Abstract
Microbial ecologists and systematists are challenged to discover the early ecological changes that drive the splitting of one bacterial population into two ecologically distinct populations. We have aimed to identify newly divergent lineages (“ecotypes”) bearing the dynamic properties attributed to species, with the rationale that discovering their ecological differences would reveal the ecological dimensions of speciation. To this end, we have sampled bacteria from the Bacillus subtilis-Bacillus licheniformis clade from sites differing in solar exposure and soil texture within a Death Valley canyon. Within this clade, we hypothesized ecotype demarcations based on DNA sequence diversity, through analysis of the clade's evolutionary history by Ecotype Simulation (ES) and AdaptML. Ecotypes so demarcated were found to be significantly different in their associations with solar exposure and soil texture, suggesting that these and covarying environmental parameters are among the dimensions of ecological divergence for newly divergent Bacillus ecotypes. Fatty acid composition appeared to contribute to ecotype differences in temperature adaptation, since those ecotypes with more warm-adapting fatty acids were isolated more frequently from sites with greater solar exposure. The recognized species and subspecies of the B. subtilis-B. licheniformis clade were found to be nearly identical to the ecotypes demarcated by ES, with a few exceptions where a recognized taxon is split at most into three putative ecotypes. Nevertheless, the taxa recognized do not appear to encompass the full ecological diversity of the B. subtilis-B. licheniformis clade: ES and AdaptML identified several newly discovered clades as ecotypes that are distinct from any recognized taxon.
The last decade has revealed unimagined levels of species diversity among the prokaryotes. Annealing experiments with environmental DNA have pushed our estimates of community diversity into the millions of species (20, 56), with a billion species estimated worldwide (15). Sequencing of the 16S rRNA gene has identified tens of thousands of species, yet to be characterized, within a single community (24). Since only about 9,000 species have been recognized and described globally, the task of characterizing the many remaining species presents a profound challenge for microbial systematists and ecologists. In particular, we need to describe the unique ecological role each species plays, the differences that allow them to coexist, and the processes by which they have originated.
These key issues may all be addressed by discovering the early ecological changes that drive the splitting of one bacterial population into two ecologically distinct populations. One might expect that a comparison of the most closely related species recognized by bacterial systematics would provide insight into the first ecological differences associated with the splitting of lineages. However, the species of bacterial systematics are often extremely broadly defined, whether measured by diversity of genome content and physiology (22), by DNA sequence (53), or, most importantly, by ecology (61). So, if we are to identify the first ecological changes distinguishing newly divergent lineages, we may need to identify taxa that are more narrowly defined and more newly divergent than the named species.
Our approach is to identify bacterial clades with the properties shared by many modern species concepts—that species should each be cohesive (subject to forces that constrain the diversity within the species), irreversibly separate and ecologically distinct from other species, and invented only once (13). These criteria are met by bacterial ecotypes, which are defined as ecologically homogeneous clades, whose diversity is constrained by a force of cohesion, either periodic selection or genetic drift (5, 10, 11, 29, 47, 60). In periodic selection, natural selection favoring each adaptive mutant within an ecotype can purge the ecotype's diversity genomewide (28, 30), owing to the low rates of recombination in bacteria, occurring usually within an order of magnitude of the mutation rate (6, 7, 33, 57). Because different ecotypes are distinct in their ecological niches, periodic selection and genetic drift cannot extend beyond the limits of an ecotype. The divergence between ecotypes is thus not constrained by periodic selection or drift; also, recombination may decelerate divergence but in the end is not sufficient to prevent adaptive divergence between ecotypes (5, 10). Ecotypes therefore have the species-like properties of cohesion, as well as ecological distinctness and irreversible separateness from other ecotypes (4).
Moreover, longstanding ecotypes can be discovered from patterns of DNA sequence diversity, provided that a particular model of bacterial evolution applies. In the Stable Ecotype model, ecotypes are formed only rarely, and there is recurrent purging of diversity within ecotypes, either by periodic selection or by genetic drift (11). In this case, each ecotype can accumulate a unique set of sequence mutations at every gene in the genome over the ecotype's long history as a distinct lineage, and diversity within each ecotype is recurrently purged, allowing ecotypes to be discovered as DNA sequence clusters for any gene shared among ecotypes (9, 11, 29, 50, 61).
We have employed two recently developed algorithms to hypothesize the sequence clusters that correspond to ecotypes and to confirm that the ecotypes so hypothesized are indeed ecologically distinct. AdaptML (25) and Ecotype Simulation (ES) (29) each analyze the evolutionary history of a clade to yield appropriate criteria for demarcating the significant clades corresponding to ecotypes (10). These algorithms have an advantage over the universal molecular thresholds used by bacterial systematics to identify species, because there is no evidence of a single molecular criterion that would be applicable for all bacteria (2, 10, 11), and the criteria used thus far by systematics have frequently led to overly diverse species (11, 53).
The ES and AdaptML algorithms have successfully identified extremely closely related ecotypes that would otherwise have been subsumed as unrecognized entities within species (25, 29). Comparisons of the habitat associations of these ecotypes have then led to discovery of the ecological dimensions of ecotype divergence: solar exposure of soil for Bacillus (29), photic zone depth and temperature for Synechococcus from hot springs (61), host range for Legionella (9), and season and particle substrate size for marine Vibrio (25). Discovering the ecological dimensions of early divergence simultaneously addresses the evolutionary origins of bacterial diversity, the basis of niche partitioning, and the unique role each ecotype plays in its community.
Here we aim to expand the set of known ecological dimensions of early divergence within the Bacillus subtilis-Bacillus licheniformis clade, which includes B. subtilis and its three subspecies (35, 41), B. vallismortis, B. mojavensis, B. atrophaeus, B. amyloliquefaciens, B. licheniformis, B. sonorensis (38), B. tequilensis (21), clades earlier described as “B. axarquiensis” and “B. malacitensis” (44) but now reclassified as B. mojavensis (59), and the clade earlier described as “B. velezensis” (43) but now reclassified as B. amyloliquefaciens (58). We collected soil following the “Evolution Canyon” paradigm developed by Eviatar Nevo and colleagues: an evolution canyon is an east-west-running canyon, which provides habitats that contrast sharply in solar exposure but are identical in geological and macroclimatic features (36). Comparisons of the biota of the two slopes of evolution canyons have revealed adaptive differences related to solar exposure in a great diversity of organisms, including bacteria, fungi, plants, arthropods, and vertebrates (36). In the present study, we collected soil from a west-running, hot-desert canyon in Death Valley, California, yielding isolates from the south-facing slope (SFS), with the most intense solar exposure; the shadier north-facing slope (NFS); and the canyon bottom's arroyo (A), with intermediate solar exposure and more access to water than either slope. In addition, we collected soil of two texture types, a sandy soil and a badland-like silty soil, which also differed in their chemical properties and in the plants they supported. We will demonstrate that differences associated with solar exposure and soil texture are among the dimensions of ecological divergence for newly divergent Bacillus ecotypes in the Death Valley canyon.
MATERIALS AND METHODS
Habitats and soil collection.
Soil was collected on June 3, 2007, from Radio Facility Wash (RFW) near Furnace Creek in Death Valley National Park. The study site is flanked by the deep alluvium-filled Death Valley floor to the west, and to the east by the Funeral Mountains, which are a fault-bounded block in the southwestern portion of the Basin and Range geologic province. The elevation is 45 m below sea level. The geology of the site includes variably consolidated mixtures of recent to late Holocene silt, clay, and fine sand, with no significant soil development. Surficial material is commonly underlain by older playa or lacustrine sequences of the middle to early Holocene and Pleistocene ages (57a). The mean annual maximum temperature for the 30-year interval from 1971 to 2000 was 45°C; the mean annual minimum temperature was 4°C; and the mean annual precipitation was 6.3 cm (39). Despite the small amount of precipitation, rainfall at higher elevations does not penetrate the ground surface, resulting in periodic flooding of the wash.
