Abstract
Enteropathogenic Escherichia coli (EPEC) is an important causal agent of diarrheal illness throughout the world. Nevertheless, researchers have only recently begun to explore its capacity to form biofilms. Strain O55:H7 (DMS9) is a clinical isolate belonging to the atypical EPEC (aEPEC) group, which displays a high degree of genetic relatedness to enterohemorrhagic E. coli. Strain DMS9 formed a robust biofilm on an abiotic surface at 26°C, but not at 37°C. It also formed a dense pellicle at the air-liquid interface and developed a red, rough, and dry (RDAR) morphotype on Congo red agar. Unlike a previously described E. coli O157:H7 strain, the aEPEC strain seems to express cellulose. Transposon mutagenesis was used to identify biofilm-deficient mutants. One of the mutants was inactivated in the csgFG genes, required for assembly and secretion of curli fimbriae, while a second mutant had a mutation in crl, a thermosensitive global regulator that modulates σS activity and downstream expression of curli and cellulose. The two mutants were deficient in their biofilm formation capabilities and did not form a pellicle at the air-liquid interface. Unlike in Salmonella, the csgFG mutant in aEPEC completely lost the RDAR phenotype, while the crl mutant displayed a unique RDAR “pizza”-like morphotype. Genetic complementation of the two mutants resulted in restoration of the wild-type phenotype. This report is the first to describe and analyze a multicellular behavior in aEPEC and support a major role for curli and the crl regulator in biofilm development at low temperatures corresponding to the nonmammalian host environment.
Diarrheal illness is a worldwide public health problem that is responsible for over 2 million deaths each year, particularly among infants and children younger than 5 years of age (16). One of the most common causes of infant diarrhea in developing countries is enteropathogenic Escherichia coli (EPEC) (16, 35, 41, 62). EPEC virulence is characterized by intimate contact between the bacterium and the apical plasma membrane of the intestinal brush border cells, followed by localized destruction of the intestinal brush border and distortion of the apical enterocyte membrane. The interactions between the bacterium and the host cell result in gross cytoskeletal rearrangements, particularly the formation of an actin-rich, cup-like pedestal at the site of bacterial contact, the so-called attaching and effacing (AE) lesion (16). The genes associated with AE histopathology are localized on a 35.6-kb pathogenicity island called the locus of enterocyte effacement (LEE). LEE contains several virulence genes, including the eae gene that encodes the adhesin intimin, which is required for the attachment of EPEC to epithelial cells, genes encoding a type III secretion system (TTSS), and tir, which encodes the translocated intimin receptor (22, 30, 39). EPEC strains also contain a large plasmid, referred to as the EPEC adherence factor (EAF) plasmid. This plasmid encodes a regulator of virulence genes called Per that regulates the expression of a chromosomal regulator, Ler, which then activates the expression of other LEE genes in a regulatory cascade (22, 30, 39). The EAF plasmid also encodes the adhesin bundle-forming pilus (BFP), which is responsible for the bacteria's localized adherence to epithelial cells (42) and has been shown to be important for initial binding to host cells (17), as well as for species-specific cell adhesion (61). BFP has also been shown to be involved in bacterium-bacterium interactions leading to aggregation, the formation of complex, three-dimensional microcolonies, and the dispersal of these microcolonies (32, 36, 59, 60). The EPEC group is subdivided into two classes referred to as typical (tEPEC) and atypical (aEPEC) EPEC. A tEPEC strain contains the EAF plasmid, while aEPEC strains lack this plasmid (62). The aEPEC strains are genetically related to the enterohemorrhagic E. coli (EHEC) strains and are both considered emerging pathogens (62). Hence, understanding their transmission mode, as well as their potential survival strategies outside the host, might enable better control of their spread.
The ability of bacteria to grow as biofilms is considered to be a major requirement for survival under harsh environmental conditions (18, 43). This trait has also been linked to the pathogenesis of several human and animal diseases, as well as to enhanced resistance to antimicrobials (18, 25, 27, 70). While biofilm formation by E. coli K-12 strains has been extensively studied (9, 19, 21, 24, 47, 56, 64), very little is known regarding biofilm formation by EPEC strains and their survival outside the host.
Recently, Moreira et al. (40) published the first description of biofilm formation by a tEPEC strain (O127:H6) under continuous flowthrough conditions. These researchers found that appendage structures, namely, type I fimbriae, antigen 43, BFP, and EspA, are expressed during the initial stages of biofilm development. Mutants that did not express BFP or EspA formed more diffuse biofilms than did the wild-type (wt) strain.
Biofilm formation in aEPEC strains has not been described so far. Since some EPEC traits, such as BFP and other pEAF-bearing genes, are not present in atypical strains, the genes required for biofilm formation in the two classes might also differ. Here, we analyze for the first time biofilm formation in an aEPEC strain belonging to the O55:H7 serotype. This strain is capable of multicellular behavior characterized by the formation of a robust biofilm on an abiotic surface, a dense pellicle at the air-liquid interface and a red, dry, and rough (RDAR) morphotype on agar plates. Initial transposon mutagenesis analysis has identified curli fibers and the Crl regulator as important participants in the formation of all three types of biofilms.
MATERIALS AND METHODS
Strains and plasmids.
Clinical isolates of the E. coli strains O55:H10 (EPEC), O55:H7 (EPEC), O7:H4 (enterotoxigenic), O153:H− (enterotoxigenic), O126:H27 (enteroaggregative), and O26:H11 (EHEC) came from the collection of the E. coli Reference Laboratory (Israel Ministry of Health, Jerusalem, Israel). EHEC O157:H7 85/170 and EPEC O127:H6 e2348/69 were gifts from I. Rosenshine (Hebrew University, Jerusalem, Israel). E. coli K-12 strain TG-1 was obtained from Amersham Biosciences (United Kingdom).
For transposon mutagenesis, a spontaneous streptomycin (Sm)-resistant variant of strain O55:H7 (from here on, designated DMS9) was isolated by sequential transfer onto Luria-Bertani (LB) agar plates (10 g Bacto-tryptone, 5 g yeast extract, and 5 g NaCl) containing increasing concentrations of the antibiotic. The antibiotic-resistant variant was able to grow on 100 μg/ml of streptomycin and was designated DMS9S. Strain DMS9S exhibited biofilm phenotypes comparable to that of the wt strain (DMS9). The plasmid pGP704 carrying mini-Tn10 (23) and its host, E. coli SM10λpir, were gifts from V. de Lorenzo (Centro Nacional de Biotecnología CSIC, Campus de Cantoblanco, Madrid, Spain). This strain is resistant to kanamycin (Km; 50 μg/ml), as well as ampicillin (Amp; 100 μg/ml). E. coli DH5 was used as a host for all cloning experiments, and plasmid pBR322 was used for complementation experiments. Strains were grown in LB or in LB without salt (LBNS), as indicated below.
