Abstract
Vibrio vulnificus is a heterogeneous species that comprises strains virulent and avirulent for humans and fish, and it is grouped into three biotypes. In this report, we describe a PCR-based methodology that allows both the species identification and discrimination of those isolates that could be considered dangerous to public health. Discrimination is based on the amplification of a variable region located within the gene pilF, which seems to be associated with potential human pathogenicity, regardless of the biotype of the strain.
Vibrio vulnificus is a marine species from warm and tropical ecosystems that can be found associated with the surface of algae or the external and internal mucous surfaces of aquatic animals (16). The species is present in high numbers in filtering organisms, such as oysters, especially in warmer months (16). V. vulnificus is highly heterogeneous and comprises strains virulent and avirulent for humans, shrimp, and fish, and it is grouped into three biotypes (5, 16, 24). Biotypes 1 and 3 are opportunistic human pathogens, while biotype 2 is pathogenic for aquatic animals such as eels (5, 16, 24). This biotype comprises a zoonotic serovar (serovar E) also involved in human infections (2).
V. vulnificus human infection follows the ingestion of undercooked seafood, particularly raw oysters, or the exposure of wounds to seawater or marine animals (16). The ingestion of V. vulnificus by healthy individuals can result in gastroenteritis, but when the bacterium is ingested by individuals with underlying chronic disease, particularly liver disease, the infection can develop into septic shock, rapidly followed by death with a probability of 50% (16). Wound infections by V. vulnificus can progress rapidly to necrotizing fasciitis, requiring fasciotomies and debridement (16), and in individuals with underlying chronic disease, it may lead to secondary septicemia with a course identical to that of primary septicemia (16).
The recent description of a plasmid encoding the virulence genes essential for fish vibriosis (13) facilitated the design of a PCR protocol for the rapid identification of the strains that pose a risk for fish culture (21). In contrast, the genetic basis of human infections is poorly understood, thereby inhibiting the design of methods targeting specific virulence genes to identify the strains posing a public health hazard, especially in oysters for human consumption. Nevertheless, several genotyping systems based on polymorphisms in some loci, such as 16S rRNA or the vcg (virulence correlated gene) locus, apparently divide V. vulnificus populations into two genotypes, one associated with an environmental origin and the other with a clinical (human) one, and they have been proposed as a means to distinguish the strains with human pathogenic potential (15, 19). However, although these systems work well with biotype 1 strains, they fail with biotype 2 and 3 isolates of human origin, which present an environmental genotype (22). This last finding suggests that new systems should be designed taking into account the entire genetic diversity of human pathogenic isolates.
The present work arises from a previous multilocus sequence typing analysis on the genetic diversity of the species in which four housekeeping genes and three virulence-associated genes were partially sequenced in a collection of more than 100 strains belonging to the three biotypes isolated worldwide (20). Results with a selection of these isolates revealed that the population apparently was divided into two groups in terms of sequence variability for the fragment of the pilF gene. One group comprised most of the human clinical strains, regardless of the biotype, together with some environmental strains, while the other group encompassed most of the environmental strains. PilF is a protein required for pilus-type IV assembly whose mutation in some bacterial pathogens implies attenuated virulence for mice (6). Taking these results into account, the objective of the present work was to develop a PCR methodology based on pilF variability that could identify the isolates that potentially are dangerous to public health, under the hypothesis that differences in the pilF gene in V. vulnificus are related to human pathogenic potential.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The V. vulnificus strains used in this study and their characteristics, including the genotyping, are listed in Table 1. Strains were routinely grown in tryptone soy broth or agar plus 5 g/liter NaCl (TSB-1 or TSA-1; Pronadisa, Spain) at 28°C for 24 h. For genomic DNA extraction, bacteria were grown overnight with shaking at 28°C. The strains were maintained both as lyophilized stocks and as frozen stocks at −80°C in marine broth (Difco) plus 20% (vol/vol) glycerol.
TABLE 1.
