Abstract
To overcome some of the deficiencies with current molecular typing schema for Campylobacter spp., we developed a prototype PCR binary typing (P-BIT) approach. We investigated the distribution of 68 gene targets in 58 Campylobacter jejuni strains, one Campylobacter lari strain, and two Campylobacter coli strains for this purpose. Gene targets were selected on the basis of distribution in multiple genomes or plasmids, and known or putative status as an epidemicity factor. Strains were examined with Penner serotyping, pulsed-field gel electrophoresis (PFGE; using SmaI and KpnI enzymes), and multilocus sequence typing (MLST) approaches for comparison. P-BIT provided 100% typeability for strains and gave a diversity index of 98.5%, compared with 97.0% for SmaI PFGE, 99.4% for KpnI PFGE, 96.1% for MLST, and 92.8% for serotyping. Numerical analysis of the P-BIT data clearly distinguished strains of the three Campylobacter species examined and correlated somewhat with MLST clonal complex assignations and with previous classifications of “high” and “low” risk. We identified 18 gene targets that conferred the same level of discrimination as the 68 initially examined. We conclude that P-BIT is a useful approach for subtyping, offering advantages of speed, cost, and potential for strain risk ranking unavailable from current molecular typing schema for Campylobacter spp.
Campylobacter species, particularly C. jejuni subsp. jejuni (hereafter C. jejuni), represent the most commonly reported bacterial cause of gastroenteritis in humans in the developed world (47), with New Zealand having one of the highest rates of infection (55). The sheer scale of infection makes concerted epidemiological studies difficult, as does the extremely wide distribution of the organism, found in all major avian and mammalian food animals, their products, and indeed environments. Moreover, many Campylobacter spp. are susceptible to spontaneous genetic change through a variety of mechanisms that can result in conflicting data for genetic typing methods aiming to establish a molecular epidemiological link between strains (reviewed by On and colleagues [47]).
The poor discrimination of phenotypic typing methods led to intense developments in molecular epidemiological tools for more accurate data. Although a wide range of genotypic methods have been described (47), two methods are now more commonly used by laboratories worldwide. The availability of standardized protocols for macrorestriction profiling with pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) have facilitated major contributions to our understanding of the epidemiology of these bacteria. Nonetheless, issues remain, notably relating to the speed, cost, and ease of data analysis from these methods. Furthermore, although MLST has proven useful in evaluating the original host of a given strain, no current methods provide information on the relative risk to human health from individual strains. Various studies, including those identifying stable clones found in humans and various animals as well as strain types only in a particular animal host (5, 13, 38, 48, 61), and whole-genome microarray-based comparisons revealing a correlation between genome content and stress survival (46) indicate that not all strains are of equal risk to humans.
In this study, we designed a range of specific PCR assays and investigated the distribution of 68 genes associated with epidemicity factors in C. jejuni, to establish the basis of a novel PCR binary typing (P-BIT) system that is inexpensive, rapid, and highly portable. We compared our data with MLST and PFGE (using restriction enzymes SmaI and KpnI) results for the same isolates of C. jejuni (n = 58), C. coli (n = 2), and C. lari (n = 1).
MATERIALS AND METHODS
Bacterial isolates.
Fifty-eight C. jejuni isolates, 2 C. coli isolates, and 1 C. lari isolate from four countries and a variety of sources were included in this study (Table 1). Isolates were grown on Columbia sheep blood agar plates for 48 h at 42°C under microaerobic conditions.
TABLE 1.
Campylobacter jejuni binary typing isolates
| Isolate no. | Species | Country | Source | Isolation date | P-BIT coded | Penner serotype(s) | PFGE type |
MLST type |
||
|---|---|---|---|---|---|---|---|---|---|---|
| SmaI | KpnI | ST | CC | |||||||
| CMB06688/2 | C. jejuni | New Zealand | Bovine | January 2007 | 005551 | 3 | Sm029 | Kp186 | 3607 | 1332 |
| CMB06690 | C. coli | New Zealand | Bovine | January 2007 | 250640 | Sm131 | Kp132 | 3072 | 828 | |
| CMB06690/1 | C. lari | New Zealand | Bovine | January 2007 | 006000 | Sm207 | Kp185 | |||
| CPH003647 | C. jejuni | New Zealand | Bovine | August 2000 | 774601 | 10 | Sm098 | Kp260 | 520 | 21 |
| CPH004049 | C. jejuni | New Zealand | Bovine | September 2000 | 774601 | 10 | Sm039 | Kp060 | 520 | 21 |
| CPH004158 | C. jejuni | New Zealand | Human | September 2000 | 774601 | 10 | Sm039 | Kp060 | 520 | 21 |
| CPH005816 | C. jejuni | New Zealand | Duck | December 2001 | 000512 | 8, 17 | Sm223 | Kp018 | 995 | Ub |
| CPH010117 | C. jejuni | New Zealand | Duck | January 2001 | 200610 | 5 | Sm147 | Kp271 | 699 | 692 |
| CPH010873 | C. jejuni | New Zealand | Bovine | February 2001 | 770601 | 10 | Sm016 | Kp262 | 520 | 21 |
| CPH0110608 | C. jejuni | New Zealand | Water | October 2001 | 000675 | 2 | Sm140 | Kp098 | 977 | 1034 |
| CPH0110747 | C. jejuni | New Zealand | Water | October 2001 | 420000 | 5 | Sm001 | Kp087 | 3581 | U |
| CPH0112578j | C. jejuni | New Zealand | Water | December 2001 | 424200 | 1 | Sm218 | Kp252 | 2381 | U |
| CPH011377 | C. jejuni | New Zealand | Bovine | March 2001 | 477600 | 1, 44 | Sm098 | Kp179 | 50 | 21 |
| CPH012121 | C. jejuni | New Zealand | Water | April 2001 | 600420 | 15 | Sm235 | Kp102 | 45 | 45 |
| CPH013864 | C. jejuni | New Zealand | Duck | June 2001 | 020550 | NTa | Sm147 | Kp129 | 1525 | U |
| CPH014045 | C. jejuni | New Zealand | Water | July 2001 | 200010 | 33 | Sm100 | Kp190 | 1457 | U |
| CPH014202 | C. jejuni | New Zealand | Water | July 2001 | 420000 | 27 | Sm001 | Kp290 | 3580 | U |
| CPH0210258 | C. jejuni | New Zealand | Human | January 2002 | 410450 | 52 | Sm001 | Kp184 | 418 | 45 |
| CPH0214000-01 | C. jejuni | New Zealand | Ovine | October 2002 | 074010 | 5 | Sm053 | Kp373 | 2392 | 52 |
| CPH0214086-01 | C. jejuni | New Zealand | Human | October 2002 | 270000 | 3 | Sm033 | Kp080 | 436 | U |