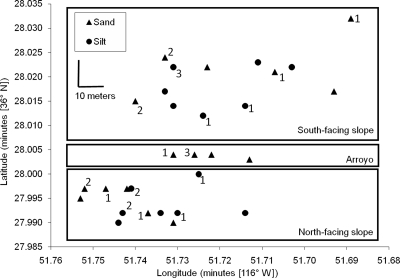
The walls of Radio Facility Wash have a slope of about 40° and are about 12 m high. RFW runs due west at the points of collection, yielding three major habitats with regard to solar exposure and access to water: the south-facing slope (36°28.012′ to 36°28.032′N, 116°51.689′ to 116°51.740′W), the north-facing slope (36°27.990′ to 36°28.000′N, 116°51.714′ to 116°51.753′W), and the arroyo at the wash bottom (36°28.003′ to 36°28.004′N, 116°51.713′ to 116°51.731′W) (Fig. 1). In addition to the solar exposure dimension, the slopes of the wash were heterogeneous in surface soil texture, with sandy and badland-like silty soil, while the arroyo surface contained only sandy soil. Soil was collected from the top 3 mm.
FIG. 1.
Collection sites within Radio Facility Wash. Next to each site is the number of strains (if not zero) that were isolated from the clade most closely associated with silty soil (i.e., that containing putative ecotypes 12 and 19). This silt-associated clade was isolated from silty and sandy sites throughout the study area, and from both slopes.
Plant-based characterization of habitats.
The five habitat types were characterized by the plants present at the peak of the flowering season (25 February 2008).
Chemical and physical analysis of soil.
Soil samples were assayed by Colorado State University's Soil, Water, and Plant Testing Lab for salinity, conductivity, pH, organic content, nitrogen, phosphorus, potassium, zinc, iron, manganese, copper, boron, and proportions of sand, silt, and clay.
Strain isolation.
Strains of the B. subtilis-B. licheniformis clade were isolated as spores from soil as previously described (12), except that the isolates were not screened for utilization of l-arabinose and mannitol. Briefly, the soil was heated in water at 80°C for 10 min, and bacteria were streaked for isolation on Luria broth plates. Strains that tested positive for citrate utilization, nitrate reduction, and gelatin hydrolysis were kept for molecular characterization. The 118 strains so isolated were confirmed by sequence analysis (as described below) to be members of the B. subtilis-B. licheniformis clade (see Fig. 2; see also Table S1 in the supplemental material).
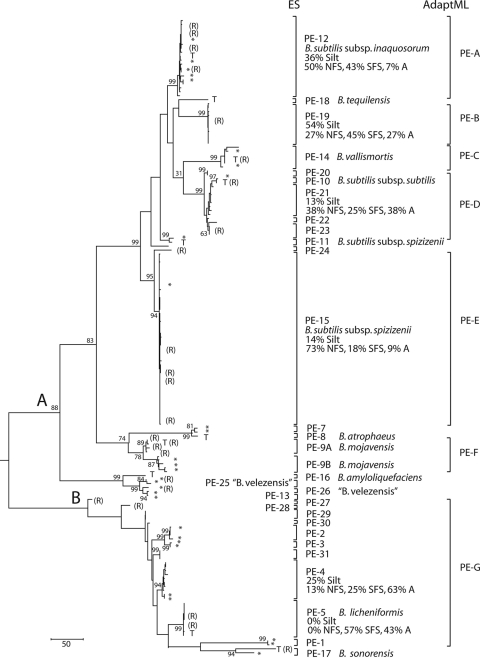
FIG. 2.
Phylogenetic, Ecotype Simulation, and AdaptML analyses of the B. subtilis-B. licheniformis clade. The phylogeny is based on a maximum-parsimony analysis of the recombination-free concatenation of dnaJ, gyrA, and rpoB, rooted by strain C-125 of B. halodurans. The B. subtilis and B. licheniformis subclades are defined as the descendants of ancestors A and B, respectively, in the figure. Bootstrap support at >50% is indicated, except for nodes within putative ecotypes. The bracketed ecotype demarcations indicate the largest clades consistent with containing a single ecotype (in ES) and containing strains of a single habitat spectrum (in AdaptML). The habitat sources of strains from RFW are indicated in Table S1 in the supplemental material. For ecotypes with ≥5 RFW strains, the percentages of isolates from different habitats are indicated. Strains not isolated from RFW are indicated by asterisks, except that type strains are indicated by a “T.” Individuals that had recombined in one of the three protein-coding genes are indicated by an “R”; their positions in the tree are based on the remaining two genes that had not recombined. Putative ecotypes 1 to 17 have been demarcated previously (29). The strains previously assigned to PE 6 were found in the present analysis to fall within PE 12; those previously assigned to PE 9 are designated here as PE 9A and 9B.
DNA extraction, PCR amplification, and sequencing.
DNA was extracted using the Puregene DNA purification kit from Gentra Systems, by following the manufacturer's protocol for Gram-positive bacteria (29). PCR amplification of rpoB and gyrA was performed as described previously (29). For dnaJ, we used forward primer 5′-GGG GTA GGT AAG AGC GCT TC-3′ and reverse primer 5′-CGG CAT TTC GCA GTA AAT ATC-3′, and PCR involved 30 cycles with annealing at 52°C.
The 16S rRNA gene was sequenced for a subset of the sequences, chosen to represent the ecotypes predicted by Ecotype Simulation (see below). Two overlapping segments of the gene were amplified and then aligned. For the region spanning gene positions 4 to 964, we used forward primer 5′-TAT CGG AGA GTT TGA TCC TGG-3′ and reverse primer 5′-GCG TTG CTT CGA ATT AAA CC-3′; for the region spanning positions 695 to 1505, we used forward primer 5′-TAG CGG TGA AAT GCG TAG AG-3′ and reverse primer 5′-GAT ACG GCT ACC TTG TTA CGA-3′. Thirty cycles of PCR were performed at an annealing temperature of 60°C.
PCR products were purified using the Qiagen QIAquick spin kit and were sent to the University of Pennsylvania DNA Sequencing Facility for sequencing. Sequences were aligned using ClustalW multiple alignment (55). After the initial alignment, all gaps and single nucleotide changes were checked in the chromatograms of both the forward and the reverse sequences. Where the forward and reverse sequences disagreed, the strand conforming to the consensus nucleotide of the strain's closest relatives was chosen to be correct, if possible; otherwise, the nucleotide site was recorded as ambiguous. The length of the concatenation of partial sequences of dnaJ, gyrA, and rpoB was 1,776 bp.
Phylogeny reconstruction.
The ES and AdaptML algorithms do not explicitly model the introduction of sequence diversity into an ecotype by recombination (25, 29), so we have aimed to analyze only the nonrecombinant sequence diversity at the three protein-coding loci. We therefore identified strains that underwent recombination, as well as the particular gene involved in each recombination event, using the “majority rules” rationale (14, 29). Using the maximum-parsimony criterion, a recombination-free phylogeny of all strains in the study was estimated with Mega (54), based on the concatenation of the three protein-coding genes, with recombinant fragments removed. The recombination-free alignment was used as input for both ES and AdaptML, with further accommodations for recombination made by ES, as discussed previously (29) (see also http://fcohan.web.wesleyan.edu).
We included in phylogenetic and ES analyses reference strains from each ecotype previously demarcated (29).
Ecotype Simulation and AdaptML analyses.
