Abstract
Virulent phage 1358 is the reference member of a rare group of phages infecting Lactococcus lactis. Electron microscopy revealed a typical icosahedral capsid connected to one of the smallest noncontractile tails found in a lactococcal phage of the Siphoviridae family. Microbiological characterization identified a burst size of 72 virions released per infected host cell and a latent period of 90 min. The host range of phage 1358 was limited to 3 out of the 60 lactococcal strains tested. Moreover, this phage was insensitive to four Abi systems (AbiK, AbiQ, AbiT, and AbiV). The genome of phage 1358 consisted of a linear, double-stranded, 36,892-bp DNA molecule containing 43 open reading frames (ORFs). At least 14 ORFs coded for structural proteins, as identified by SDS-PAGE coupled to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. The genomic organization was similar to those of other siphophages. All genes were on the same coding strand and in the same orientation. This lactococcal phage was unique, however, in its 51.4% GC content, much higher than those of other phages infecting this low-GC Gram-positive host. A bias for GC-rich codons was also observed. Comparative analyses showed that several phage 1358 structural proteins shared similarity with two Listeria monocytogenes phages, P35 and P40. The possible origin and evolution of lactococcal phage 1358 is discussed.
The first sequenced genome of a phage infecting Lactococcus lactis (bIL67) was reported in 1994 (57). Its genomic characterization was performed with the prospect of a better understanding of lactococcal phage biology. L. lactis is a Gram-positive bacterium added to milk to produce an array of fermented dairy products. In this human-made environment, substantial amounts of lactococcal cells are cultivated on a daily basis in large fermentation vats, and these added cells randomly encounter virulent phages present in heat-treated but nonsterile milk. Moreover, it is widely acknowledged that the increased use of the same bacterial strains within existing dairy facilities inevitably leads to milk fermentation failures due to the multiplication of virulent phages. This biotechnological problem reduces yields and lowers the quality of fermented products (51).
Over 700 lactococcal phage isolates have been reported in the literature (3). To date, more than 25 complete genome sequences of lactococcal phages are publicly available in the NCBI database, and the sequencing of others is under way. These numbers indicate that Lactococcus phages are among the most studied of the bacterial viruses. All lactococcal phages belong to the order Caudovirales and are included within two families according to their tail morphology: the Siphoviridae (long noncontractile tail [most lactococcal phages]) and the Podoviridae (short noncontractile tail [few lactococcal phages]) (14). Currently, phages infecting L. lactis strains have been divided into 10 genetically distinct groups (14). The complete genomic sequence is available for at least one representative of 8 of the groups.
Early sequencing efforts concentrated on the genomes of lactococcal phages belonging to the 936, c2, and P335 groups (Siphoviridae), because members of these groups were regularly isolated in dairy plants (8, 36, 50). PCR-based methods were also devised to rapidly classify these phages (41). These Siphoviridae phages pose a significant risk to the dairy industry, and their characterization is important for developing adapted antiphage strategies to limit their propagation and evolution.
In recent years, representatives of the less recognized lactococcal phage groups have been characterized, including phages Q54 (22), KSY1 (13), 1706 (23), asccφ28 of the P034 group (39), and P087 (63). Their molecular characterizations were aimed at understanding why some phage groups (936, c2, and P335) predominate while the others have remained marginal, at best. However, it was recently reported that P034-like phages may be emerging in certain regions (52). Genomic and microbiological analyses indicated that members of these rare phage groups were likely the result of recombination between different lactococcal phages and phages infecting other Gram-positive bacteria, and they may not be fit to multiply rapidly in milk. For example, lactococcal phage 1706 shares similarities with Ruminococcus and Clostridium prophages (23). Similarly, L. lactis phage P087 structural proteins share identity with gene products found in a prophage in the Enterococcus faecalis genome (63). It was also shown previously that lactococcal phage asccφ28 was related to Streptococcus pneumoniae phage Cp-1 and Bacillus subtilis φ29-like phages (39). It was suggested that phages 1706, asccφ28, and P087 acquired a receptor-binding protein complex from another lactococcal phage that enabled them to infect a L. lactis host.
Here, we report the complete genome sequence and analysis of phage 1358, a virulent representative of the 9th lactococcal phage group.
MATERIALS AND METHODS
Bacteria, phages, and culture conditions.
All phages and bacterial strains used in this study were obtained from the Félix d'Hérelle Reference Center for Bacterial Viruses (www.phage.ulaval.ca). L. lactis cells were grown statically at 30°C in M17 broth (Oxoid) supplemented with 0.5% glucose (GM17) (60), and Listeria strains were grown in tryptic soy agar (TSA) broth (Quelab). Lactococcal phages were routinely amplified at 30°C in GM17 broth supplemented with 0.01 M calcium chloride (GM17-Ca), and the lysates were stored at 4°C.
Microbiological assays.
One-step growth assays of phage 1358 were performed in triplicate, as reported elsewhere previously (49), with a multiplicity of infection (MOI) of 0.05 and with the host L. lactis SMQ-388 or L. lactis SMQ-382. These assays were also carried out at 21°C and 30°C. The burst size was calculated by dividing the average phage titer after the exponential phase by the average titer before the infected cells began to release virions (49). To measure the efficacy of natural phage defense mechanisms against phage 1358, the efficiency of plaquing (EOP) was calculated by dividing the phage titer on the tested L. lactis strain by the titer on phage-sensitive wild-type strain L. lactis SMQ-388. The bacterial strains tested were L. lactis SMQ-388 transformed with vector pNZ123 (16), containing the phage defense mechanism AbiK (19), AbiQ (18), or AbiT (11), and vector pLC5 (25), containing AbiV (25). The phage-sensitive strain contained only the pNZ123 or pLC5 vector. The host range of virulent lactococcal phage 1358 was assessed by spotting 10 μl of a 10−2 dilution of the high-titer lysate (109 PFU/ml) on top agar containing an L. lactis strain. A total of 59 industrial and laboratory L. lactis strains were tested. Moreover, phage 1358 was also tested against Lactococcus raffinolactis ATCC 43920 as well as eight Listeria strains, namely, Listeria innocua HER1030 and HER1035 and Listeria monocytogenes HER1034, HER1082, HER1083, HER1184, HER1247, and HER1394. Finally, Listeria phage P35 was tested on the host of phage 1358, Lactococcus lactis SMQ-388.
Electron microscopy.
A 7.5-μl drop of phage lysate (1010 PFU/ml) was placed onto a copper Formvar-carbon-coated grid (Ted Pella Inc.). The liquid was removed after 1 min by touching the edge of the grid with blotting paper. The stain (7.5 μl of 3% phosphotungstic acid [pH 7]) was applied in the same way. Phage morphology was observed by using a JEOL 1230 transmission electron microscope at 80 kV. Dimensions of the phage are the means for at least 20 specimens.
Phage DNA preparation and sequencing.
Phage 1358 genomic DNA was isolated by using a Maxi lambda DNA purification kit (Qiagen) with previously described modifications (15). The restriction profile of the isolated DNA was compared to the previously published profile of phage 1358 to confirm its identity (14). Restriction endonucleases (Roche Diagnostics) were used as recommended by the manufacturer. After restriction, phage DNA samples were heated for 10 min at 70°C to prevent cohesive end ligation. The terminal redundancy of the genome was also verified through a search of restricted submolar fragments. The DNA fragments were separated in a 0.8% agarose gel in 1× Tris-acetate-EDTA buffer, stained with ethidium bromide, and photographed under UV illumination. Genome sequencing was first performed by using GS-FLX shotgun sequencing (McGill University and Genome Quebec Center, Montreal, Quebec, Canada), followed by pyrosequencing of the phage DNA. Approximately 11,000 reads were generated, and this resulted in a single contig that was assembled to 58× coverage. To identify genome ends, two primer pairs were designed, and direct sequencing of the total genomic phage DNA was carried out with an ABI Prism 3130XL apparatus (Applied Biosystems) from the Laboratory of Nucleic Acids Analysis (Pavillon Charles-Eugène-Marchand, Université Laval). Standard PCR procedures were used (54).
Bioinformatic analysis.
