Abstract
We investigated the prevalence and diversity of Escherichia coli strains isolated from surface waters from multiple watersheds within the South Nation River basin in eastern Ontario, Canada. The basin is composed of mixed but primarily agricultural land uses. From March 2004 to November 2007, a total of 2,004 surface water samples were collected from 24 sampling sites. E. coli densities ranged from undetectable to 1.64 × 105 CFU 100 ml−1 and were correlated with stream order and proximity to livestock production systems. The diversity of 21,307 E. coli isolates was characterized using repetitive extragenic palindromic PCR (rep-PCR), allowing for the identification of as many as 7,325 distinct genotypes, without capturing all of the diversity. The community was temporally and spatially dominated by a few dominant genotypes (clusters of more than 500 isolates) and several genotypes of intermediary abundance (clustering between 10 and 499 isolates). Simpson diversity indices, assessed on a normalized number of isolates per sample, ranged from 0.050 to 0.668. Simpson indices could be statistically discriminated on the basis of year and stream order, but land use, discharge, weather, and water physical-chemical properties were not statistically important discriminators. The detection of Campylobacter species was associated with statistically lower Simpson indices (greater diversity; P < 0.05). Waterborne E. coli isolates from genotypes of dominant and intermediary abundance were clustered with isolates obtained from fecal samples collected in the study area over the same period, and 90% of the isolates tested proved to share genotypes with fecal isolates. Overall, our data indicated that the densities and distribution of E. coli in these mixed-use watersheds were linked to stream order and livestock-based land uses. Waterborne E. coli populations that were distinct from fecal isolates were detected and, on this basis, were possibly naturalized E. coli strains.
Escherichia coli is ubiquitously distributed in fecal material from humans and warm-blooded animals (38). The detection of E. coli in water is an implicit indicator of recent fecal contamination and therefore of the risk of cooccurrence of enteric pathogens that can cause illness in susceptible populations (62). Many jurisdictions evaluate and mandate compliance with drinking and recreational water quality standards on the basis of the presence and abundance of E. coli (14, 44). For example, Canadian recreational water quality standards stipulate that E. coli densities in excess of a geometric mean of 200 CFU per 100 ml indicate that the water is unsuitable for swimming and bathing (23).
In a background of increasing occurrence of microbial contamination of surface water, a variety of methods for elucidating the sources of fecal contamination have been developed, and these microbial source tracking (MST) methods are recommended components of fecal pollution abatement strategies (16, 57). So-called library-dependent MST methods compare environmental isolates to collections of isolates obtained from likely sources of fecal pollution in the area of investigation. The host source is distinguished on the basis of the similarity of environmental isolates to reference fecal isolates. Comparison can be undertaken on the basis of genomic fingerprinting methods, including repetitive extragenic palindromic PCR (rep-PCR), ribotyping, or pulsed-field gel electrophoresis (PFGE) (13, 17, 31, 54, 57). A variety of studies using these methods have revealed enormous diversity in the fecal and environmental E. coli populations. For example, 461 distinct PFGE genotypes and 175 distinct enterobacterial repetitive intergenic consensus (ERIC)-PCR genotypes were detected in a collection of 555 E. coli strains isolated from river water in Texas (10). As many as 291 and 94 rep-PCR genotypes were distinguished in collections of 643 river isolates and 353 beach water E. coli isolates, respectively (43). Significant diversity was also revealed using multilocus enzyme electrophoresis (MLEE) and multilocus sequence typing (MLST) on 185 E. coli isolates from freshwater beaches, where an average of 40 alleles per locus were detected (59). Almost 60% of 657 E. coli isolates in a fecal reference collection had unique (i.e., detected in only one individual) fingerprints determined by rep-PCR (32). Extensive diversity of E. coli was also observed in soils in temperate climates, where the growth and persistence of “naturalized” populations without any known fecal input have been found (7, 28, 30). Naturalized populations have been dominated by the B1 phylogroup and may have adapted in ways that enhance their survival in temperate secondary habitats (59). The temporal and spatial diversity of E. coli may not be a significant factor in coarse-source (e.g., human versus animal) classification of E. coli by means of ribotyping procedures (48). Ultimately, the characterization and understanding of the diversity of populations of selected microorganisms in surface watercourses affected by multiple sources of fecal pollution (as in agricultural watershed settings, for example) may be more critical for assessing the specific impacts of contamination-mitigating measures than previously thought. For instance, restricting the access of cattle on pasture to adjacent water by implementing vegetative buffering along watercourses creates habitat for varied wildlife, which then contribute to fecal pollution. In this context, the diversity in populations of indicator bacteria could be useful for better understanding how changes in landscape use influence fecal source inputs.
As part of a research program evaluating the impact of agriculture on water quality and the efficacy of better agricultural management practices to mitigate agricultural pollution, we have conducted a multiyear study of the microbiological water quality for a suite of different-sized watersheds in the South Nation River basin in eastern Ontario, Canada (41, 46, 61). Land use in this river basin is mixed, consisting primarily of agricultural activities, light urban development, and interspersed wildlife habitat. Surface water systems in the study region differ widely in their contributing areas and therefore in their discharges (61).
In the work undertaken here, we sought to determine the spatial and seasonal variability in the density and the structure of populations of E. coli in surface waters within the South Nation River basin. The specific objectives of the study were (i) to characterize the seasonal distribution and abundance of E. coli in different watershed settings within the river basin, (ii) to evaluate the spatial distribution of E. coli densities and diversity with respect to upstream land use activities, (iii) to use rep-PCR to elucidate the dominant E. coli genotypes and the diversity of E. coli populations and to explore linkages to pathogen presence, season, and environmental and land use variables, and (iv) using rep-PCR, to evaluate the concordance between waterborne isolates and fecal isolates obtained from within the study area. The study is distinguished by an intensive 4-year sampling of numerous (n = 24) sites that differed in their stream order and proximal land use activity; the number of E. coli isolates (≈21,000) included in the analysis; and the use of two distinct rep-PCR fingerprinting methods (ERIC and BOXA1R) to characterize the isolates. Furthermore, we used classification and Regression Tree (CART) analysis to evaluate relationships between the abundance and diversity of E. coli in water samples and environmental and land use variables.
MATERIALS AND METHODS
Characterization of sampling sites and surface water sampling.
The South Nation River basin is located in eastern Ontario, Canada. The total area of the basin is approximately 3,900 km2. The topography in the region is generally flat; subsurface tile drainage and groundwater are the primary flow contributors. Roughly 60% of the land use in the South Nation watershed is farming, consisting primarily of dairy operations and cropping systems that receive manure (Statistics Canada's 1996 agriculture census data are available at http://ceps.statcan.ca/english/profil/PlaceSearchForm1.cfm).
Surface water was sampled from 24 discrete locations within an area of approximately 200 km2 (41). The sample sites were located on tributaries to the South Nation River and on the South Nation River proper. Land use and stream order information were characterized using data from an intensive roadside survey, remote sensing imagery (e.g., LANDSAT and SPOT series), and digital elevation model (DEM) databases of the study region (9, 41, 46) (Table 1). The various watercourses (ranging in size from drainage ditches, to tributaries of various sizes, to the main reach of the South Nation River) were characterized as follows. A DEM was “filled” using ArcMap Geographic Information Systems software, version 9.1, and the Spatial Analyst package (Environmental Systems Research Institute [ESRI], Redlands, CA), removing small defects and/or sinks (areas of no outward flow), in the surface of the DEM. Next, flow direction was calculated on the filled DEM, and subsequently flow accumulation was calculated, in order to determine the direction of flow on a cell-by-cell basis and the number of upstream cells contributing to each contributing cell and coincident sampling location. Stream thresholds were identified in order to produce a representative drainage network, and the Shreve order (49) was calculated using the Stream Order routine in Spatial Analyst and was assigned to water sample site locations. The larger the watercourse, the larger the Shreve order (Table 1).
TABLE 1.
