Abstract
A molecular analysis of betaproteobacterial ammonia oxidizers and a N2O isotopomer analysis were conducted to study the sources of N2O emissions during the cow manure composting process. Much NO2−-N and NO3−-N and the Nitrosomonas europaea-like amoA gene were detected at the surface, especially at the top of the composting pile, suggesting that these ammonia-oxidizing bacteria (AOB) significantly contribute to the nitrification which occurs at the surface layer of compost piles. However, the 15N site preference within the asymmetric N2O molecule (SP = δ15Nα − δ15Nβ, where 15Nα and 15Nβ represent the 15N/14N ratios at the center and end sites of the nitrogen atoms, respectively) indicated that the source of N2O emissions just after the compost was turned originated mainly from the denitrification process. Based on these results, the reduction of accumulated NO2−-N or NO3−-N after turning was identified as the main source of N2O emissions. The site preference and bulk δ15N results also indicate that the rate of N2O reduction was relatively low, and an increased value for the site preference indicates that the nitrification which occurred mainly in the surface layer of the pile partially contributed to N2O emissions between the turnings.
The very sensitive greenhouse gas nitrous oxide (N2O) has a 296 times higher impact than CO2 (39) and is also responsible for ozone depletion (10). Agricultural activities such as the use of nitrate fertilizers, livestock production, and manure management, including composting, are known to be important sources of N2O emissions (18). To devise a strategy to mitigate N2O emissions, it is essential to understand its sources in detail. However, the sources of N2O emissions during the composting process are still largely unclear.
In the composting process, a part of NH4+-N is known to be processed through nitrification-denitrification and emitted as N2 and N2O. Nitrous oxide is known to be generated through both the nitrification and denitrification processes as intermediate products or by-products. Nitrous oxide emission is a very complex process because denitrifying bacteria are phylogenetically diverse (60), and nitrifiers are also known to utilize the denitrification process even under aerobic conditions (42). It is thus very difficult to estimate the relative contributions of nitrification and denitrification in actual N2O emissions from the environment. Until now, there has been insufficient knowledge about the relative contributions of these processes to N2O emissions during the animal manure composting process. Measurement of the actual contributions of N2O emissions from compost piles in the field is therefore critical to establishing a strategy of mitigating N2O emissions.
Recently, a high-precision analytical technique for determining intramolecular 15N site preference in asymmetric molecules of N2O was developed (47). Since N2O has two N atoms within the molecule (central and outer N), distribution of a stable isotope, 15N, results in the distribution of three isotopomers, such as 15N15NO, 15N14NO, and 14N15NO. By using this newly developed innovative technique, the latter two types of molecules, which exist abundantly in the environment, can be individually measured. The difference in δ15N between δ15Nα and δ15Nβ is the so-called site preference (SP = δ15Nα − δ15Nβ, where 15Nα and 15Nβ represent the 15N/14N ratios at the center and end sites of the nitrogen atoms, respectively). The site preference enabled us to identify the source and sinks of N2O in the environment (48, 49, 50, 56). Using this technique, Sutka et al. (44) found that the site preference for N2O from hydroxylamine oxidation (∼33‰) and nitrite reduction (∼0‰) differs in a pure culture study and noted that this difference can be used to distinguish the relative contributions of nitrification and denitrification sources to N2O emissions. There have still been only several reported studies which applied this measurement technique to field N2O samples (48, 53) or referred to the relative contributions of nitrification and denitrification. To our knowledge, the present study is the first to apply this isotopomer analysis technique to the determination of N2O sources in the composting process. We specifically used this technique to understand the actual contributions of nitrification and denitrification to N2O emissions during the cow manure composting process.
Ammonia oxidation, the conversion of ammonium to nitrite via hydroxylamine, is an initial step of the nitrification-denitrification process and is critical to the nitrogen cycle in the terrestrial environment (4, 24). In the nitrification process, N2O is generated as a by-product when ammonia oxidizers convert hydroxylamine to nitrite (35). Since NO2−-N and NO3−-N accumulate in the latter stages of the composting process (29, 30), it is obvious that nitrifiers are active in compost piles. Therefore, it is important to clarify the role and significance of ammonia oxidizers in N2O emissions during the composting process. However, since the pure culture isolation method is so difficult and time-consuming, little is known about these ammonia oxidizers. A molecular approach based on PCR has been recently developed and has to date been used to target the ammonia monooxygenase gene (amoA) or 16S rRNA gene of betaproteobacterial ammonia oxidizers in soil, wetlands, and marine sediments (2, 3, 6, 7, 13, 32, 52). Using these techniques, substantial information about uncultured ammonia-oxidizing bacteria (AOB) that are partially or wholly responsible for nitrification in the environment will become available. Since the microbial community drastically changes through the composting process (19, 29), and a high accumulation of nitrite or nitrate will occur, especially in the latter half of the process (30), we continuously sampled and analyzed the diversity and abundance of AOB throughout the process. Our objectives in this study were to elucidate the sources of N2O emissions during the cow manure composting process by combining the isotopomer analysis and molecular analysis of betaproteobacterial AOB.
MATERIALS AND METHODS
Composting experiment.
The composting experiments were performed twice at the National Agriculture Research Center for the Hokkaido Region (Sapporo City, Hokkaido, Japan) from 29 May through 11 August 2008 (pile 1) and 2 September through 14 October 2008 (pile 2). The cows were fed orchard grass silage, corn silage, oat hay, alfalfa hay, beet pulp, and two types of concentrate mixtures to meet their digestible energy requirements, as recommended by the Japanese feeding standard for dairy cattle (1). Lactating Holstein cow excrement and dried grass (Orchard grass; Dactylis glomerata) were used in this study.