Open reading frame (ORF) predictions were performed by using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), BioEdit (26), and Heuristic GeneMark (7). The prediction was bolstered by visual inspection using criteria such as the presence of a ribosome-binding site (RBS), the possible existence of short ORFs and non-AUG start codons, and codon usage analysis. The putative RBSs were determined by using the 3′ end of L. lactis IL1403 16S rRNA (10). The translated ORF products were compared with known protein sequences by using BLASTp (5) and the nonredundant public GenBank database. BLAST searches were also done by using the ACLAME database of clustered viral proteins maintained at the Service de Conformation de Macromolécules Biologiques et de Bioinformatique de l'Université Libre de Bruxelles (http://aclame.ulb.ac.be/) (43). Conserved domains were searched by using the CDD database, with an E value cutoff of <0.01 (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The isoelectric point (pI) and molecular mass (MM) of deduced phage proteins were determined by using Compute pI/Mw (http://ca.expasy.org/tools/pi_tool.html). The relationship between the GC content and genome position was calculated by using the GC Content and GC Skew diagrams of the Center for Nanostructure Technology and Biomolecular Technology at the University of Kaiserslautern (http://nbc3.biologie.uni-kl.de/).
The usage of codons was determined by using the Codon Usage program from the bioinformatics toolbox of DNA2.0 (https://www.dna20.com/). The data for the bacterial strains came from the codon usage database of the Kazusa DNA Research Institute (http://www.kazusa.or.jp/codon/). For the analysis of phage codon usage, each ORF was analyzed independently, and the entire data were added. The percentage of synonymous codon usage was calculated for each amino acid. The codon usages of one representative of each phage group infecting L. lactis and one phage of L. monocytogenes as well as three Lactococcus lactis and two Listeria monocytogenes host strains were investigated.
Analysis of phage 1358 structural proteins.
One liter of phage lysate was concentrated with polyethylene glycol (PEG) and purified on a discontinuous CsCl gradient and a one-step CsCl gradient (54). Purified phages were recovered by ultracentrifugation using a Beckman SW41 Ti rotor at 35,000 rpm (210,053 × g) for 3 h, followed by a second ultracentrifugation using a Beckman NVT65 rotor at 60,000 rpm (342,317 × g) for 18 h. The phage preparation (8 × 1010 PFU/ml) was then dialyzed against phage buffer (0.02 M Tris-HCl [pH 7.4], 0.1 M NaCl, 0.1 M MgSO4) and analyzed for structural proteins by standard Tris-glycine 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (42). Samples were mixed with 4× sample loading buffer and boiled for 5 min before loading. Protein bands were detected by Coomassie blue staining. The bands were cut out of the gel, digested with trypsin, and identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) at the Centre Protéomique de l'Est du Québec (Quebec City, Quebec, Canada).
Nucleotide sequence accession number.
A sequence was submitted to the GenBank database under accession number GQ403788.
RESULTS AND DISCUSSION
Morphology.
Virulent lactococcal phage 1358 was isolated from a dairy environment in 1981 by the New Zealand Dairy Research Institute (33). Phage 1358 is a member of the Siphoviridae family of the Caudovirales order (1), as are the majority of lactococcal phages. Based on stringent DNA-DNA hybridization studies and electron microscopy observations, this bacterial virus was previously recognized as being a unique lactococcal phage and was designated the representative of its group (14, 35). The only other reported member of this lactococcal phage group is possibly phage 1404 (35). Phages 1358 and 1404 were previously described as type c small isometric phages; both had the same host range, and they have DNA homology, as shown by DNA-DNA hybridization (33). Furthermore, their EcoRI restriction profiles differed by only one DNA fragment. Two other possible group members have been suggested, but no significant morphological or genetic characteristics are available (35).
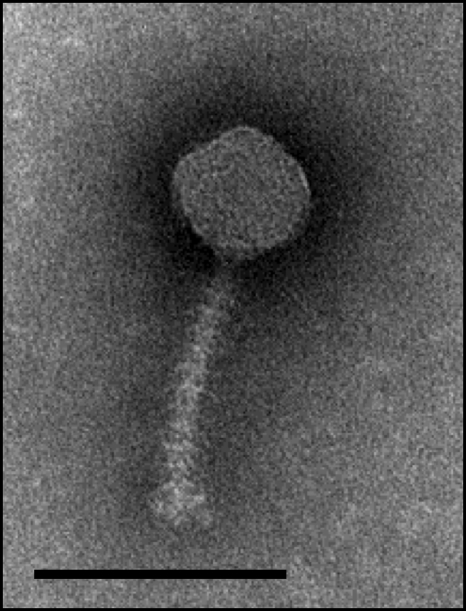
Phage 1358 has an icosahedral capsid with a diameter of 54 ± 3 nm and a noncontractile tail 103 ± 4 nm in length and 9 ± 1 nm in width (Fig. 1). Phage 1358 has one of the shortest observed tails for a lactococcal siphophage. Phages Q54 and φ50 (P335 group) have tails of a similar length, 109 nm and 105 nm, respectively, while the tail lengths of other lactococcal phages of the Siphoviridae family are between 130 and 276 nm. Interestingly, 9 to 11 horizontal bars were evenly spaced throughout the noncontractile tail of phage 1358 (Fig. 1). To our knowledge, this feature is unique among lactococcal phages (23) and is rarely seen among other bacterial viruses. It was previously observed for a limited number of phages infecting Lactobacillus, Rhizobium, and Streptomyces (2, 64). Five phages available at the Félix d'Hérelle Center for Bacterial Viruses (www.phage.ulaval.ca) also contain this type of tail structure: Acinetobacter calcoaceticus phages HER32 and HER162, Sinorhizobium meliloti phages HER112 and HER119, and Bacillus thuringiensis phage HER380. It was previously reported that the number and the position of these cross-bars are variable between different phages and sometimes even for the same phage (53). For example, Lactobacillus delbrueckii subsp. lactis phage JCL-1032 contains approximately 19 bars (21), but there are only two bars for phage SLP of Serratia marcescens. Interestingly, the latter phage can occasionally produce virions without cross-bars (64). Despite these studies, the function of this structural feature is unknown.
FIG. 1.
Electron micrograph of phage 1358. Scale bar, 100 nm.
Microbiological characterization.
Host range analysis showed that phage 1358 infected only 3 of the 60 lactococcal strains tested, including its host strain, L. lactis SMQ-388. One of the two other phage 1358-sensitive bacterial strains was L. lactis SMQ-382, the host of phage 1483. The latter virulent phage, which belongs to the P335 group (14), was also isolated in the 1980s by the New Zealand Dairy Research Institute (34). The third phage 1358-sensitive L. lactis strain was an industrial strain that was highly resistant to phages of the 936 group. In fact, phage 1358 is the first virulent phage known to infect this industrial lactococcal strain. This limited host range explains, in part, why this phage is not commonly found in dairy environments.
A single-step growth curve for phage 1358 was performed by using cells of its host, L. lactis SMQ-388, grown in GM17 medium at 30°C. The burst size was calculated to be 72 ± 2 PFU released per infected lactococcal cell, and its latent period was 90 ± 1.3 min. When the same single-step growth assay was performed at 21°C, the burst size increased to 86 ± 6 new virions per infected cell, but its latent period also increased to 132 ± 3 min. The experiment was repeated at 30°C by using L. lactis SMQ-382 as the host strain. The burst size was calculated to be 77 ± 36 PFU/cell, and the latent period was 90 ± 3 min. For other virulent lactococcal phages, the burst size varied between 42 and 400 PFU/cell, and the latent period was generally between 20 and 60 min (data not shown). Together, these data indicate that phage 1358 is similar to other lactococcal phages in its burst size, but its long latent period may also contribute to its scarcity in industrial settings. Its relatively long latent period at 21°C and 30°C suggests that phage 1358 would not be a significant concern in most milk processes such as the manufacture of buttermilk (48) and many types of cheeses (8). Of note, phage 1706, another uncommon lactococcal phage, also has a long latent period of 85 ± 2 min, but its burst size is more than twice that of phage 1358 (23).
The sensitivity of phage 1358 to four abortive infection mechanisms was also determined. First, we introduced a high-copy vector (pNZ123 or pLC5) expressing AbiK (19), AbiQ (18), AbiT (11), or AbiV (25) into L. lactis SMQ-388, and the resulting transformants were challenged with phage 1358. None of the four bacteriophage resistance mechanisms were effective against this virulent phage. To our knowledge, this is the first lactococcal phage to be insensitive to these four Abi systems.
Genome sequence and GC content.
Phage 1358 has a linear double-stranded DNA genome composed of 36,892 bp (Table 1). Its genome size is similar to those of many other lactococcal phage genomes, including phages belonging to the 936 (28) and P335 (40) groups. Sequencing of the genomic extremities revealed the absence of cohesive ends and the presence of terminal redundancy. Analyses of several restriction profiles uncovered the presence of submolar fragments (data not shown), confirming that phage 1358 is a pac-type phage. Further analyses located the pac site near or within the putative terminase subunit (data not shown). A similar location was reported previously for other phages (9, 12, 58, 69).