Transposon mutagenesis and genetic analysis.
Plasmid pGP704 was used to deliver the mini-Tn10 into E. coli strain DMS9S, as described previously (23). Individual colonies of transconjugants were replica plated onto agar plates containing LB-Amp and LB supplemented with Km and Sm. Clones which displayed the phenotype Sm+/Km+/Amp− were considered to contain the mini-Tn10 insertion.
To determine the number of Tn10 insertions within the chromosome of the transconjugants, chromosomal DNA isolated from individual clones was cut with HindIII (a restriction enzyme for which a single site is present in the mini-Tn10 element) and run on a 1% agarose gel. Southern blot hybridization, using the Km resistance gene as a probe, was used to confirm the presence of a single insertion. For identification of the insertion sites in the tested mutants, a HindIII chromosomal digest was generated and self-ligated. The ligation mixture was used as a template for inverse PCR, as described previously (28). The primers used were Tn-F and Tn-R. The resultant fragments, which were composed of the junction between the 3′ end of Tn10 and the adjacent chromosomal DNA, were sequenced (Macrogen, Seoul, South Korea). Sequence analysis was performed using BLAST (2) at the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST).
To verify the location of the transposon insertion, the identity of the DNA sequence near the 5′ end of the mini-Tn10 insertion was also tested, using one primer derived from the mini-Tn10 and another that was derived from the putative sequence of the affected gene. The primer sets A59-F/A59-R and C24-F/C24-R were used to examine the sequences in the A59 (csgFG) and C24 (crl) mutants, respectively (Table 1). The sequences of the primers were determined using GeneRunner software and the EHEC genome sequence (http://genome.gen-info.osaka-u.ac.jp/bacteria/o157/index.html).
TABLE 1.
Primers used in this study
| Primer | Sequencea | Source | nt position |
|---|---|---|---|
| Km-F | 5′-GGAGAAAACTCACCGAGGC-3′ | Km resistance cassette from Tn10 | 647-665b |
| Km-R | 5′- GTCGGGCAATCAGGTGCG-3′ | Km resistance cassette from Tn10 | 1255-1272b |
| Tn-F | 5′-AACGCAGACCGTTCCGTGG-3′ | Mini-Tn10 sequence | 1535-1553b |
| Tn-R | 5′-TCGCTCAGGCGCAATCACG-3′ | Mini-Tn10 sequence | 923-905b |
| A59-F | 5′-TGGTCGTCCGCTGGTTGATGATTA-3′ | E. coli O157:H7, ORF ECs0265 | 351-374c |
| A59-R | 5′-CGGAACATTCATCAGTGTAA-3′ | Mini-Tn10 sequence | 160-141b |
| C24-F | 5′-TGCTGCTTTCATTTGTTCCGG-3′ | E. coli O157:H7, ORF ECs1413 | 344-364c |
| C24-R | 5′-TGGCTCCCTCACTTTCTGGCT-3′ | Tn10 sequence | 332-352b |
| crl-F | 5′-GGGGATATC TGATAATAATAAGGAGTATTGATTATGACGTTACCGAGTGGACA-3′ | E. coli O157:H7, ORF ECs0267 | 1-20d |
| crl-R | 5′-GGGGTCGACTCAGTGATGGTGATGGTGATGCGCCGTTAACTTCACCGGCTCGTCA-3′ | E. coli O157:H7, ORF ECs0267 | 402-377c |
| curli-F | 5′-GGGGATATCTGATAATAATAAGGAGTATTGATTATGCGTGTCAAACATGCAGT-3′ | E. coli O157:H7, ORF ECs1415 | 1-20c |
| curli-R | 5′-GGGGTCGACGGGGCACTCACGCTTTCGCTTAAACAGTAAATGCCGGATTATT-3′ | E. coli O157:H7, intergenic region between ORF ECs1414 and ORF ECs1413 | 1459990-1459928c |
| pBR-F | 5′-AAATCTAACAATGCGCT-3′ | GenBank/EMBL accession no. J01749 | 89-105d |
| pBR-R | 5′-CACCTGTCCTACGAGTTGC-3′ | GenBank/EMBL accession no. J01749 | 751-770d |
The EcoRV restriction site is underlined. The SalI restriction site is in bold.
Tn10 minitransposon sequence (GenBank accession number AJ601386).
E. coli O157:H7 Sakai, Genome Information Research Center, Osaka University, Japan (http://genome.gen-info.osaka-u.ac.jp/bacteria/o157/viewer.html).
pBR322 sequence (GenBank accession number J01749).
Construction of recombinant plasmids.
For complementation experiments, crl and csgFG were cloned into plasmid pBR322. DMS9S chromosomal DNA was used as a template for the amplification of the crl gene with the primers crl-F/crl-R (Table 1) as follows: 95°C for 3 min; 35 cycles of 95°C for 30 s, 69°C for 1 min, and 72°C for 1.5 min. PCR amplification of the csgFG genes was performed with the primers curli-F/curli-R (Table 1) under the following conditions: 95°C for 3 min; 35 cycles of 95°C for 30 s, 49°C for 1 min, and 72°C for 1.5 min. The amplified fragments were each digested with EcoRV and SalI, and ligated to pBR322 that had been precut with the two enzymes. The recombinant plasmid was introduced into competent E. coli DH5α by electroporation by using the Invitrogen Electroporator II with electric pulses of 15 kW/cm for 7 milliseconds. Cells were then transferred into super optimal broth medium (55), incubated at 37°C for 2 h, and spread onto LB-Amp plates. Plasmids that were sensitive to tetracycline were considered to harbor recombinant genes. Confirmation of the presence of intact crl and csg genes was performed by PCR amplification of the inserts, using primers (pBR322-F/R; Table 1) external to the EcoRV and SalI sites present on pBR322, followed by sequence analysis. The plasmid containing the crl gene was designated pCRL, and the plasmid containing the csgFG genes was designated pCSG.
