Abstract
Methods for rapid detection and quantification of infectious viruses in the environment are urgently needed for public health protection. A fluorescence-activated cell-sorting (FACS) assay was developed to detect infectious adenoviruses (Ads) based on the expression of viral protein during replication in cells. The assay was first developed using recombinant Ad serotype 5 (rAd5) with the E1A gene replaced by a green fluorescent protein (GFP) gene. Cells infected with rAd5 express GFP, which is captured and quantified by FACS. The results showed that rAd5 can be detected at concentrations of 1 to 104 PFU per assay within 3 days, demonstrating a linear correlation between the viral concentration and the number of GFP-positive cells with an r2 value of >0.9. Following the same concept, FACS assays using fluorescently labeled antibodies specific to the E1A and hexon proteins, respectively, were developed. Assays targeting hexon showed greater sensitivity than assays targeting E1A. The results demonstrated that as little as 1 PFU Ads was detected by FACS within 3 days based on hexon protein, with an r2 value greater than 0.9 over a 4-log concentration range. Application of this method to environmental samples indicated positive detection of infectious Ads in 50% of primary sewage samples and 33% of secondary treated sewage samples, but none were found in 12 seawater samples. The infectious Ads ranged in quantity between 10 and 165 PFU/100 ml of sewage samples. The results indicate that the FACS assay is a rapid quantification tool for detecting infectious Ads in environmental samples and also represents a considerable advancement for rapid environmental monitoring of infectious viruses.
Waterborne viral infection is one of the most important causes of human morbidity in the world. There are hundreds of different types of human viruses present in human sewage, which, if improperly treated, may become the source of contamination in drinking and recreational waters (6, 12, 19). Furthermore, as water scarcity intensifies in the nation, so has consideration of wastewater reuse as a valid and essential alternative for resolving water shortages (31).
Currently, routine viral monitoring is not required for drinking or recreational waters, nor is it required for wastewater that is discharged into the environment. This lack of a monitoring effort is due largely to the lack of methods that can rapidly and sensitively detect infectious viruses in environmental samples. In the past 20 years, tremendous progress has been made in detection of viruses in the environment based on molecular technology (32, 33, 35). PCR and quantitative real-time PCR (qPCR) methods have improved both the speed and sensitivity of viral detection compared with detection by the traditional tissue culture method (2, 11, 17, 18). However, they provide little information on viral infectivity, which is crucial for human health risk assessment (22-24, 35). Our previous work using a real-time PCR assay to detect human adenoviruses (Ads) in sewage could not differentiate the infectious viruses in the secondary treated sewage from those killed by chlorination disinfection (15). In this research, we pursued an innovative approach to detecting infectious viruses in water using fluorescence-activated cell sorting (FACS). This method is rapid and sensitive, with an established record in microbiological research (29, 34, 39).
FACS is a specialized type of flow cytometry which provides a method for counting and sorting a heterogeneous mixture of biological cells into two or more kinds, one cell at a time, based upon the specific light-scattering and fluorescent characteristics of each cell (4, 25, 34, 38). It is a useful method since it provides fast and quantitative recording of fluorescent signals from individual cells (14, 16, 34, 47). The FACS viral assay is based on the expression of viral protein inside the recipient cell during viral replication (16). Specific antibody labeled with fluorescence is bound to the target viral protein, which results in fluorescence emission from infected cells. Viral particles outside the cell will not be captured, because the size of virus is below the detection limit of flow cytometry. Therefore, detection of cells, which can be captured with fluorescently labeled viral antibody, is a definitive indication of the presence of infectious virus.
This research used human Ads as the target for development of the FACS method. The rationale for this choice is as follows. (i) Ads are important human pathogens that may be transmitted by water consumption and water spray (aerosols) (26, 32). The health hazard associated with exposure to Ads has been demonstrated by epidemiological data and clinical research (1, 7, 9, 35, 40, 43). (ii) Ads are among the most prevalent human viruses identified in human sewage and are frequently detected in marine waters and the Great Lakes (17, 32, 33, 35). (iii) Ads are more resistant to UV disinfection than any other bacteria or viruses (3, 5, 10, 24, 41, 42, 44). Thus, they may survive wastewater treatment as increasing numbers of wastewater treatment facilities switch from chlorination to UV to avoid disinfection by-products. (iv) Some serotypes of Ads, including enteric Ad 40 and 41, are fastidious. They are difficult to detect by plaque assay, and a routine assay of infectivity takes 7 to 14 days (8, 20).
In this study, recombinant Ad serotype 5 (rAd5) with the E1A gene (the first transcribed gene after infection) replaced by a green fluorescent protein (GFP) gene was first used to test for sensitivity and speed of the assay. Two other viral proteins were then used as targets for development of FACS assays using Ad serotype 2 (Ad2) and Ad41. This study demonstrated the feasibility, sensitivity, and reliability of the assay for detection of infectious Ads in environmental samples.
MATERIALS AND METHODS
Adenoviruses, host cells, and plaque assays.
Ad2 and rAd5 were obtained from the University of Southern California (courtesy of Michael Lai). rAd5 is a recombinant virus in which the E1A gene is replaced by the GFP gene, which can express GFP during viral replication within cells. Ad41 was kindly provided to the laboratory by Shawn Tompson at the Los Angeles Sanitation District.
A human embryonic kidney cell line (HEK-293A) was also obtained from the University of Southern California (courtesy of Michael Lai) and was used in this study between passages 42 and 50. HEK-293A is a mutant of the original Graham HEK-293 cell (ATCC CRL-1573), which expresses the viral proteins from transforming genes of Ad5, including E1A and E1B (13). HEK-293A has a better ability to adhere to a petri dish surface to form a monolayer culture for plaque assay (20). Human lung carcinoma cell line A549 was obtained from the Los Angeles Sanitation District (courtesy of Shawn Thompson) at passage 108 and was used in this study between passages 115 and 123. The culture conditions for these two cell lines have been described by Jiang et al. (20).