List of landscape variables (independent criteria) used in CART analyses
| Variable namea | Variable description (unit)b |
|---|---|
| SHREVE | Stream order. Lower orders (numbers) represent smaller tributaries, while higher orders (numbers) represent larger watercourses. Shreve's method adds the order of each converging branch together regardless of size. |
| DBARN_xK | Barn density in catchment area 2, 5, 10, and 20 km upstream (obs. km−2) |
| DDEVHI_xK | High-density development in catchment area 2, 5, 10, and 20 km upstream (obs. km−2) |
| DDEVLO_xK | Low-density development in catchment area 2, 5, 10, and 20 km upstream (obs. km−2) |
| DFORE_xK | Forest density in catchment area 2, 5, 10, and 20 km upstream (obs. km−2) |
| DPAST_xK | Pasture density in catchment area 2, 5, 10, and 20 km upstream (obs. km−2) |
| CROPP-xK | Cropland coverage in catchment area 2, 5, 10, and 20 km upstream (km2 km−2) |
| DEVELP-xK | Developed land coverage in catchment area 2, 5, 10, and 20 km upstream (km2 km−2) |
| FORAGEP-xK | Forage land coverage in catchment area 2, 5, 10, and 20 km upstream (km2 km−2) |
| PASTP-xK | Pasture land coverage in catchment area 2, 5, 10, and 20 km upstream (km2 km−2) |
| SHRUBP-xK | Shrub land coverage in catchment area 2, 5, 10, and 20 km upstream (km2 km−2) |
| VEGP-xK | Forest coverage in catchment area 2, 5, 10, and 20 km upstream (km2 km−2) |
| WATWETP-xK | Water plus wetland coverage in catchment area 2, 5, 10, and 20 km upstream (km2 km−2) |
| BASIN_(landuse) | Cropland, developed land, forage land, pasture land, and forest coverage in entire catchment area (km2 km−2) |
| NUD_(landuse) | Upstream distances (km) from sample location to nearest land use observations listed above and further land use observations, including poultry barns (POULTRY BARN), pasture with livestock access to sampled tributary (PASTURE ACCESS), horse barns (HORSE BARN), hog barns (HOG BARN), dairy operations (DAIRY OP), and cattle barns (CATTLE BARN) |
x, maximum upstream distance from the sample site at which the sample site catchment area was defined.
obs., number of observations.
From March 2004 to November 2007, a total of 2,004 water samples were collected for enumeration of E. coli. With the exception of one sample site (designated MST-1), 1-liter samples of surface water were collected from within a 0.5-m depth of the surface directly into sterile containers (Systems Plus, Woodstock, ON, Canada). Samples from MST-1 were taken from a municipal drinking water intake, which draws water from a depth of about 6 m within the South Nation River.
Water isolates of E. coli for detailed genotypic analysis were obtained from 5 (MST-1, MST-5, MST-6, MST-9, and MST-15) of the 24 sample sites. These were chosen as representative of different stream orders and land use activities within the river basin. Briefly, MST-1 is located on the South Nation River (Shreve stream order, 50,846), MST-5 on the Little Castor River (Shreve stream order, 1,695), MST-6 on the Payne River (Shreve stream order, 3,942), MST-9 on Butternut Creek (Shreve stream order, 1,090), and MST-15 on a municipal drainage ditch (Shreve stream order, 54). Water samples (n = 311) were collected on a biweekly basis from March 2004 to August 2007 (Table 2).
TABLE 2.
Sample period, number of water samples collected, and number of E. coli isolates genotyped using BOX-PCR and ERIC-PCR for each site and each year
| Site | Dates of sample collection | No. of samples (no. of isolates) fingerprinted |
||||
|---|---|---|---|---|---|---|
| 2004 | 2005 | 2006 | 2007 | Total | ||
| MST-1 | 29 Mar. 2004 to 13 Aug. 2007 | 19 (1,477) | 18 (1,712) | 29 (998) | 6 (96) | 69 (4,283) |
| MST-5 | 25 Apr. 2004 to 13 Aug. 2007 | 22 (1,671) | 27 (3,489) | 23 (640) | 9 (180) | 81 (5,980) |
| MST-6 | 25 Apr. 2004 to 13 Aug. 2007 | 20 (1,552) | 26 (3,704) | 21 (652) | 9 (180) | 76 (5,458) |
| MST-9 | 25 Apr. 2005 to 13 Aug. 2007 | —a | 22 (3,326) | 21 (407) | 10 (200) | 53 (3,933) |
| MST-15 | 30 Aug. 2005 to 30 July 2007 | — | 9 (1,205) | 18 (360) | 5 (88) | 32 (1,653) |
—, the site was not sampled in 2004.
Fecal sampling.
Between April 2004 and September 2007, fecal samples (n = 150) were collected from a variety of sources within the study area (40). These included livestock and companion animals (alpaca, bovine, dog, horse, poultry, rabbit, sheep, and swine), avian wildlife (goose and gull), mammalian wildlife (beaver, deer, fox, groundhog, moose, muskrat, and raccoon), and human wastewater (septic tanks, wastewater treatment plant effluent). Wildlife fecal material was obtained directly from animals by local trappers. Livestock slurry or solid manure samples were obtained from several commercial farms either once or on several different dates.
E. coli enumeration, isolation, and confirmation.
Water and fecal samples were shipped on ice packs within 24 h to Agriculture and Agri-Food Canada (AAFC) laboratories in London, Ontario, Canada, where they were analyzed immediately. Water samples were enumerated as described elsewhere (61). Briefly, 10- to 100-ml portions of water were filtered through sterile, 0.45-μm-pore-size, 47-mm-diameter cellulose acetate filters (Pall Gelman GN-6; VWR International, Mississauga, ON, Canada), and the filters were plated onto mFC basal medium (Difco, Fisher Scientific, Ottawa, ON, Canada) supplemented with 100 mg liter−1 of 3-bromo-4-chloro-5-indolyl-β-d-glucopyranoside (BCIG) (hexylammonium salt; Inverness Medical, Ottawa, ON, Canada [11]) and then incubated overnight at 44.5°C. Fecal samples were serially diluted in sodium metaphosphate buffer (2 g liter−1) and mixed thoroughly. E. coli from fecal material was isolated on mFC-BCIG medium as described previously (39). All single colonies were restreaked twice onto LB agar (Difco, Fisher Scientific). Isolates were considered to be E. coli if they grew at 44.5°C, had a positive reaction for β-d-glucuronidase (blue color on mFC-BCIG agar), fermented lactose, and produced indole. Confirmed isolates were inoculated into sterile 96-well microtiter plates containing 100 μl well−1 of LB broth and were incubated overnight at 37°C. Sterile glycerol (Sigma-Aldrich Canada Ltd., Mississauga, ON, Canada) was then added to each well at a final concentration of 15% (vol/vol), and the plates were stored at −70°C. For this study, 21,307 water and 10,966 fecal E. coli isolates were confirmed and further genotyped by rep-PCR.
Additional microbiological analyses of water.
A subset (n = 180) of the 311 water samples from which diversity indices were determined was also analyzed for the presence of Salmonella spp., Campylobacter spp., E. coli O157:H7, Cryptosporidium oocysts, and Giardia cysts. The pathogen abundance data and detailed descriptions of the methodology used to obtain them have been published previously (46, 61).
Rep-PCR fingerprinting.
Cell suspensions of E. coli were prepared by inoculating 100 μl of fresh LB broth per well in a sterile 96-well microtiter plate with frozen stock cultures. Cells were grown statically at 37°C overnight and were centrifuged at 710 × g for 25 min (Centra CL3 microplate centrifuge; Thermo IEC, Needham Heights, MA). The supernatant was removed, and cells were resuspended in 100 μl sterile Milli-Q water and agitated at 1,000 rpm with a microplate shaker (Sarstedt, Montreal, QC, Canada) for 5 min. The cell suspension either was used directly as a template for the PCR or was frozen at −20°C until required.
Bacteria were genotyped by both ERIC-PCR and BOX-PCR, with primers described elsewhere (58). The final reaction mixture (25 μl) consisted of 1× PCR buffer (Promega, Madison, WI), 1.5 mM MgCl2, 0.1 mg ml−1 gelatin, 200 μM each deoxynucleoside triphosphate (Invitrogen, Burlington, ON, Canada), 2 μM (each) forward and reverse primers ERIC-1 and ERIC, 1 U of Taq polymerase (Promega, Madison, WI), and 2 μl of the E. coli cell suspension as a template for ERIC-PCR. The same protocol was used for BOX-PCR except that the gelatin was replaced with 1% (vol/vol) dimethyl sulfoxide, and the single primer BOXA1R was used. Amplification was performed with a Thermo MBS Satellite 0.2 thermocycler (VWR International, Mississauga, ON, Canada) as follows. After an initial denaturation at 94°C for 10 min, 34 cycles of denaturation (94°C for 3 s; 92°C for 30 s), annealing (50°C for 1 min), and extension (65°C for 1 min) were performed, followed by a final extension (65°C for 8 min). PCR products were resolved by horizontal electrophoresis in a 25-cm by 50-cm gel (Gator A3-1; Owl Separations, Portsmouth, NH) prepared with 1.5% (wt/vol) agarose (Invitrogen, Mississauga, ON, Canada) and 1× Tris-borate-EDTA buffer. Six microliters of loading dye was added to 25 μl of the PCR product, and 7 μl of this mixture was loaded into wells fitted to an 8-mm by 1-mm comb tooth size. Every eighth well received the MassRuler DNA ladder (Fermentas, Burlington, ON, Canada). Gels were subjected to 2.5 V cm−1 for 16 h in 1× Tris-borate-EDTA buffer. The gel was stained with 1 μg ml−1 ethidium bromide solution for 10 min and was destained in Milli-Q water for 10 min. Gel images were captured as 16-bit tagged-image format file (TIFF) images by using AlphaEase FC software and an Alpha Innotech digital gel documentation system (Fisher Scientific).