The plasmids were introduced into strain DMS9S by electroporation, as described above, and selection of the recombinant clones was performed on LB agar supplemented with Amp, Sm, and Km. The presence of the recombinant plasmid in each clone was confirmed by PCR with the primers pBR-F/R and sequence analysis.
Biofilm formation on polystyrene.
Detection and quantification of biofilms on polystyrene were performed using 96-well polystyrene microtiter plates (Greiner Bio-One, Frickenhausen, Germany), as described previously (15, 20), with a few modifications as detailed. Tested strains were grown overnight at 37°C in LB. The optical density of the culture was adjusted to an optical density at 600 nm (OD600) of 1.0 (corresponding to ca. 108 CFU/ml). The culture was then diluted 1:10 in fresh LB or LBNS and used to inoculate four replicate wells per microtiter plate (100 μl per well). Controls consisted of uninoculated wells containing sterile medium only. Plates were incubated statically for 72 h at 26°C to allow for biofilm development. Following this incubation, planktonic cells were removed by aspiration, and each plate was washed three times with 250 μl/well of sterile double-distilled water (SDDW). The plates were dried (30°C for 15 min), and each well was stained with 100 μl of 0.1% crystal violet (CV; Hy-Laboratory, Rehovot, Israel) for 15 min at 26°C. Wells were thoroughly rinsed three times with 150 μl SDDW and air-dried, and the CV was solubilized in 100 μl 33% acetic acid for 5 min with orbital shaking at 150 rpm. Finally, the amount of extracted CV was determined by measuring the OD595 by using an enzyme-linked immunosorbent assay (ELISA) plate reader (ELx 800 U.V; BioTec Instruments, Inc., Burlington, VT). For initial screening of the library, individual colonies were transferred into a 96-well plate containing 100 μl of LBNS and grown for 20 h at 37°C in the presence of the appropriate antibiotics. The optical density of each well was recorded to identify mutants with impaired growth, and 5 μl of each culture was transferred to 95 μl of fresh LBNS with the appropriate antibiotics. The biofilm formation assay was performed as described above.
Pellicle formation.
Strains were grown overnight in LB at 37°C, and 5 μl was transferred into 4 ml LBNS in 15-ml glass tubes. Following incubation for 3 days at 26°C, the formation of a biofilm (pellicle) at the air-liquid interface was tested visually and photographed by a digital camera. In some cases, the tubes were stained with CV (1 min), washed thoroughly with distilled water, and photographed.
Calcofluor-binding assay.
Bacterial strains were grown on LBNS plates containing 200 mg/liter of the dye calcofluor (Sigma-Aldrich Co., St. Louis, MO) for 3 days at 26°C. Calcofluor binding was detected by fluorescence under a short-UV-light lamp.
Western analysis.
Bacteria from pellicles, liquid culture, and aggregates at the bottom of tubes were collected for Western analysis. Bacteria were pelleted and resuspended in 150 μl 2× SDS loading buffer with or without pretreatment with 70 μl hexafluoro-2-propanol (HFIP). HFIP was removed prior to addition of loading buffer by spinning for 30 min in a rotary vacuum at 45°C. Without HFIP treatment, curli fibers will remain polymerized, and CsgA subunits will not migrate into the resolving part of the gel. Samples were separated by SDS-PAGE (15% polyacrylamide gel). CsgA was probed by anti-CsgA antibody and developed as described previously (66).
Microscopy.
Bacterial colonies were visualized under a stereomicroscope with a Leica MZ FLIII binocular lens (Leica Microsystems GmbH, Wetzlar, Germany). Photographs were taken with a Leica DC 200 digital camera. For confocal microscopy studies, fresh colonies were used to inoculated 4 ml LBNS in 15-ml glass tubes. Bacteria were incubated at 26°C for 72 h without shaking. The pellicle was gently removed from the tube, washed twice with phosphate-buffered saline (PBS), and fixed overnight with 2.5% glutaraldehyde in 0.1 M PBS (pH 7.0) at 4°C. The pellicle was then washed twice with PBS and stained with 0.01% acridine orange (AO) in 10 mM Tris (pH 8.0) and 1 mM EDTA for 30 min. The stain was washed with SDDW (three washes, 30 min each, with gentle shaking). Fluorescence images were collected using an Olympus IX81 confocal laser scanning microscope (Japan) equipped with 488-nm argon-ion and 543-nm helium-neon lasers, as described previously (57). Images were collected and processed using the Olympus FluoView 500 confocal software. Additional image processing and three-dimensional reconstitution of confocal stacks were carried out using Imaris 5.0.1 software (Bitplane, Zurich, Switzerland). Pellicle depths derived from FluView-500-processed image stacks of 10 randomly chosen areas in a single experiment were used to calculate the average pellicle depth.
For studies of glass-associated biofilms, bacteria were grown as described above and diluted 1:20 in LBNS. Then, 100 μl of the diluted solution was incubated statically in 16-well Chamber Slides (Nalge Nunc International, Rochester, NY) for 72 h at 26°C. The wells were then washed three times with PBS and stained with AO, as described above.
Transmission electron microscopy.
Bacterial cells (10 μl) that were collected from pellicles were placed onto Formvar-coated copper grids (Ernest F. Fullam Inc., Latham, NY) for 2 min, washed with deionized water, and negatively stained with 2% (wt/vol) uranyl acetate for 90 s. Samples were visualized under a Philips CM100 transmission electron microscope as described previously (66).
Statistical analysis.
All experiments were repeated at least three times unless otherwise stated. Data were analyzed using the General Linear Models (GLM) procedure of the SAS software system 8.02 (SAS Institute Inc., Cary, NC). Differences were considered to be significant when P was ≤0.05.
RESULTS
aEPEC strain O55:H7 forms robust biofilm and displays a multicellular behavior.
While examining biofilm formation among clinical isolates of E. coli, we observed that an EPEC isolate of serotype O55:H7 (DMS9) formed a substantially more robust biofilm than did the other tested strains (Fig. 1). Based upon its serologic traits, this strain is considered to be a member of the aEPEC group (62). In agreement with its serologic classification, a PCR analysis confirmed that the O55:H7 strain contains the eae gene, but not the bfp or the stx genes (data not shown).
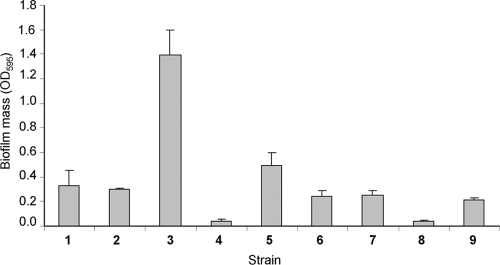
FIG. 1.
