Abstract
Although branched-chain amino acids are synthesized as building blocks of proteins, we found that the fungus Aspergillus nidulans excretes them into the culture medium under hypoxia. The transcription of predicted genes for synthesizing branched-chain amino acids was upregulated by hypoxia. A knockout strain of the gene encoding the large subunit of acetohydroxy acid synthase (AHAS), which catalyzes the initial reaction of the synthesis, required branched-chain amino acids for growth and excreted very little of them. Pyruvate, a substrate for AHAS, increased the amount of hypoxic excretion in the wild-type strain. These results indicated that the fungus responds to hypoxia by synthesizing branched-chain amino acids via a de novo mechanism. We also found that the small subunit of AHAS regulated hypoxic branched-chain amino acid production as well as cellular AHAS activity. The AHAS knockout resulted in higher ratios of NADH/NAD+ and NADPH/NADP+ under hypoxia, indicating that the branched-chain amino acid synthesis contributed to NAD+ and NADP+ regeneration. The production of branched-chain amino acids and the hypoxic induction of involved genes were partly repressed in the presence of glucose, where cells produced ethanol and lactate and increased levels of lactate dehydrogenase activity. These indicated that hypoxic branched-chain amino acid synthesis is a unique alternative mechanism that functions in the absence of glucose-to-ethanol/lactate fermentation and oxygen respiration.
Hypoxia (oxygen [O2] depletion) imposes a challenge to most eukaryotes, since O2 serves as a substrate for the biosynthesis of some essential compounds such as sterols, unsaturated fatty acids, and heme. For most eukaryotes, O2 respiration is an indispensable mitochondrial mechanism that conserves the energy required for growth. Eukaryotic cells exposed to hypoxia induce adaptation mechanisms to ensure survival. Such mechanisms include a metabolic shift from oxidative phosphorylation to fermentation, a decrease in translation and cell growth, and cell death (34). Transcription factors, such as hypoxia-inducible factor (HIF) in higher eukaryotes and Hap1p and Rox1p in Saccharomyces cerevisiae (6, 8, 9, 33), affect the expression of hypoxic genes that participate in these processes (8, 9, 33).
Eukaryotic microbes inhabit various concentrations of O2. Transient flooding often causes O2 depletion in the soil environment where soil fungi reside. Pathogenic fungi proliferate in animal tissues or cells where the O2 concentration is lower than the atmospheric condition (12). Hypoxic responses and adaptation of the budding yeast S. cerevisiae are the best characterized among eukaryotic microbes. Upon hypoxia, the yeast alters global transcription (10, 32, 39) and produces cellular proteins for energy conservation and for the biosynthesis of cellular components (2, 10). For example, S. cerevisiae downregulates the genes for respiratory complexes and the tricarboxylic acid cycle in response to hypoxia and consequently acquires most energy for growth by alcohol fermentation. The expression of some ergosterol- and heme-biosynthetic genes is upregulated for maximal O2 incorporation into these molecules (21, 40). Similar regulation has been identified in Schizosaccharomyces pombe and Candida albicans (18, 39, 47). Aspergillus nidulans, which belongs to the same phylum (Ascomycota) as these yeasts, uses nitrate as an alternative electron acceptor to O2 under hypoxic conditions. This reaction proceeds using niaD and niiA, which encode nitrate and nitrite reductases (14, 37, 38, 45, 46). Global protein production by the fungus is altered when the fungus is cultured under hypoxic conditions (36).
