Abstract
Cheese is a complex and dynamic microbial ecosystem characterized by the presence of a large variety of bacteria, yeasts, and molds. Some microorganisms, including species of lactobacilli or lactococci, are known to contribute to the organoleptic quality of cheeses, whereas the presence of other microorganisms may lead to spoilage or constitute a health risk. Staphylococcus aureus is recognized worldwide as an important food-borne pathogen, owing to the production of enterotoxins in food matrices. In order to study enterotoxin gene expression during cheese manufacture, we developed an efficient procedure to recover total RNA from cheese and applied a robust strategy to study gene expression by reverse transcription-quantitative PCR (RT-qPCR). This method yielded pure preparations of undegraded RNA suitable for RT-qPCR. To normalize RT-qPCR data, expression of 10 potential reference genes was investigated during S. aureus growth in milk and in cheese. The three most stably expressed reference genes during cheese manufacture were ftsZ, pta, and gyrB, and these were used as internal controls for RT-qPCR of the genes sea and sed, encoding staphylococcal enterotoxins A and D, respectively. Expression of these staphylococcal enterotoxin genes was monitored during the first 72 h of the cheese-making process, and mRNA data were correlated with enterotoxin production.
Staphylococcus aureus is a significant bacterial pathogen producing a variety of proteins and toxins that contribute to its ability to colonize and cause diseases (15). Some S. aureus strains are able to produce staphylococcal enterotoxins (SEs) in food matrices and are responsible for food poisoning, characterized by such symptoms as nausea, vomiting, abdominal cramps, and diarrhea (2). In France S. aureus is reported as the most frequent pathogen involved in food-borne diseases associated with dairy products (12) and especially with raw milk cheeses (9). It is generally accepted that SE production constitutes a risk when S. aureus bacteria exceed a threshold of 105 S. aureus CFU per gram of cheese during manufacture. Numerous studies have reported on S. aureus behavior during cheese manufacturing, focusing only on S. aureus growth and enterotoxin production (1, 8, 11, 18, 21, 22, 31, 32, 34-36, 42, 43, 49). To our knowledge, no study has investigated the expression of the genes encoding SEs in foods or during food production. Numerous parameters, such as pH, aeration, or temperature, could, indeed, affect the expression of these genes. During the cheese-making process, natural staphylococcal contamination is minor in the total microbial population. The initial S. aureus contamination is usually below 103 CFU/ml of raw milk, while bacterial starters are inoculated at least at 106 CFU/ml of milk. Analysis of SE gene expression in situ thus requires an efficient method of extracting bacterial RNA from cheese to ensure recovery of quantifiable amounts of staphylococcal RNA. A precise method is also needed to quantify minor transcripts in the extracted bacterial RNA.
Two main strategies were initially used to extract bacterial RNA from cheese, one based on separation of cells from the cheese matrix prior to RNA extraction (29, 45) and the other using a direct method (4, 33). The method of Bonaïti et al. (4), which was developed to analyze the surface of cheese curd, is not directly applicable for all cheese fractions. Two methods were developed on model semifat cheeses, and these either have not been applied to cheese with standard fat content (33) or gave bad results with commercial cheese (45). Makhzami et al. separated cells from the cheese matrix using a Nycodenz density gradient, but cheeses contained more than 108 CFU/g of cheese (29), which is not always the case at the beginning of cheese processing. In the absence of a reliable method, there was still a need to develop an efficient bacterial RNA extraction method usable at any time in the cheese-making processing and applicable to any fraction of the cheese.
Real-time reverse transcription-quantitative PCR (RT-qPCR) is the most sensitive method for the detection of low-abundance mRNA and is therefore the method of choice for our studies. A good normalization strategy is needed to correct for sample-to-sample variations due to the different steps of the real-time RT-qPCR process from the RNA extraction to the PCR step (5, 6). Moreover, in our studies, one drawback is that staphylococcal RNA extracted from the cheese matrix cannot be quantified. However, relative measurements of mRNA expression changes can be determined by using internal reference genes. Ideally, reference genes used in relative RT-qPCR studies are controls equally expressed under different conditions. Several housekeeping genes have already been used in S. aureus gene expression studies as single internal reference genes, including ftsZ (41), pta (37), hu (7, 17, 39), and gyrB (19, 20, 46, 50). However, previous studies have shown that the expression of many housekeeping genes is differentially regulated, depending on experimental conditions (14, 44). The use of multiple reference genes results in a more accurate and reliable normalization of gene expression data (47). A Visual Basic Application (VBA) for Microsoft Excel called geNorm makes it possible to determine the most stably expressed genes from a series of candidate reference genes and the number of genes required for accurate normalization (47).
In the present study we developed an efficient method to extract bacterial RNA accessible for RT-qPCR from cheese and adapted a simple, sensitive, and reproducible method for quantification of relative transcript levels in order to study S. aureus enterotoxin gene expression during cheese manufacture. The method was applied to semihard cheeses and soft cheeses. We investigated the expression of 10 potential reference genes in S. aureus during growth in milk and in cheese to define good internal controls for experiments. The RNA extraction method was validated for RNA purity and quality. The method was used to determine the proportion of staphylococcal RNA in total RNA extracted from cheeses, which was then compared with the proportion of staphylococcal cells in the bacterial population. Finally, we studied the expression of two SE genes over the first 72 h of cheese making and compared the transcript levels with SE protein production.
MATERIALS AND METHODS
Bacterial strains.
S. aureus strain 361F, which was used throughout this study, was isolated by De Buyser et al. in 1985 (10) from raw ewe milk cheese that was implicated in staphylococcal food poisoning. This strain can produce enterotoxins A and D. Lactococcus lactis subsp. lactis (IL-416) and L. lactis subsp. cremoris (SK11) were grown in coculture with S. aureus, respectively, in milk and in cheese.
Growth conditions in milk and cell sampling.