Quantification of biofilms formed on a 96-well polystyrene plate, using the CV method. Strains were incubated statically in LB broth for 3 days at 26°C. Data are from a representative experiment with three replicates. 1, E. coli TG-1; 2, O55:H10 (EPEC); 3, O55:H7 (EPEC DMS9); 4, O7:H4 (enterotoxigenic E. coli); 5, O153:H− (enterotoxigenic E. coli); 6, O126:H27 (enteroaggregative E. coli); 7, O26:H11 (EHEC); 8, O157:H7 (EHEC) 85/170; 9, O127:H6 e2348/69 (EPEC). The error bars represent the standard errors of the means.
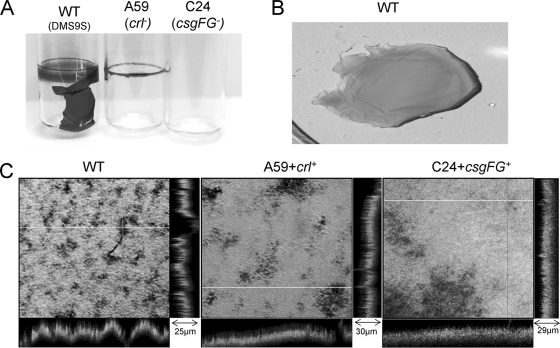
Biofilm-forming bacterial strains often develop a typical colony morphology known as the RDAR morphotype (13, 51, 54). Incubation of the DMS9 strain on Congo red plates demonstrated the development of a characteristic RDAR morphotype at 26°C, but not at 37°C. Furthermore, the size of the colony developed at 26°C was substantially larger than that developed at 37°C (Fig. 2). Following propagation on an LBNS plate supplemented with calcofluor for 3 days at 26°C, strain DMS9 displayed a bright fluorescence under UV irradiation (data not shown). These results indicated that unlike E. coli O157:H7 (63), cellulose is a component of the extracellular polymorphic substances (EPS) of the aEPEC strain. Another feature characteristic of multicellular behavior is the formation of a pellicle at the air-liquid interface (51). When grown statically at 26°C, but not at 37°C, strain DMS9 formed a dense pellicle above the liquid culture broth. In fact, bacteria were distributed at three phases, air-liquid (pellicle), liquid (planktonic), and liquid-solid, at the bottom of the tube. The pellicle was avidly bound to the glass walls of the tube, as was evident from its capacity to withstand the pressure of the culture column when the tube was held upside down. The pellicle seems to be quite elastic, as it remained intact when a tip of a plastic quad-loop inoculation needle was gently pushed against it (Fig. 3).
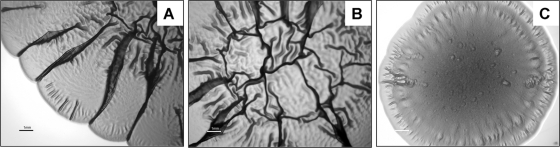
FIG. 2.
EPEC strain DMS9 displays a characteristic RDAR morphotype. Strain DMS9 was inoculated on the center of Congo red plates and incubated for 4 days at 26°C (A and B) or 37°C (C). Panel A presents a section of the peripheral region of a single representative colony, and panel B shows its central region. The bar represents 1 mm.
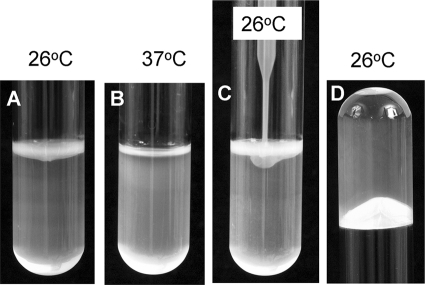
FIG. 3.
Pellicle formation at the air-liquid interface. Strain DMS9 incubated statically for 3 days in LBNS broth forms a dense pellicle at 26°C (A) but not at 37°C (B). The band at the top seen at 37°C corresponds to light reflection. The robustness and elasticity of the pellicle are illustrated by the inability of a plastic quad-loop inoculation device, pushed gently against the pellicle, to penetrate through (C) and by its capacity to hold a liquid column of 4 ml broth when the tube was held upside down (D).
Isolation of biofilm formation mutants.
To identify EPEC genes involved in biofilm formation, we constructed a mini-Tn10 library with the DMS9S strain and used a rapid screening test to identify mutations affecting biofilm formation. Clones with reduced biofilm mass were retested for their capacity to produce biofilm by using the CV assay. The biofilms formed by six such mutants were compared to the biofilm formed by the wt strain (Fig. 4). Since direct selection of mutants using the biofilm screening method might also yield mutants with impaired growth rates and lead to the erroneous classification of these mutants as biofilm mutants, the growth rates of the different mutants were tested in LB and LBNS media at 26°C and 37°C. All of the tested clones displayed growth characteristics similar to those of the wt strain (data not shown), implying that they were true biofilm mutants.
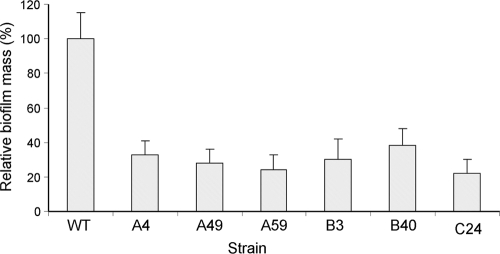
FIG. 4.
Screening of the transposon library for biofilm-deficient mutants. Strains were incubated statically in LBNS broth for 3 days at 26°C, and the biofilm was quantified by the CV method. Data represent the OD595 values of the different strains relative to that of the wt strain, which was set as 100%. Average data of three independent experiments are presented. The biofilm masses (CV values) of all mutants were significantly smaller than that of the wt (P < 0.0001). The error bars represent the standard errors of the means.
Characterization of the mini-Tn10 insertion sites.