V. vulnificus strains used in the study: source, biotype, genotype, serovar, and PCR results
| Strain(s) | Biotype/serovara | Origin | Genotypingb |
Country of isolation | PCR |
Human serum resistance | ||
|---|---|---|---|---|---|---|---|---|
| vcg | 16S rRNA | Vvha | Vvpdh | |||||
| ATCC33816, CECT 5164, CECT 5168, CECT 5169, | Bt1/NT | Human blood | C | B | United States | + | + | + |
| CECT 529Td | Bt1/NT | Human blood | E | A | United States | + | − | − |
| CECT 5166 | Bt1/NT | Wound infection | E | B | United States | + | + | + |
| CECT 5165d | Bt1/NT | Seawater | E | A | United States | + | − | − |
| E4 | Bt1/NT | Seafood | C | B | United States | + | − | − |
| JEc | Bt1/NT | Seafood | E | B | United States | + | + | − |
| MLT364, MLT362 | Bt1/NT | Environmental | C | B | United States | + | + | + |
| VV1003c, | Bt1/NT | Environmental | C | B | United States | + | + | − |
| VV352c,d, VV425c | Bt1/NT | Environmental | E | A | United States | + | + | − |
| MLT404d, MLT406d | Bt1/NT | Environmental | E | A | United States | + | − | − |
| V4d | Bt1/NT | Human blood | C | B | Australia | + | + | + |
| CS9133 | Bt1/NT | Human blood | C | B | South Korea | + | + | + |
| CECT 5167, N87, KH03 | Bt1/NT | Human blood | C | B | Japan | + | + | + |
| YN03 | Bt1/NT | Human blood | E | A | Japan | + | + | + |
| L49 | Bt1/NT | Brackish water | E | A | Japan | + | − | − |
| YJ106 | Bt1/NT | Human blood | C | B | Taiwan | + | + | + |
| CG118 | Bt1/NT | Seawater | C | B | Taiwan | + | + | + |
| CG100d | Bt1/NT | Oyster | C | B | Taiwan | + | + | + |
| CG106 | Bt1/NT | Oyster | E | A | Taiwan | + | − | − |
| CG110, CG111 | Bt1/NT | Seawater | C | B | Taiwan | + | − | − |
| CECT 4867 | Bt1/NT | Diseased eel | E | A | Sweden | + | − | − |
| 94-9-118 | Bt1/NT | Human expectoration | E | A | Denmark | + | + | + |
| 94-9-130 | Bt1/NT | Water | E | A | Denmark | + | − | − |
| 94-9-119 | Bt1/NT | Human wound | E | A | Denmark | + | + | + |
| A2, A4, A5, A6, A7, PD-1, PD-3, PD-5, PD-12, V1 | Bt1/NT | Eel tank water | E | A | Spain | + | − | − |
| PD-2-66 | Bt1/NT | Eel tank water | E | B | Spain | + | − | − |
| CECT 4606d | Bt1/NT | Healthy eel | E | A | Spain | + | − | − |
| Riu-1 | Bt1/NT | Seawater | E | A | Spain | + | − | − |
| Riu-3 | Bt1/NT | Seawater | E | AB | Spain | + | − | − |
| 94385d | Bt1/NT | Leg wound | E | B | Spain | + | + | + |
| CECT 4608d | Bt1/NT | Eel farm water | C | B | Spain | + | + | + |
| CECT 5768, A13, A21, A22, CECT 5689, CECT 5769, A15, A16, A17, A18, CECT 5198, CECT 5343, A10, A11, A14d | Bt2/SerA | Diseased eel | E | A | Spain | + | − | − |
| 4/7/17, 4/7/19, | Bt2/SerA | Diseased eel | E | A | Denmark | + | − | − |
| 21B, 22, 26, 27, 21A | Bt2/SerA | Diseased eel | ND | ND | Denmark | + | − | − |
| CECT 4864, CECT 4999d, CECT 4605, CECT 4607, CECT 4604d, CECT 4602, CECT 4603, CECT 4601 | Bt2/SerE | Diseased eel | E | A | Spain | + | + | + |
| PD-2-47, PD-2-50, PD-2-51, PD-2-55, PD-2-56, CECT 5763 | Bt2/SerE | Eel tank water | E | A | Spain | + | + | + |
| C1, CECT 5762 | Bt2/SerE | Healthy eel | E | A | Spain | + | + | + |
| Riu-2 | Bt2/SerE | Seawater | E | A | Spain | + | + | + |