TABLE 1.
ORFs deduced from the phage 1358 genomic sequence and their predicted functionsd
| ORF | Position |
Size (aa) | MM (kDa) | pI | GC % | Putative RBS and start codona | Predicted function (or domain)b | Best match in databasesc | No. of identical ORFs/overall no. of ORFs (%) | Size (aa) | E value | GenBank accession no. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | ||||||||||||
| 1 | 18 | 623 | 201 | 22.4 | 5.5 | 54.8 | GAAAGGAacgccgaaacATG | Terminase small subunit | DSY4528 (Desulfitobacterium Y51) | 22/68 (32) | 275 | 0.059 | YP_520761.1 |
| 2 | 565 | 1929 | 454 | 52.1 | 6.9 | 53.3 | AGAAAGGActtaacaccgATG | Terminase large subunit | gp2 (Listeria phage P40) | 193/426 (45) | 430 | 1.0E−100 | YP_002261418.1 |
| 3 | 1944 | 3587 | 547 | 61.2 | 4.5 | 54.7 | AAAGGAGaaagcacaATG | Portal protein | gp3 (Listeria phage P35) | 272/545 (49) | 537 | 1.0E−147 | YP_001468787.1 |
| 4 | 3927 | 4865 | 312 | 34.7 | 6.5 | 57.0 | AAAGGAGctaaaacATG | Minor head protein | gp4 (Listeria phage P40) | 77/252 (30) | 303 | 4.0E−26 | YP_002261420.1 |
| 5 | 4970 | 5602 | 210 | 22.7 | 4.5 | 50.2 | AAGGAGaaacacaATG | Structural protein | gp5 (Listeria phage P40) | 65/212 (30) | 198 | 1.0E−04 | YP_002261421.1 |
| 6 | 5635 | 6528 | 297 | 32.5 | 5.2 | 45.4 | AGAAAGAGGTgctcaataATG | Major head protein | gp6 (Listeria phage P35) | 128/291 (43) | 302 | 4.0E−57 | YP_001468790.1 |
| 7 | 6588 | 7157 | 189 | 20.0 | 5.3 | 52.6 | AAGGgGGTgcacATG | — | |||||
| 8 | 7175 | 7495 | 106 | 12.3 | 4.3 | 50.5 | AGAAaAGGAcatcgaaacaATG | — | |||||
| 9 | 7586 | 8176 | 196 | 21.8 | 5.0 | 53.1 | AGAAAGGGGcttctcgcATG | Structure protein | gp7 (Listeria phage P35) | 82/202 (40) | 178 | 2.0E−27 | YP_001468791.1 |
| 10 | 8185 | 8535 | 116 | 13.0 | 5.7 | 54.7 | AGtAGGTggctgcATG | Head-tail connector | EF0342 (Enterococcus faecalis V583) | 43/120 (35) | 112 | 1.0E−09 | NP_814134.1 |
| 11 | 8535 | 8987 | 150 | 16.6 | 9.7 | 55.0 | GAGGgGGTggcgtaATG | Structural protein | gp9 (Listeria phage P40) | 64/142 (45) | 154 | 6.0E−27 | YP_002261425.1 |
| 12 | 8984 | 9424 | 146 | 16.5 | 5.2 | 51.7 | GAAAGGgctcgggGGTggcaagaaATG | Structural protein | EF0344 (Enterococcus faecalis V583) | 35/123 (28) | 127 | 2.0E−09 | NP_814136.1 |
| 13 | 9421 | 10902 | 493 | 51.6 | 4.6 | 50.0 | AGAAAGaAGGTgctcaaATG | Structural protein | gp11 (Listeria phage P35) | 118/198 (59) | 326 | 7.0E−64 | YP_001468795.1 |
| 14 | 10987 | 11598 | 203 | 22.3 | 5.8 | 48.2 | GAAAGGAcattaaaccATG | gp12 (Listeria phage P35) | 39/153 (25) | 152 | 4.0E−07 | YP_001468796.1 | |
| 15 | 11658 | 11918 | 86 | 9.8 | 4.8 | 45.6 | GGTacgccGAcgcgcgcttcATG | gp13 (Listeria phage P40) | 29/84 (34) | 87 | 8.0E−05 | YP_002261429.1 | |
| 16 | 11915 | 13987 | 690 | 73.5 | 7.1 | 49.8 | GAAAGGGGaaagcactcaATG | Tail tape measure protein | gp14 (Listeria phage P35) | 292/691 (42) | 628 | 1.0E−132 | YP_001468798.1 |
| 17 | 13987 | 15045 | 352 | 39.6 | 5.4 | 52.4 | GAAAGGAattacaatataATG | Structural protein | gp15 (Listeria phage P40) | 50/139 (35) | 428 | 3.0E−24 | YP_002261431.1 |
| 18 | 15061 | 16683 | 540 | 60.6 | 6.3 | 50.6 | GAGGaaataaacagATG | Structural protein | gp16 (Listeria phage P35) | 40/114 (35) | 387 | 2.0E−13 | YP_001468800.1 |
| 19 | 16694 | 18484 | 596 | 66.3 | 6.5 | 53.1 | AGAAtaAGGGGgcttaaATG | Structural protein | — | ||||
| 20 | 18503 | 19684 | 393 | 43.4 | 5.3 | 51.0 | AGAAAGGAcaaataaaaatATG | Structural protein | gp17 (Listeria phage P40) | 44/135 (32) | 279 | 4.0E−06 | YP_002261433.1 |
| 21 | 19750 | 20196 | 148 | 16.4 | 8.0 | 42.7 | GAAAGGcGGaaagaGTG | Holin | gp18 (Listeria phage P40) | 60/114 (52) | 151 | 3.0E−26 | YP_002261434.1 |
| 22 | 20162 | 20863 | 233 | 25.6 | 9.7 | 54.4 | AGAAAaaGAGGTacaaaATG | Endolysin | Hydrolase (Streptococcus pyogenes MGAS315) | 77/219 (35) | 254 | 1.0E−20 | NP_665110.1 |
| 23 | 21538 | 21762 | 74 | 8.6 | 9.5 | 45.3 | GAAAGGGGcacacATG | — | |||||
| 24 | 21759 | 22034 | 91 | 10.1 | 10.9 | 51.8 | GAGGTattcataaATG | — | |||||
| 25 | 22127 | 22903 | 258 | 28.7 | 4.1 | 49.8 | GGAGaaaactcaATG | — | |||||
| 26 | 22903 | 24123 | 406 | 46.7 | 7.0 | 53.2 | GAGGaagcctaATG | Hypothetical protein CLOBOL_02547 (Clostridium bolteae ATCC BAA-613) | 143/407 (35) | 391 | 4.0E−49 | ZP_02085017.1 | |
| 27 | 24136 | 25008 | 290 | 31.6 | 4.2 | 49.6 | AAAGGAGcaaatcATG | Phage protein (Clostridium botulinum NCTC-2916) | 77/188 (40) | 206 | 1.0E−16 | ZP_02614116.1 | |
| 28 | 25011 | 26972 | 653 | 72.9 | 7.1 | 52.7 | AGAAAttGAGGacgaattctaataATG | DNA polymerase family A | DNA polymerase (Clostridium cellulolyticum H10) | 293/666 (43) | 652 | 1.0E−143 | ZP_01576560.1 |
| 29 | 27051 | 29600 | 849 | 96.3 | 5.6 | 50.0 | AAAGGAGTagaaaatcaATG | Primase/helicase | gp39 (Staphylococcus phage tp310-2) | 230/771 (29) | 815 | 3.0E−84 | YP_001429934.1 |
| 30 | 29756 | 31153 | 465 | 50.1 | 9.5 | 51.3 | AAAGGAGTagaaagccaATG | — | |||||
| 31 | 31426 | 31554 | 42 | 4.8 | 4.4 | 41.9 | AAAGGAGcgaatcaaaATG | — | |||||
| 32 | 31664 | 32071 | 135 | 14.7 | 9.0 | 52.9 | AGAAAGcAGGTagaaccATG | — | |||||
| 33 | 32056 | 32319 | 87 | 9.8 | 9.1 | 51.9 | AAAGAGGAcgcggacgcATG | — | |||||
| 34 | 32316 | 32492 | 58 | 6.6 | 10.0 | 56.5 | GAGGTaattttagATG | — | |||||
| 35 | 32492 | 32686 | 64 | 7.2 | 10.0 | 51.3 | AAAGGAGaatcggaaataATG | — | |||||
| 36 | 32683 | 32925 | 80 | 9.2 | 6.7 | 47.7 | GAGGTatgaaATG | — | |||||
| 37 | 32925 | 33152 | 75 | 8.3 | 4.2 | 50.0 | AAAGGGTtgggaataATG | — | |||||
| 38 | 33152 | 33343 | 63 | 7.3 | 6.0 | 50.0 | GAGGgcgactaATG | — | |||||
| 39 | 33345 | 33569 | 74 | 8.7 | 9.3 | 51.1 | AGAAAGAGGTggtaaaATG | — | |||||
| 40 | 33566 | 33868 | 100 | 11.0 | 5.7 | 47.5 | AGGAGataagaaaatcATG | — | |||||
| 41 | 33865 | 34677 | 270 | 30.4 | 5.0 | 51.5 | AAAGGGGcagaccgATG | SAI_0594 (Streptococcus agalactiae H36B) | 32/73 (43) | 104 | 2.0E−07 | ZP_00782379.1 | |
| 42 | 34667 | 35008 | 113 | 12.7 | 9.7 | 54.7 | AGGGGgcaaaaaATG | VRR-NUC domain | Bbp43 (Bordetella phage BPP-1) | 41/86 (47) | 87 | 2.0E−13 | NP_958712.1 |
| 43 | 35005 | 36738 | 577 | 66.4 | 6.5 | 51.1 | AGAAAGGGcataaaggactATG | Helicase | SNF2-related (C. cellulolyticum H10) | 188/539 (34) | 457 | 5.0E−71 | ZP_01576554.1 |
RBS, ribosome-binding site (AGAAAGGAGGT). Underlining indicates nucleotides identical to the RBS consensus; lowercase type indicates spacer nucleotides between the RBS and the start codon; boldface type indicates the start codon.