| CPH0214128-01 | C. jejuni | New Zealand | Human | October 2002 | 675610 | 8, 17 | Sm036 | Kp096 | 50 | 21 |
| CPH0214244-01 | C. jejuni | New Zealand | Ovine | October 2002 | 776601 | 10 | Sm037 | Kp019 | 520 | 21 |
| CPH0214392-01 | C. jejuni | New Zealand | Human | October 2002 | 020410 | 5 | Sm090 | Kp079 | 693 | U |
| CPH0214443-01 | C. jejuni | New Zealand | Human | October 2002 | 274540 | 15 | Sm068 | Kp088 | 3606 | U |
| CPH0214447-01 | C. jejuni | New Zealand | Human | October 2002 | 270010 | 3 | Sm033 | Kp081 | 436 | U |
| CPH0214589-01 | C. jejuni | New Zealand | Human | October 2002 | 400030 | 45 | Sm001 | Kp032 | 137 | 45 |
| CPH0310726-01 | C. jejuni | New Zealand | Ovine | February 2003 | 274000 | 5 | Sm053 | Kp373 | 2382 | 52 |
| CPH0310881-01 | C. jejuni | New Zealand | Human | February 2003 | 377630 | 1, 44 | Sm098 | Kp043 | 50 | 21 |
| CPH0310892-01 | C. jejuni | New Zealand | Human | February 2003 | 677610 | 1, 44 | Sm098 | Kp043 | 50 | 21 |
| CPH0311081-01 | C. jejuni | New Zealand | Human | March 2003 | 677600 | 2 | Sm246 | Kp119 | 53 | 21 |
| CPH0311417-01 | C. jejuni | New Zealand | Human | March 2003 | 005500 | 9 | Sm152 | Kp068 | 2374 | U |
| CPH0311750 | C. coli | New Zealand | Human | April 2003 | 250040 | Sm142 | Kp042 | 829 | 828 | |
| CPH0311897-01 | C. jejuni | New Zealand | Human | April 2003 | 377610 | 2 | Sm039 | Kp038 | 53 | 21 |
| CPH0312128-01 | C. jejuni | New Zealand | Human | April 2003 | 274050 | 11 | Sm021 | Kp109 | 257 | 257 |
| CPH0312133-01 | C. jejuni | New Zealand | Human | April 2003 | 677600 | 1, 44 | Sm038 | Kp037 | 50 | 21 |
| CPH0312157 | C. jejuni | New Zealand | Water | April 2003 | 377630 | 2 | Sm048 | Kp021 | 53 | 21 |
| CPH0314240 | C. jejuni | New Zealand | Chicken | October 2003 | 274050 | 11 | Sm021 | Kp178 | 257 | 257 |
| CPH0315217 | C. jejuni | New Zealand | Water | December 2003 | 410030 | 45 | Sm001 | Kp032 | 137 | 45 |
| CPH0410333 | C. jejuni | New Zealand | Chicken | January 2004 | 677600 | 1 | Sm038 | Kp037 | 50 | 21 |
| CPH0411548 | C. jejuni | New Zealand | Chicken | April 2004 | 674150 | 58 | Sm002 | Kp004 | 1517 | 354 |
| CPH0411624-04 | C. jejuni | New Zealand | Water | May 2004 | 201611 | 5 | Sm145 | Kp243 | 991 | 692 |
| CPH0512595C | C. jejuni | New Zealand | Chicken | September 2005 | 674150 | NT | Sm047 | Kp173 | 1517 | 354 |
| NCTC11168 | C. jejuni | U.K. | Human | 1977 | 777630 | 2 | Sm039 | Kp243 | 43 | 21 |
| NZRM 4128 | C. jejuni | New Zealand | Human | 2002 | 677600 | 1, 44 | Sm013 | Kp020 | 50 | 21 |
| NZRM 4136 | C. jejuni | New Zealand | Chicken | 2001 | 410552 | 27 | Sm004 | Kp002 | 474 | 48 |
| NZRM 4141 | C. jejuni | New Zealand | Human | 2003 | 411440 | 25 | Sm001 | Kp013 | 45 | 45 |
| P110b | C. jejuni | New Zealand | Chicken | August 2005 | 374610 | 4, 13 | Sm010 | Kp066 | 474 | 48 |
| P145a | C. jejuni | New Zealand | Chicken | September 2005 | 374650 | 4, 13 | Sm006 | Kp299 | 3609 | 48 |
| RM1221 | C. jejuni | USA | Chicken | 2000 | 254150 | 53 | Sm168 | Kp301 | 354 | 354 |
| RM1864 = 81-176 | C. jejuni | USA | Human | 1988 | 011013 | 23, 36 | Sm248 | Kp315 | 604 | 42 |
| SVS1099 | C. jejuni | Denmark | Chicken | 777610 | 2 | Sm037 | Kp295 | 21 | 21 | |
| SVS1425 | C. jejuni | Denmark | Chicken | 677600 | 1, 44 | Sm083 | Kp060 | 50 | 21 | |
| SVS3141 | C. jejuni | Denmark | Porcine | 377630 | 2 | Sm039 | Kp039 | 53 | 21 | |
| SVS380-827 | C. jejuni | Denmark | Chicken | 410550 | 55 | Sm001 | Kp003 | 45 | 45 | |
| SVS4039 | C. jejuni | Denmark | Bovine | 410440 | 6, 7 | Sm001 | Kp291 | 45 | 45 | |
| SVS5001 | C. jejuni | Denmark | Human | 1996 | 377630 | 2 | Sm016 | Kp039 | 53 | 21 |
| SVS5141 | C. jejuni | Denmark | Human | 1996 | 377630 | 2 | Sm016 | Kp039 | 53 | 21 |
| SVS72-64077 | C. jejuni | Denmark | Turkey | 000555 | 17 | Sm252 | Kp364 | 48 | 48 | |
| SVS72-74172 | C. jejuni | Denmark | Turkey | 270030 | NT | Sm009 | Kp294 | 48 | 48 | |
| SVS835-770 | C. jejuni | Denmark | Chicken | 410010 | 42 | NVc | NV | NV | NV | |
| SVS992 | C. jejuni | Denmark | Chicken | 777630 | 2 | Sm176 | Kp314 | 21 | 21 | |
NT, not typeable.
U, unassigned.
NV, nonviable.
To generate the P-BIT code, the genes from the 18-target P-BIT scheme were divided into six groups of three genes in the order shown in Table 2. The PCR results were recorded as 0 or 1, and the result for the first gene in the group was multiplied by 1, the second by 2, and the third by 4. The results were then added together to give a number from 0 to 7 representing the results for the three genes. The six groups’ results were then concatenated.
DNA extraction.
Between 5 and 10 colonies were transferred to a 1.5-ml tube containing phosphate-buffered saline (PBS) (BR0014G; Oxoid, Basingstoke, England) and centrifuged at 6,000 × g. The supernatant was discarded and DNA was extracted from the pellet using DNeasy blood and tissue kits (Qiagen, Hilden, Germany). The DNA quantity and purity were established using the Nanodrop 1000 (Thermo Scientific, Waltham, MA), and the DNA was stored at −20°C until required. DNA was diluted in sterile Milli-Q water as required.
Risk-based binary typing of isolates.
Selection of gene targets for potential use in a PCR binary typing (P-BIT) system was based upon several criteria. We chose predominantly those genes implicated as markers of epidemicity that were overrepresented in genotypes associated with human illness (1, 4, 14, 16, 17, 20, 22, 26-31, 35-37, 43, 44, 46, 49, 50, 52, 53, 59, 60, 64, 65, 67, 69-71) or associated with virulence factors that were not components of the core genome (33, 34, 60), to evaluate a basis for subtyping that could also be related to the risk to human health from individual strains. These genes were generally involved in cell surface, mobility, and toxin production. Primers were designed using Primer3 (58) via Geneious Pro (10; available from http://www.geneious.com/) and the sequences of the identified genes and the identified strains as loaded in GenBank. Primers were designed to produce PCR products with sizes and melting temperatures that will allow for easy multiplexing in the future. The primers were checked for sequence homology with all known sequences using the Basic Local Alignment Search Tool (BLAST; www.ncbi.nlm.nih.gov/BLAST/) and synthesized by Invitrogen (Carlsbad, CA). In addition, one primer set (Cj0265) designed by Price and colleagues (56) was also included in the risk-based binary typing system. The target genes and primer sequences are listed in Table 2.