| CIP8190 | Bt2/SerE | Human blood | E | A | France | + | + | + |
| CECT 4868 | Bt2/SerE | Diseased eel | E | A | Norway | + | + | + |
| 90-2-11 | Bt2/SerE | Diseased eel | E | A | Denmark | + | + | + |
| 4-8-112 | Bt2/SerE | Human wound | E | A | Denmark | + | + | + |
| 94-9-123 | Bt2/SerE | Seawater | E | A | Denmark | + | + | + |
| CCUG38521 | Bt2/SerE | Human blood | ND | ND | Sweden | + | + | + |
| Ö122 | Bt2/SerE | Diseased eel | ND | ND | Sweden | + | + | + |
| CECT 4866 | Bt2/SerE | Human blood | E | A | Australia | + | + | + |
| CECT 4865 | Bt2/SerE | Diseased shrimp | E | A | Taiwan | + | + | + |
| UE516 | Bt2/SerE | Diseased eel | E | A | Taiwan | + | + | + |
| CECT 898, CECT 897, CECT 4174, CECT 4862 | Bt2/SerE | Diseased eel | E | A | Japan | + | + | + |
| 95-8-161d, 95-8-162d | Bt2/SerI | Diseased eel | E | A | Denmark | + | + | + |
| CECT 4869d | Bt2/SerI | Diseased eel | E | A | Belgium | + | − | − |
| 95-8-7d | Bt2/SerI | Diseased eel | E | A | Denmark | + | + | + |
| 95-8-6d | Bt2/SerI | Diseased eel | E | A | Denmark | + | − | − |
| 97, 162d, 11028d, vv12, vv32 | Bt3/SerO | Diseased human | E | AB | Israel | + | + | + |
| V. alginolyticus CECT 521T | − | − | Not tested | |||||
| V. anguillarum CECT 522T | − | − | Not tested | |||||
| V. cholerae CECT 514T | − | − | Not tested | |||||
| V. natriegens CECT | − | − | Not tested | |||||
| V. parahaemolyticus CECT 511T | − | − | Not tested | |||||
Bt, biotype; NT, nontypeable; SerE, serovar E; SerA, serovar A; SerI, serovar I; SerO, serovar O.
Genotyping data are from Sanjuan et al. (20); E type and C type are according to Rosche et al. (19); A type, B type, and AB type are according to Lee et al. (13). ND, not done.
Translucent strain.
Strains used for whole pilF gene sequencing.
DNA isolation and manipulation.
For DNA sequencing, genomic DNA was extracted according to the Mini-Prep protocol (4). For PCR determinations from overnight cultures, DNA was extracted by the boiling method (8). Briefly, the samples were boiled for 10 min and supernatants, obtained after centrifugation, either were directly used as DNA samples (bacterial cultures) or were subjected to extraction with phenol-chlorophorm, precipitation with ethanol, and suspension in 100 μl of Tris-HCl 10 mM. All DNA samples were stored at −20°C until use.
Sequence analysis.
Primers to amplify the full-length pilF gene were designed from the published sequences of strains YJ016 and CMCP6, both from biotype 1 (Table 2) (7, 12), and were used with 20 strains representative of the genetic variability of the species (Table 1). The sequences were aligned with AlignX (Invitrogen) and were compared by using the maximum likelihood (ML) approach implemented in Modeltest V3.7 (17). Likelihood scores for each model for nucleotide substitution were estimated with PAUP*4.0b10, and the best model was determined using the Akaike information criterion (AIC) (1). ML phylogenetic trees were obtained with PHYML 2.4.4 (10) using the previously determined models of nucleotide substitution, and support for the nodes was evaluated by bootstrapping with 1,000 replicates. The 20 sequences were submitted to GenBank.
TABLE 2.
