Abstract
A marine bacterium, Hahella chejuensis, recently has attracted attention due to its lytic activity against a red-tide dinoflagellate. The algicidal function originates from its red pigment, prodigiosin, which also exhibits immunosuppressive or anticancer activity. Genome sequencing and functional analysis revealed a gene set contained in the hap gene cluster that is responsible for the biosynthesis of prodigiosin. To screen for the factors affecting the prodigiosin biosynthesis, we constructed a plasmid library of the H. chejuensis genomic DNA, introduced it into Escherichia coli strains harboring the hap cluster, and observed changes in production of the red pigment. Among the screened clones, hapXY genes whose products constitute a two-component signal transduction system were elucidated as positive regulators of the pigment production. In addition, an Hfq-dependent, noncoding region located at one end of the hap cluster was confirmed to play roles in regulation. Identification of factors involved in the regulation of prodigiosin biosynthesis should help in understanding how the prodigiosin-biosynthetic pathway is organized and controlled and also aid in modulating the overexpression of prodigiosin in a heterologous host, such as E. coli, or in the natural producer, H. chejuensis.
Harmful algal blooms (HABs), commonly called red tide, are a phenomenon in which toxin-producing marine algae rapidly proliferate in the offshore area. The HAB-causing phytoplanktons are reported to interact with other organisms such as bacteria and fungi. Among them, the marine bacteria are known to play important roles in decreasing or developing HABs (3, 5, 14). For instance, Hahella chejuensis, isolated from the coastal area of Marado in South Korea (15), is capable of killing Cochlodinium polykrikoides (12). C. polykrikoides is a major microalga that causes HABs, especially in the Northeast Pacific coastal area (8). The bacterial determinant that kills C. polykrikoides was further characterized as a red pigment referred to as prodigiosin (12). Prodigiosin belongs to a family of tripyrrole antibiotic molecules called prodiginines, which have potential as anticancer agents or immunosuppressants (24). The prodigiosin congener isolated from H. chejuensis also exerts an immunosuppressive effect (11).
Through completed genome sequencing of H. chejuensis and its functional analysis, the genomic region involved in biosynthesis of prodigiosin was elucidated (12). This complete set of prodigiosin-biosynthetic genes was named the hap gene cluster. The red pigment prodigiosin was further characterized structurally, and the biosynthetic pathway was proposed by Kim and colleagues (13, 14). Genes of the hap cluster share homology with those in the pig cluster and the red cluster which are involved in prodiginine-biosynthetic intermediates of Serratia marcescens and Streptomyces coelicolor, respectively (7, 23, 25). Enzymes encoded by the genes in the pig and red clusters have been characterized (24). However, gene expression of the hap cluster can be tightly controlled, based on the observation that heterologous expression of the hap cluster alone failed to produce the pigment in Escherichia coli. The recombinant E. coli was able to produce the pigment only when the culture filtrate of H. chejuensis was added to the growth media (12). This result indicates that another regulatory cue is needed for prodigiosin biosynthesis, which prompted us to search for regulatory factors that modulate prodigiosin biosynthesis in H. chejuensis.
In this study, regulatory factors for biosynthesis of prodigiosin in H. chejuensis were identified by functional screening. To search for such factors, a plasmid library derived from the genomic DNA of H. chejuensis was constructed and transformed into E. coli strains carrying the hap cluster. In the cases of Serratia marcescens and Streptomyces coelicolor, molecular inputs, such as cell-produced quorum-sensing signal molecules or two-component systems (TCSs) for signal transduction, have been verified as key regulatory signals for prodigiosin biosynthesis so far (4, 9, 10, 20-22). Similarly, some clones of interest uncovered in this study include molecular factors such as those that belong to the TCS. Also, we elucidated that an apparently noncoding region in the hap cluster functions as a key factor of prodigiosin biosynthesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Hahella chejuensis KCTC 2396 was originally isolated from marine sediment from Marado, located southwest of Jeju island, South Korea (15). H. chejuensis cells were cultured at 30°C in ZoBell's medium (5 g peptone, 1 g yeast extract, 0.01 g FePO4, 250 ml distilled water, and 750 ml aged and filtrated seawater). E. coli strains were cultured at 37°C in Luria broth. The antibiotic concentrations used for plasmid selection in E. coli were as follows: ampicillin (Ap) 100 μg/ml, chloramphenicol (Cm) 20 μg/ml, and kanamycin (Km) 25 μg/ml. 1× CopyControl induction solution (Epicentre, Madison, WI) was added to the medium so that an observable amount of prodigiosin was produced due to the high copy number of pCC1FOS harboring the hap cluster. To induce gene expression in the pUC vectors, 0.7 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added. The bacterial strains and plasmids used in this study are listed in Table 1 .