Adenovirus plaque assays using HEK-293A and A549 were developed and described previously (20). Briefly, serially diluted Ads were inoculated onto confluent cells in 6-well plates and incubated for 1.5 h, with gentle rocking every 20 min for viral adsorption. Then, inoculated cells were overlaid with 1.25% agarose, containing nutrients and antibiotics. A second overlay was applied at 4 to 5 days postinfection. Inoculated cell cultures were examined microscopically daily up to 2 weeks postinfection, and plaques were counted at 10 days.
FACS analyses.
FACS analysis was performed using a BD FACSCanto II flow cytometer (BD Biosciences, San Jose, CA). The instrument was set up to measure forward-angle scattered light (FSC) (483 to 493 nm), side-angle scattered light (SSC) (483 to 493 nm), green fluorescence light (FL-l) (515 to 545 nm), and red fluorescence light (FL-4) (653 to 669 nm). Data were collected for at least 10,000 events in the BD Cell Quest software program (BD Biosciences) and were analyzed offline using the free software program WinMDI (version 2.9; available at http://facs.scripps.edu/software.html).
For detection of rAd5 with GFP, HEK-293A cells grown in 12-well plates (3 × 105 cells/well) were infected with rAd5 at concentrations between 1 and 104 PFU per well for 1, 2, 3, 4, and 5 days. The cells were collected, washed with prewarmed phosphate-buffered saline (PBS) and 1× binding buffer (BB) (1.4 M NaCl, 0.025 M CaCl2, and 0.1 M HEPES), and then fixed with 2% formaldehyde in 1× BB before injection into a FACS. Infected cells emitting GFP fluorescence were detected by the FL-1 filter (515 to 545 nm).
Detection of rAd5 was also performed based on the viral hexon protein to compare with the rAd5 GFP assay. HEK-293A cells grown in 12-well plates (3 × 105 cells/well) were infected with rAd5 at concentrations between 1 and 104 PFU/well for 3 days. The cells were then collected, washed with prewarmed PBS and 1× BB, and incubated with 50 μl of permeate buffer (BD Biosciences) and 50 μl of hexon primary antibody (Natutec, Frankfurt, Germany) at a concentration of 10 μg/ml, 20 μg/ml, or 30 μg/ml for 30 min on ice or at 4°C in the dark. After two washes in PBS with 10% fetal bovine serum (FBS), the cells were incubated with 50 μl of permeate buffer and 100 μl of a 1:400 dilution (2 mg/ml) of goat antimouse secondary antibody labeled with Alexa Fluor 647 (Invitrogen, Carlsbad, CA) on ice for 30 min in the dark. After two more washes, the cells were fixed in 1× BB with 2% paraformaldehyde and analyzed for green (515 to 545 nm) and red (653 to 669 nm) fluorescence by flow cytometry.
FACS detection of rAd5, Ad2, and Ad41 by using viral hexon protein in HEK-293A cells was also tested for 2, 3, and 5 days of incubation. The experimental setup for these assays was similar to the viral hexon assay described above except that the optimized hexon antibody concentration of 20 μg/ml was used. Infected cells were analyzed for red (FL-4; 653 to 669 nm) fluorescence by flow cytometry.
For FACS assay of Ads based on viral E1A, A549 cells grown in 12-well plates (3 × 105 cells/well) were used to replace HEK-293A because HEK-293A interferes with the assay of the viral E1A protein due to the presence of the transformed viral gene. Ad2 was inoculated into A549 cells at concentrations between 1 and 104 PFU/well for 5 days. Detection of Ad2 based on the viral E1A protein was performed using an experimental setup similar to that for the hexon protein except that mouse anti-E1A monoclonal antibody (Abcam, Cambridge, MA) was used as the primary antibody. After secondary antibody binding and fixation, the cells were injected into the FACS for detection of fluorescent cells using red fluorescence (FL-4; 653 to 669 nm).
Comparison of FACS and qPCR assay of Ads.
Approximately 105 PFU/ml rAd5 in PBS was exposed to 254-nm UV light in a 6-well cell culture plate (BD Falcon, NJ). A collimated-beam UV apparatus (Spectrolinker XL-1000 UV cross-linker; Spectronics Corp., Westbury, NY) emitting nearly monochromatic UV radiation at 254 nm was used, and the UV intensity was set at 0.5 W/cm2. After periods (0, 80, 160, and 280 s) of UV exposure, samples (100 μl each) were taken and subjected to FACS assay and qPCR assay. The UV exposure doses were 0, 40, 80, and 140 mJ/cm2 in the present study, which were calculated from the exposure time and UV intensity.
A FACS assay based on rAd5 GFP after a 3-day incubation was performed as described above. For qPCR, the Ad genome was extracted from the UV-treated samples using a QIAmp viral RNA minikit (Qiagen Inc., California). PCR protocols and primers were essentially the same as previous described by He and Jiang (15). Briefly, qPCR was carried out in 25-μl reaction mixtures consisting of 12.5 μl of 2× TaqMan master mix (Invitrogen, California), 2 μl primers, 1 μl TaqMan probe, 5 μl template DNA, and 4.5 μl double-distilled water (ddH2O). The final thermocycling profile for adenoviruses was 95°C for 15 s, 56°C for 15 s, and 62°C for 30 s with 1 s of automatic increment every cycle for 45 cycles. The serially diluted rAd5 virus was used to establish the standard curve. PBS was used as the negative control.
