Abstract
Metallic copper alloys have recently attracted attention as a new antimicrobial weapon for areas where surface hygiene is paramount. Currently it is not understood on a molecular level how metallic copper kills microbes, but previous studies have demonstrated that a wide variety of bacteria, including Escherichia coli, Staphylococcus aureus, and Clostridium difficile, are inactivated within minutes or a few hours of exposure. In this study, we show that bacteria isolated from copper alloy coins comprise strains that are especially resistant against the toxic properties exerted by dry metallic copper surfaces. The most resistant of 294 isolates were Gram-positive staphylococci and micrococci, Kocuria palustris, and Brachybacterium conglomeratum but also included the proteobacterial species Sphingomonas panni and Pseudomonas oleovorans. Cells of some of these bacterial strains survived on copper surfaces for 48 h or more. Remarkably, when these dry-surface-resistant strains were exposed to moist copper surfaces, resistance levels were close to those of control strains and MICs for copper ions were at or below control strain levels. This suggests that mechanisms conferring resistance against dry metallic copper surfaces in these newly isolated bacterial strains are different from well-characterized copper ion detoxification systems. Furthermore, staphylococci on coins did not exhibit increased levels of resistance to antibiotics, arguing against coselection with copper surface resistance traits.
Copper in its ionic form is a required trace element for most pro- and eukaryotic organisms, including humans. While needed in small amounts, copper can easily become toxic when in surplus. This toxicity is caused mainly by the intrinsic properties of copper, as free copper ions undergo redox cycling reactions alternating between Cu(I) and Cu(II). This also results in the transfer of electrons to hydrogen peroxide and the concomitant generation of hydroxyl radicals that readily attack and damage cellular biomolecules. Recently, it was found that the majority of copper stress in Escherichia coli, as indicated by hydroxyl radical formation, occurs within the periplasm, away from the cytoplasmic DNA, and is thus copper-mediated oxidative stress (25). The cytoplasm might thus be better protected from copper-mediated oxidative stress, and indeed cells usually prevent accumulation of significant intracellular concentrations of free copper ions either by producing copper-binding chaperones (26, 36) or unspecific chelators such as glutathione (20, 30) or by efflux (14, 35). Nevertheless, copper ions within the cytoplasm also cause damage. Surprisingly, this damage is not related to oxidative stress but is exerted directly by the metal ions. It seems that copper ions attack and displace iron atoms from enzymes with solvent-exposed iron sulfur clusters such as those of hydratases (24). Thus, the presence of oxygen is not needed for this reaction, and there is no copper-mediated oxidative stress involved in this damage (24).
While we are now gaining a more detailed picture of why copper ions are toxic to cells, we do not understand why metallic copper surfaces kill single-celled organisms such as bacteria and yeasts. Earlier studies have demonstrated that metallic copper surfaces efficiently inactivate microbes upon contact (9, 11, 32), especially when exposed to dry surfaces (10). These beneficial properties led to the official registration of copper alloys as antimicrobials through the U.S. Environmental Protection Agency in 2008. There is now great hope that metallic copper surfaces will be able to help control hospital-acquired (nosocomial) infections. Indeed, there are ongoing trials in which dry touch surfaces in hospitals around the world are replaced by copper alloys. Results from a German hospital trial indicate that copper surfaces such as door knobs, light switches, and push plates diminished the bacterial load by up to 30% compared to stainless steel control surfaces (A. Mikolay et al., unpublished data). Similar studies in Great Britain and South Africa found that the numbers of bacteria on the surfaces of copper-containing items such as trolleys, desks, toilet seats, tap handles, or push plates were 71% (28) or 90% to 100% (5) lower than those on their stainless steel, wood, or tile control equivalents.
A potential challenge when applying metallic copper might be the probable emergence and spread of resistant bacteria, similar to what was observed after the introduction of antibiotics. The goal of this study was to investigate if bacteria that can withstand dry metallic copper surfaces can be isolated and if there is a link to multiple drug resistance. Where can potentially pathogenic bacteria that are in contact with both humans and metallic copper surfaces be found? Actually, people handle copper surfaces every day. Most coins around the world are made from copper or copper alloys. This includes the U.S. penny, which is composed of copper plated over a zinc core, and the nickel, dime, and quarter, which are cupronickel alloys (www.usmint.gov/). Coins of the European Union, such as the 50-cent coin, are made from an 89% copper alloy, as are the bicolored one- and two-Euro coins, which consist of different copper alloys (http://www.copperinfo.co.uk/coins/).
In the present study we isolated and initiated characterization of aerobic heterotrophic bacteria from copper alloy coins as an example of heavily used copper surfaces and person-to-person vectors. We believe that knowledge of the physiology and resistance mechanisms of these microbes will help us to adapt our strategies for using metallic copper surfaces in hygiene-sensitive areas. This might not only diminish total bacterial numbers but also prevent the emergence and spread of multiple-drug-resistant strains in hospitals equipped with copper surfaces.
MATERIALS AND METHODS
Bacterial strains and growth media.
The following type and collection strains were used in this study: Staphylococcus haemolyticus DSM 20263, Staphylococcus hominis DSM 20328, Staphylococcus warnerii DSM 20316, Staphylococcus epidermidis DSM 20044, Acinetobacter johnsonii DSM 6963, Micrococcus luteus DSM 20030, Bacillus anthracis Sterne 34F2 (pXO1+ pXO2−) (37), Bacillus cereus DSM 31, Pantoea stewarti DSM 30176, Brachybacterium conglomeratum DSM 1-241, Massilia timonae DSM 16850, Kocuria marina JCM 13363, Psychrobacter faecalis DSM 14664, Pseudomonas oleovorans DSM 1045, and Sphingomonas panni (DSM 15761).