Primers used in this study
| Primer | Sequence | Product size (bp) | Target | Utility |
|---|---|---|---|---|
| VvhaF | CCGCGGTACAGGTTGGCGC | 521 | Hemolysin/cytolisin gene | V. vulnificus-specific sequence |
| VvhaR | CGCCACCCACTTTCGGGCC | |||
| PilFF | CGATTGGTAGGCAATAGAC | 917 | pilF | Primers to sequence pilF gene |
| PilFR | GCAACTCAACCTCAAGACG | |||
| VvpdhF | TGTCGGTGAAAACGGCAAAGCTG | 338 | pilF | V. vulnificus strains potentially dangerous for humans |
| VvpdhR | GGTATCGATTTCCAACTTAGCGAGGTTGAGCACC |
Multiplex PCR design.
The alignment of the sequences showed a differential region of 34 nucleotides that is present in the strains isolated from humans and the strain belonging to the zoonotic serovar (Fig. 1). This region was used as a reverse primer to amplify a 338-bp fragment of pilF, which hypothetically is present only in the isolates endangering public health. Table 2 shows the Vvpdh (for V. vulnificus potentially dangerous for humans) primer set. A second primer set was included to amplify a fragment of 521 bp of the cytolysin gene vvhA, which is species specific (11) (Table 2). PCR was performed in 50-μl reaction volumes that contained 0.2 mM Vvha forward and reverse primers, 0.4 mM Vvpdh forward and reverse primers, 1.5 U of Taq DNA polymerase (GoTaq; 5 U/μl; Promega), 10 μl of 5× Taq reaction buffer (Gotaq Green; Promega), 1.125 mM MgCl2, 0.65 mM deoxynucleoside triphosphate (dNTP) mix (Promega) and 2 μl of DNA crude extracts. The PCR was performed in a Techne thermocycler (TC-412). The reaction started with 10 min of denaturation at 94°C and was followed by 45 cycles of 40 s of denaturation at 94°C, 45 s of annealing at 54°C, and 45 s of extension at 72°C. An additional extension at 72°C for 10 min completed the reaction. The detection limit of the PCR in phosphate-buffered saline-1% NaCl (PBS-1) was evaluated by extracting DNA from 10-fold serial dilutions of V. vulnificus in PBS-1 (108 to 101 CFU/ml) and subjecting the samples to the PCR.
FIG. 1.
Alignment of a segment of the sequenced pilF genes showing the region used for the design of the primer VvpdhR.
Mice virulence.
The virulence potential for humans was tested in BALB/c mice of 20 g of average weight (5 to 6 weeks old) pretreated with Desferal (Sigma) (iron-overloaded mouse model) according to previously published procedures (3). To this end, groups of six animals were injected intraperitoneally (i.p.) with 10-fold serial bacterial dilutions of V. vulnificus in PBS-1 (0.2 ml per mouse), and mortalities were recorded daily for 1 week and considered only if the bacteria were isolated in pure culture from internal organs. The 50% lethal dose (LD50) was calculated by the Reed and Muench method (18). Appropriate controls for each experiment also were included (mice inoculated with PBS-1 and mice inoculated with Desferal) (3).
Serum resistance.
The bacterial resistance to human serum (male AB; Sigma-Aldrich, Spain), was assessed in 96 microwell plates by mixing 100 μl of a bacterial suspension in PBS-1 containing 104 CFU per ml with 100 μl of fresh human serum. The mixtures were incubated at 37°C for 24 h, and aliquots were removed at 0, 1, 3, 9 and 24 h postincubation for bacterial counts on TSA-1 plates by drop-plate methodology. Experiments were done in triplicate. A strain was considered resistant if the cell survival percentage was higher than or equal to 100% ± 20% after 24 h of incubation. The strains CECT 4604 (formerly E86) and CECT 4606 (formerly E109), which are highly resistant and sensitive to human serum, respectively, were used as controls (3).
Nucleotide sequence accession numbers.
The 20 sequences determined in the course of this work were submitted to GenBank under accession numbers FJ756476 to FJ756489 and FJ899603 to FJ899608.
