Abstract
There are numerous PCR-based assays available to characterize bovine fecal pollution in ambient waters. The determination of which approaches are most suitable for field applications can be difficult because each assay targets a different gene, in many cases from different microorganisms, leading to variation in assay performance. We describe a performance evaluation of seven end-point PCR and real-time quantitative PCR (qPCR) assays reported to be associated with either ruminant or bovine feces. Each assay was tested against a reference collection of DNA extracts from 247 individual bovine fecal samples representing 11 different populations and 175 fecal DNA extracts from 24 different animal species. Bovine-associated genetic markers were broadly distributed among individual bovine samples ranging from 39 to 93%. Specificity levels of the assays spanned 47.4% to 100%. End-point PCR sensitivity also varied between assays and among different bovine populations. For qPCR assays, the abundance of each host-associated genetic marker was measured within each bovine population and compared to results of a qPCR assay targeting 16S rRNA gene sequences from Bacteroidales. Experiments indicate large discrepancies in the performance of bovine-associated assays across different bovine populations. Variability in assay performance between host populations suggests that the use of bovine microbial source-tracking applications will require a priori characterization at each watershed of interest.
The ability to discriminate between bovine and other sources of fecal contamination is necessary for the accurate evaluation of human health risks associated with agricultural runoff and focused water quality management to make waters safe for human use. Many methods have been proposed to identify bovine fecal pollution using a variety of different microbiology and molecular techniques. One of the most widely used approaches utilizes a PCR to amplify a gene target that is specifically found in a host population. Currently, there are numerous PCR-based assays for the detection and/or quantitative assessment of bovine fecal pollution available for microbial source-tracking (MST) applications (1, 5-7, 11, 14, 17, 18, 21, 23). These assays target genes ranging from mitochondrial DNA to ribosomal rRNA to other functional genes involved in microorganism-host interactions.
The majority of the reported bovine-associated PCR assays target 16S rRNA genes from the order Bacteroidales. This bacterial group constitutes a large proportion of the normal gut microbiota of most animals, including bovines (28), and contains subpopulations closely associated with other animal hosts such as swine, horse, and human (1, 3, 6, 18, 24). Host-associated PCR-based assays targeting Bacteroidales genetic markers have been used to investigate the sources and levels of fecal pollution at a number of beaches and inland watersheds, with variable levels of success (10, 13, 22, 27). Researchers have postulated that differences in host animal age, health, diet, and geographic location may influence bacterial community structures in the bovine gastrointestinal tract (2, 9, 26). Without a priori knowledge of the potential representational bias introduced by such factors, it may be difficult to use these assays with confidence as indicators of bovine fecal pollution.
Assay specificity and sensitivity and the prevalence and abundance of genetic marker determinations are typically estimated from the systematic testing of a collection of reference fecal sources collected from known animal sources. However, the characterization of assay performance has been limited, in most cases, to animal sources originating from a particular geographic region or industry, such as dairy or beef. The determination of assay performance across a range of different host populations is essential as the field moves toward the implementation of PCR-based host-associated fecal pollution assessment approaches.
We report a performance study of seven PCR and quantitative PCR (qPCR) assays targeting Bacteroidales genes reported to be associated with either ruminant (e.g., bovine, goat, sheep, deer, and others) or bovine feces. Each assay was tested against a reference collection of DNA extracts from 247 individual bovine fecal samples representing 11 different populations. Assay specificity was determined by testing 175 fecal DNA extracts from 24 different animal species. For qPCR assays, the abundance of each genetic marker was measured within each bovine population and compared to quantities of Bacteroidales 16S rRNA genetic markers. These analyses indicated large discrepancies in assay performance across different bovine populations.
MATERIALS AND METHODS
Fecal and wastewater reference collections.
A total of 431 individual fecal samples were collected for analysis as previously described (23). Target (bovine) fecal specimens (n = 247) were collected over a 2-year period from 11 different populations, including herd 1 (located in CO; n = 25), herd 2 (CO; n = 25), herd 3 (OH; n = 20), herd 4 (NE; n = 29), herd 5 (NE; n = 30), herd 6 (WA; n = 10), herd 7 (GA; n = 31), herd 8 (WY; n = 49), herd 9 (WA; n = 10), herd 10 (WA; n = 10), and herd 11 (WA; n = 8). Nontarget fecal samples (n = 175) represented a total of 24 different animal sources, including Homo sapiens (human; n = 16), Lama pacos (alpaca; n = 2), Anser sp. (Canadian goose; n = 12), Felis catus (cat; n = 10), Gallus gallus (chicken; n = 10), Odocoileus virginianus (white-tail deer; n = 9), Odocoileus hemionus (mule deer; n = 5), Cervus elaphus (elk; n = 5), Alces alces (moose; n = 1), Antilocapra american (pronghorn; n = 4), Canis familiaris (dog; n = 10), Anas sp. (duck; n = 12), Capra aegagrus (goat; n = 7), Equus caballus (horse; n = 7), Pelecanus sp. (pelican; n = 5), Sus scrofa (pig; n = 22), Laridae (gull; n = 12), Ovis aries (sheep; n = 10), Eudorcas thomsoni (gazelle; n = 1), Giraffa camelopardalis (giraffe; n = 1), Okapi johnstoni (okapi; n = 3), Budorcas taxicolor (takin; n = 1), Elaphodus cephalophus (tufted deer; n = 1), Procyon loter (raccoon; n = 2), and Meleagris sp. (turkey; n = 7). All wildlife samples were collected from feral animals except those from gazelle, giraffe, okapi, takin, and tufted deer, which originated from the Cincinnati Zoo. Each fecal sample was collected from a different individual to maximize the opportunity to observe false-positive amplifications.