Strains were grown in Luria-Bertani broth (Difco BD, Sparks, MD), nutrient broth (Difco BD, Sparks, MD), or R2A medium (Difco BD, Sparks, MD) as required at 30°C with shaking to stationary growth phase (16 to 32 h of incubation). Bacto Agar (Difco BD, Sparks, MD) was added at 15 g/liter for solid media.
Isolation procedures.
Aerobic heterotrophic bacteria were isolated from regular European 50-cent coins collected in Germany (27 coins) and Portugal (25 coins) in October 2007. Before use, coins were incubated in sterile plastic bags or petri dishes for at least 24 h at room temperature to reduce contamination with adventitious environmental germs. Two independent isolation procedures were performed. For method one, both sides of the coins were stamped on solid agar medium plates using sterile forceps. The plates were incubated at 30°C until colonies formed (1 to 3 days). Colonies were purified by streaking repeatedly on the same media. Alternatively, coins were incubated in liquid media and shaken for 2 days at 30°C. Mixed cultures were then diluted in the same growth media and plated. Colonies were purified by repeatedly streaking on the same media. Overall, each of the three growth media was used for about one-third of copper surface bacteria for selection and isolation.
DNA extraction and 16S rRNA gene amplification and sequencing.
Total DNA of the isolates was extracted from stationary-phase cultures either by the freeze-thaw method (31) or with a blood and tissue kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Amplification of the 16S rRNA gene was accomplished by PCR with the high-fidelity DNA-polymerase mix (Roche, Indianapolis, IN) or Taq polymerase (Sigma, St. Louis, MO), 1.5 mM MgCl2, 100 μM deoxynucleoside triphosphates (dNTPs), and 10 pmol of the primers 27F (5′-AGAGTTTGATCMTGGCTCAG; corresponding to E. coli 16S rRNA gene bases 8 to 27) (21) and 1525R (5′-AAGGAGGTGWTCCARCC-3′; corresponding to E. coli 16S rRNA gene bases 1525 to 1541) (21) with about 100 ng template DNA. PCRs were performed for 32 cycles, each consisting of a 30-s denaturation step at 96°C, a 30-s annealing step at 52°C, and a 1-min extension step at 72°C.
PCR products were purified and partially sequenced on an ABI 3730xl automated sequencer by GATC Biotech AG (Konstanz, Germany) in a 96-well setup using 16S rRNA gene primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) (21). Alternatively, PCR products were sequenced at the Microbiology Laboratory, Faculdade de Ciências e Tecnologia, Universidade de Coimbra, Portugal, on an ABI Prism 310 automated sequencer with the same primer.
Taxonomic and phylogenetic analyses.
The quality of 16S rRNA gene sequences was checked manually using the Bioedit editor (29) and aligned against representative reference sequences of the most closely related strains, obtained from the Ribosomal Database Project (7) and the European Molecular Biology Laboratory (EMBL), using the multiple-alignment CLUSTAL X software package (3). The method of Jukes and Cantor (22) was used to calculate evolutionary distances. Sequences were also checked for chimeric properties by using the CHECK_CHIMERA routine of the Ribosomal Database Project II (RDP-II) (27). Phylogenetic analysis was conducted using the neighbor-joining method as implemented in the computer program Molecular Evolutionary Genetics Analysis (MEGA4) (38) to determine the phylogenetic placement of each isolate relative to the type strains. Tree topologies were evaluated by performing bootstrap analysis (12) of 1,000 data sets by using the MEGA4 package (23). An isolate was considered to be a member of a particular species if the isolate and the particular type strain clustered with a bootstrap value of 90% or greater and displayed a similarity of 97% or more with respect to their 16S rRNA gene sequence divergence.
Dry metallic copper surface testing.
For identification of metallic copper surface-resistant bacteria, approximately 109 cells of the isolates were applied to 1- by 1-in. copper coupons (C110) as described previously (10). Cells were left on the coupons for 1 day, 2 days, 7 days, and 31 days under ambient conditions in sterile plastic petri dishes. In contrast to the original protocol (10), bacteria were not removed with phosphate-buffered saline (PBS) but were stamped directly on solid agar media and the coupon removed. Resistance to copper surfaces was counted as negative if 10 or fewer colonies were observed after incubation for 2 days. However, in most instances there was a clear-cut difference between resistant and sensitive isolates because resistant strains yielded a high density of colonies. For comparison, the bacteria were also tested on stainless steel (S304) surfaces to address sensitivity to desiccation. Representative strains were additionally challenged by exposure to plastic (polyvinylchloride) or aluminum surfaces to rule out access to a vital source of iron from stainless steel. Results were compared with those for type strains and strain collection bacteria of the same species. Experiments were repeated three times.
Moist metallic copper surface testing.
Frequently, bacteria are also tested for resistance on metallic copper surfaces that have been kept moist for the duration of the experiment (9, 11, 33, 42). We also tested the isolates for the occurrence of copper surface resistance under these conditions. For this, 40-μl aliquots of approximately 109 cells of each isolate in PBS were applied as a standing droplet on copper coupons in triplicates, and 10 μl was removed after 1, 3, 24, and 48 h and 7 days and plated on solid agar media. Survivors were counted as CFU. Experiments were repeated three times.