Once S. aureus strain 361F had been precultured in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, MI) at 37°C with shaking (200 rpm) for 6 h, it was inoculated into 200 ml of sterile reconstituted milk (100 g of skim milk powder [Régilait, Saint-Martin-Belle-Roche, France] per liter of distilled water, sterilized at 110°C for 10 min). For the coculture with L. lactis, milk was also inoculated with strain IL-416 grown in M17 (Difco Laboratories) broth complemented with 1% (wt/vol) lactose at 30°C without shaking for 16 h. S. aureus strain 361F cultures were performed under the following conditions: (i) at 37°C with shaking (200 rpm), (ii) at 37°C without shaking, (iii) at 30°C with shaking, (iv) at 37°C with shaking in coculture with L. lactis IL-416, and (v) at 37°C with shaking in milk acidified with lactic acid (pH 5.3). Bacterial growth was monitored for 24 h. Serial dilutions of milk cultures were prepared in sterile 1% (wt/vol) peptone water and plated using a spiral plater (Spiral System, Cincinnati, OH) on BHI medium supplemented with 5% (wt/vol) NaCl for staphylococcus cell enumeration and on M17 medium supplemented with 1% (wt/vol) lactose for lactococcus cell enumeration. Plates were incubated for 2 days at 37°C (BHI medium plates) and at 30°C (M17 medium plates). Bacterial samples were collected in two independent experiments at four growth phases for RT-qPCR analysis: at the beginning of the exponential growth phase (2 h), at mid-exponential growth phase (6 h), at the end of the exponential growth phase (10 h), and at stationary phase (24 h). After a rapid centrifugation (3,000 × g at 4°C for 5 min), cell pellets were recovered and immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Semihard cheese process and commercial cheeses.
Experimental semihard cheeses were prepared under controlled bacteriological conditions according to a protocol adapted from semihard cheese technology. Once S. aureus strain 361F and L. lactis strain SK11 had been precultured, respectively, in BHI broth at 37°C with shaking (200 rpm) for 6 h and in sterile reconstituted milk at 30°C without shaking for 16 h, 6 liters of commercial full-fat, microfiltered milk (Marguerite, Villefranche sur Saône, France) preheated at 32°C was inoculated with S. aureus to a final concentration of 103, 104, or 106 CFU/ml and with L. lactis before the addition of 40 ml of filtered rennet extract (520 mg of chymosin/liter; Berthelot, France) for 100 liters of milk. Coagulation proceeded for about 45 min, and the curd was then cut into cubes and, after a slow stirring for 20 min, prepressed in whey for 30 min. The curd was finally cut into three equal parts, poured into molds, and pressed. Then, the temperature was slowly decreased to 16°C in 4 h. Whey draining was continued for almost 24 h. After the cheeses were taken out of the molds, they were salted for 5 h in sterile brine (pH 5 at 10°C), dried for 24 h, and conditioned in a plastic bag under vacuum. Model cheeses were approximately 250 g. Ripening took place at 12°C. Each cheese was manufactured twice using two different milk batches. The other cheeses used in the present study (Vieux Pané, Roquefort, and Munster) were purchased commercially from local cheese suppliers.
Bacterial enumeration in cheese samples.
The staphylococcus and lactococcus cell growth in cheeses was estimated for the first 3 days of the cheese-making procedure. Five grams of cheese, cut into small pieces, was added to 45 ml of trisodium citrate solution (71.4 mmol/liter). After incubation at room temperature for 10 min, the samples were homogenized using an Ultra-Turrax mechanical blender (T-25 IKA; Labo Moderne, Paris, France) at 16,500 rpm for two 30-s sequences. Serial dilutions of milk (before rennet addition) or homogenized cheese (after rennet addition) and incubation conditions for bacterial enumeration were then performed as described in the protocol for growth in milk.
Bacterial cell recovery from the cheese matrix.
Homogenized cheeses in trisodium citrate solution (10 ml) were centrifuged at 4,600 × g for 10 min at 4°C. After the fat layer on the top and the supernatant had been discarded, the pellet (containing bacteria and cheese particles) was washed with 1 ml of cold Tris base-EDTA (TE; 10 mM Tris base, 1 mM EDTA, pH 8) buffer and frozen in liquid nitrogen. The sample was then stored at −80°C until the RNA extraction, which was performed within 1 week. The yield of the recovery was measured by cell counts, as described above, before the first centrifugation and just before freezing.
RNA purification and quantification.
All solutions used for this procedure were stored at 4°C, and centrifugation steps were performed at 4°C to limit RNase activity. Cell pellets were suspended in diethyl pyrocarbonate (DEPC)-treated water before being transferred into 1.5-ml microtubes containing 0.5 g of 0.1-mm diameter glass beads (Sigma Aldrich, Saint Louis, MO), 30 μl of 3 M sodium acetate (pH 5.2), 30 μl of 10% (wt/vol) SDS, and 500 μl of water-saturated phenol-chloroform (5:1) (Sigma Aldrich), as described by Hiron et al. (24). Cells were then disrupted three times using a Fast Prep FP120 system (Bio1001; Thermo Electron Corporation) at maximum speed for 45 s, with chilling on ice for 1 min between pulses. After centrifugation (at 4°C 15 000 × g for 15 min), the aqueous phase was recovered, and RNA was purified with Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After isopropanol precipitation, total RNA was suspended in DEPC-treated water.
Extracts were adjusted to about 0.2 μg/μl to perform rigorous DNase treatments using a DNA-free kit (Ambion, Austin, TX) according to the manufacturer's instructions. RNA concentration, purity (represented by the A260 nm/A230 nm and A260 nm/A280 nm ratios [30]) and integrity (RNA integrity number [RIN]) (40) were assessed using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE) and an Agilent 2100 Bioanalyzer (Agilent Technology, Palo Alto, CA).
Reverse transcription and real-time qPCR.
Oligonucleotides for real-time qPCR (Table 1) were designed using Primer Express, version 2.0, software (Applied Biosystems, Foster City, CA) and purchased from Eurogentec SA (Seraing, Belgium). Secondary structures were controlled with Oligo Calc (27), and a BLAST analysis was performed to check primer specificity.
TABLE 1.