Conserved domains were found at the NCBI database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), within the CDD database, with an E value of 0.01. Boldface type indicates that the protein was found in the virion structure.
— indicates no hit or no significant match.
pI and MM values were taken from the ExPASy website under Compute pI/MM (http://ca.expasy.org/tools/pi_tool.html). aa, amino acids.
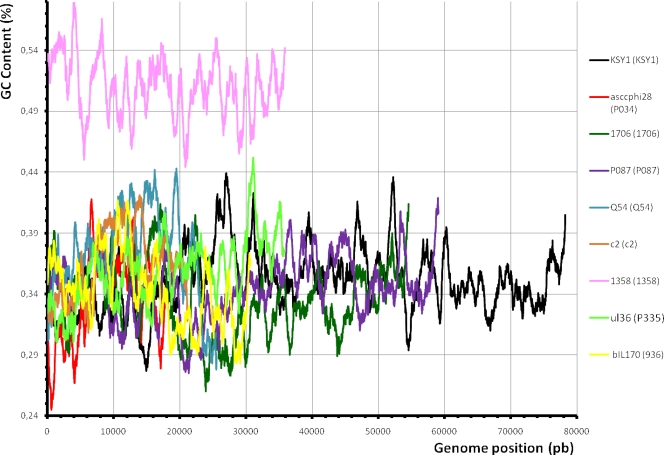
One of the most interesting features of this genome is its GC content, which was calculated to be 51.04%. This GC content is much higher than those of its L. lactis hosts (35.3%) (10, 46, 67) and all other characterized lactococcal phages (28). The lowest reported GC content for a lactococcal phage genome was 33% for phages such as phages 1706 (23) and asscφ28 (39), while the highest was 37% for KSY1 (13). Thus, the GC content of the phage 1358 genome is significantly higher, and it is not due to a specific genetic module, since the high GC content was found throughout the genomic sequence (Fig. 2). The lowest- and the highest-GC-content regions have GC contents of 44.4% and 57.9%, respectively.
FIG. 2.
Comparison of the GC content of phage 1358 with that of other lactococcal phages. The GC contents of these phages are in relation to their genome positions. pb, base pairs.
This unusual GC content is also reflected in the codon usage of phage 1358. Compared to the three bacterial strains of L. lactis for which the complete genome sequence is available, phage 1358 uses the optimal codon for only seven amino acids, including the two unique codons for methionine and tryptophan (Table 2). For comparative purposes, the codon usage was also determined for lactococcal phages from other groups. They use between 13 and 15 optimal codons (Table 2). Globally, a bias for GC-rich codons was observed for the genome of lactococcal phage 1358. This somewhat incompatible codon usage may also explain the long latent period of this lactococcal phage. Previous codon usage analyses revealed that many phages exhibit codon bias (47, 55, 56) and at different degrees across the genome (45). In these cases, this difference could be explained by the mosaic structure of some genomes that include genetic elements derived from phages infecting various hosts (29).
TABLE 2.
Analysis of the codon usage of L. lactis and L. monocytogenes bacterium and phage strains
| Amino acid | Codon | % Codon usage |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. lactis host strainsa | L. monocytogenes host strainsb | L. monocytogenes phage P35 |
L. lactis phage |
||||||||||
| 1358 | bIL170 | Q54 | P335 | P087 | asccφ28 | c2 | 1706 | KSY1 | |||||
| Ala | GCG | 11.0 | 21.7 | 21.0 | 29.5 | 7.7 | 15.4 | 12.4 | 2.2 | 20.5 | 10.5 | 2.8 | 1.9 |
| GCA | 31.6 | 36.4 | 36.5 | 37.6 | 37.9 | 33.6 | 37.4 | 34.4 | 36.4 | 37.3 | 37.6 | 46.6 | |
| GCT | 41.5 | 31.0 | 32.2 | 13.1 | 47.8 | 36.4 | 39.1 | 56.6 | 33.9 | 41.4 | 52.5 | 47.4 | |
| GCC | 16.0 | 10.9 | 10.3 | 19.8 | 6.6 | 14.7 | 11.0 | 6.8 | 9.3 | 11.0 | 7.2 | 4.2 | |
| Arg | AGG | 4.1 | 3.6 | 3.8 | 4.2 | 10.5 | 5.4 | 5.8 | 3.3 | 5.5 | 10.3 | 4.8 | 5.2 |
| AGA | 22.2 | 21.6 | 24.6 | 7.8 | 45.4 | 25.8 | 32.9 | 26.7 | 30.2 | 35.0 | 36.1 | 46.3 | |
| CGG | 6.1 | 7.8 | 7.3 | 11.8 | 3.8 | 6.7 | 4.7 | 2.6 | 11.0 | 1.7 | 1.2 | 2.8 | |
| CGA | 15.4 | 15.0 | 12.2 | 14.5 | 11.6 | 15.6 | 18.8 | 13.3 | 11.5 | 9.9 | 10.8 | 11.0 | |
| CGT | 40.5 | 33.5 | 33.5 | 16.5 | 19.8 | 27.4 | 29.6 | 48.6 | 32.4 | 33.3 | 42.6 | 31.3 | |
| CGC | 11.8 | 18.5 | 16.8 | 45.3 | 9.0 | 19.1 | 8.3 | 5.5 | 9.3 | 9.9 | 4.6 | 3.5 | |
| Asn | AAT | 79.0 | 68.4 | 67.9 | 32.9 | 62.5 | 52.0 | 70.5 | 67.5 | 75.3 | 46.4 | 77.6 | 64.3 |
| AAC | 21.0 | 31.6 | 32.1 | 67.2 | 37.5 | 48.0 | 29.5 | 32.5 | 24.7 | 53.7 | 22.4 | 35.7 | |
| Asp | GAT | 72.7 | 73.4 | 73.2 | 14.2 | 54.3 | 47.6 | 71.0 | 63.6 | 66.8 | 47.8 | 77.7 | 62.2 |
| GAC | 27.3 | 26.6 | 26.8 | 85.8 | 45.7 | 52.4 | 29.0 | 36.4 | 33.2 | 52.2 | 22.3 | 37.8 | |
| Cys | TGT | 77.3 | 63.9 | 60.5 | 27.4 | 66.1 | 73.3 | 64.2 | 84.4 | 63.4 | 73.3 | 77.6 | 74.1 |
| TGC | 22.7 | 36.1 | 39.5 | 72.6 | 33.9 | 26.7 | 35.9 | 15.6 | 36.6 | 26.7 | 22.4 | 25.9 | |
| Gln | CAG | 16.5 | 15.4 | 15.7 | 36.0 | 20.4 | 17.4 | 19.7 | 11.8 | 9.3 | 17.4 | 11.3 | 12.5 |
