Abstract
The succinic acid producer Mannheimia succiniciproducens can efficiently utilize sucrose as a carbon source, but its metabolism has not been understood. This study revealed that M. succiniciproducens uses a sucrose phosphotransferase system (PTS), sucrose 6-phosphate hydrolase, and a fructose PTS for the transport and utilization of sucrose.
Mannheimia succiniciproducens MBEL55E, a capnophilic (CO2 loving) Gram-negative facultative anaerobic rumen bacterium, efficiently produces succinic acid from a wide range of carbon sources, including pentose sugar (xylose), hexose sugars (fructose and glucose), and disaccharides (lactose, maltose, and sucrose) (3, 9). Sucrose is inexpensive, readily available, and abundant (7, 15), making it an attractive raw material for cost-effective bio-based production of succinic acid. Although M. succiniciproducens utilizes sucrose relatively faster than other carbon sources, the sucrose metabolism in M. succiniciproducens is not well understood. Here, we report the characteristics of sucrose transport and metabolism in M. succiniciproducens.
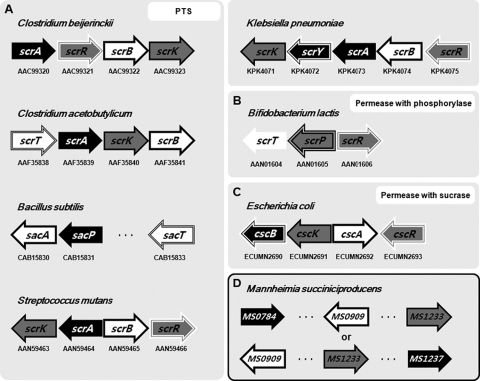
Sucrose metabolism including transport and utilization in bacteria can be categorized into three types (Fig. 1A to C and Fig. 2A). The phosphotransferase system (PTS) includes the phosphoenolpyruvate (PEP)-dependent sucrose-specific PTS, sucrose 6-phosphate hydrolase, and fructokinase, and is found in both Gram-positive and -negative bacteria. A non-PTS system includes sucrose permease and sucrose phosphorylase. Another non-PTS system involves sucrose permease and sucrase (sucrose hydrolase or invertase) (15, 16). The genes encoding each enzyme system are often clustered (Fig. 1A to C).
FIG. 1.
Genetic organization of different sucrose utilization systems in bacteria and a proposed sucrose utilization system in M. succiniciproducens. Arrows indicate genes, and the numbers below the arrows are the locus number or protein number in the NCBI (http://www.ncbi.nlm.nih.gov/) and KEGG (http://www.genome.jp/kegg/) databases. The successive numbers of the sucrose utilization genes indicate that they are clustered and often are cotranscribed. The serial three dots between the arrows imply that gene cluster for the sucrose utilization is separated by other genes. (A) Several different PTSs in Gram-positive and -negative bacteria. Clostridium beijerinckii contains scrA for PTS, scrR for the LacI family sucrose regulator, scrB for sucrose 6-phosphate hydrolase, and scrK for fructokinase. Clostridium acetobutylicum contains scrT for the transcriptional regulator, scrA, scrK, and scrB. Bacillus subtilis contains sacA, sucrose 6-phosphate hydrolase, sacP for PTS, and sacT for the transcriptional regulator. Streptococcus mutans contains scrK, scrA, scrB, and scrR. Klebsiella pneumoniae contains scrK, scrY for sucrose-specific outer membrane porin, scrA, scrB, and scrR. (B) Permease with phosphorylase system in Bifidobacterium lactis, which contains scrT for sucrose-specific permease, scrP for sucrose phosphorylase, and scrR. (C) Permease with sucrase system in Escherichia coli UMN026, which contains cscB for sucrose permease, cscK for fructokinase, cscA for sucrose 6-phosphate hydrolase, and cscR for the LacI family sucrose regulator. (D) Deduced sucrose utilization system in M. succiniciproducens. MS0784 was found to encode the sucrose PTS, with MS0909 coding for sucrose 6-phosphate hydrolase, while fructokinase is not present (see text).