Candidate reference genes and genes of interest
| Gene group and name | Molecular function | Primer name and sequencea | Amplicon length (bp) | Efficiencyb | r2 |
|---|---|---|---|---|---|
| Candidate reference genes | |||||
| gyrB | DNA gyrase, subunit B | F: CCAGGTAAATTAGCCGATTGC | 121 | 95 | 0.994 |
| R: AAATCGCCTGCGTTCTAGAG | |||||
| ftsZ | Cell division protein | F: ATCCAAATCGGTGAAAAATTAACAC | 121 | 96 | 0.998 |
| R: CCATGTCTGCACCTTGGATTG | |||||
| hu | DNA-binding protein | F: TTTACGTGCAGCACGTTCAC | 125 | 92 | 0.999 |
| R: AAAAAGAAGCTGGTTCAGCAGTAG | |||||
| rplD | 50S Ribosomal protein L4 | F: TGAAAGATAATGCTGAGCGTAAAG | 121 | 104 | 0.998 |
| R: ACAATCCGTGCTCCACAATG | |||||
| recA | Recombinase A | F: GGGAGACACTCACGTTGGTTTAC | 121 | 86 | 0.996 |
| R: AACTTTTTCACGAATTTGGTTGATG | |||||
| sodA | Superoxide dismutase | F: TTAGGTAATAAGCGTGTTCCCATAC | 125 | 83 | 0.994 |
| R: GGCTTGGTTAGTCGTAAACAATG | |||||
| gap | Glyceraldehyde-3-phosphate | F: AGTGAACCAGATGCAAGCAAATTAC | 121 | 96 | 0.999 |
| dehydrogenase | R: TTTTAGCGCCTGCTTCAATATG | ||||
| rpoB | RNA polymerase beta chain | F: GCGAACATGCAACGTCAAG | 121 | 98 | 0.999 |
| R: GACCTCTGTGCTTAGCTGTAATAGC | |||||
| pta | Phosphate acetyltransferase | F: AAAGCGCCAGGTGCTAAATTAC | 121 | 98 | 0.994 |
| R: CTGGACCAACTGCATCATATCC | |||||
| tpi | Triose phosphate isomerase | F: TTCTGAACGTCGTGAATTATTCC | 121 | 100 | 0.996 |
| R: TTCACGCTCTTCGTCTGTTTC | |||||
| Enterotoxin genes | |||||
| sea | Enterotoxin A | F: TCAATTTATGGCTAGACGGTAAACAA | 93 | 101 | 0.994 |
| R: GAAGATCCAACTCCTGAACAGTTACA | |||||
| sed | Enterotoxin D | F: AAACGTTAAAGCCAATGAAAACA | 150 | 98 | 0.997 |
| R: TGATCTCCTGTACTTTTATTTTCTCCTA |
F, forward; R, reverse.
PCR amplification efficiency was determined from the standard curves, using the formula 10(−1/slope). For the calculations, the base of the exponential amplification function is used (e.g., 1.94 means 94% amplification efficiency).
Annealing of 100 ng of total RNA extracted from S. aureus pure culture or 500 ng of total RNA extracted from coculture of S. aureus and L. lactis with random nonamers (CyScribe cDNA postlabeling kit; GE Healthcare Amersham Biosciences) was performed for 10 min at 20°C after denaturation of RNA secondary structures (5 min at 65°C). cDNA was synthesized by a 1-h reverse transcription at 42°C using PowerScript Reverse Transcriptase and Ultra pure dNTP (Clontech-Takara Bio Europe, Saint Germain en Laye, France), followed by enzyme inactivation (15 min at 70°C).
The cDNA levels were then analyzed by quantitative PCR using an ABI Prism 7900 HT Sequence Detection System (SDS) (Applied Biosystems). Each sample was tested in duplicate in a 96-well plate (Applied Biosystems). The reaction mixture (final volume, 20 μl) consisted of 10 μl of SYBR green PCR Mastermix (Applied Biosystems), 1.2 μl of each primer (300 nM final), 2.6 μl of ultrapure H2O, and 5 μl of a 1/100 (cDNA from S. aureus pure culture) or 1/10 (cDNA from coculture of S. aureus and L. lactis) dilution of cDNA. The absence of genomic DNA in RNA samples was checked by real-time qPCR before cDNA synthesis (minus RT control).
A blank (no-template control) was incorporated in each assay. The thermocycling program consisted of one hold at 50°C for 2 min and one hold at 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Melting curve data were then collected to check PCR specificity, contamination, and the absence of primer dimers. The PCR efficiency (E) of each primer pair was determined by the dilution series method using cDNA as a template. To minimize interrun variations, a calibrator sample was used to determine and fix the fluorescence threshold. The threshold cycle (CT) values calculated by the SDS software were exported to Excel for the relative quantification analysis.
Real-time RT-qPCR data analysis.
The CT values of each sample were transformed as described in the freely distributed geNorm user manual into relative quantities (Q) with a reference (ref) sample and using the gene-specific PCR efficiency, calculated as follows: Q = E(refCt − sampleCt). Normalization was applied by dividing the relative quantities of genes of interest by the geometric mean of the relative quantities of selected reference genes (normalization factor [NF]). Standard deviation of the normalized expression was calculated as described in the geNorm user manual.
The stability of mRNA expression of the potential reference genes was evaluated by using the geNorm VBA applet for Microsoft Excel (47). This program calculates the gene expression stability measure (M) for a potential reference gene as the average pairwise variation for that gene with all other tested genes. Two ideal reference genes are expected to have an identical expression ratio in all samples, whatever the experimental conditions. Therefore, candidate reference genes are ranked on the assumption that those with the lowest M value have the most stable expression. Finally, the program determines the optimal number of genes for accurate normalization by calculating the pairwise variation (Vn/n + 1) between two normalization factors, NFn (normalization factor based on the geometric mean of the n most stable genes) and NFn + 1, such that if Vn/n + 1 is superior to 0.15, the (n + 1)th gene has a significant effect on normalization and should preferably be included for calculation of a reliable normalization factor. Authors of geNorm nevertheless recommend the minimal use of the three most stable internal control genes for calculation of the normalization factor (NF3) and stepwise inclusion of more control genes until the (n + 1)th gene has no significant contribution to the newly calculated normalization factor.
The absolute quantification of the expression of the most stably expressed gene in S. aureus during growth in milk was assessed with specific primers for S. aureus and L. lactis in an RNA sample extracted from cheese to evaluate the proportions of S. aureus and L. lactis RNAs. The amplification of target gene transcript in the sample was compared to a standard curve obtained with specific primers for both species and prepared with known concentrations of RNA extracted independently from pure culture of S. aureus or L. lactis.
Analysis of enterotoxin production in cheese.