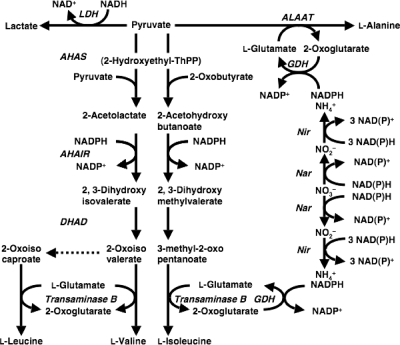
Branched-chain amino acids are essential nutrients, and their metabolic disorders can cause acidosis (4). Both eukaryotic and prokaryotic microbes metabolize them as carbon, nitrogen, and energy sources. An evolutionarily conserved mechanism synthesizes branched-chain amino acids in microbes. For example, l-Val is synthesized from pyruvate in a pathway comprising four reactions catalyzed by acetohydroxy acid synthase (AHAS), acetohydroxy acid isomeroreductase (AHAIR), dihydroxy acid dehydratase (DHAD), and transaminase B (Fig. 1), which are encoded by the S. cerevisiae genes ILV2, ILV5, ILV3, and BAT1/2 (3, 11, 13, 15, 23, 27, 30, 42). These enzymes catalyze the biosynthesis of l-Ile from pyruvate and 2-oxobutyrate. An intermediate of l-Val synthesis, 2-oxovalerate, is a precursor for l-Leu synthesis. These mechanisms have been genetically manipulated to breed bacteria producing branched-chain amino acids, which are of industrial use in nutritional and health supplements and in chemical synthesis (1, 17). However, the mechanism of eukaryotic microbes, especially the hypoxic response of the mechanism, has been relatively less characterized.
FIG. 1.
Pathway for branched-chain amino acid synthesis. AHAS, acetohydroxy acid synthase; AHAIR, acetohydroxy acid isomeroreductase; ALAAT, alanine aminotransferase; DHAD, dihydroxy acid dehydratase, GDH, glutamate dehydrogenase; Nar, nitrate reductase; Nir, nitrite reductase; ThPP, thiamine pyrophosphate.
The primary role of branched-chain amino acids is to serve as building blocks for cellular proteins. However, we found during the course of our studies of fungal metabolism under hypoxic conditions that hypoxically incubated A. nidulans cells do not incorporate a portion of de novo synthesized branched-chain amino acids into proteins but rather excrete them into the culture medium. This suggests a role for branched-chain amino acids and/or their biosynthetic mechanisms other than simply as building blocks for proteins. Here, we investigated the physiological significance of hypoxic branched-chain amino acid production by A. nidulans and investigated expression of the mechanism and properties of knockout strains of AHAS-encoding genes in this fungus. The physiological significance of branched-chain amino acid production in reoxidizing NAD(P)H to generate NAD(P)+ under hypoxic conditions is also discussed.
MATERIALS AND METHODS
Strains, culture, and media.
Aspergillus nidulans strains A26 (biA1) and A89 (biA1 argB2) were obtained from the Fungal Genetic Stock Center (University of Kansas Medical Center). Conidia (108) were transferred to 500-ml Erlenmeyer flasks containing 100 ml of GMM (1% glucose, 0.6% NaNO3, 10 mM KH2PO4, 7 mM KCl, 2 mM MgSO4, 0.2% Hutner's trace metals [vol/vol] [37]) and normoxically incubated at 30°C for 16 h at 120 rpm (preculture). For preculturing strain DAHAS-L, valine, leucine, and isoleucine (10 mM each) were added to the medium. Resultant mycelia (0.1 g [wet weight]) were collected by centrifugation, washed twice with 0.85% NaCl, and then inoculated into 500-ml Erlenmeyer flasks containing 100 ml of MMEN or MMDN (100 mM ethanol or 1% glucose, 10 mM NaNO3, 10 mM KH2PO4, 7 mM KCl, 2 mM MgSO4, 0.2% Hutner's trace metals [vol/vol]). Biotin (0.2 μg liter−1) was added to all media. The headspace in the flasks was replaced with nitrogen gas by purging the air for 15 min, and then the flasks were sealed with butyl rubber stoppers and rotated at 30°C at 120 rpm to maintain hypoxic conditions. Normoxic conditions were maintained by agitating 100 ml of MMEN or MMDN in flasks sealed with cotton plugs but without replacing the headspace air. Arginine (0.2 mg liter−1) was added when culturing the strain under arginine auxotrophy.
Determination of organic and amino acids.