| CAA | 83.5 | 84.6 | 84.3 | 64.0 | 79.6 | 82.6 | 80.3 | 88.2 | 90.7 | 82.6 | 88.7 | 87.5 | |
| Glu | GAG | 17.9 | 18.1 | 18.5 | 38.0 | 21.2 | 19.0 | 21.5 | 18.2 | 20.0 | 26.2 | 17.4 | 16.8 |
| GAA | 82.1 | 81.9 | 81.5 | 62.0 | 78.8 | 81.0 | 78.5 | 81.8 | 80.0 | 73.8 | 82.6 | 83.2 | |
| Gly | GGG | 12.4 | 13.3 | 13.1 | 20.1 | 8.8 | 21.9 | 10.7 | 3.3 | 19.6 | 15.3 | 5.0 | 5.9 |
| GGA | 37.8 | 29.4 | 30.1 | 15.0 | 33.8 | 34.8 | 40.2 | 29.3 | 37.3 | 27.6 | 29.7 | 31.0 | |
| GGT | 37.0 | 35.8 | 36.0 | 14.2 | 40.4 | 26.4 | 34.2 | 62.9 | 28.6 | 43.1 | 55.8 | 49.4 | |
| GGC | 12.6 | 21.5 | 20.9 | 50.7 | 17.0 | 16.9 | 14.9 | 4.5 | 14.5 | 14.0 | 9.5 | 13.7 | |
| His | CAT | 74.7 | 70.4 | 70.7 | 28.0 | 70.2 | 46.5 | 66.7 | 70.1 | 75.7 | 55.1 | 79.6 | 74.7 |
| CAC | 25.3 | 29.7 | 29.3 | 72.0 | 29.8 | 53.5 | 33.3 | 29.9 | 24.3 | 44.9 | 20.4 | 25.3 | |
| Ile | ATA | 11.3 | 13.2 | 14.5 | 19.5 | 25.1 | 27.7 | 19.3 | 13.6 | 17.0 | 22.1 | 22.6 | 20.4 |
| ATT | 68.2 | 62.2 | 59.8 | 29.5 | 56.9 | 57.6 | 59.9 | 61.1 | 68.0 | 55.5 | 59.3 | 56.5 | |
| ATC | 20.5 | 24.6 | 25.7 | 51.0 | 18.0 | 14.6 | 20.8 | 25.4 | 15.1 | 22.5 | 18.2 | 23.1 | |
| Leu | TTG | 21.3 | 13.2 | 13.1 | 14.8 | 16.7 | 13.3 | 19.8 | 17.0 | 14.6 | 22.0 | 12.6 | 16.1 |
| TTA | 31.6 | 39.4 | 39.8 | 15.5 | 38.7 | 47.1 | 36.1 | 38.0 | 46.2 | 36.7 | 41.3 | 31.9 | |
| CTG | 5.9 | 5.1 | 4.8 | 9.8 | 4.9 | 1.4 | 4.8 | 2.8 | 2.7 | 3.3 | 1.3 | 3.6 | |
| CTA | 7.7 | 14.0 | 14.3 | 10.0 | 14.2 | 14.5 | 9.4 | 11.2 | 9.7 | 17.1 | 12.9 | 14.8 | |
| CTT | 25.6 | 22.3 | 22.2 | 11.8 | 23.8 | 20.7 | 25.1 | 26.4 | 21.2 | 17.2 | 27.4 | 29.8 | |
| CTC | 7.9 | 6.0 | 6.0 | 38.2 | 1.7 | 3.1 | 4.9 | 4.6 | 5.8 | 3.7 | 4.5 | 3.8 | |
| Lys | AAG | 17.2 | 14.3 | 13.9 | 36.0 | 23.6 | 20.2 | 22.3 | 19.8 | 18.0 | 20.4 | 22.4 | 26.5 |
| AAA | 82.8 | 85.7 | 86.1 | 64.0 | 76.4 | 79.8 | 77.7 | 80.2 | 82.0 | 79.6 | 77.6 | 73.5 | |
| Phe | TTT | 75.5 | 67.6 | 66.9 | 30.0 | 69.1 | 65.4 | 73.8 | 64.1 | 83.7 | 64.0 | 71.1 | 61.5 |
| TTC | 24.5 | 32.5 | 33.1 | 70.0 | 30.9 | 34.6 | 26.2 | 35.9 | 16.3 | 36.0 | 28.9 | 38.5 | |
| Pro | CCG | 8.7 | 19.9 | 18.9 | 41.0 | 8.6 | 28.1 | 12.2 | 2.0 | 10.7 | 11.4 | 2.8 | 2.2 |
| CCA | 46.5 | 52.0 | 52.5 | 12.8 | 38.3 | 30.7 | 47.0 | 52.9 | 22.0 | 32.3 | 53.4 | 58.2 | |
| CCT | 36.4 | 23.2 | 23.6 | 15.1 | 49.6 | 32.0 | 33.0 | 42.3 | 53.1 | 51.5 | 41.5 | 37.4 | |
| CCC | 8.4 | 5.0 | 5.0 | 31.0 | 3.6 | 9.2 | 7.9 | 2.8 | 14.1 | 4.8 | 2.3 | 2.2 | |
| Ser | AGT | 22.3 | 23.8 | 23.7 | 6.1 | 26.1 | 27.2 | 23.8 | 26.4 | 22.0 | 30.6 | 22.0 | 21.9 |
| AGC | 9.0 | 15.4 | 16.0 | 33.3 | 17.4 | 24.1 | 14.2 | 4.4 | 6.7 | 20.3 | 6.2 | 7.6 | |
| TCG | 5.6 | 11.0 | 11.2 | 26.7 | 4.2 | 7.7 | 5.2 | 3.6 | 5.6 | 4.1 | 2.2 | 2.0 | |
| TCA | 33.0 | 17.4 | 17.7 | 19.2 | 32.6 | 22.6 | 31.0 | 33.8 | 35.7 | 25.3 | 35.2 | 33.6 | |
| TCT | 25.4 | 21.5 | 20.7 | 10.4 | 18.6 | 15.0 | 21.5 | 27.9 | 26.5 | 18.7 | 31.6 | 33.2 | |
| TCC | 4.7 | 11.0 | 10.7 | 4.4 | 1.1 | 3.3 | 4.2 | 3.9 | 3.6 | 1.0 | 2.8 | 1.7 | |
| Thr | ACG | 11.8 | 20.9 | 20.1 | 33.5 | 9.9 | 11.2 | 10.9 | 4.6 | 16.5 | 11.8 | 4.8 | 2.7 |
| ACA | 39.2 | 41.9 | 42.9 | 35.4 | 46.8 | 45.1 | 39.0 | 40.8 | 40.9 | 44.9 | 44.8 | 45.9 | |
| ACT | 36.5 | 26.0 | 26.4 | 12.9 | 37.5 | 32.5 | 39.6 | 45.7 | 29.1 | 37.3 | 46.1 | 47.0 | |
| ACC | 12.5 | 11.2 | 10.6 | 19.3 | 5.7 | 11.2 | 10.5 | 8.9 | 13.5 | 6.0 | 4.2 | 4.4 | |
| Tyr | TAT | 78.2 | 68.0 | 67.4 | 45.3 | 73.0 | 65.8 | 75.4 | 75.7 | 78.9 | 67.0 | 78.8 | 72.3 |
| TAC | 21.8 | 32.0 | 32.6 | 54.7 | 27.0 | 34.2 | 24.6 | 24.3 | 21.1 | 33.0 | 21.2 | 27.7 | |
| Val | GTG | 13.5 | 19.7 | 20.0 | 20.7 | 7.5 | 7.0 | 13.0 | 6.0 | 12.4 | 11.0 | 6.7 | 7.1 |
| GTA | 19.7 | 30.9 | 31.7 | 18.4 | 40.3 | 44.4 | 24.8 | 40.0 | 20.4 | 37.2 | 37.8 | 53.0 | |
| GTT | 48.5 | 37.0 | 36.5 | 14.9 | 39.8 | 39.6 | 47.9 | 47.2 | 50.7 | 40.6 | 48.2 | 30.0 | |
| GTC | 18.3 | 12.4 | 11.8 | 46.0 | 12.4 | 9.1 | 14.4 | 6.8 | 16.5 | 11.3 | 7.3 | 9.6 | |
| Stop | TGA | 19.0 | 18.7 | 19.7 | 27.9 | 25.0 | 27.7 | 26.5 | 21.6 | 14.3 | 30.8 | 18.4 | 21.4 |
| TAG | 12.0 | 9.9 | 8.0 | 16.3 | 9.4 | 10.6 | 20.4 | 14.8 | 21.4 | 12.8 | 18.4 | 18.3 | |
| TAA | 69.0 | 71.4 | 72.2 | 55.8 | 65.6 | 61.7 | 53.1 | 63.6 | 64.3 | 56.4 | 63.2 | 60.3 | |
These data represent the means for three L. lactis strains.
