. Contam. Toxicol. 173:85-116. [PubMed] [Google Scholar]
- 57.Helm, P. A., M. L. Diamond, R. Semkin, and T. F. Bidleman. 2000. Degradation as a loss mechanism in the fate of α-hexachlorocyclohexane in Arctic watersheds. Environ. Sci. Technol. 34:812-818. [Google Scholar]
- 58.Heritage, A. D., and I. C. MacRae. 1977. Degradation of lindane by cell-free preparations of Clostridium sphenoides. Appl. Environ. Microbiol. 34:222-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horne, I., X. Qiu, R. J. Russell, and J. G. Oakeshott. 2003. The phosphotriesterase gene opdA in Agrobacterium radiobacter P230 is transposable. FEMS Microbiol. Lett. 222:1-8. [DOI] [PubMed] [Google Scholar]
- 60.Hynkova, K., Y. Nagata, M. Takagi, and J. Damborsky. 1999. Identification of the catalytic triad in the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. FEBS Lett. 446:177-181. [DOI] [PubMed] [Google Scholar]
- 61.Imai, R., Y. Nagata, M. Fukuda, M. Takagi, and K. Yano. 1991. Molecular cloning of a Pseudomonas paucimobilis gene encoding a 17-kilodalton polypeptide that eliminates HCl molecules from γ-hexachlorocyclohexane. J. Bacteriol. 173:6811-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ito, M., Z. Prokop, M. Klvana, Y. Otsubo, M. Tsuda, J. Damborsky, and Y. Nagata. 2007. Degradation of beta-hexachlorocyclohexane by haloalkane dehalogenase LinB from gamma-hexachlorocyclohexane-utilizing bacterium Sphingobium sp. MI1205. Arch. Microbiol. 188:313-325. [DOI] [PubMed] [Google Scholar]
- 63.Iwata, H., S. Tanabe, N. Sakai, A. Nishimura, and R. Tatsukawa. 1994. Geographical distribution of persistent organochlorines in air, water and sediments from Asia and Oceania, and their implications for global distribution from lower latitudes. Environ. Pollut. 85:15-33. [DOI] [PubMed] [Google Scholar]
- 64.Jagnow, G., K. Haider, and P. C. Ellwardt. 1977. Anaerobic dechlorination and degradation of hexachlorocyclohexane isomers by anaerobic and facultative anaerobic bacteria. Arch. Microbiol. 115:285-292. [DOI] [PubMed] [Google Scholar]
- 65.Janssen, D. B. 2004. Evolving haloalkane dehalogenases. Curr. Opin. Chem. Biol. 8:150-159. [DOI] [PubMed] [Google Scholar]
- 66.Johri, A. K., M. Dua, D. Tuteja, D. M. Saxena, and R. Lal. 1998. Degradation of alpha, beta, gamma and delta-hexachlorocyclohexanes by Sphingobium paucimobilis. Biotechnol. Lett. 20:2347-2353. [Google Scholar]
- 67.Jornvall, H., B. Persson, M. Krook, S. Atrian, R. Gonzalez-Duarte, J. Jeffery, and D. Ghosh. 1995. Short-chain dehydrogenases/reductases (SDR). Biochemistry 34:6003-6013. [DOI] [PubMed] [Google Scholar]
- 68.Jürgen, H. J., and R. Roth. 1989. Case study and proposed decontamination steps of the soil and groundwater beneath a closed herbicide plant in Germany. Chemosphere 18:1163-1169. [Google Scholar]
- 69.Kmunicek, J., K. Hynkova, T. Jedlicka, Y. Nagata, A. Negri, F. Gago, R. C. Wade, and J. Damborsky. 2005. Quantitative analysis of substrate specificity of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26. Biochemistry 44:3390-3401. [DOI] [PubMed] [Google Scholar]
- 70.Kumar, M., P. Chaudhary, M. Dwivedi, R. Kumar, D. Paul, R. K. Jain, S. K. Garg, and A. Kumar. 2005. Enhanced biodegradation of β- and δ-hexachlorocyclohexane in the presence of α- and γ-isomers in contaminated soils. Environ. Sci. Technol. 39:4005-4011. [DOI] [PubMed] [Google Scholar]
- 71.Kumari, B., V. K. Madan, and T. S. Kathpal. 2007. Status of insecticide contamination of soil and water in Haryana, India. Environ. Monit. Assess. 136:239-244. [DOI] [PubMed] [Google Scholar]
- 72.Kumari, R., S. Subudhi, M. Suar, G. Dhingra, V. Raina, C. Dogra, S. Lal, J. R. van der Meer, C. Holliger, and R. Lal. 2002. Cloning and characterization of lin genes responsible for the degradation of hexachlorocyclohexane isomers by Sphingomonas paucimobilis strain B90. Appl. Environ. Microbiol. 68:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kutz, F. W., P. H. Wood, and D. P. Bottimore. 1991. Organochlorine pesticides and polychlorinated biphenyls in human adipose tissue. Rev. Environ. Contam. Toxicol. 120:1-82. [DOI] [PubMed] [Google Scholar]
- 74.Lal, R., M. Dadhwal, K. Kumari, P. Sharma, A. Singh, H. Kumari, S. Jit, S. K. Gupta, A. Nigam, D. Lal, M. Verma, J. Kaur, K. Bala, and S. Jindal. 2008. Pseudomonas sp. to Sphingobium indicum: a journey of microbial degradation and bioremediation of hexachlorocyclohexane. Indian J. Microbiol. 48:3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lal, R., C. Dogra, S. Malhotra, P. Sharma, and R. Pal. 2006. Diversity, distribution and divergence of lin genes in hexachlorocyclohexane-degrading sphingomonads. Trends Biotechnol. 24:121-130. [DOI] [PubMed] [Google Scholar]
- 76.Lammel, G., Y. S. Ghimb, A. Gradosa, H. Gaod, H. Hühnerfussc, and R. Lohmann. 2007. Levels of persistent organic pollutants in air in China and over the Yellow Sea. Atmos. Environ. 41:452-464. [Google Scholar]
- 77.Langenhoff, A. A. M., A. A. M. Nipshagen, C. Bakker, J. Krooneman, and G. Visscher. 2001. Monitoring stimulated reductive dechlorination at the Rademarkt in Groningen, The Netherlands. Battelle Press, San Diego, CA.