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Derivation, description, and/or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| Hahella chejuensis KCTC 2396 | Wild type | 15 |
| Escherichia coli strains | ||
| EPI300 | Commercial strain used for cloning | Epicentre |
| HC81008E02 | EPI300 transformed with pHC81008E02 | 12 |
| HC81002H12 | EPI300 transformed with pHC81002H12 | 12 |
| HC81006F09R4 | EPI300 transformed with pHC81006F09R4 | 14 |
| DH5α | Commercial strain used for cloning | Invitrogen |
| Plasmids | ||
| pCC1FOS | High-copy-inducible fosmid cloning vector, Cmr | Epicentre |
| pHC81008E02 | pCC1FOS containing a 34.8-kb genomic fragment of the H. chejuensis hap cluster | 12 |
| pHC81002H12 | pCC1FOS containing a 32.7-kb genomic fragment of the H. chejuensis hap cluster | 12 |
| pHC81006F09R4 | pCC1FOS containing a 36.4-kb genomic fragment of the H. chejuensis hap cluster | 14 |
| pUC18 | Cloning vector, 2.69 kb, Apr | Invitrogen |
| pUC19 | Cloning vector, 2.69 kb, Apr | Invitrogen |
| p8E02-G3 | pUC19 containing a 3.2-kb genomic fragment of H. chejuensis | This study |
| p8E02-G3ΔHK | Variant of p8E02-G3 in which the histidine kinase gene is partially deleted | This study |
| p8E02-G3ΔRR | Variant of p8E02-G3 in which the response regulator gene is partially deleted | This study |
| pSKK1 | pUC19 containing a 1.2-kb insert of the hap region | This study |
| pSKK2 | pUC18 containing a 1.2-kb insert of the hap region | This study |
| pSKK2-EAB150 | pUC18 with a fragment derived from the serial deletion of pSKK2 | This study |
| pKD4 | Contains Kmr flanked with FRT sites, Apr | 2 |
| pKD46-RecA | recA+ derivative of pKD46 containing genes involved in λ Red recombination | Ji Hoon Shim |
FRT, FLP recombination target.
Construction of a genomic DNA library of H. chejuensis.
The genomic DNA of H. chejuensis KCTC 2396 was prepared by using the Wizard SV DNA purification system purchased from Promega (Madison, WI) and was randomly sheared by sonication for 0.2 s. The sheared genomic fragments of 3 to 5 kb in size were selected and ligated into the high-copy-number plasmid pUC19, which was digested with SmaI and then dephosphorylated. Ligation reaction was performed with a 1:3 ratio of pUC19 to H. chejuensis genomic DNA fragments overnight at 16°C. Ligates were transformed into E. coli DH5α, and the resulting transformants were pooled to construct a genomic library.
Phenotypic screening for alteration in pigment production.
E. coli strains HC81008E02, HC81002H12, and HC81006F09 R4 each harbor fosmids pHC81008E02, pHC81002H12, and pHC81006F09 R4 that contain the hap cluster of H. chejuensis KCTC 2396. HC81008E02 and HC81002H12 were determined not to produce the red pigment prodigiosin under normal growth conditions, even when the harboring fosmids are induced to high copy number (12), whereas HC81006F09 R4 is a variant that constitutively produces the red pigment if the fosmid is at high copy number (14). Plasmids that contain the H. chejuensis genomic DNA were isolated from the pooled E. coli transformants and were transformed into the E. coli strains that contain the hap cluster to monitor the changes in colony color. Clones that make HC81008E02 and HC81002H12 produce the red pigment constitutively without the need to add the culture filtrate of H. chejuensis into the growth medium or that make HC81006F09 R4 produce little or no red pigment under normal growth conditions were picked up as candidates that may positively or negatively modulate prodigiosin biosynthesis. Red pigments from the clones of interest were confirmed to be prodigiosin through liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) analysis (13).
Nucleotide sequencing and data analysis.
For sequencing, plasmids were isolated from the clones that were selected through the phenotypic screening according to the protocols described in Molecular Cloning: a Laboratory Manual by Joseph Sambrook and D. W. Russell (16). M13F and M13R primers were used to uncover the genomic region of H. chejuensis embedded in the plasmid vectors by sequencing the ends of the inserts. DNA sequencing was performed at Genotech Co. Ltd. (Daejeon, South Korea). Similarity searches of nucleotide sequences were performed by using BLAST at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) server, and information on the whole insert carried on the plasmids was recovered by using Artemis (downloaded from http://www.sanger.ac.uk/Software/Artemis) with the genome sequence and annotation data of H. chejuensis (12).
Prodigiosin production assay.
Cultures of H. chejuensis incubated for 48 h and those of the hap cluster-containing E. coli strains incubated for 12 h were inoculated into the fresh ZoBell and LB media, respectively, with or without antibiotics and inducers to dilute 100-fold. Cells were incubated for 24 h at 30°C, with vigorous shaking at 200 rpm, to produce prodigiosin. Prodigiosin exhibits a characteristic maximum absorption spectrum at 534 nm in acidified ethanol. Cells were harvested by centrifugation at 13,000 rpm for 5 min, and then intracellular prodigiosin was extracted from the cells by suspending them in an acidified ethanol solution (4% [vol/vol] 1 M HCl in ethanol) (18). The relative prodigiosin concentration expressed per cell was determined as the ratio of absorbance of the extracted prodigiosin solution at 534 nm to the turbidity of the culture suspension at 600 nm (A534/A600).
Serial deletion of pSKK1 and pSKK2.
Unidirectional deletion was accomplished by the Erase-a-Base kit obtained from Promega (Madison, WI). pSKK1 and pSKK2 that contained a 1.2-kb insert of the hap region (Table 1) were digested with PstI and XbaI to generate a 3′ overhang resistant to ExoIII digestion and a 5′ overhang labile to ExoIII, respectively. Treatment of ExoIII nuclease lasted for 150 s. ExoIII-digested plasmids were collected at every 10-s interval and treated with S1 nuclease to remove single-stranded tails from the sample DNA and then with Klenow DNA polymerase to release blunt ends by filling single-stranded sequences. Finally, each sample was ligated to circularize the deletion-containing plasmids and transformed into E. coli carrying the hap cluster.