About 2.5 metric tons of dairy cow excrement [total solids (TS), 18.9% ± 0.4% (pile 1) and 20.2% ± 0.6% (pile 2); volatile solids (VS), 80.6% ± 1.0% TS (pile 1) and 82.4% ± 0.8% (pile 2)] and 250 kg of dried grass [TS, 83.0% ± 0.2% (pile 1) and 87.6% ± 0.1% (pile 2); VS, 84.5% ± 0.1% TS (pile 1) and 93.6% ± 0.1% (pile 2)] were mixed. TS and VS were measured using a gravimetric method (see “Chemical analysis of the compost” below). About 2.5 metric tons of the mixture was piled up on a waterproof concrete floor. Each pile had a volume of 3 m3, a diameter of 2.8 m, and a height of 1.4 m at the start of the experiment. The compost piles were turned with a front loader and a manure spreader once every 2 weeks. The temperatures of the compost piles and ambient air were measured hourly using a Thermo Recorder RTW-30S (Espec, Japan).
N2O emission measurement.
Nitrous oxide emissions were measured using a dynamic chamber system and an IPD (infrared photoacoustic detector; INNOVA, Denmark), as described previously (29). The chamber system was designed to estimate the total gas emissions from compost piles, with a PVC (polyvinyl chloride) chamber equipped with blower ventilation and a gas sampling port on the ventilation exhaust. The chamber used in this study was 4 m in width, 6 m in depth, and 4 m in height. Four vent holes 10 cm in diameter were installed in the upper part of the chamber and connected to the ventilation blower, installed outside, with PVC pipe. The airflow was controlled using the inverter and was at 271 m3/h constantly throughout the experimental period. Fresh air was introduced under the skirt of the chamber. The air was subsampled using a Teflon tube (4 mm in diameter) inserted just before the in-line fan. The N2O concentrations of exhaust air were measured every 30 min, with two replications. According to the technical data of the IPD, the detection limit of N2O is 0.03 ppm at a pressure of 1 atm and a temperature of 25°C, and this can be translated to 0.06 mg/m3.
Analysis of N2O isotopomer ratios.
Gas samples for N2O isotopomer analysis were collected at 2- to 7-day intervals at the sampling port of the IPD, using a sampling system which consisted of an ammonium trap (300 ml of 2 mM H2SO4 solution in a scrubbing bottle), water- and CO2-absorbing columns (7 mm inside diameter [i.d.]), 25-cm glass tubes packed with Mg(ClO4)2 (8/24 and 20/40 mesh; Wako Pure Chemical Industries, Osaka, Japan) and with Ascarite (NaOH on support, 8/20 and 20/30 mesh; Thomas Scientific, Swedesboro, NJ), a 1-liter glass bottle equipped with two stopcocks, a bellows pump (MB-21; Senior Aerospace Metal Bellows, Sharon, MA), and a flow monitor. During the sampling, concentration monitoring by the IPD was interrupted, and the Teflon tube was replaced with a Tygon tube (3/8-in. i.d., 3 m). The chamber air was allowed to flow through the bottle at 0.5 liters min−1 for 15 min. Ambient air was also sampled at 2 m above ground in an evacuated 1- to 2-liter stainless steel canister.
The N2O isotopomer ratios were measured using a gas chromatography-isotope ratio mass spectrometry (GC-IRMS) (MAT 252; Thermo Fisher Scientific K.K., Yokohama, Japan) system described elsewhere (46). Site-specific nitrogen isotope analysis in N2O was conducted using ion detectors that had been modified for mass analysis of fragment ions of N2O (NO+) containing N atoms in the center positions of N2O molecules, whereas the bulk (average) nitrogen and oxygen isotope ratios were determined from molecular ions (47). Pure N2O (purity, >99.999%; Showa Denko K.K., Japan) was calibrated with international standards and used as a working standard for the isotopomer ratios. The notation of the isotopomer ratios is shown below. The measurement precision was typically better than 0.1‰ for δ15Nbulk (where 15Nbulk represents the average 15N/14N isotope ratio) and δ18O and better than 0.5‰ for δ15Nα and δ15Nβ.
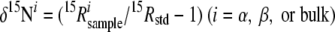 |
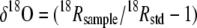 |
Here, 15Rα and 15Rβ represent the 15N/14N ratios at the center and end sites of the nitrogen atoms, respectively; 15Rbulk and 18R show average isotope ratios for 15N/14N and 18O/16O, respectively. Subscripts “sample” and “std” indicate isotope ratios for the sample and the standard, respectively, N for atmospheric N2, and O for Vienna Standard Mean Ocean Water (V-SMOW). We also define the 15N site preference (SP) as an illustrative parameter of the intramolecular distribution of 15N, as follows.
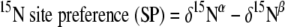 |
The N2O concentration was measured simultaneously with isotopomer ratios by comparing the peak area of the major ion (mass of 44 and 30 in molecular ion analysis and fragment ion analysis, respectively) obtained with sample gas and with a reference gas (349 ppb N2O in air; Japan Fine Products Co., Ltd.) (46).
Isotopomer ratios for compost-derived N2O (δcompost) were calculated from those for chamber gas samples (δchamber) and ambient air samples (δair) using the following mass balance equation:
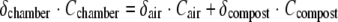 |
where C is the N2O concentration and Ccompost equals Cchamber minus Cair.
Chemical analysis of the compost.
A fresh sample of about 1 kg from each zone (Fig. 1) was collected just before each turning and at the start and end of the experiment. The level of total solids was measured after the samples dried overnight at 105°C. The dried samples were processed at 600°C for 1 h, and the level of volatile solids was calculated using the following equation (9):
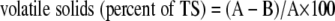 |
where A is the weight of the dried residue and B is the weight of the residue.
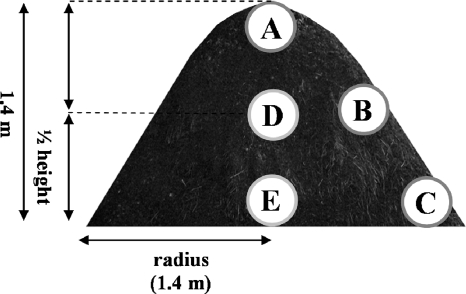
FIG. 1.
Sampling points of the piles. Samples were taken just before each turning.