- 78.Law, S. A., T. F. Bidleman, M. J. Martin, and M. V. Ruby. 2004. Evidence of enantioselective degradation of α-hexachlorocyclohexane in groundwater. Environ. Sci. Technol. 38:1633-1638. [DOI] [PubMed] [Google Scholar]
- 79.Li, J., T. Zhu, F. Wang, X. H. Qiu, and W. L. Lin. 2006. Observation of organochlorine pesticides in the air of the Mt. Everest region. Ecotoxicol. Environ. Safe. 63:33-41. [DOI] [PubMed] [Google Scholar]
- 80.Li, Y. F. 1999. Global technical hexachlorocyclohexane usage and its contamination consequences in the environment: from 1948 to 1997. Sci. Total Environ. 232:121-158. [Google Scholar]
- 81.Liu, H., J. J. Zhang, S. J. Wang, X. E. Zhang, and N. Y. Zhou. 2005. Plasmid-borne catabolism of methyl parathion and p-nitrophenol in Pseudomonas sp. strain WBC-3. Biochem. Biophys. Res. Commun. 334:1107-1114. [DOI] [PubMed] [Google Scholar]
- 82.Lodha, B., P. Bhat, M. S. Kumar, A. N. Vaidya, S. Mudliar, D. J. Killedar, and T. Chakrabarti. 2007. Bioisomerization kinetics of γ-HCH and biokinetics of Pseudomonas aeruginosa degrading technical HCH. Biochem. Eng. J. 35:12-19. [Google Scholar]
- 83.Ma, A. Z., J. Wu, G. S. Zhang, T. Wang, and S. P. Li. 2005. Isolation and characterization of a HCH degradation Sphingomanas sp. strain BHC-A. Acta Microbiol. Sin. 45:728-732. [PubMed] [Google Scholar]
- 84.MacRae, I. C., K. Raghu, and E. M. Bautista. 1969. Anaerobic degradation of the insecticide lindane by Clostridium sp. Nature 221:859-860. [DOI] [PubMed] [Google Scholar]
- 85.MacRae, I. C., K. Raghu, and T. F. Castro. 1967. Persistence and biodegradation of four common isomers of benzene hexachloride in submerged soils. J. Agric. Food Chem. 15:911-914. [Google Scholar]
- 86.Malhotra, S., P. Sharma, H. Kumari, A. Singh, and R. Lal. 2007. Localization of HCH catabolic genes (lin genes) in Sphingobium indicum B90A. Indian J. Microbiol. 47:271-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Malik, A., K. P. Singh, and P. Ojha. 2007. Residues of organochlorine pesticides in fish from the Gomti river, India. Bull. Environ. Contam. Toxicol. 78:335-340. [DOI] [PubMed] [Google Scholar]
- 88.Manickam, N., M. Mau, and M. Schlömann. 2006. Characterization of the novel HCH-degrading strain, Microbacterium sp. ITRC1. Appl. Microbiol. Biotechnol. 69:580-588. [DOI] [PubMed] [Google Scholar]
- 89.Manickam, N., R. Misra, and S. Mayilraj. 2007. A novel pathway for the biodegradation of γ-hexachlorocyclohexane by a Xanthomonas sp. strain ICH12. J. Appl. Microbiol. 102:1468-1478. [DOI] [PubMed] [Google Scholar]
- 90.Manickam, N., M. K. Reddy, H. S. Saini, and R. Shanker. 2007. Isolation of hexachlorocyclohexane-degrading Sphingomonas sp. by dehalogenase assay and characterization of genes involved in γ-HCH degradation. J. Appl. Microbiol. 104:952-960. [DOI] [PubMed] [Google Scholar]
- 91.Manonmani, H. K., D. H. Chandrashekaraiah, N. S. Reddy, C. D. Elcey, and A. A. Kunhi. 2000. Isolation and acclimation of a microbial consortium for improved aerobic degradation of α-hexachlorocyclohexane. J. Agric. Food Chem. 48:4341-4351. [DOI] [PubMed] [Google Scholar]
- 92.Marek, J., J. Vevodova, I. K. Smatanova, Y. Nagata, L. A. Svensson, J. Newman, M. Takagi, and J. Damborsky. 2000. Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. Biochemistry 39:14082-14086. [DOI] [PubMed] [Google Scholar]
- 93.Mars, A. E., T. Kasberg, S. R. Kaschabek, M. H. van Agteren, D. B. Janssen, and W. Reineke. 1997. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J. Bacteriol. 179:4530-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Masai, E., S. Shinohara, H. Hara, S. Nishikawa, Y. Katayama, and M. Fukuda. 1999. Genetic and biochemical characterization of a 2-pyrone-4,6-dicarboxylic acid hydrolase involved in the protocatechuate 4,5-cleavage pathway of Sphingomonas paucimobilis SYK-6. J. Bacteriol. 181:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mencía, A. M., A. I. Martínez-Ferri, M. Alcalde, and V. De Lorenzo. 2006. Identification of a γ-hexachlorocyclohexane dehydrochlorinase (LinA) variant with improved expression and solubility properties. Biocat. Biotrans. 24:223-230. [Google Scholar]