Site-directed mutagenesis.
Disruption of the putative histidine kinase gene in p8E02-G3 (see Table S1 in the supplemental material) was accomplished by fusion PCR. To knock out the dimerization domain of the histidine kinase, four primers were designed. Primers HKlinkR (5′-GATTGAGTCCATGCTTAACGCCGAAAGGCTGGAAACAGGTC) and HKlinkF (5′-GACCTGTTTCCAGCCTTTCGGCGTTAAGCATGGACTCAATC) with flanking sequences of the deletion target region of the histidine kinase gene share overlapping 20-base oligonucleotides. Primers OutEco (5′-ATTGAATTCGAGCTCGGTACCCG) and OutBam (5′-ATGGATCCCCTGATAGTTATTAATATG) (restriction sites are underlined) contain the end sequences of the p8E02-G3 insert with the restriction sites of EcoRI and BamHI for cloning into pUC19. First, PCR products were generated by using p8E02-G3 as the PCR template and amplified with primer sets of either OutEco/HKlinkR or HKlinkF/OutBam. These PCR products were combined for the second-round PCR by using them as templates with primers OutEco/OutBam. The final PCR products were digested with EcoRI and BamHI, ligated into pUC19, and treated with the same enzymes to generate p8E02-G3ΔHK. p8E02-G3ΔRR, a plasmid containing an in-frame deletion in the putative response regulator gene, was constructed by using the same strategy with primers RRlinkR (5′-GGAAATTAACCTGACGACGCGCTACCGATTTGTCGGAGAAG), RRlinkF (5′-CTTCTCCGACAAATCGGTAGCGCGTCGTCAGGTTAATTTCC), OutEco, and OutBam. Oligonucleotide primers were synthesized by Genotech Co. Ltd. (Daejeon, South Korea). The PCR conditions are as follows: denaturation for 5 min at 95°C; 25 cycles of 30 s at 95°C, 30 s at 55°C, and 3 min at 72°C; additional extension for 10 min at 72°C; and storage at 4°C. Taq polymerase obtained from Bioneer Co. (Daejeon, South Korea) was used.
hfq gene disruption.
The hfq gene deletion in E. coli EPI300 was conducted by using the λ Red system reported by Datsenko and Wanner (2). The PCR product for the recombination reaction that carries a kanamycin resistance cassette was amplified with primers containing 50-bp homology extensions flanking the hfq gene and 20-bp priming sequences for pKD4 as a template (HfqKmH1, 5′-GTACAAATAAGCATATAAGGAAAAGAGAGAATGGCTAAGGGGCAATCTTTGTGTAGGCTGGAGCTGCTTC; HfqKmH2, 5′-CTTCGCTGTCCTGTTGCGCGGAAGTATTCTGCGCGCTGCTACCATGATGGCATATGAATATCCTCCTTAG). The PCR fragment was transformed into E. coli harboring the Red helper plasmid pKD46-RecA. Recombinants that show kanamycin resistance were selected, and then replacement of the hfq gene with the cassette was validated by PCR and sequencing.
RESULTS
Screening of factors modulating prodigiosin biosynthesis.
To search for factors in H. chejuensis that may positively or negatively affect biosynthesis of prodigiosin, three clones of E. coli EPI300 that carry pHC81008E02, pHC81002H12, or pHC81006F09 R4 were utilized. These fosmids, pHC81008E02, pHC81002H12, and pHC81006F09 R4, contain the hap gene cluster of H. chejuensis KCTC 2396 (12). HC81008E02 and HC81002H12 are defined as clones of E. coli that carry pHC81008E02 and pHC81002H12, respectively. They produce no red pigment under normal growth conditions and did so only when the culture filtrate of H. chejuensis was added to the medium. In contrast, HC81006F09 R4, a variant E. coli clone carrying pHC81006F09, constitutively produces the pigment (14).
Plasmids pooled from the genomic DNA library of H. chejuensis were transformed into HC81008E02 and HC81002H12, and 22 transformants that were able to synthesize the pigment without the presence of the culture filtrate based on changes in colony color (data not shown) were selected for further characterization. The transformed plasmids should carry certain genomic regions that positively modulate prodigiosin biosynthesis. A similar experiment was conducted with HC81006F09 R4 to identify clones that negatively regulate the biosynthesis of prodigiosin. Thirty plasmids transformed into HC81006F09 R4 constitutively repressed the production of the red pigment and resulted in colonies that were pale red or white (data not shown). These candidate plasmids that either positively or negatively modulate the pigment production were verified by retransformation of each plasmid into E. coli EPI300 with an appropriate fosmid.
The genomic regions of H. chejuensis contained in these plasmids were elucidated by determining the end sequences of the inserts and by matching them to the genome sequence of H. chejuensis (see Table S1 in the supplemental material). They could be divided into four classes based on their locations in the genome and the functions of the gene products. Eight clones containing the genomic fragments in the hap locus that encodes the prodigiosin-biosynthetic gene cluster were categorized as class 1. Nine clones were placed in class 2, and they contain genes possibly having regulatory functions; five of them have genes coding the histidine kinase or the response regulator that are related to the TCS. In class 3, nine clones carry genes encoding membrane-associated proteins. Class 4 contains the remaining clones having candidate regulators of prodigiosin biosynthesis that do not belong to class 1, 2, or 3. Clones represented by two genomic regions, one that contains genes for a TCS and the other that is a small fragment in the hap cluster, were selected for further study.
