Abstract
Quantification and sizing of filamentous cyanobacteria in environmental samples or cultures are time-consuming and are often performed by using manual or semiautomated microscopic analysis. Automation of conventional image analysis is difficult because filaments may exhibit great variations in length and patchy autofluorescence. Moreover, individual filaments frequently cross each other in microscopic preparations, as deduced by modeling. This paper describes a novel approach based on object-oriented image analysis to simultaneously determine (i) filament number, (ii) individual filament lengths, and (iii) the cumulative filament length of unbranched cyanobacterial morphotypes in fluorescent microscope images in a fully automated high-throughput manner. Special emphasis was placed on correct detection of overlapping objects by image analysis and on appropriate coverage of filament length distribution by using large composite images. The method was validated with a data set for Planktothrix rubescens from field samples and was compared with manual filament tracing, the line intercept method, and the Utermöhl counting approach. The computer program described allows batch processing of large images from any appropriate source and annotation of detected filaments. It requires no user interaction, is available free, and thus might be a useful tool for basic research and drinking water quality control.
Automated quantification and sizing of single cells by microscopy and image analysis are routinely used to determine microbial biomass (2, 6, 18). In contrast, fully automated quantification and measurement of the length of filamentous organisms are considerably more difficult, and not all problems have been solved satisfactorily yet, even though there is a strong demand for such approaches both in microbial ecology and in drinking water quality control (5). The filamentous cyanobacterium Planktothrix rubescens is a prominent organism in this context. It produces a variety of potent toxins (3, 4, 11, 12, 14) that may threaten animal and human health. Its ability to form blooms in some freshwater habitats makes this organism highly relevant in terms of public health and drinking water control (8, 9, 13, 23). Moreover, P. rubescens has recently been reported to unexpectedly invade drinking water reservoirs (15). Not surprisingly, monitoring of filamentous cyanobacteria is a standard task in many laboratories. Cyanobacterial biomass is often used as a proxy for estimating toxin concentration, as determination of this biomass is less costly than chemical analysis. Determination of abundance and biomass is routinely performed by microscopy using either the Utermöhl sedimentation method (20) or collection of cells on filters (22). These methods involve manual microscopy and are thus labor-intensive and time-consuming.
A major improvement in filament quantification occurred when charge-coupled device (CCD) cameras and image analysis software became available. Several automated or semiautomated methods have been proposed for measuring total filament length, either indirectly (e.g., the line-intercept method) (16, 17) or directly by filament detection via image analysis of either fluorescence (10, 22) or bright-field images (1). However, the filamentous nature of P. rubescens (i.e., its remarkable length-to-width ratio) poses problems for determination of the number and length of individual filaments by conventional image analysis. P. rubescens filaments are around 5 to 8 μm wide (7, 23), whereas their length ranges from less than 50 μm to more than 2,500 μm. If the length of individual filaments is measured, filaments have to be located entirely within a field of view (FOV), which means that large areas have to be imaged. Larger FOV areas may be viewed by using lower magnifications, but this comes at the cost of lower levels of fluorescence intensity and resolution, which are important for both efficient imaging and precise measurement. A second problem results from the fact that filamentous organisms often overlap in a microscopic preparation. Overlapping objects in a two-dimensional image, like the images generated by wide-field microscopy, cannot be correctly separated by conventional image analysis that is based on thresholding and binarization; two overlapping objects typically are merged into a single object. While the effect of this artifact on the automated determination of abundance is obvious, the corresponding error in the length measurements depends greatly on the method used. These problems have been discussed in great detail by Walsby and Avery (22), but no automated solution has been proposed yet.
The aim of this work was to develop a computer program that (i) counts filaments, (ii) measures the length of individual filaments, and (iii) determines the cumulative lengths of filaments in large composite images generated by epifluorescence microscopy. The probability that a filament is located only partially in an FOV and thus cannot be measured and the probability of filament overlaps were assessed theoretically by using a Monte Carlo simulation approach. Here we specifically address the problem of overlapping filaments and describe a strategy to recognize and correctly count them. The performance of the algorithm was tested by using measurements for filaments in images obtained by the manual filament tracing, line-intercept (16), and Utermöhl methods.
MATERIALS AND METHODS
Monte Carlo simulation.
The number of filaments on a membrane filter that were located partially outside an FOV, depending on the filament length and FOV size, and the number of filaments overlapping in an FOV as a function of filament density and filament length were estimated by using Monte Carlo simulation. The simulation was written in VB.Net and featured a graphical user interface. The simulation assumes that filaments are straight lines and uses a random number generator to create and position filaments in an FOV.
Microscopy system and software development environment.