Cheese samples (25 g) were tested for the presence of enterotoxins (A to E) 72 h after milk inoculation by using a European Union extraction method for screening, followed by dialysis concentration coupled to a Vidas SET2 (bioMérieux, Marcy l'Etoile, France) detection kit. Samples found positive for enterotoxins were further analyzed by using the European Union Community Reference Laboratory with the coagulase-positive staphylococci confirmatory method described by Hennekinne et al. (23) to determine the type of enterotoxin produced.
RESULTS
Bacteria growth in milk and cheese.
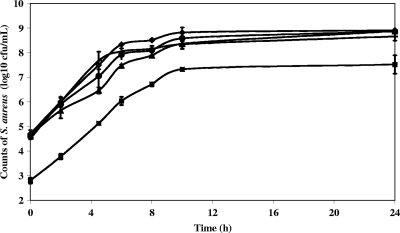
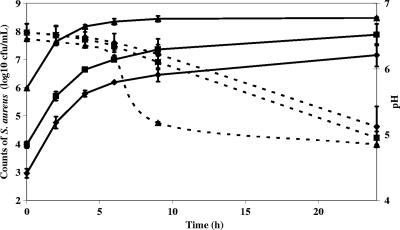
Potential reference gene transcript levels were measured during growth of S. aureus strain 361F in milk under conditions that might occur during the cheese-making process and during the first 72 h of cheese manufacture. Cell counts in milk for 24 h are reported in Fig. 1. The increase in staphylococcal cell numbers and the pH reduction in semihard cheeses manufactured with L. lactis strain SK11 during the first 24 h are reported in Fig. 2. Although pHs finally reached the same values under all conditions (5 ± 0.1), the decrease in cheeses inoculated with 106 S. aureus CFU/ml was faster than in other cheeses, suggesting that S. aureus in that concentration may facilitate the acidifying activity and growth of L. lactis (data not shown). We noticed, however, an early growth slowdown in milk cultures inoculated with 106 S. aureus CFU/ml, which may be due to faster acidification and/or to nutritional competition between L. lactis and S. aureus. Bacterial RNA was extracted, and microbiological analysis was performed on molding samples until 72 h after inoculation of the milk, using the method developed in the present study.
FIG. 1.
Growth of S. aureus strain 361F in reconstituted milk. Data are expressed as the means of two biological repetitions at 37°C with shaking (filled diamond), 30°C with shaking (filled triangle), 37°C without shaking (filled horizontal bar), 37°C with shaking and lactic acid (filled circle), and 37°C with shaking and L. lactis strain IL-416 (filled square). When interval bars are not visible they fall within plot symbols.
FIG. 2.
Growth of S. aureus strain 361F and pH evolution in cheese. Data are expressed as the means of two biological repetitions. S. aureus bacteria were inoculated at 103 CFU/ml (⧫), 104 CFU/ml (▪), and 106 CFU/ml (▴). The dashed line indicates pH, and the solid line indicates the S. aureus count. When interval bars are not visible, they fall within plot symbols.
RNA extraction from cheese.
The first step in the development of tools to quantify enterotoxin gene expression in cheese was the cheese RNA extraction strategy. After many attempts to directly extract bacterial RNA from cheeses (data not shown), we succeeded in separating the bacterial cells from the cheese matrix before RNA extraction. During the first step, casein and fat were eliminated from the cheese matrix by combining mechanical and chemical treatments known to destabilize the cheese matrix. Next, bacteria were recovered and washed. Bacterial recovery yield was around 100%. The RNA extraction yield was (6.1 ± 1.6) × 10−5 ng/cell when the total bacterial population was around 107 CFU/g of cheese and (2.9 ± 0.9) × 10−5 ng/cell when the population was greater than 107 CFU/g of cheese. Better yields [(8.1 ± 1.7) × 10−5 ng/bacterial cell] were obtained with cheese inoculated with 106 S. aureus CFU/ml of milk when the population reached more than 107 CFU/g. The average A260/A230 and A260/A280 ratios were, respectively, 1.80 ± 0.50 and 2.06 ± 0.14. The RIN of the RNA samples was around 9.5 ± 0.5, which means that they were not degraded and could be used for real-time RT-qPCR. From Vieux Pané, Roquefort, and Munster we obtained, respectively, 76 μg of RNA/g of cheese with a RIN of 8.2, 39 μg of RNA/g with a RIN of 7.6, and 190 μg of RNA/g with a RIN of 8.7.
Selection of potential internal controls for RT-qPCR experiments on S. aureus RNA.
A total of 10 housekeeping genes known to be involved in different aspects of cellular functions were evaluated for their potential as good internal controls for S. aureus gene expression experiments (Table 1). Primers were designed, and their efficiency (E) was controlled using sample standard curves, generated by serial 10-fold dilutions and calculated according to the equation E = 10−1/slope. Their specificity was checked using BLAST against NCBI databases and confirmed on RNA extracted from a complex ecosystem (data not shown).
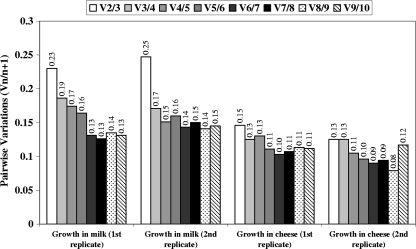
Transcription profiling from real-time RT-qPCR assays on the 82 samples of this study was carried out for the 10 candidate reference genes. Using geNorm VBA (see the Materials and Methods section) for each independent series and combination of cultures, the 10 genes were ranked according to their stability of gene expression measure (M) (Table 2), with the lowest M value indicating the greatest stability. All the genes presented M values below 1.5, which is the default limit defined by Vandesompele et al. (47) and means that they all presented rather good expression stability. ftsZ, recA, pta, and rpoB were the four most stably expressed genes in S. aureus during growth in milk under the different conditions of pH, temperature, and aeration that might occur during the cheese-making process. During growth in cheese, ftsZ, pta, and gyrB showed the greatest stability. Finally, to determine the optimal number of reference genes required for accurate normalization, geNorm provided the pairwise variation (Vn/n + 1) values between each combination of sequential normalization factors (Fig. 3). Based on criteria of Vandesompele (47), the use of the three most stably expressed genes, namely, gyrB, ftsZ, and pta, as internal controls for samples extracted from cheese would suffice for reliable data normalization.
TABLE 2.