In ES, sequence data were analyzed to estimate the number of ecotypes (n) and the rates of ecotype formation (Ω) and periodic selection (σ), which were then used to infer the ecotype demarcations (see Fig. 2) (29). Given the extremely large population sizes and the high rates of worldwide migration in this clade (34, 40), we excluded genetic drift from the analysis, as previously discussed (29).
AdaptML used as input the habitat source of each organism, as well as the sequence-based phylogeny of the organisms (25). The algorithm yielded the rate of evolutionary transition in ecological adaptation (corresponding to the ecotype formation rate in ES), the demarcation of organisms into ecotypes (Fig. 2), and the habitat spectrum of each ecotype (the probability that a member of an ecotype is isolated from each habitat) (Table 1).
TABLE 1.
Habitat spectruma of each class of ecotypes identified by AdaptML
| Class of ecotypes | Habitat spectrum |
Ecotypes convergent on this habitat spectrum | ||||
|---|---|---|---|---|---|---|
| SFS |
NFS |
Arroyo, sandy | ||||
| Silty | Sandy | Silty | Sandy | |||
| Silt-arroyo | 0.19 | 0.25 | 0.10 | 0.06 | 0.40 | B, C, G |
| Sand-NFS | 0.06 | 0.16 | 0.12 | 0.52 | 0.14 | A, D, E, F |
The habitat spectrum represents the fraction of time that a lineage from a class of ecotypes is estimated to be in each habitat type.
Each ecotype demarcated by either algorithm represents a clade putatively adapted to a particular set of environments. In some cases, different ecotypes have independently converged on the same niche, at least as defined by adaptation to solar exposure and soil texture (Table 1).
FA assays.
Strains were grown on Trypticase soy broth agar (Difco) for 24 h at 28°C. Harvesting of the cells, saponification, methylation, and extraction were performed as recommended (45) for fatty acid (FA) evaluation with the Sherlock microbial identification system (MIDI, Inc., Newark, DE). The samples were analyzed on an Agilent Technologies 6890N gas chromatograph. The fatty acid content for each strain is reported as the percentage of each fatty acid among all fatty acids present.
Nucleotide sequence accession numbers.
All sequences produced for this study are available through GenBank. Accession numbers are GQ488877 to GQ489007 for dnaJ, GQ488617 to GQ488746 for gyrA, and GQ488747 to GQ488876 for rpoB.
RESULTS
Phylogeny.
We constructed a maximum-parsimony tree of the RFW isolates and reference strains, based on the recombination-free concatenation of the three protein-coding genes (dnaJ, gyrA, and rpoB) (Fig. 2). The tree confirms the previous result (38) that the B. subtilis-B. licheniformis clade is clearly divided into two subclades, one containing B. subtilis and its close relatives and the other containing B. licheniformis and B. sonorensis. The tree indicates several clades, unrecognized by systematics, that are closely related to named species, including those clades labeled in Fig. 2 as putative ecotypes (PE) 2 to 4, 15, 19 to 23, 25 to 26, and 29 to 31.
Bacterial systematists have used divergence in the 16S rRNA gene as a guide to demarcating species (51, 52), and so we measured 16S rRNA divergence to determine whether the unrecognized clades appear to be outside the boundaries of existing recognized species. Recognized species within the B. subtilis-B. licheniformis clade are frequently as little as 0.3% divergent in 16S rRNA (Table 2), which was about the level of divergence of clades PE 2 and PE 4 from each other and from the clade containing the type strain of B. licheniformis (PE 5). Other unrecognized clades were much more closely related than recognized species, e.g., PE 4 and PE 28, with a divergence of 0.07%.
TABLE 2.
Divergence in 16S rRNA sequence among putative ecotypes and recognized taxa
| Putative ecotype | Divergencea |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
B. subtilis subcladeb |
B. licheniformis subcladec |
|||||||||||||||
| PE 12 | PE 18 | PE 19 | PE 14 | PE 10 | PE 21 | PE 23 | PE 15 | PE 11 | PE 9B | PE 9A | PE 5 | PE 2 | PE 31 | PE 29 | PE 4 | |
| PE 12 | — | 0.18 | 0.08 | 0.28 | 0.08 | 0.08 | 0.15 | 0.15 | 0.08 | 0.11 | 0.17 | 1.56 | 1.23 | 1.58 | 1.44 | 1.51 |
| PE 18 | — | 0.18 | 0.67 | 0.18 | 0.18 | 0.18 | 0.36 | 0.18 | 0.36 | 0.36 | 1.46 | 1.46 | 1.46 | 1.46 | 1.46 | |
| PE 19 | — | 0.19 | 0.14 | 0.14 | 0.21 | 0.07 | 0.00 | 0.21 | 0.25 | 1.71 | 1.30 | 1.64 | 1.51 | 1.58 | ||
| PE 14 | 0.09 | 0.34 | 0.32 | 0.39 | 0.25 | 0.19 | 0.39 | 0.43 | 1.90 | 1.49 | 1.83 | 1.69 | 1.76 | |||
| PE 10 | — | 0.00 | 0.07 | 0.21 | 0.14 | 0.21 | 0.25 | 1.71 | 1.30 | 1.64 | 1.51 | 1.58 | ||||
| PE 21 | — | 0.07 | 0.21 | 0.14 | 0.21 | 0.25 | 1.71 | 1.30 | 1.64 | 1.51 | 1.58 | |||||
| PE 23 | — | 0.27 | 0.21 | 0.27 | 0.32 | 1.65 | 1.37 | 1.71 | 1.58 | 1.64 | ||||||
| PE 15 | — | 0.07 | 0.14 | 0.19 | 1.78 | 1.37 | 1.71 | 1.58 | 1.64 | |||||||
| PE 11 | — | 0.14 | 0.19 | 1.63 | 1.36 | 1.71 | 1.57 | 1.64 | ||||||||
| PE 9b | — | 0.05 | 1.78 | 1.37 | 1.71 | 1.58 | 1.64 | |||||||||
| PE 9a | 0.04 | 1.83 | 1.42 | 1.76 | 1.62 | 1.69 | ||||||||||
| PE 5 | 0.04 | 0.55 | 0.20 | 0.34 | 0.28 | |||||||||||
| PE 2 | — | 0.34 | 0.48 | 0.27 | ||||||||||||
| PE 28 | — | 0.14 | 0.07 | |||||||||||||
| PE 27 | — | 0.21 | ||||||||||||||
| PE 4 | — | |||||||||||||||
Comparisons within the B. subtilis subclade and within the B. licheniformis subclade are given in boldface. —, only one strain was sequenced from the ecotype, and therefore, the within-ecotype divergence is not indicated. From RFW isolates, 1,460 bp was sequenced; from other strains, 1,466 bp was available from the All Species Living Tree Project, with the exceptions of 1,169 bp for B. subtilis subsp. inaquosorum and 548 bp for B. tequilensis (63).
PE 12 is B. subtilis subsp. inaquosorum; PE 18 is B. tequilensis; PE 14 is B. vallismortis; PE 10 is B. subtilis subsp. subtilis; PE 15 and PE 11 are B. subtilis subsp. spizizenii; and PE 9B and PE 9A are B. mojavensis.
PE 5 is B. licheniformis.
We performed ES and AdaptML to determine which of these unrecognized clades and which of the recognized species and subspecies appear to be ecotypes.
Ecotype Simulation analysis.