Image acquisition was performed with a motorized microscope (AxioImager Z.1; Zeiss, Germany) using a CCD camera (AxioCam MRm; 12-bit grayscale; 1,388 by 1,040 pixels [px]; Zeiss, Germany), a 10× EC Plan-Neofluar (NA 0.3) objective, and a light-emitting diode (LED) epifluorescence illumination device (Colibri, Zeiss, Germany) with four LEDs (365 nm, 450 to 700 nm, 470 nm, and 590 nm) in combination with a multifilter cube (filter set 62 HE; Zeiss, Germany) and a filter cube for green excitation (filter set 43; Zeiss, Germany). For extension of the FOV size, multiple single adjacent images were acquired with green excitation and assembled automatically using the MosaiX module of the AxioVision software (Zeiss, Germany). The composite image was 11 by 15 single images and covered an area of approximately 1 cm2 (∼15,000 by 15,000 px; 0.645 μm pixel−1). Images were stored in the JPG format (grayscale, 8 bit). For internal validation by manual filament tracing, smaller images (5,000 by 5000 px) were acquired.
The filament counting and sizing program and the Monte Carlo simulation were developed in Microsoft Visual Basic.Net (VB.Net). Both programs are available free (http://www.technobiology.ch).
Program and algorithm description. (i) Fixel (filament element) concept.
The pixel (picture element) depicts the smallest element of an image in conventional image processing. Therefore, objects that are measured are represented by groups of connected pixels. As a consequence, overlapping objects (i.e., objects sharing the same pixels) are detected as a single object. As filaments are expected to frequently overlap as a function of their density and length (Fig. 1A), a new concept had to be introduced to correctly handle overlapping objects (for examples, see the left panels in Fig. 2C to F). We used a model-based object-oriented image analysis approach. The assumptions for our model were as follows: (i) filaments appear in an image as bright objects on a dark background; (ii) filaments are approximately 10 px wide; and (iii) filaments are unbranched. The basic objects in the algorithm are “filaments” and “fixels,” and filaments consist of multiple fixels. The term fixel, analogous to pixel, was used to describe the smallest element of a filament. A fixel is a rectangle that covers a small part of a filament and has the same orientation as the filament at its location.
FIG. 1.
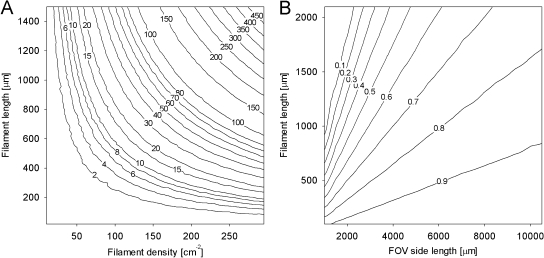
Monte Carlo simulations. (A) Predicted number of filament intersections as a function of filament density (filaments cm−2) in a field of view (FOV) and filament length. (B) Proportion of filaments of a given length located entirely within an FOV of a given size. For example, approximately 75% of 2-mm-long filaments are expected to be entirely in an FOV with an area of 1 cm2.
FIG. 2.
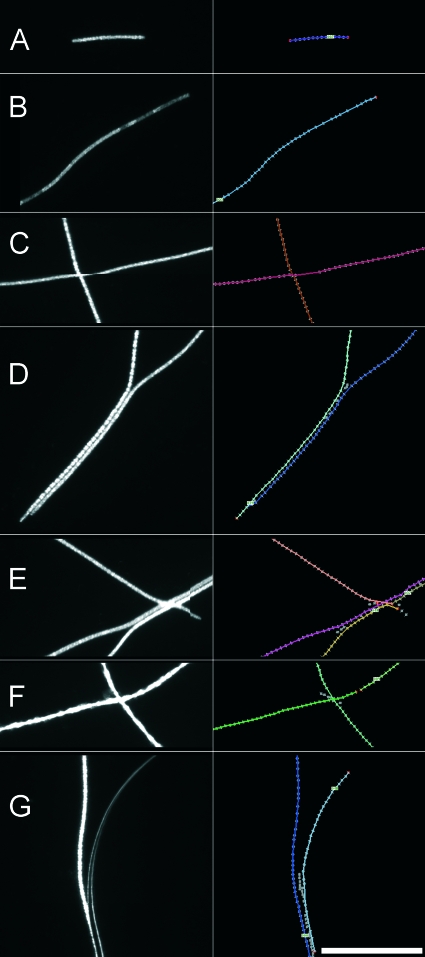
Sections from typical fluorescent microscope images of P. rubescens. The images on the left are the original images, and the images on the right are the annotated images automatically produced by the program for validation purposes. Bar = 100 μm. Detected filaments are shown as colored overlays and are enumerated. Detected fixels are indicated by gray squares, and endpoint fixels are indicated by red squares. (A to D) Correctly detected filaments. (A) Typical short filament. (B) Filament exhibiting a patchy fluorescent signal. (C) Two crossing filaments. Note the horizontal shift depicting an artifact created by stitching of the composite image. (D) Two parallel filaments. (E) Two parallel filaments crossed by a third filament. The filaments were wrongly assembled by the program. (F) Two crossing filaments. One filament has an incorrect break. (G) Filament in close proximity to an air bubble. An air bubble is incorrectly detected as a filament.