The Ad genome reduction was calculated based on qPCR results using log10 (Nt/N0), where N0 and Nt are the concentration of rAd5 before and after exposure to UV irradiation, respectively. The Ad infectivity reduction was calculated based on the FACS-GFP assay using changes in GFP fluorescence of cells before and after exposure to UV irradiation.
Environmental samples.
Primary sewage effluent and secondary effluent samples were collected from two southern California domestic wastewater treatment facilities. Sewage samples (20 ml) were centrifuged at 288,000 × g, 4°C, for 90 min using a Beckman ultracentrifuge (Beckman L8-ML) with an SW 41 Ti rotor. Pellets were resuspended in 200 μl PBS (pH 7.4). Seawater samples were collected over a weeklong period in February 2009 at the coastal Pacific Ocean near Los Angeles, CA. Twelve different samples were tested for the presence of infectious Ads using FACS. Seawater samples (500 ml) were concentrated ∼1,000-fold to a final volume of ∼500 μl, using a Centricon Plus-80 ultrafiltration system with a 100-kDa-molecular-mass-cutoff membrane (Millipore Inc.) as previously described (17, 18). Concentrated sewage and seawater samples were stored frozen at −80°C immediately and used later for infection of cells. Before inoculation onto confluent HEK-293A in 12-well plates; the concentrated sewage and seawater samples were clarified by filtering through 0.2-μm-pore-size filters (PES [4 mm, low protein binding]; Whatman, Piscataway, NJ). The FACS assay targeting the hexon protein after 3 days of incubation on HEK-293A cells was used for environmental samples. Highly diluted rAd5 was used as a positive control, while PBS was used as a negative control.
RESULTS
FACS assay of rAd5 based on GFP.
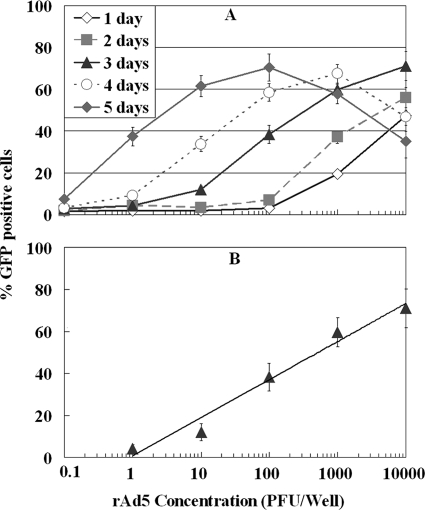
Figure 1A shows the sensitivity of FACS detection of rAd5 based on GFP at different incubation times on HEK-293A cells. A dose-response relationship was observed between the number of PFU of rAd5 used for inoculation and the proportion of GFP-positive cells. The GFP-positive cells were detected within 24 h of incubation at an inoculation density of 100 PFU per well. For incubation times of 3 and 4 days, GFP-positive cells were detected at inoculation densities of 1 and 0.1 PFU per well of rAd5, respectively. Prolonging the incubation time to 4 to 5 days resulted in a decrease in the GFP signal in samples with a viral density of >100 PFU per well. This is due to cell death at the high level of infection. Thus, the results suggest that a 3-day incubation may be the optimal time for assay of environmental samples when viral density is generally in the low range. A linear correlation with an r2 value of 0.97 was obtained by plotting the percentages of GFP-positive cells versus the initial numbers of rAd5 inoculated at concentrations ranging from 1 to 104 PFU/well after 3 days of incubation (Fig. 1B).
FIG. 1.
(A) Effects of the initial viral concentration and incubation time on FACS detection of rAd5 GFP on HEK-293A cells. (B) Dose-response curve for rAd5 by FACS-GFP assay after 3-day incubation on HEK-293A cells. The linear correlation is expressed as follows: y = 18.1 log10x + 0.81; r2 = 0.97. Each data point represents replication of three or more wells.
FACS assay of rAd5 based on hexon protein.
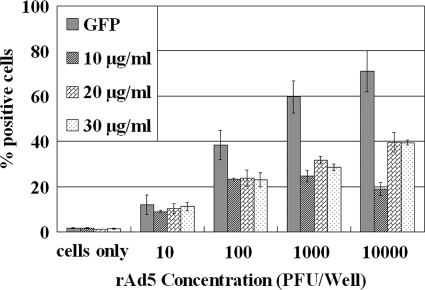
Hexon is the most abundant structural protein in Ads, with 240 copies of the trimeric molecule in the capsid, accounting for 63% of the total protein mass (30, 37). Optimization of the FACS assay based on the hexon protein was performed using a serial dilution of rAd5 incubated on HEK-293A cells for 3 days and with different concentrations of Ad hexon antibody. As shown in Fig. 2, the efficiencies of rAd5 detection based on the hexon protein were lower than those using GFP, especially in samples with the higher rAd5 inoculation density. In comparing the assay sensitivity and the amount of hexon antibody used in FACS assay, the results showed that 50 μl of 20 μg/ml hexon antibody was optimal for the FACS-hexon assay under the study conditions. The nonspecific adsorption of hexon antibody increased with the increase in the amount of hexon antibody, which only made the background fluorescence of HEK-293A cells higher. The final concentration of 20 μg/ml was used for subsequent experiments owing to the higher sensitivity and lower antibody consumption.
FIG. 2.
Optimization of the hexon primary antibody concentration (10 μg/ml, 20 μg/ml, and 30 μg/ml) for FACS-hexon assay using rAd5 incubated on HEK-293A cells for 3 days. In comparison with the FACS-GFP assay, the FACS-hexon assay showed a lower sensitivity. Each data point represents replication of three or more wells.