TABLE 2.
Target genes for Campylobacter jejuni binary typing
| Gene/ORFc | Primera | Product size (bp) | Reference(s) |
|---|---|---|---|
| Cj0008 | For: AGTCCTGAAGCGTTTGCTGT | 245 | 8 |
| Rev: CATTTTTCATTTACCCATTGCTT | |||
| cdtB (Cj0078c) | For: AGCCACAGAAAGCAAATGGA | 550 | 59 |
| Rev: TGCTTGAGTTGCGCTAGTTG | |||
| Cj0122 | For: TCCTTTGCAAACACTTACCAA | 250 | 46 |
| Rev: CCAACGAGGGTGTTTAGCAT | |||
| exbD1 (Cj0180) | For: AGAGTTCGCAGCAAGCAGAG | 248 | 46, 70 |
| Rev: TGAAAGAGCTATTTTTCCATAGCC | |||
| tonB1 (Cj0181) | For: TTTTAAACAAGGAGCGGAATTT | 547 | 46, 70 |
| Rev: TTTTGGCAATGGAGAAGCTC | |||
| Cj0265b | For: AAGCGAAAATAACAGGGTTTTGC | 175 | 56 |
| Rev: GCTTACCTTATCCCATTTGCCA | |||
| cheW (Cj0283c) | For: TGCAAAAACAGCAAACTCAGA | 398 | 31, 69 |
| Rev: AAGTTTCAGGTGGTGGATCG | |||
| panB (Cj0298c) | For: TTTCTCGCGGAGCTAAAAAG | 150 | 8 |
| Rev: CGATTTCTATACCCCCTTCCA | |||
| modC (Cj0300) | For: GCCTAAATTGGGGCGTATAGA | 299 | 46 |
| Rev: GCAAGATTTTTGGTTCTCTTGC | |||
| fdxA (Cj0333c) | For: TGCTTGTGGTTCTTGTATTGATG | 245 | 69 |
| Rev: CTGGAGTGTCCCCACTTCTC | |||
| cmeB (Cj0366c) | For: CAACCCGCACCAGGTTATAC | 546 | 35, 36, 50 |
| Rev: TCCACCAATAAGCCCTGTTC | |||
| Cj0423 | For: GCAAGTGGCGGGTCTATTT | 148 | 46 |
| Rev: CAAGCCAAACAATAATAGTCCAA | |||
| Cj0484 | For: AAATGGGAGTTCCAATTGCTT | 445 | 46 |
| Rev: ATTGGCAAGGCGATAAAAAC | |||
| cft (Cj0612c) | For: TAAACAAAGAAATGTATGCAGCAA | 445 | 27, 70 |
| Rev: GCAATATTTTTGATGTATTGATCAGC | |||
| Cj0659 | For: TGYCTTTGTGCTTTGTTTTTAGC | 101 | 46 |
| Rev: TCATTTAATGCAAGGTAATTATGTTCT | |||
| Cj0727 | For: TTATGCCTTARGYGGGGATG | 345 | 46 |
| Rev: ATTGGCCTTGCATAACCTTTT | |||
| CJE0837 | For: ATGCTGTACCACGAGGAGGT | 148 | 12 |
| Rev: TTTACACTTTCCCCCTGAGC | |||
| cfrA (Cj0755) | For: AGCAGGGATAAGCCCTCTTG | 203 | 46, 49, 69, 70 |
| Rev: AGCGATCTATTTGCCAYTCG | |||
| tpx (Cj0779) | For: GCAGCGGGTAAAACTCAAAT | 353 | 49 |
| Rev: TTTGCAATATCTGGCATTTCTG | |||
| flgS (Cj0793) | For: CTTGCAAGCATGGGTAGTGT | 404 | 71 |
| Rev: GGCTTTTCACATTCGCTTTC | |||
| serA (Cj0891c) | For: GGGGTGGGCGTAGATAATGT | 450 | 49 |
| Rev: TTTTGCAATTTCTTGCTTTCC | |||
| peb1A (Cj0921c) | For: TTGATCAAGCAACAGGTGAAA | 503 | 14, 27, 28, 69 |
| Rev: TGGTTCAAAACTATCTGGCAAA | |||
| cheB (Cj0924c) | For: GGATCTTCAACAGGTGGTCCT | 354 | 31 |
| Rev: TCCCATGCCCGTTAAAATAA | |||
| jlpA (Cj0983) | For: AGCTTGCGGAAATTCCATAG | 147 | 14 |
| Rev: TCACCATCTGCATTGCATTTA | |||
| csrA (Cj1103) | For: GTCGTTCAAACAGGGAAAGG | 152 | 50, 51 |
| Rev: TTTTTGCTTAAGTCATCAAGTTTAATA | |||
| pglB-s/wlaF (Cj1126c) | For: CAAAGCAATGTGGATTTGTTTC | 247 | 65 |
| Rev: CGCTTAAAACCACTCCGTTG | |||
| pglB-g/wlaF (Cj1126c) | For: GCCCGCTAGAATGTCTTTGA | 153 | 65 |
| Rev: AAAACCACTCCGTTGCTAAGAT | |||
| Cj1135 | For: TTAGAATTTGCAAAACGCAAAG | 303 | 8, 9 |
| Rev: ATCTTCCCTACCCCAGCCTA | |||
| Cj1136 | For: TGGTTTGACCATGCTTGTATTT | 252 | 8, 9, 46 |
| Rev: CAAAACCATGCCAAAATGAA | |||
| wlaN (Cj1139) | For: AGCATTAGAAAGTTGCATTAACCA | 252 | 8, 9, 30, 43, 52 |
| Rev: CATTCTTCGCAAGCATTAAGTTC | |||
| neuA1 (Cj1143) | For: GAATGTGGTTTTATCGATGGAG | 446 | 8, 9 |
| Rev: GGCCTATCATGCAAAAGAGC | |||
| CJE1500 | For: ACGATTTCGTTTTCCCTAAACTC | 545 | 12 |
| Rev: AGGACTAAAATCCCCATAAGGATAA | |||
| neuB3 (Cj1317) | For: CAGGTGCTGATGCGATAAAA | 349 | 4, 22, 69 |
| Rev: GTAGCAATGCCCGTTGAAA | |||
| Cj1321 | For: TTTATACATCCTCAATCTTTTGTTTCA | 249 | 3 |
| Rev: GCAAGGCAACAGGCTAACTC | |||
| neuC2/ptmD (Cj1328) | For: GCCCTGAGTTTGGACTCACT | 253 | 37, 53 |
| Rev: CCGCCACATAAATGCACTAAA | |||
| neuA3/ptmB (Cj1331) | For: CGCAAGTGTAGCCCAAAAAT | 447 | 20, 22, 37, 53, 64, 65 |
| Rev: TCCATTACAAAAAGCCCTGTG | |||
| maf5/pseE (Cj1337) | For: TTGGGGCTAGACGATTATGG | 146 | 22, 26 |
| Rev: TGAGAATGTGTTTGCCCATC | |||
| pldA (Cj1351) | For: GCTTTGGCTAATTATCTTGGTGA | 455 | 70 |
| Rev: CAAAGGCGTGGAACAAATAAA | |||
| Cj1357 | For: TGGTGGAATGGAACTCAGAA | 552 | 46 |
| Rev: TCCATCGCTTTGGAATTTTT | |||
| Cj1365 | For: CTGATACAGCTGGGAAACAGC | 150 | 3 |
| Rev: GAAACACCCCAGCTCCATAA | |||
| gmhA2 (Cj1424) | For: TAGCAGGAAATGGTGGCAGT | 450 | 9, 46 |
| Rev: GCTTGCAAGAAAAACCTTTACC | |||
| fcl (Cj1428) | For: CGCAGGTACAGCATTGGTAG | 546 | 9 |
| Rev: CCCTCGCTTAACAATTTTGC | |||
| Cj1435 | For: TCCTTATCCTCGTTATGAATGGTT | 154 | 9 |
| Rev: TAACGATTGTGTGGCCTTCA | |||
| glf (Cj1439) | For: TTGGTTCTGGATTGTTTGGT | 555 | 9 |
| Rev: CGCCTTTAGGGATACCTTGAT | |||
| cadF (Cj1439) | For: ATGGTTTAGCAGGTGGAGGA | 348 | 1, 14, 28, 50, 51, 59, 69 |