DNA purification.
All DNA extractions were performed with a FastDNA Kit for Soils (Q-Biogene, Carlsbad, CA) as described previously (23). DNA extraction yields were determined with a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). All DNA purifications were diluted to a test concentration of 1 ng/μl and stored at −20°C in 50-μl aliquots in GeneMate Slick Low-Adhesion microtubes (ISC BioExpress) until time of analysis. Extraction controls, with purified water substituted for fecal material, were performed each day samples were extracted to monitor for potential extraneous DNA contamination.
Primers and probes.
Seven PCR-based assays reported to be specific for either ruminant or bovine feces and the GenBac3 qPCR assay that targets the rRNA genes of Bacteroidales were utilized in this study (Table 1). For qPCR assays, TaqMan fluorogenic probes were 5′ labeled with 6-FAM (6-carboxyfluorescein) or TET (tetrachloro derivative of carboxyfluorescein) and 3′ labeled with TAMRA (6-carboxytetramethylrhodamine).
TABLE 1.
End-point PCR and qPCR assay primers, probes, reaction conditions, and DNA targets
| Assay | Platform | Primer name, primer sequence, and/or probe sequence (5′ to 3′) | Annealing temp (°C) | No. of cycles | Reported host target(s) | DNA target | Reference(s) |
|---|---|---|---|---|---|---|---|
| CF128 | PCR | CF128: CCAACYTTCCCGWTACTC | 62 | 35 | Ruminants | 16S rRNA | 1, 22 |
| 708R: CAATCGGAGTTCTTCGTG | |||||||
| CF193 | PCR | CF193: TATGAAAGCTCCGGCG | 62 | 35 | Ruminants | 16S rRNA | |
| 708R: CAATCGGAGTTCTTCGTG | |||||||
| Bac2 | PCR | Bac2F: GCTTGTTGCGTTCCTTGAGATAAT | 60 | 35 | Bovine | HDIG domain protein | 23 |
| Bac2R: ACAAGCCAGGTGATACAGAAAG | |||||||
| Bac3 | PCR | Bac3F: CTAATGGAAAATGGATGGTATCT | 62 | 35 | Bovine | Sialic acid-specific 9-O-acetylesterase secretory protein homolog | |
| Bac3R: GCCGCCCAGCTCAAATAG | |||||||
| BoBac | qPCR | BoBac367f: GAAGRCTGAACCAGCCAAGTA | 60 | 40 | Bovine | 16S rRNA | 11 |
| BoBac467r: GCTTATTCATACGGTACATACAAG | |||||||
| BoBac402f: 6-FAM-TGAAGGATGAAGGTTCTATGGATTGTAAACTT-TAMRA | |||||||
| CowM2 | qPCR | CowM2F: CGGCCAAATACTCCTGATCGT | 60 | 40 | Bovine | HDIG domain protein | 21 |
| CowM2R: GCTTGTTGCGTTCCTTGAGATAAT | |||||||
| CowM2P: 6-FAM-AGGCACCTATGTCCTTTACCTCATCAACTACAGACA-TAMRA | |||||||
| CowM3 | qPCR | CowM3F: CCTCTAATGGAAAATGGATGGTATCT | 60 | 40 | Bovine | Sialic acid-specific 9-O-acetylesterase secretory protein homolog | |
| CowM3R: CCATACTTCGCCTGCTAATACCTT | |||||||
| CowM3P: 6-FAM-TTATGCATTGAGCATCGAGGCC-TAMRA | |||||||
| GenBac3 | qPCR | GenBactF3: GGGGTTCTGAGAGGAAGGT | 60 | 40 | No data | 16S rRNA | 20 |
| GenBactR4: CCGTCATCCTTCACGCTACT | |||||||
| GenBactP2: 6FAM-CAATATTCCTCACTGCTGCCTCCCGTA-TAMRA |
End-point PCR amplification.
Four end-point PCR assays were used in this study, including CF128, CF193, Bac2, and Bac3 (Table 1). Reaction conditions and thermal cycling parameters for all end-point PCR assays are described elsewhere (22, 23) with the exception that TaKaRa Ex Taq Hot Start (Takara Bio, Inc., Japan) DNA polymerase was used for all assays. A volume of 25 ml was used for reactions performed with a DNA Engine Tetrad2 Peltier thermal cycler (Bio-Rad, Hercules, CA) for 35 cycles. All reactions were performed in duplicate in 96-well polypropylene plates. Results were visualized by loading 5 μl of PCR product onto either a 2% (CF128 and CF193) or 3% (Bac2 and Bac3) agarose gel containing a 1× final concentration of GelStar nucleic acid stain (Cambrex BioScience, Rockland, ME). Agarose gels were photographed with a Gel Logic 100 Imaging system (Carestream Health Inc.) and an UV transilluminator (model 614A; Fisher Scientific) at the maximum setting and an exposure time of 2.5 s. For specificity and prevalence testing, all replicate reactions must elicit the appropriate size band to be considered a positive reaction, except for sensitivity experiments. To monitor for potential sources of extraneous DNA during laboratory analyses, a minimum of three no-template amplifications with purified water substituted for template DNA were performed for each 96-well end-point PCR and qPCR.
Preparation of qPCR standards.