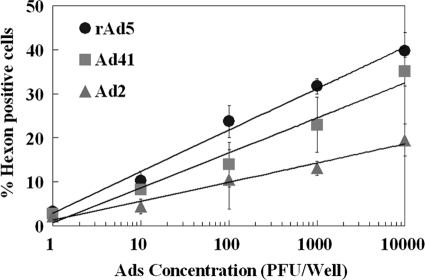
Comparison of FACS assay of Ad2, Ad5, and Ad41 based on hexon protein.
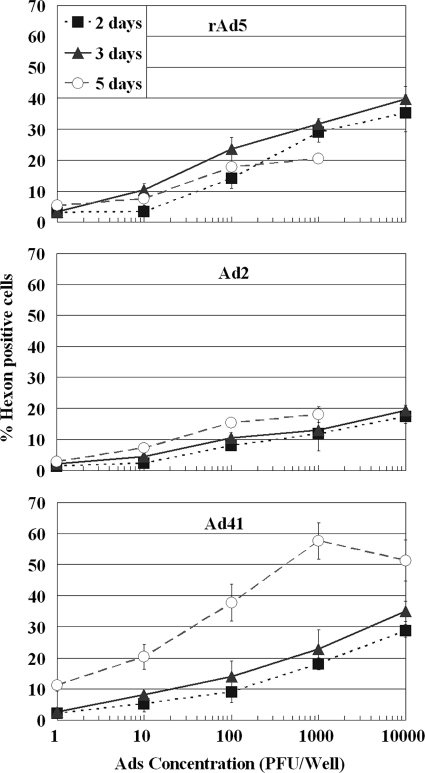
Figure 3 compares the efficiencies of FACS detection of different Ad serotypes based on the viral hexon protein. Although the hexon antibody used in the study has affinity toward a broad spectrum of Ad serotypes, the binding efficiency may differ by serotype. The results demonstrated that all three serotypes could be detected in 3 days at an inoculation concentration of 1 PFU/well. The fluorescence signals at 3 days of incubation (about 2.5 to 3.5% positive), although relatively low, were significantly above the cell-only background level (set at 1.5%) (Fig. 3). Prolonging the incubation time to 5 days did not increase the efficiency of detection for Ad5 but improved the efficiency for Ad41. A significant amount of cell death was detected at 5 days of incubation at inoculation concentrations of 1,000 PFU/well and greater.
FIG. 3.
Effects of the initial viral density and incubation time on FACS-hexon detection of rAd5, Ad2, and Ad41 on HEK-293A cells. Each data point represents replication of two or more wells.
FACS assay of Ad2 based on E1A protein.
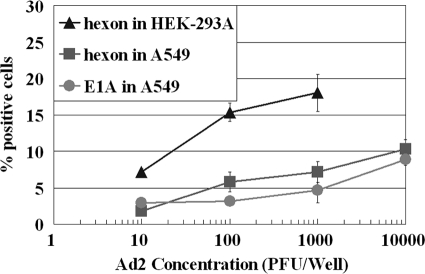
This experiment explored using the Ad E1A protein as the target for a FACS assay to improve the speed of detection because E1A is the early protein expressed during viral infection. The cell line A549 was used with Ad2 because HEK-293A is unsuited for an E1A assay (see Materials and Methods) and previous studies in our laboratory showed that A549 supported the replication of Ad2 at a higher rate (20). Figure 4 shows the sensitivity of the FACS assay based on E1A at 5 days of incubation was greater than that based on the hexon protein at an inoculation density of 10 PFU/well. However, at an inoculation density of ≥100 PFU/well of Ad2, the signal of the hexon protein was greater than that of the E1A protein (Fig. 4). Comparing the FACS Ad assay using A549 with that using the HEK-293A cell line, the sensitivity and efficiency with the A549 cell line were significantly lower (Fig. 4). More than 15% of fluorescent cells were detected using HEK-293A cells with hexon antibody at the inoculation density of 100 PFU/well for a 5-day incubation, while only less than 6% fluorescent cells were present when A549 was used for the same inoculation density and incubation period (Fig. 4). These results suggest that the sensitivity of the cell line is a more important factor for the FACS assay than the viral protein used as a target. Thus, the HEK-293A cell line in combination with the hexon antibody is the best choice for a FACS assay of a broad spectrum of Ad serotypes (hereinafter referred to as the FACS-hexon assay).
FIG. 4.
Comparison of FACS assay of Ad2 based on E1A and hexon protein and FACS-hexon assay using A549 cells and HEK-293A cells after 5-day incubation. Each data point represents replication of two or more wells.
Quantification of Ads by FACS assays.
Based on the above results, standard curves for quantification of infectious Ads by FACS-hexon assay were developed for the 3-day incubation on HEK-293A cells. A linear correlation was obtained by plotting the percentages of hexon-positive cells versus the logarithm of initial Ads inoculated at concentrations ranging from 1 to 104 PFU/well (Fig. 5). The correlation efficiencies were greater than 0.9 for all three Ad serotypes over the 4-log concentration range, indicating that the FACS assay based on the hexon protein could be used as a quantification tool for detecting infectious Ads. However, as shown in Fig. 5, the slope of the equation was different for each serotype due to the different affinities and replication rates of each serotype with the cell line (20, 48).
FIG. 5.
The standard curves for quantification of infectious Ads by FACS-hexon assay after 3 days of incubation on HEK-293A cells. The correlations are as follows: y = 9.43 log10x + 2.89 [r2 = 0.99]; y = 7.95 log10x + 0.66 [r2 = 0.97]; and y = 4.33 log10x + 1.26 [r2 = 0.97] for Ad5, Ad41, and Ad2, respectively. Each data point represents replication of two or more wells.