Computer-assisted image analysis and cluster assignment.
Gel images were normalized, and fingerprints were assigned to isolates, with the BioNumerics software package (version 4.5; Applied Maths, Kortrijk, Belgium) as described elsewhere (15). Filtering and background subtraction were optimized for each image independently according to the methodology available at http://www.ecolirep.umn.edu/addinggelimages.shtml. The positions of fingerprints on gels were normalized using the MassRuler DNA ladder as the external standard in the range of 300 bp to 6,000 bp. Strains were assigned to different clusters by calculating the similarity coefficients with the curve-based Pearson similarity coefficient. Similarity trees were generated using the unweighted-pair group method using average linkages. Repeated experiments where the same isolate fingerprint was run on different gels under similar conditions consistently showed an average similarity of 85%. Hence, clusters were initially assigned using the software on the basis of 85% similarity, and the final assignments were determined on the basis of careful and laborious visual inspection.
Fingerprint data analysis.
The diversity captured in the E. coli collections was estimated by rarefaction analysis using the analytical approximation algorithm of reference 27, and 95% confidence intervals were estimated (24). Calculations were performed with the freeware program Analytical Rarefaction 1.3, available at http://www.uga.edu/strata/software/. Curves were plotted using SigmaPlot (version 9.1; SPSS Inc., Chicago, IL). The asymptotes of the rarefaction curves were estimated using the Michaelis-Menten equation, which is available in SigmaPlot as the one-site saturation ligand model (26). The asymptote is a measure of richness at sampling saturation and was used to estimate the fraction of total community diversity captured within the E. coli collection and the number of isolates required to capture half of the predicted diversity. The SigmaPlot curve fitter uses the Marquardt-Levenberg algorithm to find the coefficients that give the best fit between the equation and the data (42). The Simpson diversity indices for populations of E. coli were calculated with the software calculator available at the Chang Bioscience website, and confidence intervals were calculated (21, 50). The Wilcoxon signed-rank test (a nonparametric form of the paired Student t test) was carried out using SPSS software for Windows (version 15.0; SPSS Inc., Chicago, IL) and was used to determine whether Simpson index values calculated from water samples with decreasing numbers of randomly chosen isolates were coming from the same distribution (P, 0.05).
E. coli abundance and Simpson diversity index linkages with season and environmental and land use variables.
We determined if the abundance of E. coli varied coherently with season, stream order, and land use (e.g., the density of agricultural, human, or wildlife fecal sources in proximity to each sampling location) as follows. Potential landscape sources of seasonal E. coli pollution in the study area were determined by predicting seasonal E. coli densities on the basis of land use and stream order (Table 1). The statistical approach used to make these determinations is regression tree analysis in the data-mining software CART version 6.0 (Salford Systems, San Diego, CA) (5, 53). CART is a well-established automated nonparametric binary recursive partitioning (decision tree) methodology. These CART-based exploratory analyses can help identify spatial and temporal hot spots for E. coli contamination and, thus, potential landscape sources of fecal contamination. Data on the densities of E. coli in water (the “dependent variable” or target variable”; expressed in CFU 100 ml−1) were grouped into “data nodes” defined on the basis of land use and stream order attributes (“predictor variables”; described in Table 1) by using CART. Due to the sensitivity of least-square regression methods to outliers and heteroscedasticity properties not uncommon in E. coli density data, the least absolute deviation (LAD) regression criteria were employed for these data in CART. Thus, the mean absolute deviation (MAD) rather than the standard deviation (SD) was used to describe within-node data dispersion. The general assumptions and overall CART modeling approach used here, except for the use of LAD rather than least-squares methods, have been described previously (36). For purposes of brevity, we present only CART results reflecting, at a maximum, two (secondary) tree levels.
CART was also used to determine if the diversity of E. coli populations was related to land use, climate, season, and environmental varibles. The Simpson diversity indices (dependent variable) were evaluated in terms of the independent land use variables (Table 1), as well as the year, the season, and additional water physical/chemical variables (Table 3). For these CART analyses, the least-squares approach was used because of the relatively small impact of outliers. In addition, the association of the occurrence of pathogens (Campylobacter, Salmonella, Cryptosporidium oocysts, Giardia cysts [61]) with CART-produced Simpson index groups was summarized.
TABLE 3.
Season, climate, and water physical and chemical data used to predict Simpson indexes using CARTa
| Variable name(s) | Variable description (unit) |
|---|---|
| SEASON | Season, determined by solstice and equinox dates |
| AMIA_AMN | NH3 (ammonia) + ammonium concn in sample water (mg liter−1) |
| NITRITE | NO2− (nitrite) concn in sample water (mg liter−1) |
| NITRATE | NO3− (nitrate) concn in sample water (mg liter−1) |
| REA_PHOS | Reactive phosphorus concn in sample water |
| TOTKN | Total Kjeldahl nitrogen concn in sample water (mg liter−1) |
| TOTPHO | Total phosphorus concn in sample water (mg liter−1) |
| TEMP | Temp of water at time of sample collection (°C) |
| pH | pH of water at time of sample collection |
| CONDUCTIVITY | Electrical conductivity of water at time of sample collection (mS cm−1) |
| DISS_OXYGEN_P, DISS_OXYGEN_MGL | Gaseous O2 dissolved in sample water (mg liter−1), measured as a percentage of saturation and in mg liter−1 |
| ORP | Oxidation reduction potential (mV) |
| TURBIDITY | Cloudiness of sample water as measured with a nephelometer sensor (nephelometric turbidity units) |
| DIS_SPC, DIS_PAY, DIS_RUS | River discharge at Spencerville, Payne, and Russell hydrometric stations, respectively (m3 s−1) |
| RUS_TOTALRAIN, RUS_TOTALRAINXD | Total rainfall on day of sampling (mm); total rainfall x = 1, 2, 3, and 7 days in advance of sampling at the Russell meteorological station (mm) |
| RUS_MAXTEMP, RUS_MINTEMP, | |
| RUS_MEANTEMP | Daily maximum, minimum, and mean temp (°C) at Russell and daily avg temp (°C) |
Physical and chemical analyses were undertaken on the same water samples as those used for microbiological analyses.
RESULTS
Seasonal and spatial variations in densities of E. coli.
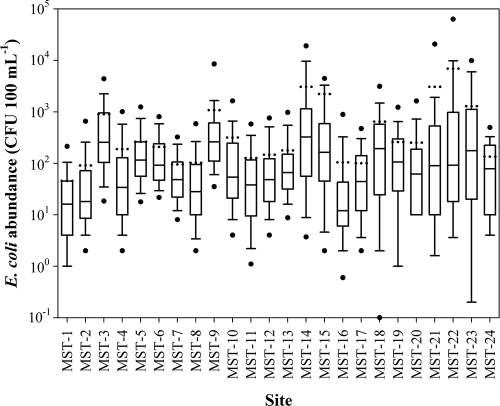
The abundance of E. coli in surface waters was determined on 2,004 water samples collected from 24 discrete surface water sample sites over a 45-month period (Fig. 1). Counts ranged from undetectable (detection limit, 1 CFU 100 ml−1) to 1.64 × 105 CFU 100 ml−1 (median, 64 CFU 100 ml−1). A total of 1,206 water samples (60%) had E. coli counts at or below the Ontario Provincial Water Quality Objectives of 100 CFU 100 ml−1, and 1,461 samples (73%) had counts at or below the Canadian Recreational Water Quality guidelines of 200 CFU 100 ml−1. The sampling sites differed consistently in their degree of contamination, with sites MST-1, MST-2, and MST-16 having the lowest (16, 18, and 12 CFU 100 ml−1, respectively) and sites MST-3, MST-9, and MST-14 having the highest (255, 260, and 325 CFU 100 ml−1, respectively) median viable E. coli counts over the 45-month study period. The Shreve stream order variable was negatively correlated with E. coli density, presumably because of dilution in the larger watercourses (Table 4). The strongest correlations were found in the summer, with an R of −0.61 (P < 0.05).
FIG. 1.
Box plots of E. coli counts (CFU 100 ml−1) in surface waters for each of the 24 sites of the South Nation River watershed. A box and whiskers represent the first and third quartiles (bottom and top lines of the box), the median (middle line of the box), and the smallest and largest observations (bottom and top whiskers) of a data distribution. Dots below and above the whiskers represent the 5th and 95th percentiles, respectively.
TABLE 4.