Mycelia were separated from the medium by filtration, washed five times with chilled water, and then lyophilized. Culture filtrate (10 ml) was immediately frozen in liquid nitrogen and lyophilized. Freeze-dried mycelia (100 mg) were transferred into liquid nitrogen, ground into a fine powder, and then suspended in 3 ml of methanol/water (50:50, vol/vol). After incubation at room temperature for 16 h, the extract was centrifuged at 11,000 × g for 5 min at room temperature, and then the supernatant was transferred to 15-ml tubes. The culture filtrate and the cell extracts were lyophilized, methylated using methylchlorocarbonate as described previously (41), and analyzed by gas chromatography-electron impact mass spectrometry using a gas chromatograph-mass spectrometer (QP2010; Shimadzu, Kyoto, Japan) equipped with a capillary column (DB-5, 30 m by 0.32 mm; J & W Scientific, CA). The injection and ion-source temperatures were 250°C and 210°C, respectively. Helium gas (1 ml min−1) was utilized as a carrier. The oven temperature was controlled at 80°C for 2 min and then increased to 330°C at 8°C min−1 and maintained for 10 min. Metabolites were identified by their retention times and mass fragments. Thereafter, p-hydroxybenzoic acid was added as an internal control. Peak areas of total ion currents were integrated and used to quantify those of authentic compounds.
Disruption of the AHAS and the GDH genes.
Plasmids for disrupting the genes for AHAS-L, AHAS-S, and GDH were constructed by inserting DNA fragments encoding the 5′ and 3′ regions of each gene into the up- and downstream regions of the argB gene cloned within the SmaI site of pBluescript KS+ (pBSarg1) (37), respectively. The DNA fragments encoding the 5′ regions fused with appropriate restriction sites were amplified using various primers (see Table S1 in the supplemental material), digested with restriction enzymes (Table S1), and ligated with pBSarg1 that had already been spliced with the same restriction enzymes. The 3′ region of each gene was amplified using various primers (Table S1), digested with restriction enzymes (Table S1), and inserted into the same restriction sites of the resulting plasmid to generate pDAHASL, pDAHASS, and pDGDH.
A. nidulans strain A89 was transformed using the plasmids as described previously (37). Total fungal DNA prepared as described by Takasaki et al. (37) was Southern blotted using a DIG DNA labeling and detection kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions.
Quantitative PCR.
After incubating for 6 and 12 h under normoxic or hypoxic conditions, total RNA was extracted from the mycelia and applied (2.0 μg) to first-strand cDNA synthesis as described previously (35, 36). Real-time quantitative PCR proceeded using a MiniOpticon version 3.1 (Bio-Rad Laboratories Inc., CA). Gene-specific primers were designed based on sequence data (http://www.broad.mit.edu/annotation/genome/aspergillus_nidulans) so that the lengths of the PCR products ranged between 160 and 190 bp (see Table S1 in the supplemental material). Real-time PCR proceeded in a final volume of 50 μl. The SYBR Premix Ex Taq (Bio-Rad) was applied according to the manufacturer's instructions, and then PCR proceeded as follows: initial denaturation at 94°C for 1 min, followed by 40 cycles of denaturation at 94°C for 10 s, annealing at 62°C for 30 s, and elongation at 72°C for 20 s. Specific amplification was confirmed by analyzing melting curves from 65°C to 95°C. No fluorescence was increased in a control experiment without a template.
Enzyme assays.
Fungal cells were collected by filtration, washed twice with 0.7% NaCl, suspended in buffer A (20 mM potassium phosphate [pH 7.2], 10% glycerol, 0.3 mM N-tosyl-l-phenylalanine, 0.3 mM phenylmethylsulfonyl fluoride), and homogenized as described previously (34). Cellular debris was sedimented by centrifugation at 1,500 × g for 10 min, and then the supernatant was separated by centrifugation at 10,000 × g for 15 min followed by 100,000 × g for 60 min. After a centrifugation at 100,000 × g for 60 min, cytosolic protein extracts are produced.