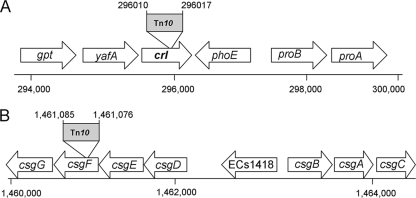
Southern blot hybridization, using the Km resistance cassette as a probe, displayed two hybridization bands in each of the tested mutants, consistent with the presence of a single insertion (data not shown). Two insertional mutants, designated A59 and C24, were chosen for further characterization based upon their poor biofilm phenotype (Fig. 4). Identification of the transposon insertion sites was performed using inverse PCR and sequence analysis. The DNA sequence for the A59 mutant displayed 100% and 99% similarities to the crl genes of EHEC (Sakai strain) and E. coli K-12 MG1655, respectively. The crl gene product is a thermosensor that is involved in the modulation of σS activity in E. coli and Salmonella (48, 49). To confirm the location of the transposon, PCR analysis and sequencing of the chromosomal 5′-flanking region of the transposon were performed. The insertion site is shown in Fig. 5A, suggesting the generation of a small deletion of 6 nucleotides (nt) when the transposon is inserted. The mini-Tn10 insertion site in mutant C24 was mapped to an open reading frame (ORF) similar to that of the gene csgF (95% similarity to the EHEC ortholog). The product of csgF, CsgF (Fig. 5B), is required for efficient curli assembly in vivo, together with CsgG and CsgE (7, 31, 50). Since the Tn10 insertion in csgF might have also affected transcription of the downstream gene, csgG (Fig. 5), the mutant was designated the csgFG mutant. To verify that the insertions did not result in additional unintended changes in the DNA near the insertion sites, we performed a PCR analysis of the DNA region adjacent to the 5′ end of the transposon, using the primer pairs A59-F/Km-R and C24-F/Km-R (Table 1). Sequencing of the resulting DNA fragments confirmed the location of the transposon and showed that this insertion also resulted in a small (7-nt) deletion (Fig. 5B). These findings confirmed that the insertions did not cause any major deletion or reshuffling of the DNA sequence near the insertion sites in the two mutants.
FIG. 5.
Schematic representation of the ORFs located near the mini-Tn10 insertion in two selected mutants: A59 (A) and C24 (B). Chromosomal localization of the insertion sites and coordinates are based on the genome map for E. coli O157:H7 Sakai (http://genome.gen-info.osaka-u.ac.jp/bacteria/o157/index.html).
In order to verify that the two mutations are responsible for the observed phenotypic changes, we constructed recombinant DMS9S clones expressing crl and csgFG on the pBR322 vector.
Effects of Crl and curli on biofilm formation.
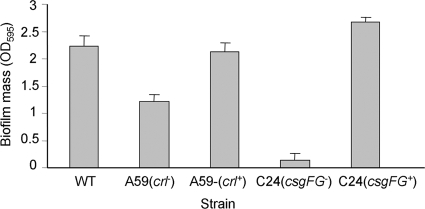
The wt strain formed a substantial amount of biofilm on a polystyrene surface when grown in LBNS medium under static conditions for 72 h at 26°C, whereas the two mutants formed smaller amounts of biofilm, with the C24 csgFG mutant forming a very poor biofilm. Complementation of the mutations resulted in the formation of biofilm masses comparable to that of the wt (Fig. 6). Confocal microscopy analysis of biofilms formed under similar conditions on a glass surface revealed that the two mutants produced impaired biofilms, which were arrested at the microcolony stage. The CsgFG− mutant adhered rather poorly to the glass surface compared to the Crl− mutant. Following the complementation of the mutations, the two mutants regained their respective biofilm formation capacities. They each developed robust biofilms (Fig. 7), with the C24 CsgFG strain forming an even denser biofilm.
FIG. 6.
Biofilm quantification on 96-well polystyrene plates. Bacteria were grown in static LBNS cultures for 3 days at 26°C. The biofilm mass was determined using the CV method. The average OD595 values from three separate experiments are presented. The wt strain tested was DMS9S. The biofilm masses of the A59 and C24 mutants were significantly smaller than that of the wt (P < 0.0001). The error bars represent the standard errors of the means.
FIG. 7.
Biofilm formation on a glass surface. Cells were incubated for 3 days at 26°C in chamber slides, washed, and then stained with AO. Confocal microscopy image stacks were collected using FluoView 500 software, and three-dimensional reconstitution was performed using Imaris software. The pictures were transformed into black-and-white images and processed using Microsoft Office Picture Manager software to enhance their clarity.
Curli expression and air-liquid pellicle.
Strain DMS9S, but not the crl or the csgFG mutants, formed a thick pellicle at the air-liquid interface when grown statically in LBNS for 72 h at 26°C (Fig. 8A and B). Interestingly, the crl mutant preserved some of its biofilm-producing traits, as it was found to adhere to the glass tube wall in a ring-like form, while no such attachment was observed for the csgFG mutant. Curli expression was examined in DMS9S and in the two mutants by Western blot analysis as well as by electron microscopy (Fig. 9). Expression of curli was abolished in the csgFG mutant either in planktonic cells or in aggregates collected from the bottom of the tube (Fig. 9A). However, cells of the crl mutant which formed a ring in the tube appeared to express curli, as can be judged by the faint band of CsgA subunits (Fig. 9B). The introduction of pCSG and pCRL into the csgFG and crl mutants, respectively, resulted in regaining the capacity to form pellicles and express curli (Fig. 9A). Confocal microscopy of the wt pellicle revealed highly heterogenic structures with variable depths and bacterial densities, whereas the topologies of the pellicles formed by the two complemented mutants were more homogeneous (Fig. 8C).
FIG. 8.
Pellicle formation at the air-liquid interface. Bacteria were incubated in LBNS for 3 days at 26°C in glass tubes, and their pellicles were visualized by staining with CV (A). The wt pellicle was removed from the tube and stained with AO (B). AO-stained pellicles of the wt and complemented mutants were examined by confocal microscopy (C). Stacks were collected and processed using FluoView 500 software. The z-section values of representative areas are denoted. Pictures were processed using Microsoft Office Picture Manager software to enhance their clarity.
FIG. 9.
Curli expression during pellicle formation. Bacteria were grown in LBNS broth for 3 days at 26°C. (A) Western blots of the wt (lanes 1 and 2), the A59 crl+ strain (lanes 3 and 4), the C24 csgFG+ strain (lanes 5 and 6), and the A59 crl mutant (lanes 7 and 8). Since the C24 csgFG mutant did not form a pellicle, planktonic bacteria from the liquid culture and aggregates at the bottom were collected for Western analysis (lanes 11 and 12). A mixture of pellicle, planktonic cells, and aggregates of the wt strain was used as the control (lanes 9 and 10). Samples were pretreated with (+) or without (−) HFIP before being loaded onto an SDS-polyacrylamide gel and probed with CsgA antibody. (B) Transmission electron microscopy of the wt, A59 crl+, C24 csgFG+, and A59 crl strains collected from pellicles. Scale bars are equal to 500 nm.