A two-component system affects the pigment production.
p8E02-G3 is a plasmid that induces the red-pigment production of HC81008E02. The red pigment turned out to be prodigiosin according to LC-tandem MS (LC-MS/MS) analysis (data not shown). Analysis of the nucleotide sequence of the p8E02-G3 insert revealed the presence of three genes: a histidine kinase gene and a response regulator gene of the TCS and a truncated gene containing the FOG: tetratricopeptide repeat domain. Based on BLASTP searches of the SwissProt database, the product of HCH_01435, hereinafter referred to as HapX, appears most similar to the YclK/AfsQ2/MtrB/EnvZ/PhoR class of histidine kinases, whereas the HCH_01436 product, referred to as HapY, is most related to OmpR/YclJ/PhoB/MprA/AfsQ1 response regulators.
The histidine kinase HapX is composed of three domains. They are the histidine kinase-like ATPase domain that belongs to the family of several ATP-binding proteins, the HAMP domain that is widely distributed as a putative regulatory element in transmembrane and other signaling proteins (1), and the histidine kinase A domain that functions in dimerization and accepting phosphates. Domains for ATP-binding and dimerization form the kinase core. The activation signal generated by histidine kinases are transmitted to response regulators. Like most bacterial response regulators, HapY consists of a receiver domain that accepts the phosphorylation signal and the transcriptional regulatory domain for DNA binding (Fig. 1A).
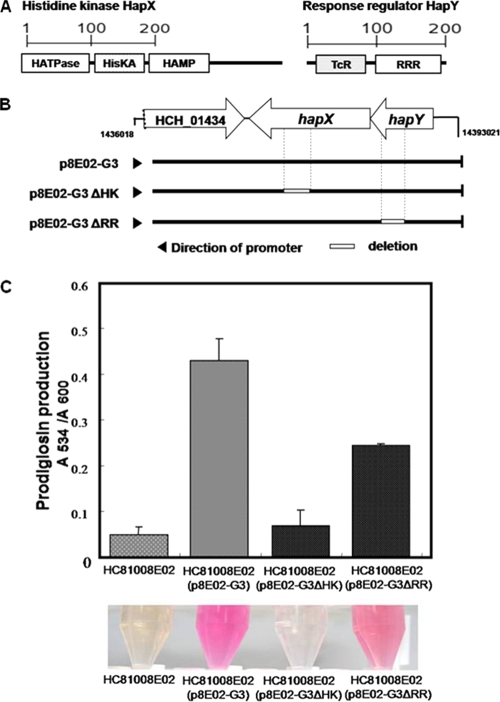
FIG. 1.
Effects of two-component signal transduction system genes on prodrigiosin biosynthesis. (A) Domains of the histidine kinase HapX and the response regulator HapY. (B) Construction of in-frame deletion mutants. p8E02-G3 contains genes involved in a two-component signal transduction system that consists of a histidine kinase and a response regulator. A mutant, p8E02-G3ΔHK, lacking the His kinase A domain of the histidine kinase was constructed. Transcriptional regulatory domain for DNA binding of the response regulator was deleted to produce another mutant, p8E02-G3ΔRR. Arrowheads, directions of the plasmid-encoded lac promoters. (C) Spectrophotometric and colorimetric analysis for prodigiosin production by HC81008E02 containing p8E02-G3, p8E02-G3ΔHK, or p8E02-G3ΔRR. Prodigiosin production assay was conducted as described in Materials and Methods. Samples were prepared in three replicates, and error bars indicate the standard errors of the means. HC81008E02 does not produce prodigiosin, based on LC-MS/MS analysis (S.-K. Kwon and J. F. Kim, unpublished observation). HATPase, histidine kinase-like ATPase domain; HisKA, histidine kinase A domain; HAMP, histidine kinases, adenylyl cyclases, methyl binding proteins, and phosphatases domain; TcR, transcriptional regulatory protein, C terminal; RRR, response regulator receiver domain.
To determine whether the mutations of these two genes can prevent the positive regulation of p8E02-G3 in prodigiosin biosynthesis, in-frame deletions of some essential domains of the TCS were introduced. To eliminate the activities of the histidine kinase or the response regulator, p8E02-G3ΔHK and p8E02-G3ΔRR lacking the histidine kinase A domain or the transcriptional regulatory domain (Fig. 1B) were constructed and transformed into HC81008E02. The resulting transformants were cultured, and then the red pigment produced was extracted. Whereas production of the red pigment was slightly reduced for HC81008E02 with p8E02-G3ΔRR, HC81008E02 carrying p8E02-G3ΔHK could synthesize a barely detectable amount of the pigment, if any, as shown in Fig. 1C.
A noncoding region in the hap cluster modulates prodigiosin biosynthesis.