The plasmid DNA construct used as an absolute DNA standard for the CowM2 and CowM3 qPCR assays is described elsewhere (21). A custom plasmid DNA standard sequence was constructed for the BoBac assay that consists of the primer sequences and unique probe hybridization site (5′-VIC-TAGGAACAGGCGGCGACGA-TAMRA-3′). To build the BoBac plasmid DNA construct, long oligonucleotides including Frag1 (5′-GGGTTTAAAGGGAGCGTAGGGTGAGTCGTATTACAATTCACTGGCC GTCGAAGRCTGAACCAGCCAAGTAATCGTAGGAACAGGCGGCGAC GATA-3′) and Frag2 (5′-GCTTATTCATACGGTACATACAAGGAGAGGC GGCTACACCACGAATCCGCCTTATATCGTCGCCGCCTGTTCCTA CG-3′) containing the primer sequences were designed such that their 3′ ends overlapped. The two overlapping fragments were then combined into a single DNA molecule using overlap extension PCR (4). The plasmid construct was then inserted into the pCR4 TOPO plasmid vector (Invitrogen), and the resulting recombinant plasmids were purified from transformed Escherichia coli cell cultures using a Qiagen Plasmid Purification Kit (Qiagen, Valencia, CA). Plasmid DNA was linearized by NotI restriction digestion (New England BioLabs, Beverly, MA), quantified with a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies), and diluted in 10 mM Tris-0.1 mM EDTA (pH 8.0) to generate samples ranging from approximately 25 to 2.5 × 106 molecules of template DNA. Calibration curve equations for the GenBac3 assay were generated from qPCR analyses of serially diluted genomic DNA preparations from cultured cell suspensions of Bacteroides thetaiotaomicron (ATCC 29741). Total DNA concentrations in these purified stock solutions were spectrophotometrically determined, and ribosomal DNA target sequence copy concentrations were estimated from reported mean genome sizes and rRNA gene copy numbers per genome of respective species (8).
qPCR amplification.
Four qPCR assays were used in this study, including BoBac, CowM2, CowM3, and GenBac3 (Table 1). All amplifications were performed in a 7900 HT Fast Real Time Sequence Detector (Applied Biosystems). Reaction conditions and thermal cycling parameters for all qPCR assays were used as originally reported by respective laboratories and are described elsewhere with the exception of the BoBac qPCR assay (Table 1). The BoBac assay was run for 40 cycles rather than 50, and the annealing temperature was increased from 57°C to 60°C. All reactions were performed in duplicate in MicroAmp Optical 96-well reaction plates with MicroAmp 96-well Optical Adhesive Film (Applied Biosystems). Data were initially analyzed with Sequence Detector Software (version 2.2.2) at a threshold determination of 0.08 for host-specific assays and 0.03 for the general fecal indicator bacteria assay (GenBac3). Threshold cycle (CT) values were exported to Microsoft Excel in preparation for further statistical analysis.
Monitoring for PCR inhibition in DNA extracts.
DNA isolation from fecal samples may not remove all substances that can interfere with end-point and qPCR assays, and the degree of interference may vary between samples. Therefore, internal controls designed to evaluate the suitability of isolated DNA for quantitative analysis were included for each DNA extract. All fecal DNA extracts were screened for inhibition utilizing either the CowM2 or Entero1 internal amplification control (IAC) multiplex assays (21). The criterion for concluding that no significant PCR inhibition occurred with the CowM2 IAC assay for fecal DNA composite preparations was established as a CT of 34.6 ± 1, based on 58 repeated experiments measuring the simplex mean CT values for control reaction mixtures containing 50 copies of the CowM2 IAC in buffer. Individual bovine fecal extracts were also tested for inhibition using the previously reported multiplex Entero1 IAC assay (21). The criterion for concluding no significant PCR inhibition in these assays was defined as a CT of 33.3 ± 1, based on 50 repeated experiments measuring the simplex mean CT values for control reaction mixtures containing 25 copies of the Entero1 IAC in buffer.
Sensitivity, specificity, and prevalence.
Presence or absence data generated by end-point PCR assays was used to estimate the sensitivity and specificity of the CF128, CF193, Bac2, and Bac3 assays. The sensitivity of each end-point PCR assay was determined by testing a separate composite DNA preparation for each bovine population (n = 11) at four different concentrations, including 1 × 10−9 g, 1 × 10−10 g, 1 × 10−11 g, and 1 × 10−12 g of total DNA. Each composite consisted of an equal ratio of DNA from 10 randomly selected individual fecal samples with the exception of the herd 11 population (n = 8). Four replicates were tested for each composite DNA concentration. Sensitivity was defined as the total number of positive samples that tested positive correctly (TPC) divided by the sum of TPC and the total number of amplifications that tested negative incorrectly (TPI) [sensitivity = TPC/(TPC+TNI)]. Prevalence was estimated by testing all 247 individual bovine fecal samples for the presence of DNA target at a 1 ng/μl concentration of total DNA. Specificity is the ability of an assay to discriminate between the target animal host and other animal sources and was mathematically expressed as follows: specificity = TNC/(TNC+TPI), where TNC represents the total number of negative samples that tested negative correctly, and TPI is the total number of samples that tested positive incorrectly. Specificity was characterized by testing the 175 nontarget fecal DNA samples for the presence of target DNA at a 1 ng/μl concentration of total DNA.
Estimating abundance of DNA targets in bovine populations.