- 96.Mertens, B., N. Boon, and W. Verstraete. 2006. Slow-release inoculation allows sustained biodegradation of gamma-hexachlorocyclohexane. Appl. Environ. Microbiol. 72:622-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Middeldorp, P. J., W. van Doesburg, G. Schraa, and A. J. Stams. 2005. Reductive dechlorination of hexachlorocyclohexane (HCH) isomers in soil under anaerobic conditions. Biodegradation 16:283-290. [DOI] [PubMed] [Google Scholar]
- 98.Middeldorp, P. J., M. Jaspers, A. J. B. Zehnder, and G. Schraa. 1996. Biotransformation of α-, β-, γ-, and δ-hexachlorocyclohexane under methanogenic conditions. Environ. Sci. Technol. 30:2345-2349. [Google Scholar]
- 99.Mishra V., R. Lal, and Srinivasan. 2001. Enzymes and operons mediating xenobiotic degradation in bacteria. Crit. Rev. Microbiol. 27:133-166. [DOI] [PubMed] [Google Scholar]
- 100.Miyauchi, K., Y. Adachi, Y. Nagata, and M. Takagi. 1999. Cloning and sequencing of a novel meta-cleavage dioxygenase gene whose product is involved in degradation of γ-hexachlorocyclohexane in Sphingomonas paucimobilis. J. Bacteriol. 181:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Miyauchi, K., H. S. Lee, M. Fukuda, M. Takagi, and Y. Nagata. 2002. Cloning and characterization of linR, involved in regulation of the downstream pathway for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 68:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Miyauchi, K., S. K. Suh, Y. Nagata, and M. Takagi. 1998. Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene whose product is involved in degradation of γ-hexachlorocyclohexane by Sphingomonas paucimobilis. J. Bacteriol. 180:1354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Miyazaki, R., Y. Sato, M. Ito, Y. Ohtsubo, Y. Nagata, and M. Tsuda. 2006. Complete nucleotide sequence of an exogenously isolated plasmid, pLB1, involved in γ-hexachlorocyclohexane degradation. Appl. Environ. Microbiol. 72:6923-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mohn, W. W., J. Garmendia, T. C. Galvao, and V. de Lorenzo. 2006. Surveying biotransformations with a la carte genetic traps: translating dehydrochlorination of lindane (γ-hexachlorocyclohexane) into lacZ-based phenotypes. Environ. Microbiol. 8:546-555. [DOI] [PubMed] [Google Scholar]
- 105.Mohn, W. W., B. Mertens, J. D. Neufeld, W. Verstraete, and V. de Lorenzo. 2006. Distribution and phylogeny of hexachlorocyclohexane-degrading bacteria in soils from Spain. Environ. Microbiol. 8:60-68. [DOI] [PubMed] [Google Scholar]
- 106.Nagasawa, S., R. Kikuchi, and M. Matsuo. 1993. Indirect identification of an unstable intermediate in γ-HCH degradation by Pseudomonas paucimobilis UT26. Chemosphere 26:2279-2288. [Google Scholar]
- 107.Nagasawa, S., R. Kikuchi, Y. Nagata, M. Takagi, and M. Matsuo. 1993. Aerobic mineralization of γ-HCH by Pseudomonas paucimobilis UT26. Chemosphere 26:1719-1728. [Google Scholar]
- 108.Nagasawa, S., R. Kikuchi, Y. Nagata, M. Takagi, and M. Matsuo. 1993. Stereochemical analysis of γ-HCH degradation by Pseudomonas paucimobilis UT26. Chemosphere 26:1187-1201. [Google Scholar]
- 109.Nagata, Y., R. Endo, M. Ito, Y. Ohtsubo, and M. Tsuda. 2007. Aerobic degradation of lindane (γ-hexachlorocyclohexane) in bacteria and its biochemical and molecular basis. Appl. Microbiol. Biotechnol. 76:741-752. [DOI] [PubMed] [Google Scholar]
- 110.Nagata, Y., A. Futamura, K. Miyauchi, and M. Takagi. 1999. Two different types of dehalogenases, LinA and LinB, involved in γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26 are localized in the periplasmic space without molecular processing. J. Bacteriol. 181:5409-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nagata, Y., T. Hatta, R. Imai, K. Kimbara, M. Fukuda, K. Yano, and M. Takagi. 1993. Purification and characterization of γ-hexachlorocyclohexane (γ-HCH) dehydrochlorinase (LinA) from Pseudomonas paucimobilis. Biosci. Biotechnol. Biochem. 59:1582-1583. [DOI] [PubMed] [Google Scholar]
- 112.Nagata, Y., R. Imai, A. Sakai, M. Fukuda, K. Yano, and M. Takagi. 1993. Isolation and characterization of Tn5-induced mutants of Pseudomonas paucimobilis UT26 defective in γ-hexachlorocyclohexane dehydrochlorinase (LinA). Biosci. Biotechnol. Biochem. 57:703-709. [DOI] [PubMed] [Google Scholar]