Determination of CuCl2 MICs.
Cultures were grown until stationary growth phase (24 to 48 h) at 30°C with shaking. Cultures were diluted 1:100 in fresh media, streaked onto solid media containing increasing CuCl2 concentrations, and incubated at 30°C. Growth was examined after 2 days. Experiments were repeated three times.
Evaluation of antibiotic resistance levels among staphylococci.
The antibiotic susceptibility of all staphylococcal isolates was determined by the agar diffusion method, using bioMérieux antibiotic disks (Bio-discs; bioMérieux, Marcy l'Etoile, France) (see Table S1 in the supplemental material for details). In short, staphylococci were streaked on fresh solid agar media and grown at 30°C for 24 h. With the help of a sterile loop, cells were removed and resuspended in sterile deionized water, and 100 μl of the cell suspension was transferred and spread on Mueller-Hinton agar plates (Difco BD, Sparks, MD). After excess moisture was dried from the plates, antibiotic disks were applied on the surface of the agar. Plates were incubated at 30°C for 24 h. The growth inhibition diameters were measured and bacteria classified as sensitive, intermediate, or resistant according to established French national guidelines (8).
RESULTS
Bacteria can be isolated from copper coins.
We reasoned that copper coins are ideal starting materials for the natural selection of metallic copper surface-resistant bacteria. Also, copper coins from general circulation are a suitable contact interface with the human skin microbiome because coins are handled by a wide variety of people with diverse sets of personal skin microbes. Thus, we hypothesized that there is significant overlap between human-associated bacteria and bacteria exposed to copper coins. These germs in turn could be potentially transferred to antimicrobial copper surfaces in hospitals.
For isolation of copper surface-resistant bacteria, we employed two different methods. Both the liquid and the solid medium selection procedures yielded an assortment of (facultative) aerobic heterotrophic bacteria. The advantage of the solid media was that the number of bacteria from each face of the coins could be directly counted. This amounted to an average of about 6 CFU per coin (independent of the origin of the coins). Including the liquid procedure, a total of 294 isolates were isolated from 52 coins (Table 1). As the goal was to isolate copper surface-resistant bacteria, complete coverage of the bacterial coin population was not paramount. Therefore, colonies obtained from the liquid medium procedure were selected by colony morphology, whereas all isolates from the solid medium procedure were used for further analysis. This might have resulted in a small bias toward a higher incidence of species from different groups of bacteria and toward fewer species from the same genus.
TABLE 1.
Strains isolated from copper coins
| Species | Family | No. of isolates | No. of isolates surviving at day: |
||
|---|---|---|---|---|---|
| 1 | 2 | 7 | |||
| Brachybacterium conglomeratum | Dermabacteraceae | 2 | 1 | 1 | 1 |
| Dermacoccus nishinomiyaensis | 2 | 0 | 0 | 0 | |
| Agrococcus jenensis | Microbacteriaceae | 1 | 0 | 0 | 0 |
| Curtobacterium flaccumfaciens | 2 | 0 | 0 | 0 | |
| Frigoribacterium faeni | 1 | 0 | 0 | 0 | |
| Microbacterium insulae, Microbacterium lacticum, Microbacterium oxydans, Microbacterium ulmi | 4 | 1 | 0 | 0 | |
| Micrococcus luteus, Micrococcus lylae | Micrococcaceae | 59 | 23 | 8 | 5 |
| Pseudoclavibacter helvolus | 1 | 1 | 0 | 0 | |
| Arthrobacter chlorophenolicus, Arthrobacter oxydans | 2 | 0 | 0 | 0 | |
| Kocuria marina, Kocuria palustris, Kocuria rhizophila, Kocuria sp. | 10 | 4 | 1 | 0 | |
| Aeromicrobium sp. | Nocardioidaceae | 1 | 0 | 0 | 0 |
| Propioniferax innocua | Propionibacteriaceae | 1 | 0 | 0 | 0 |
| Bacillus anthracis, Bacillus benzoevorans, Bacillus cereus, Bacillus circulans, Bacillus insolitus, Bacillus licheniformis, Bacillus macroides, Bacillus megaterium, Bacillus mycoides, Bacillus nealsonii, Bacillus phychrodurans, Bacillus pumilus, Bacillus silvestris, Bacillus simplex, Bacillus sp., Bacillus thuringiensis, Bacillus weihenstephanensis | Bacillaceae | 50 | 0 | 0 | 0 |
| Paenibacillus cineris, Paenibacillus favisporus, Paenibacillus rhizosphaerae | Paenibacillaceae | 1 | 0 | 0 | 0 |
| Lysinibacillus fusiformis, Lysinibacillus sphaericus | Planococcaceae | 1 | 1 | 0 | 0 |