Comparison of FACS and qPCR assay of Ads.
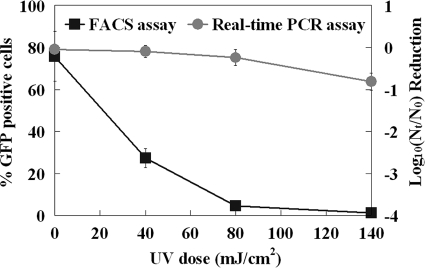
As shown in Fig. 6, the viral infectivity detected by FACS declined rapidly for rAd5 exposed to increasing UV doses from 0 to 80 mJ/cm2. Infectious rAd5 was nearly undetectable at the highest UV dose (140 mJ/cm2). Taking into account that the detection limit of the GFP-based FACS assay was 1 PFU/well after a 3-day incubation, the reductions were approximately a 4 log10 inactivation for a UV dose of 140 mJ/cm2 at an initial concentration of 104 PFU/well (Fig. 6). However, the qPCR assay indicated that less than 1 log10 reduction of genome copy was achieved with the same UV dose of 140 mJ/cm2 (Fig. 6).
FIG. 6.
Comparison of FACS and qPCR assays of adenovirus after UV irradiation. rAd5 before and after UV exposure was detected by FACS-GFP after 3 days of incubation and qPCR assay. Each data point represents replication of two or more wells.
Validation of FACS assay for Ads in environmental waters.
The application of the FACS-hexon assay to concentrated environmental water samples demonstrated positive detection of Ads in 3 of 6 trials using primary sewage effluents and in 3 of 12 trials for the secondary treated sewage samples (Table 1) . The percentages of hexon-positive cells were 5 to 8% and 4 to 6% after a 3-day incubation for primary and secondary effluent, respectively, which were significantly above the background fluorescence threshold of 1.5%. None of the 12 concentrated seawater samples showed positive detection of infectious Ads by FACS-hexon assay (Table 1), nor did any samples test positive by PCR assay (data not shown). Computation of Ads in sewage samples using standard curves estimated 10 to 165 PFU/100 ml for primary sewage and 10 to 105 PFU/100 ml for secondary sewage. The range of estimation of infectious viruses was due to the unknown serotype of Ad present in sewage samples and the difference in the standard curve for each serotype.
TABLE 1.
Detection of infectious adenoviruses in environmental samples by FACS based on viral hexon protein
| Sampling sourcea | Sampling dateb | Sample type | No. positive by FACS | Hexon-positive cells (%) | Estimated Ads in water samples (PFU/100 ml) |
|---|---|---|---|---|---|
| Coastal Pacific, Los Angeles | 02/26/09-02/13/09 | Seawater | 0 of 10 | ||
| S. Cal. sewage treatment facility | 03/31/09-04/08/09 | Primary effluents | 4 of 6 | 5 to 8 | 10-165 |
| S. Cal. sewage treatment facility | 03/31/09-04/08/09 | Secondary effluents | 4 of 12 | 4 to 6 | 10-105 |
S. Cal., southern California.
Month/day/year.
DISCUSSION
The results of the present study demonstrated that FACS is a rapid and quantitative method by which to detect infectious Ads in water and clearly distinguish them from inactivated viruses. This assay has shortened the viral detection time from 7 to 14 days using a traditional plaque assay or cell cytopathic effect (CPE) assay to 1 to 3 days (Table 2). In comparison with the qPCR assay, it is shown that although qPCR has a proven record of sensitivity and speed for viral detection (Table 2), past reports and this study have also demonstrated its deficiency in separating infectious viruses from inactivated ones (22-24). The qPCR is especially insensitive to viral genome damage during UV disinfection, as has been demonstrated in this study, because the qPCR assay requires a short nucleic acid sequence as the target to achieve high efficiency (11, 22, 35). UV exposure breaks the viral genome in multiple locations but does not necessary completely degrade the short DNA fragments. We have shown that the qPCR assay led to a 3-log overestimation of rAd5 after UV exposure. FACS, on the other hand, detects only infectious viruses because viral proteins outside the cell will not be captured by the flow cytometer (their sizes are beyond the detection limit).
TABLE 2.
Speeds and sensitivities of adenovirus detection by different methods
| Detection time | Detection limit for method |
||||||
|---|---|---|---|---|---|---|---|
| qPCR (PFU) | Epifluorescence microscopy based on rAd5 GFP on HEK-293A cellsa (PFU) | FACS based on rAd5 GFP on HEK-293A cells (PFU) | FACS based on hexon protein on HEK-293A cells (PFU) | FACS based on hexon protein on A549 cells (PFU) | Plaque assay on HEK-293A cells (PFU) | ICC qRT-PCR (IU) | |
| 3-∼4 h | 1-10 | ||||||
| 1 day | 1 | 100 | |||||
| 2 days | 0.1 | 10 | |||||
| 3 days | 0.01 | 1 | 1-10 | 5 | |||
| 5 days | 0.01 | 0.1 | 1 | 10 | |||
| 7-14 days | 3-10 | 0.1 | |||||
| Reference | This study | This study | This study | This study | This study | Jiang et al., 2009 (20)b | Ko et al., 2005 (23)c |
Detection of infectious rAd5 by epifluorescence microscopy is based on the expression of GFP in HEK-293A cells. This method reveals the rAd5 infection by observing GFP by microscopy but cannot quantify the number of infectious Ads.
In the work of Jiang et al., the detection limit for mixed primary/secondary sewage was 10 PFU Ads, and that for primary sewage was 3 PFU Ads.