These data represent the means for two L. monocytogenes strains.
Genes and gene products.
A total of 43 open reading frames (ORFs) longer than 40 codons were predicted (Table 1) from the genome sequence. Only orf12 and orf15 were not preceded by a putative ribosomal binding site, and only orf21 had an alternative start codon (GTG). All genes were on the same DNA strand, all had the same orientation, and 19 gene sequences overlapped. The sizes of the gene products varied from 42 amino acids (orf31) to 849 amino acids (orf29). The coding sequence represented approximately 93% of the complete genome sequence, which is typical of most phages (4). The longest noncoding region was located between orf22 and orf23 (674 bp).
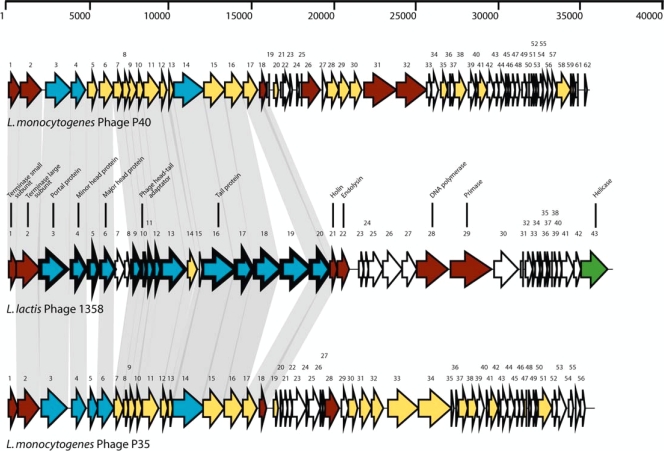
The genome organization of phage 1358 was typical of lactococcal phages with two expected gene clusters, early- and late-expressed genes. The late-expressed gene region was the more discernible, and it contained genes coding for proteins involved in packaging (orf1 and orf2), morphogenesis (orf3 to orf20), and lysis (orf21 and orf22) (Fig. 3). In agreement with the virulent nature of phage 1358, no lysogeny module was found in the genome.
FIG. 3.
Genomic organizations of virulent lactococcal phage 1358 and L. monocytogenes phages P40 and P35. The scale above the map is in base pairs. Each arrow represents a putative ORF, and the numbering refers to Table 1. The putative functions inferred from bioinformatic or structural analyses are indicated above the ORFs. For the phage 1358 genome, arrows with thick outlines represent gene products detected by LC-MS/MS analyses. Gray shadows linking ORFs with the same color indicate more than 23% amino acid identity. White arrows represent ORFs for which no putative function can be attributed. Red arrows represent ORFs sharing identity with at least one phage shown here, and green arrows represent ORFs that do not share identity. Blue arrows represent ORFs sharing identity with proteins for which a function was attributed.
Bioinformatic analyses.
Comparative analyses with sequences in public databases such as GenBank (5) or ACLAME (43) revealed that 17 of the ORFs (39.5%) of phage 1358 had no significant matches, confirming that the phage gene pool is still largely unexplored. Most of the unknown phage proteins are likely encoded by early-expressed genes (Table 1). Conversely, similarities were found between deduced ORFs of phage 1358 and gene products from phages infecting the food-borne pathogen Listeria monocytogenes, particularly phages P40 and P35 (Table 1 and Fig. 3). In total, 15 of phage 1358's 43 ORFs (34.9%) were best matched with proteins of either Listeria phage P35 or P40 (Table 1). All of these Listeria-related phage proteins (ORF2 to ORF6, ORF9, ORF11, ORF13 to ORF18, ORF20, and ORF21) were predicted to be structural proteins or to be involved in cell lysis (ORF21/holin) (Table 1 and Fig. 3). The amino acid identities were between 25% and 49%. Four of the phage 1358 ORFs (ORF26, ORF27, ORF28, and ORF43) shared identity (35 to 43%) with proteins found in Clostridium, two ORFs (ORF10 and ORF12) shared identity (35% and 28%, respectively) with hypothetical proteins of Enterococcus, and two ORFs (ORF22/endolysin and ORF41) shared identity (35% and 43%, respectively) with streptococcal proteins.
Although the matches were not highly significant, the gene products resulting from the expression of the clusters orf26 to orf29 and orf42 and orf43 shared similarity with deduced proteins found in bacterial strain L. monocytogenes HCC23 (GenBank accession number NC_011660) (data not shown). These phage 1358-associated genes in L. monocytogenes HCC23 were contiguous and corresponded to the locus tags LMHCC_2574 to LMHCC_2576 and LMHCC_2586 to LMHCC_2588. Some of these ORFs were related to conserved phage-associated proteins. Taken together, these data suggest that lactococcal phage 1358 shares similarity with Listeria phages. Also of note, no best matches occurred with lactococcal proteins, confirming the uniqueness of this L. lactis phage. Only ORF13 (structural protein), ORF16 (putative tape measure protein), and ORF22 (endolysin) could be associated with lactococcal phage gene products, from phages KSY1 (gp055), bIL285 (gp52), and asccφ28 (gp12), respectively.
Structural proteome of phage 1358.
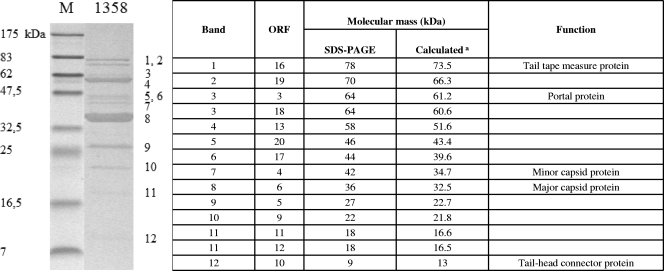
The structural proteome of phage 1358 was characterized by SDS-PAGE coupled with LC-MS/MS analysis. A total of 12 protein bands were analyzed, and two of them were found to contain two structural proteins (bands 3 and 11) (Fig. 4). Thus, a total of 14 structural proteins could be linked to a deduced proteome of phage 1358. The genes coding for these proteins (orf3 to orf6, orf9 to orf13, and orf16 to orf20) were clustered within a genomic region considered to be the morphogenesis module, as determined by bioinformatic analysis. Although the functions for the proteins encoded by orf7 and orf8 are not clear, the orf14 and orf15 gene products are likely tail chaperone proteins (59) and, thus, not found as part of the virion structure.
FIG. 4.
LC-MS/MS analysis of phage 1358 structural proteins. (Left) Coomassie blue staining of a 12% SDS-polyacrylamide gel showing phage 1358 structural proteins. Letters on the right indicate bands cut out of the gel and identified by LC-MS/MS. The sizes (in kDa) of the proteins in the broad-range molecular mass standard (M) are indicated on the left. (Right) Identification of phage 1358 proteins from corresponding bands shown in the left panel. Numbers at the right correspond to the numbers indicated in the left panel. aCalculated from the gene sequence.
Band 8 was the principal structural protein of phage 1358 and was associated with orf6. Considering the position of orf6 in the phage 1358 genome and the concentration of ORF6 in the phage structure, it is likely to be the major capsid protein. Analogous observations coupled to conserved domains suggested that ORF4 is a minor capsid protein. ORF16 is likely the tape measure protein based on its size and the location of the gene. The MMs of structural proteins estimated by SDS-PAGE were in agreement with the calculated masses from the corresponding gene sequences. For all phage 1358 structural proteins except orf19, corresponding deduced proteins could be found in L. monocytogenes phages P35 and P40.
Origin of lactococcal phage 1358.