BoBac, CowM2, CowM3, and GenBac3 qPCR assay abundance of DNA targets in host animals was determined by estimating the log10 mean copy number of DNA targets in composite fecal samples representing each bovine population. DNA composites for each bovine population consisted of 10 randomly selected individual samples with the exception of herd 11 (n = 8). For each population composite, select DNA sample extracts were normalized to equal concentrations of total DNA prior to pooling to generate a final test concentration of 1 ng per reaction mixture. The abundance of DNA targets for PCR assays CF128, CF193, Bac2, and Bac3 was not determined. Instead, the frequency of detection at a 1-ng test concentration was calculated for each assay using all 247 individual bovine fecal samples.
Calculations and statistical analysis.
For all qPCR assays, amplification efficiency [10(1/−slope)/2], linearity (R2 value), analytical precision, and range of quantification were calculated from 7 to 32 independently generated standard curves. The analytical precision of CT measurements determined from plasmid or genomic DNA standards was expressed as a percent coefficient of variation (CV; standard deviation expressed as a percentage of the mean). Master calibration curves, unknown DNA concentration estimates, and credible intervals were determined using a Monte Carlo Markov chain (MCMC) approach (25). MCMC calculations were performed using the publicly available software WinBUGS, version 1.4.1 (http://www.mrc-bsu.cam.ac.uk/bugs) (15). To estimate within-population variance (σ2) and differences between each qPCR assay and bovine population, two-way nested analysis of variance (ANOVA) was used. In this analysis, bovine population was a fixed factor, and fecal samples were the random factor nested within a respective bovine population. ANOVA tests were performed using SAS software (Cary, NC) with the procedures PROC MIXED and PROC GLM (19).
RESULTS
qPCR calibration curves, range of quantification, and precision.
Calibration curves used to generate estimates of DNA target concentrations in fecal samples are reported in Table 2. Bovine-associated qPCR assays exhibited a range of quantification (ROQ) of 10 to 25 to 2.5 × 106 copies, while the ROQ GenBac3 ranged from 10 to 1 × 104 copies (in all cases, the entire range of copy numbers was tested). Precision of CT measurements across defined ROQs for all assays was less than a 4.0% CV, amplification efficiencies ranged from 93% to 97.5%, and all R2 values were ≥0.998 (Table 2).
TABLE 2.
Calibration curve equations and performance characteristics of host-associated and general fecal indicator qPCR assays
| Assay | No. of curvesa | Calibration equation | R2b | Amplification efficiency (%)c | ROQ (copies) for target DNAd | %CV across ROQe |
|---|---|---|---|---|---|---|
| BoBac | 32 | Y = 40.4 − 3.45X | 0.998 | 97.5 | 10-1 × 106 | 3.9 |
| CowM2 | 24 | Y = 41.9 − 3.67X | 0.999 | 93.5 | 25-2.5 × 106 | 2.8 |
| CowM3 | 24 | Y = 42.7 − 3.69X | 0.999 | 93 | 25-2.5 × 106 | 3.1 |
| GenBac3 | 7 | Y = 37.1 − 3.49X | 0.999 | 96.5 | 10-4 × 104 | 1.2 |
Number of individual standard curves used to determine a respective calibration equation.
R2 denotes the coefficient of determination representing the proportion of variability in the data set accounted for by the linear model.
Amplification efficiency = 10(1/−slope)/2.
Range of quantification measured in copies of target DNA for each respective qPCR assay.
Data represent the qPCR analytical precision of measuring standard concentrations expressed as the mean percent coefficient of variation.
Specificity and prevalence.
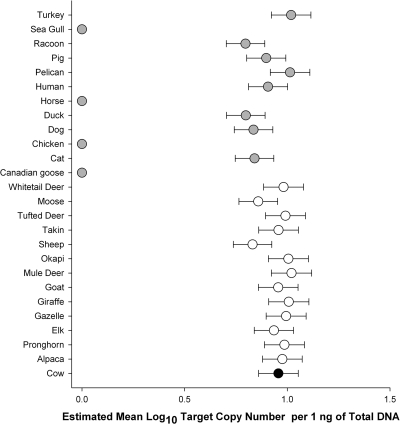
Specificity values were 76%, 99.9%, 100%, and 98.9% with PCR assays CF128, CF193, Bac2, and Bac3, respectively (Table 3). For qPCR assays, specificity ranged from 47.4% for the BoBac assay to 100% for the CowM2 and CowM3 assays. The estimated mean log10 target copy concentrations per 1 ng of total DNA from fecal DNA extracts from nonbovine reference samples that elicited false-positive detection with the BoBac assay are compared with the mean log10 target copy concentration from all bovine samples in Fig. 1.
TABLE 3.