Internal control gene stability value M for all potential reference genes
| Growth condition and replicate no. |
M for the indicated genea |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| gyrB | ftsZ | hu | rplD | recA | sodA | gap | rpoB | pta | tpi | |
| In milk | ||||||||||
| Replicate 1 | 0.923 | 0.481 | 1.174 | 0.834 | 0.642 | 1.094 | 1.020 | 0.734 | 0.481 | 0.972 |
| Replicate 2 | 0.814 | 0.595 | 1.242 | 0.904 | 0.595 | 1.063 | 1.144 | 0.766 | 0.729 | 0.972 |
| Both replicatesb | 0.859 | 0.613 | 1.213 | 0.930 | 0.721 | 1.128 | 1.058 | 0.779 | 0.613 | 0.984 |
| In cheese | ||||||||||
| Replicate 1 | 0.370 | 0.370 | 0.636 | 0.438 | 0.920 | 0.842 | 0.758 | 0.693 | 0.497 | 0.581 |
| Replicate 2 | 0.309 | 0.511 | 0.459 | 0.664 | 0.698 | 0.851 | 0.607 | 0.373 | 0.309 | 0.561 |
| Both replicatesb | 0.392 | 0.464 | 0.609 | 0.681 | 0.790 | 0.876 | 0.730 | 0.556 | 0.392 | 0.647 |
A low M value corresponds to high expression stability of the particular control gene.
Analysis performed on both replicates.
FIG. 3.
Pairwise variations (Vn/n + 1) for all the samples during growth in milk and in cheese (two replicates). Pairwise variations between every combination of sequential normalization factors were calculated to determine the minimum number of housekeeping genes required for accurate normalization in the different samples. The cutoff value, below which the inclusion of an additional reference gene does not result in a significant improvement of normalization, was set at 0.15.
Proportion of staphylococcal RNA in total RNA.
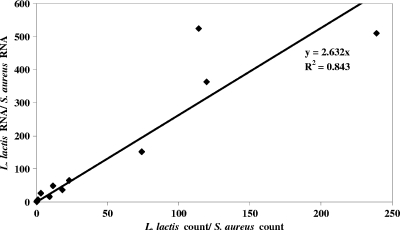
We studied the expression of one of the candidate reference genes, recA (among the four most stably expressed genes during S. aureus growth in milk) in S. aureus and L. lactis RNAs extracted from cheese samples to check that proportions of each species are the same in RNA samples and microbiological counts. Starting from equal amounts of RNA as the reaction template, standard curves for each species-specific primer were plotted. The efficiency of L. lactis primers was 91% (r2 = 0.997), and that of S. aureus primers was 89% (r2 = 0.999). Using standard curves made it possible to calculate recA expression levels in each species and thus to estimate the ratio of L. lactis to S. aureus RNA in samples extracted from cheese by real-time RT-qPCR. The ratio based on the recA standard was 2.6 times greater than what was determined by microbiological counts in the first 9 h of manufacture (Fig. 4). After that time, this ratio continued to increase. It is therefore of major concern to use good reference genes to normalize this kind of variation.
FIG. 4.
Correlation between L. lactis and S. aureus microbiological counts and RNA proportions in samples extracted from cheese during the first 9 h of manufacture.
sea and sed expression in S. aureus during cheese manufacture.
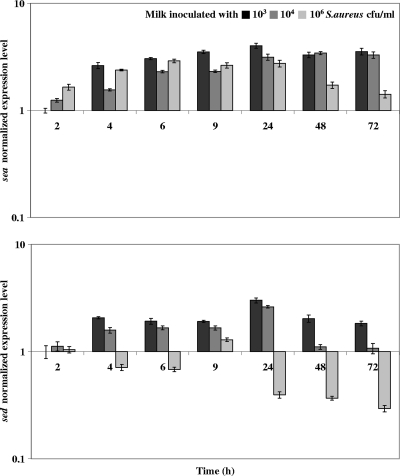
We examined sea and sed RNA and protein levels during cheese manufacture. Normalized expression levels of sea and sed genes in S. aureus strain 361F during the first 72 h of cheese manufacture are reported in Fig. 5. Regardless of the inoculation level of S. aureus in milk or bacterial growth phase, sea expression levels remained essentially unchanged, while sed expression was statistically lower in cheese inoculated with 106 S. aureus CFU/ml of milk than in cheeses inoculated with 103 (P < 0.001) or 104 (P < 0.05) S. aureus CFU/ml.
FIG. 5.
sea and sed expression levels in cheese. Data are expressed as the means of two biological repetitions. Gene expression levels were quantified using real-time RT-qPCR. Normalization was provided using the geometric mean expression levels of three reference genes (ftsZ, pta, and gyrB). The expression level at 2 h after inoculation of milk with 103 S. aureus CFU/ml was used as the calibrator. Vertical bars indicate standard deviations.
Relative to the staphylococcal counts in cheese, SEA and SED production rates are lower in cheese inoculated with 106 S. aureus CFU/ml of milk than in cheeses inoculated with 103 or 104 S. aureus CFU/ml (Table 3). In the case of SED, we attribute this difference to lower expression of the sed gene in cheese in the higher-density inoculation. However, low-level production of SEA does not seem to be explained at the sea-transcriptional level.
TABLE 3.
SEA and SED production in cheese
| S. aureus inoculation level (CFU/ml of milk) | Amt of protein produced (ng/g of cheese)a |
|
|---|---|---|
| SEA | SED | |
| 103 | 0.065 | 0.26 |
| 104 | 0.325 | 1.425 |
| 106 | 0.445 | 1.235 |
Data are the means of two biological repetitions.