The structural proteins of virulent lactococcal phage 1358 bear considerable similarity to those of two L. monocytogenes phages. Based on amino acid sequence similarities and the restricted number of phage 1358 genes associated with lactococcal phages, it is tempting to speculate that phage 1358 is derived, at least in part, from a phage infecting another low-GC Gram-positive bacterium, namely, Listeria. Although not identified by bioinformatic analysis, it is likely that phage 1358 acquired genes that allow it to recognize L. lactis hosts. Alternatively, the Listeria phages noted above may be derived from a lactococcal phage. We tested whether lactococcal phage 1358 could infect eight Listeria strains, including the host of Listeria phage P35 (HER1247). We also tested whether Listeria phage P35 could infect the host of phage 1358, Lactococcus lactis SMQ-388. No plaques were observed in these assays.
L. monocytogenes is found in environments such as soil, water, sewage, silage, farms, animals, humans, and foods including milk and cheeses (20, 27, 32, 37, 65, 66, 68). Listeria phages are also found in similar environments (38, 44). For example, phage P35 was isolated from silage (30). Consequently, it is possible that L. monocytogenes phages have been in contact with L. lactis phages, leading to genetic exchange.
Moreover, it was previously shown that Listeria genes can be expressed and proteins can be produced in Lactococcus (6, 31, 61). It is not the first time that a phage infecting a generally-recognized-as-safe (GRAS) bacterium (L. lactis) shared structural similarities with phages infecting pathogenic bacteria (20, 62). The recently described rare virulent lactococcal phage P087 had structural relationships with an E. faecalis prophage (63).
The most striking finding of the analysis of phage 1358 was the high GC content (51%) throughout the genome, which was much higher than those of other lactococcal phages and hosts (10, 46, 67) as well as Listeria phages P35 (40%) and P40 (39%) and hosts (24). Moreover, this unusual GC content was reflected in the codon usage. Since GC content is sometimes used to classify organisms, this uniformly higher GC content suggests that phage 1358 may not be derived directly from a Listeria phage, as suggested by data from the proteomic analysis. Instead, they may share an unknown ancestor.
In that regard, a recent study of six Listeria phages clearly showed that virulent phages P35 and P40 also form a distinct group among listerial phages, as they were clustered in a separate branch of a phylogenetic tree (17). Among others, they differ in genome size and organization and have a rather broad host range and a higher GC content (17). About half of their deduced proteins are without a significant match in the databases, while some ORFs shared similarities with proteins found in Clostridium and Enterococcus. Thus, similarly to lactococcal phage 1358, Listeria phages P35 and P40 are rather unique.
We have characterized the reference member of the 9th lactococcal phage group, phage 1358. As illustrated by its long latent period, phage 1358 is not particularly well adapted to proliferate in rapid industrial fermentation processes, probably due to its high GC content. As far as we are aware, this rare group of lactococcal phages is limited to two members, both isolated in New Zealand. Bioinformatic analyses suggest that phage 1358 likely infected another bacterial genus or species before infecting L. lactis and has not subsequently experienced enough time to equilibrate its GC content to that of its current host (49). It remains unclear why these rare phages have not evolved to thrive in industrial fermentations. As other phage genomes become available, additional links may be made that could further explain the origin of phage 1358.
Acknowledgments
We thank Barbara-Ann Conway for editorial assistance. We are grateful to Hélène Deveau, Geneviève Rousseau, Denise Tremblay, and Manuela Villion for helpful discussion.
Marie-Ève Dupuis was the recipient of a Natural Sciences and Engineering Research Council (NSERC) undergraduate student research award and a FQRNT/Novalait graduate scholarship. This work was funded by a strategic grant from NSERC of Canada.
Footnotes
Published ahead of print on 8 January 2010.
REFERENCES
- 1.Ackermann, H. W. 1998. Tailed bacteriophages: the order Caudovirales. Adv. Virus Res. 51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann, H.-W., G. Brochu, and H. P. E. Konjin. 1994. Classification of Acinetobacter phages. Arch. Virol. 135:345-354. [DOI] [PubMed] [Google Scholar]
- 3.Ackermann, H.-W., and A. M. Kropinski. 2007. Curated list of prokaryote viruses with fully sequenced genomes. Res. Microbiol. 158:555-566. [DOI] [PubMed] [Google Scholar]
- 4.Allison, G. E., and T. R. Klaenhammer. 1998. Phage resistance mechanisms in lactic acid bacteria. Int. Dairy J. 8:207-226. [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Millerand, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahey-El-Din, M., P. G. Casey, B. T. Griffin, and C. G. Gahan. 2008. Lactococcus lactis-expressing listeriolysin O (LLO) provides protection and specific CD8(+) T cells against Listeria monocytogenes in the murine infection model. Vaccine 26:5304-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bissonnette, F., S. Labrie, H. Deveau, M. Lamoureux, and S. Moineau. 2000. Characterization of mesophilic mixed starter cultures used for the manufacture of aged cheddar cheese. J. Dairy Sci. 83:620-627. [DOI] [PubMed] [Google Scholar]
- 9.Black, L. W. 1989. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43:267-292. [DOI] [PubMed] [Google Scholar]
- 10.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouchard, J. D., É. Dion, F. Bissonnette, and S. Moineau. 2002. Characterization of the two-component abortive phage infection mechanism AbiT from Lactococcus lactis. J. Bacteriol. 184:6325-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casjens, S., L. Sampson, S. Randall, K. Eppler, H. Wu, J. B. Petri, and H. Schmieger. 1992. Molecular genetic analysis of bacteriophage P22 gene 2 product, a protein involved in the initiation of heedful DNA packaging. J. Mol. Biol. 227:1086-1099. [DOI] [PubMed] [Google Scholar]
- 13.Chopin, A., H. Deveau, S. D. Ehrlich, S. Moineau, and M.-C. Chopin. 2007. KSY1, a lactococcal phage with a T7-like transcription. Virology 365:1-9. [DOI] [PubMed] [Google Scholar]
- 14.Deveau, H., S. J. Labrie, M.-C. Chopin, and S. Moineau. 2006. Biodiversity and classification of lactococcal phages. Appl. Environ. Microbiol. 72:4338-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deveau, H., M. R. van Calsteren, and S. Moineau. 2002. Effect of exopolysaccharides on phage-host interactions in Lactococcus lactis. Appl. Environ. Microbiol. 68:4364-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeVos, W. M. 1987. Gene cloning and expression in lactic streptococci. FEMS Microbiol. Rev. 46:281-295. [Google Scholar]
- 17.Dorscht, J., J. Klumpp, R. Bielmann, M. Schmelcher, Y. Born, M. Zimmer, R. Calendar, and M. J. Loessner. 2009. Comparative genome analysis of Listeria bacteriophages reveals extensive mosaicism, programmed translational frameshifting and a novel prophage insertion site. J. Bacteriol. 191:7206-7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Émond, É., É. Dion, S. A. Walker, E. R. Vedamuthu, J. K. Kondo, and S. Moineau. 1998. AbiQ, an abortive infection mechanism from Lactococcus lactis. Appl. Environ. Microbiol. 64:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Émond, É., B. J. Holler, I. Boucher, P. A. Vandenbergh, E. R. Vedamuthu, J. K. Kondo, and S. Moineau. 1997. Phenotypic and genetic characterization of the bacteriophage abortive infection mechanism AbiK from Lactococcus lactis. Appl. Environ. Microbiol. 63:1274-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsman, P. 1993. Characterization of a prolate-headed bacteriophage of Lactobacillus delbrueckii subsp. lactis, and its DNA homology with isometric-headed phages. Arch. Virol. 132:321-330. [DOI] [PubMed] [Google Scholar]
- 22.Fortier, L.-C., A. Bransi, and S. Moineau. 2006. Genome sequence and global gene expression of Q54, a new phage species linking the 936 and c2 phage species of Lactococcus lactis. J. Bacteriol. 188:6101-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garneau, J. E., D. M. Tremblay, and S. Moineau. 2008. Characterization of 1706, a virulent phage from Lactococcus lactis with similarities to prophages from other Firmicutes. Virology 373:298-309. [DOI] [PubMed] [Google Scholar]