| Rev: CAAAACCAAAATGACCTTCCA | |||
| Cj1549 | For: AACCCAATACGGCTTTGATG | 153 | 8, 9 |
| Rev: CTTTCAATTGGCGTTCCTGT | |||
| Cj1551 | For: GGTAGTGTTTTTGGCTCGGTA | 145 | 8, 9 |
| Rev: GGGTTTTGTTTTGCTCGTGT | |||
| iamA (Cj1647) | For: TTAGTGGCGGTATGCAAAAA | 254 | 43, 59 |
| Rev: ATAAGTGCCGCAAAATCCAA | |||
| p19 (Cj1659) | For: CGGCGATCCAAAAGAACTTA | 252 | 49, 70 |
| Rev: GCCACCATAGGCATCAAAGT | |||
| flgE2 (Cj1729c) | For: TAACGCGAAAGGACAAGGAC | 555 | 22, 29, 43 |
| Rev: GCTTGAGGAGGTTGTGGTGT | |||
| cst-II | For: ATTGCTGGAAATGGACCAAG | 546 | 1, 16, 17 |
| Rev: TCCGATATAGTGTGAATTATCATTTTT | |||
| CJE1733 | For: GATGTATCCGCCTTTGGCTA | 447 | 12 |
| Rev: GGGTTCTTGTTATAAAGCCCATT | |||
| cgtA | For: CCTTTGAATCCTTGGGCTTT | 555 | 1, 16, 17, 44 |
| Rev: TGAAGAAAAACTTTGTTTTGTTGCT | |||
| cgtB | For: GCAAGGGCAATAGAAAGCTG | 550 | 1, 16, 17, 30, 44 |
| Rev: TTTGAGCTTGACTTAACATTGGA | |||
| virB8/comB1 (Cjp1) | For: AATGGATATGAGAGCCGATGA | 345 | 33 |
| Rev: AGCTCCATTGGTATAAAATCAAAAA | |||
| virB9/comB2 (Cjp2) | For: AACACAGGCGAAAGCATTTT | 349 | 33 |
| Rev: CCATTGCAAAGCGTGTTCT | |||
| virB10 (Cjp3) | For: TTAGCTGATCGTGGCAACAC | 452 | 33 |
| Rev: GCGTAAGGCATTCCGTATCT | |||
| virB11 (Cjp5) | For: GAAATGGAAGCAAACGAGCTT | 452 | 1, 30, 33 |
| Rev: TTTGGATCAATTTCACCCATT | |||
| virD4 (Cjp6) | For: TCAGAGCTAAAATGCCCCTAAG | 351 | 33 |
| Rev: TCGCACTTGTAACCCAATGA | |||
| virB4/comB4 (Cjp53) | For: ACAGGAGCATTTGCACCAC | 345 | 33 |
| Rev: GTTCGCCCCAATCATTTTT | |||
| tetO | For: TTTCCCGTTTATCACGGAAG | 448 | 60, 67 |
| Rev: CAAGCAATATTTCCCGCTGT | |||
| HS1.07 | For: TGAACTTGCTTGGGAAATAAATC | 455 | 24, 25 |
| Rev: CAATCATCGGAGCATTAGCA | |||
| HS19.09 | For: ACCTTAGAGAAATTTTTACCCTTGG | 345 | 24, 25 |
| Rev: ATCAGAATTTTCCAAATCGATAGC | |||
| HS23/36.17 | For: TCTTCTTTTTGGTTTGGATGG | 355 | 24, 25 |
| Rev: GCAAATCTTGATGGGTTTGTG | |||
| HS41.06 | For: TTACTTGGTGGAAGCAATTCG | 249 | 24, 25 |
| Rev: TTCTTTTTCCCAAGTGTGTGC | |||
| fhuA | For: GCCGGTGATCTGTTCTTCAT | 547 | 15, 69, 70 |
| Rev: TGTTACCGAGGAAGGTGTCC | |||
| aphA3 | For: ACCGCTGCGTAAAAGATACG | 154 | 67 |
| Rev: TCCAGCCATAGCATCATGTC | |||
| CAT | For: AAAGCCTAATCCTCCGGAAA | 148 | 67 |
| Rev: GTATTTTCCGCCCTCCTCAT |
For, forward; Rev, reverse.
Primers designed by Price and colleagues (56).
Genes selected for the 18-target P-BIT system are shown in boldface.
DNeasy-extracted DNA from each isolate was tested using each gene target in PCRs containing 2.5 mM MgCl2, 1× PCR buffer II (50 mM KCl, 10 mM Tris, pH 8.3; Applied Biosystems, Foster City, CA), 0.2 mg/ml bovine serum albumin (Sigma-Aldrich, St. Louis, MO), 250 μM each deoxynucleoside triphosphate (dNTP), 12.5 pmol each primer, 1.25 U AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA), and 0.5 ng or 5 ng of extracted DNA. PCRs were run in ABI 9700 (Applied Biosystems, Foster City, CA) thermal cyclers using the following cycling conditions: 5 min at 95°C followed by 40 cycles of 1 min at 95°C, 1 min at 59°C, and 1 min at 74°C and then a final extension at 74°C for 8 min.
PCR products were run in 2% agarose gels with 1× Tris-borate-EDTA (TBE) for 70 min at 110 V and visualized using ethidium bromide. The presence or absence of target PCR products was loaded into a BioNumerics version 5.10 (Applied Maths, Ghent, Belgium) database. Interstrain relationships were assessed by numerical analysis of the P-BIT data using the simple matching coefficient and Ward's clustering.
Typing of isolates.
The Penner serotyping system was used to determine the heat-stable serotypes of the isolates using the passive hemagglutination technique and antisera produced in house according to the method of Penner and Hennessy (54).
All isolates except SVS835-770, which had become nonviable, were analyzed by PFGE using the standardized PulseNet protocol (57), and Salmonella enterica serovar Braenderup H9812 restricted with XbaI as a size standard (21). Samples were digested using SmaI and KpnI. Gels were prepared with 1% (wt/vol) SeaKem Gold agarose (Lonza, Rockland, ME) and electrophoresed for 18 h using respective initial and final switch times of 6.8 s and 38.4 s for SmaI and 5.2 s and 42.3 s for KpnI. PFGE profiles were analyzed and compared using BioNumerics version 5.10 and submitted to the PulseNet Aotearoa (New Zealand) Campylobacter database, where SmaI and KpnI pattern designations were assigned.