RESULTS
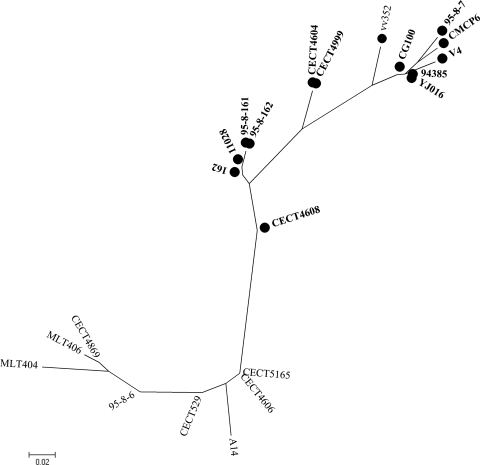
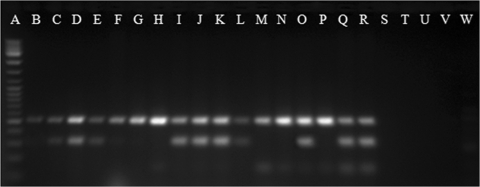
We analyzed the sequence variability of the full-length pilF gene in 20 V. vulnificus strains from different biotypes and origins, thus trying to cover most of the genetic diversity of the species (Table 1). The ML tree grouped the human isolates of biotypes 1 and 3 together with isolates belonging to the zoonotic serovar of V. vulnificus and some environmental isolates (Fig. 2). Part of the pilF polymorphism resided in a variable region of 34 nucleotides, which was identified after sequence alignment (Fig. 1). This region was used as the PCR reverse primer (VvpdhR) to amplify a pilF fragment of 338 bp in the isolates of the species believed to be dangerous to public health, under the hypothesis that all of them shared this fragment. The primers VvpdhR and VvpdhF were combined with a species-specific primer, and the specificity of the multiplex PCR was tested on a collection of 112 V. vulnificus strains belonging to all three biotypes, which were recovered worldwide from environmental and clinical (human and animal) sources, together with five strains of other Vibrio species (Table 1). As expected, no amplification product was obtained from the other Vibrio species, whereas all V. vulnificus strains gave the species-specific PCR product (Table 1, Fig. 3). All of the V. vulnificus human isolates of biotype 1 amplified with the Vvpdh primers, with the exception of the type strain of the species (CECT 529T) (Table 1, Fig. 3). This isolate was confirmed to lack human virulence potential after determining its virulence degree in iron-overloaded mice (50% LD50 > 108 CFU per mouse) and its survival in human serum (0% survival after 24 h of incubation). In addition, we found positive results from some biotype 1 environmental isolates (Table 1). Since the species is an autochthonous member of the marine ecosystem where virulent and avirulent strains can coexist, the possibility that these Vvpdh-positive biotype 1 environmental isolates could be pathogenic for humans was evaluated by assessing their susceptibility to human serum. Results of human serum resistance tests showed that approximately 80% of these isolates resisted the bactericidal activity of human serum (survival rate of more than 90% ± 10%) (Table 1, Fig. 2). The serum-sensitive isolates either lacked capsule or were less capsulated, since they developed translucent colonies on agar plates (data not shown).
FIG. 2.
Maximum likelihood phylogenetic tree of 20 V. vulnificus isolates obtained from the alignment of the pilF gene. Isolates that resisted the action of the human sera are shown in boldface. A circle represents Vvpdh-positive isolates.
FIG. 3.
Electrophoresis of amplified products by the multiplex PCR. Lane A, DNA ladder mix (Fermentas); lanes B to D, human clinical strains of biotype 1 (CECT 529T, YJ016, and CECT 5168); lanes E to H, environmental isolates of biotype 1 (MLT 362, CECT 4606, CECT 5165, and CECT 4867); lanes I to L, eel and human isolates of biotype 2 serovar E (CECT 4999, CECT 4866, CCUG 38521, and CIP 8190); lanes M and N, eel isolates of biotype 2 serovar A (CECT 5768 and CECT 5689); lanes O and P, eel isolates of serovar I (95-8-161 and 95-8-6); lanes Q and R, human isolates of biotype 3 (vv12 and 11028); and lanes S to W, other vibrio species (V. alginolyticus, V. anguillarum, V. cholerae, V. natriegens, and V. parahaemolyticus).