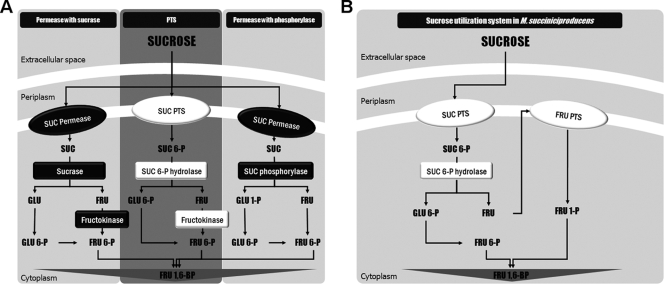
FIG. 2.
Schematic diagram of sucrose utilization systems. (A) Three kinds of sucrose utilization systems in bacteria. From the left are shown a permease with sucrase system, a sucrose PTS, and a permease with phosphorylase system. (B) Sucrose utilization system in M. succiniciproducens identified in this study. SUC, sucrose; FRU, fructose; GLU, glucose; P, phosphate; BP, bisphosphate.
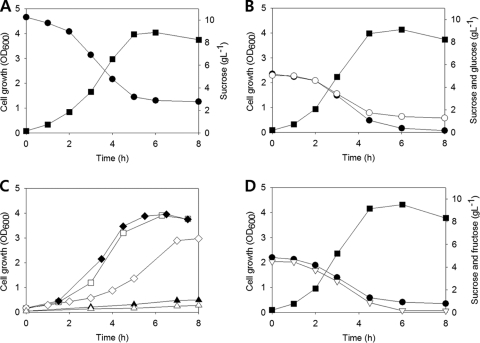
M. succiniciproducens was able to grow on sucrose very efficiently with a relatively high sucrose uptake rate of 11.18 ± 0.31 mmol g (dry cell weight)−1 h−1 (Fig. 3A). In order to identify the proteins responsible for the uptake and utilization of sucrose in M. succiniciproducens, the most probable proteins for sucrose uptake and utilization were first deduced by using protein BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to compare the sequences of proteins in M. succiniciproducens with those of other bacteria capable of utilizing sucrose (see Table S1 in the supplemental material). A list of candidate genes involved in sucrose metabolism was obtained (Table 1). Then these genes were individually deleted from the chromosome, creating strains that were examined for the utilization of sucrose in M. succiniciproducens. The genes, MS0784, MS0807, MS0909, MS1233, and MS1237, were knocked out from the chromosome of the wild-type M. succiniciproducens MBEL55E strain as previously described (5, 6), and the resulting mutants were named MBEL55EΔ0784, MBEL55EΔ0807, MBEL55EΔ0909, MBEL55EΔ1233, and MBEL55EΔ1237, respectively. The deletion of a sucrose permease gene candidate (MS0807) in M. succiniciproducens did not affect cell growth on sucrose. Also, BLAST searches suggested that M. succiniciproducens does not possess sucrose phosphorylase. These observations suggest that M. succiniciproducens most likely uses PTS rather than a permease system for sucrose uptake. Thus, we initially assumed that M. succiniciproducens sucrose utilization system involves MS0784 or MS1237 for sucrose PTS, MS0909 for sucrose 6-phosphate hydrolase, and MS1233 for fructokinase (Fig. 1D).
FIG. 3.
