Abstract
Biodegradation of synthetic compounds has been studied extensively, but the metabolic diversity required for catabolism of many natural compounds has not been addressed. 5-Nitroanthranilic acid (5NAA), produced in soil by Streptomyces scabies, is also the starting material for synthetic dyes and other nitroaromatic compounds. Bradyrhizobium JS329 was isolated from soil by selective enrichment with 5NAA. When grown on 5NAA, the isolate released stoichiometric amounts of nitrite and half of the stoichiometric amounts of ammonia. Enzyme assays indicate that the initial step in 5NAA degradation is an unusual hydrolytic deamination for formation of 5-nitrosalicylic acid (5NSA). Cloning and heterologous expression revealed the genes that encode 5NAA deaminase (naaA) and the 5NSA dioxygenase (naaB) that cleaves the aromatic ring of 5NSA without prior removal of the nitro group. The results provide the first clear evidence for the initial steps in biodegradation of amino-nitroaromatic compounds and reveal a novel deamination reaction for aromatic amines.
The research on biodegradation/biotransformation of nitro compounds has focused on synthetic chemicals, but there are a substantial number of natural nitro-substituted compounds whose metabolism has not been explored. The biodegradation pathways for natural nitro compounds probably provided the metabolic diversity that enabled the rapid and recent evolution of pathways for degradation of synthetic nitro compounds.
5-Nitroanthranilic acid (5-NAA), a natural nitroaromatic compound, is produced by Streptomyces scabies, but its physiological role is unclear (15). Synthetic 5NAA is used as the starting material for various nitroaromatic compounds and dyes (3). Substitution of the aromatic ring with amino, nitro, and carboxyl functional groups creates an interesting challenge for catabolic enzymes because any of the three groups could serve as a point of attack for dioxygenase enzymes prior to ring cleavage.
Synthetic nitroanilines are toxic and used for the synthesis of pharmaceuticals, dyes, and pigments (27). In sewage, nitroanilines can be formed from the corresponding dinitroaromatic compounds under aerobic or anaerobic conditions (11). Early reports indicated that nitroanilines were resistant to biodegradation (1, 9, 11), but 4-nitroaniline was degraded by Pseudomonas sp. strain P6 (32) and Stenotrophomonas strain HPC 135 (26). Saupe reported that 3-nitroaniline could be degraded aerobically (27). The biodegradation pathways of nitroanilines are unknown, and they are typically classified as nondegradable or poorly degradable compounds (27).
As part of a search for novel metabolic diversity and an effort to study the degradation pathway for recalcitrant nitroanilines, we report here the biodegradation of 5NAA as the sole carbon, nitrogen, and energy source by Bradyrhizobium JS329. The degradation pathway involves an unusual hydrolytic removal of the amino group and subsequent ring fission without prior removal of the nitro group.
(A preliminary report of this work was presented at the 109th General Meeting of the American Society for Microbiology, 2009 [25].)
MATERIALS AND METHODS
Isolation and growth of 5NAA-degrading bacteria.
Soils were suspended (10%, wt/vol) in nitrogen-free minimal medium (BLK, pH 7.2) (4) containing 5NAA (50 μM) and incubated at 30°C with shaking. When 5NAA disappeared, subcultures were transferred to fresh medium with increasing concentrations of 5NAA. After repeated subculture into media with increasing levels of 5-NAA, a pure culture was isolated on agar plates containing BLK and 5NAA (500 μM).
Isolates were routinely grown on BLK agar plates or in BLK liquid medium supplemented with 5NAA (500 μM). When large amounts of cells were required, cultures were grown in BLK supplemented with succinate (7.5 mM) and 5NAA (500 μM) as the carbon and nitrogen sources. The noninduced cultures were grown with succinate (7.5 mM) and NH4Cl (500 μM). Exponential-phase cells were harvested by centrifugation and washed twice with phosphate buffer (20 mM, pH 7.0) prior to use.
Analytical methods.
5NAA, 5-nitrosalicylic acid (5NSA), gentisate and, salicylate were analyzed by high-performance liquid chromatography (HPLC) with an Agilent Eclipse XDB-C18 column (4.6 mm by 150 mm; 5 μm), using the method described previously (21). 5NAA was monitored at 370 nm (retention time [RT], 7.1 min), 5NSA at 305 nm (RT, 7.5 min), gentisate at 330 nm (RT, 5.4 min), and salicylate at 305 nm (RT, 7.3 min).