Total nitrogen (T-N) was measured using the Kjeldahl method (5). The C/N ratio was measured using vario Max CNS (Elementar, Germany).
To measure inorganic N, pH, and electrical conductivity (EC), 7.5 g of fresh compost was placed into a 50-ml polypropylene tube with 30 ml of deionized water, shaken (200 rpm, 30 min), and then centrifuged (1,500 × g, 20 min). The supernatant was collected, and NH4-N, NO2-N, and NO3-N were measured using ion chromatography (DX-AQ 2211; Dionex); pH and EC were determined with calibrated electrodes (Horiba, Japan).
DNA extraction.
DNA extraction from the compost samples was performed using the commercially available DNA extraction kit Isofecal (Nippon Gene, Japan). The extraction was done according to the manufacturer's instructions, and the concentrations of DNA samples were measured by NanoDrop (Thermo Scientific). The purified DNA samples were stored at −20°C until further analysis.
AOB community structure analysis.
Among the ammonia oxidizers in the environment, we analyzed only the betaproteobacterial AOB community using the PCR-denaturing gradient gel electrophoresis (DGGE) method targeting the amoA and 16S rRNA genes. The nested PCR procedure was used to obtain a highly specific PCR product. Thermal cycler TP400 (Takara, Japan) and DNA polymerase PrimeStar (Takara, Japan) were used in this study. To amplify the amoA fragment, the primer set of amoA-1F and amoA-2R was used as described previously (40). To amplify the betaproteobacterium-specific 16S rRNA gene, the CTO189f-GC and CTO654r primer pair was used as described previously (13). The reaction mixture was prepared with template DNA (ca. 20 ng), 5 μM of each primer, 5× PCR buffer for PrimeStar (included in the kit), 0.2 mM of each deoxynucleoside triphosphate (dNTP), and 0.5 U of PrimeStar DNA polymerase, at a final volume of 20 μl. The thermal profile for the amoA gene was as follows: initial denaturation at 98°C for 5 min; 30 cycles of denaturation at 95°C for 10 s, annealing at 55°C for 5 s, and extension at 72°C for 1 min; final extension at 72°C for 7 min; and cooling at 4°C. The thermal profile for the 16S rRNA gene was as follows: initial denaturation at 98°C for 5 min; 30 cycles of denaturation at 95°C for 10 s, annealing at 57°C for 5 s, and extension at 72°C for 1 min; final extension at 72°C for 7 min; and cooling at 4°C. The PCR product was purified using the commercial kit MonoFas (GL Science, Japan) and used for the second PCR, using the same primer pairs. A GC clamp (5′-CGC CCG CCG CGC CCC GCG CCC GGC CCG CCG CCC CCG CCC C-3′) was attached to the 5′ end of the forward primer to improve the separation of the PCR fragments. DGGE analysis of the amplified bacterial amoA gene was performed on the DCode universal mutation detection system (Bio-Rad), according to the manufacturer's instructions. Polyacrylamide gels (7%, wt/vol) containing a linear formamide/urea gradient ranging from 25% to 65% denaturant were used. The gels were run for 15 h at 100 V and 60°C and stained with SYBR green for 30 min. The bands were visualized with a transilluminator (AE-6911FXFD; ATTO, Japan).
Real-time PCR.
Real-time PCR was performed using the commercially available kit SYBR Premix Ex Taq II (Takara, Japan), with a 20-μl reaction mix that consisted of 40 ng of template DNA. The primer pair used to amplify the amoA gene is described above. The primer pair used to amplify the 16S rRNA gene was 341F and 517R, as described previously (23). The PCR protocol for amoA quantification was as follows: 10 s at 95°C and 40 cycles consisting of 10 s at 95°C, 10 s at 55°C, and 34 s at 72°C. The PCR protocol for bacterial 16S rRNA gene quantification was as follows: 10 s at 95°C and 40 cycles consisting of 10 s at 95°C and 34 s at 60°C. Reactions were carried out in an ABI 7500 real-time PCR system (ABI). An external standard curve was prepared using serial dilutions of a known copy number of the plasmid pGEM-T Easy vector (Promega) containing the amoA gene of Nitrosomonas europaea (NBRC 14298). The standard curve for the bacterial 16S rRNA gene was prepared using the 16S rRNA gene of Paracoccus denitrificans (NCIMB 16712), and the same vector was used for the amoA gene.
Cloning and sequencing.
The excised DGGE bands were reamplified with Ex Taq (Takara, Japan) with the primer set without a GC clamp. The PCR product was purified using a commercial kit, as mentioned above, and cloned using pGEM-T Easy vector systems, according to the instruction manual (Promega). Cells from randomly picked colonies (3 colonies per sample) were resuspended in 20 μl of prepared PCR mixtures, and the inserts were amplified as mentioned above. The clones with the correct inserts were chosen for sequencing. The plasmid DNA was purified using the Wizard Plus minipreps DNA purification system (Promega), according to the instruction manual, and sequenced with the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems) and the ABI Prism 3100 genetic analyzer.
Statistical analysis.
The chemical analysis and gaseous concentration data were analyzed by analysis of variance (ANOVA), using the general linear model procedure described by the SAS Institute (41). Tukey's multiple range comparison tests were used to separate the means. A P value of <0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
The amoA and 16S rRNA gene sequences reported in this study have been deposited in the DNA Data Bank of Japan (DDBJ) under accession numbers AB495025 to AB495032 and AB496413 to AB496419.
RESULTS
Composting, N2O emissions, and isotopomer analysis.
In both piles 1 and 2, a significant reduction of weight, moisture content, and volatile solids occurred, indicating the active degradation of organic contents (Table 1). The maximum temperatures of the center zones of the piles were 74.9°C (pile 1) and 73.8°C (pile 2). These maximum temperatures indicate the high rate of degradation and microbial activity in both piles. However, the NO2−-N and NO3−-N content tended to be higher in pile 2 than in pile 1.
TABLE 1.