| Staphylococcus capitis, Staphylococcus equorum, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus hominis, Staphylococcus lugdunensis, Staphylococcus pasteuri, Staphylococcus sp., Staphylococcus vitulinus, Staphylococcus warneri | Staphylococcaceae | 115 | 27 | 11 | 0 |
| Leuconostoc citreum | Leuconostocaceae | 2 | 1 | 0 | 0 |
| Brevundimonas bullata | Caulobacteraceae | 4 | 0 | 0 | 0 |
| Roseomonas pecunia | Acetobacteraceae | 1 | 0 | 0 | 0 |
| Sphingomonas panni, Sphingomonas sp. | Sphingomonadaceae | 2 | 1 | 1 | 0 |
| Cupriavidus metallidurans | Burkholderiaceae | 1 | 0 | 0 | 0 |
| Massilia aurea, Massilia timonae | Oxalobacteraceae | 2 | 0 | 0 | 0 |
| Enterobacter cowanii | Enterobacteriaceae | 1 | 1 | 0 | 0 |
| Erwinia persicina | 1 | 1 | 0 | 0 | |
| Pantoea agglomerans, Pantoea ananatis, Pantoea stewartii, Pantoea vagans | 13 | 5 | 0 | 0 | |
| Acinetobacter iwoffii, Acinetobacter johnsonii, Acinetobacter ursingii | Moraxellaceae | 4 | 1 | 0 | 0 |
| Moraxella osloensis | 2 | 1 | 0 | 0 | |
| Psychrobacter faecalis | 3 | 1 | 0 | 0 | |
| Pseudomonas asplenii, Pseudomonas fragi, Pseudomonas oleovorans, Pseudomonas putida | Pseudomonadaceae | 5 | 1 | 1 | 0 |
| Stenotrophomonas maltophilia | Xanthomonadaceae | 1 | 0 | 0 | 0 |
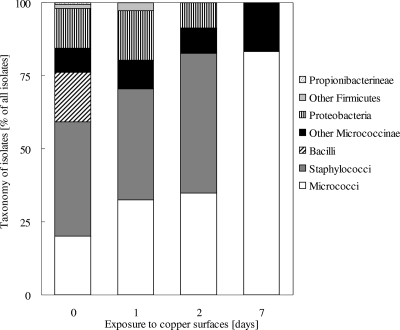
Gram-positive bacteria are the predominant group of bacteria from copper coins and are the most resistant to copper surfaces.
The majority of the isolates were Gram-positive bacteria (Fig. 1 and Table 1). The largest group comprised the staphylococci (115 isolates), followed by micrococci (59 isolates) and bacilli (50 isolates). The remainder belonged to the Actinobacteria (except micrococci), including other Micrococcineae (24 isolates) and Propionibacterineae (2 isolates). One species, Roseomonas pecunia, has not been characterized before; a complete species description has been submitted for publication (A. Lopes et al., submitted for publication). Finally, four non-Bacillus Firmicutes isolates were identified. Surprisingly, no corynebacteria or flavobacteria were among the isolates from coins.
FIG. 1.
Relative abundances of bacteria isolated from copper coins and their resistance to metallic copper surfaces. The relative abundances of 294 bacterial strains isolated from European copper coins at time zero and the relative abundances of copper surface-resistant isolates after exposure to experimental pure metallic copper surfaces for 1, 2, and 7 days are shown.
Only 40 isolates were Gram negative (Fig. 1). The biggest group among these were the gammaproteobacteria (30 isolates), including Pseudomonas oleovorans, Pantoea stewartii, and Acinetobacter johnsonii. Other proteobacteria were from the alphaproteobacteria subgroup (seven isolates), including two species of the genus Sphingomonas (two isolates), and a few betaproteobacteria (three isolates), with two Massilia species.
Upon testing for survival on pure copper surfaces (99% Cu, C110), a total of 71 isolates were found to survive 1 day of exposure, 23 survived 2 days of exposure, and 6 survived 1 week of exposure, but none survived 1 month of exposure (Fig. 1). The predominant genus of the 1-day survivors was Staphylococcus (27 strains), followed by Micrococcus (23 strains), the Gram-negative bacteria (proteobacteria) (12 strains), and Micrococcineae other than micrococci (7 strains). A similar trend was observed after 2 days of exposure, but the staphylococci (11 strains) and the micrococci (8 strains) had similar numbers of surviving strains. Only two actinomycetales other than micrococci, (Kocuria palustris and Brachybacterium conglomeratum) and two proteobacteria (Sphingomonas panni and Pseudomonas oleovorans) remained among the survivors. Finally, after 7 days only four micrococcal strains (all M. luteus) and one Brachybacterium conglomeratum strain survived. In general, the isolates survived from 16 times (M. luteus) up to 5,760 times (Pseudomonas oleovorans) longer on copper surfaces than their type strains or controls (Table 2). Remarkably, most but not all control and type strains of the respective copper coin-resistant isolates were killed much faster than the isolates. There were two exceptions to this trend. One was Kocuria marina, which survived 48 h, whereas the coin strain of K. marina survived for only 24 h. The other exception was Sphingomonas panni. Each S. panni strain survived for 48 h on metallic copper (Table 2). This suggests that resistance against metallic copper surfaces is rare but not absent from bacteria, especially among other strains of species that were found on copper coins.
TABLE 2.