Ammonia and nitrite were measured as reported previously (21). Protein was measured with a Pierce (Rockford, IL) BCA protein assay reagent kit.
Chemicals.
5-Nitroanthranilic acid, 4-nitrocatechol, salicylic acid, and gentisic acid were from Sigma-Aldrich (Milwaukee, WI). 5-Nitrosalicylic acid was from Eastman Kodak (Rochester, NY). 5-Hydroxyanthranilic acid was from Acros Organics (Geel, Belgium).
Identification of bacteria.
A genomic DNA purification system (Promega, Madison, WI) was used to extract genomic DNA, which was the template for 16S rRNA gene amplification by PCR with 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) (18) and 1489R (5′-TACCTTGTTACGACTTCA-3′) (30). The 16S rRNA gene sequence (794 bp) was compared to published DNA sequences deposited in GenBank (http://www.ncbi.nlm.nih.gov/) by using BLASTN (2).
Respirometry.
Oxygen uptake was measured polarographically at 25°C with a Clark-type oxygen electrode connected to a YSI model 5300 biological oxygen monitor.
Cell extracts and enzyme assays.
Cells were passed three times through a French pressure cell (20,000 lb/in2). The cell lysate was clarified by centrifugation (20,000 × g, 4°C, 20 min). The supernatant was concentrated by ultrafiltration (molecular mass cutoff, 10 kDa; Microcon Centrifugal Filter Devices) at 4°C and washed twice with cold phosphate buffer, and then the rententate was suspended in cold phosphate buffer. The 5NAA deaminase and 5NSA 1,2-dioxygenase enzyme assays were performed at 30°C with potassium phosphate buffer (pH 7.0, 20 mM). Typical assays contained 0.6 to 3.6 mg protein and 0.05 to 0.5 μmol substrate in a total volume of 1 ml. Trifluoroacetic acid (TFA) was added to the mixture (1:100, vol/vol) to stop the reaction. The acidified mixture was clarified by centrifugation before HPLC analysis.
Gene library construction and screening.
Genomic DNA (3 μg) from 5NAA-grown Bradyrhizobium JS329 cells was extracted with an UltraClean microbial DNA isolation kit (Carlsbad, CA). DNA was randomly sheared by vortexing for 2 min. The sheared DNA fragments (approximately 30 kb in size) were end repaired to generate blunt, 5′-phosphorylated ends. The blunt-end fragments were ligated into CopyControl vector pCC1FOS and transfected into phage T1-resistant Escherichia coli strain Epi300-T1R as described in the manufacturer's protocol (Epicentre Biotechnologies, Madison, WI) to create a genomic library.
Clones were screened for 5NAA-degrading activity after 40 h of growth in 96-well plates containing LB medium supplemented with chloramphenicol (12.5 μg/ml), 1× Fosmid CopyControl induction solution (Epicentre Biotechnologies, Madison, WI), and 5NAA (100 μM). Because 5NAA is yellow, a clone designated pJS800, able to catalyze 5NAA transformation, was detected by loss of the yellow color (Table 1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| Bradyrhizobium sp. strain JS329 | 5-Nitroanthranilic acid degrader | This study |
| Escherichia coli EPI300 | Host strain for pJS800, pJS801, and pJS802 | Epicentre (Madison, WI) |
| Plasmids | ||
| pCC1FOS | Cmr; 8.1-kb fosmid vector for construction of the genomic library | Epicentre (Madison, WI) |
| pJS800 | Cmr; 40-kb pCC1FOS containing naaA and naaB from JS329 (5NAA+ 5NSA+) | This study |
| pJS801 | Cmr Kmr; transposon Tn5 insertion into naaA of pJS800 (5NAA− 5NSA+) | This study |
| pJS802 | Cmr Kmr; transposon Tn5 insertion into naaB of pJS800 (5NAA+ 5NSA−) | This study |
Identification of key genes.