PCR and qPCR assay specificity results
| Animala | Locale | No. of samplesb | End-point PCR resultc |
qPCR resultc |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Bac2 | Bac3 | CF128d | CF193d | CowM2 | CowM3 | BoBac | |||
| Alpaca* | WV | 2 | - | 2 | SO | SO | - | - | 2 |
| Pronghorn* | WY | 4 | - | - | SO | SO | - | - | 4 |
| Elk* | WY | 5 | - | - | SO | SO | - | - | 4 |
| Gazelle* | OH | 1 | - | - | SO | SO | - | - | 1 |
| Giraffe* | OH | 1 | - | - | SO | SO | - | - | 1 |
| Goat* | WA | 7 | - | - | SO | SO | - | - | 7 |
| Mule deer* | WY | 5 | - | - | SO | SO | - | - | 5 |
| Okapi* | OH | 3 | - | - | SO | SO | - | - | 3 |
| Sheep* | DE | 10 | - | - | SO | SO | - | - | 10 |
| Takin* | OH | 1 | - | - | SO | SO | - | - | 1 |
| Tufted deer* | OH | 1 | - | - | SO | SO | - | - | 1 |
| Moose* | WY | 1 | - | - | SO | SO | - | - | 1 |
| White-tailed deer* | WY | 9 | - | - | SO | SO | - | - | 9 |
| Canadian goose | GA | 12 | - | - | - | - | - | - | - |
| Cat | WV | 10 | - | - | - | - | - | - | 9 |
| Chicken | GA | 10 | - | - | 4 | - | - | - | - |
| Dog | WV | 10 | - | - | 3 | - | - | - | 7 |
| Duck | GA | 12 | - | - | 3 | - | - | - | 7 |
| Horse | WA | 7 | - | - | - | 1 | - | - | - |
| Human | WV | 16 | - | - | - | - | - | - | 6 |
| Pelican | FL | 5 | - | - | - | - | - | - | - |
| Pig | OH | 22 | - | - | 20 | - | - | - | 11 |
| Racoon | WA | 2 | - | - | - | - | - | - | 1 |
| Sea gull | GA | 12 | - | - | - | - | - | - | - |
| Turkey | OH | 7 | - | - | - | - | - | - | 2 |
| Total | 175 | 0 | 2 | 30 | 1 | 0 | 0 | 92 | |
| Specificity (%) | 100.0 | 98.9 | 76.0 | 99.9 | 100.0 | 100.0 | 47.4 | ||
Ruminant animal sources are indicated with an asterisk.
Number of individual fecal samples tested from a particular animal source.
The numbers of individual fecal sources that yielded a false-positive result are indicated. −, all test samples were negative.
Samples from ruminant animals were omitted (SO) from the CF128 and CF193 data sets because these assays were originally reported to target all ruminant animal sources.
FIG. 1.
Simple scatter plot indicating the estimated mean log10 target number of BoBac genetic marker concentrations in 1 ng of total DNA from respective fecal DNA extracts from reference animal sources. Black indicates a bovine source. White and gray denote ruminant and nonruminant animal sources, respectively. Mean estimated log10 target copy number values of zero indicate no amplification from the respective animal source. Error bars represent the standard deviation of the posterior distribution of the estimated mean log10 target copy number per nanogram of total DNA.
The prevalence of each PCR assay DNA target was estimated by testing DNA extracts from 247 individual bovine fecal samples and calculating a frequency of detection (Table 4). Prevalence ranged from 54% to 85% when samples from all bovine populations were included.
TABLE 4.
Prevalence of PCR assay genetic markers
| Assay | Overall prevalence (%)a | Prevalence (%) by herd no. |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
| CF128 | 85 | 96 | 92 | 70 | 100 | 33 | 100 | 97 | 100 | 100 | 100 | 100 |
| CF193 | 68 | 76 | 0 | 0 | 100 | 10 | 100 | 97 | 100 | 100 | 100 | 100 |
| Bac2 | 54 | 0 | 0 | 0 | 100 | 0 | 90 | 80 | 87 | 100 | 100 | 87 |
| Bac3 | 69 | 16 | 80 | 0 | 100 | 0 | 90 | 100 | 100 | 100 | 100 | 100 |
Prevalence value for all DNA extracts from 247 individual bovine fecal samples.
PCR sensitivity.
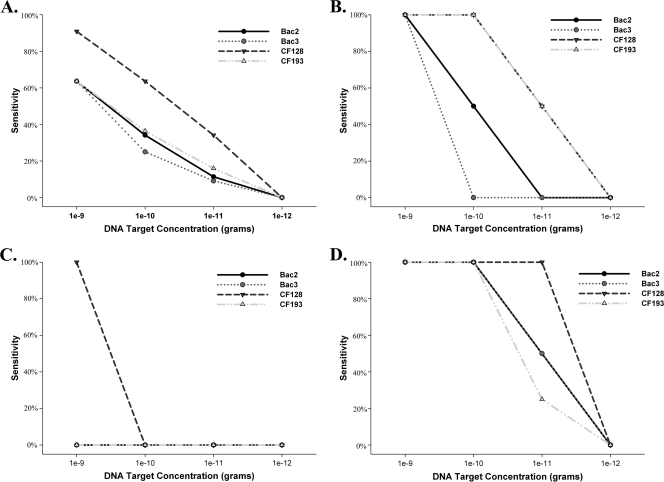
The sensitivity of each PCR assay (CF128, CF193, Bac2, and Bac3) was determined at four different total DNA concentrations. Sensitivity for each total DNA concentration was then plotted to compare sensitivity trends of each PCR assay for each bovine population and across all populations (Fig. 2). All PCR assays indicated a 0% sensitivity score at a concentration of 1 × 10−12 g of total DNA, regardless of the bovine population.
FIG. 2.
Simple straight-line and scatter plots indicating the sensitivity of host-associated PCR assays at four different concentrations ranging from 1 × 10−9 to 1 × 10−12 g of total DNA. Panel A represents the overall sensitivity of each PCR assay including fecal samples from all bovine feeding management groups. Panels B, C, and D indicate sensitivity for herd 8, herd 5, and herd 9 populations, respectively.
Abundance of fecal bacterial genes in bovine populations.