Spearman's rank correlations between Shreve stream order and abundance
| Season | No. of samples | Ra |
|---|---|---|
| All | 2,004 | −0.36 |
| Spring | 595 | −0.15 |
| Summer | 750 | −0.61 |
| Fall | 575 | −0.25 |
| Winter | 84 | −0.35 |
P < 0.05 for all correlations.
Relationships of land use and stream order with E. coli abundance.
Relationships between E. coli densities and landscape attributes related to potential fecal sources and stream order were explored using CART. Overall, the results indicate that higher median E. coli densities in the water were associated with areas where cattle barns were closer to observation sites, the proportion of upstream forage lands was greater, the stream order was smaller, and pasture density within a surface water catchment area to a maximum distance of 2 km upstream of a sample site was relatively greater. In detail, CART was run using seasonal data sets; data split definitions and terminal node (defined as a data grouping where no further splitting of data occurs) statistics are given in Table 5. All CART models, except those employed on the winter data sets, cross-validated using the 10-fold default 90% learning and 10% data testing approach, effectively indicating the “structural significance” of the resulting models. For the 595 samples collected in the spring, the E. coli abundance data were stratified into terminal node groups on the basis of the primary splitting variables: NUD_CATTLE BARN, SHREVE, and DPAST_2K (Table 1). For summer, the data split trends and the variables used to define primary splits were for the most part similar. NUD_PAST (upstream distance to nearest pasture) was, however, important for partitioning the lower-stream-order groups into relatively “higher” (median ± MAD, 468 ± 3,650 CFU 100 ml−1) and “lower” (174 ± 521 CFU 100 ml−1) median E. coli terminal nodes. For fall data (n = 575), the highest median abundance was found where the proportion of forage lands was greater than 0.39 km2 km−2 within catchment areas defined as being up to 2 km upstream of the sample site. The lowest median groupings were found where that proportion was ≤0.39 km2 km−2, and where NUD_FORAGE (upstream distance to nearest forage land) was greater than 2.95 km upstream (24 ± 84 CFU 100 ml−1). Stream order was not important for the fall data set. For the winter data set (n = 84), CART did not provide an internally cross-validated model, and therefore, no results are reported.
TABLE 5.
| Data set (total no. of samples)c | Root node split criterion | Secondary split criteriond | Median E. coli density (CFU 100 ml−1) ± MAD | No. of samples |
|---|---|---|---|---|
| E. coli densities for spring (595) | NUD_CATTLE BARN, >2.29 km | DPAST_2K, >1.26 obs. km−2 | 58 ± 1,040 | 190 |
| NUD_CATTLE BARN, >2.29 km | DPAST_2K, ≤1.26 obs. km−2 | 20 ± 206 | 315 | |
| NUD_CATTLE BARN, ≤2.29 km | SHREVE, >5.0 | 108 ± 258 | 61 | |
| NUD_CATTLE BARN, ≤2.29 km | SHREVE, ≤5.0 | 230 ± 516 | 29 | |
| E. coli densities for summer (750) | SHREVE, >15.5 | NUD_CATTLE BARN, ≤3.65 km | 112 ± 114 | 87 |
| SHREVE, >15.5 | NUD_CATTLE BARN, >3.65 km | 20 ± 88 | 383 | |
| SHREVE, ≤15.5 | NUD_PAST, ≤1.16 km | 468 ± 3,650 | 141 | |
| SHREVE, ≤15.5 | NUD_PAST, >1.16 km | 174 ± 521 | 139 | |
| E. coli densities for fall (575) | FORAGEP_2K, >0.39 km2 km−2 | NA | 230 ± 608 | 64 |
| FORAGEP_2K, ≤0.39 km2 km−2 | NUD_FORAGE, >2.95 km | 24 ± 84 | 73 | |
| FORAGEP_2K, ≤0.39 km2 km−2 | NUD_FORAGE, ≤2.95 km | 78 ± 526 | 438 |
Root node split criterion, the variable and condition by which all the data were divided into two nodal groupings (child nodes); secondary split criterion, the variable and condition by which the child nodes derived from the root nodal split were divided (for purposes of brevity, we present only results up to this tree level in this study). The variables that define the E. coli split criteria are described in Table 1.
Terminal nodes are data groupings where no further splitting occurs.
All data sets are for 2004 to 2007. Note that winter data are not included, since CART could not cross-validate that seasonal data set, as a result of lack of data structure.
obs., observations; NA, not applicable, since there was no terminal node of data at that level in the tree model.
Seasonal and spatial variation in E. coli population structure.
A total of 21,307 E. coli strains were isolated from 311 water samples taken at 5 discrete sampling sites over a 4-year period. These were genotyped using BOX-PCR and ERIC-PCR. The number of isolates genotyped per water sample ranged from 7 to 368 (mean ± SD, 67 ± 60) for BOX-PCR and from 7 to 412 (mean ± SD, 67 ± 62) for ERIC-PCR. Water samples that were relatively pristine yielded fewer isolates.
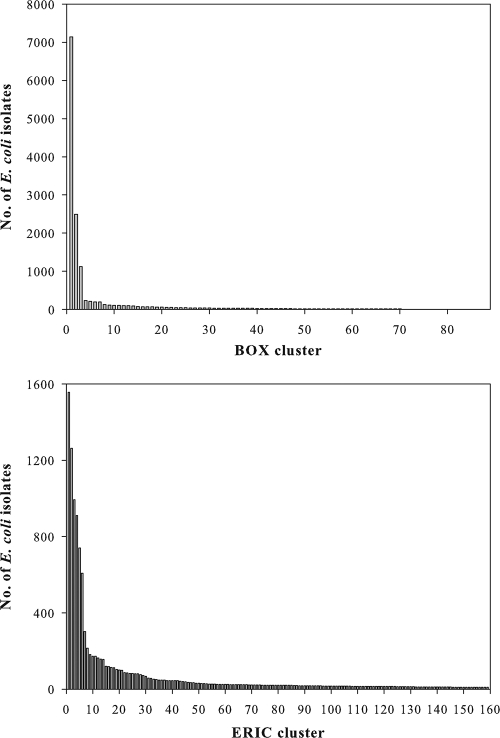
A total of 4,930 distinct genotypes were detected using BOX-PCR, and 7,235 distinct genotypes were detected using ERIC-PCR. With both methods, the population comprised a very few genotypes clustering more than 500 isolates, several genotypes clustering between 10 and 499 isolates, and numerous genotypes clustering fewer than 10 isolates (Fig. 2). Using BOX-PCR genotyping, 3 dominant genotypes (0.06% of all genotypes) clustered 10,752 isolates (50.5% of the total collection). Using ERIC-PCR genotyping, 6 dominant genotypes (0.08% of all genotypes) clustered 6,071 isolates (28.5% of the total collection). A total of 4,418 isolates were clustered both within BOX and ERIC dominant genotypes, representing 41% of the isolates within dominant BOX clusters and 73% of the isolates within dominant ERIC clusters. These dominant isolates were consistently found at each sampling site, in every sampling year, and in every season studied. Overall, although the diversity of genotypes within the collection of E. coli isolates obtained in this study was enormous, the majority of the isolates belonged to “dominant” genotypes, defined here as representing at least 500 individual isolates within the total collection.
FIG. 2.
Rank abundance curves for genotypes detected by rep-PCR using the BOXA1R (top) or ERIC (bottom) primers.
Many genotypes clustered between 10 and 499 E. coli isolates, defined here as genotypes with “intermediate abundance” (Fig. 2). There were 81 genotypes of intermediate abundance by BOX-PCR (clustering 3,264 E. coli isolates) and 153 by ERIC-PCR (clustering 5,697 E. coli isolates), and their spatial, seasonal, and annual distributions differed widely within the watershed.
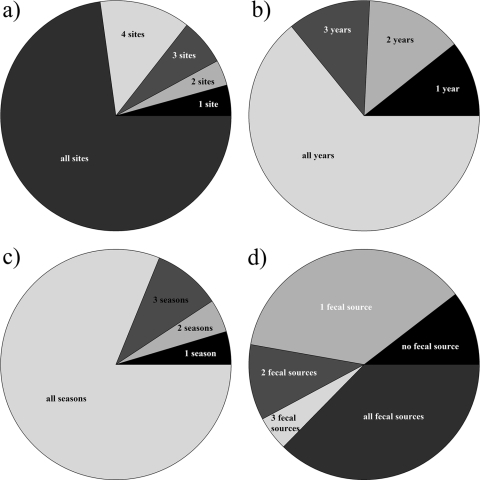
Isolates that clustered to the dominant and intermediate-abundance ERIC genotypes (n = 11,768) were pooled, and spatial (Fig. 3a), annual (Fig. 3b), and seasonal (Fig. 3c) variations were determined. The majority of the isolates (72.8%) belonged to genotypes that were detected at all 5 sampling sites. Progressively smaller portions of the collection consisted of isolates that were detected at 4, 3, or 2 sites or only 1 site. Those isolates that were detected at only 1 site (n = 507) were not consistently detected at the same site. Most isolates belonged to genotypes that were detected in all seasons. Only 4.6% belonged to genotypes that were detected in only 1 season, and these were distributed among all 4 seasons.