In the first step of the analysis, an image derived from epifluorescence microscopy is loaded and transferred into a two-dimensional pixel array. The background of the image is estimated by using the mean pixel intensities of the first scan line in the x direction and assuming that most of the pixels present are background pixels. Subsequently, the array is processed by a kernel to detect all fixels in the image.
(ii) Fixel detection.
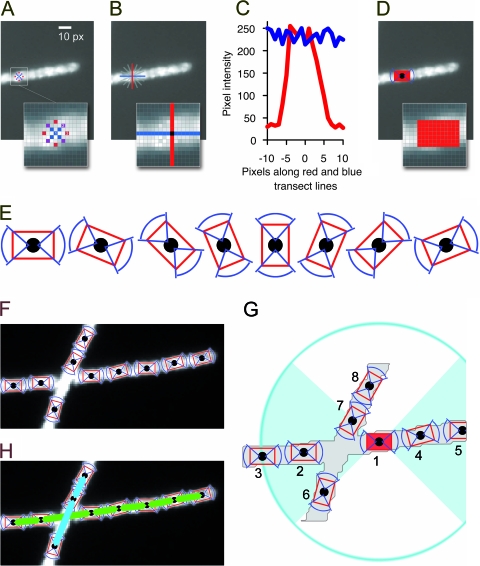
The primary kernel for fixel detection is shown in Fig. 3A. It is not a kernel in the classical sense since it analyzes only a subset of selected pixels in close proximity to a center pixel. The gray-level intensities of the pixels are compared to three empirically determined thresholds (T1 = 16, T2 = 13, and T3 = 10), and the result is positive if all of the intensities are greater than the background-corrected threshold intensity. If a potential fixel is detected, its orientation is determined. A cross-like structure consisting of two orthogonal transect lines (Fig. 3A and B) through the central pixel is used to sum pixel intensities. This structure is rotated in 22.5° steps, and the difference of the sum of the two transect lines is analyzed (Fig. 3B). If one of the lines is parallel to the filament and the other line is perpendicular to the filament, the difference of the integrated intensities of the lines is maximal (Fig. 3C). Once the orientation of the fixel is established, a rectangular field with the same orientation is moved along a line perpendicular to the fixel's orientation, and the precise placement of the fixel is locally optimized by maximizing the sum of the gray-level intensities in the rectangle. Finally, the fixel location is set, and the corresponding area is locked so that no other fixel can be placed in it (Fig. 3D). All fixels detected in the image are stored in a list.
FIG. 3.
Concept of the program illustrated using a sample image section. (A) The image is processed by a circular kernel to detect filamentous structures (fixels) by applying three brightness thresholds. (B) Potential fixels are checked for high gray-level intensity on the filament and low gray-level intensity on the sides. A cross-like structure (red and blue lines) is rotated at the location, and gray-level profiles are measured. (C) The orientation of a fixel is determined by comparing orthogonal gray-level profiles. (D) Once a fixel is set, its area is locked in order not to set other fixels in it. (E) All possible orientations of a fixel. (F) Detected fixels on two crossing filaments. (G) Filament assembly based on allocating possible neighbors to each fixel. Defined criteria have to be met. In this example, possible neighbors for fixel 1 were evaluated. Fixels 2 to 8 are within range, but fixel 8 is outside the 90° angle of both ends of fixel 1. Fixels 6 and 7 differ too much in their orientation with respect to fixel 1. Fixels 2 to 5 are thus the only valid neighbors. The best neighbors on both ends are fixels 2 and 4, as they are most closely situated. (H) Assembled fixels.
(iii) Assignment of neighboring fixels.
After detection of all fixels and determination of their orientations in the image, filaments are reconstructed by connecting neighboring fixels. In addition to its orientation and spatial position in the image, a fixel also has two sides that can connect to a neighboring fixel. Figure 3E illustrates all possible fixel orientations, and Fig. 3F shows fixels detected in two crossing filaments. For every fixel, two lists of suitable neighbors are generated, one for each side of the fixel. Potential neighbors have to be within a defined distance (6 to 70 px), and the difference in orientation must not exceed 45°. From all potential neighbors of a fixel, the best neighbor on each side is selected by an empirical optimization function that considers distance, orientation, and gray-level intensity profile for fixels (Fig. 3G). There are three possibilities: (i) a fixel may have no neighbors and is not considered for filament assembly, (ii) a fixel may have two neighbors and can be a part of a filament, or (iii) a fixel may have only one neighbor and can be a starting point for filament assembly.
(iv) Filament assembly.
Starting from the ends, filaments are assembled by joining each fixel to its best-scoring neighbor on the other side in a recursive way. As there are twice as many “end fixels” as filaments, too many filaments are assembled. As filament assembly from the two different ends does not necessarily have to result in the same filament (e.g., in the case of ambiguous crossings), a score for each filament assembly is generated, and the version of a filament with the better score is kept (Fig. 3H).
(v) Iteration of neighbor assignment and filament assembly.