In the work of Ko et al., the detection limit was 5 IU for Ad41 with 3 days' incubation and 0.1 IU with 7 days' incubation.
Table 2 also compares the FACS assay with epifluorescence microscopy direct observation for virally encoded GFP during infection. Although microscopy can easily identify rAd5 infection based on GFP expression on HEK-293A cells with greater sensitivity and speed, it cannot quantify the infected cells.
The detection time and sensitivity of FACS were similar to those of the integrated cell culture with reverse transcription-PCR (ICC-RT-PCR) assay, which detects viral mRNA during replication in cells (Table 2). ICC-RT-PCR has been shown to be a viable method for detecting infectious Ads in environmental samples (22, 23). However, ICC-RT-PCR involved complicated procedures of cell culture, RNA extraction, and RT-PCR assay, while the FACS assay relies on the fluorescent cells detected by flow cytometry, which has the potential for developing into an online sensing system.
The speed and sensitivity of the FACS assay depend on the expression rate and level of target viral protein in infected cells and the efficiency of capture of target protein by primary and secondary antibodies. Ad replication starts as soon as the viral DNA enters the nucleus, about 30 min after virus adsorption to host cells (22). The first protein to be made is the E1A protein, but the hexon protein is the most abundant of the structural proteins, accounting for 63% of the total protein mass (37). Transcription of the E1A gene is known to persist throughout infection, whereas the hexon is gene involved in the assembly of virions and is transcribed preferentially at later times after infection, reaching higher copy numbers during infection (22, 31). Thus, both the hexon protein and E1A are logical targets for development of the FACS assay.
The FACS assay based on rAd5 GFP showed a higher sensitivity and greater speed than the assay based on viral hexon (Table 2). This is likely due to the efficiency of capture of the target protein by primary and secondary antibodies. Although hexon expresses at a higher level than GFP (which replaced E1A in rAd5) during viral replication in cells (22), the fluorescence emission from hexon also depends on antibody binding. The hexon primary antibody has to get into the host cell and captures the hexon inside HEK-293A cells. The secondary antibody also has to enter the cell to bind with primary antibody. The efficiency with which the hexon primary antibody captures the hexon protein inside the infected cells is affected by the specificity of primary antibody, the ratio of antibodies to cells, the rate of penetration, and the condition of cell membrane. As a result, the percentages of GFP-positive cells were higher than those of hexon-positive cells. However, both GFP-based and hexon-based FACS showed adequate sensitivity for detection of low concentrations of infectious Ads in infected cells after 3 days of incubation.
The comparison of FACS based on E1A and that based on the hexon protein using Ad2 on A549 cells showed that hexon-based FACS was more sensitive than E1A-based FACS after 5 days of incubation at an inoculation density of >10 PFU/well. This result is likely due to the accumulation of the hexon protein in cells 5 days after infection. Previous studies also showed primers designed based on hexon were more sensitive than those designed based on E1A in the ICC-RT-PCR assay, which detects viral mRNA in host cells (22).
In addition to a good viral target, a high efficiency of antibody binding, and fluorescence capture, the cell line is another critical factor that determines the speed and sensitivity of the assay. The comparison of the FACS-hexon assay on A549 and HEK-293A cells indicated greater sensitivity on HEK-293A cells (Table 2). The reason may be that the HEK-293A cells can express E1A and E1B, which can “jump start” viral replication in cells. The previous study in our laboratory showed that the HEK-293A cell line was more sensitive than A549 in detection of Ads by 50% tissue culture infective dose (TCID50) (20).
Various Ad serotypes have different replication rates in HEK-293A cells (20, 22), and their affinities to hexon antibody may also differ. These differences result in variability of FACS assay efficiency for different serotypes. Separate quantification curves may be used to accurately determine infectious Ads of different serotypes. However, this issue is not limited to the FACS assay. Previous studies in our laboratory also showed that Ad5, Ad2, and Ad40 required different incubation times to produce CPE. The ICC-RT-PCR method also requires longer incubation times after infection for some serotypes than for others (22). As a result, a FACS assay of environmental samples can give a range of Ad numbers using the standard curves of different serotypes, because serotypes of Ads in environmental samples are still largely unknown. Nevertheless, the Ad numbers in sewage samples estimated by FACS were similar to those in a previous study using a plaque assay (20).
In conclusion, we have developed a new method for rapid detection of infectious Ads in environmental water samples. This method has the potential for application in detection of infectious Ads in environments and evaluation of viral stability and inactivation during the water treatment and disinfection process.
Acknowledgments
We are grateful to Michael Lai at the University of Southern California and Shawn Thompson at Los Angeles Sanitation District for providing cell lines and adenoviruses for this study. We thank Luis Villarreal and David Camerini at UC Irvine for discussion and input on FACS detection methods. Special thanks go to Rosina Girones at the University of Barcelona for visiting our laboratory and for the inspirational discussion of viruses in the water environment. We also thank Weiping Chu at UC Irvine for her technical assistance with operation of the flow cytometer.
Financial support for this research was provided by the UCI Urban Water Research Center, Environmental Institute, and California Sea Grant. Financial and travel support for D. Li are partially provided by a Chinese Scholarship Council Postgraduate Scholarship and a special fund from the State Key Joint Laboratory of Environment Simulation and Pollution Control (08Y01ESPCT).
We are grateful to Han-chang Shi and Miao He at Tshinghua University for their support for the Ph.D. joint training program.
Footnotes
Published ahead of print on 15 January 2010.