- 113.Nagata, Y., M. Kamakura, R. Endo, R. Miyazaki, Y. Ohtsubo, and M. Tsuda. 2006. Distribution of γ-hexachlorocyclohexane-degrading genes on three replicons in Sphingobium japonicum UT26. FEMS Microbiol. Lett. 256:112-118. [DOI] [PubMed] [Google Scholar]
- 114.Nagata, Y., K. Miyauchi, J. Damborsky, K. Manova, A. Ansorgova, and M. Takagi. 1997. Purification and characterization of a haloalkane dehalogenase of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 63:3707-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nagata, Y., K. Miyauchi, and M. Takagi. 1999. Complete analysis of genes and enzymes for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. J. Ind. Microbiol. Biotechnol. 23:380-390. [DOI] [PubMed] [Google Scholar]
- 116.Nagata, Y., K. Mori, M. Takagi, A. G. Murzin, and J. Damborsky. 2001. Identification of protein fold and catalytic residues of γ-hexachlorocyclohexane dehydrochlorinase LinA. Proteins 45:471-477. [DOI] [PubMed] [Google Scholar]
- 117.Nagata, Y., T. Nariya, R. Ohtomo, M. Fukuda, K. Yano, and M. Takagi. 1993. Cloning and sequencing of a dehalogenase gene encoding an enzyme with hydrolase activity involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 175:6403-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nagata, Y., R. Ohtomo, K. Miyauchi, M. Fukuda, K. Yano, and M. Takagi. 1994. Cloning and sequencing of a 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase gene involved in the degradation of γ-hexachlorocyclohexane in Pseudomonas paucimobilis. J. Bacteriol. 176:3117-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nagata, Y., Z. Prokop, Y. Sato, P. Jerabek, A. Kumar, Y. Ohtsubo, M. Tsuda, and J. Damborsky. 2005. Degradation of β-hexachlorocyclohexane by haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 71:2183-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nalin, R., P. Simonet, T. M. Vogel, and P. Normand. 1999. Rhodanobacter lindaniclasticus gen. nov., sp. nov., a lindane-degrading bacterium. Int. J. Syst. Bacteriol. 49:19-23. [DOI] [PubMed] [Google Scholar]
- 121.Nawab, A., A. Aleem, and A. Malik. 2003. Determination of organochlorine pesticides in agricultural soil with special reference to γ-HCH degradation by Pseudomonas strains. Bioresour. Technol. 88:41-46. [DOI] [PubMed] [Google Scholar]
- 122.Negri, A., E. Marco, J. Damborsky, and F. Gago. 2007. Stepwise dissection and visualization of the catalytic mechanism of haloalkane dehalogenase LinB using molecular dynamics simulations and computer graphics. J. Mol. Graph. Model. 26:643-651. [DOI] [PubMed] [Google Scholar]
- 123.Oakley, A. J., Z. Prokop, M. Bohac, J. Kmunicek, T. Jedlicka, M. Monincova, I. Kuta-Smatanova, Y. Nagata, J. Damborsky, and M. C. Wilce. 2002. Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product- and water-mediated inhibition. Biochemistry 41:4847-4855. [DOI] [PubMed] [Google Scholar]
- 124.Ogram, A. V., Y. P. Duan, and S. L. Trabue. 2000. Carbofuran degradation mediated by three related plasmid systems. FEMS Microbiol. Ecol. 32:197-203. [DOI] [PubMed] [Google Scholar]
- 125.Ohisa, N., and M. Yamaguchi. 1978. Gamma-BHC degradation accompanied by the growth of Clostridium rectum isolated from paddy field soil. Agric. Biol. Chem. 42:1819-1823. [Google Scholar]
- 126.Ohisa, N., M. Yamaguchi, and N. Kurihara. 1980. Lindane degradation by cell-free extracts of Clostridium rectum. Arch. Microbiol. 125:221-225. [DOI] [PubMed] [Google Scholar]
- 127.Okai, M., K. Kubota, M. Fukuda, Y. Nagata, K. Nagata, and M. Tanokura. 2009. Crystallization and preliminary X-ray analysis of γ-hexachlorocyclohexane dehydrochlorinase LinA from Sphingobium japonicum UT26. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:822-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Okeke, B. C., T. Siddique, M. C. Arbestain, and W. T. Frankenberger. 2002. Biodegradation of γ-hexachlorocyclohexane (lindane) and α-hexachlorocyclohexane in water and a soil slurry by a Pandoraea species. J. Agric. Food Chem. 50:2548-2555. [DOI] [PubMed] [Google Scholar]
- 129.Osterreicher-Cunha, P., T. Langenbach, J. P. Torres, A. L. Lima, T. M. de Campos, E. A. Vargas, Jr., and A. R. Wagener. 2003. HCH distribution and microbial parameters after liming of a heavily contaminated soil in Rio de Janeiro. Environ. Res. 93:316-327. [DOI] [PubMed] [Google Scholar]
