Abstract
Arthrobacter sp. strain JBH1 was isolated from nitroglycerin-contaminated soil by selective enrichment. Detection of transient intermediates and simultaneous adaptation studies with potential intermediates indicated that the degradation pathway involves the conversion of nitroglycerin to glycerol via 1,2-dinitroglycerin and 1-mononitroglycerin, with concomitant release of nitrite. Glycerol then serves as the source of carbon and energy.
Nitroglycerin (NG) is manufactured widely for use as an explosive and a pharmaceutical vasodilator. It has been found as a contaminant in soil and groundwater (7, 9). Due to NG's health effects as well as its highly explosive nature, NG contamination in soils and groundwater poses a concern that requires remedial action (3). Natural attenuation and in situ bioremediation have been used for remediation in soils contaminated with certain other explosives (16), but the mineralization of NG in soil and groundwater has not been reported.
To date, no pure cultures able to grow on NG as the sole carbon, energy, and nitrogen source have been isolated. Accashian et al. (1) observed growth associated with the degradation of NG under aerobic conditions by a mixed culture originating from activated sludge. The use of NG as a source of nitrogen has been studied in mixed and pure cultures during growth on alternative sources of carbon and energy (3, 9, 11, 20). Under such conditions, NG undergoes a sequential denitration pathway in which NG is transformed to 1,2-dinitroglycerin (1,2DNG) or 1,3DNG followed by 1-mononitroglycerin (1MNG) or 2MNG and then glycerol, under both aerobic and anaerobic conditions (3, 6, 9, 11, 20), and the enzymes involved in denitration have been characterized in some detail (4, 8, 15, 21). Pure cultures capable of completely denitrating NG as a source of nitrogen when provided additional sources of carbon include Bacillus thuringiensis/cereus and Enterobacter agglomerans (11) and a Rhodococcus species (8, 9). Cultures capable of incomplete denitration to MNG in the presence of additional carbon sources were identified as Pseudomonas putida, Pseudomonas fluorescens (4), an Arthobacter species, a Klebsiella species (8, 9), and Agrobacterium radiobacter (20).
Here we describe the isolation of bacteria able to degrade NG as the sole source of carbon, nitrogen, and energy. The inoculum for selective enrichment was soil historically contaminated with NG obtained at a facility that formerly manufactured explosives located in the northeastern United States. The enrichment medium consisted of minimal medium prepared as previously described (17) supplemented with NG (0.26 mM), which was synthesized as previously described (18). During enrichment, samples of the inoculum (optical density at 600 nm [OD600] ∼ 0.03) were diluted 1/16 in fresh enrichment medium every 2 to 3 weeks. Isolates were obtained by dilution to extinction in NG-supplemented minimal medium. Cultures were grown under aerobic conditions in minimal medium at pH 7.2 and 23°C or in tryptic soy agar (TSA; 1/4 strength).
Early stages of enrichment cultures required extended incubation with lag phases of over 200 h and exhibited slow degradation of NG (less than 1 μmol substrate/mg protein/h). After a number of transfers over 8 months, the degradation rates increased substantially (2.2 μmol substrate/mg protein/h). A pure culture capable of growth on NG was identified based on 16S rRNA gene analysis (504 bp) as an Arthrobacter species with 99.5% similarity to Arthrobacter pascens (GenBank accession no. GU246730). Purity of the cultures was confirmed microscopically and by formation of a single colony type on TSA plates. 16S gene sequencing and identification were done by MIDI Labs (Newark, DE) and SeqWright DNA Technology Services (Houston, TX). The Arthrobacter cells stained primarily as Gram-negative rods with a small number of Gram-positive cocci (data not shown); Gram variability is also a characteristic of the closely related Arthrobacter globiformis (2, 19). The optimum growth temperature is 30°C, and the optimum pH is 7.2. Higher pH values were not investigated because NG begins to undergo hydrolysis above pH 7.5 (data not shown). The isolated culture can grow on glycerol, acetate, succinate, citrate, and lactate, with nitrite as the nitrogen source. Previous authors described an Arthrobacter species able to use NG as a nitrogen source in the presence of additional sources of carbon. However, only dinitroesters were formed, and complete mineralization was not achieved (9).