After the neighbor detection and filament assembly procedure, there may still be unassigned fixels. Therefore, up to four iterations of neighbor detection and filament assembly are performed, in which fixels already assigned to filaments are progressively removed from the fixel list after each iteration.
(vi) Filament annotation and length measurement.
For manual verification of filament detection, an annotated image is created for each image analyzed, in which all fixels and filaments detected are imprinted and enumerated. Filament length is measured by adding the distances between the fixels of a filament. As the end regions are systematically underestimated by this method, an empirically determined value (24 px) is added to each filament length, as determined by correlation analysis (see Results). Finally, a report is generated as a text file comprising the lengths of the individual filaments.
Image set for internal validation.
Seven representative images (5,000 by 5,000 px; approximately 0.1 cm2) of P. rubescens filaments on polycarbonate membrane filters were analyzed (see below). Manual measurement of filaments by tracing single filaments in AxioVision was performed independently by four researchers. The results of the researchers were compared with each other and with the output of the automated routine. Additionally, the images were evaluated by the line-intercept method described by Nedoma et al. (16), using 16 horizontal test bars of the length (5,000 px each). To assess the rate of correct recognition and assembly of parallel or overlapping filaments, the annotated images generated by the program were manually compared to the original images. In order to verify the accuracy of measurement of the lengths of individual filaments, 100 filaments that were correctly detected by the program and by all four researchers were selected. For each filament, the means of the four replicate length measurements determined by the researchers were compared to the automated length measurement. The seven test images are available online at http://www.technobiology.ch.
Sampling and evaluation of joint data set with an external laboratory.
Depth profile samples (0, 1, 2.5, 5, 7.5, 10, 12.5, 15, 20, 30, 40, 80, 120, and 135 m) were obtained from Lake Zürich on 9 July 2008 during a routine sampling conducted by the staff of the Zürich Water Supply. The samples (50 ml) were fixed with paraformaldehyde (final concentration, 2%). Volumes between 1.5 and 12 ml, depending on the filament density, were filtered onto polycarbonate membrane filters (diameter, 25 mm; pore size, 5 μm; Sterico). Filters were imaged as described above to obtain large composite images (∼15,000 by 15,000 px; 1 cm2) using green light fluorescence illumination to detect the autofluorescence of P. rubescens. Four replicate preparations were obtained for the depth at which the P. rubescens abundance was maximal. Images were then analyzed by the program described here and in parallel by manual tracing of filaments and drawing of poly-lines using AxioVision. Additionally, the samples were independently analyzed by the WVZ using the Utermöhl method (25 ml per sample; fixation with Lugol's solution [final concentration, approximately 0.1%] for at least 5 days; sedimentation time of at least 24 h; analyzed area, 1 cm2; measurement of 20 to 30 individual filaments for determination of the average length).
RESULTS
Monte Carlo simulation.
The need for a strategy to handle overlapping filaments and the importance of analyzing large FOV areas for correct measurement of individual filaments lengths were demonstrated by a computational simulation using the Monte Carlo method. Figure 1A shows the results of prediction of the number of expected filament crossings as a function of filament density and filament length. By performing the simulation using the total number of filaments and the average filament length (487 μm) for the seven images (total area, 0.7 cm2) (Table 1) as input, 36 intersections were predicted. For the internal validation set of seven images, 44 intersections were counted.
TABLE 1.
Data for the internal validation set of seven imagesa
| Image | Manual count | Automated count | No. of incorrectly handled overlaps and touches/total no. | No. of incorrectly handled parallel filaments/total no. | No. of air bubbles | No. of premature breaks |
|---|---|---|---|---|---|---|
| 1 | 25.8 ± 1.5 | 27 | 1/5 | 0/0 | 1 | 0 |
| 2 | 18.5 ± 0.5 | 23 | 1/3 | 0/0 | 3 | 2 |
| 3 | 27.5 ± 1.5 | 27 | 0/1 | 0/1 | 0 | 0 |
| 4 | 44.0 ± 0.0 | 44 | 1/10 | 2/3 | 0 | 0 |
| 5 | 36.8 ± 0.4 | 40 | 3/8 | 1/3 | 0 | 2 |
| 6 | 21.8 ± 0.4 | 21 | 0/2 | 1/1 | 0 | 0 |
| 7 | 17.5 ± 1.1 | 18 | 0/5 | 1/2 | 0 | 1 |
| Total | 191.8 ± 3.6 | 200 | 6/44 | 5/11 | 4 | 5 |
Manual and automated counts are given, and typical issues such as overlaps, parallel filaments, air bubbles, and filament breaks on images were quantified. The number before a slash indicates the count of incorrectly handled events; the number after the slash indicates the total number of handled events.
Internal validation.