The class 1 clones (see Table S1 in the supplemental material) contain hap genes and adjacent intergenic sequences located in a single locus of the hap cluster. Plasmids positively and negatively modulating pigment production contain the same region, but these two types of clones have opposite directions for the plasmid-encoded promoter. Plasmids having the promoter in the direction of hap transcription negatively affected the pigment production; on the other hand, those having the promoter opposite the direction of hap transcription positively affected production (Fig. 2A). A 1-kb fragment of the overlapping region common to all of these plasmids was cloned into pUC18 at its BamHI and EcoRI sites and was designated pSKK2. As a consequence of the transformation of pSKK2 into HC81008E02 and HC81002H12, cells became constitutively red. pSKK1 was obtained by incorporation of the 1-kb fragment into pUC19, in which the insert was integrated in the opposite orientation at the point of the lac promoter in the vector, with respect to pSKK2. HC81006F09 R4 transformed with pSKK1 produced no more pigment (Fig. 2B). The same region of the genomic fragments showed different activities toward pigment production according to the coded direction of the lac promoter of the vector.
FIG. 2.
A regulatory region in the hap cluster that affects prodigiosin biosynthesis. (A) Candidate clones containing a region in the hap cluster that alters prodigiosin biosynthesis. Six plasmid clones contain hap genes or genes located in the hap cluster. Plasmids positively or negatively modulating pigment production share the common overlapping region, but these two types of clones have opposite directions of the plasmid-encoded lac promoters (shown by arrowheads). Plasmids having the promoter in the direction of hap transcription negatively affected pigment production; those having the promoter opposite the direction of hap transcription positively affected it. Filled arrows indicate the hap genes homologous to the pig genes of Serratia marcescens and to the red genes of Streptomyces coelicolor A3(2). (B) Plasmid maps of pSKK1 and pSKK2 and pigment production of HC81006F09 R4 (pSKK1), HC81008E02 (pSKK2), and HC81002H12 (pSKK2). The overlapping region common to all of these plasmids was cloned into pUC19 and pUC18 to produce pSKK1 and pSKK2, respectively. HC81006F09 R4 transformed with pSKK1 was turned off in pigment production. HC81008E02 or HC81002H12 carrying pSKK2 became constitutively red.
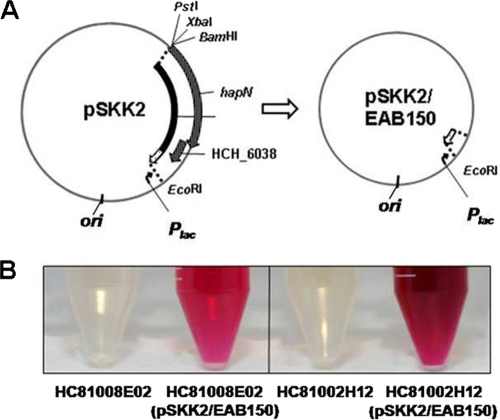
pSKK2 thus was suspected to contain factors which positively affect prodigiosin biosynthesis. The 1,072-bp insert of pSKK2 has two potential protein coding sequences and an intergenic region. Two protein coding regions, hapN annotated as a putative oxidoreductase gene and a hypothetical gene HCH_06038, were integrated in the opposite direction of the lac promoter. To specify the positive regulatory factor, a serial deletion of the insert cloned in pUC18 was conducted from the hapN gene side as described in the experimental procedures. The plasmids created at each time point were transformed into HC81002H12, and it was observed whether they produced the red pigment. The minimal effective plasmid was elucidated by this serial deletion process and named pSKK2/EAB150 (Fig. 3A). Its insert was only 124 bp long, and no apparent functional genes exist according to the annotation (Fig. 3B).
FIG. 3.
Effect of a noncoding region part of the hap cluster on prodigiosin biosynthesis. (A) Plasmid map of pSKK2/EAB150; (B) pigment production of HC81008E02 (pSKK2/EAB150) and HC81002H12 (pSKK2/EAB150). Through serial deletions of pSKK2, the minimum region that affects prodigiosin biosynthesis was elucidated. pSKK2/EAB150 carries this genomic region, and its effect on prodigiosin biosynthesis was confirmed by pigment production after transformation of this plasmid into HC81008E02 and HC81002H12.
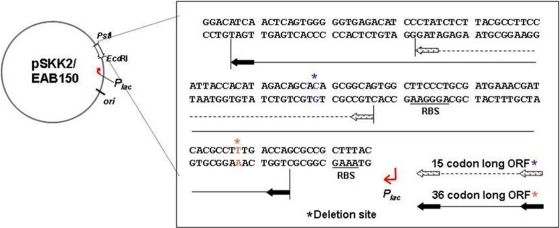
No homologue of the 124-bp sequence was detectable with BLASTN/BLASTX searches against the nonredundant nucleotide collection (nr/nt) database or even in the specialized sequence read archive (SRA) transcript libraries at NCBI (http://www.ncbi.nlm.nih.gov). Also, there was no significant match retrieved from the CAMERA database (http://camera.calit2.net/) that houses environmental metagenomic sequences. This suggests that the region may function at the level of transcription as a noncoding RNA (ncRNA) molecule or may encode a small peptide that is either novel or evaded homology searches due to insufficient sequence length. In this region, several small open reading frames (ORFs) with potential initiation codons (CTG and GTG) were found. Those beginning with CTG were of the lengths of 36 and 15 codons. The putative ORFs beginning with GTG were 32 codons in length. Of these, only 36- and 15-codon-long ORFs were preceded by appropriate sequences that resemble the ribosome-binding sites (Fig. 4).
FIG. 4.
Open reading frames present in the sequence of the pSKK2/EAB150 insert. Two ORFs that start with the CTG initiation codon, preceded by appropriate sequences that resemble the ribosome-binding site, were found in this intergenic region of 124 bases. Those ORFs were 36 and 15 codons long. Frame shift mutations were introduced into these ORFs to test the possibility that either of the two ORFs encodes a small protein (asterisks represent the bases deleted to release a mutant of each ORF).