FIG. 3.
(a to c) Detection of specific ERIC-PCR genotypes representing isolates (n = 11,768) within the 5 sites sampled during the study (a), during the 4 years of the study (b), and during each season (c). (d) Genotypes of waterborne isolates were clustered with a reference collection (n = 10,966) of fecal E. coli strains, and their identities with the genotypes of isolates from the fecal sources (livestock and companion animals, avian wildlife, mammalian wildlife, human) were established.
The predicted richness for collections obtained over the entire study period from each of the 5 sampling sites was estimated by rarefaction analysis (Table 6). The Michaelis-Menten fit with the experimental data was excellent (r2, >0.98). None of the rarefaction curves reached an asymptote, indicating that further acquisition of isolates would be required to capture all of the diversity within these populations. The collections are estimated to have captured only 17% to 27% of the predicted richness, and the predicted asymptotes for richness at sampling saturation ranged widely, from 1,220 to 6,964 genotypes for BOX-PCR and from 2,083 to 10,374 genotypes for ERIC-PCR.
TABLE 6.
Estimated richness of E. coli populations obtained from 5 representative sampling sites over the study perioda
| Method | Site | r2b | No. of genotypes: |
% of predicted genotypes that were detected | No. of isolates required to capture 50% of predicted genotypes (mean ± SD)e | |
|---|---|---|---|---|---|---|
| Predicted (mean ± SD)c | Detectedd | |||||
| BOX-PCR | MST-1 | 0.9999 | 5,794 ± 196 | 1,196 | 21 | 14,384 ± 578 |
| MST-5 | 0.9999 | 6,964 ± 207 | 1,621 | 23 | 19,007 ± 685 | |
| MST-6 | 0.9999 | 6,138 ± 190 | 1,479 | 24 | 17,282 ± 654 | |
| MST-9 | 0.9999 | 3,970 ± 133 | 888 | 22 | 13,697 ± 549 | |
| MST-15 | 0.9998 | 1,220 ± 69 | 325 | 27 | 4,589 ± 325 | |
| ERIC-PCR | MST-1 | 0.9999 | 10,295 ± 514 | 1,755 | 17 | 19,614 ± 1,123 |
| MST-5 | 0.9998 | 9,599 ± 385 | 2,152 | 22 | 19,390 ± 937 | |
| MST-6 | 0.9998 | 10,374 ± 413 | 2,093 | 20 | 20,985 ± 985 | |
| MST-9 | 0.9998 | 6,491 ± 311 | 1,470 | 23 | 13,294 ± 769 | |
| MST-15 | 0.9998 | 2,083 ± 172 | 509 | 24 | 5,050 ± 509 | |
The total numbers of water samples and E. coli isolates obtained from each site are given in Table 2.
Coefficient of determination of the goodness of fit to the Michaelis-Menten equation.
The Vmax parameter in the Michaelis-Menten equation.
The number of genotypes detected in the rarefaction analysis is used to estimate the percentage of total community diversity that was captured in the collection.
The Km parameter in the Michaelis-Menten equation.
Diversity of E. coli within water samples, variability, and relationships with key drivers.
A Simpson index for E. coli populations within each of 311 water samples was estimated using ERIC-PCR fingerprints. Since different numbers of isolates were collected for different samples, a crucial issue was the relationship between the sample size (i.e., the number of E. coli isolates from a given water sample) and the diversity index determined for that water sample. If, as expected, apparent diversity increased with sample size, then diversity indices for water samples would not be comparable unless the number of isolates treated from each water sample was normalized. We determined if normalization was required as follows. Collections of E. coli isolates (n, 178 to 368) were obtained from 10 different water samples that were chosen because they were relatively heavily contaminated. First, a Simpson index was obtained, and estimates of sampling saturation (where the estimate of “true” diversity is expected to occur) were determined by rarefaction analysis of the 10 collections by using all of the available isolates obtained from each water sample. Then subsets of isolates (n, 20, 50, 100, 150, and 200, etc.) were chosen randomly from each water sample, and diversity analyses were performed using each normalized discrete sample size. In all cases in this study, decreasing the number of isolates in the diversity analysis reduced the number of genotypes that were predicted to be in the sample by rarefaction analysis. When the genotypes of all the available isolates were considered, the predicted number of genotypes in each water sample was 109 ± 48 (average ± SD) (n = 10) for BOX-PCR and 142 ± 56 for ERIC-PCR (data not shown). When the sample size was reduced and normalized to 20 randomly selected isolates from each water sample, the predicted number of genotypes decreased to 16 ± 7 for BOX-PCR and 46 ± 27 for ERIC-PCR. In contrast, the Simpson indices estimated with BOX-PCR fingerprints for the 10 samples were independent of sample size, with average values (± SD) of 0.257 ± 0.132 when all isolates were considered, and 0.264 ± 0.120 when only 20 randomly chosen isolates were considered (P = 0.88). With the ERIC-PCR fingerprints, however, the estimated Simpson indices were responsive to sample size. The index increased from 0.079 ± 0.045 when the entire collection was considered to 0.118 ± 0.039 when only 20 isolates per sample were considered (P < 0.05). On the basis of these results, the diversity index for a given water sample was estimated using 20 E. coli isolates randomly chosen from the sample. ERIC-PCR data were chosen, rather than BOX-PCR data, because of the greater discriminatory power of the former. Simpson diversity index values ranged between 0.050 and 0.668, with a median value of 0.120.
Potential relationships between diversity (Simpson index determined with ERIC-PCR data) and the various variables given in Tables 1 and 3 were explored using the CART approach. Overall, among the various variables tested, only the sampling year and stream order were identified as important (Table 7). The modeling results in Table 7 met the default 10-fold 90% learning and 10% data testing cross-validation criteria (therefore, the resulting model is structurally significant in that capacity). The lowest-average CART-produced Simpson index group, representing data from the years 2004 and 2006 to 2007 and Shreve stream orders of >572, had an average Simpson index ± SD of 0.121 ± 0.069 (Table 7). For the group of Simpson index data from 2005, the average was 0.193 ± 0.104. The group of Simpson index data defined by the years 2004 and 2006 to 2007 and a Shreve stream order of ≤572 had the highest average (0.209 ± 0.127).
TABLE 7.
Results of CART analyses for the Simpson index
| Root node split criteriona | Secondary split criteriona | Simpson index (mean ± SD) | No. of samples | % of samples positive for the following pathogen: |
||||
|---|---|---|---|---|---|---|---|---|
| Salmonella | Campylobacter | E. coli O157:H7 | Cryptosporidium | Giardia | ||||
| Yr = 2004, 2006-2007 | SHREVE, ≤572 | 0.209 ± 0.127 | 23 | 5 | 10 | 0 | 45 | 30 |
| SHREVE, >572 | 0.121 ± 0.069 | 181 | 13 | 32 | 0 | 47 | 27 | |
| Yr = 2005 | NA | 0.193 ± 0.104 | 102 | 11 | 10 | 0 | 52 | 12 |
Regression tree split criteria for terminal nodes (data groupings where no further splitting occurs) are given. Root node split criterion, the variable and condition by which all the data were divided into two nodal groupings (child nodes); secondary split criterion, the variable and condition by which the child nodes derived from the root nodal split were divided. NA, not applicable. Within-nodal data descriptions are given as Simpson indices.
In a parallel study, we determined the distributions of pathogens in the South Nation River basin study area and their associations with indicator microorganisms and hydrologically based pollution drivers (61). In this study, the presence and absence of pathogens in water samples were evaluated with respect to the diversity of E. coli populations within the water samples (Tables 7 and 8). The Simpson index was not associated with the probability of detecting Salmonella, E. coli O157:H7, Cryptosporidium, or Giardia in a given water sample (Table 8). In contrast, Campylobacter was found at higher frequencies in larger tributaries and streams when the E. coli diversity was high (Table 8).
TABLE 8.