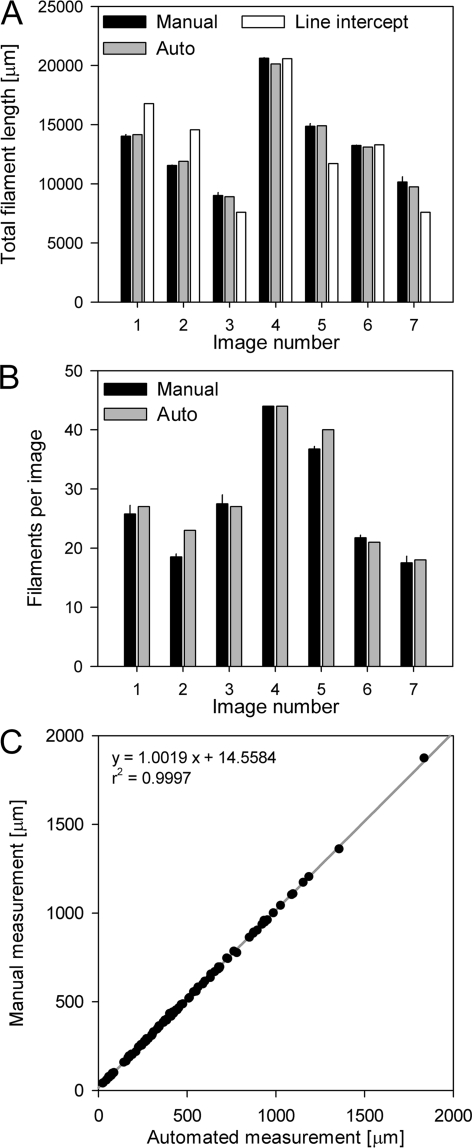
A high correlation was found between manual and automated measurements for both numbers of filaments (linear regression, r2 = 0.9591, P = 0.001) and cumulative filament length (linear regression, r2 = 0.9946, P < 0.001) for the seven images tested (Fig. 4A and B). In addition, the automated measurement of cumulative length was more accurate than the previously described line-intercept method (linear regression of manual measurements and line-intercept estimates, r2 = 0.7433, P = 0.0216).
FIG. 4.
Comparison of the different methods for an internal validation set of seven composite images. (A) Measurement of cumulative filament length by manual filament tracing, the automated method, and the line-intercept method. The error bars for the manual measurements indicate the standard deviations of four replicates. (B) Filament numbers as quantified by manual counting and by the automated method. The error bars for the manual measurements indicate the standard deviations of four replicates. (C) Linear regression of 100 selected filaments of different lengths from the seven images that were individually measured manually and by the automated approach.
For the four manual determination replicates there were only small variations in filament counts (coefficient of variation [CV], 0.02), and the automated estimates of filament numbers tended to be slightly higher than the manually determined values. However, the means of the data for manual replicates and automated counting were not statistically significantly different from one another (one-way analysis of variance, P = 0.997, post hoc test by Tukey's method).
The numbers of incorrectly handled events for parallel and overlapping filaments for automated filament detection, as well as the numbers of frequently observed artifacts, such as air bubbles below the coverslip or premature filament breaks, are shown in Table 1. Most importantly, overlapping filaments were handled correctly in >85% of the cases. The image sections shown in Fig. 2 as examples were derived from the seven images and illustrate some of the artifacts observed.
To prove the ability of our method to accurately measure the lengths of individual filaments independent of length or curvature, 100 randomly chosen filaments that were correctly identified in the seven images by both manual and automatic detection were compared individually (Fig. 4C). A high linear correlation was found between the automated and manual length determinations (r2 = 0.9997), and the slope of the regression was close to 1 (Fig. 4C). The observed offset of 14.6 μm resulted from the fact that the centers of the first and last fixels of a filament are not located exactly at the ends of the filament. Based on this regression analysis, a mean of 12 px per filament end was added for correction (as described in Materials and Methods).
Comparative evaluation of a data set.
To validate the method, joint sampling with the Zürich Water Supply was conducted on 9 July 2008. The automated method described here, manual filament tracing of microscopic images, and independently validated and certified analysis using the Utermöhl approach by the Zürich Water Supply were used to estimate filament concentrations, as well as cumulative filament lengths, in the water samples from the depth profile (Fig. 5).
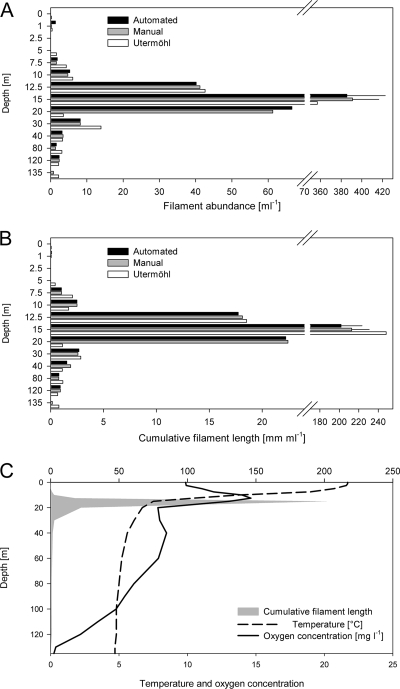
FIG. 5.