DISCUSSION
The RNA extraction method we developed was applied to semihard cheeses and soft cheeses during the first 3 days of cheese manufacture and after ripening. The RNA extraction method we developed gave the necessary quality for transcriptional analysis studies by real-time RT-qPCR. As explained by Imbeaud et al. (25), an A260/280 ratio above 1.8 indicates limited protein contamination, whereas the A260/230 ratio below 1.8 in some samples is indicative of the presence of residual organic compounds. Studies by Imbeaud et al. demonstrated that samples presenting organic compound contamination did not cause inhibition in downstream applications such as cDNA synthesis (25). In this study, RNA samples were of high quality, as reflected by high RIN values (above 9 when extractions were during the first 3 days of cheese manufacture and above 7 for commercial cheeses). Moreover, the RNA extraction method provided a sufficient amount of good-quality RNA for transcriptional analysis at all stages of the cheese-making process, from the molding until the end of ripening, even when the bacterial population was below 107 CFU/g of cheese. We checked that the RNA level of the minor species (S. aureus) in RNA extracted from cheese corresponded to levels measured by microbiological analysis of the cheese. The proportions of staphylococci to lactococci by RNA comparisons and by microbiological counts were comparable for the first 9 h of cheese manufacture and then differed. It is thus important to use a normalization strategy for transcript quantification. Compared to previously reported cheese RNA extraction strategies (4, 29, 33, 45), our method presents the advantage of being efficient at any stage of cheese manufacture, and cell pellet preparation can be performed quickly together with microbiological analysis of the cheese sample. The procedure also allows a larger amount of cheese to be processed if greater amounts of RNA are needed.
The RT-qPCR protocol we present contains the necessary information to repeat the experiments and was validated by checking primer specificity and efficiency. The limit of detection was not calculated, but we always worked within the range of standard curves developed to determine primer PCR efficiency. We selected and used multiple reference genes to achieve an accurate and reliable normalization of RT-qPCR data (47). We thus fulfilled criteria recently reported by Bustin et al. in the MIQE (minimum information for publication of quantitative real-time PCR experiments) guidelines (6).
We investigated the expression stability of 10 potential reference genes under different environmental conditions. While many studies used the amount of 16S rRNA as an internal control (28, 48, 52), this was not our choice, as the rRNA molecules are not representative of the mRNA fraction. Among the 10 candidates, according to geNorm analysis of expression (47), ftsZ, recA, pta, and rpoB were the four most stably expressed genes in S. aureus during growth in milk under different conditions, and ftsZ, recA, and rpoB were found to be stably expressed in BHI broth (13). However, during growth in cheese, transcriptional stability of the reference genes differed substantially, indicating that expression has to be validated under each experimental condition (6). Each of the three genes chosen to normalize our RT-qPCR data during S. aureus growth in cheese (ftsZ, pta, and gyrB) had already been used separately in previous studies (37, 41, 50). We used the geometric mean of expression results using all three genes to normalize expression of our gene of interest (47).
The tools we developed allowed us to follow enterotoxin A and D gene expression levels during cheese manufacture for the first time. A reliable measure of their expression was possible when the ratio of S. aureus to L. lactis starter culture was below 10−3, i.e., with at least 103 S. aureus per ml of milk at the beginning of cheese manufacture. At a higher ratio of S. aureus to L. lactis, the CT values obtained with RT-qPCR were too high and resulted in unusable data for evaluation of transcript abundance (data not shown). SE gene expression has already been explored in tryptic soy or BHI broth to compare the expression levels of these genes in different strains (13, 28). Studies of S. aureus 361F in BHI broth (13) showed that, as in the cheese matrix, the S. aureus growth phase has no significant impact on the sea expression pattern. During growth in BHI broth, the sed expression pattern presented a modest postexponential induction ratio (<10-fold) (13) that we did not observe during growth in cheese. SED production is maximal during the transition from the exponential to the stationary phases of growth, a consequence of its regulation by the staphylococcal two-component regulatory element known as the agr (accessory gene regulator) system that responds to a quorum-sensing octapeptide (3, 26, 53). In a model of continuous culture technique, Wright et al. showed that the minimum cell density predicted to promote specific agr expression was 1.2 × 108 CFU/ml (51). In cheeses the staphylococcal population rarely reaches this value, and this threshold may vary when we work under different conditions. This situation can thus explain why we did not see any induction, compared to results in BHI medium, when the population reached 4 × 108 to 2 × 109 CFU/ml (13).
When S. aureus was inoculated at 106 CFU/ml, the population reached the threshold of 108 CFU/g of cheese, and sed expression was not induced. An effect of the starter culture on staphylococcal gene expression might be hypothesized, as suggested by a recent study reporting that L. lactis was able to dramatically impair S. aureus expression of several virulence genes (16). Under mixed cultures performed in a chemically defined medium held at a constant pH, the induction of the agr system, as well as of the agr-regulated enterotoxin gene sec, was strongly impaired (38), while sea, which is not under the control of the agr system, was not affected (16). The reason for agr downregulation resulting from the interaction between the two species (S. aureus and L. lactis inoculated at the same level, i.e., 106 CFU/ml) was not clearly elucidated (16), but this observation might partly explain the curiously low sed expression level monitored in cheeses inoculated with 106 S. aureus CFU/ml of milk compared to those inoculated with smaller amounts. Our data suggest that the interaction between the two species in cheese could be more important at equal inoculation ratios (106 CFU/ml) than under other conditions. Growth of both species was indeed faster, and the acidifying activity was higher under these conditions (Fig. 2). Whether such interactions with L. lactis could have an impact on agr-regulated sed gene expression in cheese remains to be confirmed.
Results of this study make it possible to correlate enterotoxin D gene expression and production for the first time. This correlation should be further investigated by following enterotoxin production together with gene expression in cheeses to determine whether enterotoxin D regulation occurs at the transcriptional or posttranscriptional level. A correlation between enterotoxin A production and gene expression was not observed in this study, suggesting that enterotoxin A regulation may occur at the posttranscriptional level.
In conclusion, this study highlights the importance of investigating and characterizing enterotoxin gene regulation. Understanding the dynamics of SE gene expression in cheese could help cheese manufacturers to anticipate the conditions under which SE production is activated during the cheese-making process. Moreover, this work presents an easily transferable set of tools to study gene expression of minor species in a complex ecosystem, providing RNA extraction methods and a means to accurately calculate expression of the gene of interest. The approach can be applied to other microorganisms by designing specific primers and, in particular, to other complex ecosystems containing S. aureus. We stress the importance of validating reference gene expression stability under the specific test conditions.
Acknowledgments
We thank the PICT platform at INRA Jouy-en-Josas for the use of the ABI Prism SDS 7900 HT and the 2100 BioAnalyzer Agilent. We are grateful to M. L. De Buyser for providing strain 361F, P. Quénée for L. lactis strains IL-416 and SK11, J. A. Hennekinne and I. Papinaud for enterotoxin quantification, and D. White and A. Gruss for reading the manuscript.