Comparison of logarithmically transformed (base 10) and raw Simpson index diversity values for water samples in which specific pathogens were present or absent
| Pathogen (no. of samples examined) | Mean Simpson index value ± 95% confidence interval of meana (no. of samples) |
Effect size index (d)b |
Pc |
|||||
|---|---|---|---|---|---|---|---|---|
| Log transformed |
Raw |
Log transformed | Raw | Log transformed | Raw | |||
| Presence | Absence | Presence | Absence | |||||
| Salmonella spp. (186) | 0.920 ± 0.089 (21) | 0.847 ± 0.034 (165) | 0.140 ± 0.048 (21) | 0.164 ± 0.015 (165) | −0.33 | −0.24 | 0.144 | 0.359 |
| Campylobacter spp. (182) | −0.937 ± 0.053 (38) | 0.832 ± 0.038 (144) | 0.126 ± 0.019 (38) | 0.171 ± 0.017 (144) | −0.48 | −0.46 | 0.002 | 0.001 |
| Cryptosporidium oocysts (153) | 0.833 ± 0.055 (74) | 0.870 ± 0.050 (79) | 0.174 ± 0.027 (74) | 0.156 ± 0.021 (79) | 0.16 | 0.17 | 0.317 | 0.290 |
| Giardia cysts (153) | 0.890 ± 0.078 (34) | 0.841 ± 0.042 (119) | 0.149 ± 0.028 (34) | 0.169 ± 0.020 (119) | −0.21 | −0.19 | 0.293 | 0.247 |
Simpson index diversity values for samples in which pathogens were present (Presence) or absent (Absence) were obtained with ERIC-PCR data. The 95% confidence interval of the mean was calculated by multiplying the standard error of the mean by 1.96.
Calculated as (μ1 − μ2)/σpooled. An effect size of 0.20 is considered small; 0.50, medium; 0.80, large (12).
By Welch's t test (for samples of unequal size and unequal variance) utilizing Satterthwaite's approximation for degrees of freedom. P values are 2-sided.
Concordance between water genotypes and fecal genotypes.
The distributions of specific ERIC genotypes in the water and in the fecal collections of E. coli were compared (Fig. 3d). The 6 dominant ERIC genotypes (genotypes with 500 or more isolates each, representing a total of 6,071 isolates) and the 153 ERIC genotypes with intermediate abundance (genotypes with 10 to 499 isolates each, representing a total of 5,697 isolates) were clustered together with the 10,996 E. coli isolates obtained from fecal samples collected in the study area between 2004 and 2007 by using Bionumerics software (Fig. 3d). Of the 159 water genotypes, 110, including the 6 dominant genotypes (representing 10,539 isolates [89.5% of the isolates tested]) were shared with isolates found in the fecal collection. The remaining 49 genotypes (detected in 1,229 isolates [10.5% of the isolates tested]) were not detected in the fecal collection. Thirty-three (8,410 water isolates) of the 36 ERIC genotypes that were detected at all 5 sampling sites were also detected in the fecal collection. Three of the 36 ERIC genotypes that were detected at all 5 sampling sites (107 water isolates) were absent from the fecal collection. Twenty three (496 water isolates) of the 44 genotypes detected at only 1 or at 2 sampling sites were not detected in the fecal collection. A total of 4,328 isolates, representing 53 genotypes, were detected in only 1 fecal source; 1,251 isolates, representing 22 genotypes, were detected in 2 fecal sources; 567 isolates, representing 10 genotypes, were found in 3 fecal sources; and 4,393 isolates, representing 25 genotypes, were found in all fecal sources. Of the 6 waterborne ERIC genotypes clustering more than 500 E. coli isolates, 4 were found in all fecal sources whereas 2 (clustering 1,556 and 993 isolates) were detected only in livestock.
DISCUSSION
Abundance and distribution of E. coli.
In the present study, we characterized the distribution and the community structure of E. coli in surface waters from multiple watersheds within the South Nation River basin in eastern Ontario, Canada, over a 45-month period. The surface watercourses represented a suite of different-sized, mixed-land-use catchments characterized by various degrees of urban development, livestock and crop production systems, and wildlife habitats. E. coli was isolated from the 24 discrete sampling sites with comparable densities in different years and seasons, but with average densities consistently over Canadian guidelines for recreational water quality (23). The CART analyses suggested that for spring and summer, denser E. coli populations were found at sites proximal to livestock production systems. However, stream order was also found to be important for spring and summer, perhaps reflecting the significance of dilution in attenuating pollution. For instance, in spring samplings, at sites where a cattle barn was located in the catchment area approximately 2.3 km upstream, median densities differed by 122 CFU 100 ml−1 depending on whether the Shreve order of the watercourse was greater than or equal to 5 (the higher the stream order, the lower the density). However, the associations of the stream order with the densities of microorganisms will vary seasonally, and the results obtained here suggest that this hydrological variable should be used cautiously (Table 3). In fall samplings, the greatest E. coli densities were found where proportions of forage lands were relatively greater within the sample site catchment area defined as being up to approximately 2 km upstream of the sampling site. Manure application on forage fields over the entire growing season is common in the region; moreover, forage fields are often near livestock operations. The area of investigation is heavily tile drained, and thus, contaminants in manure at the time of application can quickly be transported to adjacent watercourses (37). We previously showed with the same data set that densities of E. coli (and Enterococcus spp.) were strongly correlated to rainfall variables, suggesting that precipitation-induced transport through tile drainage systems or surface runoff is a potentially important exposure pathway (61). Significantly, the CART analyses also indicated that landscape indicators of fecal pollution were constrained to upstream distances less than approximately 3 km. This finding is important, since it means that the observations required for understanding pollution drivers in these kinds of landscapes may only need to be acquired within a short distance upstream.
E. coli diversity and drivers.
E. coli genotypic and phenotypic diversity is thought to be very large (30), and it has been suggested that collections of as many as 40,000 isolates might be necessary in order to capture all of the E. coli diversity based on rep-PCR DNA fingerprinting (32). Consistent with this notion, the more than 20,000 E. coli isolates analyzed here were estimated by rarefaction analysis to capture only as much as 27% of the predicted genotypes in the area studied. Overall, at the level of discrimination afforded by rep-PCR, it was completely impractical to saturate sampling in this freshwater drainage system.
Sampling sites MST-1, MST-5, MST-6, and MST-9 were more diversified in E. coli richness than was site MST-15. The latter, a drainage ditch, is likely under the influence of local fecal inputs, as opposed to other sites, which drain and integrate fecal inputs from larger areas and likely from diversified fecal sources. Otherwise, the similarity of E. coli communities between sites, years, and seasons, despite the potential for different fecal sources in the different sampling areas, is likely due to integration as water flows through the drainage basin. This lack of spatial variation in E. coli populations in surface freshwater was also observed in populations obtained from 6 beaches of Lake Huron and the St. Clair River (59). In another lake study conducted on 11 sites over a 9-month period, rep-PCR genotyping of E. coli isolates showed that a few E. coli genotypes consistently dominated populations recovered from the area.
At the temporal scale, changes in the composition of the E. coli community in surface water could be the consequence of seasonal fecal population structure change, with summer populations derived from more numerous sources than winter populations (60). Moreover, fecal E. coli communities are known to change during the lifetime of individual animals or to be influenced by the diet, and such changes could also be responsible for the year-to-year differentiation of water communities observed here (15, 34, 47).
For community studies, diversity is usually assessed using three criteria (i.e., richness, evenness, and composition [18, 45]), which usually generate considerable amounts of data. The use of diversity indices as composite estimators of diversity enables data reduction and comparison of several samples but is likely influenced by sample size: increasing the sample size will likely increase the specific richness and affect species composition up to the point where all species are captured. For instance, Shannon index and evenness are under- and overestimated by low coverage, whereas the Simpson index tends to be less sensitive to sample size (25, 52). An accurate estimation of diversity would require determining the best compromise given the sample size. With E. coli densities differing widely between our samples, using a normalization approach, we estimated that rep-PCR analyses on 20 to 50 isolates per sample would allow the capture of a representative fraction of the diversity: Simpson index values proved to reach a plateau around 50 isolates, but specific richness, estimated by rarefaction analysis fitted to the Michaelis-Menten equation, increased continuously with the acquisition of more isolates. However, both Simpson index and predicted richness values based on 20 isolates were significantly correlated to values obtained using the full range of isolates, and the use of normalized sample sizes allowed diversity estimation to be independent of the level of contamination. We thus determined that 20 isolates would represent a good sample size for the study of E. coli diversity in the environment, and we used this number of isolates (chosen randomly from all of the isolates of a given sample) to characterize diversity in the present work.
The diversity in waterborne E. coli populations was not related to the likelihood of detecting pathogenic microorganisms, except for Campylobacter spp., where higher diversity was associated with a significantly higher probability of detection (Tables 7 and 8). The least diverse E. coli populations were detected in smaller reaches, where fecal contamination would have been predominantly of bovine origin (dairy operations). Overall, E. coli diversity was not a robust indicator of the probability of pathogen detection.
Relationship between waterborne and fecal E. coli strains within the watershed.