Growth profiles of the wild-type M. succiniciproducens strain and its mutant strains in MH5S medium. (A) Cell growth (▪) and sucrose consumption (•) profiles of M. succiniciproducens MBEL55E cultured in MH5S medium. OD600, optical density at 600 nm. (B) Nondiauxic growth of M. succiniciproducens MBEL55E (▪) in the presence of sucrose (•) and glucose (○). In this experiment, 5 g liter−1 sucrose and 5 g liter−1 glucose were used instead of 10 g liter−1 sucrose in MH5S medium. (C) Cell growth profiles of MBEL55EΔ0784 (▴), MBEL55EΔ1237 (□), MBEL55EΔ0909 (▵), MBEL55EΔ1233 (⧫), and MBEL55EΔ2178 (⋄) in MH5S medium. (D) Nondiauxic growth of M. succiniciproducens MBEL55E (▪) in the presence of sucrose (•) and fructose (▿). In this experiment, 5 g liter−1 sucrose and 5 g liter−1 fructose were used instead of 10 g liter−1 sucrose in MH5S medium.
TABLE 1.
Predicted sucrose utilization-associated enzymes, phenotypes of each mutant, and specific enzyme activities
| Related enzyme in sucrose utilization systems | Corresponding gene in M. succiniciproducens | Deletion mutant (sucrose utilization phenotype)a | Enzyme sp act (mU mg of protein−1)b |
||||
|---|---|---|---|---|---|---|---|
| Wild type |
Mutant |
||||||
| MH5S | MH5G | BHI | MH5S | BHI | |||
| Sucrose PTS | MS0784c | MBEL55EΔ0784 (Scr−) | 3.70 ± 0.15d | 1.06 ± 0.10d | 1.40 ± 0.08d | ND | 0.10 ± 0.0d |
| MS1237c | MBEL55EΔ1237 (Scr+) | ||||||
| Sucrose 6-phosphate hydrolase | MS0909c | MBEL55EΔ0909 (Scr−) | 18.3 ± 3.2 | 17.4 ± 0.1 | 20.4 ± 0.6 | ND | 1.7 ± 0.3 |
| Fructokinase | MS1233c | MBEL55EΔ1233 (Scr+) | 0.032 ± 0.002 | ND | ND | 0.054 ± 0.006 | ND |
| Fructose PTS | MS2178 | MBEL55EΔ2178 (Scr+) | 119.3 ± 2.9 | ND | 38.8 ± 1.4 | 80.9 ± 1.1 | 22.9 ± 1.07 |
| MS0784 | MBEL55EΔ0784 (Scr−) | ND | ND | 38.8 ± 1.4 | 29.1 ± 1.23 | ||
Scr+ and Scr−, respectively, indicate that each mutant grows or rarely grows (μ ≤ 0.15 h−1) in MH5S medium.
BHI is a rich complex medium containing 6 g liter−1 brain and heart (infusion form), 6 g liter−1 peptic digest of animal tissue, 5 g liter−1 sodium chloride, 3 g liter−1 glucose, 14.5 g liter−1 pancreatic digest of gelatin, and 2.5 g liter−1 disodium phosphate. BHI is used for cultivating mutant strains such as MBEL55EΔ0784 and MBEL55EΔ0909, which rarely grow in MH5S medium. ND, not determined.
Initially predicted by BLAST searches with enzymes listed in Table S1 in the supplemental material. MS0807 for sucrose permease was also predicted by the homology searches, and the explanation for this can be found in the text.
Sucrose PTS activity was measured in cpm and converted to mU/mg.
It has been shown that the genes encoding proteins responsible for sucrose uptake and utilization exist as an operon or a gene cluster in most bacteria, which is indicated by sequential accession or locus numbers as listed in Fig. 1A to C and Table S1. However, in M. succiniciproducens, the genes responsible for sucrose metabolism were not clustered together in its genome and a distinct sucrose-specific repressor was not found (Fig. 1D and Table S1). The absence of a sucrose repressor might be one of the reasons why sucrose is simultaneously metabolized with glucose without showing glucose-sucrose diauxic growth in M. succiniciproducens (Fig. 3B).