Chemical component profiles of compost piles
| Compost pile | Wt (kg) | Chemical component profilea |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TS (%) | VS (% TS) | NO2− (mg/kg TS) | NO3− (mg/kg TS) | NH4+ (mg/kg TS) | TKN (g/kg TS) | C/N ratio | pH | EC (mS/cm) | ||
| 1, initial | 2,790 | 27.0 (0.7)c | 82.7 (0.2)a | 17.8 (0.7)c | 8.3 (10.3)c | 512.2 (107.3)b | 28.9 (1.0)b | 18.8 (0.9)a | 9.3 (0.1)b | 2.2 (0.0)d |
| 1, final | 840 | 50.4 (0.9)b | 68.8 (0.1)c | 38.6 (7.4)b | 1,783.8 (41.7)a | 47.4 (7.8)c | 34.2 (0.6)a | 10.9 (0.1)b | 8.9 (0.1)c | 5.3 (0.1)b |
| 2, initial | 2,770 | 24.7 (0.3)d | 74.8 (0.6)b | 2.3 (2.2)c | 7.9 (5.6)c | 843.1 (48.9)a | 26.9 (0.3)b | 19.6 (0.3)a | 8.6 (0.1)d | 2.7 (0.3)c |
| 2, final | 970 | 53.9 (0.7)a | 62.1 (1.4)d | 138.4 (11.6)a | 243.8 (5.7)b | 121.6 (15.4)c | 33.5 (0.7)a | 12.3 (0.0)b | 9.6 (0.0)a | 5.9 (0.2)a |
TS, total solids (n = 3); VS, volatile solids (n = 3); TKN, total Kjeldahl nitrogen (n = 3); EC, electrical conductivity (n = 3). Values followed by different letters indicate significant difference (P < 0.05). The values in parentheses indicate standard errors.
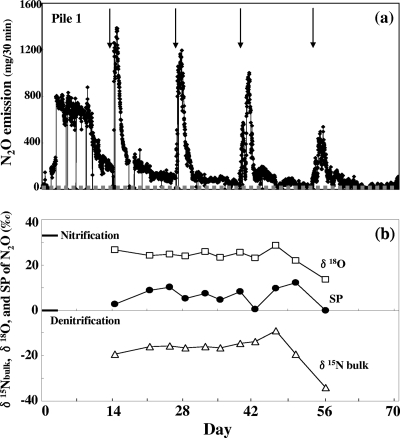
The peak N2O emissions from pile 1 occurred just after the turning, as we reported previously (Fig. 2a) (28). Figure 2b shows the variation of site preference, δ15N, and δ18O during the composting period of pile 1. The site preference of N2O was low and ranged from 0.0‰ to 12.0‰ throughout the composting period, indicating that the relative contribution of denitrification in N2O emissions is high during the cow manure composting process. In particular, the average site preference of N2O just after the turning (2.0‰ ± 2.3‰, n = 4) was significantly (P < 0.01) lower than that of the N2O samples between the turnings (8.7‰ ± 2.4‰, n = 7). Both the bulk δ15N and δ18O values did not show significant variation, except for the last sample on day 56, and ranged between approximately −34.2 and −9.1‰ in δ15N and approximately 13.7 and 28.7‰ in δ18O, respectively (see Table S1 in the supplemental material). These values were distinct from the values reported for other field samples such as seawater (δ15N, 1.7 to 22.7‰; δ18O, 46 to 105‰ [55]) or soil (δ15N, approximately −46 to −5‰; δ18O, approximately −3 to 9‰ [36]).
FIG. 2.
(a) N2O emission profile of pile 1. The arrows indicate the turnings. The gray dotted line indicates the detection limit. (b) δ15Nbulk, δ18O, and site preference (SP) of N2O. Open triangles indicate δ15Nbulk, open squares indicate δ18O, and closed circles indicate the site preference. The standard site preferences of N2O from nitrification (33‰) and denitrification (0‰) are also indicated (44).
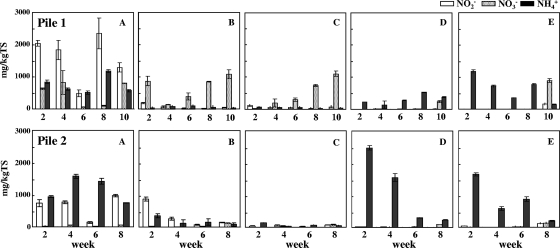
The distribution of inorganic nitrogen (NH4+-N, NO2−-N, and NO3−-N) in each part of the compost pile is shown in Fig. 3. Interestingly, remarkable NO2−-N and NH4+-N accumulations were detected in sample A at the top of the pile. In addition, the average total solid value obtained for sample A from pile 1 throughout the composting period was 27.7%a ± 5.3%, significantly lower (P < 0.05) than those obtained for the surface samples B (42.8%b ± 6.8%) and C (41.7%b ± 8.4%). This tendency was similar for samples from pile 2 (sample A, 29.7%ab ± 6.7%; sample B, 48.9%ab ± 13.4%; and sample C, 60.1%b ± 11.7%). (Values followed by different letters indicate significant difference [P < 0.05].) Although NO3−-N was detected in the surface samples (B and C) from pile 1, little was detected in those from pile 2. However, only NH4+-N was detected in the center and bottom samples (D and E) from both piles. These phenomena were observed at each turning, which was done every 2 weeks. From these results, active nitrification appeared to occur at the surfaces of the piles, especially at the top. The accumulated NO2−-N and NO3−-N which was transferred to the center or bottom zone by turning was not detected in the samples taken at the next turning.
FIG. 3.
NH4+, NO2−, and NO3−-N profiles of the compost piles. Open bars indicate nitrite, dotted bars indicate nitrate, and closed bars indicate ammonium. The error bars indicate the standard deviations (n = 3). A to E indicate the sampled zones of the pile, described in Fig. 1.
Distribution and abundance of the amoA gene.