DNeasy-extracted DNA from each isolate, except SVS835-770, which had become nonviable, was analyzed using multilocus sequence typing (MLST) as described by Dingle and colleagues (7). Amplification was performed in a 25-μl reaction mixture using AmpliTaq Gold master mixture (Applied Biosystems, Foster City, CA) and 5 pmol of each primer. Products were sequenced with an ABI Genetic Analyser 3130XL (Applied Biosystems, Foster City, CA). The sequence information was collated and alleles were assigned using the Campylobacter PubMLST database (http://pubmlst.org/campylobacter/) (23). Novel alleles and sequence types (ST) were submitted for allele and ST and clonal complex (CC) designations when appropriate.
RESULTS
Risk-based binary typing.
Sixty-eight PCRs were designed for 67 prospective risk-based binary typing genes. Two primer sets for pglB were designed using the sequence from positions 81 to 176 (accession no. AF108897) (66). Both sequences aligned only with C. jejuni, but one primer set contained mismatches with some strains (pglB-s) while the other showed 100% alignment for both (pglB-g). Five targets produced negative results for all 61 isolates tested, fhuA, aphA-3, CAT, HS41.06, and HS19.09, of which the latter two were serotype-specific targets (24) subsequently shown to produce positive results for isolates of the target serotype (data not shown). Four targets (cheW, cft, pglB-g, and p19) were positive for all C. jejuni and C. coli isolates tested and negative for C. lari. One target (csrA) was positive for all C. jejuni isolates, negative for C. lari, and variable for C. coli isolates. Eleven targets (fdxA, cmeB, Cj0659, flgS, serA, peb1A, cheB, pglB-s, pldA, Cj1357, and iamA) were positive for all C. jejuni isolates and negative for C. coli and C. lari isolates. The remaining 49 targets were detected in at least 1, and at most 57, of the 58 C. jejuni isolates studied. The risk-based binary typing system produced 47 types from 61 isolates with a diversity index (62) of 98.5%.
A subset of 18 targets was selected to yield the same discriminatory potential as the entire scheme. Their selection was based upon several criteria, including discriminatory potential among the strain set examined, position in the genome, and known or predicted function.
Numerical analysis of P-BIT data.
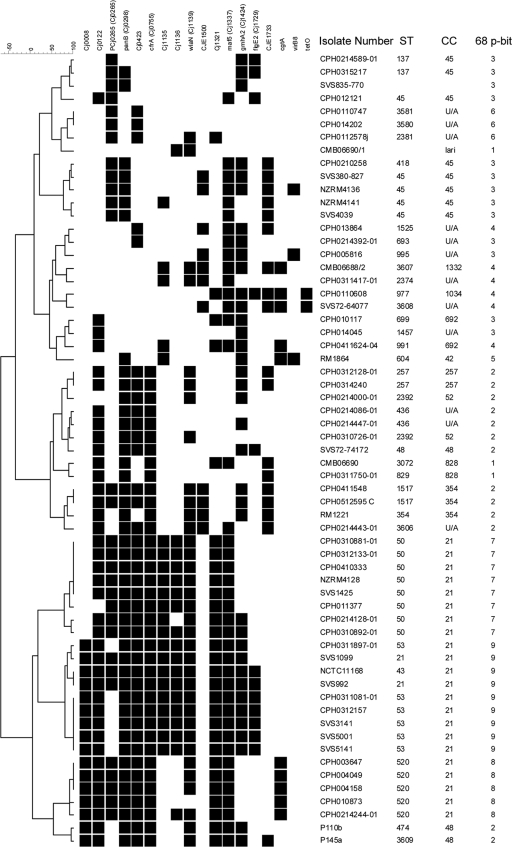
A dendrogram of the cluster analysis (Fig. 1) revealed nine clusters at the 67% similarity level (S-level). Cluster 1 comprised the two C. coli isolates and one C. lari isolate studied, distinguished at the species level at the 68% S-level. Cluster 2 contained 13 C. jejuni isolates, comprised of several highly related clusters assigned to CC 48, 257, 354, and 52, in addition to three sequence types yet to be assigned to a CC. Cluster 3 contained 13 isolates assigned to CC 45 and 692 (in which each of these types clustered closely with isolates unassigned to a CC) and one isolate (SVS835-770) for which MLST data was not available. Cluster 4 contained six isolates, three of which were unassigned to a CC and one each of CC 692, 1034, and 1332. Cluster 5 was represented by a single isolate of CC 42. Cluster 6 comprised three isolates unassigned to any CC. Clusters 7, 8, and 9 contained a total of 22 isolates all assigned to CC 21, but representing ST 50 (cluster 7), ST 520 (cluster 8), and ST 21, ST 43, and ST 53 (cluster 9), respectively. A cluster analysis based on the suggested 18-target P-BIT set yielded a similar topology (Fig. 2).
FIG. 1.
Cluster analysis of P-BIT data based on 68 gene targets using the simple matching coefficient and Ward's clustering.
FIG. 2.
Cluster analysis of P-BIT data based on the suggested minimal set of 18 gene targets using the simple matching coefficient and Ward's clustering. “68 p-bit” refers to the clusters illustrated in Fig. 1.
A minimum spanning tree was prepared for the 18-target P-BIT subset using the binary coefficient, maximum neighbor distance of 2 changes, and minimum size of 2 types. The tree is displayed in Fig. 3 with ST labels. Six clusters were revealed. Cluster 1 comprised isolates assigned to CC 48 and 21, and cluster 2 contained the ST 520 isolates from CC 21. Cluster 3 comprised isolates of CC 48, 52, 257, 354, and 828 (C. coli) and isolates that have yet to be assigned a CC. Cluster 4 contained isolates of CC 692 and one unassigned isolate. Cluster 5 contained isolates assigned to CC 45 and one isolate for which MLST was unavailable (SVS835-770). Clusters 6 and 7 contained only isolates that had not been assigned a CC.
FIG. 3.
Minimum spanning tree based on 18 P-BIT targets. The tree was created using BioNumerics 5.1 software, using the binary coefficient. The MST is presented with logarithmic branch lengths; complexes are shaded based on a maximum neighbor distance of 2 changes and minimum of 2 types. Thick lines between nodes indicate one change, thinner lines two changes, dark dotted lines three to four changes, and gray dotted lines five or more changes. Mix 1 contains ST 3580 and 3581, and mix 2 contains ST 21 and 43. nv, nonviable; lari, C. lari isolate.
The wide range of sources and wide variation in detection rates between target genes in this study make investigation of source bias problematic. However chi-square analysis of the P-BIT data revealed that, at the 95% confidence level, CJE1500 was detected more frequently in poultry and wild bird isolates and less frequently in animal, human, and water samples than would be expected if isolates from all sources had an equal probability of carrying this gene (data not shown). However, we do not know how many of the human isolates were derived from diarrheal cases that arose from consumption of contaminated poultry but assume the proportion is not insignificant (47).
Typing.