Some E. coli strains can survive and replicate in some secondary habitats, and strains that are repeatedly isolated from environmental matrices and that are distinct from those obtained from known fecal sources may be environmentally adapted, or “naturalized” (28, 29). Naturalized E. coli populations have been detected in a variety of environments, such as soil or water, and in tropical, temperate, or cold regions (4, 7, 8, 29). About a quarter of the waterborne collection clustered in genotypes that were not detected in the fecal collection (Fig. 3d). These isolates could therefore represent a sizable “naturalized” population. Alternatively, the fecal collection may have significantly underrepresented the total diversity of E. coli within the area during the experiment, and the ability to source all environmental isolates is therefore not complete. Finally, in spite of the supposed robustness of the method (33), rearrangements that modify the abundance or the location of rep-PCR primer binding sites within the genome could potentially create new apparent “genotypes” not detected in fecal populations.
About one-third of the water isolates clustered with the livestock and companion animal fecal isolates (Fig. 3d). This is in agreement with the spatial relationships between E. coli abundance in the water and proximity to sources of cattle manure pollution from the numerous dairy farms in the area (Table 4). About half of the isolates belonged to genotypes that were detected in multiple fecal sources. Perhaps the digestive system of ruminants is a unique niche selecting for distinct genotypes within the area of study, whereas monogastrics tend to harbor a larger diversity of shared types (20). Overall, the study identified a few very dominant genotypes that were detected in one fecal source and many genotypes that were detected in multiple sources.
Based on the 22 complete genome sequences of E. coli that are currently available, the E. coli pangenome (the complete complement of noncore genes distributed within individuals within the species) has been estimated to comprise at least 42,500 gene families (51), significantly more than the human genome. Studies have revealed enormous diversity in E. coli populations in primary and various secondary habitats. These include feces from various mammalian and avian hosts (2, 22), stored dairy and swine manure (39), home septic systems (19), fresh and marine surface water (35, 43, 56), beach sand (59), soils (7), and the freshwater macrophytic green alga Cladophora (8). Many aspects of this enormous diversity have importance for the assessment and management of water quality. First, rep-PCR is a microbial source tracking (MST) method commonly employed to elucidate the source of fecal pollution of surface water (16, 32, 57). Environmental isolates are compared to reference isolates from potential fecal sources (e.g., human, livestock, wildlife) in the area of investigation, and the likely host source is assigned on the basis of similarity of rep-PCR fingerprints. The tractability of this library-dependent MST method is contingent on the ability to undertake representative sampling, the host specificity of the fingerprints obtained, and the persistence of fecal genotypes in secondary habitats. Second, the diversity of environmental populations may reflect how impacted they are by point or diffuse sources of fecal pollution. For example, isolated collections of E. coli were obtained from a marine coastal area that was exposed to storm water sewerage following significant precipitation events (6). The diversity of populations determined by rep-PCR was distinctly higher before than after precipitation events, indicating that populations dominated by sewage contamination were significantly less diverse than background populations of E. coli in this saline aquatic environment. Third, the structure of surviving populations is in part due to differential survival in secondary habitats. Different strains of E. coli differ widely in their abilities to persist or proliferate in manured soils (55) or subtropical waters and sediments (1), potentially confounding quantitative relative source assignation by MST (3). Finally, environmental isolates of E. coli that are distinct from fecal isolates may represent types that have become uniquely adapted for extraintestinal survival and proliferation (7, 28, 30).
In summary, we demonstrated here that E. coli abundance and distribution in mixed-use watersheds were linked to the size of the watercourse, as well as agricultural land uses associated with livestock production systems. E. coli populations proved to be genotypically diverse, although dominant genotypes were observed across sites, seasons, and sampling years. A comparison of waterborne and fecal E. coli populations suggested the occurrence of naturalized E. coli populations in the watershed studied and detected dominant genotypes that were shared among multiple sources, or were found in livestock only. Temporal changes in E. coli water populations were likely to be related to changes in fecal inputs over time, emphasizing the necessity of constructing temporally integrative fecal libraries when microbial source tracking approaches are considered.
Acknowledgments
This work was funded in part by the AAFC National Water Quality Surveillance Research Program and the Watershed Evaluation of Beneficial Management Practice (WEBs) program. E. Lyautey and Z. Lu were supported by the NSERC Visiting Fellowship in Government Laboratories program.
We thank the South Nation Conservation Authority for assistance in obtaining samples.
Footnotes
Published ahead of print on 28 December 2009.
REFERENCES
- 1.Anderson, K. L., J. E. Whitlock, and V. J. Harwood. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. A., J. E. Whitlock, and V. J. Harwood. 2006. Diversity and distribution of Escherichia coli genotypes and antibiotic resistance phenotypes in feces of humans, cattle, and horses. Appl. Environ. Microbiol. 72:6914-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes, B., and D. M. Gordon. 2004. Coliform dynamics and the implications for source tracking. Environ. Microbiol. 6:501-509. [DOI] [PubMed] [Google Scholar]
- 4.Beversdorf, L. J., S. M. Bornstein-Forst, and S. L. McLellan. 2007. The potential for beach sand to serve as a reservoir for Escherichia coli and the physical influences on cell die-off. J. Appl. Microbiol. 102:1372-1381. [DOI] [PubMed] [Google Scholar]
- 5.Breiman, L., J. Freidman, R. Olshen, and C. Stone. 1984. Classification and regression trees. Wadsworth International, Pacific Grove, CA.
- 6.Brownell, M. J., V. J. Harwood, R. C. Kurz, S. M. McQuaig, J. Lukasik, and T. M. Scott. 2007. Confirmation of putative stormwater impact on water quality at a Florida beach by microbial source tracking methods and structure of indicator organism populations. Water Res. 41:3747-3757. [DOI] [PubMed] [Google Scholar]
- 7.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, M. J. Sadowsky, and S. Ishii. 2006. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 8:504-513. [DOI] [PubMed] [Google Scholar]
- 8.Byappanahalli, M. N., R. L. Whitman, D. A. Shively, J. Ferguson, S. Ishii, and M. J. Sadowsky. 2007. Population structure of Cladophora-borne Escherichia coli in nearshore water of Lake Michigan. Water Res. 41:3649-3654. [DOI] [PubMed] [Google Scholar]
- 9.Canadian Soil Information System (CanSIS). 2004. Ontario Soils Datalayer (version 2004 and 2005). Agriculture and Agri-Food Canada, Eastern Cereal and Oilseeds Research Centre, Ottawa, Ontario, Canada.
- 10.Casarez, E. A., S. D. Pillai, and G. D. Di Giovanni. 2007. Genotype diversity of Escherichia coli isolates in natural waters determined by PFGE and ERIC-PCR. Water Res. 41:3643-3648. [DOI] [PubMed] [Google Scholar]
- 11.Ciebin, B. W., M. H. Brodsky, R. Reddington, G. Horsnell, A. Choney, G. Palmateer, A. Ley, R. Joshi, and G. Shears. 1995. Comparative evaluation of modified m-FC and m-TEC media for membrane filter enumeration of Escherichia coli in water. Appl. Environ. Microbiol. 61:3940-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, J. 1992. A power primer. Quantitative methods in psychology. Psychol. Bull. 11:155-159. [DOI] [PubMed] [Google Scholar]
- 13.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dufour, A. P. 1984. Health effects criteria for fresh recreational waters. Office of Research and Development, U.S. Environmental Protection Agency, Research Triangle Park, NC.
- 15.Duriez, P., and E. Topp. 2007. Temporal dynamics and impact of manure storage on antibiotic resistance patterns and population structure of Escherichia coli isolates from a commercial farm. Appl. Environ. Microbiol. 73:5486-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edge, T. A., and K. A. Schaefer (ed.). 2006. Microbial source tracking in aquatic ecosystems: the state of the science and an assessment of needs. National Water Research Institute, Burlington, Ontario, Canada.
- 17.Farber, J. M. 1996. An introduction of the hows and whys of molecular typing. J. Food Prot. 59:1091-1101. [DOI] [PubMed] [Google Scholar]
- 18.Forney, L. J., X. Zhou, and C. J. Brown. 2004. Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7:210-220. [DOI] [PubMed] [Google Scholar]
- 19.Gordon, D. M., S. Bauer, and J. R. Johnson. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513-1522. [DOI] [PubMed] [Google Scholar]
- 20.Gordon, D. M., and A. Cowling. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575-3586. [DOI] [PubMed] [Google Scholar]
- 21.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen, D. L., S. Ishii, M. J. Sadowsky, and R. E. Hicks. 2009. Escherichia coli populations in Great Lakes waterfowl exhibit spatial stability and temporal shifting. Appl. Environ. Microbiol. 75:1546-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Health and Welfare Canada. 1992. Guidelines for Canadian recreational water quality. H49-70/1991E. Ministry of Supply and Services Canada, Ottawa, Ontario, Canada.