Resistance of representative copper coin isolates to dry or moist copper surfaces and ionic copper
| Type or control strain or coin isolatea | Survival time on: |
MIC (mM CuCl2) | |
|---|---|---|---|
| Dry copper surfaces | Moist copper surfaces | ||
| Escherichia coli W3110b | >30 s | >1 h | 3.5 |
| Pantoea stewartii DSM 30176 | >30 s | >48 h | 1.5 |
| P. stewartii L10 | >24 h | >1 h | 4.5 |
| Acinetobacter johnsonii DSM 6963 | >1 min | >1 h | 2.5 |
| A. johnsonii L18 | >24 h | >1 h | 3.0 |
| Pseudomonas oleovorans DSM 1045 | >30 s | >1 h | 2.5 |
| P. oleovorans L19 | >48 h | >24 h | 3.5 |
| Sphingomonas panni DSM 15761 | >48 h | >48 h | 2.0 |
| S. panni R65P | >48 h | >1 h | 0.75 |
| Staphylococcus haemolyticus DSM 20263 | >1 h | >3 h | 3.5 |
| S. haemolyticus L70 | >48 h | >3 h | 2.0 |
| Staphylococcus epidermidis DSM 20044 | >1 h | >1 h | 1.0 |
| S. epidermidis L77 | >24 h | >24 h | 1.5 |
| Staphylococcus warnerii DSM 20316 | >1 min | >1 h | 2.5 |
| S. warnerii L47 | >48 h | >24 h | 2.0 |
| Brachybacterium conglomeratum DSM 10241 | >10 min | >1 h | 0.5 |
| B. conglomeratum N96 | >7 days | >1 h | 0.5 |
| Micrococcus luteus DSM 20030 | >3 h | >24 h | 2.0 |
| M. luteus L51 | >48 h | >3 h | 1.5 |
| Kocuria marina JCM 13363 | >48 h | >1 h | 2.0 |
| K. marina L73 | >24 h | >24 h | 2.0 |
| K. palustris R40 | >48 h | >1 h | 0.75 |
In each group, the first strain is a type or control strain and the second and/or third strain is a coin isolate.
Lab strain of E. coli, included as an example of a copper-sensitive bacterium.
All strains that were resistant to metallic copper surfaces survived for at least 1 month on stainless steel surfaces (data not shown). Most of the representative strains shown in Table 2 also survived for 1 month on plastic or aluminum, surfaces that cannot serve as a vital iron source. Unexpectedly, some Gram-negative strains were sensitive to aluminum or plastic surfaces. The type strain of Pseudomonas oleovorans was inactivated within 1 day upon exposure to plastic and aluminum but not to stainless steel (data not shown). The Pantoea stewartii type strain was inactivated on aluminum after 1 day but not on plastic or stainless steel surfaces. In general, all strains shown in Table 2 survived longer on plastic, stainless steel, or aluminum than on copper surfaces. Thus, the differences in survival time on dry metallic copper observed among the strains cannot be attributed to resistance against desiccation but are specific to dry metallic copper.
Coin bacteria are sensitive to wet copper surfaces and to copper ions.
Recent studies demonstrated that the toxicity exerted by dry metallic copper surfaces is different from that of wet copper surfaces and that the mechanism of killing differs from that of copper ions (9, 10). We therefore reasoned that the isolates found to be resistant to dry metallic copper (Fig. 1 and Table 1) might carry new resistance mechanisms unrelated to defense systems against moist copper surfaces or ionic copper. When strains that survived dry metallic copper for 2 days were tested on moist copper coupons, some of them were more sensitive than their control strains. The coin isolate of P. stewartii, strain L10, was dead after 1 h of exposure, but the control survived for 1 week of exposure (Table 2). Similarly, S. panni R65P from coins was killed by moist exposure after 1 h, but the type strain survived for 48 h. The type strain of M. luteus survived eight times longer on moist copper than the M. luteus L51 isolate. Other coin isolates were as resistant to moist exposure as their controls (A. johnstonii, S. haemolyticus, and B. conglomeratum). This lower resistance against moist metallic copper suggests that the resistance mechanisms that have evolved to withstand dry copper surfaces are different from those needed for survival on moist copper.
Likewise, there was no correlation between the dry copper surface resistance of the 2-day survivors and their CuCl2 MICs on solidified agar media (Table 2). Virtually all dry copper surface-resistant isolates were as sensitive to copper ions as their control strains and exhibited resistance levels comparable to those of E. coli (Table 2). Exceptions were S. panni R65P and K. palustris R40, which were about three times less resistant against CuCl2, and P. stewartii L10, which was three times more resistant than its type strain (Table 2).
Staphylococci isolated from coins are no more antibiotic resistant than their type strains.
It might be argued that prolonged use of metallic copper surfaces not only would select for resistance traits against this challenge but also would favor coselection with innate antibiotic resistance genes. We investigated if our staphylococcal strains from copper coins already had undergone such an evolutionary process by testing these bacteria in the bioMérieux antibiotic disk assay. Overall, the isolates did not exhibit an increased resistance to the antibiotics tested (Table 1). Most of the strains were scored as sensitive. This suggests that at least in the case of copper coins, there has been no coselection of metallic copper resistance and resistance against antibiotics. However, this possibility might become an issue once copper surfaces are widely used in health-related areas, making constant evaluation of antibiotic resistance in exposed microbes obligatory.
DISCUSSION
Bacteria from coins and copper surface resistance.
Surprisingly little attention has been paid to the microbial load of copper coinage. The antimicrobial properties of coins have been demonstrated, and coins have been found to have a lower bacterial load than paper currency (19, 34). Nevertheless, coins have been shown to carry opportunistic pathogens, such as a variety of species of the genera Staphylococcus, Bacillus, and Corynebacterium (34, 44). An earlier study identified S. aureus, E. coli, and P. aeruginosa on coins (1). The most comprehensive study on the bacterial flora from coins was published in 2005, comprising a total of 25 isolates from coinage collected from 17 countries (44).