- 130.Otyepka, M., P. Banás, A. Magistrato, P. Carloni, and J. Damborský. 2008. Second step of hydrolytic dehalogenation in haloalkane dehalogenase investigated by QM/MM methods. Proteins 70:707-717. [DOI] [PubMed] [Google Scholar]
- 131.Padma, T. V., R. M. Dickhut, and H. Ducklow. 2003. Variations in α-hexachlorocyclohexane enantiomer ratios in relation to microbial activity in a temperate estuary. Environ. Toxicol. Chem. 22:1421-1427. [PubMed] [Google Scholar]
- 132.Pal, R., S. Bala, M. Dadhwal, M. Kumar, G. Dhingra, O. Prakash, S. R. Prabagaran, S. Shivaji, J. Cullum, C. Holliger, and R. Lal. 2005. Hexachlorocyclohexane-degrading bacterial strains Sphingomonas paucimobilis B90A, UT26 and Sp+, having similar lin genes, represent three distinct species, Sphingobium indicum sp. nov., Sphingobium japonicum sp. nov. and Sphingobium francense sp. nov., and reclassification of [Sphingomonas] chungbukensis as Sphingobium chungbukense comb. nov. Int. J. Syst. Evol. Microbiol. 55:1965-1972. [DOI] [PubMed] [Google Scholar]
- 133.Persson, B., M. Krook, and H. Jornvall. 1991. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur. J. Biochem. 200:537-543. [DOI] [PubMed] [Google Scholar]
- 134.Phillips, T. M., H. Lee, J. T. Trevors, and A. G. Seech. 2004. Mineralization of hexachlorocyclohexane in soil during solid-phase bioremediation. J. Ind. Microbiol. Biotechnol. 31:216-222. [DOI] [PubMed] [Google Scholar]
- 135.Phillips, T. M., H. Lee, J. T. Trevors, and A. G. Seech. 2006. Full-scale in situ bioremediation of hexachlorocyclohexane-contaminated soil. J. Chem. Technol. Biotechnol. 81:289-298. [Google Scholar]
- 136.Phillips, T. M., A. G. Seech, H. Lee, and J. T. Trevors. 2001. Colorimetric assay for lindane dechlorination by bacteria. J. Microbiol. Methods 47:181-188. [DOI] [PubMed] [Google Scholar]
- 137.Phillips, T. M., A. G. Seech, H. Lee, and J. T. Trevors. 2005. Biodegradation of hexachlorocyclohexane (HCH) by microorganisms. Biodegradation 16:363-392. [DOI] [PubMed] [Google Scholar]
- 138.Popp, P., L. Bruggemann, P. Keil, U. Thuss, and H. Weiss. 2000. Chlorobenzenes and hexachlorocyclohexanes (HCHs) in the atmosphere of Bitterfeld and Leipzig (Germany). Chemosphere 41:849-855. [DOI] [PubMed] [Google Scholar]
- 139.Prakash, O., M. Suar, V. Raina, C. Dogra, R. Pal, and R. Lal. 2004. Residues of hexachlorocyclohexane isomers in soil and water samples from Delhi and adjoining areas. Curr. Sci. 87:73-77. [Google Scholar]
- 140.Quintero, J. C., M. T. Moreira, G. Feijoo, and J. M. Lema. 2005. Effect of surfactants on the soil desorption of hexachlorocyclohexane (HCH) isomers and their anaerobic biodegradation. J. Chem. Technol. Biotechnol. 80:1005-1015. [Google Scholar]
- 141.Quintero, J. C., M. T. Moreira, G. Feijoo, and J. M. Lema. 2005. Anaerobic degradation of hexachlorocyclohexane isomers in liquid and soil slurry systems. Chemosphere 61:528-536. [DOI] [PubMed] [Google Scholar]
- 142.Raina, V., A. Hauser, H. R. Buser, D. Rentsch, P. Sharma, R. Lal, C. Holliger, J. T. Poiger, M. D. Muller, and H. P. E. Kohler. 2007. Hydroxylated metabolites of β- and δ-hexachlorocyclohexane: bacterial formation, stereochemical configuration, and occurrence in groundwater at a former production site. Environ. Sci. Technol. 41:4292-4298. [DOI] [PubMed] [Google Scholar]
- 143.Raina, V., D. Rentsch, T. Geiger, P. Sharma, H. R. Buser, C. Holliger, R. Lal, and H. P. E. Kohler. 2008. New metabolites in the degradation of α- and γ-hexachlorocyclohexane (HCH): pentachlorocyclohexenes are hydroxylated to cyclohexenols and cyclohexenediols by the haloalkane dehalogenase LinB from Sphingobium indicum B90A. J. Agric. Food Chem. 56:6594-6603. [DOI] [PubMed] [Google Scholar]
- 144.Raina, V., M. Suar, A. Singh, O. Prakash, M. Dadhwal, S. K. Gupta, C. Dogra, K. Lawlor, S. Lal, J. R. van der Meer, C. Holliger, and R. Lal. 2008. Enhanced biodegradation of hexachlorocyclohexane (HCH) in contaminated soils via inoculation with Sphingobium indicum B90A. Biodegradation 19:27-40. [DOI] [PubMed] [Google Scholar]
- 145.Ricking, M., and J. Schwarzbauer. 2008. HCH residues in point-source contaminated samples of the Teltow Canal in Berlin, Germany. Environ. Chem. Lett. 6:83-89. [Google Scholar]