Comparison of different methods (automated, manual filament tracing, and Utermöhl methods) for enumeration of filaments for a depth profile for Lake Zürich on 9 July 2008. (A) Filament abundance for the depth profile. (B) Cumulative filament length for the depth profile. The error bars for the automated and manual measurements for a depth of 15 m indicate the standard deviations for four experimental replicates. Note that the depths are not originally scaled in panels A and B but are categories for better visibility. (C) Cumulative filament length (gray area) for the depth gradient (data from panel B).
Great variation in the filament concentration in the depth profile was observed. Almost no filaments were present in the upper half of the epilimnion (depth, <7.5 m) and in the hypolimnion (>40 m). In contrast, in the metalimnion there was a pronounced peak below the depth where the maximum oxygen concentration was observed at 15 m, where the density was approximately 400 filaments ml−1 (Fig. 5A). The distribution of the cumulative filament length was similar, and the maximal value was roughly 200 m liter−1 at a depth of 15 m (Fig. 5B). The pattern observed is in accordance with previous observations (24) and confirmed the ability of Planktothrix to form dense layers in the thermocline and eventually to be responsible for a steep oxygen gradient in its layer (Fig. 5C).
The three different methods for determination of filament density and cumulative length produced similar results, and no significant difference was observed, with the exception of one measurement obtained by the Utermöhl approach for a depth of 20 m, which was right below the depth where the filament concentration was maximal. A statistical analysis to compare the methods was performed by using linear regression. Automated and manual filament measurements for images showed high correlations for both filament numbers (r2 = 1.000, P < 0.0001) and cumulative filament length (r2 = 1.000, P < 0.0001). For four replicate preparations from 15 m, the manual measurements yielded coefficients of variations of 0.07 for filament number and 0.08 for cumulative filament length, whereas the CVs for the automated method were 0.10 and 0.11, respectively. After exclusion of the data for 20 m (for justification, see Discussion), a comparison of the automated method and the Utermöhl approach also yielded high correlations for filament number (r2 = 0.9995, P < 0.0001) and cumulative length (r2 = 0.9998, P < 0.0001).
DISCUSSION
We describe a model-based, object-oriented image analysis approach for detection and sizing of individual filamentous cyanobacteria in fluorescence microscope images. A central problem for determining the length distributions of these organisms results from the great length of some filaments and from the great variation in length in a sample. Determination of the length of a single filament is possible only if the filament is completely located in the field of view (FOV) observed. The probability that a filament is partially out of an FOV is a function of the filament length and the FOV size, as shown by Monte Carlo simulation (Fig. 1B). This implies that large FOV areas need to be observed (approximately 1 cm2 for P. rubescens) in order to avoid underestimation of the mean filament length. This can be accomplished by stitching single FOVs together to form large composite images, as described in this study. Image stitching is a technique to produce large high-resolution images (e.g., in microphotography of tissue samples), and most microscopy suppliers provide hardware and software for this purpose. Processing of such large images requires a powerful computer, preferably a computer equipped with a 64-bit operating system and 4 GB of RAM. Still, even in such large images, only approximately 75% of P. rubescens filaments that are 2,000 μm long would be correctly represented. Thus, it might be advisable to use a mathematical correction when filament length distributions are determined in detail, based on the simulation results described here. However, if the focus of an analysis is determination of the cumulative length or biovolume, image stitching is not necessary, and equally precise results can be obtained using smaller single images.
Filamentous organisms are difficult to analyze by conventional image processing, as they frequently overlap (Fig. 1A). The extent of overlapping in an image can be predicted and is a function of filament length and filament density, as shown by a comparison of a computational simulation with the actual frequencies of overlaps. Overlap frequency may be addressed by reducing the filament density, but the cost is analysis of fewer individuals per sample, which compromises the precision of the measurement. For reliable quantification, particularly reliable quantification of the mean length, or for reliable determination of the length distribution of filaments, a large number of individuals have to be measured and overlapping cannot be avoided. Therefore, we decided to use a model-based, object-oriented image analysis approach explicitly designed for analyzing unbranched, overlapping filaments. This approach does not primarily detect individual whole filaments but detects small elements of filaments (fixels). These elements are then iteratively assembled to reconstruct entire filaments by using a probabilistic model (i.e., they are linked with their most likely neighbors). Our method is robust with respect to several critical issues related to detection of filaments by image analysis, such as patchy fluorescent signals (1) or the shifting artifacts frequently observed when individual FOVs are stitched together to produce a large composite image (Fig. 2C).
The automated image analysis method was compared to the “gold standard,” which is manual tracing of filaments in images by experienced researchers. A comparison of the results of manual analysis by four individuals showed that there were only small differences between evaluations, both in filament counts and in cumulative length measurements. Despite its accuracy and robustness with respect to artifacts, the manual method is time-consuming and tedious (it takes up to several hours for each composite image of a 1-cm2 area).