In the present study we isolated 294 strains from two countries, Germany and Portugal. Though the goal of the study was not an inventory of the bacterial diversity of copper coins, our results generally follow those of previous studies. Xu et al. reported 100% Gram-positive isolates from coins, with the majority belonging to the genera Bacillus (40%) and Staphylococcus (28%) (44). Other studies also found predominantly staphylococci and bacilli but also corynebacteria (34). The predominant groups in the present study were the staphylococci (115 isolates) and bacilli (50 isolates), but a significant number of other bacteria, including Gram-negative strains (40 isolates) were also identified. However, corynebacteria, which are typical skin symbionts, were not observed. Compared to studies assessing the bacterial diversity of human skin, significant overlap with our isolates can be observed. A recent inventory of the human skin microbiome also identified a mixed population of bacteria from the dry sites of the palm of the hand proximal to the little finger, with a higher prevalence of betaproteobacteria and Flavobacteriales (15). There were no flavobacterial isolates derived from our copper coins. Bacteria of this phylum may have not been able to grow on the media used. Alternatively, flavobacteria might be especially sensitive to exposure to copper surfaces and consequently could not be selected.
Another study found propionibacteria to predominate human palms, followed by Streptococcaceae and Staphylococcaceae (13). Streptococci were also absent from our isolates, suggesting a high sensitivity against metallic copper. Thus, overall the bacteria derived from copper coins constitute a subgroup of the typical skin surface bacteria, with a bias toward staphylococci, bacilli, and betaproteobacteria.
Previous work by us (10) and others (9, 11, 32) has clearly demonstrated that metallic copper surfaces have strong antimicrobial properties against both Gram-positive and -negative bacteria. While coins probably provide a strong selective force for bacteria accidently exposed to these copper surfaces, 76% of our isolates were copper surface sensitive upon retesting on copper coupons; i.e., cells were killed in less than 1 day. The coins used for this study were regular coinage from general circulation and therefore probably were soiled with organic matter. Soiling has previously been demonstrated to enable bacteria to withstand copper surfaces for an extended time, rendering the biocidal surfaces inactive (2, 39, 42). The isolated coin bacteria that failed to exhibit copper surface resistance were thus probably protected by soiled patches or particles from the surface. Nevertheless, retesting of the strains surviving the initial selection provided evidence that dry copper surface resistance is not rare among bacteria related to the human skin.
One unexpected complication involved the bacilli. While scoring resistant to copper surfaces in our initial screens (data not shown), a closer examination of these Firmicutes elucidated that survival was because of the production of endospores. Endospores of Clostridium difficile have been demonstrated to withstand contact with copper surfaces (42). Another study, however, found a >5-log-unit reduction of dormant endospores of C. difficile following 24 to 48 h of exposure to copper surfaces (41). In our research, some of the Bacillus spores from the copper coin isolates were able to germinate and form colonies even after 1 month of exposure on pure copper surfaces (data not shown), but vegetative cells of all coin-derived bacilli proved to be sensitive (Table 1).
Dry copper surfaces provide different antimicrobial properties than ionic copper, with potential implications for the challenge posed by multiple-drug-resistant germs.
The aim of this study was to identify bacteria able to withstand dry copper surfaces. On first glance these bacteria might pose a future complication for a more general application of metallic copper surfaces. However, knowledge of the taxonomic identity of these bacteria is the first step in adapting existing hygiene procedures to deal with this challenge. Dry copper surface resistance and resistance against copper ions do not go hand in hand. Most of the isolates that were resistant to surfaces were as sensitive to copper ions as their respective type strains. However, resistance systems that confer an increased level of resistance against copper ions compared to strains of the same species lacking these determinants have been studied frequently. The most prominent representatives are Pco from an E. coli strain isolated from a piggery (43), Cop from Pseudomonas syringae (6), and Tcr from enterococci (16). These very efficient copper ion resistance systems fail, however, in protecting cells from exposure to dry metallic copper, as exemplified by Pco. An E. coli strain harboring the pco genes was almost as sensitive to copper surfaces as a strain lacking pco or a strain lacking all major copper ion resistance systems, i.e., CopA, CueO, and Cus (10).
Frequently, the antimicrobial properties of metallic copper are tested with aqueous bacterial cultures exposed to the surfaces (9, 11, 33, 42). We believe that this experimental setup does not reflect the situation in a clinical environment where most touch surfaces are dry. Therefore, we have recently adapted a method for studying bacteria exposed to dry copper surfaces (10). Bacteria in a protocol mimicking dry touch surfaces were killed within minutes, in contrast to about 1 to 8 h under wet conditions (9, 33). Recent results from hospital trials conducted in three countries (South Africa, Great Britain, and Germany) confirm the efficient antimicrobial properties of copper surfaces in a real-life setting (5, 28; Mikolay et al., unpublished data). Therefore, studying copper surface-resistant bacteria and their mechanisms of survival will probably strengthen our comprehension for use of copper surfaces and their further development. The results reported in this present work suggest that resistance mechanisms against dry metallic copper differ from those responsible for defense of bacteria against wet surfaces or dissolved copper ions. Thus, it is unsurprising that our isolates where as sensitive as their respective type or control strains when survival on copper ion-containing solid agar media was examined (Table 2). There is further indication that dry copper surfaces pose a significantly different kind of stress to bacteria than copper surfaces that are left wet. Under moist conditions, the dry copper surface-resistant coin isolates exhibited inactivation rates more similar to those of the sensitive controls (Table 2).