To evaluate the coding capacity of the 124-bp region to be translated into small peptides, we created frameshift mutants of the 36- and 15-codon ORFs for functional investigation by transforming these mutants into HC81002H12. When the colony colors of the transformants were examined, levels of the red-pigment biosynthesis by the 36-codon ORF mutants were similar to that for the corresponding 124-bp insert, but the 15-codon ORF mutant lost the ability to synthesize the red pigment. Thus, we sought more direct proof to confirm the translation of these ORFs. Even if they are in fact synthesized, the small sizes of these putative peptides would make it difficult for them to be detected. We tagged them by introducing the GFPuv coding sequence without the start codon after the 36- and 15-codon ORFs in order to facilitate their detection. As a result of observed in-frame GFPuv fusions to the ORFs, neither could direct the synthesis of significant levels of GFPuv (data not shown). Even for the frameshift mutant of the 15-codon ORF that caused the halt of red-pigment biosynthesis, no direct proof of translation of this ORF was obtained.
Biosynthesis of prodigiosin depends on the Hfq function.
If part of the 124-bp sequence is transcribed into a small ncRNA, its function may depend on Hfq. Hfq is involved in the expression of many genes not only by affecting mRNA stability but by interacting with ncRNA (19). An hfq mutant of EPI300 was constructed by utilizing the λ Red recombinase, into which the fosmids pHC81008E02, pHC81002H12, and pHC81006F09 R4 carrying the prodigiosin biosynthesis gene cluster were transformed. pSKK1 or pSKK2 containing the 124-bp sequence was then transformed into one of these fosmid-carrying hfq mutants to result in the Δhfq HC81008E02 (pSKK2), Δhfq HC81002H12 (pSKK2), and Δhfq HC81006F09 R4 (pSKK1) mutants. Δhfq HC81008E02 (pUC18), Δhfq HC81002H12 (pUC18), and Δhfq HC81006F09 R4 (pUC19) were also constructed to be used as controls.
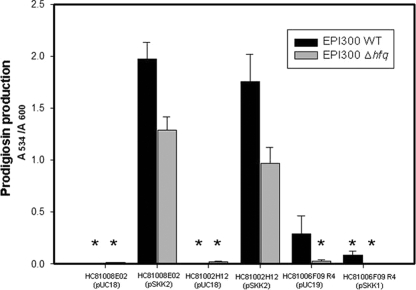
As presented in Fig. 5, pigment production by the hfq mutants of HC81008E02 (pSKK2) and HC81002H12 (pSKK2) was decreased 34.7% and 44.9% compared to those of wild-type HC81008E02 (pSKK2) and HC81002H12 (pSKK2), respectively. In contrast, pigment production was not observed for the wild-type and hfq mutant strains of HC81008E02 (pUC18) and HC81002H12 (pUC18). This indicates that the function of the 124-bp noncoding region that is transcribed by the plasmid-encoded lac promoter depends on intact Hfq. In the case of HC81006F09 R4 (pUC19), pigment production was abolished in the hfq mutant, whereas the wild-type HC81006F09 R4 was red. Pigment production was not detectable for both wild-type and hfq mutant strains of HC81006F09 R4 (pSKK1).
FIG. 5.
Prodigiosin production by wild type and hfq mutants of HC81008E02, HC81002H12, and HC81006F09 R4 with or without the region carried by pSKK2 or pSKK1. Prodigiosin production assay was conducted as described in Materials and Methods (asterisks represent the samples whose ethanol extracts showed no indication of pigment production). Samples were prepared in three replicates, and error bars indicate the standard errors of the means.
Additionally, an investigation to identify the molecular determinant of this 124-bp noncoding region was conducted based on its potential as an untranslated ncRNA. This 124-bp sequence was compared with known ncRNA sequences of the Rfam database by using Infernal (http://infernal.janelia.org/). These sequences showed no significant bit score with ncRNA when the cutoff value was 24.
DISCUSSION
Prodigiosin, possibly the pigment of the medieval transubstantiation miracles of bloody bread, is a “marvelous” compound with medical and industrial significance thanks to antibiotic, anticancer, and immunosuppressive activities (24). For the purpose of mitigating HABs, prodigiosin itself may be more desirable than direct application of its producers, such as H. chejuensis or Serratia spp., to the natural environment, since live organisms might unwillingly influence the balance of the marine ecosystem or pose a potential problem to unidentified nontarget organisms, such as some invertebrates or other lower eukaryotes. Thus, understanding the regulatory mechanisms of prodigiosin biosynthesis is important to overcoming the limitations of prodigiosin overproduction and to paving the way to better production of prodigiosin in its natural hosts or in a stabilized heterologous expression system.
In this study, an E. coli-based heterologous system was established to search for the regulatory factors that are involved in prodigiosin biosynthesis of H. chejuensis. Fifty-two clones that altered the pigment production were identified from a genomic library of H. chejuensis. The genetic information of these selected clones delivers reasonable cues for further investigation of the regulation of the biosynthesis of a tripyrrole antibiotic pigment, prodigiosin.