- 24.Heck, K. L. J., G. van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 25.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 26.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurlbert, S. H. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 28.Ishii, S., W. B. Ksoll, R. E. Hicks, and M. J. Sadowsky. 2006. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 72:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii, S., D. L. Hansen, R. E. Hicks, and M. J. Sadowsky. 2007. Beach sand and sediments are temporal sinks and sources of Escherichia coli in Lake Superior. Environ. Sci. Technol. 41:2203-2209. [DOI] [PubMed] [Google Scholar]
- 30.Ishii, S., and M. J. Sadowsky. 2008. Escherichia coli in the environment: implications for water quality and human health. Microbes Environ. 23:101-108. [DOI] [PubMed] [Google Scholar]
- 31.Ishii, S., and M. J. Sadowsky. 2009. Applications of the rep-PCR DNA fingerprinting technique to study microbial diversity, ecology and evolution. Environ. Microbiol. 11:733-740. [DOI] [PubMed] [Google Scholar]
- 32.Johnson, L. K., M. B. Brown, E. A. Carruthers, J. A. Ferguson, P. E. Dombek, and M. J. Sadowsky. 2004. Sample size, library composition, and genotypic diversity among natural populations of Escherichia coli from different animals influence accuracy of determining sources of fecal pollution. Appl. Environ. Microbiol. 70:4478-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang, H. P., and W. M. Dunne. 2003. Stability of repetitive-sequence PCR patterns with respect to culture age and subculture frequency. J. Clin. Microbiol. 41:2694-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katouli, M., A. Lund, P. Wallgren, I. Kuhn, O. Soderlind, and R. Mollby. 1995. Phenotypic characterization of intestinal Escherichia coli of pigs during suckling, postweaning, and fattening periods. Appl. Environ. Microbiol. 61:778-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kon, T., S. C. Weir, E. T. Howell, H. Lee, and J. T. Trevors. 2007. Genetic relatedness of Escherichia coli isolates in interstitial water from a Lake Huron (Canada) beach. Appl. Environ. Microbiol. 73:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lapen, D. R., G. C. Topp, E. G. Gregorich, H. N. Hayhoe, and W. E. Curnoe. 2001. Divisive field-scale associations between corn yields, management, and soil information. Soil Tillage Res. 58:193-206. [Google Scholar]
- 37.Lapen, D. R., E. Topp, C. D. Metcalfe, H. Li, M. Edwards, N. Gottschall, P. Bolton, W. Curnoe, M. Payne, and A. Beck. 2008. Pharmaceutical and personal care products in tile drainage following land application of municipal biosolids. Sci. Total Environ. 399:50-65. [DOI] [PubMed] [Google Scholar]
- 38.Leclerc, H., D. A. A. Mossel, S. C. Edberg, and C. B. Struijk. 2001. Advances in the bacteriology of the Coliform group: their suitability as markers of microbial water safety. Annu. Rev. Microbiol. 55:201-234. [DOI] [PubMed] [Google Scholar]
- 39.Lu, Z., D. R. Lapen, A. Scott, A. Dang, and E. Topp. 2005. Identifying host sources of fecal pollution: diversity of Escherichia coli in confined dairy and swine production systems. Appl. Environ. Microbiol. 71:5992-5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyautey, E., A. Hartmann, F. Pagotto, K. Tyler, D. R. Lapen, G. Wilkes, P. Piveteau, A. Rieu, W. J. Robertson, D. T. Medeiros, T. A. Edge, V. Gannon, and E. Topp. 2007. Characteristics and frequency of detection of fecal Listeria monocytogenes shed by livestock, wildlife, and humans. Can. J. Microbiol. 53:1158-1167. [DOI] [PubMed] [Google Scholar]
- 41.Lyautey, E., D. R. Lapen, G. Wilkes, K. McCleary, F. Pagotto, K. Tyler, A. Hartmann, P. Piveteau, A. Rieu, W. J. Robertson, D. T. Medeiros, T. A. Edge, V. Gannon, and E. Topp. 2007. Distribution and characteristics of Listeria monocytogenes isolates from surface waters of the South Nation River watershed, Ontario, Canada. Appl. Environ. Microbiol. 73:5401-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marquardt, D. W. 1963. An algorithm for least squares estimation of parameters. J. Soc. Ind. Appl. Math. 11:431-441. [Google Scholar]
- 43.McLellan, S. L. 2004. Genetic diversity of Escherichia coli isolated from urban rivers and beach water. Appl. Environ. Microbiol. 70:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.National Research Council and Committee on Indicators for Waterborne Pathogens (ed.). 2004. Indicators for waterborne pathogens. National Academies Press, Washington, DC.
- 45.Purvis, A., and A. Hector. 2000. Getting the measure of biodiversity. Nature 405:212-219. [DOI] [PubMed] [Google Scholar]
- 46.Ruecker, N. J., S. L. Braithwaite, E. Topp, T. Edge, D. R. Lapen, G. Wilkes, W. Robertson, D. Medeiros, C. W. Sensen, and N. F. Neumann. 2007. Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment. Appl. Environ. Microbiol. 73:3945-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell, J. B., F. Diez-Gonzalez, and G. N. Jarvis. 2000. Effects of diet shifts on Escherichia coli in cattle. J. Dairy Sci. 83:863-873. [DOI] [PubMed] [Google Scholar]
- 48.Scott, T. M., J. Caren, G. R. Nelson, T. A. Jenkins, and J. Lukasik. 2004. Tracking sources of fecal pollution in a South Carolina watershed by ribotyping Escherichia coli: a case study. Environ. Forensics 5:15-19. [Google Scholar]
- 49.Shreve, R. L. 1966. Statistical law of stream numbers. J. Geol. 74:17-37. [Google Scholar]
- 50.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 51.Snipen, L., T. Almoy, and D. Ussery. 2009. Microbial comparative pan-genomics using binomial mixture models. BMC Genomics 10:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soetaert, K., and C. Heip. 1990. Sample-size dependence of diversity indexes and the determination of sufficient sample-size in a high-diversity deep-sea environment. Mar. Ecol. Prog. Ser. 59:305-307. [Google Scholar]
- 53.Steinberg, D., and M. Golovnya. 2007. CART 6.0 user's guide. Salford Systems, San Diego, CA.
- 54.Stoeckel, D. M., M. V. Mathes, K. E. Hyer, C. Hagedorn, H. Kator, J. Lukasik, T. L. O'Brien, T. W. Fenger, M. Samadpour, K. M. Strickler, and B. A. Wiggins. 2004. Comparison of seven protocols to identify fecal contamination sources using Escherichia coli. Environ. Sci. Technol. 38:6109-6117. [DOI] [PubMed] [Google Scholar]
- 55.Topp, E., M. Welsh, Y. C. Tien, A. Dang, G. Lazarovits, K. Conn, and H. Zhu. 2003. Strain-dependent variability in growth and survival of Escherichia coli in agricultural soil. FEMS Microbiol. Ecol. 44:303-308. [DOI] [PubMed] [Google Scholar]
- 56.Udenika Wijesinghe, R., Y. A. Feng, C. W. Wood, D. M. Stoeckel, and J. N. Shaw. 2009. Population dynamics and genetic variability of Escherichia coli in a mixed land-use watershed. J. Water Health 7:484-496. [DOI] [PubMed] [Google Scholar]
- 57.U.S. Environmental Protection Agency. June 2005. Microbial source tracking guide document. EPA/600/R-05/064. National Risk Management Research Laboratory, Office of Research and Development, U.S. EPA, Cincinnati, OH.
- 58.Versalovic, J., M. Schneider, F. J. De Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 59.Walk, S. T., E. W. Alm, L. M. Calhoun, J. M. Mladonicky, and T. S. Whittam. 2007. Genetic diversity and population structure of Escherichia coli isolated from freshwater beaches. Environ. Microbiol. 9:2274-2288. [DOI] [PubMed] [Google Scholar]
- 60.Whitman, R. L., K. Przybyla-Kelly, D. A. Shively, M. B. Nevers, and M. N. Byappanahalli. 2008. Sunlight, season, snowmelt, storm, and source affect E. coli populations in an artificially ponded stream. Sci. Total Environ. 390:448-455. [DOI] [PubMed] [Google Scholar]
- 61.Wilkes, G., T. Edge, V. Gannon, C. Jokinen, E. Lyautey, D. Medeiros, N. Neumann, N. Ruecker, E. Topp, and D. R. Lapen. 2009. Seasonal relationships among indicator bacteria, pathogenic bacteria, Cryptosporidium oocysts, Giardia cysts, and hydrological indices for surface waters within an agricultural landscape. Water Res. 43:2209-2223. [DOI] [PubMed] [Google Scholar]
- 62.Yates, M. V. 2007. Classical indicators in the 21st century—far and beyond the coliform. Water Environ. Res. 79:279-286. [DOI] [PubMed] [Google Scholar]