The activity of AHAS was measured by the method of Leyval et al. (25) with the following modification. The reaction mixture contained 200 mM potassium phosphate (pH 7.4), 50 mM sodium pyruvate, 10 mM MgCl2, 0.1 mM thiamine pyrophosphate, and 0.1 mM flavin adenine dinucleotide (FAD), and the reaction was initiated by adding cell extract. After incubation at 37°C for 20 min, the reaction was acidified with 0.1 ml of 50% H2SO4 and incubated at 37°C for 25 min to form acetoin from α-acetolactate. The amount of acetoin formed was determined using the Voges-Proskauer method (44) by measuring A535. Transaminase B was assayed in a reaction mixture (200 μl) containing 100 mM Tris-HCl (pH 9.0), 0.25 mM pyridoxal-5′-phosphate, 5 mM 2-oxoisovalerate, 10 mM potassium glutamate, and cell extract. After incubation at 30°C for 30 min, the reaction was terminated with 60 μl of 21% perchloric acid, and the mixture was centrifuged at 15,000 × g for 5 min at 4°C. The supernatant was neutralized by adding trace amounts of 5 M KOH and centrifuged at 15,000 × g for 5 min at 4°C, and then l-Val in the supernatants was quantified by high-performance liquid chromatography (HPLC).
The activity of GDH was assayed in reaction mixtures containing 100 mM potassium phosphate (pH 7.2), 10% glycerol, 5 mM ammonium chloride, 5 mM 2-oxoglutarate, and 0.2 mM NADPH. The activity of LDH was assayed in reaction mixtures containing 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid-KOH (pH 7.5), 5 mM sodium pyruvate, and 0.2 mM NADPH. These reactions were initiated by adding cell extract, and then A340 was monitored at 25°C using a Beckman DU-7500 spectrophotometer.
The activity of NAD(P)+ transhydrogenase was measured by the method of Kaplan (20). The reaction mixture contained 50 mM sodium phosphate buffer (pH 7.0), 1 mM KCN, 1 mM dithiothreitol, 1 mM EDTA, 0.4 mM 3-acetylpyridine-NAD+, and 0.4 mM NADPH. The reduction of 3-acetylpyridine-NAD+ by NADPH was measured by determining the increase in A375. An extinction coefficient of 5.1 mM−1cm−1 was used to calculate specific activity.
Determination of the intracellular pyridine nucleotides.
Mycelia (20 mg [wet weight]) were washed twice with chilled water, immediately transferred into liquid nitrogen, ground into a fine powder, and then suspended with 400 μl of extraction buffer supplied in the NAD(P)+ and NAD(P)H quantitation kit (Bio Version, Mountain View, CA). Pyridine nucleotides were quantified using the same kit according to the manufacturer's instructions.
Other methods.
Ammonium, nitrate, glucose, lactate, ethanol, and O2 were quantified as described previously (45, 46). The protein concentrations were determined using a kit (Bio-Rad) according to the manufacturer's instructions. Proteins resolved by two-dimensional polyacrylamide gel electrophoresis were identified as described in the supplemental material.
RESULTS
Hypoxic production of amino acids in culture medium.
We investigated metabolic changes in A. nidulans FGSC A26 (wild type [WT]) cultured under normoxic and hypoxic conditions and measured amino acids and organic acids in the culture media. Ethanol served as a carbon and energy source, and nitrate was the nitrogen source. The O2 concentration was maintained at <2 μM during hypoxic culture. After a 12-h incubation the fungus produced malonate, maleate, and succinate in the medium under normoxic conditions (Table 1). Culture under hypoxia generated smaller amounts of these organic acids but considerable amounts of lactate. The nine amino acids shown in Table 1 were undetectable in normoxic culture media, whereas 2.1 to 61 μmol of them was detectable in hypoxic media. Intracellular levels of these amino acids were also somewhat higher in hypoxic cells. These results indicated that hypoxic cells produced more of these amino acids. The most abundantly produced amino acids were Ala and Glu (Table 1), which are also produced by other eukaryotes in a hypoxic environment (28, 29). Smaller quantities of branched-chain amino acids (Val, Leu, and Ile) than Ala and Glu accumulated, but the amounts were still fairly large (10 to 15 μmol).
TABLE 1.