Fosmid pJS800 was purified from the E. coli host with a FosmidMAX DNA purification kit (Epicentre Biotechnologies, WI) and subjected to mutagenesis in vitro using an Ez-Tn5 <KAN-2> insertion kit (Epicentre Biotechnologies, Madison, WI). The randomly mutated fosmids were reintroduced into E. coli (TranforMax EC100; Epicentre Biotechnologies, Madison, WI) by electroporation. Tn5 transposon mutants were selected on LB agar plates supplemented with chloramphenicol (12.5 μg/ml) and kanamycin (50 μg/ml). The resistant mutants were screened for the ability to transform 5NAA or 5NSA by HPLC. Clones pJS801 and pJS802 (Table 1) were sequenced to confirm the transposon insertion site.
The mutants that lost the ability to transform 5NAA or 5NSA were sequenced by primer walking, starting with Ez-Tn5 <KAN-2> specific outward reading primers (Ez-Tn5 <KAN-2> FP-1 forward primer and Ez-Tn5 <KAN-2> RP-1 reverse primer; Epicentre Biotechnologies, Madison, WI). DNA was sequenced by Nevada Genomics Center (Reno, NV). Pairwise comparison against GenBank sequences was performed using BLAST. The promoter was predicted using the promoter prediction tool on the Berkeley Drosophila Genome Project website (http://www.fruitfly.org/seq_tools/promoter.html).
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rRNA genes of Bradyrhizobium JS329 and the sequences of genes involved in 5NAA degradation were deposited in GenBank under accession numbers GU188568 and GU188569, respectively.
RESULTS
Isolation and identification of 5NAA-degrading bacteria.
Biodegradation of 5NAA was detected in enrichment culture after 8 days with 5NAA as the sole source of carbon, nitrogen, and energy. Among several soil samples, only soil from a potato farm yielded a bacterial isolate able to grow on 5NAA. The partial 16S rRNA gene sequence (794 bp) of the isolate was 99% identical to the 16S rRNA genes of Bradyrhizobium elkanii, but because 16S rRNA genes do not discriminate among Bradyrhizobium species (31), the isolate was designated Bradyrhizobium sp. JS329.
Growth of bacteria.
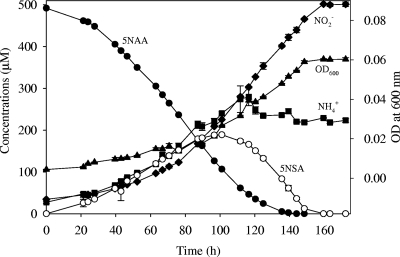
JS329 grew best at pH 7.2 and 30°C on concentrations of 5NAA up to 5 mM. During growth on 5NAA, JS329 released stoichiometric amounts of nitrite along with some ammonia, which indicates that nitrite is released without prior reduction or assimilation and that the ammonia serves as the source of nitrogen. 5-Nitrosalicylic acid (5NSA) accumulated transiently during 5NAA degradation (Fig. 1). The release of 5NSA suggested that the amino group was released before the nitro group.
FIG. 1.
Growth of Bradyrhizobium sp. JS329 on 5NAA as the sole carbon, nitrogen, and energy source. •, 5NAA; ♦, NO2−; ○, 5NSA; ▴, optical density at 600 nm (OD600); ▪, NH4+.
JS329 also grew on 5NSA as the sole carbon, nitrogen, and energy source with the accumulation of nitrite (0.5 mol NO2−/mol 5NSA) in the culture medium. We hypothesized that the initial attack might be on the adjacent amino and carboxyl groups, similar to the initial step in the anthranilic acid degradation pathway, to yield 4-nitrocatechol, but 4-nitrocatechol did not serve as a growth substrate, which suggested that 4-nitrocatechol is not involved in the degradation of 5NAA. The growth yields were similar for growth on 5NAA and 5NSA (data not shown). Either ammonium or nitrite could serve as the nitrogen source for growth. JS329 grew well with 5NAA as the nitrogen source and glucose, succinate, pyruvate, or acetate as the carbon source.
Respirometry.