One of the isolates (SVS835-770) became nonviable before PFGE and MLST could be undertaken. In our laboratory, Penner serotyping is only available for C. jejuni and MLST is only available for C. jejuni and C. coli isolates. Three isolates were untypeable by Penner serotyping, and the remaining 55 isolates had 22 Penner serotypes and a diversity index of 92.8%. The 60 isolates analyzed by PFGE produced 38 SmaI and 52 KpnI types with diversity indices of 97.0% and 99.4%, respectively. Combining SmaI and KpnI produced 54 types and a diversity index of 99.7%. The 59 isolates analyzed by MLST produced 32 sequence types and a diversity index of 96.1%.
DISCUSSION
Typing systems for microorganisms may be assessed by a number of criteria to establish their fitness for purpose (68; R. J. Meinersmann, presented at Campylobacter, Helicobacter and Related Organisms, Cape Town, South Africa, 1997). Some C. jejuni strains resist digestion with certain restriction enzymes, making characterization by PFGE typing problematic (13, 18), while MLST target genes may not always provide data appropriate for the scheme (40). P-BIT relies on a code derived from positive or negative results of PCR analyses for a wide range of genes widely distributed in C. jejuni genomes and extrachromosomal elements and thus offers complete typeability. In addition, our results indicate the P-BIT approach to be more discriminatory than MLST and SmaI-based PFGE typing, another important feature. We found KpnI-based PFGE typing to be the most discriminatory of the methods used in our study, notwithstanding that not all strains prove typeable with this enzyme (18).
Although the more widespread use of standardized methodologies for PFGE-based typing of bacterial pathogens has proven valuable for epidemiological studies (2, 11, 18, 19, 32), determining and using normalization parameters to enable a meaningful comparison of PFGE macrorestriction profiles can be challenging. Furthermore, the PFGE apparatus is of moderate cost. The increased availability of DNA sequencing facilities has made MLST a popular option and offers excellent portability, enabling researchers to readily compare their data with results obtained worldwide. The MLST approach also offers phylogenetically meaningful data that have proven invaluable for population genetic (6, 41) and source attribution (13, 42) studies. However, while technological advances continue to reduce the cost of performing the analysis, MLST still requires substantive capital outlay for implementation and running costs can be prohibitive for small routine laboratories. Our P-BIT approach requires only the most basic molecular biology laboratory equipment for implementation (approximately one-tenth the cost of PFGE equipment, for example) and no more than e-mail for the exchange of data between laboratories. The PCRs used have been designed to enable multiplexing, to further reduce running costs. We have identified a core set of 18 gene targets that conferred the same discriminatory potential in this study as the complete range of 68 targets employed. We believe these features, together with its high discriminatory power and complete typeability, render P-BIT a worthy tool for laboratories engaged in epidemiological studies of C. jejuni. The distinctiveness of profiles obtained from the few C. coli and C. lari strains included suggest P-BIT data may also provide a basis for species identification between closely related Campylobacter species (itself a challenging task [45]), but further work is required to substantiate this.
The P-BIT approach to typing yields a simple binary code (albeit one based on whole-genomic polymorphism) that is not suited for analysis with algorithms aimed at predicting phylogenetic relationships. The bases for determining strain relatedness with MLST and P-BIT are also very different, with the former evaluating change between housekeeping gene sequences and the latter determining, by PCR, the presence of diverse, widely distributed genes that may be subject to selective pressure. Nevertheless, some congruence between these methods was seen when comparing the interstrain relationships inferred. In particular, isolates assigned to CC 21, 45, 52, 257, and 354 formed or dominated well-defined clusters in the P-BIT analyses (Fig. 1 to 3). With the population genetic structure of C. jejuni believed to be significantly shaped through recombination events and horizontal gene transfer (7, 63), the correlation of P-BIT and MLST is perhaps not surprising, with P-BIT potentially offering additional insight into the genomic evolution of strains that may be useful for relatively uncommon MLST types or those that cannot be assigned to a clonal complex.
Subtyping using PFGE depends on the distribution of restriction sites within the organismal cellular DNA. As with P-BIT, the data are not aimed at determining strain phylogeny unless banding patterns are indistinguishable, in which case strains are assumed to be related. It has become common practice to confirm strain relationships inferred with one restriction enzyme with the use of a second (47). Despite these caveats, we compared the clustering of C. jejuni strains in a dendrogram based on SmaI and KpnI PFGE profiles (data not shown) with the P-BIT and MLST results, given the agreement between the latter two methods. The PFGE analysis agreed with some of the P-BIT and MLST results, with homogeneous clusters of ST 257, 436, and 2392 each formed at S-levels of 80%; other clusters were dominated (but not exclusive to other ST) by strains assigned to ST 137, 50, and 520 at S-levels between 70 and 92%. Five of 6 ST 53 strains also clustered together (S-level of 75%), and two highly related ST 45 strains (NZRM 4141 and SVS 4039) gave PFGE profiles determined as 90% similar. However, although all CC 21 strains formed a distinct cluster in the PFGE analysis, the infraspecific relationships between the constituent ST differed from that of the P-BIT and MLST analyses and also included the three CC 48 strains examined. Strains assigned to CC 45 and 354 were divergently distributed in the PFGE analysis. Although the discriminatory potential of PFGE typing can exceed that of both P-BIT and MLST, our results support the need for caution when using PFGE data to infer clonal relationships, for reasons described in more detail elsewhere (47).
One feature we had in mind when developing this approach was the possibility of quantifying the risk to human health from individual strains through generation of a simple binary code relating to epidemicity factors. There have been various observations of “stable clones” present in human clinical specimens and various host animals (39, 47, 48, 61) as well as whole-genomic comparative studies indicating that host-specific types are less well adapted for stress survival than those found more widely (46). The selection of initial marker genes in this study was on a metagenomic basis but focused on genes that conferred, or were implicated in, some aspect of strain virulence. Many aspects of pathogenicity in Campylobacter are poorly understood, but examination of some of the strains classified as “high risk” and “low risk” by On and colleagues (46) showed them to cluster distinctly (Fig. 1). Notably, all “high-risk” strains (NCTC11168, SVS992, SVS1099, SVS1425, SVS3141, and SVS5001) were found to belong to CC 21, a globally common type, with four of the five “low-risk” strains (SVS380-827, SVS4039, SVS72-64077, and SVS835-770) appearing in related clusters 3 and 4 (Fig. 1). We recognize that our P-BIT system uses PCR primers that are designed to amplify specific sequence orientations of the target genes and that genes may be present in particular strains with different flanking regions that would therefore not be detected. Thus, P-BIT data provides indicative but not absolute data regarding the presence or absence of a gene. Nevertheless, for the establishment of a simple, inexpensive, and effective typing method, we feel this is an acceptable compromise. For genes carried on plasmids, there is a risk in long-term studies that strains will be cured of such extrachromosomal DNA and thereby lose a specific marker. This appears to be the case with one strain examined here, 81-176 (RM1864), in which the plasmid-borne tetO gene conferring tetracycline resistance was not detected. Our example of this strain was a kind gift from colleagues, and we have no information on the number of passages it may have experienced before it was received. However, the isolate's first description was in 1988 in another laboratory, and it is likely to have been subcultured many times before we received it, allowing ample opportunity for the plasmid to be cured. We examined a heated lysate of 81-176 to ascertain if the result may have been due to the DNA preparation method used being suited for genomic DNA extraction, but still obtained no tetO-derived amplicon (data not shown). In the context of the P-BIT system, we do not consider the inclusion of tetO inappropriate since it represents an important virulence marker (found in two of the strains examined here: see Fig. 2) and potentially valuable for short-term epidemiological studies such as outbreaks, where plasmid loss is unlikely to occur. Further studies of the relationship between P-BIT profile, stress survival, and strain type frequency in human populations are also warranted.