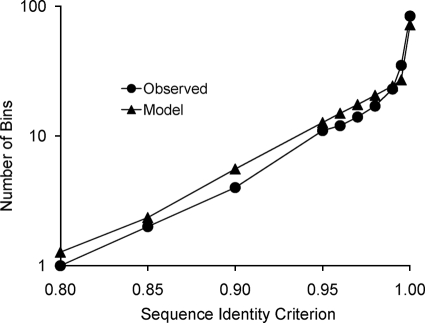
ES was performed on the three-gene concatenation of the RFW isolates and reference strains within the B. subtilis-B. licheniformis clade, excluding strains that contained a recombinant segment. To begin ES, the recombinant-free sequence diversity was characterized by the “clade sequence diversity” curve of Fig. 3 (3, 29, 32). This curve represents the history of cladogenesis among lineages of this clade that were included in our sample. The clade sequence diversity curve was the input data used by ES to estimate the rates of ecotype formation and periodic selection, as well as the number of ecotypes (n) (29).
FIG. 3.
Clade sequence diversity pattern for the B. subtilis-B. licheniformis clade. For both the observed and model curves, sequences for the concatenation of genes were binned by complete linkage clustering into clusters with different levels of minimum pairwise sequence identity, to yield the history of lineage splitting (29). The observed curve was based on the actual sequences from our survey of diversity, and the model curve was based on the average number of bins at each criterion level over 1,000 replicate runs of ES, using the parameter trio representing the maximum-likelihood solution (i.e., n = 28, Ω = 0.019, σ = 1.1).
ES estimated the rate of periodic selection (1.1; confidence interval [CI], 0.16 to 5.6) to be much greater than the rate of ecotype formation (0.019; CI, 0.013 to 0.029) (all rates are per nucleotide substitution in the concatenation), which is consistent with the Stable Ecotype model. ES estimated a total of 28 ecotypes in the B. subtilis-B. licheniformis clade (CI, 13 to 44). The fit of the ES-estimated parameters to the observed clade sequence diversity curve is shown in Fig. 3.
Ecotypes were then demarcated by finding the most inclusive subclades that were each consistent with containing a single ecotype (i.e., n = 1), as previously described (29) (Fig. 2). ES hypothesized 14 putative ecotypes beyond those previously identified (29): 11 putative ecotypes were discovered in RFW (PE 19 to 24 and PE 27 to 31), and 3 were recognized (or formerly recognized) taxa not included in our previous ES analysis but included here as reference strains: B. tequilensis (PE 18) and “B. velezensis” (PE 25 and 26).
For the most part, the ecotypes demarcated by ES corresponded very closely to the taxa recognized by systematics. Among previously classified strains, several taxa corresponded to putative ecotypes, including Bacillus subtilis subsp. inaquosorum (PE 12), Bacillus subtilis subsp. subtilis (PE 10), B. vallismortis (PE 14), B. atrophaeus (PE 8), B. sonorensis (PE 17), and B. licheniformis (PE 5). Exceptions included the more diverse taxon Bacillus subtilis subsp. spizizenii, which was split into two ecotypes (PE 11 and PE 15), and B. mojavensis, which was previously identified as PE 9 but is now split into PE 9A and 9B. Also, three strains of “B. velezensis,” currently classified as B. amyloliquefaciens, were split into two ecotypes (PE 25 and 26) separate from the type strain of B. amyloliquefaciens (Fig. 2).
Ecological distinctness of ecotypes hypothesized by ES.
Putative ecotypes demarcated by ES must be independently confirmed to be ecologically distinct, since under various evolutionary models (other than the Stable Ecotype model), a single ecotype may contain multiple sequence clusters (11, 29). We assayed for ecological distinctness by testing whether ecotypes were heterogeneous in their microgeographic associations with different habitat types, as defined by solar exposure and soil texture. We found a significant association between ecotypes and solar exposure (χ2 = 38.5, 12 df, P < 0.0001, including those ecotypes with ≥5 members sampled [Fig. 2]). Two ecotypes appeared highly specialized by solar exposure: PE 5 was found only on the more exposed south-facing slope and in the arroyo, and PE 15 was found largely on the north-facing slope.
Putative ecotypes demarcated by ES were also different in terms of their associations with soil texture (χ2 = 13.0; 6 df, P = 0.044), but specialization with soil texture was not as extreme as for solar exposure. The clade of PE 12 and PE 19 contained a much higher fraction of isolates from silty soil (45%) than the B. subtilis-B. licheniformis clade at large (22%).
Another approach to testing the ES-demarcated ecotypes for ecological distinctness involved the AdaptML algorithm, as we next explain.
AdaptML analysis.
The concatenation-based, recombination-free phylogeny (Fig. 2) and the habitat source of each strain (see Table S1 in the supplemental material) were the inputs for AdaptML. When both ecological dimensions (solar exposure and soil texture) were used as habitat source inputs, AdaptML recognized two classes of ecotypes, one relatively specialized to the arroyo and the silty soil of the slopes, and the other relatively specialized to sandy soil and the north-facing slope (Table 1). There was much overlap between the habitat spectra of these ecotype classes, especially along the soil texture dimension. For example, the two habitat spectra differed only by a factor of 1.5 in their affinities with silty soil (i.e., strains from the silt-arroyo-preferring ecotypes had a 29% probability of isolation from silty habitats, while strains from the sand-NFS ecotypes had a 19% probability of isolation from silty habitats [Table 1]), but the habitat spectra differed by a factor of 4.0 in their affinities with the south-facing slope (Table 1).
AdaptML hypothesized seven ecotypes among RFW isolates, four convergent on the sand-NFS ecotype class and three convergent on the silt-arroyo ecotype class (Fig. 2; Table 1). AdaptML and ES agreed on the demarcations of four putative ecotypes from RFW (PE 12, 19, 14, and 15). In other cases, a clade that was demarcated as a single ecotype by AdaptML was split by ES into multiple ecotypes, as seen for AdaptML ecotypes D, F, and G (Fig. 2).
While AdaptML found seven ecotypes when both ecological dimensions were included in the analysis (Table 1), the algorithm found only a single ecotype within the entire B. subtilis-B. licheniformis clade when either ecological dimension was analyzed alone (data not shown).
Physical, chemical, and biotic differences among habitats.
The “silty” soils contained about 50% silt and about 45% clay, with very little sand. The “sandy” soils contained about 55 to 80% sand and about 15 to 40% clay, with very little silt (see Table S2 in the supplemental material). The SFS sandy soil contained less sand than the sandy soils of the NFS and arroyo. Overall, the soil from RFW showed low organic content (<2% in all soils), high pH (ranging from 8.4 to 9.1), and low nutrient content, with most soils containing nitrogen at <8 ppm. One exception to the low nitrogen level was the south-facing silty soil (with 25.3 ppm); possibly there is more nitrogen-fixing activity in this soil. Boron, nitrogen, and organic content levels were highest in the silty soils.
Also, these habitats differed in the plants growing in each, with far fewer plant species inhabiting the SFS than either the arroyo or the NFS (see Table S3 in the supplemental material). The soil texture dimension had a profound impact on plant distribution; not a single plant was found on the silty soils of either slope.
Fatty acid content.
We analyzed the levels of the 11 major fatty acids in six ecotypes (or pools of ecotypes in cases where sample sizes were low) from the B. subtilis subclade. These fatty acids included four isomethyl-branched FAs, previously shown to contribute to adaptation to warmer temperatures, and two anteisomethyl-branched and three unsaturated FAs, previously shown to contribute to adaptation to cooler temperatures (19) (Table 3). For each isolate we calculated the sums of high-temperature-adapting (isomethyl-branched) and low-temperature-adapting (anteisomethyl-branched and unsaturated) FA levels, and we produced a heat adaptation index by dividing the high- by the low-temperature-adapting FA levels.
TABLE 3.