In the case of biotype 2, all isolates belonging to the zoonotic serovar positively amplified with the Vvpdh primers regardless of their origin, while the isolates belonging to serovar A did not, and the isolates belonging to serovar I gave variable results (Table 1, Fig. 3). All of the biotype 2 isolates that were positive for Vvpdh resisted the bactericidal action of human serum (survival rate of 90% ± 10%) (Table 1). This result is in agreement with previous studies, demonstrating that biotype 2 is formed by three distinct clonal complexes (20), one of which is able to infect humans (2), another is unable to do so (9), and the third (a minority) has yet to be studied. Finally, all biotype 3 strains, all of human origin, amplified with the Vvpdh primers and resisted the bactericidal activity of human serum (Table 1).
DISCUSSION
In a previous work on multilocus sequence typing (20), we found evidence that the human isolates of V. vulnificus of the three biotypes could be separated from the others on the basis of variations in the pilF gene sequence. This result leads us to hypothesize that pilF is a good genetic marker for human virulence potential in this species. To test this hypothesis, we sequenced the entire gene in 20 human and nonhuman isolates, and the sequences were analyzed. The ML tree grouped the human isolates together with those of the zoonotic serovar, and the sequence alignment identified a variable region that facilitated the design of a primer set. A multiplex PCR method was developed and tested on a wide range of V. vulnificus isolates that were representative of the genetic diversity of the species to assess its ability to differentiate V. vulnificus isolates that pose a risk to public health. All of the V. vulnificus human isolates of the three biotypes gave the expected amplification products, with the exception of the type strain of the species (CECT 529T), a blood isolate lacking the Vvpdh band. Despite its human clinical origin, this strain proved avirulent for iron-overloaded mice, the animal model used to predict human virulence in V. vulnificus, and to be sensitive to human serum. The type strain of the species was isolated from human blood in the 1970s and has been kept under laboratory conditions since then. It is possible that spontaneous mutations have affected this gene, with some diminishing its virulence. Alternatively, it is possible that this strain was isolated from an immunosuppressed host and was weakly virulent in origin. In fact, this isolate was grouped together with environmental strains by other typing methods for V. vulnificus, such as typing the vcg locus or 16S rRNA (Table 1 and reference 22). Thus, the negative result obtained by PCR for the type strain of the species does not necessarily invalidate the usefulness of this PCR as a method to discriminate strains that may be dangerous to public health. Furthermore, a number of environmental biotype 1 and 2 isolates also gave a positive result for both primers in the PCR. Most of these isolates were resistant to human serum and, therefore, seem to belong to the group of V. vulnificus isolates dangerous to public health. The serum sensitivity of the remaining Vvpdh-positive environmental isolates could be related to the lack of capsule, as those strains developed translucent colonies on agar plates. In fact, capsule is reported to be essential for V. vulnificus resistance to human serum and virulence (3, 14, 23). Thus, the encapsulated environmental Vvpdh-positive isolates could constitute a risk to public health. As expected, no correspondences between the results in the Vvpdh PCR and vcg/rRNA typing were found (Table 1). In this sense, although 91% of the Vvpdh-negative strains showed the profile usually associated with nondangerous strains (vcg E type and rRNA A type [15, 19, 25]), 62% of the Vvpdh-positive strains also were vcg E type and rRNA A type (Table 1). This result is due mainly to the biotype 2 and 3 isolates that were positive for the Vvpdh PCR.
In conclusion, two genotypes can be identified within V. vulnificus strains on the basis of pilF variability. One encompasses most of the strains that potentially are dangerous for public health (Vvpdh positive) regardless of their biotype, and the other comprises all other strains (Vvpdh negative). PilF is a protein required for pilus type IV assembly whose mutation in other bacterial pathogens is involved in attenuated virulence for mice (6). The exact role of pilF in virulence for mice/humans in V. vulnificus strains has yet to be determined. Additional studies using natural samples of molluscan shellfish would clarify the potential of multiplex PCR for V. vulnificus risk assessment.
Acknowledgments
This work has been financed by grants AGL2008-03977/ACU and CSD2009-00006 from the Spanish Ministry for Education and Science and grant MTKD-CT-2004-0145019 from the European Union.
We thank SCSIE of the University of Valencia for technical support in determining the sequences.
Footnotes
Published ahead of print on 28 December 2009.