Dry metallic copper surfaces are not a habitat on which bacteria actually can grow and propagate. This is in sharp contrast to environments such as piggeries or orchards, where both antibiotics and copper compounds are applied to an actively growing and interacting microbial community, including microbial biofilms. Animal guts, biofilms, and clinical environments are recognized as settings where coselection of antibiotic and metal resistances may or may not occur (reviewed in reference 4). Therefore, a major concern when applying metal ions such as copper salts as antimicrobials is the potential of very efficient copper ion resistance systems related to Pco, Cop, or Tcr driving the coselection with resistances against antimicrobial agents and antibiotics. In fact, Tcr-like systems were found quite frequently when copper ions were used as a growth supplement in husbandry. These copper ion-detoxifying systems were also found to be genetically linked to genetic determinants conferring multiple drug resistance (to macrolides and glycopeptides) in livestock (4, 18). However, the appearance of copper and antibiotic resistance in bacteria isolated from pigs could not be correlated, and the available data did not support coselection of these traits (17).
Given that dry copper surfaces do not support growth of microbes, coselection of different resistance mechanisms is expected to be of minor concern. Nevertheless, antibiotic and copper surface resistance systems could co-occur, facilitating intra- and interspecies spread. Yet, at least for the staphylococci that we have isolated, copper surface resistance and resistance against common antibiotics do not seem to be intrinsically coupled (Table 1). Because dry copper surfaces also inactivate bacteria expressing efficient copper ion resistance determinants such as pco, previous reports of the co-occurrence of metal (ion) and antibiotic resistances in staphylococci likely have little consequence for the safe use of metallic copper (40). Further work is currently in progress to unravel the mechanisms that enable some bacteria to withstand the toxic properties of metallic copper surfaces. We expect to find mechanisms that go beyond resistance against copper ions. This knowledge might then be applied to develop new strategies involving the use of copper surfaces in the battle against bacteria responsible for nosocomial infections. Ideally, these efforts will lead to minimization, if not prevention, of the spread of such pathogens in hospitals and other places where human health is at risk.
Supplementary Material
Acknowledgments
This work was supported by the International Copper Association (ICA) and the Copper Development Association (CDA), by the UNL Research Council, and by the Nebraska Redox Biology Center (to G.G.). C.E.S. was supported by a Fundação para a Ciência e Tecnologia, Portugal, graduate fellowship. Research in the lab of P.V.M. was funded by the Fundação para a Ciência e Tecnologia under project PPCDT/AMB/60909/2004. Equipment for this project was purchased with Nebraska Tobacco Settlement Biomedical Research Development Funds.
We thank Paul D. Fey (UNMC), William A. Day (UofA), and Greg A. Somerville (UNL) for the generous gifts of strains; Filipa Sobreira Pires Salavessa Fontes, Maria Inês Simões Brandão, Nadine Taudte, Travis Krans, Corinna Hermsen, and Ee Wen Lam for skillful technical assistance; and Jennifer Quaranta for critically reading the manuscript.
Footnotes
Published ahead of print on 4 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abrams, B. L., and N. G. Waterman. 1972. Dirty money. JAMA 219:1202-1203. [PubMed] [Google Scholar]
- 2.Airey, P., and J. Verran. 2007. Potential use of copper as a hygienic surface; problems associated with cumulative soiling and cleaning. J. Hosp. Infect. 67:271-277. [DOI] [PubMed] [Google Scholar]
- 3.Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132:221-241. [DOI] [PubMed] [Google Scholar]
- 4.Baker-Austin, C., M. S. Wright, R. Stepanauskas, and J. V. McArthur. 2006. Co-selection of antibiotic and metal resistance. Trends Microbiol. 14:176-182. [DOI] [PubMed] [Google Scholar]
- 5.Casey, A. L., D. Adams, T. J. Karpanen, P. A. Lambert, B. D. Cookson, P. Nightingale, L. Miruszenko, R. Shillam, P. Christian, and T. S. J. Elliott. 2009. Role of copper in reducing hospital environment contamination. J. Hosp. Infect. doi: 10.1016/j.jhin.2009.08.018. [DOI] [PubMed]
- 6.Cha, J. S., and D. A. Cooksey. 1991. Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 88:8915-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The Ribosomal Database Project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comité de l'Antibiogramme de la Société Française de Microbiologie. 1998. Communiqué du Comité de l'Antibiogramme de la Société Française de Microbiologie. Bull. Soc. Fr. Microbiol. 13:243-258. [Google Scholar]
- 9.Elguindi, J., J. Wagner, and C. Rensing. 2009. Genes involved in copper resistance influence survival of Pseudomonas aeruginosa on copper surfaces. J. Appl. Microbiol. 106:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espirito Santo, C., N. Taudte, D. H. Nies, and G. Grass. 2008. Contribution of copper ion resistance to survival of Escherichia coli on metallic copper surfaces. Appl. Environ. Microbiol. 74:977-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faundez, G., M. Troncoso, P. Navarrete, and G. Figueroa. 2004. Antimicrobial activity of copper surfaces against suspensions of Salmonella enterica and Campylobacter jejuni. BMC Microbiol. 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 13.Fierer, N., M. Hamady, C. L. Lauber, and R. Knight. 2008. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:17994-17999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke, S., G. Grass, C. Rensing, and D. H. Nies. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grice, E. A., H. H. Kong, S. Conlan, C. B. Deming, J. Davis, A. C. Young, G. G. Bouffard, R. W. Blakesley, P. R. Murray, E. D. Green, M. L. Turner, and J. A. Segre. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasman, H. 2005. The tcrB gene is part of the tcrYAZB operon conferring copper resistance in Enterococcus faecium and Enterococcus faecalis. Microbiology 151:3019-3025. [DOI] [PubMed] [Google Scholar]
- 17.Hasman, H., and F. M. Aarestrup. 2005. Relationship between copper, glycopeptide, and macrolide resistance among Enterococcus faecium strains isolated from pigs in Denmark between 1997 and 2003. Antimicrob. Agents Chemother. 49:454-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasman, H., and F. M. Aarestrup. 2002. tcrB, a gene conferring transferable copper resistance in Enterococcus faecium: occurrence, transferability, and linkage to macrolide and glycopeptide resistance. Antimicrob. Agents Chemother. 46:1410-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havas, F. 2000. About the bacteriological state of notes and coins. Magyar Allatorvosok Lapja 122:501-503. [Google Scholar]
- 20.Helbig, K., C. Bleuel, G. J. Krauss, and D. H. Nies. 2008. Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 190:5431-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, J. L. 1994. Similarity analysis of rRNAs, p. 683-700. In P. Gerhardt, W. A. Wood, N. r. Krieg, and R. Murray (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC.