Fatty acid levels for each putative ecotype
| Putative ecotype | Mean (SD) level of fatty acida |
Heat adaptation indexb | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High-temp adapting |
Low-temp adapting |
|||||||||||||
| 14:0 | 16:0 | Iso 14:0 | Iso 15:0 | Iso 16:0 | Iso 17:0 | Sum | Anteiso 15:0 | Anteiso 17:0 | 16:1 ω11c | 16:1 ω7c alcohol | Iso 17:1 ω10c | Sum | ||
| PE 12 | 0.51 (0.39) | 3.33 (0.62) | 1.32 (0.31) | 28.77 (4.66) | 3.18 (1.07) | 9.13 (2.01) | 42.40 (4.07) | 39.06 (4.01) | 9.43 (1.78) | 1.93 (3.08) | 0.51 (0.19) | 1.24 (0.73) | 52.16 (3.13) | 0.82 (0.12) C |
| PE 19 | 0.29 (0.04) | 2.93 (0.45) | 0.95 (0.22) | 23.39 (1.72) | 3.31 (0.65) | 11.71 (1.04) | 39.35 (1.66) | 39.17 (1.80) | 12.96 (0.96) | 1.16 (0.13) | 0.55 (0.11) | 2.10 (0.28) | 55.92 (1.78) | 0.71 (0.05) BD |
| PE 20 to 23 | 0.36 (0.07) | 3.48 (0.89) | 1.13 (0.15) | 24.13 (3.18) | 3.21 (0.44) | 8.86 (1.02) | 37.33 (3.43) | 42.00 (2.11) | 12.16 (1.72) | 1.49 (0.26) | 0.49 (0.12) | 1.31 (0.51) | 57.45 (3.09) | 0.65 (0.10) BD |
| PE 14 | 0.30 (0.04) | 3.38 (0.35) | 1.32 (0.11) | 23.86 (2.09) | 4.10 (0.35) | 11.67 (1.31) | 40.95 (2.89) | 39.31 (1.75) | 11.63 (0.92) | 1.07 (0.20) | 0.67 (0.10) | 1.52 (0.28) | 54.20 (2.75) | 0.76 (0.09) BC |
| PE 15 | 0.24 (0.03) | 2.68 (0.53) | 0.97 (0.19) | 19.15 (2.56) | 3.47 (0.73) | 9.21 (1.01) | 32.81 (2.10) | 44.74 (1.44) | 15.10 (1.40) | 1.01 (0.17) | 0.57 (0.10) | 1.44 (0.38) | 62.87 (1.99) | 0.52 (0.05) A |
| PE 9A and 9B | 0.28 (0.11) | 2.72 (0.51) | 0.88 (0.16) | 20.71 (6.67) | 3.00 (0.51) | 8.54 (0.54) | 33.14 (6.85) | 41.81 (4.23) | 14.95 (2.51) | 1.60 (0.57) | 0.71 (0.08) | 2.31 (0.56) | 61.38 (6.69) | 0.56 (0.19) AB |
Expressed as a fraction of the total fatty acid content. In the case of ecotypes for which few isolates were sampled, clades of closely related putative ecotypes were pooled. The sums of high-temperature-adapting and low-temperature-adapting fatty acids are reported.
Ratio of high- to low-temperature-adapting fatty acids, given as mean (standard deviation). Capital letters A to D indicate the ecotypes and ecotype pools that are significantly different according to multcomp analysis (23); two ecotypes are significantly different if they do not share a capital letter.
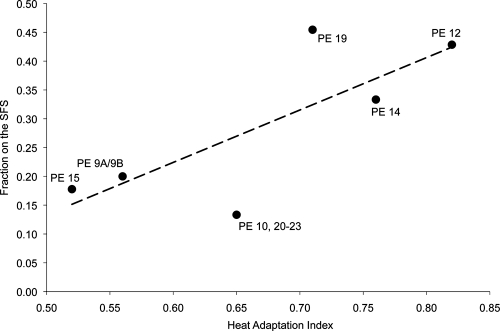
Putative ecotype 15 and the pool of PE 9A and 9B showed lower heat adaptation indices than other ecotypes; the heat adaptation index of PE 15 was significantly different from that of every other ecotype except the pool of PE 9A and 9B (Table 3). Also, the two very closely related ecotypes PE 12 and PE 19 were significantly different in their heat adaptation indices. Those ecotypes with higher heat adaptation indices tended to have a higher fraction of strains isolated from the south-facing slope (r = 0.77, 4 df, P = 0.036 by a one-tailed test) (Fig. 4).
FIG. 4.
Relationship between the mean heat adaptation index of a putative ecotype and the fraction of strains isolated from the south-facing slope. The heat adaptation index of each strain was quantified as the sum of the isomethyl-branched fatty acid levels divided by the sum of the anteisomethyl-branched and unsaturated fatty acid levels (Table 3).
We tested for possible ecological heterogeneity within the ecotypes demarcated by ES and AdaptML by analyzing the diversity of heat adaptation indices within ecotypes. Interestingly, in four of the five ecotypes with at least one individual isolated per slope, the strains isolated from the south-facing slope had a greater median heat adaptation index than the strains isolated from the north-facing slope. While not significant, this result suggests the possibility of ecological divergence within the hypothesized ecotypes.
Geography of isolates from silty soil.
The clade containing PE 12 and 19 was much more frequently isolated from silty soil than all other ecotypes. As shown in Fig. 1, strains of this silt-associated clade were isolated from silty and sandy sites throughout the study area.
DISCUSSION
We have aimed to identify the ecological dimensions of speciation in the B. subtilis-B. licheniformis clade. Our first step was to demarcate newly divergent, ecologically distinct clades (ecotypes) within the clade, with the goal of identifying the ecological differences between the young ecotypes. Rather than accept a universal molecular threshold of divergence for hypothesizing clades of biological significance, as in the established systematics of bacteria (11, 42, 51), we have utilized two algorithms (Ecotype Simulation and AdaptML) that analyze a clade's evolutionary history to determine the appropriate demarcations for the particular clade. Through Ecotype Simulation, we identified 31 sequence clusters within the B. subtilis-B. licheniformis clade as putative ecotypes, with 19 of these present in RFW; AdaptML identified 7 ecotypes in RFW. We next consider to what extent the putative ecotypes we identified represent the ecological diversity within the B. subtilis-B. licheniformis clade.
Ecological distinctness of the putative ecotypes.
Sequence clusters identified by ES are most likely to correspond to ecotypes if the Stable Ecotype model applies (11). However, under some alternative models of bacterial evolution, the clusters hypothesized to be ecotypes under ES might actually be ecologically interchangeable members of the same ecotype (11). This is the case, for example, under the Geotype-plus-Boeing model, where formerly geographically isolated populations (geotypes) have been mixed recently by human transport (11), possibly the case for geotypes of Yersinia pestis (1, 62). To accommodate the Geotype-plus-Boeing and other models, the ecotypes hypothesized by ES must be independently confirmed as ecologically distinct (8, 11, 29).
We followed three steps to confirm the ecological distinctness of ecotypes. First, we tested whether the ecotypes indicated by sequence-based analysis were significantly different in their habitat associations within the Death Valley canyon, with the rationale that heterogeneity in habitat associations would indicate the ecotypes’ ecological distinctness. As noted by Hunt et al. (25), confirming ecological distinctness through habitat differences is a conservative process: statistical power to distinguish ecotypes is reduced when ecotypes are only quantitatively different in their habitat preferences (i.e., when they share the same habitats); likewise, in the case of Bacillus, statistical power is reduced when ecotypes are occasionally isolated as spores that have randomly settled on a habitat that the ecotype does not prefer. We show below that, despite overlap in habitats, the ecotypes hypothesized by sequence-based analyses were significantly different in their habitat associations. Second, we examined whether the habitat associations we discovered were due to intrinsic physiological differences among ecotypes. Finally, we examined whether the ecotypes identified by ES and AdaptML were each ecologically homogeneous.