- 24.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvé, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. García-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gómez-López, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vázquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 25.Haaber, J., S. Moineau, L.-C. Fortier, and K. Hammer. 2008. AbiV, a novel abortive phage infection mechanism on the chromosome of Lactococcus lactis subsp. cremoris MG1363. Appl. Environ. Microbiol. 74:6528-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 27.Hayes, P. S., J. C. Feeley, L. M. Graves, G. W. Ajello, and D. W. Fleming. 1986. Isolation of Listeria monocytogenes from raw milk. Appl. Environ. Microbiol. 51:438-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hejnowicz, M. S., M. Golebiewski, and J. Bardowski. 2009. Analysis of the complete genome sequence of the lactococcal bacteriophage bIBB29. Int. J. Food Microbiol. 131:52-61. [DOI] [PubMed] [Google Scholar]
- 29.Hendrix, R. W. 2002. Bacteriophages: evolution of the majority. Theor. Popul. Biol. 61:371-480. [DOI] [PubMed] [Google Scholar]
- 30.Hodgson, D. A. 2000. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 35:312-323. [DOI] [PubMed] [Google Scholar]
- 31.Innocentin, S., V. Guimarães, A. Miyoshi, V. Azevedo, P. Langella, J. M. Chatel, and F. Lefèvre. 2009. Lactococcus lactis expressing either Staphylococcus aureus fibronectin-binding protein A or Listeria monocytogenes internalin A can efficiently internalize and deliver DNA in human epithelial cells. Appl. Environ. Microbiol. 75:4870-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivanek, R., Y. T. Gröhn, and M. Wiedmann. 2006. Listeria monocytogenes in multiple habitats and host populations: review of available data for mathematical modeling. Foodborne Pathog. Dis. 3:319-336. [DOI] [PubMed] [Google Scholar]
- 33.Jarvis, A. W. 1984. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl. Environ. Microbiol. 47:343-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarvis, A. W. 1987. Sources of lactic streptococcal phages in cheese plants. N. Z. J. Dairy Sci. Technol. 22:93-103. [Google Scholar]
- 35.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 36.Josephsen, J., N. Andersen, H. Behrndt, E. Brandsborg, G. Christiansen, M. B. Hansen, S. Hansen, E. W. Nielsen, and F. K. Vogensen. 1994. An ecological study of lytic bacteriophages of Lactococcus lactis subsp. cremoris isolated in a cheese plant over a five year period. Int. Dairy J. 4:123-140. [Google Scholar]
- 37.Kathariou, S. 2002. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 65:1811-1829. [DOI] [PubMed] [Google Scholar]
- 38.Kim, J. W., R. M. Siletzky, and S. Kathariou. 2008. Host ranges of Listeria-specific bacteriophages from the turkey processing plant environment in the United States. Appl. Environ. Microbiol. 74:6623-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotsonis, S. E., I. B. Powell, C. J. Pillidge, G. K. Limsowtin, A. J. Hillier, and B. E. Davidson. 2008. Characterization and genomic analysis of phage asccphi28, a phage of the family Podoviridae infecting Lactococcus lactis. Appl. Environ. Microbiol. 74:3453-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labrie, S., J. Josephsen, H. Neve, F. K. Vogensen, and S. Moineau. 2008. Morphology, genome sequence, and structural proteome of the type phage P335 from Lactococcus lactis. Appl. Environ. Microbiol. 74:4636-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labrie, S., and S. Moineau. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 43.Leplae, R., A. Hebrant, S. J. Wodak, and A. Toussaint. 2004. ACLAME: a classification of mobile genetic elements. Nucleic Acids Res. 32:D45-D49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loessner, M. J., and C. E. D. Rees. 2005. Listeria phages: basics and applications, p. 362-379. In M. K. Waldor, D. I. Friedman, and S. L. Adhya (ed.), Phages: their role in bacterial pathogenesis and biotechnology, 1st ed. ASM Press, Washington, DC.
- 45.Lucks, J. B., D. R. Nelson, G. R. Kudla, and J. B. Plotkin. 2008. Genome landscapes and bacteriophage codon usage. PLoS Comput. Biol. 4:e1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McEwan, N. R. 2005. Codon utilization, DNA landscaping and fractal analysis in bacteriophage phi(adh). Acta Virol. 49:169-176. [PubMed] [Google Scholar]
- 48.Moineau, S., M. Borkaev, B. J. Holler, S. A. Walker, J. K. Kondo, E. R. Vedamuthu, and P. A. Vandenbergh. 1996. Isolation and characterization of lactococcal phages from U.S. buttermilk plants. J. Dairy Sci. 79:2104-2111. [Google Scholar]
- 49.Moineau, S., E. Durmaz, S. Pandian, and T. Klaenhammer. 1993. Differentiation of two abortive mechanisms by using monoclonal antibodies directed toward lactococcal bacteriophage capsid proteins. Appl. Environ. Microbiol. 59:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moineau, S., J. Fortier, H.-W. Ackermann, and S. Pandian. 1992. Characterization of lactococcal bacteriophages from Quebec cheese plants. Can. J. Microbiol. 38:875-882. [Google Scholar]
- 51.Moineau, S., and C. Lévesque. 2005. Control of bacteriophages in industrial fermentation, p. 286-296. In E. Kutter and A. Sulakvelidze (ed.), Bacteriophages: biology and application. CRC Press, Boca Raton, FL.
- 52.Raiski, A., and N. Belyasova. 2009. Biodiversity of Lactococcus lactis bacteriophages in the Republic of Belarus. Int. J. Food Microbiol. 130:1-5. [DOI] [PubMed] [Google Scholar]
- 53.Sally, E. P., and G. Stanley. 1978. Partial characterization of a bacteriophage of Lactobacillus bulgaricus isolated from yoghurt. J. Appl. Bacteriol. 44:321-323. [Google Scholar]
- 54.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 55.Sau, K., and A. Deb. 2009. Temperature influences synonymous codon and amino acid usage biases in the phages infecting extremely thermophilic prokaryotes. In Silico Biol. 9:1-9. [PubMed] [Google Scholar]
- 56.Sau, K., S. K. Gupta, S. Sau, and T. C. Ghosh. 2005. Synonymous codon usage bias in 16 Staphylococcus aureus phages: implication in phage therapy. Virus Res. 113:123-131. [DOI] [PubMed] [Google Scholar]
- 57.Schouler, C., S. D. Ehrlich, and M.-C. Chopin. 1994. Sequence and organization of the lactococcal prolate-headed bIL67 phage genome. Microbiology 140:3061-3069. [DOI] [PubMed] [Google Scholar]
- 58.Schwudke, D., A. Ergin, K. Michael, S. Volkmar, B. Appel, D. Knaber, A. Konietzny, and E. Strauch. 2008. Broad-host-range Yersinia phage PY100: genome sequence, proteome analysis of virions, and DNA packaging strategy. J. Bacteriol. 190:332-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siponen, M., G. Sciara, M. Villion, S. Spinelli, J. Lichière, C. Cambillau, S. Moineau, and V. Campanacci. 2009. Crystal structure of ORF12 from Lactococcus lactis phage p2 identifies a tape measure protein chaperone. J. Bacteriol. 191:728-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Turner, M. S., F. Waldherr, M. J. Loessner, and P. M. Giffard. 2007. Antimicrobial activity of lysostaphin and a Listeria monocytogenes bacteriophage endolysin produced and secreted by lactic acid bacteria. Syst. Appl. Microbiol. 30:58-67. [DOI] [PubMed] [Google Scholar]
- 62.Vázquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Domínguez-Bernal, W. Goebel, B. González-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Villion, M., M.-C. Chopin, H. Deveau, S. D. Ehrlich, S. Moineau, and A. Chopin. 2009. P087, a lactococcal phage with a morphogenesis module similar to an Enterococcus faecalis prophage. Virology 388:49-56. [DOI] [PubMed] [Google Scholar]
- 64.Vinas, M. C., D. Gargallo, J. G. Loren, and J. Guinea. 1984. Morphological characterization of the Serratia marcescens bacteriophage SLP. J. Basic Microbiol. 25:285-288. [DOI] [PubMed] [Google Scholar]
- 65.Waak, E., W. Tham, and M. L. Danielsson-Tham. 2002. Prevalence and fingerprinting of Listeria monocytogenes strains isolated from raw whole milk in farm bulk tanks and in dairy plant receiving tanks. Appl. Environ. Microbiol. 68:3366-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watkins, J., and K. P. Sleath. 1981. Isolation and enumeration of Listeria monocytogenes from sewage sludge and river water. J. Appl. Bacteriol. 50:l-9. [DOI] [PubMed] [Google Scholar]
- 67.Wegmann, U., M. O'Connell-Motherway, A. Zomer, G. Buist, C. Shearman, C. Canchaya, M. Ventura, A. Goesmann, M. J. Gasson, O. P. Kuipers, S. D. van Sinderen, and J. Kok. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:3256-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Welshimer, H. J., and J. Donker-Voet. 1971. Listeria monocytogenes in nature. Appl. Microbiol. 21:516-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu, H., L. Sampson, R. Parr, and S. Casjens. 2002. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol. Microbiol. 45:1631-1646. [DOI] [PubMed] [Google Scholar]