The deletion of MS0784 or MS1237 (candidate genes for sucrose PTS) in M. succiniciproducens MBEL55E was performed next. Cells were grown in MH5S medium (containing per liter 2.5 g of yeast extract, 2.5 g of polypeptone, 1 g of NaCl, 8.7 g of K2HPO4, 10 g of NaHCO3, 0.02 g of CaCl2·2H2O, and 0.2 g of MgCl2·6H2O) supplemented with 10 g liter−1 sucrose. Compared to the wild-type strain (μ of 0.72 h−1), the MBEL55EΔ1237 strain grew normally (μ of 0.68 h−1), but the MBEL55EΔ0784 strain did not grow well (μ of 0.15 h−1) (Fig. 3A and C); the slow growth was possibly due to the presence of polypeptone and yeast extract. This result indicates that MS0784 encodes the sucrose PTS (Fig. 3C). Deletion of the candidate gene for sucrose 6-phosphate hydrolase, MS0909, also slowed growth (μ of 0.13 h−1) (Fig. 3C).
In order to examine whether the gene products of MS0784 and MS0909 truly encode the sucrose PTS and sucrose 6-phosphate hydrolase, respectively, enzyme assays were performed for MBEL55EΔ0784 and MBEL55EΔ0909 and the results were compared with those of the wild-type strain (Table 1). As two deletion mutants, MBEL55EΔ0784 and MBEL55EΔ0909, poorly grew in MH5S medium, we used the cells cultured in a rich complex medium, BHI (Bacto brain heart infusion; Becton Dickinson and Company, Sparks, MD), for a better comparison of enzyme activities. The activity of sucrose PTS was measured using radioactive [14C]sucrose. Permeabilized cells prepared by adding toluene were suspended and incubated in the assay buffer with or without PEP and were filtered through DEAE-cellulose DE81 filter paper (Whatman International, Ltd., Maidstone, England) (4, 10). Its radioactivity was counted by using an LS 6500 scintillation counter (Beckman Coulter, Fullerton, CA). The PTS activity was calculated as the difference in the radioactivity between mixtures containing PEP and those lacking it. One unit of enzyme activity was defined as the amount of enzyme necessary to catalyze the conversion of 1 μmol of substrate per minute into specific products. The Km value and Vmax of the sucrose PTS were 55 μM and 17.64 mU mg of protein−1, respectively. The specific PTS enzyme activity was 0.10 ± 0.01 mU mg of protein−1 in the MBEL55EΔ0784 strain cultured in BHI medium. On the other hand, the activities were 3.70 ± 0.15 and 1.40 ± 0.08 mU mg of protein−1 in the wild-type strain cultured in MH5S medium and BHI medium, respectively (Table 1). Thus, it is confirmed that MS0784 encodes a PEP-dependent sucrose PTS and the growth of M. succiniciproducens on sucrose is much retarded without this gene and its activity.
The sucrose 6-phosphate hydrolase activity was measured by following the formation of glucose 6-phosphate and subsequent NADP-linked glucose 6-phosphate dehydrogenase activity (11). The Km value and Vmax of sucrose 6-phosphate hydrolase were 13.6 mM and 33.52 mU mg of protein−1, respectively. The specific sucrose 6-phosphate hydrolase activity was 1.7 ± 0.3 mU mg of protein−1 in the MBEL55EΔ0909 strain cultured in BHI medium, while the activities were 18.3 ± 3.2 and 20.4 ± 0.6 mU mg of protein−1 in the wild-type strain cultured in MH5S medium and BHI medium, respectively (Table 1). Thus, MS0909 encodes a sucrose 6-phosphate hydrolase, and M. succiniciproducens uses this protein as a major enzyme for the hydrolysis of sucrose 6-phosphate. Furthermore, negligible cell growth and the presence of little enzyme activity observed in the mutants MBEL55EΔ0784 and MBEL55EΔ0909 suggest that MS0784 and MS0909 are the major genes encoding the sucrose PTS and sucrose 6-phosphate hydrolase, respectively, in M. succiniciproducens. To examine whether both enzymes are inducible by sucrose, the enzyme activities were measured using cells cultured in the MH5G medium, which is MH5S medium that contains glucose instead of sucrose (Table 1). As shown in Table 1, the activities of the sucrose PTS and sucrose 6-phosphate hydrolase were 1.06 ± 0.1 and 17.4 ± 0.1 mU mg of protein−1, respectively. Comparison of these activities with those obtained with the cells cultured in the sucrose medium (MH5S) suggested that the sucrose PTS is inducible by sucrose while sucrose 6-phosphate hydrolase is not.