Ecological distinctness of the putative ecotypes hypothesized by ES was suggested by differences in their habitat associations within the Death Valley canyon. In contingency tests, the putative ecotypes hypothesized by ES were significantly heterogeneous in both the solar exposure and soil texture dimensions (Fig. 2). Also, an AdaptML analysis implicated both ecological dimensions in the divergence of ecotypes. While AdaptML analysis of both environmental dimensions together identified multiple ecotypes, separate analyses of each environmental dimension alone yielded no evidence of ecological specialization (i.e., the whole B. subtilis-B. licheniformis clade was deemed a single ecotype). We may conclude that ecological specialization has involved both solar exposure and soil texture, perhaps through an interaction of the two.
AdaptML and ES identified some of the same ecotypes within the B. subtilis clade, confirming that these ecotypes are distinct in one or both of the ecological dimensions analyzed. For example, two very closely related clades, identified as PE 12 and PE 19, were assigned as distinct ecotypes by both ES and AdaptML, with PE 12 assigned to the sand-NFS habitat spectrum and PE 19 assigned to the silt-arroyo habitat spectrum. In other cases, AdaptML pooled into a single ecotype several clades that were hypothesized to be separate ecotypes by ES; this was the case for the clades labeled as putative ecotypes D, F, and G by AdaptML (Fig. 2).
The greater splitting of ecotypes by ES than by AdaptML is consistent with differences in approach between these algorithms. ES does not utilize as an input any ecological data about the organisms (e.g., habitat source data) and so works independently of the investigators’ knowledge of the ecological dimensions of divergence. It can therefore hypothesize ecotypes whose ecological divergence is beyond the imagination of the investigators; the downside is that the ecological distinctness of putative ecotypes identified by ES must be independently confirmed (10). Because AdaptML utilizes habitat source data, it provides the advantage that it can hypothesize the demarcations of ecotypes and confirm the ecological distinctness of ecotypes, all in one step; however, AdaptML can identify only those ecotypes that are divergent in the ecological dimensions specified by the investigator (10). Thus, our results suggest that the multiple ecotypes hypothesized by ES within an AdaptML ecotype (e.g., within AdaptML's ecotype D) are divergent in some other ecological dimensions not included in our analysis. Testing this could involve sampling along other ecological axes (e.g., the rhizospheres of different plant species, or associations with different bacterial communities [10]), or by physiological comparisons, aided perhaps by genome content analyses (16, 17) or by genomewide gene expression analyses (E. Becraft, F. M. Cohan, A. F. Koeppel, and D. M. Ward, unpublished data).
Even where ES and AdaptML agree in their ecotype demarcations, they suggest the existence of additional, unidentified ecological dimensions of early ecotype divergence. This is because different demarcated ecotypes have converged on the same ecological niche, as defined by solar exposure and soil texture. For example, within the B. subtilis subclade, the clades labeled by AdaptML as ecotypes B and C represent convergences onto the silt-arroyo habitat spectrum, and ecotypes A, D, and E have converged on the sand-NFS spectrum (Table 1). The long-term coexistence of these ecotypes is likely to require ecological divergence along some other, currently unknown ecological dimension(s) (26).
We next consider whether the differences in habitat association among putative ecotypes could be based on accidents of colonization history rather than on ecological specialization. For example, one might argue that the difference in habitat association between PE 15 and PE 12 was caused by initial settling of these clades on RFW's north- and south-facing slopes, respectively, with very little subsequent dispersal. Such a biogeographic explanation is unlikely, because Bacillus has been shown to migrate at extremely high rates, even between continents (34, 40), and because Bacillus spores are known to resist environmental stresses (37).
In the case of ecotype associations with silty versus sandy soil, we may further rule out colonization history in explaining habitat associations by considering the geography of isolates. A model of habitat association by colonization history alone would predict that the outlier, the silt-associated clade containing PE 12 and 19, would be found only in a small number of silty collection sites. However, the clade was isolated from multiple silty and sandy collection points throughout our study site in RFW (Fig. 1). It would appear that the predominance of this clade in silty soil, and more generally the significant heterogeneity of ecotype associations with soil texture, is due to ecological specialization and not to colonization history. Eventual identification of the physiological differences underlying these habitat associations may include adaptations to the physical soil differences described in Table S2 in the supplemental material and/or the plant resource differences described in Table S3 in the supplemental material.
In the case of ecotype associations with solar exposure, our fatty acid data provide direct evidence of adaptation to warmer temperatures for those putative ecotypes most tightly associated with the SFS. As seen in previous studies (29, 48), those ecotypes most frequently isolated from the SFS of Radio Facility Wash had significantly higher heat adaptation index levels (based on fatty acid content) than ecotypes less frequently isolated from the SFS (Fig. 4). In contrast to the previous association between solar exposure association and fatty acid content, the RFW case is especially interesting, because here the association is not between nearly absolute specialists for particular slopes (49) but rather among slope generalists that are more subtly different in their slope associations.
Are the ecotypes identified ecologically homogeneous?
We next consider whether the ecotypes we have resolved represent the most immediate products of speciation. This requires not just that the putative ecotypes be ecologically distinct but also that the membership of each ecotype be ecologically homogeneous. If an ecotype we identify contains strains specialized to different environments, the identified “ecotype” may actually contain several newly divergent ecotypes that have not yet had time to accumulate neutral nucleotide substitutions in the marker loci.
This possibility is suggested by a previous survey of genomic diversity within PE 10, which corresponds to B. subtilis subsp. subtilis (Fig. 2) (16). Members of this ecotype contain different sets of genes involved in the uptake and metabolism of carbohydrates, indicating possible niche specialization to different food resources, as well as different sets of genes involved in sporulation and germination, suggesting life in different environments with different optimal cues for successful germination. However, no ecological differences among the strains studied have been confirmed (16). The present study offers some suggestive evidence for extremely newly divergent ecotypes beyond the resolution of nucleotide substitutions at our three loci. We found that within the ecotypes identified, the strains isolated from the SFS tended (not significantly) to have a higher heat adaptation index than isolates from the NFS. This is consistent with the existence of multiple temperature-distinguished ecotypes within the ES-identified ecotypes. This hypothesis could be tested through analysis of a more rapidly evolving molecular marker (10), such as variable-number tandem repeat (VNTR) loci (27), or by a genomewide single-nucleotide polymorphism (SNP) analysis (18). Here we would expect to find two or more molecular clusters within a given ES-identified ecotype, one with a significantly higher mean heat adaptation index and greater SFS association than the other.
Nevertheless, until the diversity within ES-identified ecotypes is confirmed, we will consider the ecotypes ES has identified to be our best approximation for the immediate products of speciation and our best clue for identifying the ecological dimensions of speciation. This study thus adds soil texture and further confirms solar exposure as ecological dimensions of speciation in the B. subtilis-B. licheniformis clade.
Implications for Bacillus systematics.
The recognized species and subspecies of the B. subtilis-B. licheniformis clade were nearly identical to ecotypes demarcated by ES and AdaptML, with a few exceptions in which a recognized taxon was split into two or three ecotypes. This close correspondence between hypothesized ecotypes and recognized species and subspecies contrasts with the findings for other taxa, where a single species may contain many ecotypes discernible through ES or AdaptML analysis of multiple loci, as seen within B. simplex (29), Vibrio splendidus (25), and hot spring Synechococcus (61).