- 22.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 23.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 24.Macomber, L., and J. A. Imlay. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106:8344-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macomber, L., C. Rensing, and J. A. Imlay. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189:1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magnani, D., O. Barre, S. D. Gerber, and M. Solioz. 2008. Characterization of the CopR regulon of Lactococcus lactis IL1403. J. Bacteriol. 190:536-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marais, F., S. Mehtar, and L. Chalkley. 2009. Antimicrobial efficacy of copper touch surfaces in reducing environmental bioburden in a South African community healthcare facility. J. Hosp. Infect. doi: 10.1016/j.jhin.2009.07.010. [DOI] [PubMed]
- 29.McArthur, J. V., and R. C. Tuckfield. 2000. Spatial patterns in antibiotic resistance among stream bacteria: effects of industrial pollution. Appl. Environ. Microbiol. 66:3722-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miras, R., I. Morin, O. Jacquin, M. Cuillel, F. Guillain, and E. Mintz. 2008. Interplay between glutathione, Atx1 and copper. 1. Copper(I) glutathionate induced dimerization of Atx1. J. Biol. Inorg. Chem. 13:195-205. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen, P., D. Fritze, and F. G. Priest. 1995. Phenetic diversity of alkaliphilic Bacillus strains: proposal for nine new species. Microbiology 141:1745-1761. [Google Scholar]
- 32.Noyce, J. O., H. Michels, and C. W. Keevil. 2006. Potential use of copper surfaces to reduce survival of epidemic meticillin-resistant Staphylococcus aureus in the healthcare environment. J. Hosp. Infect. 63:289-297. [DOI] [PubMed] [Google Scholar]
- 33.Noyce, J. O., H. Michels, and C. W. Keevil. 2006. Use of copper cast alloys to control Escherichia coli O157 cross-contamination during food processing. Appl. Environ. Microbiol. 72:4239-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pachter, B., L. Kozer, S. Pachter, and M. Weiner. 1997. Dirty money? A bacteriologic investigation of US currency. Infect. Med. 14:574. [Google Scholar]
- 35.Rensing, C., B. Fan, R. Sharma, B. Mitra, and B. P. Rosen. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc. Natl. Acad. Sci. U. S. A. 97:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singleton, C., and N. E. Le Brun. 2007. Atx1-like chaperones and their cognate P-type ATPases: copper-binding and transfer. Biometals 20:275-289. [DOI] [PubMed] [Google Scholar]
- 37.Sterne, M. 1937. The effects of different CO concentrations on the growth of virulent anthrax strains. Pathogenicity and immunity tests on guinea pigs and sheep with anthrax variants derived from virulent strains. Onderstepoort J. Vet. Sci. Anim. Ind. 9:49-67. [Google Scholar]
- 38.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 39.Tolba, O., A. Loughrey, C. E. Goldsmith, B. C. Millar, P. J. Rooney, and J. E. Moore. 2007. Survival of epidemic strains of nosocomial- and community-acquired methicillin-resistant Staphylococcus aureus on coins. Am. J. Infect. Control 35:342-346. [DOI] [PubMed] [Google Scholar]
- 40.Ugur, A., and O. Ceylan. 2003. Occurrence of resistance to antibiotics, metals, and plasmids in clinical strains of Staphylococcus spp. Arch. Med. Res. 34:130-136. [DOI] [PubMed] [Google Scholar]
- 41.Weaver, L., H. T. Michels, and C. W. Keevil. 2008. Survival of Clostridium difficile on copper and steel: futuristic options for hospital hygiene. J. Hosp. Infect. 68:145-151. [DOI] [PubMed] [Google Scholar]
- 42.Wheeldon, L. J., T. Worthington, P. A. Lambert, A. C. Hilton, C. J. Lowden, and T. S. Elliott. 2008. Antimicrobial efficacy of copper surfaces against spores and vegetative cells of Clostridium difficile: the germination theory. J. Antimicrob. Chemother. 62:522-525. [DOI] [PubMed] [Google Scholar]
- 43.Williams, J. R., A. G. Morgan, D. A. Rouch, N. L. Brown, and B. T. Lee. 1993. Copper-resistant enteric bacteria from United Kingdom and Australian piggeries. Appl. Environ. Microbiol. 59:2531-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, J., J. E. Moore, and B. C. Millar. 2005. Ribosomal DNA (rDNA) identification of the culturable bacterial flora on monetary coinage from 17 currencies. J. Environ. Health 67:51-55. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.