This work was carried out as part of the SEFRO project, with financial support from the Agence Nationale de la Recherche (the French National Research Agency) under the Programme National de Recherche en Alimentation et Nutrition Humaine, project ANR06-PNRA029. The P2-level experimental cheese plant acquisition was supported by Ile-de-France.
Footnotes
Published ahead of print on 8 January 2010.
REFERENCES
- 1.Aoyama, K., C. Takahashi, Y. Yamauchi, F. Sakai, H. Igarashi, S. Yanahira, and H. Konishi. 2008. Examination of Staphylococcus aureus survival and growth during cheese-making process. Shokuhin Eiseigaku Zasshi. 49:116-123. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 2.Balaban, N., and A. Rasooly. 2000. Staphylococcal enterotoxins. Int. J. Food Microbiol. 61:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Bayles, K. W., and J. J. Iandolo. 1989. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J. Bacteriol. 171:4799-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonaïti, C., S. Parayre, and F. Irlinger. 2006. Novel extraction strategy of ribosomal RNA and genomic DNA from cheese for PCR-based investigations. Int. J. Food Microbiol. 107:171-179. [DOI] [PubMed] [Google Scholar]
- 5.Bustin, S. A. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29:23-39. [DOI] [PubMed] [Google Scholar]
- 6.Bustin, S. A., V. Benes, J. A. Garson, J. Hellemans, J. Huggett, M. Kubista, R. Mueller, T. Nolan, M. W. Pfaffl, G. L. Shipley, J. Vandesompele, and C. T. Wittwer. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611-622. [DOI] [PubMed] [Google Scholar]
- 7.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 8.Cords, B. R., and S. R. Tatini. 1973. Applicability of heat-stable deoxyribonuclease assay for assessment of staphylococcal growth and the likely presence of enterotoxin in cheese. J. Dairy Sci. 56:1512-1519. [DOI] [PubMed] [Google Scholar]
- 9.De Buyser, M.-L., B. Dufour, M. Maire, and V. Lafarge. 2001. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int. J. Food Microbiol. 67:1-17. [DOI] [PubMed] [Google Scholar]
- 10.De Buyser, M. L., F. Janin, and F. Dilasser. 1985. Contamination of ewe cheese with Staphylococcus aureus: study of an outbreak of food poisoning. Zentralbl. Bakteriol. Mikrobiol. Hyg. 1(Suppl. 14):677-678. [Google Scholar]
- 11.Delbes, C., J. Alomar, N. Chougui, J. F. Martin, and M. C. Montel. 2006. Staphylococcus aureus growth and enterotoxin production during the manufacture of uncooked, semihard cheese from cows’ raw milk. J. Food Prot. 69:2161-2167. [DOI] [PubMed] [Google Scholar]
- 12.Delmas, G., A. Gallay, E. Espié, S. Haeghebaert, N. Pihier, F. X. Weill, H. De Valk, V. Vaillant, and J. C. Désenclos. 2006. Les toxi-infections alimentaires collectives en France entre 1996 et 2005. BEH 51-52:418-422. [Google Scholar]
- 13.Derzelle, S., F. Dilasser, M. Duquenne, and V. Deperrois. 2009. Differential temporal expression of the staphylococcal enterotoxins genes during cell growth. Food Microbiol. 26:896-904. [DOI] [PubMed] [Google Scholar]
- 14.Dheda, K., J. F. Huggett, S. A. Bustin, M. A. Johnson, G. Rook, and A. Zumla. 2004. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37:112-114, 116,118-119. [DOI] [PubMed] [Google Scholar]
- 15.Dinges, M. M., P. M. Orwin, and P. M. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Even, S., C. Charlier, S. Nouaille, N. L. Ben Zakour, M. Cretenet, F. J. Cousin, M. Gautier, M. Cocaign-Bousquet, P. Loubiere, and Y. Le Loir. 2009. Staphylococcus aureus virulence expression is impaired by Lactococcus lactis in mixed cultures. Appl. Environ. Microbiol. 75:4459-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 18.Gaya, P., M. Medina, L. Bautista, and M. Nunez. 1988. Influence of lactic starter inoculation, curd heating and ripening temperature on Staphylococcus aureus behaviour in Manchego cheese. Int. J. Food Microbiol. 6:249-257. [DOI] [PubMed] [Google Scholar]
- 19.Goerke, C., M. G. Bayer, and C. Wolz. 2001. Quantification of bacterial transcripts during infection using competitive reverse transcription-PCR (RT-PCR) and LightCycler RT-PCR. Clin. Diagn. Lab Immunol. 8:279-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez-Lucia, E., J. Goyache, J. A. Orden, A. Domenech, F. Javier Hernandez, J. A. Ruiz-Santa Quiteria, B. Lopez, J. L. Blanco, and G. Suarez. 1992. Growth of Staphylococcus aureus and synthesis of enterotoxin during ripening of experimental Manchego-type cheese. J. Dairy Sci. 75:19-26. [DOI] [PubMed] [Google Scholar]
- 22.Hamama, A., N. El Hankouri, and M. El Ayadi. 2002. Fate of enterotoxigenic Staphylococcus aureus in the presence of nisin-producing Lactococcus lactis strain during manufacture of Jben, a Moroccan traditional fresh cheese. Int. Dairy J. 12:933-938. [Google Scholar]
- 23.Hennekinne, J. A., F. Guillier, S. Perelle, M. L. De Buyser, S. Dragacci, S. Krys, and B. Lombard. 2007. Intralaboratory validation according to the EN ISO 16 140 standard of the Vidas SET2 detection kit for use in official controls of staphylococcal enterotoxins in milk products. J. Appl. Microbiol. 102:1261-1272. [DOI] [PubMed] [Google Scholar]
- 24.Hiron, A., E. Borezee-Durant, J. C. Piard, and V. Juillard. 2007. Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 189:5119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imbeaud, S., E. Graudens, V. Boulanger, X. Barlet, P. Zaborski, E. Eveno, O. Mueller, A. Schroeder, and C. Auffray. 2005. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 33:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. U. S. A. 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kibbe, W. A. 2007. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 35:W43-W46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, Y. D., B. Y. Moon, J. H. Park, H. I. Chang, and W. J. Kim. 2007. Expression of enterotoxin genes in Staphylococcus aureus isolates based on mRNA analysis. J. Microbiol. Biotechnol. 17:461-467. [PubMed] [Google Scholar]