Systematists of the B. subtilis-B. licheniformis clade thus appear to have effectively recognized the ecological diversity among close relatives. Perhaps one element of the successful approach of Bacillus systematists has been to demarcate species with very little divergence in the 16S rRNA gene, with closely related species frequently divergent by as little as 0.3% (Table 2). While many of the ecotypes within this clade are formally recognized by systematics, we are only beginning to identify the ecological dimensions on which the named taxa have diverged; four of the five named species and subspecies isolated from RFW were adapted to the same sand-NFS habitat spectrum (Fig. 2; Table 1).
The recognized taxa do not appear to encompass the full diversity of the B. subtilis-B. licheniformis clade. The present study and our previous study on Negev Desert isolates within this clade (29) have discovered numerous putative ecotypes that are each closely related to a recognized taxon. In one case (PE 19 versus B. subtilis subsp. inaquosorum), the ecological dimensions of divergence have been identified, but the ecological dimensions of other divergences have not been worked out. Based on these results, we suggest that the B. subtilis-B. licheniformis clade is replete with newly divergent ecotypes waiting to be confirmed as ecologically distinct. Bacillus ecologists and systematists are challenged to discover the yet unknown ecological dimensions of speciation leading to the putative ecotypes identified here.
Toward a universal approach to discovering a clade's ecological diversity.
This study has provided a general protocol for identifying the ecological dimensions of speciation by using theory-based algorithms to identify newly divergent, ecologically distinct clades. Demarcating these clades does not require experience or intuition with the taxon, since ES utilizes only sequence data, although for the moment, confirming the ecological distinctness of putative ecotypes does require intuition and knowledge informing us of the likely ecological dimensions of early divergence. This requirement for experience would change if systematists of a focus clade were to characterize habitats by the other organisms that grow there. Just as large terrestrial biomes may be characterized by their dominant plants (e.g., grassland), a bacterial habitat may be characterized by its dominant bacteria, through assays of 16S rRNA sequence diversity (31). Biotic characterization of communities may allow putative ecotypes within a focus clade to be confirmed as distinct by their associations with other bacteria. The ecological dimensions of divergence could then be identified by previous knowledge of the preferred habitats of taxa outside the focus clade (10). This general approach will allow us to move all the way from surveying sequence diversity within a focus clade, to hypothesizing and confirming ecotypes, and then to identifying the dimensions of ecological divergence within the focus clade, all by using universal methods and leveraging what is already known from other taxa.
Supplementary Material
Acknowledgments
We thank Will Cohan for assistance in collecting soil in Death Valley.
We are grateful to the National Park Service for supporting our research in Death Valley. This research was supported by funds awarded to J.S. from a DFG grant (SI 1352/1-2) and to F.M.C. from an NSF FIBR grant (EF-0328698), as well as by research funds from Wesleyan University.
Footnotes
Published ahead of print on 4 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. U. S. A. 101:17837-17842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., and M. Wagner. 2008. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6:431-440. [DOI] [PubMed] [Google Scholar]
- 3.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 4.Cohan, F. M. 2001. Bacterial species and speciation. Syst. Biol. 50:513-524. [DOI] [PubMed] [Google Scholar]
- 5.Cohan, F. M. 1994. The effects of rare but promiscuous genetic exchange on evolutionary divergence in prokaryotes. Am. Nat. 143:965-986. [DOI] [PubMed] [Google Scholar]
- 6.Cohan, F. M. 2005. Periodic selection and ecological diversity in bacteria, p. 78-93. In D. Nurminsky (ed.), Selective sweep. Landes Bioscience, Georgetown, TX.
- 7.Cohan, F. M. 1996. The role of genetic exchange in bacterial evolution. ASM News 62:631-636. [Google Scholar]
- 8.Cohan, F. M. 2006. Towards a conceptual and operational union of bacterial systematics, ecology, and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1985-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohan, F. M., A. Koeppel, and D. Krizanc. 2006. Sequence-based discovery of ecological diversity within Legionella, p. 367-376. In N. P. Cianciotto, Y. Abu Kwaik, P. H. Edelstein, B. S. Fields, D. F. Geary, T. G. Harrison, C. Joseph, R. M. Ratcliff, J. E. Stout, and M. S. Swanson (ed.), Legionella: state of the art 30 years after its recognition. ASM Press, Washington, DC.
- 10.Cohan, F. M., and A. F. Koeppel. 2008. The origins of ecological diversity in prokaryotes. Curr. Biol. 18:R1024-R1034. [DOI] [PubMed] [Google Scholar]
- 11.Cohan, F. M., and E. B. Perry. 2007. A systematics for discovering the fundamental units of bacterial diversity. Curr. Biol. 17:R373-R386. [DOI] [PubMed] [Google Scholar]
- 12.Cohan, F. M., M. S. Roberts, and E. C. King. 1991. The potential for genetic exchange by transformation within a natural population of Bacillus subtilis. Evolution 45:1393-1421. [DOI] [PubMed] [Google Scholar]
- 13.de Queiroz, K. 2005. Ernst Mayr and the modern concept of species. Proc. Natl. Acad. Sci. U. S. A. 102(Suppl. 1):6600-6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Didelot, X., and D. Falush. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dykhuizen, D. E. 1998. Santa Rosalia revisited: why are there so many species of bacteria? Antonie Van Leeuwenhoek 73:25-33. [DOI] [PubMed] [Google Scholar]
- 16.Earl, A. M., R. Losick, and R. Kolter. 2007. Bacillus subtilis genome diversity. J. Bacteriol. 189:1163-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earl, A. M., R. Losick, and R. Kolter. 2008. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, J. T., S. M. Beckstrom-Sternberg, T. Pearson, J. S. Beckstrom-Sternberg, P. S. Chain, F. F. Roberto, J. Hnath, T. Brettin, and P. Keim. 2009. Whole-genome-based phylogeny and divergence of the genus Brucella. J. Bacteriol. 191:2864-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fulco, A. J. 1983. Fatty acid metabolism in bacteria. Prog. Lipid Res. 22:133-160. [DOI] [PubMed] [Google Scholar]
- 20.Gans, J., M. Wolinsky, and J. Dunbar. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387-1390. [DOI] [PubMed] [Google Scholar]
- 21.Gatson, J. W., B. F. Benz, C. Chandrasekaran, M. Satomi, K. Venkateswaran, and M. E. Hart. 2006. Bacillus tequilensis sp. nov., isolated from a 2000-year-old Mexican shaft-tomb, is closely related to Bacillus subtilis. Int. J. Syst. Evol. Microbiol. 56:1475-1484. [DOI] [PubMed] [Google Scholar]
- 22.Goris, J., K. T. Konstantinidis, J. A. Klappenbach, T. Coenye, P. Vandamme, and J. M. Tiedje. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57:81-91. [DOI] [PubMed] [Google Scholar]
- 23.Hothorn, T., F. Bretz, and P. Westfall. 2008. Simultaneous inference in general parametric models. Biom. J. 50:346-363. [DOI] [PubMed] [Google Scholar]
- 24.Huber, J. A., D. B. Mark Welch, H. G. Morrison, S. M. Huse, P. R. Neal, D. A. Butterfield, and M. L. Sogin. 2007. Microbial population structures in the deep marine biosphere. Science 318:97-100. [DOI] [PubMed] [Google Scholar]