We conclude that P-BIT is a useful approach for subtyping, offering advantages of speed, cost, and potential for strain risk ranking unavailable elsewhere. We hope the wider scientific community will be encouraged to investigate further its usage in national and global surveillance studies and outbreak investigations.
Acknowledgments
This project was funded by the New Zealand Ministry of Research, Science and Technology through an ESR-administered Capability Fund project.
We acknowledge Aruni Premaratane and Stephanie Brandt for technical assistance and Beverley Horn for statistical assistance. We also thank the anonymous reviewers for constructive comments on the manuscript.
Footnotes
Published ahead of print on 18 December 2009.
REFERENCES
- 1.Bereswill, S., and M. Kist. 2003. Recent developments in Campylobacter pathogenesis. Curr. Opin. Infect. Dis. 16:487-491. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2009. Campylobacter jejuni infection associated with unpasteurized milk and cheese—Kansas, 2007. MMWR Morb. Mortal. Wkly. Rep. 57:1377-1379. [PubMed] [Google Scholar]
- 3.Champion, O. L., M. W. Gaunt, O. Gundogdu, A. Elmi, A. A. Witney, J. Hinds, N. Dorrell, and B. W. Wren. 2005. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. U. S. A. 102:16043-16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou, W. K., S. Dick, W. W. Wakarchuk, and M. E. Tanner. 2005. Identification and characterization of NeuB3 from Campylobacter jejuni as a pseudaminic acid synthase. J. Biol. Chem. 280:35922-35928. [DOI] [PubMed] [Google Scholar]
- 5.Devane, M. L., C. Nicol, A. Ball, J. D. Klena, P. Scholes, J. A. Hudson, M. G. Baker, B. J. Gilpin, N. Garrett, and M. G. Savill. 2005. The occurrence of Campylobacter subtypes in environmental reservoirs and potential transmission routes. J. Appl. Microbiol. 98:980-990. [DOI] [PubMed] [Google Scholar]
- 6.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. A. Wareing, and M. C. J. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorrell, N., O. L. Champion, and B. W. Wren. 2005. Advances in Campylobacter jejuni comparative genomics through whole genome DNA microarrays, p. 79-99. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 9.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drummond, A. J., B. Ashton, M. Cheung, J. Heled, M. Kearse, R. Moir, S. Stones-Havas, T. Thierer, and A. Wilson. 2009. Geneious v4.6. Biomatters, Auckland, New Zealand.
- 11.Fitzgerald, C., L. O. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerdlow, R. Flahart, J. Sexton, and P. I. Fields. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Binkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dobson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:0072-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.French, N. P., A. Midwinter, B. Holland, J. Collins-Emerson, R. Pattison, F. Colles, and P. Carter. 2009. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children's playgrounds. Appl. Environ. Microbiol. 75:779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friis, L. M., C. Pin, B. M. Pearson, and J. M. Wells. 2005. In vitro cell culture methods for investigating Campylobacter invasion mechanisms. J. Microbiol. Methods 61:145-160. [DOI] [PubMed] [Google Scholar]
- 15.Galindo, M. A., W. A. Day, B. H. Raphael, and L. A. Joens. 2001. Cloning and characterization of a Campylobacter jejuni iron-uptake operon. Curr. Microbiol 42:139-143. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert, M., J.-R. Brisson, M.-F. Karwaski, J. Michniewicz, A.-M. Cunningham, Y. Wu, N. M. Young, and W. W. Wakarchuk. 2000. Biosynthesis of ganglioside mimics in Campylobacter jejuni OH4384. J. Biol. Chem. 275:3896-3906. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert, M., P. C. R. Godschalk, C. T. Parker, H. P. Endtz, and W. W. Wakarchuk. 2005. Genetic basis for the variation in the lipooligosaccharide outer core of Campylobacter jejuni and possible association of glycosyltransferase genes with post-infectious neuropathies, p. 219-248. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 18.Gilpin, B., A. Cornelius, B. Robson, N. Boxall, A. Ferguson, C. Nicol, and T. Henderson. 2006. Application of pulsed-field gel electrophoesis to identify potential outbreaks of campylobacteriosis in New Zealand. J. Clin. Microbiol. 44:406-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graham, C., R. Whyte, B. Gilpin, A. Cornelius, J. A. Hudson, D. Morrison, H. Graham, and C. Nicol. 2005. Outbreak of campylobacteriosis following pre-cooked sausage consumption. Aust. N. Z. J. Public Health 29:507-510. [DOI] [PubMed] [Google Scholar]
- 20.Guerry, P., P. Doig, R. A. Alm, D. H. Burr, N. Kinsella, and T. J. Trust. 1996. Identification and characterization of genes required for post-translational modification of Campylobacter coli VC167 flagellin. Mol. Microbiol. 19:369-378. [DOI] [PubMed] [Google Scholar]
- 21.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. M. Ribot. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jagannathan, A., and C. Penn. 2005. Motility, p. 331-347. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 23.Jolley, K. A., M.-S. Chan, and M. C. J. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlyshev, A. V., O. L. Champion, C. Churcher, J.-R. Brisson, H. C. Jarrell, M. Gilbert, D. Brochu, F. St. Michael, J. Li, W. W. Wakarchuk, I. Goodhead, M. Sanders, K. Stevens, B. White, J. Parkhill, B. W. Wren, and C. M. Szymanski. 2005. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 55:90-103. [DOI] [PubMed] [Google Scholar]
- 25.Karlyshev, A. V., O. L. Champion, G. W. P. Joshua, and B. W. Wren. 2005. The polysaccharide capsule of Campylobacter jejuni, p. 249-258. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 26.Karlyshev, A. V., D. Linton, N. A. Gregson, and B. W. Wren. 2002. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148:473-480. [DOI] [PubMed] [Google Scholar]
- 27.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5-21. [DOI] [PubMed] [Google Scholar]
- 28.Konkel, M. E., L. A. Joens, and P. F. Mixter. 2000. Molecular characterization of Campylobacter jejuni virulence determinants, p. 217-240. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. American Society for Microbiology, Washington, DC.