A common regulatory theme of the red-pigment production in the species of Serratia and Streptomyces is the use of a TCS for signal transduction (6, 17, 24). We showed here that H. chejuensis also utilizes a TCS for prodigiosin biosynthesis to respond to environmental cues. A putative histidine kinase gene, hapX, uncovered from one of the two genomic regions selected for further study, was confirmed to act as a positive regulatory factor via molecular deletion and biochemical analysis. Deletion of the sensor kinase resulted in a complete abolishment of red-pigment production, while that of a response regulator gene, hapY, affected it only partially. This observation enables one to hypothesize that the histidine kinase presumably has another yet identified cognate response regulator playing a more direct role in prodigiosin production. An orphan response regulator gene flanking the front of the hap cluster, HCH_06018, is suspected to encode another cognate response regulator interacting with HapX. Since the interaction between the HCH_06018 product and HapX has not been elucidated yet, investigation of the interaction between the purified proteins, as well as further genetic studies, is needed.
Screening of the regulatory factors revealed that 6 out of 52 clones were located in the same genomic locus of H. chejuensis. This suggested that the same locus located in the hap cluster is probably playing a crucial role in the regulation of prodigiosin biosynthesis. According to the serial-deletion experiment of pSKK2, a 124-bp sequence located in an intergenic region between HCH_06038 and HCH_6039 is sufficient to affect prodigiosin biosynthesis. pSKK1 and pSKK2 harbor the same 124-bp intergenic region with the directions of the plasmid-encoded lac promoters opposite to each other. pSKK1 negatively modulated prodigiosin biosynthesis, whereas pSKK2 acted positively. The 124-bp region might encode a small peptide, but its translational capacity could not be demonstrated by an experiment that utilized the green fluorescent protein (GFP) fusion. Another possibility is that this small region encodes a noncoding regulatory RNA. Regulation of gene expression mediated by small ncRNA, in many cases, depends on the RNA chaperone Hfq, which binds to the ncRNA (19). Based on the available evidence that is provided in the following paragraphs, the molecular determinant present in this region is believed most likely to function as hitherto unknown, novel ncRNA.
pSKK2 induced the production of the red pigment in E. coli containing prodigiosin-biosynthetic genes but not producing the pigment otherwise. This effect was reduced in the hfq mutant of E. coli. From these observations and the results of the serial deletion of pSKK2, we hypothesize that the 124-bp intergenic sequence in pSKK2/EAB150 that is transcribed by the lac promoter in the vector functions as an Hfq-dependent ncRNA that suppresses the repression mechanism for the hap gene expression in HC81008E02 or HC81002H12. This derepression function results in the transcription of the hap cluster and the production of prodigiosin. The partial reduction of prodigiosin production in the hfq mutant background could be due to the excess amount of ncRNA produced from the multicopy-number pUC plasmid and by the relatively strong lac promoter.
On the other hand, prodigiosin production in the variant E. coli that constitutively expresses the prodigiosin-biosynthetic genes was abolished when pSKK1 was introduced. This can be explained as the transcript expressed from the lac promoter of pSKK1 functioning as an antisense RNA that binds to the Hfq-dependent ncRNA that is transcribed in the opposite direction from the fosmid and inhibits the derepression function of the ncRNA. The pigment production was not detected with the hfq mutant of HC81006F09 R4 either, even in the absence of pSKK1. This could be due to the lowered levels of the Hfq-dependent ncRNA in the hfq mutant background, resulting in the repression of hap gene expression and no pigment production.
Even though this is not a complete study of the whole regulation system of prodigiosin biosynthesis in H. chejuensis, decisive regulatory factors of prodigiosin biosynthesis were elucidated by the screening efforts with the heterologous expression system. These factors suggest that a complex circuit of regulatory mechanisms involving a TCS and a noncoding region internal to the hap cluster exist for biosynthesis of the secondary metabolite prodigiosin in H. chejuensis.
Supplementary Material
Acknowledgments
We are indebted to an anonymous reviewer who suggested the Hfq experiment. We also thank Seong Keun Kim and Min Jung Kwak for their help in introducing the hfq mutation into EPI300, Ji Hoon Shim for providing pKD46-RecA, Jong Suk Lee for technical support to LC-MS/MS analysis, Dockyu Kim for critical comments on the manuscript, Choong Hoon Lee and Choong-Min Ryu for helpful discussions, and Yong-Jae Suh for aid in text editing.
This work was supported by a grant from the 21C Frontier Microbial Genomics and Applications Center Program, Ministry of Education, Science and Technology, Republic of Korea.