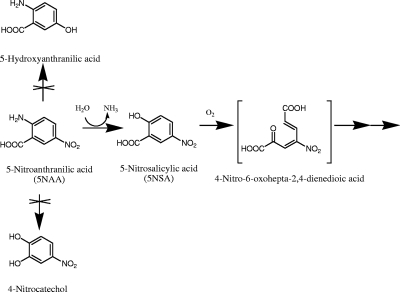
The above-described preliminary experiments suggested that 5NSA was produced from 5NAA and that nitrite was formed by subsequent removal of the nitro group. Possible transformation pathways of 5NAA could also involve the formation of 4-nitrocatechol (17) or 5-hydroxyanthranilic acid along with release of nitrite (5) (Fig. 2). The three potential intermediates were tested for the ability to stimulate oxygen uptake by 5NAA- or succinate-grown cells (see Table 3). Negative results with 4-nitrocatechol and 5-hydroxyanthranilic acid suggested that the two compounds are not involved in the 5NAA degradation pathway. Stimulation of oxygen uptake by 5NAA and 5NSA in 5NAA-grown cells, but not in succinate-grown cells, suggested that inducible enzymes catalyzing transformation of 5NAA and 5NSA were involved in the pathway.
FIG. 2.
Hypothetical initial reactions in the biodegradation of 5NAA. The middle pathway was supported by this study.
TABLE 3.
O2 uptake by 5NAA- or succinate-grown cellsa
| Test substrate | Oxygen uptake (nmol O2/min/mg of protein) by cells grown in: |
|
|---|---|---|
| 5-Nitroanthranilic acid | Succinic acid | |
| 5-Nitroanthranilic acidb | 5.7 ± 0.42 | ND |
| 5-Nitrosalicylic acidc | 6.6 ± 0.85 | ND |
| Gentisic acid | 2 ± 0.84 | ND |
| 5-Hydroxyanthranilic acid | ND | ND |
| 4-Nitrocatechol | ND | ND |
| Succinic acid | NT | 5.4 |
The reaction mixtures contained 1.8 ml of oxygen-saturated phosphate buffer (0.02 M; pH 7.0; 25°C), cells, and test substrates (20 μM). The experiments were done in duplicate except for the succinate control. ND, not detected; NT, not tested.
The stoichiometry was 1.9 ± 0.1 mol O2/mol substrate.
The stoichiometry was 1.7 ± 0.1 mol O2/mol substrate.
Cloning of the genes involved in 5NAA degradation.
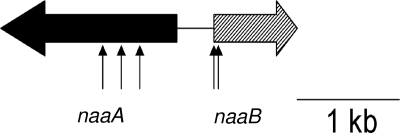
When the fosmid library was screened for clones with the ability to transform 5NAA, 3 out of 5,000 clones were able to transform both 5NAA and 5NSA to non-UV-absorbing metabolites. Tn5 mutants were prepared from pJS800. One group of mutants exemplified by pJS801 lost the ability to transform 5NAA but not 5NSA. Mutants (pJS802) that lost the ability to transform 5NSA transformed 5NAA stoichiometrically to 5NSA (data not shown). Primer walking yielded about 2.2 kb of sequence containing two adjacent and divergently transcribed genes (Fig. 3). The 5NAA deaminase (encoded by naaA) (Fig. 3) is distantly (34% amino acid identity) related to an uncharacterized M20/M25/M40 family peptidase from Hyphomonas neptunium ATCC 15444. 5NSA 1,2-dioxygenase (encoded by naaB) (Fig. 3) is distantly (29% amino acid identity) related to gentisate 1,2-dioxygenase from Oligotropha carboxidovorans OM5 (23) and salicylate 1,2-dioxygenase (13) from Pseudaminobacter salicylatoxidans (22% amino acid identity). The deduced 5NSA 1,2-dioxygenase amino acid sequence contained a conserved domain in common with salicylate dioxygenase and gentisate 1,2-dioxygenase (TIGR02272 and COG3435) (19). The promoter sequence for the naaB gene, found in the 400-bp intergenic region, is TCGTTCTGTAGAACGAACGACGATCTGTATATTGTTACTGGGAGGGTGAC, and the promoter sequence for the naaA gene is TGAATTTTGGCTGTTGACCGTAGACAGAACATCGTTTAATACCCCGAACA. The transcription starts at G or A (underlined bold character). The results for 5NAA and 5NSA transformation by the clones are consistent with hydrolytic deamination of 5NAA to 5NSA followed by ring cleavage catalyzed by the dioxygenase.
FIG. 3.
Identification of key genes. naaA, gene encoding 5NAA deaminase; naaB, gene encoding 5NSA 1,2-dioxygenase. Sites of Tn5 transposon insertion are marked by arrows.
Enzyme assays.