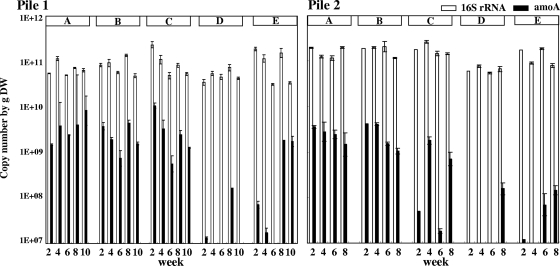
The gene copy numbers of betaproteobacterial ammonia oxidizers and all bacteria were determined by real-time PCR quantification, targeting the 16S rRNA and amoA genes, respectively (Fig. 4). In all samples (A to E), about 1011 copies/g (dry weight) of the 16S rRNA gene were detected. In contrast, the amoA gene was detected abundantly only in the surface samples (A to C), at about 109 copies/g (dry weight). In the initial to middle stage of the composting process, amoA was not detected from either the center samples (D) or the bottom ones (E). Although significant NO2−-N was accumulated in the top samples, the amoA copy numbers among the surface samples (A to C) showed no significant differences. In the latter stage of the process, in which the organic contents were mostly degraded and nitrification had actively occurred, the amoA gene was detected even from the center (D) or bottom (E) samples. Similar results were obtained in both pile 1 and pile 2.
FIG. 4.
16S rRNA (open bars) and amoA (closed bars) copy numbers g−1 (dry weight [DW]) compost for each zone of the compost piles. Error bars indicate the standard deviation for three replicate DNA extractions.
Diversity of the betaproteobacterial amoA gene.
The phylogenetic trees for the amoA and 16S rRNA genes of betaproteobacteria detected in surface samples from both piles are shown in Fig. S2 in the supplemental material. Sequences reported by previous papers (37, 38) were used to construct the phylogenetic trees. All sequences for both amoA and 16S rRNA specific for betaproteobacteria obtained in this study belong to the Nitrosomonas europaea cluster. These results suggest that the ammonia oxidizers working in the composting pile are not diverse but are instead a closely related group contributing to the ammonia oxidation.
The results obtained from PCR-DGGE for the amoA gene are shown in Fig. S1 in the supplemental material. Since the primer used in this study was a degenerate primer, multiple bands were visualized from the same sequence. Eight amoA sequences were detected in this study, and clones CMC 4 to 6 were detected from both piles 1 and 2. Some sequences were detected only in pile 1, making the band pattern of pile 2 somewhat simpler than that of pile 1.
DISCUSSION
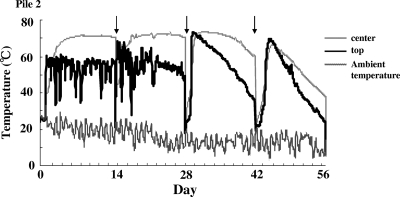
Nitrous oxide has a strong greenhouse effect, and its emissions must be mitigated. To devise a strategy for mitigation, it is necessary to understand its sources in detail. Our previous report showed the N2O emissions during the cow manure composting process occurred just after the piles were turned (28), and the N2O emissions in this study occurred in the same manner (Fig. 2a). The site preference ranged from 0.0 to 12.0, suggesting the relative importance of the denitrification process in the contribution to N2O emissions (Fig. 2b) (44). The site preference of the N2O samples released just after the turning ranged from 0.0 to 5.0, which indicates that their entire source originated from denitrification. In the top zone or surface zones of the pile, a large accumulation of NO2−-N and NO3−-N was observed. The accumulated nitrite and nitrate seem to have been moved inside the pile by the turning and consumed by subsequent denitrification. We also measured methane emissions by using an IPD and detected significant methane production within a few hours after the turning (data not shown), which shows that the core zone of the pile, even immediately after turning, was under an anoxic condition. The temperature just after the turnings ranged from 20 to 40°C inside the piles, and it took 1 or 2 days to reach maximum temperature (>65°C) (Fig. 5). These results show that the inorganic nitrogen species in the oxidized state that accumulate in the surface are transferred to the anoxic zone in the pile by turning, and the denitrification process that occurs after turning under the mesophilic condition is the main source of the N2O emissions during the composting process. Therefore, the suppression of nitrification, which actively occurs in the surface and top of the pile, may lead to significant reduction of the N2O emissions. Some previous studies reported the use of several types of nitrification inhibitors in pure culture (57), enriched nitrifying biomass (14), or arable soil (15, 51, 58). Dicyandiamide (DCD), nitrapyrin, 3,4-dimethylpyrazole phosphate (DMPP), or allylthiourea is frequently used; these chemicals all, to some extent, specifically inhibit the ammonia oxidation pathway. Soils amended with these chemicals reduce N2O emissions and nitrate reaching, with improving nitrogen uptake by the crops. Therefore, the use of these chemicals may lead to a reduction in N2O emissions from the compost. The effect of these chemical uses is not known because nitrogen removal is one of the main purposes in waste management.
FIG. 5.
Temperature profiles at the top and core of pile 2 and the ambient air. The arrows indicate the turnings.
There have been several studies on the effect of N2O reduction by bacteria on isotopomer ratios, and a simultaneous increase in the δ15N, δ18O, and site preference values was reported (20, 33, 55). Although the reported enrichment factors for site preference range from 2 to 16‰, depending on soil moisture or other environmental conditions, a Rayleigh model with an enrichment factor of 6‰ predicts that 50 percent of N2O consumption results in a 4‰ increase in site preference. Therefore, the N2O reduction would be an important factor for interpretation of the stable isotope analysis. In our study, the site preference of the samples just after the turnings with relatively high concentrations were very close to the value previously reported by pure culture study (0‰), as mentioned above, and it increased to some extent (4.5 to 12.0‰) in the samples collected between the turnings with low concentrations (see Table S1 in the supplemental material). On the other hand, both the bulk δ15N and δ18O values did not show statistically significant change throughout the process, ranging between approximately −34.2 and −9.1‰ in δ15N and approximately 13.7 and 28.7‰ in δ18O, respectively (Fig. 2b). If the increase in site preference originated from N2O reduction, the significant enrichment in both 15N and 18O should also be observed in the samples collected between the turnings. However, neither was observed (Fig. 2b), indicating the contribution of N2O reduction to the increase in site preference between the turnings was relatively low. The accumulation of NO2−-N and NO3−-N in the surface or top samples indicates that significant nitrification occurred between the turnings. These results indicate that nitrification is partially responsible for this increase in site preference.