Footnotes
Published ahead of print on 28 December 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 2.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doucette, G. J. 1995. Interactions between bacteria and harmful algae: a review. Nat. Toxins 3:65-74. [DOI] [PubMed] [Google Scholar]
- 4.Fineran, P. C., H. Slater, L. Everson, K. Hughes, and G. P. Salmond. 2005. Biosynthesis of tripyrrole and β-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol. Microbiol. 56:1495-1517. [DOI] [PubMed] [Google Scholar]
- 5.Fleming, L. E., K. Broad, A. Clement, E. Dewailly, S. Elmir, A. Knap, S. A. Pomponi, S. Smith, H. Solo Gabriele, and P. Walsh. 2006. Oceans and human health: emerging public health risks in the marine environment. Mar. Pollut. Bull. 53:545-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gristwood, T., P. C. Fineran, L. Everson, N. R. Williamson, and G. P. Salmond. 2009. The PhoBR two-component system regulates antibiotic biosynthesis in Serratia in response to phosphate. BMC Microbiol. 9:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris, A. K., N. R. Williamson, H. Slater, A. Cox, S. Abbasi, I. Foulds, H. T. Simonsen, F. J. Leeper, and G. P. Salmond. 2004. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology 150:3547-3560. [DOI] [PubMed] [Google Scholar]
- 8.Hasui, M., M. Matsuda, K. Okutani, and S. Shigeta. 1995. In vitro antiviral activities of sulfated polysaccharides from a marine microalga (Cochlodinium polykrikoides) against human immunodeficiency virus and other enveloped viruses. Int. J. Biol. Macromol. 17:293-297. [DOI] [PubMed] [Google Scholar]
- 9.Horinouchi, S. 2003. AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). J. Ind. Microbiol. Biotechnol. 30:462-467. [DOI] [PubMed] [Google Scholar]
- 10.Horng, Y. T., S. C. Deng, M. Daykin, P. C. Soo, J. R. Wei, K. T. Luh, S. W. Ho, S. Swift, H. C. Lai, and P. Williams. 2002. The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol. Microbiol. 45:1655-1671. [DOI] [PubMed] [Google Scholar]
- 11.Huh, J. E., J. H. Yim, H. K. Lee, E. Y. Moon, D. K. Rhee, and S. Pyo. 2007. Prodigiosin isolated from Hahella chejuensis suppresses lipopolysaccharide-induced NO production by inhibiting p38 MAPK, JNK and NF-κB activation in murine peritoneal macrophages. Int. Immunopharmacol. 7:1825-1833. [DOI] [PubMed] [Google Scholar]
- 12.Jeong, H., J. H. Yim, C. Lee, S. H. Choi, Y. K. Park, S. H. Yoon, C. G. Hur, H. Y. Kang, D. Kim, H. H. Lee, K. H. Park, S. H. Park, H. S. Park, H. K. Lee, T. K. Oh, and J. F. Kim. 2005. Genomic blueprint of Hahella chejuensis, a marine microbe producing an algicidal agent. Nucleic Acids Res. 33:7066-7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, D., J. S. Lee, Y. K. Park, J. F. Kim, H. Jeong, T. K. Oh, B. S. Kim, and C. H. Lee. 2007. Biosynthesis of antibiotic prodiginines in the marine bacterium Hahella chejuensis KCTC 2396. J. Appl. Microbiol. 102:937-944. [DOI] [PubMed] [Google Scholar]
- 14.Kim, D. K., Y. K. Park, J. S. Lee, J. F. Kim, H. Y. Jeong, B. S. Kim, and C. H. Lee. 2006. Analysis of a prodigiosin biosynthetic gene cluster from the marine bacterium Hahella chejuensis KCTC 2396. J. Microbiol. Biotechnol. 16:1912-1918. [Google Scholar]
- 15.Lee, H. K., J. Chun, E. Y. Moon, S. H. Ko, D. S. Lee, H. S. Lee, and K. S. Bae. 2001. Hahella chejuensis gen. nov., sp. nov., an extracellular-polysaccharide-producing marine bacterium. Int. J. Syst. Evol. Microbiol. 51:661-666. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Shu, D., L. Chen, W. Wang, Z. Yu, C. Ren, W. Zhang, S. Yang, Y. Lu, and W. Jiang. 2009. afsQ1-Q2-sigQ is a pleiotropic but conditionally required signal transduction system for both secondary metabolism and morphological development in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 81:1149-1160. [DOI] [PubMed] [Google Scholar]
- 18.Slater, H., M. Crow, L. Everson, and G. P. Salmond. 2003. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 47:303-320. [DOI] [PubMed] [Google Scholar]
- 19.Storz, G., J. A. Opdyke, and A. Zhang. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7:140-144. [DOI] [PubMed] [Google Scholar]
- 20.Takano, E., R. Chakraburtty, T. Nihira, Y. Yamada, and M. J. Bibb. 2001. A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41:1015-1028. [DOI] [PubMed] [Google Scholar]
- 21.Takano, E., T. Nihira, Y. Hara, J. J. Jones, C. J. Gershater, Y. Yamada, and M. Bibb. 2000. Purification and structural determination of SCB1, a γ-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2). J. Biol. Chem. 275:11010-11016. [DOI] [PubMed] [Google Scholar]
- 22.Thomson, N. R., M. A. Crow, S. J. McGowan, A. Cox, and G. P. Salmond. 2000. Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol. Microbiol. 36:539-556. [DOI] [PubMed] [Google Scholar]
- 23.Tsao, S. W., B. A. Rudd, X. G. He, C. J. Chang, and H. G. Floss. 1985. Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J. Antibiot. 38:128-131. [DOI] [PubMed] [Google Scholar]
- 24.Williamson, N. R., P. C. Fineran, F. J. Leeper, and G. P. Salmond. 2006. The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4:887-899. [DOI] [PubMed] [Google Scholar]
- 25.Williamson, N. R., H. T. Simonsen, R. A. Ahmed, G. Goldet, H. Slater, L. Woodley, F. J. Leeper, and G. P. Salmond. 2005. Biosynthesis of the red antibiotic, prodigiosin, in Serratia: identification of a novel 2-methyl-3-n-amyl-pyrrole (MAP) assembly pathway, definition of the terminal condensing enzyme, and implications for undecylprodigiosin biosynthesis in Streptomyces. Mol. Microbiol. 56:971-989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.