A faster approach that is based on manual image analysis but is limited to determining only the cumulative length is the line-intercept method (16). Although this approach has the potential to be performed in a fully automated manner, it was less precise than our automated method when the results were compared to manual measurement results (Fig. 4A). This may be due to the fact that the line-intercept method is dependent on a homogeneous random distribution of filaments in an image as a consequence of the underlying statistical model. It is unclear whether this assumption is legitimate for filtered preparations, as filtration is prone to artifacts, such as higher cell densities in the border zones of preparations. Notably, our method does not rely on a homogeneous random distribution of filaments.
A comparison of our approach with the Utermöhl approach also showed that there was a tight correlation. The only exception was the data for a depth of 20 m (Fig. 5A and B), for which the Utermöhl approach yielded approximately 20-fold fewer filaments than both filtration-based evaluation methods. Planktothrix filaments are able to produce gas vesicles (21) to adjust their buoyancy. Filaments containing large numbers of gas vesicles may compromise the Utermöhl approach as it is based on sedimentation. The layer at 20 m was below the depth preferred by Planktothrix (24), as deduced from ambient light measurements (data not shown). It is conceivable that filaments in this layer had increased buoyancy to ascend to the depth where the light conditions were preferable (15 m) and thus were more resistant to sedimentation.
Besides this discrepancy it seems that the variation for sample preparation, as shown by the data for four replicates from the 15-m water sample, was much greater than the variation between manual analysis and automated analysis by either approach (Fig. 5). Still, our automated method sometimes also produced artifacts (Table 1 and Fig. 2E to G), either due to incorrect assembly of fixels or image artifacts such as air bubbles. For the internal validation set, 86% of the overlap events and 55% of the parallel filaments were handled correctly. These percentages could be increased by further optimization of the program code (e.g., fine adjustment of the probability weighting for fixel assembly) and by improving the filtration technique to generate preparations with more homogeneously distributed filaments. In general, the reliability of any image analysis method depends heavily on the quality of the preparation and the nature and composition of the water sample. Notably, a low density of filaments and the presence of other large particles (e.g., organic aggregates or other algae) may negatively affect the analysis. However, as the program produces annotated output images, possible artifacts are readily identified and can be excluded from the data set.
Altogether, we describe here a fast, accurate, and revisable method for high-throughput quantification of filamentous cyanobacteria by fluorescence microscopy and image analysis. The automated image analysis method is significantly faster than manual image evaluation. While manual tracing of filaments on a 15,000- by 15,000-px image may take up to several hours (depending on the filament density), automated analysis is performed within a few minutes. The easy-to-use program is available free and can be applied to microscopic images (composite or not composite) with different dimensions. Thus, it may be a useful tool both in basic research and for monitoring toxic filamentous cyanobacteria in freshwater.
Limitations and outlook.
The program described here was designed to correctly count and measure unbranched filaments, especially filaments of Planktothrix and other species with similar morphotypes. Using it with images of branched filaments would result in incorrect prediction of numbers and lengths of single filaments (as branched morphotypes would be split into multiple “pseudofilaments” at the branching points). Still, it is possible to measure the total length (and thus biomass) of branched filaments in a sample by using our program.
The main purpose of this work was to quantify and measure the length of the dominant filamentous cyanobacterial species in Lake Zurich based on its autofluorescence. A possible extension of our approach might include other preparation protocols to distinguish between cooccurring genotypes (e.g., by fluorescence in situ hybridization [19]) or direct calculation of biovolumes.
Acknowledgments
We thank Judith Blom, Thomas Posch, and Ferdinand Schanz for valuable comments. Angela Mechsner is gratefully acknowledged for manual measurement of filaments and laboratory support. Joint sampling and provision of data by the Zürich Water Supply (WVZ) are acknowledged. Michael Zeder thanks Anthony Walsby for introduction to image analysis and for motivation to work on Planktothrix quantification.
Silke Van den Wyngaert was supported by project SNF 31003A-11765.
Footnotes
Published ahead of print on 4 January 2010.