All of the amoA- and betaproteobacterium-specific 16S rRNA sequences obtained in this study comprised the same cluster as the Nitrosomonas europaea lineage sequences. The other betaproteobacterium-like amoA or 16S rRNA gene sequences were not detected at all (see Fig. S2 in the supplemental material). These sequences were detected abundantly in the surface samples (Fig. 4); the contribution of AOB with these sequences may be related to or responsible for the accumulation of NO2−-N or NO3−-N in the compost surface. Some of our amoA sequences were very close (99% similarity) to the other amoA sequences available in the online database (34, 59). In these papers, the clones were detected from sites with high nitrogen loads or high organic content, such as a municipal solid waste disposal site or a batch reactor of animal wastewater treatment—in other words, environments with conditions similar to those of a compost pile. Therefore, the closely related species of AOB reported in these studies were actively working in our cow manure composting process, which contained a large amount of organic nitrogen and easily degradable organic content.
Recently, it was recognized that ammonia-oxidizing archaea (AOA) play an even more important role than AOB under various environmental conditions, such as soil, sediments, and seawater (12, 17, 26, 45, 54). In this study, we attempted to detect AOA in the compost pile using the primers reported in these previous studies. We did not detect any AOA sequences in the compost samples, even though AOA were detected in soil samples as a positive control (data not shown). Because of the difficulty in isolating AOA, there is only one report which has succeeded in developing a pure culture of AOA (22), and only limited information is available about AOA sequences. The accumulation of knowledge about AOA is necessary to design primers which can broadly detect AOA in the environment, including in compost. Further study is needed to find out whether or not AOA exist in compost.
Significant amounts of NO2−-N were detected in the top zone of the composting piles (Fig. 3). In terms of the determining characteristics of the top zone of the pile, two factors can be considered. First, the temperature was always around 50°C, much higher than the other surface points (samples B and C) (Fig. 5). Second, the moisture content was high compared to those of the other zones. The average total solid value in sample A of pile 1 throughout the composting period was significantly lower (P < 0.05) than those in surface samples B and C. This tendency was similar in pile 2. This difference in moisture content might affect the accumulation of NO2−-N and NO3−-N.
In the manure composting process, the organic content is degraded into water, CO2, and NH4+-N, and the heat moves upward inside the pile. As a result, the top zone is the exit point of both steam and gaseous NH3 (about 50°C) (Fig. 5). The NO2−-N accumulation and high rate of amoA copy numbers under such conditions suggest an adequate supply of oxygen and the existence of active thermophilic nitrifiers with tolerance to high ammonium concentrations. Since free ammonia is known to inhibit nitrite oxidation (8, 21, 27, 43), its presence can be one of the reasons why such NO2-N accumulation occurs. Lebedeva et al. (25) isolated the thermophilic Nitrosospira from a hot spring with low organic content. While other researchers have reported the existence of thermophilic nitrifiers in hot springs or seawater with low organic content (11, 16, 31), there is no report about thermophilic nitrifiers in an organic-rich environment, like a waste treatment system. Our data suggest the possibility of the existence of unknown thermophilic nitrifiers which are active in the high-organic-content environment of a compost pile. Efforts to isolate these unknown thermophilic nitrifiers should be made in future studies.
From the results obtained in this study, the following N2O emission model is proposed. (i) In the surface layer of the piles, nitrification occurs by the significant contribution of Nitrosomonas europaea-like betaproteobacterial ammonia oxidizers, especially at the top of the pile. The nitrification partially contributes to the N2O emissions between the turnings. (ii) Denitrification is the dominant source of nitrous oxide emissions, especially in the significant emissions which occur just after the turnings. (iii) Nitrous oxide reduction seemed to have occurred sparsely throughout the process.
Supplementary Material
Acknowledgments
We thank Atsuko Kobayashi and Kazuha Azumaya for providing laboratory-based technical assistance.
This work was supported by a grant from the National Agriculture and Food Research Organization (NARO), Japan, to K.M. and partly supported by a grant-in-aid for Scientific Research A (grant 19201004) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, to N.Y. and S.T.
Footnotes
Published ahead of print on 4 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Agriculture, Forestry and Fisheries Research Council Secretariat. 1999. Japanese feeding standard for dairy cattle. Central Association of Livestock Industry, Tokyo, Japan.
- 2.Avrahami, S., R. Conrad, and G. Braker. 2002. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 68:5685-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avrahami, S., W. Liesack, and R. Conrad. 2003. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ. Microbiol. 5:691-705. [DOI] [PubMed] [Google Scholar]
- 4.Bothe, H., G. Jost, M. Schloter, B. B. Ward, and K. Witzel. 2000. Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol. Rev. 24:673-690. [DOI] [PubMed] [Google Scholar]
- 5.Bremner, J. M. 1965. Total nitrogen; inorganic forms of nitrogen, p. 1149-1255. In C. A. Black (ed.), Methods of soil analysis, part 2. Soil Science Society of America, Madison, WI.
- 6.Cebron, A., M. Coci, J. Garnier, and H. J. Laanbroek. 2004. Denaturing gradient gel electrophoretic analysis of ammonia-oxidizing bacterial community structure in the lower seine river: impact of Paris wastewater effluents. Appl. Environ. Microbiol. 70:6726-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu, H., T. Fujii, S. Morimoto, X. Lin, K. Yagi, J. Hu, and J. Zhang. 2007. Community structure of ammonia-oxidizing bacteria under long-term application of mineral fertilizer and organic manure in a sandy loam soil. Appl. Environ. Microbiol. 73:485-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, J., H. Shim, S. Park, S. Kim, and W. Bae. 2006. Optimization of free ammonia concentration for nitrite accumulation in shortcut biological nitrogen removal process. Bioprocess Biosyst. Eng. 28:275-282. [DOI] [PubMed] [Google Scholar]
- 9.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington DC.