- 25.Hunt, D. E., L. A. David, D. Gevers, S. P. Preheim, E. J. Alm, and M. F. Polz. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081-1085. [DOI] [PubMed] [Google Scholar]
- 26.Hutchinson, G. E. 1959. Homage to Santa Rosalia or why are there so many kinds of animals? Am. Nat. 93:145-159. [Google Scholar]
- 27.Kenefic, L. J., J. Beaudry, C. Trim, R. Daly, R. Parmar, S. Zanecki, L. Huynh, M. N. Van Ert, D. M. Wagner, T. Graham, and P. Keim. 2008. High resolution genotyping of Bacillus anthracis outbreak strains using four highly mutable single nucleotide repeat markers. Lett. Appl. Microbiol. 46:600-603. [DOI] [PubMed] [Google Scholar]
- 28.Koch, A. L. 1974. The pertinence of the periodic selection phenomenon to prokaryote evolution. Genetics 77:127-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koeppel, A., E. B. Perry, J. Sikorski, D. Krizanc, W. A. Warner, D. M. Ward, A. P. Rooney, E. Brambilla, N. Connor, R. M. Ratcliff, E. Nevo, and F. M. Cohan. 2008. Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc. Natl. Acad. Sci. U. S. A. 105:2504-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin, B. R. 1981. Periodic selection, infectious gene exchange and the genetic structure of E. coli populations. Genetics 99:1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lozupone, C. A., and R. Knight. 2007. Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U. S. A. 104:11436-11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McVean, G., P. Awadalla, and P. Fearnhead. 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160:1231-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merrill, L., J. Dunbar, J. Richardson, and C. R. Kuske. 2006. Composition of Bacillus species in aerosols from 11 U.S. cities. J. Forensic Sci. 51:559-565. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura, L. K., M. S. Roberts, and F. M. Cohan. 1999. Relationship of Bacillus subtilis clades associated with strains 168 and W23: a proposal for Bacillus subtilis subsp. subtilis subsp. nov. and Bacillus subtilis subsp. spizizenii subsp. nov. Int. J. Syst. Bacteriol. 49:1211-1215. [DOI] [PubMed] [Google Scholar]
- 36.Nevo, E. 1995. Asian, African and European biota meet at ‘Evolution Canyon’ Israel: local tests of global biodiversity and genetic diversity patterns. Proc. R. Soc. Lond. B Biol. Sci. 262:149-155. [Google Scholar]
- 37.Nicholson, W. L., and A. C. Schuerger. 2005. Bacillus subtilis spore survival and expression of germination-induced bioluminescence after prolonged incubation under simulated Mars atmospheric pressure and composition: implications for planetary protection and lithopanspermia. Astrobiology 5:536-544. [DOI] [PubMed] [Google Scholar]
- 38.Palmisano, M. M., L. K. Nakamura, K. E. Duncan, C. A. Istock, and F. M. Cohan. 2001. Bacillus sonorensis sp. nov., a close relative of Bacillus licheniformis, isolated from soil in the Sonoran Desert, Arizona. Int. J. Syst. Evol. Microbiol. 51:1671-1679. [DOI] [PubMed] [Google Scholar]
- 39.PRISM Climate Group. 2009. Mean minimum and maximum temperatures. http://www.prism.oregonstate.edu/.
- 40.Roberts, M. S., and F. M. Cohan. 1995. Recombination and migration rates in natural populations of Bacillus subtilis and Bacillus mojavensis. Evolution 49:1081-1094. [DOI] [PubMed] [Google Scholar]
- 41.Rooney, A. P., N. Price, C. Ehrhardt, J. L. Swezey, and J. D. Bannan. 2009. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int. J. Syst. Evol. Microbiol. 59:2429-2436. [DOI] [PubMed] [Google Scholar]
- 42.Rosselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 43.Ruiz-García, C., V. Bejar, F. Martinez-Checa, I. Llamas, and E. Quesada. 2005. Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Velez in Malaga, southern Spain. Int. J. Syst. Evol. Microbiol. 55:191-195. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz-García, C., E. Quesada, F. Martinez-Checa, I. Llamas, M. C. Urdaci, and V. Bejar. 2005. Bacillus axarquiensis sp. nov. and Bacillus malacitensis sp. nov., isolated from river-mouth sediments in southern Spain. Int. J. Syst. Evol. Microbiol. 55:1279-1285. [DOI] [PubMed] [Google Scholar]
- 45.Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. Technical note 101. Microbial ID, Inc., Newark, DE.
- 46.Reference deleted.
- 47.Sikorski, J. 2008. Populations under microevolutionary scrutiny: what will we gain? Arch. Microbiol. 189:1-5. [DOI] [PubMed] [Google Scholar]
- 48.Sikorski, J., E. Brambilla, R. M. Kroppenstedt, and B. J. Tindall. 2008. The temperature adaptive fatty acid content in Bacillus simplex strains from “Evolution Canyon”, Israel. Microbiology 154:2416-2426. [DOI] [PubMed] [Google Scholar]
- 49.Sikorski, J., and E. Nevo. 2007. Patterns of thermal adaptation of Bacillus simplex to the microclimatically contrasting slopes of ‘Evolution Canyons’ I and II, Israel. Environ. Microbiol. 9:716-726. [DOI] [PubMed] [Google Scholar]
- 50.Smith, N. H., K. Kremer, J. Inwald, J. Dale, J. R. Driscoll, S. V. Gordon, D. van Soolingen, R. G. Hewinson, and J. M. Smith. 2006. Ecotypes of the Mycobacterium tuberculosis complex. J. Theor. Biol. 239:220-225. [DOI] [PubMed] [Google Scholar]
- 51.Stackebrandt, E., and J. Ebers. 2006. Taxonomic parameters revisited: tarnished gold standards. Microbiol. Today 33:152-155. [Google Scholar]
- 52.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. Grimont, P. Kampfer, M. C. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 53.Staley, J. T. 2006. The bacterial species dilemma and the genomic-phylogenetic species concept. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1899-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Torsvik, V., F. L. Daae, R. A. Sandaa, and L. Ovreas. 1998. Novel techniques for analysing microbial diversity in natural and perturbed environments. J. Biotechnol. 64:53-62. [DOI] [PubMed] [Google Scholar]
- 57.Vos, M., and X. Didelot. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3:199-208. [DOI] [PubMed] [Google Scholar]
- 57a.Wahl, R. R., D. A. Sawyer, S. A. Minor, M. D. Carr, J. C. Cole, W. C. Swadley, R. J. Laczniak, R. G. Warren, K. S. Green, and C. M. Engle. 1997. Digital geologic map database of the Nevada Test Site area, Nevada. U.S. Geological Survey open-file report 97-140. U.S. Geological Survey, Denver, CO.
- 58.Wang, L. T., F. L. Lee, C. J. Tai, and H. P. Kuo. 2008. Bacillus velezensis is a later heterotypic synonym of Bacillus amyloliquefaciens. Int. J. Syst. Evol. Microbiol. 58:671-675. [DOI] [PubMed] [Google Scholar]
- 59.Wang, L. T., F. L. Lee, C. J. Tai, A. Yokota, and H. P. Kuo. 2007. Reclassification of Bacillus axarquiensis Ruiz-Garcia et al. 2005 and Bacillus malacitensis Ruiz-Garcia et al. 2005 as later heterotypic synonyms of Bacillus mojavensis Roberts et al. Int. J. Syst. Evol. Microbiol. 57:1663-1667. [DOI] [PubMed] [Google Scholar]
- 60.Ward, D. M. 1998. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1:271-277. [DOI] [PubMed] [Google Scholar]
- 61.Ward, D. M., M. M. Bateson, M. J. Ferris, M. Kühl, A. Wieland, A. Koeppel, and F. M. Cohan. 2006. Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1997-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward, D. M., F. M. Cohan, D. Bhaya, J. F. Heidelberg, M. Kuhl, and A. Grossman. 2008. Genomics, environmental genomics and the issue of microbial species. Heredity 100:207-219. [DOI] [PubMed] [Google Scholar]
- 63.Yarza, P., M. Richter, J. Peplies, J. Euzeby, R. Amann, K. H. Schleifer, W. Ludwig, F. O. Glockner, and R. Rossello-Mora. 2008. The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31:241-250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.