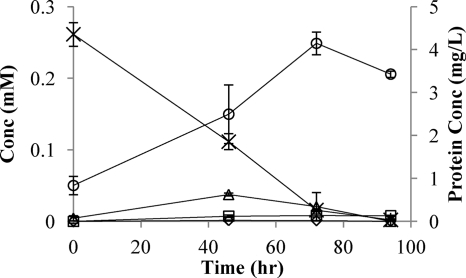
To determine the degradation pathway, cultures of the isolated strain (5 ml of inoculum grown on NG to an OD600 of 0.3) were grown in minimal medium (100 ml) supplemented with NG at a final concentration of 0.27 mM. Inoculated bottles and abiotic controls were continuously mixed, and NG, 1,2DNG, 1,3DNG, 1MNG, 2MNG, nitrite, nitrate, CO2, total protein, and optical density were measured at appropriate intervals. Nitroesters were analyzed with an Agilent high-performance liquid chromatograph (HPLC) equipped with an LC-18 column (250 by 4.6 mm, 5 μm; Supelco) and a UV detector at a wavelength of 214 nm (13). Methanol-water (50%, vol/vol) was used as the mobile phase at a flow rate of 1 ml/min. Nitrite and nitrate were analyzed with an ion chromatograph (IC) equipped with an IonPac AS14A anion-exchange column (Dionex, CA) at a flow rate of 1 ml/min. Carbon dioxide production was measured with a Micro Oxymax respirometer (Columbus Instruments, OH), and total protein was quantified using the Micro BCA protein assay kit (Pierce Biotechnology, IL) according to manufacturer's instructions. During the degradation of NG the 1,2DNG concentration was relatively high at 46 and 72 h (Fig. 1). 1,3DNG, detected only at time zero, resulted from trace impurities in the NG stock solution. Trace amounts of 1MNG appeared transiently, and trace amounts of 2MNG accumulated and did not disappear. Traces of nitrite at time zero were from the inoculum. The concentration of NG in the abiotic control did not change during the experiment (data not shown).
FIG. 1.
Growth of strain JBH1 on NG. ×, NG; ▵, 1,2DNG; ⋄, 1MNG; □, 2MNG; ○, protein.
Results from the experiment described above were used to calculate nitrogen and carbon mass balances (Tables 1 and 2). Nitrogen content in protein was approximated using the formula C5H7O2N (14). Because all nitrogen was accounted for throughout, we conclude that the only nitrogen-containing intermediate compounds are 1,2DNG and 1MNG, which is consistent with previous studies (6, 9, 20). The fact that most of the nitrogen was released as nitrite is consistent with previous reports of denitration catalyzed by reductase enzymes (4, 8, 21). The minor amounts of nitrate observed could be from abiotic hydrolysis (5, 12) or from oxidation of nitrite. Cultures supplemented with glycerol or other carbon sources assimilated all of the nitrite (data not shown).
TABLE 1.
Nitrogen mass balance
| Time (h) | % of total initial nitrogen by mass recovered ina: |
Total recovery (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1MNG | 2MNG | 1,2DNG | 1,3DNG | NG | Protein | Nitrite | Nitrate | ||
| 0 | NDb | ND | 0.9 ± 0.7 | 0.8 ± 0.6 | 82 ± 5.2 | 0.8 ± 0.2 | 14 ± 0.7 | 0.8 ± 0.3 | 100 ± 5.3 |
| 46 | 0.1 ± 0.0 | 0.8 ± 0.2 | 7.9 ± 0.4 | ND | 35 ± 3.6 | 2.0 ± 0.5 | 49 ± 1.1 | 1.7 ± 0.0 | 96 ± 4.2 |
| 72 | 0.1 ± 0.0 | 0.9 ± 0.2 | 4.3 ± 4.2 | ND | 5.0 ± 0.4 | 3.3 ± 0.2 | 81 ± 4.2 | 3.9 ± 1.9 | 98 ± 6.8 |
| 94 | ND | 0.6 ± 0.4 | ND | ND | 0.6 ± 0.4 | 3.2 ± 0.0 | 95 ± 10 | 2.6 ± 1.6 | 102 ± 10 |
Data represent averages of four replicates ± standard deviations.
ND, not detected.
TABLE 2.
Carbon mass balance
| Time (h) | % of total initial carbon by mass recovered in: |
Total recovery (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| 1MNGa | 2MNGa | 1,2DNGa | 1,3DNGa | NGa | Proteina | CO2b | ||
| 0 | NDc | ND | 1.6 ± 1.2 | 1.9 ± 0.4 | 92 ± 5.8 | 4.4 ± 0.9 | 100 ± 8.4 | |
| 46 | 0.5 ± 0.2 | 2.6 ± 0.6 | 13 ± 0.7 | ND | 39 ± 3.9 | 13 ± 3.0 | 28 ± 5.7 | 96 ± 14.1 |
| 72 | 0.4 ± 0.0 | 2.9 ± 0.7 | 7.3 ± 7.0 | ND | 5.6 ± 0.4 | 22 ± 1.2 | 59 ± 8.3 | 97 ± 17.6 |
| 94 | ND | 2.8 ± 0.3 | ND | ND | 0.8 ± 0.5 | 18 ± 0.3 | 71 ± 4.5 | 93 ± 5.6 |
Data represent averages of four replicates ± standard deviations.