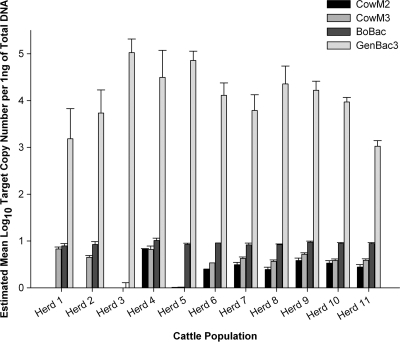
Individual bovine fecal samples were randomly collected from 11 populations and analyzed to estimate the abundance and variability of qPCR target DNA concentrations within and between populations (Fig. 3). A comparison of target DNA relative abundances between all assays was achieved by normalizing data sets to 1 ng of total DNA. The log10 mean copy number of target DNA was measured using 5 ng of total DNA for each bovine-specific assay, and 2 ng of total DNA was used for the GenBac3 qPCR assay. Within-population variance (σ2) of bovine-associated genetic markers was estimated based on CT measurements from individual bovine fecal DNA extracts from each population and fluctuated from 0.22 to 23.4 (BoBac), 0.61 to 7.25 (CowM2), and 0.65 to 6.78 (CowM3). GenBac3 variances ranged from 0.10 to 2.99. Pairwise comparisons of CT measurements from bovine populations indicated significant differences in genetic marker abundance (P < 0.05) for 13 to 37 pairings for the different qPCR assays. Only two bovine population pairings, those of herd 11 with herd 6 and of herd 11 with herd 10, were similar (P > 0.81) for all qPCR assays.
FIG. 3.
Quantification of bacterial genes from each bovine population. Mean log10 target copy number estimates are normalized per 1 ng of total DNA from respective fecal DNA extracts to compare relative abundance. Error bars represent the standard deviation of the posterior distribution of the estimated mean log10 target copy number per nanogram of total DNA.
Experiment controls.
IAC detection levels from CowM2 (expected mean CT of 34.6 ± 1.0) and Entero1 (expected mean CT of 33.8 ± 1.0) multiplex qPCR assays indicated the absence of PCR inhibitors in all DNA preparations. No-template controls indicated the absence of extraneous DNA molecules in 99% (7 false positives of 992 reactions) of PCR and qPCR experiments in this study. All extraction blank controls tested negative.
DISCUSSION
Assay specificity.
The ability to discriminate between bovine and other sources of fecal pollution is central to the concept of an MST approach. Incomplete host specificity can be a significant source of error and can confound even the most well-designed MST studies. The specificity of each host-associated assay was estimated using a broad range of animals including humans; agriculturally important animals such as swine, poultry, horse, and sheep; human companion animals (dogs and cats); and wildlife. Twelve different sources of ruminant animals were also included to characterize the degree of cross-reactivity for each assay to animals that share similar digestive physiologies. Experiments indicate a broad range of specificity values (Table 3). The broad range of observed specificities raises an important question: What level of specificity is acceptable? A recent study reporting a stochastic model that can simulate different MST application scenarios, including the use of nonspecific assays, suggests that any level of nonspecific detection can be overcome as long the frequency of the genetic marker between host populations is accurately measured (12). In principal, a correction method may suffice, but in practice it may prove too difficult to accurately characterize genetic marker occurrence among all host animals. Characterization would be especially challenging for genetic markers associated with multiple hosts such as the gene target for the BoBac qPCR assay, where more than half of all animals in a given watershed may harbor the genetic marker. Thus, specificity remains an important component in the selection of an assay for field study applications. Specificity data also suggest that genetic marker selection for novel MST assay development is paramount. CF128, CF193, and BoBac assays that target Bacteroidales 16S rRNA gene sequences exhibited variable specificity levels ranging from 47.4% to 99.9%, while Bac2, Bac3, CowM2, and CowM3 targeting genes predicted to be directly involved in host-bacteria interactions were consistently greater than 98.9%. This observation suggests that the high levels of DNA sequence conservation associated with 16S rRNA genes can be restrictive for animal-associated assays and that the search for alternative markers encoding proteins directly involved in host-specific processes is warranted.
It should be noted that the BoBac qPCR assay was originally reported as 93.3% specific for bovine feces based on DNA extracts from 15 reference samples collected from human (n = 3), pig (n = 4), horse (n = 4), and dog (n = 4), where a positive result was defined as an estimated DNA target concentration greater than 1 × 106 copies per gram of feces (10). It is unlikely that the 3°C increase in annealing temperature (60°C instead of 57°C) is responsible for these differences because a higher temperature should increase primer hybridization stringency, leading to better specificity values. Instead, the larger number of reference fecal samples (175 compared to 15), the inclusion of other ruminant samples (50 compared to 0), and the absence of a threshold of 1 × 106 copies for a positive result are more than likely responsible for the differences observed in specificity. In addition, a TAMRA quenching molecule was used instead of black hole quencher 1 (BHQ1) on the BoBac 402f probe (Table 1). Previous studies on TAMRA and BHQ1 quenchers suggest that these molecules share similar fluorescence resonance energy transfer quenching efficiencies (16) and that assay specificity is not altered by the use of either quencher (29). It is also evident that the use of a TAMRA quencher allowed the quantification of DNA targets ranging from 10 to 1 × 106 copies with a high level of precision (Table 2).
Performance of bovine-associated PCR and qPCR assays across 11 bovine populations.