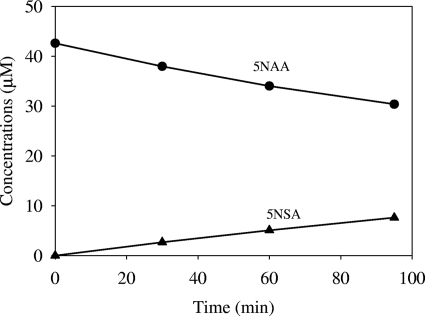
Ultrafiltered extracts from 5NAA-grown cells of Bradyrhizobium JS329 catalyzed stoichiometric transformation of 5NAA to 5NSA (specific activity, 0.8 ± 0.32 nmol/min/mg protein). No added cofactors were required for the transformation (Fig. 4). The results strongly suggest that the initial reaction involves hydrolytic elimination of the amino group. The equal stoichiometry of oxygen consumption versus substrate for 5NAA and 5NSA in intact cells clearly shows that no oxygen is involved in the conversion of 5NAA to 5NSA and strongly supports the above conclusions (see Table 3).
FIG. 4.
Stoichiometric accumulation of 5-nitrosalicylate (5NSA) from 5NAA when ultrafiltered extract from 5NAA-grown cells was incubated with 5NAA at 30°C. •, 5NAA; ▴, 5NSA.
Enzymes in crude extracts of 5NAA-grown cells of Bradyrhizobium JS329 catalyzed the oxidation of 5NSA (specific activity, 1.2 ± 0.2 nmol/min/mg protein). Gentisate was also oxidized at rates (19 ± 1.7 nmol/min/mg protein) higher than those in intact cells, which suggests that transport was limiting for gentisate. However, enzymes in cell extracts of the E. coli-containing fosmid clone pJS800, harboring naaB, did not attack gentisate but did transform salicylate (specific activity, 3.9 ± 0.9 nmol/min/mg protein) and 5NSA (specific activity, 11 ± 0.5 nmol/min/mg protein). The results indicate that gentisate dioxygenation activity in JS329 was not catalyzed by the enzyme transforming 5NSA, but the expression of the gentisate dioxygenase gene(s) in another locus was induced during growth on 5NAA.
Ultrafiltration abolished the enzymatic activity against 5NSA, which was restored by addition of ferrous but not ferric ions. Addition of flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), dithiothreitol (DTT), or NAD(P)H had no effect on the activity (Table 2). Nitrite was not detected during the transformation of 5NSA, and no UV-absorbing metabolites were detected by HPLC.
TABLE 2.
Effect of additions on transformation of 5NSA by ultrafiltered extract from 5NAA-grown cellsa
| Addition(s) | Relative rate (%) |
|---|---|
| Fe2+ | 100 |
| NADH | 23.3 ± 1.7 |
| Fe3+ | 9.4 ± 1.2 |
| Fe2+, NADH | 87.7 ± 2.2 |
| Fe2+, NADH, FAD | 102.1 ± 0.6 |
| Fe2+, NADH, FMN | 90.4 ± 0.8 |
| Fe2+, NADH, DTT | 70 ± 14 |
| No cofactor | 17 ± 2.4 |
Cells were incubated at 30°C with shaking. The concentrations were as follows: for Fe2+, 50 μM; for Fe3+, 50 μM; for NADH, 150 μM; for FMN, 10 μM; for FAD, 10 μM; and for DTT, 1 mM.
5NSA oxidation catalyzed by cell extracts required 0.97 ± 0.09 mol O2/mol 5NSA. Upon incubation with 2,2′-dipyridyl (1 mM) or o-phenanthroline (1 mM), the enzymatic activity was inhibited by 97.7% or 100%, respectively. The requirement for oxygen and Fe2+ but no external electron donor suggested that 5NSA transformation is catalyzed by a ring cleavage dioxygenase.
The properties of the enzyme catalyzing 5NSA oxidation were similar to those of salicylate 1,2-dioxygenase: 1 mol of oxygen was consumed per mol of substrate, 40% to 50% of original activity was lost after 1 week of storage at −20°C, Fe2+ was required, NAD(P)H was not required, and the enzyme was active against salicylate and inhibited by iron chelators (12).