- 10.Crutzen, P. 1981. Atmospheric chemical processes of the oxides of nitrogen, including nitrous oxide, p. 17-44. In C. C. Delwiche (ed.), Denitrification, nitrification and atmospheric nitrous oxide. John Wiley & Sons, Inc., New York, NY.
- 11.de la Torre, J., C. Walker, A. Ingalls, M. Konneke, and D. Stahl. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810-818. [DOI] [PubMed] [Google Scholar]
- 12.Francis, C., K. Roberts, J. Beman, A. Santoro, and B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freitag, T. E., and J. I. Prosser. 2003. Community structure of ammonia-oxidizing bacteria within anoxic marine sediments. Appl. Environ. Microbiol. 69:1359-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginestet, P., J. Audic, V. Urbain, and J. Block. 1998. Estimation of nitrifying bacterial activities by measuring oxygen uptake in the presence of the metabolic inhibitors allylthiourea and azide. Appl. Environ. Microbiol. 64:2266-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatch, D., H. Trindade, L. Cardenas, J. Carneiro, J. Hawkins, D. Scholefield, and D. Chadwick. 2005. Laboratory study of the effects of two nitrification inhibitors on greenhouse gas emissions from a slurry-treated arable soil: impact of diurnal temperature cycle. Biol. Fertil. Soils 41:225-232. [Google Scholar]
- 16.Hatzenpichler, R., E. Lebedeva, E. Spieck, K. Stoecker, A. Richter, H. Daims, and M. Wagner. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, J., J. Shen, L. Zhang, Y. Zhu, Y. Zheng, M. Xu, and H. Di. 2007. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ. Microbiol. 9:2364-2374. [DOI] [PubMed] [Google Scholar]
- 18.Intergovernmental Panel on Climate Change. 2001. Climate change 2001, radiative forcing of climate change, the scientific basis. Cambridge University Press, Cambridge, United Kingdom.
- 19.Ishii, K., M. Fukui, and S. Takii. 2000. Microbial succession during a composting process as evaluated by denaturing gradient gel electrophoresis analysis. J. Appl. Microbiol. 89:768-777. [DOI] [PubMed] [Google Scholar]
- 20.Jinuntuya-Nortman, M., R. Sutka, P. Ostrom, H. Gandhi, and N. Ostrom. 2008. Isotopologue fractionation during microbial reduction of N2O within soil mesocosms as a function of water-filled pore space. Soil Biol. Biochem. 40:2273-2280. [Google Scholar]
- 21.Kim, D., D. Lee, and J. Keller. 2006. Effect of temperature and free ammonia on nitrification and nitrite accumulation in landfill leachate and analysis of its nitrifying bacterial community by FISH. Bioresour. Technol. 97:459-468. [DOI] [PubMed] [Google Scholar]
- 22.Könneke, M., A. Bernhard, R. José, C. Walker, J. Waterbury, and D. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 23.Kowalchuk, G., Z. Naoumenko, P. Derikx, A. Felske, J. Stephen, and I. Arkhipchenko. 1999. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl. Environ. Microbiol. 65:396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kowalchuk, G., and J. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 25.Lebedeva, E., M. Alawi, C. Fiencke, B. Namsaraev, E. Bock, and E. Spieck. 2005. Moderately thermophilic nitrifying bacteria from a hot spring of the Baikal rift zone. FEMS Microbiol. Ecol. 54:297-306. [DOI] [PubMed] [Google Scholar]
- 26.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. Nicol, J. Prosser, S. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Y., and B. Capdeville. 1994. Some observations on free ammonia inhibition to Nitrobacter in nitrifying biofilm reactor. Biotechnol. Lett. 16:309-314. [Google Scholar]
- 28.Maeda, K., R. Morioka, D. Hanajima, and T. Osada. 2010. The impact of using mature compost on nitrous oxide emission and the denitrifier community in the cattle manure composting process. Microb. Ecol. 59:25-36. [DOI] [PubMed] [Google Scholar]
- 29.Maeda, K., R. Morioka, and T. Osada. 2009. Effect of covering composting piles with mature compost on ammonia emission and microbial community structure of composting process. J. Environ. Qual. 38:598-606. [DOI] [PubMed] [Google Scholar]
- 30.Mahimairaja, S., N. S. Bolan, and M. J. Hedley. 1995. Denitrification losses of N from fresh and composted manures. Soil Biol. Biochem. 27:1223-1225. [Google Scholar]
- 31.Mével, G., and D. Prieur. 1998. Thermophilic heterotrophic nitrifiers isolated from Mid-Atlantic Ridge deep-sea hydrothermal vents. Can. J. Microbiol. 44:723-733. [Google Scholar]
- 32.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 33.Ostrom, N., A. Pitt, R. Sutka, P. Ostrom, A. Grandy, K. Huizinga, and G. Robertson. 2007. Isotopologue effects during N2O reduction in soils and in pure cultures of denitrifiers. J. Geophys. Res. 112:G02005. [Google Scholar]
- 34.Otawa, K., R. Asano, Y. Ohba, T. Sasaki, E. Kawamura, F. Koyama, S. Nakamura, and Y. Nakai. 2006. Molecular analysis of ammonia-oxidizing bacteria community in intermittent aeration sequencing batch reactors used for animal wastewater treatment. Environ. Microbiol. 8:1985-1996. [DOI] [PubMed] [Google Scholar]
- 35.Otte, S., J. Schalk, J. Kuenen, and M. Jetten. 1999. Hydroxylamine oxidation and subsequent nitrous oxide production by the heterotrophic ammonia oxidizer Alcaligenes faecalis. Appl. Microbiol. Biotechnol. 51:255-261. [DOI] [PubMed] [Google Scholar]
- 36.Pérez, T., S. Trumbore, S. Tyler, P. Matson, I. Ortiz-Monasterio, T. Rahn, and D. Griffith. 2001. Identifying the agricultural imprint on the global N2O budget using stable isotopes. J. Geophys. Res. 106:9869-9878. [Google Scholar]
- 37.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. Schmid, H. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purkhold, U., M. Wagner, G. Timmermann, A. Pommerening-Roser, and H. Koops. 2003. 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485-1494. [DOI] [PubMed] [Google Scholar]
- 39.Rahn, T., and M. Wahlen. 1997. Stable isotope enrichment in stratospheric nitrous oxide. Science 278:1776-1778. [DOI] [PubMed] [Google Scholar]
- 40.Rotthauwe, J., K. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.SAS Institute. 2001. SAS/STAT user's guide. SAS Institute, Cary, NC.