Intracellular and extracellular metabolites from A. nidulans cultured under normoxic and hypoxic conditionsa
| Aeration | Metabolites (μmol) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organic acids |
Amino acids |
||||||||||||||
| Lactate | Malonate | Maleate | Succinate | Citrate | Malate | Ala | Glu | Val | Leu | Ile | Thr | Pro | Asp | Phe | |
| Extracellular fraction | |||||||||||||||
| Normoxia | <0.1 | 28 | 10 | 11 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 |
| Hypoxia | 19 | 7.4 | <0.1 | <0.1 | <0.1 | <0.1 | 61 | 34 | 10 | 18 | 15 | 5.2 | 6.4 | 2.3 | 2.1 |
| Intracellular fraction | |||||||||||||||
| Normoxia | <0.1 | 5.6 | 3.6 | 4.3 | 5.2 | 7.5 | 34 | 40 | 5.3 | 8.1 | 7.9 | 2.6 | 3.6 | 2.6 | 11 |
| Hypoxia | 2.3 | 6.1 | 3.2 | 4.1 | 6.2 | 9.4 | 43 | 47 | 7.0 | 11 | 9.2 | 3.0 | 4.4 | 2.9 | 10 |
Mycelia (1 g wet weight) were incubated in 500-ml Erlenmeyer flasks containing 100 ml of MMEN medium using ethanol as the sole carbon source for 12 h at 30°C under normoxic and hypoxic conditions, and intracellular and extracellular metabolites were measured. Pyruvate was undetectable. Data are means of results of three experiments. Standard errors were <20%.
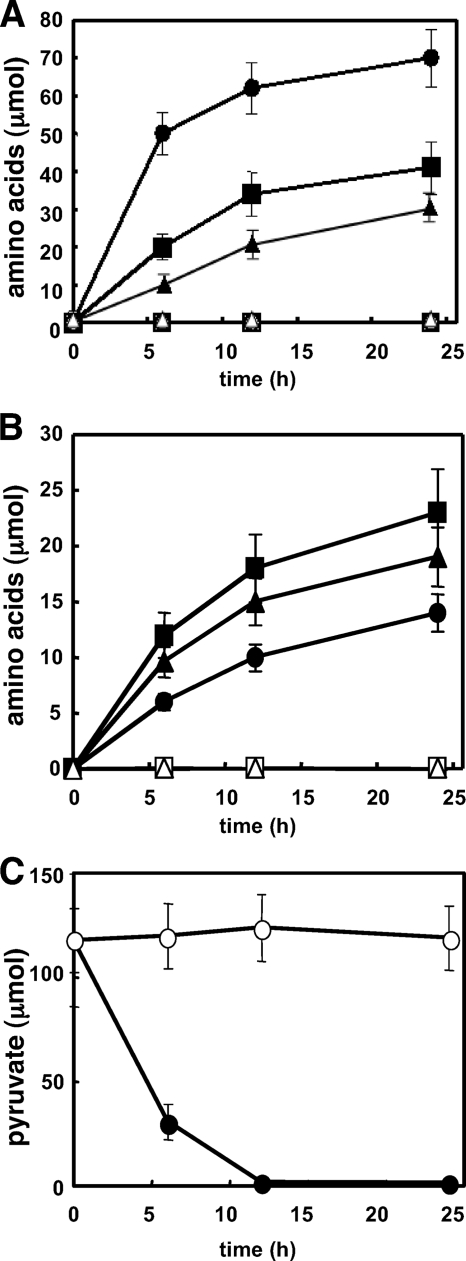
We investigated the time-dependent production of Ala, Glu, lactate, and the branched-chain amino acids by A. nidulans (Fig. 2A and B). Figure 2B shows that the fungus accumulated Val, Leu, and Ile in culture medium under hypoxic incubation conditions. Intracellular pyruvate was detected in mycelia that were precultured in normoxic medium containing glucose (Fig. 2C, time zero). After hypoxic incubation for 12 h, the intracellular pyruvate completely disappeared (Fig. 2C). Pyruvate is a direct precursor of the branched-chain amino acids (Fig. 2), Ala, and lactate. The calculated sum of the produced Val (10 μmol), Leu (18 μmol), Ile (15 μmol), Ala (68 μmol), and lactate (19 μmol) was equivalent to 156 μmol of pyruvate considering 1 mol of pyruvate generates 1 mol of each of these compounds, except for Val and Leu, which are produced from two pyruvate molecules (Fig. 1). The value after a 24-h incubation was much higher (189 μmol) and more than the amounts of consumed pyruvate (120 μmol), indicating that the branched-chain amino acids, Ala, and lactate were not only produced from intracellular pyruvate but also synthesized de novo from the carbon source.