Little is known regarding the performance of host-associated PCR and qPCR assays between different bovine populations. Useful host-associated genetic markers should ideally be uniformly distributed within and between populations. Analyses of fecal samples collected from 11 bovine populations were performed to characterize the sensitivity of each PCR assay and the prevalence of the respective DNA target and to compare the abundance of qPCR assay genetic markers relative to Bacteroidales 16S rRNA gene concentrations (Fig. 3). The general Bacteroidales assay (GenBac3) yielded the highest gene concentrations in all samples, which supports previous research reporting that members of the Bacteroidales make up a large portion of the bovine fecal bacterial community (28). The relative abundance of host-associated genetic markers followed the general trend of BoBac > CowM3 > CowM2, indicating that the less specific BoBac rRNA gene targets are more abundant than the CowM2 and CowM3 gene targets (Fig. 3).
Pairwise comparisons of host-associated and Bacteroidales CT measurements and within-population variance estimates indicated that there are minimal differences within almost all populations but significant differences between most populations tested. Pairwise comparisons indicated only three populations (herd 11, herd 6, and herd 10) where genetic markers were not significantly different (P > 0.84). These three populations are all from bovine dairy operations situated in the same geographic region with rations consisting of 70% or more forage. The pairing of these bovine populations may have occurred by chance, or similarities in geographic location and/or management practices may be responsible. qPCR abundance experiments also indicated relatively high levels of genetic markers in some populations and the complete absence in others. For example, the CowM2 qPCR genetic marker was present in seven populations but absent in herd 1, herd 2, herd 5, and herd 3 fecal samples (Fig. 3). This same trend was repeated for PCR assay sensitivity experiments (Fig. 2) and prevalence estimates, where frequencies ranged from 0% to 100% for different populations (Table 4). Prevalence frequencies for some populations are in strong disagreement from previously reported trends with the CF128, CF193, Bac2, and Bac3 assays (100%, 100%, 80%, and 91%, respectively [1, 23]). Differences between previously reported and observed values in this study suggest that more extensive testing of the performance of these assays across multiple bovine populations is needed before widespread implementation. It remains unclear what factors are responsible for these dramatic shifts in PCR and qPCR assay performance. However, data generated from the 11 populations tested in this study suggest that it is unlikely that the observed differences between populations can be attributed to animal health or age as fecal samples were collected from healthy adult animals directly from the rectum or immediately after a bowel movement. It is also unlikely that differences in geographic location are solely accountable as herd 4 and herd 5 populations are from the same facility and exhibit dramatic shifts in genetic marker prevalence (Table 4) and abundance (Fig. 3). An alternative hypothesis could be that the observed differences between bovine populations may be the result of different management practices such as the amount of forage and grain rations fed to the animals.
State of the science and implications for future research.
Our experiments suggest that some assays are more suitable than others for the characterization of fecal contamination originating from a particular bovine population. CF128 and BoBac are the most sensitive and geographically robust assays. They also amplify the most prevalent and abundant genetic markers, but both of these assays exhibit the lowest specificity levels, 76% and 47.4%, respectively. Bac2, Bac3, CowM2, and CowM3 are more than 98.9% specific but appear to exhibit lower sensitivity and genetic marker abundance in some bovine populations. Results of this study have several implications for the design of future MST case studies. The variability of genetic marker abundance and prevalence between populations suggests that the use of bovine MST applications will require a priori characterization of assay performance at each watershed of interest. The same measures may also be necessary for MST assays designed to identify other relevant agriculture animal sources such as swine and poultry where management practices can vary significantly from one facility to the next. It is important to note that the bovine populations sampled in this study represent only a small number of the possible geographic locales and management practices employed by the bovine industry. Depending on the animal facility, bovines may be exposed to various feeding regimes, antibiotics, and supplements to increase weight for beef production or milk for dairy applications. Experiments also provide support for new directions in host-associated genetic marker identification and development of novel MST assays. It is interesting that all assays evaluated in this study were originally developed from reference bovine fecal samples collected from dairy farm operations. It is reasonable to assume that experiments designed to develop novel assays for the detection of fecal bacteria from different types of bovine populations could achieve similar levels of performance.
Acknowledgments
We are grateful to Stephanie Harris, Shawn Archibeque, Elaine Berry, and Dawn King for fecal sample collection efforts.
The U.S. Environmental Protection Agency, through its Office of Research and Development, funded and managed, or partially funded and collaborated in, the research described herein.
This paper has been subjected to the Agency's peer and administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the author(s) and do not necessarily reflect the views of the Agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print on 8 January 2010.