This sucrose PTS enzyme contains an EIIB domain and a typical membrane-bound EIIC domain. However, the EIIA domain was not found. Thus, it seems that the EIIA for glucose PTS (encoded by nagE; MS1508) showing 72% homology to E. coli K-12 EIIA for glucose PTS is used for sucrose as well. The glucose PTS activities of MBEL55E and MBEL55EΔ0784 cultured in the MH5G medium were measured using radiolabeled [14C]glucose and compared. The glucose PTS activities of MBEL55E and MBEL55EΔ0784 were 7.98 ± 1.20 and 2.37 ± 1.20 mU mg of protein−1, respectively. Thus, the sucrose PTS enzyme can also function as a glucose PTS.
When cells use the sucrose PTS system, fructokinase plays an essential role in converting fructose to fructose 6-phosphate (Fig. 2A). However, when MS1233, which is a putative fructokinase in M. succiniciproducens, was disrupted, there was little apparent difference in cell growth between the wild-type strain (μ of 0.72 h−1) and MBEL55EΔ1233 strain (μ of 0.68 h−1) (Fig. 3A and C). This result suggests that the protein encoded by MS1233 might not be a fructokinase. To confirm the result, fructokinase assays were performed as described by Helanto et al. (2). Escherichia coli strain W (KCTC 1039; Korean Collection for Type Cultures, Daejeon, Korea), one of the well-known sucrose-utilizing bacteria possessing fructokinase, was used as a positive control; its fructokinase activity was 25.6 mU mg of protein−1. The fructokinase activities of the wild-type M. succiniciproducens and MBEL55EΔ1233 strains were 0.03 and 0.05 mU mg of protein−1, respectively (Table 1), which were almost negligible compared with that of the E. coli W control strain. Also, these values are 3 to 5 orders of magnitude lower than those of other sucrose-utilizing bacteria, which range from 10 to 1,000 mU mg of protein−1 (1, 2, 12, 17). Thus, it can be concluded that MS1233 does not encode fructokinase and that M. succiniciproducens does not possess a specific fructokinase.
However, no fructose was detected in the culture broth during the whole period of fermentation in sucrose medium, which implies that M. succiniciproducens is able to efficiently consume fructose. One possible option was the use of a fructose PTS, as reported earlier in Corynebacterium glutamicum (13), which also does not possess fructokinase, but instead uses a fructose PTS to reuptake the fructose secreted after the hydrolysis of sucrose 6-phosphate. The enzyme encoded by the MS2178 gene showed 58% and 36% homologies to E. coli K-12 and C. glutamicum fructose PTS IIBC protein. When the MS2178 gene was knocked out to make the MBEL55EΔ2178 strain as described earlier (5, 6), considerable growth retardation (μ of 0.40 h−1) was observed (Fig. 3C). This implies that M. succiniciproducens utilizes a fructose PTS for the metabolism of fructose as C. glutamicum does. The most distinctive feature in M. succiniciproducens, however, is that sucrose and fructose can be simultaneously utilized at similar uptake rates (Fig. 3D). According to the previous report by Moon et al. (13), fructose was detected during sucrose uptake even in the wild-type strain as well as in the fructose-PTS deletion mutant of C. glutamicum. This seems to be because the overall fructose uptake rate was innately slower than the sucrose uptake rate in C. glutamicum. On the other hand, since M. succiniciproducens can uptake both carbon sources simultaneously, almost at the same rates, and even the uptake rate of fructose seems to be faster at low concentrations (Fig. 3D), fructose might not be detected in its culture medium.