- 146.Rijnaarts, H. H. M., A. Bachmann, J. C. Jumelet, and A. J. B. Zehnder. 1990. Effect of desorption and intraparticle mass transfer on the aerobic biomineralization of α-hexachlorocyclohexane in a contaminated calcareous soil. Environ. Sci. Technol. 24:1349-1354. [Google Scholar]
- 147.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rubinos, D. A., R. Villasuso, S. Muniategui, M. T. Barral, and F. Diaz Fierros. 2007. Using the landfarming technique to remediate soils contaminated with hexachlorocyclohexane isomers. Water Air Soil Pollut. 181:385-399. [Google Scholar]
- 149.Russell, R. J., R. L. Harcourt, and J. G. Oakeshott. 1998. Bioremediation of pesticides using enzymes. ACIAR Proc. 85:341-348. [Google Scholar]
- 150.Sahu, S. K., K. K. Patnaik, S. Bhuyan, B. Sreedharan, N. Kurihara, T. K. Adhya, and N. Sethunathan. 1995. Mineralization of α-, γ- and β-isomers of hexachlorocyclohexane by a soil bacterium under aerobic conditions. J. Agric. Food Chem. 43:833-837. [Google Scholar]
- 151.Sahu, S. K., K. K. Patnaik, and N. Sethunathan. 1992. Dehydrochlorination of δ-isomer of hexachlorocyclohexane by a soil bacterium, Pseudomonas sp. Bull. Environ. Contam. Toxicol. 48:265-268. [DOI] [PubMed] [Google Scholar]
- 152.Sahu, S. K., K. K. Patnaik, M. Sharmila, and N. Sethunathan. 1990. Degradation of alpha-, beta-, and gamma-hexachlorocyclohexane by a soil bacterium under aerobic conditions. Appl. Environ. Microbiol. 56:3620-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sanghi, R., M. K. Pillai, T. R. Jayalekshmi, and A. Nair. 2003. Organochlorine and organophosphorus pesticide residues in breast milk from Bhopal, Madhya Pradesh, India. Hum. Exp. Toxicol. 22:73-76. [DOI] [PubMed] [Google Scholar]
- 154.Scott, C., G. Pandey, C. J. Hartley, C. J. Jackson, M. J. Cheesman, M. C. Taylor, R. Pandey, J. L. Khurana, M. Teese, C. W. Coppin, K. M. Weir, R. K. Jain, R. Lal, R. J. Russell, and J. G. Oakeshott. 2008. The enzymatic basis of pesticide bioremediation. Indian J. Microbiol. 48:65-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Senoo, K., and H. Wada. 1989. Isolation and identification of an aerobic γ-HCH decomposing bacterium from soil. Soil Sci. Plant Nutr. 35:79-87. [Google Scholar]
- 156.Shapir, N., E. F. Mongodin, M. J. Sadowsky, S. C. Daugherty, K. E. Nelson, and L. P. Wackett. 2007. Evolution of catabolic pathways: genomic insights into microbial s-triazine metabolism. J. Bacteriol. 189:674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shapir, N., C. Pedersen, O. Gil, L. Strong, J. Seffernick, M. J. Sadowsky, and L. P. Wackett. 2006. TrzN from Arthrobacter aurescens TC1 is a zinc amidohydrolase. J. Bacteriol. 188:5859-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sharma, P., V. Raina, R. Kumari, S. Malhotra, C. Dogra, H. Kumari, H. P. E. Kohler, H. R. Buser, C. Holliger, and R. Lal. 2006. Haloalkane dehalogenase LinB is responsible for β- and δ-hexachlorocyclohexane transformation in Sphingobium indicum B90A. Appl. Environ. Microbiol. 72:5720-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sievers, S., and P. Friesel. 1989. Soil contamination patterns of chlorinated organic compounds: looking for the source. Chemosphere 19:691-698. [Google Scholar]
- 160.Singh, A., and R. Lal. 2008. Sphingobium ummariense sp. nov., a hexachlorocyclohexane (HCH)-degrading bacterium, isolated from HCH-contaminated soil. Int. J. Syst. Evol. Microbiol. 59:162-166. [DOI] [PubMed] [Google Scholar]
- 161.Singh, A. K., P. Chaudhary, A. S. Macwan, U. N. Diwedi, and A. Kumar. 2007. Selective loss of lin genes from hexachlorocyclohexane-degrading Pseudomonas aeruginosa ITRC-5 under different growth conditions. Appl. Microbiol. Biotechnol. 76:895-901. [DOI] [PubMed] [Google Scholar]
- 162.Singh, K. P., A. Malik, and S. Sinha. 2007. Persistent organochlorine pesticide residues in soil and surface water of northern Indo-Gangetic alluvial plains. Environ. Monit. Assess. 125:147-155. [DOI] [PubMed] [Google Scholar]
- 163.Slade, R. E. 1945. The gamma isomer of hexachlorocyclohexane (gammexane). An insecticide with outstanding properties. Chem. Ind. 40:314-319. [Google Scholar]
- 164.Smets, B. F., H. Yin, and A. Esteve-Nuρez. 2007. TNT biotransformation: when chemistry confronts mineralization. Appl. Microbiol. Biotechnol. 76:267-277. [DOI] [PubMed] [Google Scholar]