- 29.Konkel, M. E., J. D. Klena, V. Rivera-Amill, M. R. Monteville, D. Biswas, B. Raphael, and J. Mickelson. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 186:3296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kordinas, V., C. Nicolaou, A. Ioannidis, E. Papavasileiou, N. J. Legakis, and S. Chatzipanagiotou. 2005. Prevalence of four virulence genes in Campylobacter jejuni determined by PCR and sequence analysis. Mol. Diagn. 9:211-215. [DOI] [PubMed] [Google Scholar]
- 31.Korolik, V., and J. Ketley. 2005. Campylobacter chemotaxis, p. 349-367. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 32.Kuusi, M., P. Klemets, I. Miettinen, I. Laaksonen, H. Sarkkinen, M.-L. Hanninen, H. Rautelin, E. Kela, and J. P. Nuorti. 2004. An outbreak of gastroenteritis from a non-chlorinated community water supply. J. Epidemiol. Community Health 58:273-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen, J. C., and P. Guerry. 2005. Plasmids of Campylobacter jejuni 81-176, p. 181-192. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 34.Larsen, J. C., C. Szymanski, and P. Guerry. 2004. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81-176. J. Bacteriol. 186:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, J., M. Akiba, and Q. Zhang. 2005. Multidrug efflux systems in Campylobacter, p. 205-218. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 36.Lin, J., L. O. Michel, and Q. Zhang. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Logan, S. M., J. F. Kelly, P. Thibault, C. P. Ewing, and P. Guerry. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificty of Campylobacter flagellins. Mol. Microbiol. 46:587-597. [DOI] [PubMed] [Google Scholar]
- 38.Manning, G., C. G. Dowson, M. C. Bagnall, I. H. Ahmed, M. West, and D. G. Newell. 2003. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl. Environ. Microbiol. 69:6370-6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manning, G., B. Duim, T. M. Wassenaar, J. A. Wagenaar, A. Ridley, and D. G. Newell. 2001. Evidence for a genetically stable strain of Campylobacter jejuni. Appl. Environ. Microbiol. 67:1185-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McTavish, S. M., C. E. Pope, C. Nicol, D. Campbell, N. French, and P. E. Carter. 2009. Multilocus sequence typing of Campylobacter jejuni, and the correlation between clonal complex and pulsed-field gel electrophoresis macrorestriction profile. FEMS Microbiol. Lett. 298:149-156. [DOI] [PubMed] [Google Scholar]
- 41.Mickan, L., R. Doyle, M. Valcanis, K. E. Dingle, L. Unicomb, J. Lanser, and the Australian Campylobacter Subtyping Study Group. 2007. Multilocus sequence typing of Campylobacter jejuni isolates from New South Wales, Australia. J. Appl. Microbiol. 102:144-152. [DOI] [PubMed] [Google Scholar]
- 42.Miller, W. G., M. D. Englen, S. Kathariou, I. V. Wesley, G. Wang, L. Pittenger-Alley, R. M. Siletz, W. Muraoka, P. J. Fedorka-Cray, and R. E. Mandrell. 2006. Identification of host-associated alleles by multilocus sequence typing of Campylobacter coli strains from food animals. Microbiology 152:245-255. [DOI] [PubMed] [Google Scholar]
- 43.Muller, J., F. Schulze, W. Muller, and I. Hanel. 2006. PCR detection of virulence-associated genes in Campylobacter jejuni strains with differential ability to invade Caco-2 cells and to colonize the chick gut. Vet. Microbiol. 113:123-129. [DOI] [PubMed] [Google Scholar]
- 44.Nachamkin, I., J. Liu, M. Li, H. Ung, A. P. Moran, M. M. Prendergast, and K. Sheikh. 2002. Campylobacter jejuni from patients with Guillain-Barre syndrome preferentially expresses a GD1a-like epitope. Infect. Immun. 70:5299-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.On, S. L. W. 2005. Taxonomy, phylogeny, and methods for the identification of Campylobacter species, p. 13-42. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 46.On, S. L. W., N. Dorrell, L. Petersen, D. D. Bang, S. Morris, S. J. Forsythe, and B. W. Wren. 2006. Numerical analysis of DNA microarray data of Campylobacter jejuni strains correlated with survival, cytolethal distending toxin and haemolysin analyses. Int. J. Med. Microbiol. 296:353-363. [DOI] [PubMed] [Google Scholar]
- 47.On, S. L. W., N. McCarthy, W. G. Miller, and B. J. Gilpin. 2008. Molecular epidemiology of Campylobacter species, p. 191-211. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 48.On, S. L. W., E. M. Neilsen, J. Engberg, and M. Madsen. 1998. Validity of Sma-defined genotypes of Campylobacter jejuni examined by SalI, KpnI, and BamHI polymorphisms: evidence of identical clones infecting humans, poultry, and cattle. Epidemiol. Infect. 120:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park, S. F. 2005. Campylobacter jejuni stress responses during survival in the food chain and colonization, p. 311-330. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 51.Park, S. F. 2002. The physiology of Campylobacter species and its relevance to their role as foodborne pathogens. Int. J. Food Microbiol. 74:177-188. [DOI] [PubMed] [Google Scholar]
- 52.Parker, C. T., S. T. Horn, M. Gilbert, W. G. Miller, D. L. Woodward, and R. E. Mandrell. 2005. Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J. Clin. Microbiol. 43:2771-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M.-A. Rajandream, K. M. Rutherford, A. H. M. Van Vliet, A. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 54.Penner, J. L., and J. N. Hennessy. 1980. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble heat-stable antigens. J. Clin. Microbiol. 12:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Population and Environmental Health Group. 2009. Notifiable and other diseases in New Zealand 2008 annual surveillance report FW09074. Institute of Environmental Science and Research, Christchurch, New Zealand.
- 56.Price, E. P., F. Huygens, and P. M. Giffard. 2006. Fingerprinting of Campylobacter jejuni by using resolution-optimized binary gene targets derived from comparative genome hybridization studies. Appl. Environ. Microbiol. 72:7793-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 59.Rozynek, E., K. Dzierzanowska-Fangrat, P. Jozwiak, J. Popowski, D. Korsak, and D. Dzierzanowska. 2005. Prevalence of potential virulence markers in Polish Campylobacter jejuni and Campylobacter coli isolates obtained from hospitalized children and from chicken carcasses. J. Med. Microbiol. 54:615-619. [DOI] [PubMed] [Google Scholar]
- 60.Schmidt-Ott, R., S. Pohl, S. Burghard, M. Weig, and U. Grob. 2005. Identification and characterization of a major subgroup of conjugative Campylobacter jejuni plasmids. J. Infect. 50:12-21. [DOI] [PubMed] [Google Scholar]
- 61.Siemer, B. L., C. S. Harrington, E. M. Nielsen, B. Borck, N. L. Nielsen, J. Engberg, and S. L. W. On. 2004. Genetic relatedness among Campylobacter jejuni serotyped isolates of diverse origin as determined by numerical analysis of amplified fragment length polymorphism (AFLP) profiles. J. Appl. Microbiol. 96:795-802. [DOI] [PubMed] [Google Scholar]
- 62.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 63.Suerbaum, S., M. Lohrengel, A. Sonnevend, F. Ruberg, and M. Kist. 2001. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 183:2553-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szymanski, C. M., S. Goon, B. Allan, and P. Guerry. 2005. Protein glycosylation in Campylobacter, p. 259-273. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 65.Szymanski, C. M., S. M. Logan, D. Linton, and B. W. Wren. 2003. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol. 11:233-238. [DOI] [PubMed] [Google Scholar]
- 66.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 67.Taylor, D. E., and D. M. Tracz. 2005. Mechanisms of antimicrobial resistance in Campylobacter, p. 193-204. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 68.van Belkum, A., P. T. Tassios, L. Dijkshoorn, S. Haeggman, B. Cookson, N. K. Fry, V. Fussing, J. Green, E. Feil, P. Gerner-Smidt, S. Brisse, and M. Struelens. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13:1-46. [DOI] [PubMed] [Google Scholar]
- 69.Van Vliet, A. H. M., and J. M. Ketley. 2001. Pathogenesis of enteric Campylobacter infection. J. Appl. Microbiol. 90:45S-56S. [DOI] [PubMed] [Google Scholar]
- 70.Wooldridge, K. G., and A. H. M. Van Vliet. 2005. Iron transport and regulation, p. 293-310. In J. M. Ketley and M. E. Konkel (ed.), Campylobacter molecular and cellular biology. Horizon Bioscience, Wymondham, United Kingdom.
- 71.Wosten, M. M. S. M., J. A. Wagenaar, and J. P. M. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279:16214-16222. [DOI] [PubMed] [Google Scholar]