Effect of Crl and curli on the RDAR morphotype.
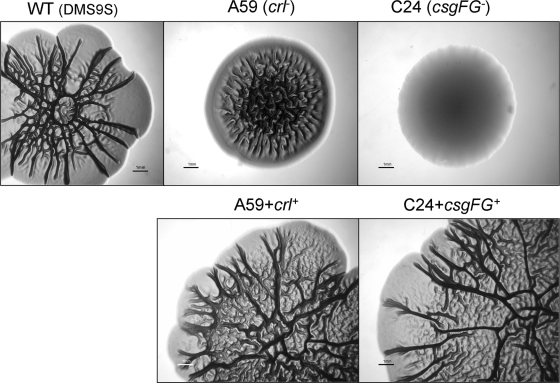
Both crl and csgFG mutants displayed a unique morphotype different from that of the wt strain, and their colonies were smaller than those of the wt strain. The csgFG mutant displayed a smooth and white (SAW) morphotype, while the crl mutant displayed a distinctive “pizza”-like morphotype, with a SAW-like region along the perimeter of the colony and a dense RDAR region in the center (Fig. 9). Complementation of the two mutations restored the typical wt morphotype, including CR binding (Fig. 10).
FIG. 10.
Comparison of the RDAR morphotypes of different strains. Strains were seeded in the center of LBNS agar plates supplemented with Congo red, and the plates were incubated for 5 days at 26°C. Photographs were taken through a binocular microscope with a digital camera. Each bar represents 1 mm.
DISCUSSION
In this study, we demonstrate for the first time that an aEPEC strain of the O55:H7 serotype displays a multicellular behavior characterized by the development of a robust biofilm on an abiotic surface (polystyrene) at 26°C, but not at 37°C, under static conditions. This strain forms a dense pellicle at the air-liquid interface and has an RDAR morphotype when grown under low-temperature and low-osmolarity conditions.
Two out of the six Tn10 mutants we initially analyzed had insertions in genes associated with curli regulation (crl) or assembly (csgFG). Curli are amyloid fibers produced by many members of the Enterobacteriaceae family. Together with cellulose, these fibers form major components of extracellular matrices in this family (10, 71). Curli have been implicated in many physiological and pathogenic processes of E. coli and Salmonella spp., including host cell adhesion and invasion (6). Yet, curli fibers were also shown to be involved in attachment to abiotic surfaces, cell aggregation, and biofilm formation (4, 5, 26, 29, 63, 65). In S. enterica serovar Enteritidis, curli was shown to allow adherence to Teflon and stainless steel, which can lead to biofilm formation and contamination of surfaces that are often used in the food industry (4).
Curli expression requires two divergently transcribed operons (Fig. 5B), the csgBAC operon, encoding the structural components of curli, and the csgDEFG operon. CsgD is a global regulator that controls the transcription of multiple genes that collectively determine transition from planktonic to sessile mode of growth, including the csgBAC operon and the cellulose biosynthesis operons (14, 29). CsgE and CsgF are two periplasmic chaperon-like proteins that facilitate efficient curli assembly, whereas CsgG is an outer membrane lipoprotein that interacts with CsgE and CsgF and forms an outer membrane channel that stabilizes the curli subunit proteins by mediating their translocation (50).
The presence of curli did not influence the attachment of the closely related pathogenic E. coli O157:H7 cells to nonhost surfaces, namely, alfalfa sprouts and lettuce leaves (12, 34). However, in the case of aEPEC, curli seems to play an important role in the interactions with abiotic surfaces, as inactivation of the csgFG genes, which impaired curli expression, also significantly reduced the capacity of this strain to form mature biofilms.
In S. enterica serovar Typhimurium, a mutant defective in the expression of csgBA genes (formerly agfBA), encoding curli (aka Tafi) subunit proteins, still maintained long-range intercellular adhesion, characterized by a colony morphotype called pink, dry, and rough and formation of a ring (but not pellicle) at the air-liquid interface (53). In contrast, the aEPEC curli-deficient mutant (csgFG) completely lost the characteristic multicellular traits; it forms a small and smooth colony and does not form a ring. These findings imply that curli proteins have a more critical role in the development of multicellular behavior in the tested aEPEC strain.
Crl was initially identified in E. coli as a cryptic regulator of curli expression (3). Later on, Crl was shown to modulate expression of curli in E. coli and Salmonella through regulation of CsgD (11, 14, 48). Crl is considered to be a thermosensor, since it modulates σS activity in E. coli and Salmonella only at temperatures below 30°C (11, 48). Crl stimulates σS-dependent transcription of numerous other genes in stationary phase and in response to different stress conditions by direct interaction with the RpoS holoenzyme (11, 44).
The crl mutant produced a smaller amount of biofilm than that produced by the wt, which was comparable to that produced by the csgFG mutant, when the two strains were grown in LB broth (Fig. 4). However, when grown in LBNS, a low-osmolarity medium, the effect of the crl mutation on biofilm formation was smaller than that of the csgFG mutation (Fig. 6). These results suggest that other osmoregulated components in aEPEC take part in controlling biofilm development.
Similar to the csgFG mutant, the crl mutant did not form a pellicle, yet it formed a ring at the air-liquid interface in the glass tube, which might be consistent with a low level of curli expression. Yet, since Crl modulates the expression of a considerable portion of the RpoS regulon in E. coli (11, 38, 44, 48), it is possible that the crl mutation has affected the expression of numerous other genes, which are also necessary for biofilm formation. Ring formation by the crl mutant is reminiscent of the Salmonella csgBA phenotype (53), further supporting variation in the mechanisms involved in the multicellular behavior of these two pathogens.
The crl mutant bound CR on agar plates, which is consistent with curli expression (31). However, it displayed a unique RDAR “pizza”-like morphotype, which is distinguished from that of either the wt or the csgFG mutant, supporting the notion regarding the involvement of other genes of the Crl-RpoS regulon in the multicellular behavior of aEPEC.