Data represent averages of duplicates ± standard deviations.
ND, not detected.
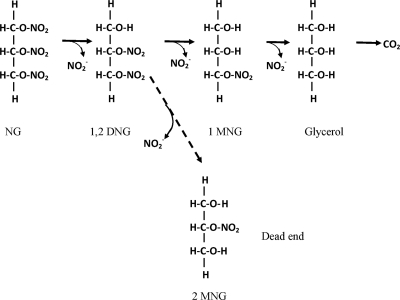
In a separate experiment cells grown on NG were added to minimal media containing 1,3DNG, 1,2DNG, 1MNG, or 2MNG and degradation over time was measured. 1,2DNG, 1,3DNG, and 1MNG were degraded at rates of 6.5, 3.8, and 8 μmol substrate/mg protein/hour. No degradation of 2MNG was detected (after 250 h), which indicates that 2MNG is not an intermediate in a productive degradation pathway. Because 1,3DNG was not observed at any point during the degradation of NG and its degradation rate is approximately one-half the degradation rate of 1,2DNG, it also seems not to be part of the main NG degradation pathway used by Arthrobacter sp. strain JBH1. The above observations indicate that the degradation pathway involves a sequential denitration of NG to 1,2DNG, 1MNG, and then glycerol, which serves as the source of carbon and energy (Fig. 2). The productive degradation pathway differs from that observed by previous authors using both mixed (1, 3, 6) and pure cultures (4, 9, 11, 20), in which both 1,3- and 1,2DNG were intermediates during NG transformation. Additionally, in previous studies both MNG isomers were produced regardless of the ratio of 1,2DNG to 1,3DNG (3, 4, 6, 9, 20). Our results indicate that the enzymes involved in denitration of NG in strain JBH1 are highly specific and catalyze sequential denitrations that do not involve 1,3DNG or 2MNG. Determination of how the specificity avoids misrouting of intermediates will require purification and characterization of the enzyme(s) involved.
FIG. 2.
Proposed NG degradation pathway.
Mass balances of carbon and nitrogen were used to determine the following stoichiometric equation that describes NG mineralization by Arthrobacter sp. strain JBH1: 0.26C3H5(ONO2)3 + 0.33O2 → 0.03C5H7O2N + 0.63CO2 + 0.75NO2− + 0.75H+ + 0.17H2O. The result indicates that most of the NG molecule is being used for energy. The biomass yield is relatively low (0.057 mg protein/mg NG), with an fs (fraction of reducing equivalents of electron donor used for protein synthesis) of 0.36 (10), which is low compared to the aerobic degradation of other compounds by pure cultures, for which fs ranges between 0.4 and 0.6 (10, 14). The results are consistent with the requirement for relatively large amounts of energy during the initiation of the degradation mechanism (each denitration probably requires 1 mole of NADH or NADPH [21]).
Although NG degradation rates were optimal at pH 7.2, they were still substantial at values as low as 5.1. The results suggest that NG degradation is possible even at low pH values typical of the subsurface at sites where explosives were formerly manufactured or sites where nitrite production lowers the pH.
NG concentrations above 0.5 mM are inhibitory, but degradation was still observed at 1.2 mM (data not shown). The finding that NG can be inhibitory to bacteria at concentrations that are well below the solubility of the compound is consistent with those of Accashian et al. (1) for a mixed culture.
The ability of Arthrobacter sp. strain JBH1 to grow on NG as the carbon and nitrogen source provides the basis for a shift in potential strategies for natural attenuation and bioremediation of NG at contaminated sites. The apparent specificity of the denitration steps raises interesting questions about the evolution of the pathway.
Acknowledgments
This work was supported by the DuPont Corporate Remediation Group. Additional support was provided by Defense Threat Reduction Agency/U.S. Army Research Office.
We thank Shirley Nishino for her assistance with laboratory techniques.
Footnotes
Published ahead of print on 8 January 2010.