- 29.Makhzami, S., P. Quénée, E. Akary, C. Bach, M. Aigle, A. Delacroix-Buchet, J. C. Ogier, and P. Serror. 2008. In situ gene expression in cheese matrices: application to a set of enterococcal genes. J. Microbiol. Methods 75:485-490. [DOI] [PubMed] [Google Scholar]
- 30.Manchester, K. L. 1996. Use of UV methods for measurement of protein and nucleic acid concentrations. Biotechniques 20:968-970. [DOI] [PubMed] [Google Scholar]
- 31.Meyrand, A., S. Boutrand-Loei, S. Ray-Gueniot, C. Mazuy, C. E. Gaspard, G. Jaubert, G. Perrin, C. Lapeyre, and C. Vernozy-Rozand. 1998. Growth and enterotoxin production of Staphylococcus aureus during the manufacture and ripening of Camembert-type cheeses from raw goats’ milk. J. Appl. Microbiol. 85:537-544. [DOI] [PubMed] [Google Scholar]
- 32.Meyrand, A., and C. Vernozy-Rozand. 1999. Croissance et entérotoxinogenèse de staphylococcus aureus dans différents fromages. Rev. Med. Vet. 150:601-616. [Google Scholar]
- 33.Monnet, C., V. Ulve, A. S. Sarthou, and F. Irlinger. 2008. Extraction of RNA from cheese without prior separation of microbial cells. Appl. Environ. Microbiol. 74:5724-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nunez, M., L. Bautista, M. Medina, and P. Gaya. 1988. Staphylococcus aureus, thermostable nuclease and staphylococcal enterotoxins in raw ewes’ milk Manchego cheese. J. Appl. Bacteriol. 65:29-34. [DOI] [PubMed] [Google Scholar]
- 35.Otero, A., M. C. Garcia, M. L. Garcia, M. Prieto, and B. Moreno. 1988. Behaviour of Staphylococcus aureus strains, producers of enterotoxins C1 or C2, during the manufacture and storage of Burgos cheese. J. Appl. Bacteriol. 64:117-122. [DOI] [PubMed] [Google Scholar]
- 36.Otero, A., M. C. García, M. L. García, J. A. Santos, and B. Moreno. 1993. Behaviour of Staphylococcus aureus strains FRI 137 and FRI 361 during the manufacture and ripening of Manchego cheese. Int. Dairy J. 3:85-96. [Google Scholar]
- 37.Pereira, S. F., A. O. Henriques, M. G. Pinho, H. de Lencastre, and A. Tomasz. 2007. Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 189:3525-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Regassa, L. B., J. L. Couch, and M. J. Betley. 1991. Steady-state staphylococcal enterotoxin type C mRNA is affected by a product of the accessory gene regulator (agr) and by glucose. Infect. Immun. 59:955-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt, K. A., A. C. Manna, and A. L. Cheung. 2003. SarT influences sarS expression in Staphylococcus aureus. Infect. Immun. 71:5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroeder, A., O. Mueller, S. Stocker, R. Salowsky, M. Leiber, M. Gassmann, S. Lightfoot, W. Menzel, M. Granzow, and T. Ragg. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwan, W. R., L. Lehmann, and J. McCormick. 2006. Transcriptional activation of the Staphylococcus aureus putP gene by low-proline-high osmotic conditions and during infection of murine and human tissues. Infect. Immun. 74:399-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tatini, S. R., J. J. Jezeski, H. A. Morris, J. C. Olson, Jr., and E. P. Casman. 1971. Production of staphylococcal enterotoxin A in cheddar and Colby cheese. J. Dairy Sci. 54:815-825. [DOI] [PubMed] [Google Scholar]
- 43.Tatini, S. R., W. D. Wesala, J. J. Jezeski, and H. A. Morris. 1973. Production of staphylococcal enterotoxin A in blue, brick, mozzarella, and Swiss cheeses. J. Dairy Sci. 56:429-435. [DOI] [PubMed] [Google Scholar]
- 44.Thellin, O., W. Zorzi, B. Lakaye, B. De Borman, B. Coumans, G. Hennen, T. Grisar, A. Igout, and E. Heinen. 1999. Housekeeping genes as internal standards: use and limits. J. Biotechnol. 75:291-295. [DOI] [PubMed] [Google Scholar]
- 45.Ulve, V. M., C. Monnet, F. Valence, J. Fauquant, H. Falentin, and S. Lortal. 2008. RNA extraction from cheese for analysis of in situ gene expression of Lactococcus lactis. J. Appl. Microbiol. 105:1327-1333. [DOI] [PubMed] [Google Scholar]
- 46.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 47.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. http://genomebiology.com/2002/3/7/RESEARCH/0034. [DOI] [PMC free article] [PubMed]
- 48.Vaudaux, P., P. Francois, C. Bisognano, W. L. Kelley, D. P. Lew, J. Schrenzel, R. A. Proctor, P. J. McNamara, G. Peters, and C. Von Eiff. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect. Immun. 70:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vernozy-Rozand, C., A. Meyrand, C. Mazuy, M. L. Delignette-Muller, G. Jaubert, G. Perrin, C. Lapeyre, and Y. Richard. 1998. Behaviour and enterotoxin production by Staphylococcus aureus during the manufacture and ripening of raw goats’ milk lactic cheeses. J. Dairy Res. 65:273-281. [DOI] [PubMed] [Google Scholar]
- 50.Wolz, C., C. Goerke, R. Landmann, W. Zimmerli, and U. Fluckiger. 2002. Transcription of clumping factor A in attached and unattached Staphylococcus aureus in vitro and during device-related infection. Infect. Immun. 70:2758-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright, J. D., and K. T. Holland. 2003. The effect of cell density and specific growth rate on accessory gene regulator and toxic shock syndrome toxin-1 gene expression in Staphylococcus aureus. FEMS Microbiol. Lett. 218:377-383. [DOI] [PubMed] [Google Scholar]
- 52.Yarwood, J. M., J. K. McCormick, M. L. Paustian, V. Kapur, and P. M. Schlievert. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, S., and G. C. Stewart. 2000. Characterization of the promoter elements for the staphylococcal enterotoxin D gene. J. Bacteriol. 182:2321-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]