- 165.Smidt, H., and W. M. de Vos. 2004. Anaerobic microbial dehalogenation. Annu. Rev. Microbiol. 58:43-73. [DOI] [PubMed] [Google Scholar]
- 166.Streltsov, V. A., Z. Prokop, J. Damborsky, Y. Nagata, A. Oakley, and M. C. Wilce. 2003. Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates. Biochemistry 42:10104-10112. [DOI] [PubMed] [Google Scholar]
- 167.Suar, M., A. Hauser, T. Poiger, H. R. Buser, M. D. Muller, C. Dogra, V. Raina, C. Holliger, J. R. van der Meer, R. Lal, and H. P. E. Kohler. 2005. Enantioselective transformation of alpha-hexachlorocyclohexane by the dehydrochlorinases LinA1 and LinA2 from the soil bacterium Sphingomonas paucimobilis B90A. Appl. Environ. Microbiol. 71:8514-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Suar, M., J. R. van der Meer, K. Lawlor, C. Holliger, and R. Lal. 2004. Dynamics of multiple lin gene expression in Sphingomonas paucimobilis B90A in response to different hexachlorocyclohexane isomers. Appl. Environ. Microbiol. 70:6650-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Subramaniam, K., and R. D. J. Solomon. 2006. Organochlorine pesticides BHC and DDE in human blood in and around Madurai, India. Indian J. Clin. Biochem. 21:169-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sundin, G. W., and C. L. Bender. 1995. Expression of the strA-strB streptomycin resistance genes in Pseudomonas syringae and Xanthomonas campestris and characterization of IS6100 in X. campestris. Appl. Environ. Microbiol. 61:2891-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sutherland, T. D., I. Horne, K. M. Weir, C. W. Coppin, M. R. Williams, M. Selleck, R. J. Russell, and J. G. Oakeshott. 2004. Enzymatic bioremediation: from enzyme discovery to applications. Clin. Exp. Pharmacol. Physiol. 31:817-821. [DOI] [PubMed] [Google Scholar]
- 172.Tauch, A., S. Götker, A. Puhler, J. Kalinowski, and G. Thierbach. 2002. The 27.8-kb R-plasmid pTET3 from Corynebacterium glutamicum encodes the aminoglycoside adenyltransferase gene cassette aadA9 and the regulated tetracycline efflux system Tet 33 flanked by active copies of the widespread insertion sequence IS6100. Plasmid 48:117-129. [DOI] [PubMed] [Google Scholar]
- 173.Thomas, J. C., F. Berger, M. Jacquier, D. Bernillon, F. Baud-Grasset, N. Truffaut, P. Normand, T. M. Vogel, and P. Simonet. 1996. Isolation and characterization of a novel gamma-hexachlorocyclohexane-degrading bacterium. J. Bacteriol. 178:6049-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Top, E. M., and D. Springael. 2003. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 14:262-269. [DOI] [PubMed] [Google Scholar]
- 175.Toteja, G. S., A. Mukherjee, S. Diwakar, P. Singh, and B. N. Saxena. 2003. Residues of DDT and HCH pesticides in rice samples from different geographical regions of India: a multicentre study. Food Addit. Contam. 20:933-939. [DOI] [PubMed] [Google Scholar]
- 176.Trantirek, L., K. Hynkova, Y. Nagata, A. Murzin, A. Ansorgova, V. Sklenar, and J. Damborsky. 2001. Reaction mechanism and stereochemistry of γ-hexachlorocyclohexane dehydrochlorinase LinA. J. Biol. Chem. 276:7734-7740. [DOI] [PubMed] [Google Scholar]
- 177.Tu, C. M. 1976. Utilization and degradation of lindane by soil microorganisms. Arch. Microbiol. 108:259-263. [DOI] [PubMed] [Google Scholar]
- 178.van Doesburg, W., M. H. van Eekert, P. J. Middeldorp, M. Balk, G. Schraa, and A. J. Stams. 2005. Reductive dechlorination of β-hexachlorocyclohexane (β-HCH) by a Dehalobacter species in coculture with a Sedimentibacter sp. FEMS Microbiol. Ecol. 54:87-95. [DOI] [PubMed] [Google Scholar]
- 179.van Eekert, M. H. A., N. J. P. van Ras, G. H. Mentink, H. H. M. Rijnaarts, A. J. M. Stams, J. A. Field, and G. Schraa. 1998. Anaerobic transformation of β-HCH by methanogenic granular sludge and soil microflora. Environ. Sci. Technol. 32:3299-3304. [Google Scholar]
- 180.van Liere, H., S. Staps, C. Pijls, G. Zwiep, R. Lassche, and A. Langenhoff. 2003. Full scale case: successful in situ bioremediation of a HCH contaminated industrial site in central Europe (The Netherlands), p. 128-132. In J. Vijgen (ed.), Forum book; 7th International HCH and Pesticides Forum. Sustainable Development and Ecological Research Center, Kiev, Ukraine.