FIG. 2.
Metabolites in culture fluid of A. nidulans grown under normoxic and hypoxic conditions. A. nidulans WT mycelia (1 g [wet weight]) precultured in MMDN medium were transferred to 500-ml Erlenmeyer flasks containing 100 ml of MMEN medium using ethanol as the sole carbon source and incubated at 30°C under normoxic (open) and hypoxic (closed) conditions. Amino acids in the medium: (A) Ala (circles), Glu (squares), and lactate (triangles); (B) Val (circles), Leu (squares), and Ile (triangles). (C) Intracellular pyruvate cultured under normoxic (open) and hypoxic (closed) conditions. Pyruvate levels in the medium were very low. Data are means of results of three experiments. The standard deviation was <20%.
A. nidulans mechanism of de novo branched-chain amino acid synthesis.
Although little has been described about the de novo biosynthetic pathways of branched-chain amino acids in A. nidulans, we searched the fungal genome to identify highly conserved genes among filamentous fungi that are similar to the genes involved in branched-chain amino acid synthesis in S. cerevisiae (3, 11, 13, 15, 23, 27, 30, 42). We found genes for the large subunit of acetohydroxy acid synthase (AHAS-L, AN4956.3), the small subunit of acetohydroxy acid synthase (AHAS-S, AN4430.3), and acetohydroxy acid isomeroreductase (AHAIR, AN2526.3), with deduced amino acid sequences of 77%, 77%, and 89% similarity, respectively, to their yeast orthologs. The deduced amino acid sequence of AHAS-L was 82% similar to that of AHAS-L from Neurospora crassa encoded by ilv-3 (5). No other genes with more than 40% similarity to each gene product were found in the A. nidulans genome. The third step in synthesizing branched-chain amino acids is catalyzed by dihydroxy acid dehydratase (DHAD). Four predicted genes (AN6346.3, AN4058.3, AN5138.3, and AN7358.3) for DHAD were found in the genome database, of which the deduced amino acid sequences are 76%, 68%, 50%, and 46% similar, respectively, to that of ILV-3, the yeast ortholog of the DHAD gene. The last step is catalyzed by transaminase B, which is involved in the aminotransferase IV family. Six predicted genes (AN4323.3, AN7878.3, AN5957.3, AN0385.3, AN7876.3, and AN8511.3) for transaminase B were identified in the fungal genome, with deduced amino acid sequences that were 74%, 67%, 56%, 46%, 44%, and 40% similar, respectively, to that of BAT-2, the yeast ortholog of the transaminase B gene. These genes might constitute a biosynthetic pathway of branched-chain amino acids in A. nidulans.
Regulation of the genes for branched-chain amino acid synthesis.
Transcripts of the predicted genes for the branched-chain amino acid biosynthesis were quantified using PCR. Culture under hypoxic conditions for 6 h resulted in 1.4- to 6.0-fold more AHAS-L, AHAS-S, AHAIR, and DHAD1, -3, and -4 transcripts than those under normoxic conditions (Table 2). A longer incubation (12 h) increased the ratio of the transcripts, indicating that the expression of these genes was upregulated by hypoxia. The exception was that of the DHAD3 gene (AN5138.3), which was slightly decreased from 6.0 to 5.3. Among the six paralogs of transaminase B genes, the expression of the genes of isozymes 2 to 5 (AN7878.3, AN5957.3, AN0385.3, and AN7876) was increased from 3.1- to 6.0-fold after culture for 12 h under hypoxic conditions, suggesting that these gene products are involved in the production of branched-chain amino acids under hypoxic conditions.