- 42.Shaw, L., G. Nicol, Z. Smith, J. Fear, J. Prosser, and E. Baggs. 2006. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ. Microbiol. 8:214-222. [DOI] [PubMed] [Google Scholar]
- 43.Smith, R., L. Burns, R. Doyle, S. Lennox, B. Kelso, R. Foy, and R. Stevens. 1997. Free ammonia inhibition of nitrification in river sediments leading to nitrite accumulation. J. Environ. Qual. 26:1049-1055. [Google Scholar]
- 44.Sutka, R. L., N. E. Ostrom, P. H. Ostrom, J. A. Breznak, H. Gandhi, A. J. Pitt, and F. Li. 2006. Distinguishing nitrous oxide production from nitrification and denitrification on the basis of isotopomer abundances. Appl. Environ. Microbiol. 72:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tourna, M., T. Freitag, G. Nicol, and J. Prosser. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357-1364. [DOI] [PubMed] [Google Scholar]
- 46.Toyoda, S., H. Mutobe, H. Yamagishi, N. Yoshida, and Y. Tanji. 2005. Fractionation of N2O isotopomers during production by denitrifier. Soil Biol. Biochem. 37:1535-1545. [Google Scholar]
- 47.Toyoda, S., and N. Yoshida. 1999. Determination of nitrogen isotopomers of nitrous oxide on a modified isotope ratio mass spectrometer. Anal. Chem. 71:4711-4718. [Google Scholar]
- 48.Toyoda, S., N. Yoshida, T. Miwa, Y. Matsui, H. Yamagishi, U. Tsunogai, Y. Nojiri, and N. Tsurushima. 2002. Production mechanism and global budget of N2O inferred from its isotopomers in the western North Pacific. Geophys. Res. Lett. 29:7-11. [Google Scholar]
- 49.Toyoda, S., N. Yoshida, T. Urabe, S. Aoki, T. Nakazawa, S. Sugawara, and H. Honda. 2001. Fractionation of N2O isotopomers in the stratosphere. J. Geophys. Res. Atmos. 106:7515-7522. [Google Scholar]
- 50.Toyoda, S., N. Yoshida, T. Urabe, Y. Nakayama, T. Suzuki, K. Tsuji, K. Shibuya, S. Aoki, T. Nakazawa, S. Ishidoya, K. Ishijima, S. Sugawara, T. Machida, G. Hashida, S. Morimoto, and H. Honda. 2004. Temporal and latitudinal distributions of stratospheric N2O isotopomers. J. Geophys. Res. Atmos. 109:D08308. [Google Scholar]
- 51.Watanabe, T. 2006. Influence of 2-chloro-6 (trichloromethyl) pyridine and dicyandiamide on nitrous oxide emission under different soil conditions. Soil Sci. Plant Nutr. 52:226-232. [Google Scholar]
- 52.Webster, G., T. M. Embley, and J. I. Prosser. 2002. Grassland management regimens reduce small-scale heterogeneity and species diversity of β-proteobacterial ammonia oxidizer populations. Appl. Environ. Microbiol. 68:20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westley, M., H. Yamagishi, B. Popp, and N. Yoshida. 2006. Nitrous oxide cycling in the Black Sea inferred from stable isotope and isotopomer distributions. Deep Sea Res. II 53:1802-1816. [Google Scholar]
- 54.Wuchter, C., B. Abbas, M. Coolen, L. Herfort, J. van Bleijswijk, P. Timmers, M. Strous, E. Teira, G. Herndl, and J. Middelburg. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamagishi, H., M. B. Westley, B. N. Popp, S. Toyoda, N. Yoshida, S. Watanabe, K. Koba, and Y. Yamanaka. 2007. Role of nitrification and denitrification on the nitrous oxide cycle in the eastern tropical North Pacific and Gulf of California. J. Geophys. Res. Biogeosci. 112:15. [Google Scholar]
- 56.Yoshida, N., and S. Toyoda. 2000. Constraining the atmospheric N2O budget from intramolecular site preference in N2O isotopomers. Nature 405:330-334. [DOI] [PubMed] [Google Scholar]
- 57.Zacherl, B., and A. Amberger. 1990. Effect of the nitrification inhibitors dicyandiamide, nitrapyrin and thiourea on Nitrosomonas europaea. Nutr. Cycl. Agroecosys. 22:37-44. [Google Scholar]
- 58.Zerulla, W., T. Barth, J. Dressel, K. Erhardt, K. Horchler von Locquenghien, G. Pasda, M. Rädle, and A. Wissemeier. 2001. 3,4-Dimethylpyrazole phosphate (DMPP)—a new nitrification inhibitor for agriculture and horticulture. Biol. Fertil. Soils 34:79-84. [Google Scholar]
- 59.Zhu, S., G. Chan, K. Cai, L. Qu, and L. Huang. 2007. Leachates from municipal solid waste disposal sites harbor similar, novel nitrogen-cycling bacterial communities. FEMS Microbiol. Lett. 267:236-242. [DOI] [PubMed] [Google Scholar]
- 60.Zumft, W. 1992. The denitrifying prokaryotes, p. 554-582. In A. Balows (ed.), The prokaryotes: a handbook of the biology of bacteria: ecophysiology, isolation, identification, applications. Springer-Verlag, New York, NY.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.