- 181.Vega, F. A., E. F. Covelo, and M. L. Andrade. 2007. Accidental organochlorine pesticide contamination of soil in Porrino, Spain. J. Environ. Qual. 36:272-279. [DOI] [PubMed] [Google Scholar]
- 182.Vijgen, J. 2006. The legacy of lindane HCH isomer production—main report. A global overview of residue management, formulation and disposal. International HCH and Pesticides Association, Holte, Denmark. http://www.cluin.org/download/misc/Lindane_Main_Report_DEF20JAN06.pdf.
- 183.Vijgen, J., L. F. Yi, M. Forter, R. Lal, and R. Weber. 2006. The legacy of lindane and technical HCH production. Organohalog. Comp. 68:899-904. [Google Scholar]
- 184.Waliszewski, S. M., R. Villalobos-Pietrini, S. Gomez-Arroyo, and R. M. Infanzon. 2003. Persistent organochlorine pesticides in Mexican butter. Food Addit. Contam. 20:361-367. [DOI] [PubMed] [Google Scholar]
- 185.Walker, K., D. A. Vallero, and R. G. Lewis. 1999. Factors influencing the distribution of lindane and other hexachlorocyclohexanes in the environment. Environ. Sci. Technol. 33:4373-4378. [Google Scholar]
- 186.Ware, G. W. (ed.). 1989. The pesticide book. Thomson Publications, Fresno, CA.
- 187.Weber, R., C. Gaus, M. Tysklind, P. Johnston, M. Forter, H. Hollert, H. Heinisch, I. Holoubek, M. Lloyd-Smith, S. Masunaga, P. Moccarelli, D. Santillo, N. Seike, R. Symons, J. P. M. Torres, M. Verta, G. Varbelow, J. Vijgen, A. Watson, P. Costner, J. Woelz, P. Wycisk, and M. Zennegg. 2008. Dioxin- and POP-contaminated sites—contemporary and future relevance and challenges. Environ. Sci. Pollut. Res. 15:363-393. [DOI] [PubMed] [Google Scholar]
- 188.Weir, K. M., T. D. Sutherland, I. Horne, R. J. Russell, and J. G. Oakeshott. 2006. A single monooxygenase, Ese, is involved in the metabolism of the organochlorides endosulfan and endosulfate in an Arthrobacter sp. Appl. Environ. Microbiol. 72:3524-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Willett, K. L., E. M. Ulrich, and R. A. Hites. 1998. Differential toxicity and environmental fates of hexachlorocyclohexane isomers. Environ. Sci. Technol. 32:2197-2207. [Google Scholar]
- 190.Wu, J., Q. Hong, P. Han, J. He, and S. Li. 2007. A gene linB2 responsible for the conversion of β-HCH and 2,3,4,5,6-pentachlorocyclohexanol in Sphingomonas sp. BHC-A. Appl. Microbiol. Biotechnol. 73:1097-1105. [DOI] [PubMed] [Google Scholar]
- 191.Wu, J., Q. Hong, Y. Sun, Y. Hong, Q. Yan, and S. Li. 2007. Analysis of the role of LinA and LinB in biodegradation of δ-hexachlorocyclohexane. Environ. Microbiol. 9:2331-2340. [DOI] [PubMed] [Google Scholar]
- 192.Wu, W. Z., Y. Xu, K.-W. Schramm, and A. Kettrup. 1997. Study of sorption, biodegradation and isomerization of HCH in stimulated sediment/water system. Chemosphere 35:1887-1894. [Google Scholar]
- 193.Yamamoto, S., S. Otsuka, Y. Murakami, M. Nishiyama, and K. Senoo. 2009. Genetic diversity of gamma-hexachlorocyclohexane-degrading sphingomonads isolated from a single experimental field. Lett. Appl. Microbiol. 49:472-477. [DOI] [PubMed] [Google Scholar]
- 194.Yang, H., P. D. Carr, S. Y. McLoughlin, J. W. Liu, I. Horne, X. Qiu, C. M. Jeffries, R. J. Russell, J. G. Oakeshott, and D. L. Ollis. 2003. Evolution of an organophosphate-degrading enzyme: a comparison of natural and directed evolution. Protein Eng. 16:135-145. [DOI] [PubMed] [Google Scholar]
- 195.Yen, K. M., and C. M. Serdar. 1988. Genetics of naphthalene catabolism in pseudomonads. Crit. Rev. Microbiol. 15:247-268. [DOI] [PubMed] [Google Scholar]
- 196.Yule, W. N., M. Chiba, and H. V. Morely. 1967. Fate of insecticide residues. Decomposition of lindane in soil. J. Agric. Food Chem. 15:1000-1004. [Google Scholar]
- 197.Zhao, G., Y. Xu, W. Li, G. Han, and B. Ling. 2007. PCBs and OCPs in human milk and selected foods from Luqiao and Pingqiao in Zhejiang, China. Sci. Total Environ. 378:281-292. [DOI] [PubMed] [Google Scholar]
- 198.Zhu, Y., H. Liu, Z. Xi, H. Cheng, and X. Xu. 2005. Organochlorine pesticides (DDTs and HCHs) in soils from the outskirts of Beijing, China. Chemosphere 60:770-778. [DOI] [PubMed] [Google Scholar]