REFERENCES
- 1.Avery, R. M., A. P. Shelton, G. M. Beards, O. O. Omotade, O. C. Oyejide, and D. O. Olaleye. 1992. Viral agents associated with infantile gastroenteritis in Nigeria—relative prevalence of adenovirus serotype-40 and serotype-41, astrovirus, and rotavirus serotype-1 to serotype-4. J. Diarrheal Dis. Res. 10:105-108. [PubMed] [Google Scholar]
- 2.Baert, L., C. E. Wobus, E. Van Coillie, L. B. Thackray, J. Debevere, and M. Uyttendaele. 2008. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appl. Environ. Microbiol. 74:543-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxter, C. S., R. Hofmann, M. R. Templeton, M. Brown, and R. C. Andrews. 2007. Inactivation of adenovirus types 2, 5, and 41 in drinking water by UV light, free chlorine, and monochloramine. J. Environ. Eng. 133:95-103. [Google Scholar]
- 4.Bottley, G., J. R. Holt, N. J. James, and G. E. Blair. 2007. Flow cytometric detection of adenoviruses and intracellular adenovirus proteins. Methods Mol. Med. 130:205-213. [DOI] [PubMed] [Google Scholar]
- 5.Chang, J. C. H., S. F. Ossoff, D. C. Lobe, M. H. Dorfman, C. M. Dumais, R. G. Qualls, and J. D. Johnson. 1985. UV inactivation of pathogenic and indicator microorganisms. Appl. Environ. Microbiol. 49:1361-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costan-Longares, A., M. Montemayor, A. Payan, J. Mendez, J. Jofre, R. Mujeriego, and F. Lucena. 2008. Microbial indicators and pathogens: removal, relationships and predictive capabilities in water reclamation facilities. Water Res. 42:4439-4448. [DOI] [PubMed] [Google Scholar]
- 7.Crabtree, K. D., C. P. Gerba, J. B. Rose, and C. N. Haas. 1997. Waterborne adenovirus: a risk assessment. Water Sci. Technol. 35:1-6. [Google Scholar]
- 8.Cromeans, T. L., X Lu, D. D. Erdman, C. D. Humphrey, and V. R. Hill. 2008. Development of plaque assays for adenoviruses 40 and 41. J. Virol. Methods 151:140-145. [DOI] [PubMed] [Google Scholar]
- 9.Cruz, J. R., P. Caceres, F. Cano, J. Flores, A. Bartlett, and B. Torun. 1990. Adenovirus type 40 and type 41 and rotaviruses associated with diarrhea in children from Guatemala. J. Clin. Microbiol. 28:1780-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eischeid, A. C., J. N. Meyer, and K. G. Linden. 2009. UV disinfection of adenoviruses: molecular indications of DNA damage efficiency. Appl. Environ. Microbiol. 75:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Espinosa, A. C., M. Mazari-Hiriart, R. Espinosa, L. Maruri-Avidal, E. Mendez, and C. F. Arias. 2008. Infectivity and genome persistence of rotavirus and astrovirus in groundwater and surface water. Water Res. 42:2618-2628. [DOI] [PubMed] [Google Scholar]
- 12.Fong, T. T., and E. K. Lipp. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 69:357-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type-5. J. Gen. Virol. 36:59-72. [DOI] [PubMed] [Google Scholar]
- 14.Hammes, F., M. Berney, Y. Y. Wang, M. Vital, O. Koster, and T. Egli. 2008. Flow-cytometric total bacterial cell counts as a descriptive microbiological parameter for drinking water treatment processes. Water Res. 42:269-277. [DOI] [PubMed] [Google Scholar]
- 15.He, J. W., and S. Jiang. 2005. Quantification of enterococci and human adenoviruses in environmental samples by real-time PCR. Appl. Environ. Microbiol. 71:2250-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hitt, D. C., J. L. Booth, V. Dandapani, L. R. Pennington, J. M. Gimble, and J. Metcalf. 2000. A flow cytometric protocol for titering recombinant adenoviral vectors containing the green fluorescent protein. Mol. Biotechnol. 14:197-203. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, S., H. Dezfulian, and W. P. Chu. 2005. Real-time quantitative PCR for enteric adenovirus serotype 40 in environmental waters. Can. J. Microbiol. 51:393-398. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, S., R. Noble, and W. P. Chui. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, S. C. 2006. Human adenoviruses in water: occurrence and health implications: a critical review. Environ. Sci. Technol. 40:7132-7140. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, S. C., J. J. Han, J. W. He, and W. P. Chu. 2009. Evaluation of four cell lines for assay of adenoviruses in water samples. J. Water Health 7:650-656. [DOI] [PubMed] [Google Scholar]
- 21.Kalyuzhniy, O., N. C. Di Paolo, M. Silvestry, S. E. Hofherr, M. A. Barry, P. L. Stewart, and D. M. Shayakhmetov. 2008. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl. Acad. Sci. U. S. A. 105:5483-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko, G., T. L. Cromeans, and M. D. Sobsey. 2003. Detection of infectious adenovirus in cell culture by mRNA reverse transcription-PCR. Appl. Environ. Microbiol. 69:7377-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko, G., N. Jothikumar, V. R. Hill, and M. D. Sobsey. 2005. Rapid detection of infectious adenoviruses by mRNA real-time RT-PCR. J. Virol. Methods 127:148-153. [DOI] [PubMed] [Google Scholar]
- 24.Ko, G., T. L. Cromeans, and M. D. Sobsey. 2005. UV inactivation of adenovirus type 41 measured by cell culture mRNA RT-PCR. Water Res. 39:3643-3649. [DOI] [PubMed] [Google Scholar]