REFERENCES
- 1.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, M., and M. J. Wolin. 1979. Effect of monensin and lasalocid-sodium on the growth of methanogenic and rumen saccharolytic bacteria. Appl. Environ. Microbiol. 38:72-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jimenez-Clavero, M., E. Escribano-Romero, C. Mansilla, N. Gomez, L. Cordoba, N. Roblas, F. Ponz, V. Ley, and J. Saiz. 2005. Survey of bovine enterovirus in biological and environmental samples by a highly sensitive real-time reverse transcription-PCR. Appl. Environ. Microbiol. 71:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kildare, B. J., C. M. Leutenegger, B. S. McSwain, D. G. Bambic, V. B. Rajal, and S. Wuertz. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 41:3701-3715. [DOI] [PubMed] [Google Scholar]
- 7.Kirs, M., and D. C. Smith. 2007. Multiplex quantitative real-time reverse transcriptase PCR for F+-specific RNA coliphages: a method for use in microbial source tracking. Appl. Environ. Microbiol. 73:808-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klieve, A. V., D. Hennessy, D. Ouwerkerk, R. J. Forster, R. I. Mackie, and G. T. Attwood. 2003. Establishing populations of Megasphaera elsdenni YE 34 Butyrivibrio fibrisolvens YE 44 in the rumen of bovine fed high grain diets. J. Appl. Microbiol. 95:3248-3258. [DOI] [PubMed] [Google Scholar]
- 10.Lamendella, R., J. W. Santo Domingo, D. B. Oerther, J. R. Vogel, and D. M. Stoeckel. 2006. Assessment of fecal pollution sources in a small northern-plains watershed using PCR and phylogenetic analysis of Bacteroidetes 16S rRNA gene. FEMS Microbiol. Ecol. 59:651-660. [DOI] [PubMed] [Google Scholar]
- 11.Layton, A., L. McKay, D. Williams, V. Garrett, R. Gentry, and G. Sayler. 2006. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 72:4214-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leach, M. D., S. L. Broschat, and D. R. Call. 2008. A discrete, stochastic model and correction method for bacterial source tracking. Environ. Sci. Technol. 42:524-529. [DOI] [PubMed] [Google Scholar]
- 13.Lee, Y., M. Molina, J. W. Santo Domingo, J. D. Willis, M. Cysterski, D. M. Endale, and O. C. Shanks. 2008. Temporal assessment of the impact of exposure to cow feces in two watersheds by multiple host-specific PCR assays. Appl. Environ. Microbiol. 74:6839-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ley, V., J. Higgins, and R. Fayer. 2002. Bovine enteroviruses as indicators of fecal contamination. Appl. Environ. Microbiol. 68:3455-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lunn, D. J., A. Thomas, N. Best, and D. Spiegelhalter. 2000. WinBUGS-A Bayesian modeling framework: concepts, structure, and extensibility. Stat. Comput. 10:325-337. [Google Scholar]
- 16.Marras, S. A. E., F. R. Kramer, and S. Tyagi. 2002. Efficiencies of fluorescence resonance energy transfer and contact-mediated quenching in oligonucleotide probes. Nucleic Acids Res. 30:e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martellini, A., P. Payment, and R. Villemur. 2005. Use of eukaryotic mitochondrial DNA to differentiate human, bovine, porcine and ovine sources in fecally contaminated surface water. Water Res. 39:541-548. [DOI] [PubMed] [Google Scholar]
- 18.Okabe, S., N. Okayama, O. Savichtcheva, and T. Ito. 2007. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 74:890-901. [DOI] [PubMed] [Google Scholar]
- 19.SAS Institute, Inc. 1990. SAS/STAT user's guide, version 6, 4th ed. SAS Institute Inc., Cary, NC.
- 20.Seifring, S. C., M. Varma, E. Atikovic, L. J. Wymer, and R. A. Haugland. 2008. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J. Water Health 6:225-237. [DOI] [PubMed] [Google Scholar]
- 21.Shanks, O. C., E. Atikovic, A. D. Blackwood, J. Lu, R. T. Noble, J. Santo Domingo, S. Siefring, M. Sivaganesan, and R. P. Haugland. 2008. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution. Appl. Environ. Microbiol. 74:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shanks, O. C., C. Nietch, M. T. Simonich, M. Younger, D. Reynolds, and K. G. Field. 2006. Basin-wide analysis of the dynamics of fecal contamination and fecal source identification in Tillamook Bay, Oregon. Appl. Environ. Microbiol. 72:5537-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shanks, O. C., J. W. Santo Domingo, R. Lamendella, C. A. Kelty, and J. E. Graham. 2006. Competitive metagenomic DNA hybridization identifies host-specific microbial genetic markers in cow fecal samples. Appl. Environ. Microbiol. 72:4054-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shanks, O. C., J. W. Santo Domingo, J. Lu, C. A. Kelty, and J. E. Graham. 2007. Identification of bacterial DNA markers for the detection of human fecal pollution in water. Appl. Environ. Microbiol. 73:2416-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sivaganesan, M., S. Seifring, M. Varma, R. A. Haugland, and O. C. Shanks. 2008. A Bayesian method for calculating real-time quantitative PCR calibration curves using absolute plasmid DNA standards. BMC Bioinformatics 9:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slyter, L. L. 1976. Influence of acidosis on rumen function. J. Anim. Sci. 43:910-929. [DOI] [PubMed] [Google Scholar]
- 27.Walters, S. P., V. P. G. Gannon, and K. G. Field. 2007. Detection of Bacteroidales fecal indicators and the zoonotic pathogens E. coli O157:H7, Salmonella, and Campylobacter in river water. Environ. Sci. Technol. 41:1856-1862. [DOI] [PubMed] [Google Scholar]
- 28.Wood, J., K. P. Scott, G. Avgustin, C. J. Newbold, and H. J. Flint. 1998. Estimation of the relative abundance of different Bacteroides and Prevotella ribotypes in gut samples by restriction enzyme profiling of PCR-amplified 16S rRNA gene sequences. Appl. Environ. Microbiol. 64:3683-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang, G. P., D. D. Erdman, M. L. Tondella, and B. S. Fields. 2009. Evaluation of tetramethylrhodamine and black hole quencher 1 labeled probes and five commercial amplification mixes in TaqMan real-time RT-PCR assays for respiratory pathogens. J. Virol. Methods 162:288-290. [DOI] [PubMed] [Google Scholar]