REFERENCES
- 1.Almesjo, L., and C. Rolff. 2007. Automated measurements of filamentous cyanobacteria by digital image analysis. Limnol. Oceanogr. Methods 5:217-224. [Google Scholar]
- 2.Blackburn, N., A. Hagstrom, J. Wikner, R. Cuadros-Hansson, and P. K. Bjornsen. 1998. Rapid determination of bacterial abundance, biovolume, morphology, and growth by neural network-based image analysis. Appl. Environ. Microbiol. 64:3246-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blom, J. F., B. Bister, D. Bischoff, G. Nicholson, G. Jung, R. D. Sussmuth, and F. Juttner. 2003. Oscillapeptin J, a new grazer toxin of the freshwater cyanobacterium Planktothrix rubescens. J. Nat. Products 66:431-434. [DOI] [PubMed] [Google Scholar]
- 4.Blom, J. F., J. A. Robinson, and F. Juttner. 2001. High grazer toxicity of [d-Asp(3) (E)-Dhb(7)]microcystin-RR of Planktothrix rubescens as compared to different microcystins. Toxicon 39:1923-1932. [DOI] [PubMed] [Google Scholar]
- 5.Chorus, I., I. R. Falconer, H. J. Salas, and J. Bartram. 2000. Health risks caused by freshwater cyanobacteria in recreational waters. J. Toxicol. Environ. Health Part B Crit. Rev. 3:323-347. [DOI] [PubMed] [Google Scholar]
- 6.Daims, H., and M. Wagner. 2007. Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis. Appl. Microbiol. Biotechnol. 75:237-248. [DOI] [PubMed] [Google Scholar]
- 7.Davis, P., J. Beard, and A. Walsby. 2003. Variation in filament width and gas vesicles of red and green isolates of Planktothrix spp. Arch. Hydrobiol. (Suppl.) Algol. Stud. 108:15-30. [Google Scholar]
- 8.Ernst, B., B. Hitzfeld, and D. Dietrich. 2001. Presence of Planktothrix sp. and cyanobacterial toxins in Lake Ammersee, Germany and their impact on whitefish (Coregonus lavaretus L.). Environ. Toxicol. 16:483-488. [DOI] [PubMed] [Google Scholar]
- 9.Ernst, B., S. J. Hoeger, E. O'Brien, and D. R. Dietrich. 2009. Abundance and toxicity of Planktothrix rubescens in the pre-alpine Lake Ammersee, Germany. Harmful Algae 8:329-342. [Google Scholar]
- 10.Ernst, B., S. Neser, E. O'Brien, S. J. Hoeger, and D. R. Dietrich. 2006. Determination of the filamentous cyanobacteria Planktothrix rubescens in environmental water samples using an image processing system. Harmful Algae 5:281-289. [Google Scholar]
- 11.Fastner, J., M. Erhard, W. W. Carmichael, F. Sun, K. L. Rinehart, H. Ronicke, and I. Chorus. 1999. Characterization and diversity of microcystins in natural blooms and strains of the genera Microcystis and Planktothrix from German freshwaters. Arch. Hydrobiol. 145:147-163. [Google Scholar]
- 12.Fastner, J., U. Neumann, B. Wirsing, J. Weckesser, C. Wiedner, B. Nixdorf, and I. Chorus. 1999. Microcystins (hepatotoxic heptapeptides) in German fresh water bodies. Environ. Toxicol. 14:13-22. [Google Scholar]
- 13.Hoeger, S. J., B. C. Hitzfeld, and D. R. Dietrich. 2005. Occurrence and elimination of cyanobacterial toxins in drinking water treatment plants. Toxicol. Appl. Pharmacol. 203:231-242. [DOI] [PubMed] [Google Scholar]
- 14.Kurmayer, R., and M. Gumpenberger. 2006. Diversity of microcystin genotypes among populations of the filamentous cyanobacteria Planktothrix rubescens and Planktothrix agardhii. Mol. Ecol. 15:3849-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naselli-Flores, L., R. Barone, I. Chorus, and R. Kurmayer. 2007. Toxic cyanobacterial blooms in reservoirs under a semiarid Mediterranean climate: the magnification of a problem. Environ. Toxicol. 22:399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nedoma, J., J. Vrba, T. Hanzl, and L. Nedbalova. 2001. Quantification of pelagic filamentous microorganisms in aquatic environments using the line-intercept method. FEMS Microbiol. Ecol. 38:81-85. [Google Scholar]
- 17.Olson, F. 1950. Quantitative estimates of filamentous algae. Trans. Am. Microsc. Soc. 69:272-279. [Google Scholar]
- 18.Pernthaler, J., A. Pernthaler, and R. Amann. 2003. Automated enumeration of groups of marine picoplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2631-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schönhuber, W., B. Zarda, S. Eix, R. Rippka, M. Herdman, W. Ludwig, and R. Amann. 1999. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 65:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Utermöhl, H. 1958. Zur Vervollkommnung der quantitativen Phytoplankton-Methodik. Mitt. Intern. Verein. Limnol. 9:1-38. [Google Scholar]
- 21.Walsby, A. E. 1994. Gas vesicles. Microbiol. Rev. 58:94-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsby, A. E., and A. Avery. 1996. Measurement of filamentous cyanobacteria by image analysis. J. Microbiol. Methods 26:11-20. [Google Scholar]
- 23.Walsby, A. E., A. Avery, and F. Schanz. 1998. The critical pressures of gas vesicles in Planktothrix rubescens in relation to the depth of winter mixing in Lake Zurich, Switzerland. J. Plankton Res. 20:1357-1375. [Google Scholar]
- 24.Walsby, A. E., and F. Schanz. 2002. Light-dependent growth rate determines changes in the population of Planktothrix rubescens over the annual cycle in Lake Zurich, Switzerland. New Phytol. 154:671-687. [DOI] [PubMed] [Google Scholar]