TABLE 2.
Expression of genes involved in branched-chain amino acid and glutamate biosynthesisa
| Predicted gene product | Gene IDb | Relative expression ratio |
||
|---|---|---|---|---|
| Ethanol |
Glucose (12 h) | |||
| 6 h | 12 h | |||
| Acetohydroxy acid synthase small subunit (AHASS) | AN4430.3 | 2.6 | 4.3 | 1.2 |
| Acetohydroxy acid synthase large subunit (AHASL) | AN4956.3 | 1.4 | 2.1 | 1.2 |
| Acetohydroxy acid isomeroreductase (AHAIR) | AN2526.3 | 3.9 | 4.4 | 2.2 |
| Dihydroxy acid dehydratase (DHAD1) | AN6346.3 | 2.4 | 2.2 | 1.5 |
| Dihydroxy acid dehydratase (DHAD2) | AN4058.3 | 1.1 | 1.3 | 1.2 |
| Dihydroxy acid dehydratase (DHAD3) | AN5138.3 | 6.0 | 5.3 | 1.1 |
| Dihydroxy acid dehydratase (DHAD4) | AN7358.3 | 1.4 | 1.6 | 2.0 |
| Transaminase B1 | AN4323.3 | 1.2 | 1.3 | 1.1 |
| Transaminase B2 | AN7878.3 | 2.3 | 5.2 | 1.0 |
| Transaminase B3 | AN5957.3 | 1.9 | 3.1 | 1.2 |
| Transaminase B4 | AN0385.3 | 13.7 | 6.0 | 1.1 |
| Transaminase B5 | AN7876.3 | 2.3 | 4.1 | 2.3 |
| Transaminase B6 | AN8511.3 | 1.1 | 1.1 | 1.0 |
| Glutamate dehydrogenase | AN4376.3 | 1.7 | 6.9 | 2.5 |
| NAD(P) transhydrogenase | AN8130.3 | 3.3 | 3.9 | 3.1 |
Transcripts were quantified by real-time PCR using total RNA prepared from A. nidulans cultured at 30°C for 6 and 12 h under normoxic and hypoxic conditions. Ethanol and glucose served as carbon sources. Data are normalized to the β-actin (actA) transcript and are shown as relative expression ratios (hypoxia versus normoxia). Data are means of results of three experiments. Standard deviations were <25%. P < 0.05.
Gene IDs are according to the A. nidulans genome database (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html).
Intracellular activities of AHAS and transaminase B were measured. We detected 1.8- and 3.4-fold more activity in the extract prepared from the hypoxic than the normoxic cells (Table 3), which agreed with upregulation of the respective genes noted above. We detected AHAIR by a proteomic differential display analysis of proteins from A. nidulans cultured under normoxic and hypoxic conditions (see Fig. S1 in the supplemental material). Quantitation of gel images indicated that AHAIR was 2.8- and 2.0-fold more abundant in cells cultured for 12 and 24 h, respectively, under hypoxic conditions than in cells cultured under similar normoxic conditions, confirming the upregulation of this protein under hypoxia. These results indicated that a de novo biosynthetic mechanism for branched-chain amino acid synthesis was upregulated under hypoxic conditions, which is consistent with hypoxic accumulation of the branched-chain amino acids.
TABLE 3.
Enzyme activities in wild type and mutants grown under normoxic and hypoxic conditionsa
| Strain | Sp act (nmol min−1 mg−1) |
|||||
|---|---|---|---|---|---|---|
| AHAS |
Transaminase B |
GDH |
||||
| N | H | N | H | N | H | |
| WT | 40 ± 8 | 73 ± 10 | 14 ± 3 | 48 ± 6 | 23 ± 4 | 57 ± 12 |
| DAHAS-L | <1 | <1 | 11 ± 2 | 38 ± 5 | 20 ± 4 | 48 ± 11 |
| DAHAS-S | 30 ± 6 | 39 ± 7 | 15 ± 4 | 52 ± 4 | 26 ± 4 | 64 ± 11 |
| DGDH | 43 ± 9 | 73 ± 13 | 17 ± 4 | 56 ± 7 | <0.1 | <0.1 |
Mycelia were incubated in MM medium for 12 h under normoxic (N) and hypoxic (H) conditions. Valine, leucine, and isoleucine (10 mM each) were added to the medium to preculture DAHAS-L. Data are means of results of three experiments ± standard errors.