- 25.Koepke, J. A., and A. L. Landay. 1989. Precision and accuracy of absolute lymphocyte counts. Clin. Immunol. Immunopathol. 52:19-27. [DOI] [PubMed] [Google Scholar]
- 26.Kukkula, M., P. Arstila, M. L. Klossner, L. Maunula, C. H. vonBonsdorff, and P. Jaatinen. 1997. Waterborne outbreak of viral gastroenteritis. Scand. J. Infect. Dis. 29:415-418. [DOI] [PubMed] [Google Scholar]
- 27.Macartney, L., H. M. A. Cavanagh, and N. Spibey. 1988. Isolation of canine adenovirus-2 from the feces of dogs with enteric disease and its unambiguous typing by restriction endonuclease mapping. Res. Vet. Sci. 44:9-14. [PubMed] [Google Scholar]
- 28.Maiti, N. K., G. K. Baruah, S. N. Sharma, and B. Singh. 1987. A study on fowl adenovirus type-I infection in the chicken. Acta Vet. Beograd. 37:293-298. [Google Scholar]
- 29.Mcsharry, J. J., R. Costantino, M. B. Mcsharry, R. A. Venezia, and J. M. Lehman. 1990. Rapid detection of herpes simplex virus in clinical samples by flow cytometry after amplification in tissue culture. J. Clin. Microbiol. 28:1864-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nemerow, G. R., L. Pache, V. Reddy, and P. L. Stewart. 2009. Insights into adenovirus host cell interactions from structural studies. Virology 384:380-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neubert, S. 2009. Wastewater reuse: how “integrated” and sustainable is the strategy? Water Policy 11:37-53. [Google Scholar]
- 32.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pusch, D., D. Y. Oh, S. Wolf, R. Dumke, U. Schroter-Bobsin, M. Hohne, I. Roske, and E. Schreier. 2005. Detection of enteric viruses and bacterial indicators in German environmental waters. Arch. Virol. 150:929-947. [DOI] [PubMed] [Google Scholar]
- 34.Quiros, C., M. Herrero, L. A. Garcia, and M. Diaz. 2007. Application of flow cytometry to segregated kinetic modeling based on the physiological states of microorganisms. Appl. Environ. Microbiol. 73:3993-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez, R. A., I. L. Pepper, and C. P. Gerba. 2009. Application of PCR-based methods to assess the infectivity of enteric viruses in environmental samples. Appl. Environ. Microbiol. 75:297-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez, W. J., H. W. Kim, C. D. Brandt, J. Arrobio, M. K. Gardner, B. Jeffries, and R. H. Parrott. 1985. Enteric adenovirus 40 and 41 (Eads) in gastroenteritis of infants and young children. Pediatr. Res. 19:A208-A208. [Google Scholar]
- 37.Rux, J. J., and R. M. Burnett. 2004. Adenovirus structure. Hum. Gene Ther. 15(12):1167-1176. [DOI] [PubMed] [Google Scholar]
- 38.Saag, M. S., M. Holodniy, D. R. Kuritzkes, W. A. OBrien, R. Coombs, M. E. Poscher, D. M. Jacobsen, G. M. Shaw, D. D. Richman, and P. A. Volberding. 1996. HIV viral load markers in clinical practice. Nat. Med. 2:625-629. [DOI] [PubMed] [Google Scholar]
- 39.Sandhu, K. S., and M. Al-Rubeai. 2008. Monitoring of the adenovirus production process by flow cytometry. Biotechnol. Progr. 24:250-261. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu, H., T. G. Phan, S. Nishimura, S. Okitsu, N. Maneekarn, and H. Ushijima. 2007. An outbreak of adenovirus serotype 41 infection in infants and children with acute gastroenteritis in Maizuru City, Japan. Infect. Genet. Evol. 7:279-284. [DOI] [PubMed] [Google Scholar]
- 41.Sirikanchana, K., J. L. Shisler, and B. J. Marinas. 2008. Effect of exposure to UV-C irradiation and monochloramine on adenovirus serotype 2 early protein expression and DNA replication. Appl. Environ. Microbiol. 74:3774-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skraber, S., B. Gassilloud, and C. Gantzer. 2004. Comparison of coliforms and coliphages as tools for assessment of viral contamination in river water. Appl. Environ. Microbiol. 70:3644-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Svraka, S., E. Duizer, H. Vennema, E. de Bruin, B. van der Veer, B. Dorresteijn, and M. Koopmans. 2007. Etiological role of viruses in outbreaks of acute gastroenteritis in the Netherlands from 1994 through 2005. J. Clin. Microbiol. 45:1389-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, and C. P. Gerba. 2003. Chlorine inactivation of adenovirus type 40 and feline calicivirus. Appl. Environ. Microbiol. 69:3979-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiemessen, C. T., M. J. Nel, and A. H. Kidd. 1996. Adenovirus 41 replication: cell-related differences in viral gene transcription. Mol. Cell. Probes 10:279-287. [DOI] [PubMed] [Google Scholar]
- 46.Videla, C., G. Carballal, A. Misirlian, and M. Aguilar. 1998. Acute lower respiratory infections due to respiratory syncytial virus and adenovirus among hospitalized children from Argentina. Clin. Diagn. Virol. 10:17-23. [DOI] [PubMed] [Google Scholar]
- 47.Weaver, L. S., and M. J. Kadan. 2000. Evaluation of adenoviral vectors by flow cytometry. Methods 21:297-U8. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, L. O., Y. F. Mei, and G. Wadell. 2003. Human adenovirus serotypes 4 and 11 show higher binding affinity and infectivity for endothelial and carcinoma cell lines than serotype 5. J. Gen. Virol. 84:687-695. [DOI] [PubMed] [Google Scholar]