The role of Crl in the multicellular behavior is complex, since it is involved in a network of multifactorial regulation circuits including OmpR/EnvZ (52, 65), CpxR/A (26), and RcsAB (26), as well as several EAL and GGDEF-EAL domain proteins that act through modulation of cyclic di-GMP concentration (58).
Expression of multicellular behavior under low temperatures (<30°C) and low osmolarity was observed for E. coli K-12 and S. enterica serovar Typhimurium (1, 29, 45, 46, 52). Yet, different E. coli strains respond differentially to environmental cues. For example, it was reported that urinary-tract-infection E. coli strains typically expressed curli and cellulose at 28°C but not at 37°C, while commensal E. coli isolates coexpressed the two EPS components at both temperatures (10). Interestingly, the fecal isolates formed a pellicle also in the absence of curli, while our aEPEC strain failed to do so. These findings point to the existence of different mechanisms in pellicle formation among pathogenic and nonpathogenic E. coli strains.
A recent study has reported that biofilm formation and multicellular behavior in the E. coli O157:H7 strain were not related to cellulose production (63). The aEPEC strain used in our study produced a red color on agar plates containing Congo red and bound calcofluor, implying that it does produce cellulose as part of its EPS. These findings support the existence of variations in the two genetically related E. coli strains.
While tEPEC strains have been isolated only from infected humans, atypical strains have been found in several animal hosts, including birds, dogs, and cows (1, 33, 37, 41, 62). The prevalence of aEPEC strains among different animal hosts is largely unknown (33), as is their capacity to survive outside their host. The finding that aEPEC may present in healthy cattle examined at slaughter (8) may suggest that fecal transport among livestock animals is involved in the dissemination of this pathogen in the environment and its transmission to an uninfected animal or human host.
Once excreted from the host, enteric pathogens are confronted with nonhost environments characterized by limited nutrient availability, osmotic stress, large variations in temperature and pH, and predation (69). It was recently suggested that the RDAR morphotype allows Salmonella to survive in the environment and pass between hosts (68). In agreement with this notion, the researchers have noticed that Tafi (curli) expression was repressed during infection in an S. enterica serovar Typhimurium mouse model, while expression occurred only when the pathogen was passed out of the animal into the feces (67). Our findings regarding the presence of multicellular behavior in aEPEC during growth at a low temperature (26°C) may imply that this pathogen also exploits similar survival strategies when residing in the environment outside the mammalian host.
Finally, characterization of additional mini-Tn10 mutants may potentially reveal other constituents and mechanisms involved in the multicellular behavior of aEPEC strains and may extend our understanding regarding the fate of this pathogen in the nonhost environment.
Acknowledgments
We thank V. de Lorenzo (Centro Nacional de Biotecnología, Madrid, Spain) for providing plasmid pGP704, I. Rosenshine (Hebrew University, Jerusalem, Israel) for technical advice and discussions, and I. Shomer (The Volcani Center) for sharing his stereomicroscope.
M.R.C. acknowledges support from NIH grant RO1AI073847.
Footnotes
Published ahead of print on 15 January 2010.
REFERENCES
- 1.Aidar-Ugrinovich, L., J. Blanco, M. Blanco, J. E. Blanco, L. Leomil, G. Dahbi, A. Mora, D. L. Onuma, W. D. Silveira, and A. F. P. de Castro. 2007. Serotypes, virulence genes, and intimin types of Shiga toxin-producing Escherichia coli (STEC) and enteropathogenic E. coli (EPEC) isolated from calves in Sao Paulo, Brazil. Int. J. Food Microbiol. 115:297-306. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arnqvist, A., A. Olsen, J. Pfeifer, D. G. Russell, and S. Normark. 1992. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol. Microbiol. 6:2443-2452. [DOI] [PubMed] [Google Scholar]
- 4.Austin, J. W., G. Sanders, W. Kay, and S. Collinson. 1998. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol. Lett. 162:295. [DOI] [PubMed] [Google Scholar]
- 5.Barak, J. D., L. Gorski, P. Naraghi-Arani, and A. O. Charkowski. 2005. Salmonella enterica virulence genes are required for bacterial attachment to plant tissue. Appl. Environ. Microbiol. 71:5685-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnhart, M. M., and M. R. Chapman. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bian, Z., and S. Normark. 1997. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 16:5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco, M., S. Schumacher, T. Tasara, C. Zweifel, J. Blanco, G. Dahbi, J. Blanco, and R. Stephan. 2005. Serotypes, intimin variants and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-eta2). BMC Microbiol. 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumer, C., A. Kleefeld, D. Lehnen, M. Heintz, U. Dobrindt, G. Nagy, K. Michaelis, L. Emody, T. Polen, R. Rachel, V. F. Wendisch, and G. Unden. 2005. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 151:3287-3298. [DOI] [PubMed] [Google Scholar]
- 10.Bokranz, W., X. Wang, H. Tschape, and U. Romling. 2005. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 54:1171-1182. [DOI] [PubMed] [Google Scholar]
- 11.Bougdour, A., C. Lelong, and J. Geiselmann. 2004. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J. Biol. Chem. 279:19540-19550. [DOI] [PubMed] [Google Scholar]
- 12.Boyer, R. R., S. S. Sumner, R. C. Williams, M. D. Pierson, D. L. Popham, and K. E. Kniel. 2007. Influence of curli expression by Escherichia coli O157:H7 on the cell's overall hydrophobicity, charge, and ability to attach to lettuce. J. Food Prot. 70:1339-1345. [DOI] [PubMed] [Google Scholar]
- 13.Branda, S. S., A. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 14.Brombacher, E., A. Baratto, C. Dorel, and P. Landini. 2006. Gene expression regulation by the curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 188:2027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Biofilms 310:20-42. [DOI] [PubMed] [Google Scholar]
- 16.Clarke, S. C., R. D. Haigh, P. P. E. Freestone, and P. H. Williams. 2003. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin. Microbiol. Rev. 16:365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cleary, J., L.-C. Lai, R. K. Shaw, A. Straatman-Iwanowska, M. S. Donnenberg, G. Frankel, and S. Knutton. 2004. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 150:527-538. [DOI] [PubMed] [Google Scholar]
- 18.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 19.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 20.Danese, P. N., L. A. Pratt, and R. Kolter. 2001. Biofilm formation as a developmental process. Methods Enzymol. 336:19-26. [DOI] [PubMed] [Google Scholar]
- 21.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dean, P., M. Maresca, and B. Kenny. 2005. EPEC's weapons of mass subversion. Curr. Opin. Microbiol. 8:28-34. [DOI] [PubMed] [Google Scholar]