REFERENCES
- 1.Accashian, J. V., R. T. Vinopal, B. J. Kim, and B. F. Smets. 1998. Aerobic growth on nitroglycerin as the sole carbon, nitrogen, and energy source by a mixed bacteria culture. Appl. Environ. Microbiol. 64:3300-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beveridge, T. J. 1990. Mechanism of gram variability in select bacteria. J. Bacteriol. 172:1609-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaumik, S., C. Christodoulatos, G. P. Korfiatis, and B. W. Brodman. 1997. Aerobic and anaerobic biodegradation of nitroglycerin in batch and packed bed bioreactors. Water Sci. Technol. 36:139-146. [Google Scholar]
- 4.Blehert, D. S., K. L. Knoke, B. G. Fox, and G. H. Chambiliss. 1997. Regioselectivity of nitroglycerin denitration by flavoprotein nitroester reductase purified from two Pseudomonas species. J. Bacteriol. 179:6912-6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cappellos, C., W. J. Fisco, C. Ribaudo, V. D. Hogan, J. Campisi, F. X. Murphy, T. C. Castorina, and D. H. Rosenblatt. 1984. Basic hydrolysis of glyceryl nitrate esters. III. Trinitroglycerin. Int. J. Chem. Kinet. 16:1027-1051. [Google Scholar]
- 6.Cristodoulatos, C., S. Bhaumik, and B. W. Brodman. 1996. Anaerobic biodegradation of nitroglycerin. Water Res. 31:1426-1470. [Google Scholar]
- 7.Environmental Protection Agency. 2007. National priorities list. Environmental Protection Agency, Washington, DC.
- 8.Marshall, S. J., D. Krause, D. K. Blencowe, and G. F. White. 2004. Characterization of glycerol trinitrate reductase (NerA) and the catalytic role of active-site residues. J. Bacteriol. 186:1802-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marshall, S. J., and G. F. White. 2001. Complete denitration of nitroglycerin by bacteria isolated from a washwater soakaway. Appl. Environ. Microbiol. 67:2622-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarty, P. L. 2007. Thermodynamic electron equivalents model for bacterial yield prediction. Modifications and comparative evaluations. Biotechnol. Bioeng. 97:377-388. [DOI] [PubMed] [Google Scholar]
- 11.Meng, M., W. Sun, L. A. Geelhaar, G. Kuma, A. R. Partel, G. F. Payne, M. K. Speedie, and J. R. Stacy. 1995. Denitration of glycerol trinitrate by resting cells and cell extracts of Bacillus thuringiensis/cereus and Enterobacter agglomerans. Appl. Environ. Microbiol. 61:2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirecki, J. E., B. Porter, and C. A. Weiss. 2006. Environmental transport and fate process descriptors for propellant compounds. U.S. Army Corps of Engineers, Vicksburg, MS.
- 13.Oh, S., D. K. Chan, B. J. Kim, and P. C. Chiu. 2004. Reduction of nitroglycerin with elemental iron: pathway, kinetics, and mechanisms. Environ. Sci. Technol. 38:3723-3730. [DOI] [PubMed] [Google Scholar]
- 14.Rittman, B. E., and P. L. McCarty. 2001. Environmental biotechnology. McGraw-Hill, Inc., New York, NY.
- 15.Snape, J. R., N. A. Walkley, A. P. Morby, S. Nicklin, and G. F. White. 1997. Purification, properties, and sequence of glycerol trinitrate reductase from Agrobacterium radiobacter. J. Bacteriol. 179:1796-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spain, J. C., J. B. Hughes, and H.-J. Knackmuss. 2000. Biodegradation of nitroaromatic compounds and explosives. CRC Press LLC, Boca Raton, FL.
- 17.Spanggord, R. J., J. C. Spain, S. F. Nishino, and K. E. Mortelmans. 1991. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl. Environ. Microbiol. 57:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.U.S. Army. 1984. Military explosives, TM 9-1300-214. U.S. Army, Washington, DC.
- 19.Ward, C. M., and G. W. Claus. 1973. Gram characteristics and wall ultrastructure of Arthrobacter crystallopoietes during coccus-rod morphogenesis. J. Bacteriol. 114:378-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White, G. F., J. R. Snape, and S. Nicklin. 1996. Biodegradation of glycerol trinitrate and pentaerythritol tetranitrate by Agrobacterium radiobacter. Appl. Environ. Microbiol. 62:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams, R. E., and N. C. Bruce. 2002. “New uses of an old enzyme”—the old yellow enzyme family of flavoenzymes. Microbiology 148:1607-1614. [DOI] [PubMed] [Google Scholar]