DISCUSSION
Our study provides the first clear evidence for the initial steps in biodegradation of nitroanilines. The enzymatic removal of either the amino group or the nitro group might be expected to initiate biodegradation of such molecules. Amino groups attached to aromatic rings are usually removed in reactions catalyzed by dioxygenases. For example, aniline is converted to catechol by aniline dioxygenase, a multicomponent dioxygenase (10, 29). Anthranilate (2-aminobenzoate) is transformed to catechol through the simultaneous removal of the adjacent amino and carboxyl groups by a dioxygenase in Pseudomonas (17) or Burkholderia cepacia DBO1 (6). A flavoprotein hydroxylase in the yeast Trichosporon cutaneum (24) converts anthranilate into 2,3-dihydroxybenzoate, whereas enzymes in Nocardia opaca convert it to 5-hydroxyanthranilate (5). The difference in deamination mechanisms among 5NAA-, anthranilate-, and aniline-degrading enzymes might be caused by steric or electronic effects of the nitro group in the 5NAA molecule. Aminohydrolases are commonly involved in elimination of amino groups from aliphatic and heterocyclic compounds (16, 28) but have not been reported to act on aromatic compounds. Purification and characterization of the novel enzyme will be required to determine its substrate range and the mechanism of the unusual reaction.
The fact that gentisate can stimulate the oxygen uptake by 5NAA-grown cells (Table 3) at first suggested that 5NSA was converted to gentisate by a flavoprotein analogous to nitrophenol monooxygenase (14). 5NSA transformation in cell extracts without an added electron donor and no nitrite accumulation established clearly that another mechanism must be involved. Our results indicate that the reaction is a ring cleavage catalyzed by a dioxygenase that is active against both 5NSA and salicylate but not gentisate. The finding is consistent with the report that an enzyme from Pseudaminobacter salicylatoxidans with 28% identity to gentisate 1,2-dioxygenase from Pseudomonas alcaligenes NCIMB 9867 can oxidize salicylate and a variety of substituted salicylates but not 5NSA (13). Our results indicate that the ring cleavage dioxygenase has evolved the ability to oxidize 5NSA specifically.
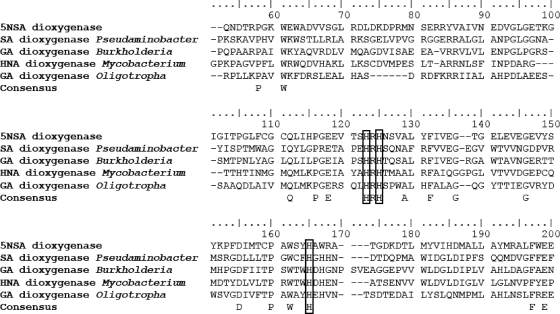
The conservation of a histidine pair is essential for the anchoring of ferrous ion in the catalytic center of salicylate and gentisate 1,2-dioxygenases (7, 13). Alignment of the sequences of salicylate 1,2-dioxygenase from Pseudaminobacter salicylatoxidans, gentisate 1,2-dioxygenase from Burkholderia multivorans CGD2M, gentisate 1,2-dioxygenase from Oligotropha carboxidovorans OM5, 1-hydroxy-2-naphthoic acid dioxygenase from Mycobacterium sp. CH1, and 5NSA 1,2-dioxygenase reveals that the histidine pair is highly conserved (Fig. 5).
FIG. 5.
Multiple sequence alignment of 5NSA 1,2-dioxygenase from Bradyrhizobium JS329, salicylate-1,2-dioxygenase from Pseudaminobacter salicylatoxidans BN12 (GenBank accession number AY323951.1), 1-hydroxy-2-naphthoic acid dioxygenase from Mycobacterium sp. CH1 (GenBank accession number ACN38281.1), gentisate 1,2-dioxygenase from Burkholderia multivorans CGD2M (GenBank accession number ZP_03569724.1), and gentisate 1,2-dioxygenase from Oligotropha carboxidovorans OM5 (GenBank accession number YP_002287389.1). The conserved histidine pair is present at positions 123 and 125. The alignment in the two ends was trimmed due to the absence of conserved sites. SA, salicylate; GA, gentisate; 5NSA, 5-nitrosalicylic acid; HNA, 1-hydroxy-2-naphthoic acid.
Inducible activities of 5NAA deaminase and 5NSA 1,2-dioxygenase were detected in both intact cells and cell extracts at specific activities sufficient to support the growth of JS329. The results indicate that the pathway is initiated by a novel hydrolytic deaminase to produce 5NSA, which in turn is oxidized by a ring fission dioxygenase (Fig. 2).