REFERENCES
- 1.Akaike, H. 1974. A new look at the statistical model identification. Automatic control, IEEE. Trans. Biomed. Eng. 19:716-723. [Google Scholar]
- 2.Amaro, C., and E. G. Biosca. 1996. Vibrio vulnificus biotype 2, pathogenic for eels, is also an opportunistic pathogen for humans. Appl. Environ. Microbiol. 62:1454-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaro, C., E. G. Biosca, B. Fouz, A. E. Toranzo, and E. Garay. 1994. Role of iron, capsule, and toxins in the pathogenicity of Vibrio vulnificus biotype 2 for mice. Infect. Immun. 62:759-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2007. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 5.Bisharat, N., V. Agmon, R. Finkelstein, R. Raz, G. Ben-Dror, L. Lerner, S. Soboh, R. Colodner, D. N. Cameron, D. L. Wykstra, D. L. Swerdlow, and J. J. Farmer III. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Israel Vibrio Study Group. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 6.Chakraborty, S., M. Monfett, T. M. Maier, J. L. Benach, D. W. Frank, and D. G. Thanassi. 2008. Type IV pili in Francisella tularensis: roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect. Immun. 76:2852-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Medici, D., L. Croci, E. Delibato, P. S. Di, E. Filetici, and L. Toti. 2003. Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype Enteritidis in poultry. Appl. Environ. Microbiol. 69:3456-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fouz, B., F. J. Roig, and C. Amaro. 2007. Phenotypic and genotypic characterization of a new fish-virulent Vibrio vulnificus serovar that lacks potential to infect humans. Microbiology 153:6-34. [DOI] [PubMed] [Google Scholar]
- 10.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 11.Hill, W. E., S. P. Keasler, M. W. Trucksess, P. Feng, C. A. Kaysner, and K. A. Lampel. 1991. Polymerase chain reaction identification of Vibrio vulnificus in artificially contaminated oysters. Appl. Environ. Microbiol. 57:707-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, C. T., C. Amaro, K. M. Wu, E. Valiente, Y. F. Chang, S. F. Tsai, C. H. Chang, and L. I. Hor. 2008. A common virulence plasmid in biotype 2 Vibrio vulnificus and its dissemination aided by a conjugal plasmid. J. Bacteriol. 190:1638-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linkous, D. A., and J. D. Oliver. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174:207-214. [DOI] [PubMed] [Google Scholar]
- 15.Nilsson, W. B., R. N. Paranjpye, A. DePaola, and M. S. Strom. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 41:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliver, J. D. 2006. Vibrio vulnificus, p. 349-366. In F. L. Thompson, B. B. Austin, and J. G. Swings (ed.), Biology of vibrios. American Society for Microbiology Press, Washington, DC.
- 17.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 18.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. (London) 27:493-497. [Google Scholar]
- 19.Rosche, T. M., Y. Yano, and J. D. Oliver. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49:381-389. [DOI] [PubMed] [Google Scholar]
- 20.Sanjuan, E. 2008. Epidemiology and phylogeny of Vibrio vulnificus biotype 2. Ph.D. thesis. University of Valencia, Valencia, Spain.
- 21.Sanjuan, E., and C. Amaro. 2007. Multiplex PCR assay for detection of Vibrio vulnificus biotype 2 and simultaneous discrimination of serovar E strains. Appl. Environ. Microbiol. 73:2029-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanjuan, E., B. Fouz, J. D. Oliver, and C. Amaro. 2009. Evaluation of genotypic and phenotypic methods to distinguish clinical from environmental Vibrio vulnificus strains. Appl. Environ. Microbiol. 75:1604-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strom, M. S., and R. N. Paranjpye. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2:177-188. [DOI] [PubMed] [Google Scholar]
- 24.Tison, D. L., M. Nishibuchi, J. D. Greenwood, and R. J. Seidler. 1982. Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl. Environ. Microbiol. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickery, M. C., W. B. Nilsson, M. S. Strom, J. L. Nordstrom, and A. DePaola. 2007. A real-time PCR assay for the rapid determination of 16S rRNA genotype in Vibrio vulnificus. J. Microbiol. Methods 68:376-384. [DOI] [PubMed] [Google Scholar]