- 23.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 24.Domka, J., J. Lee, T. Bansal, and T. K. Wood. 2007. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9:332-346. [DOI] [PubMed] [Google Scholar]
- 25.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeune. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169-175. [DOI] [PubMed] [Google Scholar]
- 27.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 28.Genevaux, P., P. Bauda, M. S. DuBow, and B. Oudega. 1999. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172:1-8. [DOI] [PubMed] [Google Scholar]
- 29.Gerstel, U., and U. Romling. 2003. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 154:659-667. [DOI] [PubMed] [Google Scholar]
- 30.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowlkes, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71:384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammar, M., A. Arnqvist, Z. Bian, A. Olsen, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 32.Hicks, S., G. Frankel, J. B. Kaper, G. Dougan, and A. D. Phillips. 1998. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect. Immun. 66:1570-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishii, S., K. P. Meyer, and M. J. Sadowsky. 2007. Relationship between phylogenetic groups, genotypic clusters, and virulence gene profiles of Escherichia coli strains from diverse human and animal sources. Appl. Environ. Microbiol. 73:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeter, C., and A. G. Matthysse. 2005. Characterization of the binding of diarrheagenic strains of E. coli to plant surfaces and the role of curli in the interaction of the bacteria with alfalfa sprouts. Mol. Plant-Microbe Interact. 18:1235-1242. [DOI] [PubMed] [Google Scholar]
- 35.Kaper, J. B. 1996. Defining EPEC. Rev. Microbiol. 27:130-133. [Google Scholar]
- 36.Knutton, S., R. K. Shaw, R. P. Anantha, M. S. Donnenberg, and A. A. Zorgani. 1999. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol. Microbiol. 33:499-509. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi, H., T. Pohjanvirta, and S. Pelkonen. 2002. Prevalence and characteristics of intimin- and Shiga toxin-producing Escherichia coli from gulls, pigeons and broilers in Finland. J. Vet. Med. Sci. 64:1071-1073. [DOI] [PubMed] [Google Scholar]
- 38.Lelong, C., K. Aguiluz, S. Luche, L. Kuhn, J. Garin, T. Rabilloud, and J. Geiselmann. 2007. The Crl-RpoS regulon of Escherichia coli. Mol. Cell. Proteomics 6:648-659. [DOI] [PubMed] [Google Scholar]
- 39.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 40.Moreira, C. G., K. Palmer, M. Whiteley, M. P. Sircili, L. R. Trabulsi, A. F. P. Castro, and V. Sperandio. 2006. Bundle-forming pili and EspA are involved in biofilm formation by enteropathogenic Escherichia coli. J. Bacteriol. 188:3952-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nougayrède, J.-P., P. J. Fernandes, and M. S. Donnenberg. 2003. Adhesion of enteropathogenic Escherichia coli to host cells. Cell. Microbiol. 5:359-372. [DOI] [PubMed] [Google Scholar]
- 43.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial developement. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 44.Pratt, L. A., and T. J. Silhavy. 1998. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29:1225-1236. [DOI] [PubMed] [Google Scholar]
- 45.Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejeune, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-464. [DOI] [PubMed] [Google Scholar]
- 47.Reisner, A., J. A. J. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 48.Robbe-Saule, V., V. Jaumouille, M.-C. Prevost, S. Guadagnini, C. Talhouarne, H. Mathout, A. Kolb, and F. Norel. 2006. Crl activates transcription initiation of RpoS-regulated genes involved in the multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:3983-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robbe-Saule, V., M. D. Lopes, A. Kolb, and F. Norel. 2007. Physiological effects of Crl in Salmonella are modulated by σS level and promoter specificity. J. Bacteriol. 189:2976-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson, L. S., E. M. Ashman, S. J. Hultgren, and M. R. Chapman. 2006. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59:870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Römling, U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62:1234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Römling, U., Z. Bian, M. Hammar, W. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Römling, U., M. Rohde, A. Olsen, S. Normark, and J. Reinkoster. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 54.Römling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 56.Schembri, M. A., L. Hjerrild, M. Gjermansen, and P. Klemm. 2003. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J. Bacteriol. 185:2236-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sela, S., S. Frank, E. Belausov, and R. Pinto. 2006. A mutation in the luxS gene influences Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 72:5653-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simm, R., A. Lusch, A. Kader, M. Andersson, and U. Romling. 2007. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spears, K. J., A. J. Roe, and D. L. Gally. 2006. A comparison of enteropathogenic and enterohaemorrhagic Escherichia coli pathogenesis. FEMS Microbiol. Lett. 255:187. [DOI] [PubMed] [Google Scholar]
- 60.Tobe, T., and C. Sasakawa. 2001. Role of bundle-forming pilus of enteropathogenic Escherichia coli in host cell adherence and in microcolony development. Cell. Microbiol. 3:579-585. [DOI] [PubMed] [Google Scholar]
- 61.Tobe, T., and C. Sasakawa. 2002. Species-specific cell adhesion of enteropathogenic Escherichia coli is mediated by type IV bundle-forming pili. Cell. Microbiol. 4:29-42. [DOI] [PubMed] [Google Scholar]
- 62.Trabulsi, L. R., R. Keller, and T. A. Tardelli Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uhlich, G. A., P. H. Cooke, and E. B. Solomon. 2006. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl. Environ. Microbiol. 72:2564-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Houdt, R., and C. W. Michiels. 2005. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156:626-633. [DOI] [PubMed] [Google Scholar]
- 65.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, X., N. D. Hammer, and M. R. Chapman. 2008. The molecular basis of functional bacterial amyloid polymerization and nucleation. J. Biol. Chem. 283:21530-21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White, A. P., D. L. Gibson, G. A. Grassl, W. W. Kay, B. B. Finlay, B. A. Vallance, and M. G. Surette. 2008. Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium. Infect. Immun. 76:1048-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.White, A. P., and M. G. Surette. 2006. Comparative genetics of the rdar morphotype in Salmonella. J. Bacteriol. 188:8395-8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winfield, M. D., and E. A. Groisman. 2003. Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yildiz, F. H. 2007. Processes controlling the transmission of bacterial pathogens in the environment. Res. Microbiol. 158:195-202. [DOI] [PubMed] [Google Scholar]
- 71.Zogaj, X., W. Bokranz, M. Nimtz, and U. Romling. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]