The mechanism of removal of the nitro group represents a particularly interesting question. For most nitrophenols, nitro group removal is catalyzed by NAD(P)H-dependent monooxygenase enzymes prior to cleavage of the aromatic ring (22). Biodegradation of 5NAA, 2,6-DNT (2,6-dinitrotoluene), and picric acid are exceptions to the general rule (8, 20). In such cases, the nitrophenol serves as the ring fission substrate. The mechanism of release of the nitro group from the ring fission product is unknown (20).
The failure to detect 5NAA-degrading bacteria in the majority of the soil samples suggests that the distribution of 5NAA degraders is limited, as did the earlier work by Hallas and Alexander (11). It seems likely that ecosystems that harbor 5NAA producers would also harbor 5NAA degraders, but the relationship between 5NAA production and degradation remains to be determined.
Acknowledgments
We thank Alok Shrestha, Youn Kim, Jill Seladi, and Se na Yoo for technical assistance. We thank Andreas Stolz for helpful suggestions and Shirley Nishino for reviewing the manuscript.
This work was supported by the Defense Threat Reduction Agency and U.S. Army Research Office grant W911NF-07-1-0077.
Footnotes
Published ahead of print on 15 January 2010.
REFERENCES
- 1.Alexander, M., and B. K. Lustigma. 1966. Effect of chemical structure on microbial degradation of substituted benzenes. J. Agric. Food Chem. 14:410-413. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Baxter, R. A., and D. Bennion. May 1956. Process for the separation of mixtures of 4- and 5-nitroanthranilic acid and their salts. U.S. patent 749,320.23.
- 4.Bruhn, C., H. Lenke, and H. J. Knackmuss. 1987. Nitrosubstituted aromatic compounds as nitrogen source for bacteria. Appl. Environ. Microbiol. 53:208-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cain, R. B. 1968. Anthranilic acid metabolism by microorganisms. Formation of 5-hydroxyanthranilate as an intermediate in anthranilate metabolism by Norcardia opaca. Antonie Van Leeuwenhoek 34:417-432. [PubMed] [Google Scholar]
- 6.Chang, H. K., P. Mohseni, and G. J. Zylstra. 2003. Characterization and regulation of the genes for a novel anthranilate 1,2-dioxygenase from Burkholderia cepacia DBO1. J. Bacteriol. 185:5871-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chua, C. H., Y. Feng, C. C. Yeo, H. E. Khoo, and C. L. Poh. 2001. Identification of amino acid residues essential for catalytic activity of gentisate 1,2-dioxygenase from Pseudomonas alcaligenes NCIB 9867. FEMS Microbiol. Lett. 204:141-146. [DOI] [PubMed] [Google Scholar]
- 8.Ebert, S., P. G. Rieger, and H. J. Knackmuss. 1999. Function of coenzyme F420 in aerobic catabolism of 2,4,6-trinitrophenol and 2,4-dinitrophenol by Nocardioides simplex FJ2-1A. J. Bacteriol. 181:2669-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehring, B., J. Prassler, P. R. Wallnoefer, and W. Ziegler. 1992. Microbial transformation of o-nitroaniline and p-nitrophenol. Umweltwiss. Schadst. Forsch. 4:81-85. [Google Scholar]
- 10.Fukumori, F., and C. P. Saint. 1997. Nucleotide sequences and regulational analysis of genes involved in conversion of aniline to catechol in Pseudomonas putida UCC22(pTDN1). J. Bacteriol. 179:399-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallas, L. E., and M. Alexander. 1983. Microbial transformation of nitroaromatic compounds in sewage effluent. Appl. Environ. Microbiol. 45:1234-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hintner, J. P., C. Lechner, U. Riegert, A. E. Kuhm, T. Storm, T. Reemtsma, and A. Stolz. 2001. Direct ring fission of salicylate by a salicylate 1,2-dioxygenase activity from Pseudaminobacter salicylatoxidans. J. Bacteriol. 183:6936-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hintner, J. P., T. Reemtsma, and A. Stolz. 2004. Biochemical and molecular characterization of a ring fission dioxygenase with the ability to oxidize (substituted) salicylate(s) from Pseudaminobacter salicylatoxidans. J. Biol. Chem. 279:37250-37260. [DOI] [PubMed] [Google Scholar]
- 14.Kadiyala, V., and J. C. Spain. 1998. A two-component monooxygenase catalyzes both the hydroxylation of p-nitrophenol and the oxidative release of nitrite from 4-nitrocatechol in Bacillus sphaericus JS905. Appl. Environ. Microbiol. 64:2479-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King, R. R., C. H. Lawrence, and L. A. Calhoun. 1998. Unusual production of 5-nitroanthranilic acid by Streptomyces scabies. Phytochemistry 49:1265-1267. [Google Scholar]
- 16.Klein, M., H. Kaltwasser, and T. Jahns. 2002. Isolation of a novel, phosphate-activated glutaminase from Bacillus pasteurii. FEMS Microbiol. Lett. 206:63-67. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi, S., S. Kuno, N. Itada, O. Hayaishi, S. Kozuka, and S. Oae. 1964. O-18 studies on anthranilate hydroxylase—a novel mechanism of double hydroxylation. Biochem. Biophys. Res. Commun. 16:556-561. [DOI] [PubMed] [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY.