The fructose PTS activity was also measured in both the wild-type strain and MBEL55EΔ2178 strain according to the method described previously (2). The apparent difference in the fructose PTS activities of the cells cultured in the MH5S medium between the wild type (119.3 ± 2.9 mU mg of protein−1) and MBEL55EΔ2178 (80.9 ± 1.1 mU mg of protein−1) suggested that deletion of the MS2178 gene affected the fructose PTS activity (Table 1). However, although the MS2178 gene encoding the fructose PTS was deleted, significant residual activity was still present in MBEL55EΔ2178.
The sucrose PTS encoded by MS0784 was also examined as a candidate of another fructose PTS. As the mutant (MBEL55EΔ0784) cannot grow in the MH5S medium, the fructose PTS activities were measured using the cells cultivated in the BHI medium (Table 1). As shown in Table 1, the fructose PTS activities of MBEL55E, MBEL55EΔ2178, and MBEL55EΔ0784 were 38.8 ± 1.4, 22.9 ± 1.07, and 29.1 ± 1.23 mU mg of protein−1, respectively. These results suggest that the sucrose PTS encoded by MS0784 also transports fructose as the fructose PTS (MS2178) does. However, considering the remaining fructose PTS activities in both mutants (MBEL55EΔ2178 and MBEL55EΔ0784), other fructose PTS systems seem to be present in M. succiniciproducens.
In many bacteria, including E. coli, the mannose PTS can transport fructose as well as mannose (14). M. succiniciproducens also has MS0616-to-MS0618 and MS2377-to-MS2379 gene clusters annotated as manXYZ encoding mannose PTS. By analyzing the membrane proteome, MS0618 and MS2379 were identified in glucose medium (8) and also constitutively expressed in either sucrose or fructose medium at similar levels (data not shown). This carbon source-independent constitutive expression might be related to the absence of the Mlc regulator, a negative regulator of the manXYZ operon, in M. succiniciproducens. Hence, it is likely that the mannose PTS operon is constitutively transcribed and translated in sucrose medium. Consequently, the two mannose PTSs might also function as an alternative fructose PTS.
In conclusion, M. succiniciproducens has some unique features in its sucrose uptake and utilization system (Fig. 2B), which can be summarized as follows. First, the genes encoding proteins involved in sucrose metabolism exist separately in the genome, forming neither an operon nor a gene cluster. Second, differently from other sucrose transport and utilization systems (Fig. 1), only sucrose PTS and sucrose 6-phosphate hydrolase take part in sucrose metabolism. Third, no fructokinase and sucrose repressor exist. Fourth, instead of fructokinase, M. succiniciproducens uses a different fructose utilization system including a fructose PTS. The results of this study not only will be useful to construct an efficient sucrose catabolism for the succinic acid production in M. succiniciproducens but also will be helpful in understanding sucrose metabolism in other microorganisms having a similar type of sucrose metabolism.
Supplementary Material
Acknowledgments
We thank Hyohak Song for helpful discussion.
This work was supported by the Genome-Based Integrated Bioprocess Development Project of the Ministry of Education, Science and Technology (MEST) through the National Research Foundation of Korea (#20090065578). Further support by the LG Chem Chair Professorship, Microsoft, and WCU (World Class University) program by MEST (R32-2008-000-10142-0) is appreciated.