- 19.Marchler-Bauer, A., J. B. Anderson, F. Chitsaz, M. K. Derbyshire, C. DeWeese-Scott, J. H. Fong, L. Y. Geer, R. C. Geer, N. R. Gonzales, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, A. Tasneem, N. Thanki, R. A. Yamashita, D. Zhang, N. Zhang, and S. H. Bryant. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205-D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishino, S. F., G. C. Paoli, and J. C. Spain. 2000. Aerobic degradation of dinitrotoluenes and pathway for bacterial degradation of 2,6-dinitrotoluene. Appl. Environ. Microbiol. 66:2139-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino, S. F., and J. C. Spain. 2006. Biodegradation of 3-nitrotyrosine by Burkholderia sp. strain JS165 and Variovorax paradoxus JS171. Appl. Environ. Microbiol. 72:1040-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishino, S. F., and J. C. Spain. 2004. Catabolism of nitroaromatic compounds, p. 575-608. In J.-L. Ramos (ed.), Pseudomonas, vol III. Biosynthesis of macromolecules and molecular metabolism. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 23.Paul, D., S. Bridges, S. C. Burgess, Y. Dandass, and M. L. Lawrence. 2008. Genome sequence of the chemolithoautotrophic bacterium Oligotropha carboxidovorans OM5T. J. Bacteriol. 190:5531-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powlowski, J. B., S. Dagley, V. Massey, and D. P. Ballou. 1987. Properties of anthranilate hydroxylase (deaminating), a flavoprotein from Trichosporon cutaneum. J. Biol. Chem. 262:69-74. [PubMed] [Google Scholar]
- 25.Qu, Y., and J. C. Spain. 2009. Abstr. 109th Gen. Meet. Am. Soc. Microbiol., abstr. Q-268. American Society for Microbiology, Washington, DC.
- 26.Qureshi, A., V. Verma, A. Kapley, and H. J. Purohit. 2007. Degradation of 4-nitroaniline by Stenotrophomonas strain HPC 135. Int. Biodeterior. Biodegradation 60:215-218. [Google Scholar]
- 27.Saupe, A. 1999. High-rate biodegradation of 3- and 4-nitroaniline. Chemosphere 39:2325-2346. [DOI] [PubMed] [Google Scholar]
- 28.Seffernick, J. L., M. L. de Souza, M. J. Sadowsky, and L. P. Wackett. 2001. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J. Bacteriol. 183:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urata, M., E. Uchida, H. Nojiri, T. Omori, R. Obo, N. Miyaura, and N. Ouchiyama. 2004. Genes involved in aniline degradation by Delftia acidovorans strain 7N and its distribution in the natural environment. Biosci. Biotechnol. Biochem. 68:2457-2465. [DOI] [PubMed] [Google Scholar]
- 30.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willems, A., R. Coopman, and M. Gillis. 2001. Phylogenetic and DNA-DNA hybridization analyses of Bradyrhizobium species. Int. J. Syst. Evol. Microbiol. 51:111-117. [DOI] [PubMed] [Google Scholar]
- 32.Zeyer, J., and P. C. Kearney. 1983. Microbial metabolism of [14C] nitroanilines to [14C] carbon dioxide. J. Agric. Food Chem. 31:304-308. [Google Scholar]