Footnotes
Published ahead of print on 15 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Caescu, C. I., O. Vidal, F. Krzewinski, V. Artenie, and S. Bouquelet. 2004. Bifidobacterium longum requires a fructokinase (Frk; ATP:d-fructose 6-phosphotransferase, EC 2.7.1.4) for fructose catabolism. J. Bacteriol. 186:6515-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helanto, M., J. Aarnikunnas, A. Palva, M. Leisola, and A. Nyyssola. 2006. Characterization of genes involved in fructose utilization by Lactobacillus fermentum. Arch. Microbiol. 186:51-59. [DOI] [PubMed] [Google Scholar]
- 3.Hong, S. H., J. S. Kim, S. Y. Lee, Y. H. In, S. S. Choi, J. K. Rih, C. H. Kim, H. Jeong, C. G. Hur, and J. J. Kim. 2004. The genome sequence of the capnophilic rumen bacterium Mannheimia succiniciproducens. Nat. Biotechnol. 22:1275-1281. [DOI] [PubMed] [Google Scholar]
- 4.Jacobson, G. R., C. A. Lee, and M. H. Saier, Jr. 1979. Purification of the mannitol-specific enzyme II of the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. J. Biol. Chem. 254:249-252. [PubMed] [Google Scholar]
- 5.Jang, Y. S., Y. R. Jung, S. Y. Lee, J. M. Kim, J. W. Lee, D. B. Oh, H. A. Kang, O. Kwon, S. H. Jang, H. Song, S. J. Lee, and K. Y. Kang. 2007. Construction and characterization of shuttle vectors for succinic acid-producing rumen bacteria. Appl. Environ. Microbiol. 73:5411-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim, J. M., K. H. Lee, and S. Y. Lee. 2008. Development of a markerless gene knock-out system for Mannheimia succiniciproducens using a temperature-sensitive plasmid. FEMS Microbiol. Lett. 278:78-85. [DOI] [PubMed] [Google Scholar]
- 7.Koutinas, A. A., R. Wang, and C. Webb. 2004. Evaluation of wheat as generic feedstock for chemical production. Ind. Crop Prod. 20:75-88. [Google Scholar]
- 8.Lee, J. W., S. Y. Lee, H. Song, and J. S. Yoo. 2006. The proteome of Mannheimia succiniciproducens, a capnophilic rumen bacterium. Proteomics 6:3550-3566. [DOI] [PubMed] [Google Scholar]
- 9.Lee, P. C., S. Y. Lee, S. H. Hong, and H. N. Chang. 2002. Isolation and characterization of a new succinic acid-producing bacterium, Mannheimia succiniciproducens MBEL55E, from bovine rumen. Appl. Microbiol. Biotechnol. 58:663-668. [DOI] [PubMed] [Google Scholar]
- 10.Lodge, J., and G. R. Jacobson. 1988. Starvation-induced stimulation of sugar uptake in Streptococcus mutans is due to an effect on the activities of preexisting proteins of the phosphotransferase system. Infect. Immun. 56:2594-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin, S. A., and J. B. Russell. 1987. Transport and phosphorylation of disaccharides by the ruminal bacterium Streptococcus bovis. Appl. Environ. Microbiol. 53:2388-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Barajas, E., B. M. Krohn, D. M. Stark, and D. D. Randall. 1997. Purification and characterization of recombinant tomato fruit (Lycopersicon esculentum Mill.) fructokinase expressed in Escherichia coli. Protein Expr. Purif. 11:41-46. [DOI] [PubMed] [Google Scholar]
- 13.Moon, M. W., H. J. Kim, T. K. Oh, C. S. Shin, J. S. Lee, S. J. Kim, and J. K. Lee. 2005. Analyses of enzyme II gene mutants for sugar transport and heterologous expression of fructokinase gene in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol. Lett. 244:259-266. [DOI] [PubMed] [Google Scholar]
- 14.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid, S. J., and V. R. Abratt. 2005. Sucrose utilisation in bacteria: genetic organisation and regulation. Appl. Microbiol. Biotechnol. 67:312-321. [DOI] [PubMed] [Google Scholar]
- 16.Sahin-Toth, M., S. Frillingos, M. C. Lawrence, and H. R. Kaback. 2000. The sucrose permease of Escherichia coli: functional significance of cysteine residues and properties of a cysteine-less transporter. Biochemistry 39:6164-6169. [DOI] [PubMed] [Google Scholar]
- 17.Thompson, J., D. L. Sackett, and J. A. Donkersloot. 1991. Purification and properties of fructokinase I from Lactococcus lactis. Localization of scrK on the sucrose-nisin transposon Tn5306. J. Biol. Chem. 266:22626-22633. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.