AHAS is involved in hypoxic production of branched-chain amino acids.
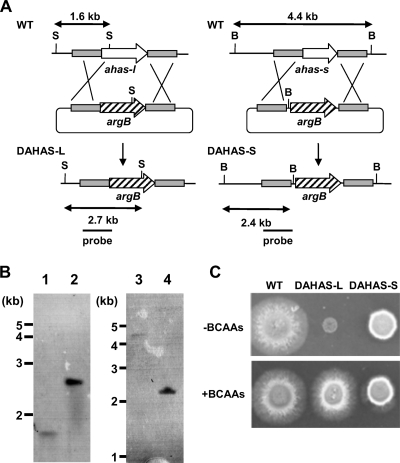
We constructed disruptants of the genes for AHAS-L and AHAS-S involved in the initial step of branched-chain amino acid synthesis. We constructed the plasmids pDAHASL and pDAHASS that are designed for double crossover with the fungal chromosome at the 5′ and 3′ regions of each gene and introduced them into A. nidulans. Total DNA samples from the transformants obtained with pDAHSL, designated DAHAS-L, were Southern blotted and amplified by PCR. The results showed that the gene for AHAS-L was deleted in the strain (Fig. 3A). Similarly, disruption of the gene for AHAS-S was confirmed in strain DAHAS-S, obtained with pDAHASS (Fig. 3B). The AHAS activity in cell extracts of strain DAHAS-L cultured under normoxic conditions was below the detection limit, whereas that of the WT was 40 nmol min−1 mg−1 (Table 3). In contrast, DAHAS-S produced 75% of the AHAS activity generated by the WT. These results indicated that AN4956.3 encodes the AHAS in this fungus and that the gene product of AN4430.3 enhances cellular AHAS activity. This is consistent with the observation that the AHAS-L of S. cerevisiae (19) and N. crassa (5) is critical for cellular AHAS activity. The activation of AHAS activity of AHAS-L by recombinant AHAS-S in S. cerevisiae has been reported (30, 31).
FIG. 3.
Effect of mutation on cell growth. (A) Strategy for homologous recombination into ahas-l and ahas-s loci to construct ahas-l and ahas-s deletion mutants. S, SacI; B, BamHI. (B) Southern blot analysis of A. nidulans wild type (FGSC A26) (lanes 1 and 3), DAHAS-L (lane 2), and DAHAS-S (lane 4). Total DNA from strains was digested with SacI and BamHI prior to blotting and hybridization. (C) Conidia (105 in 5 μl) of A. nidulans strains were spotted onto MMDN medium (upper panel) supplemented with 10 mM valine, leucine, and isoleucine (lower panel) and incubated at 37°C for 60 h under normoxic conditions.
Under normoxic conditions, DAHAS-L formed small poorly growing colonies on minimum agar plates. The addition of Val (Fig. 3C), Ile, or Leu (data not shown) to the medium restored the growth defect, and the morphology of the colonies was indistinguishable from that of the WT, indicating that AHAS-L is critical for de novo synthesis of the branched-chain amino acids. Strain DAHAS-S grew at a slightly lower rate than the WT on minimum agar plates (Fig. 3C). In contrast to results for strain DAHAS-L, adding either Val (Fig. 3C), Ile, or Leu (data not shown) had no effect on the hyphal growth of DAHAS-S, indicating that de novo synthesis of these amino acids does not limit fungal growth. This is consistent with the notion that sufficient amounts of these amino acids are synthesized in the absence of AHAS-S under normoxic conditions (see the Discussion section).
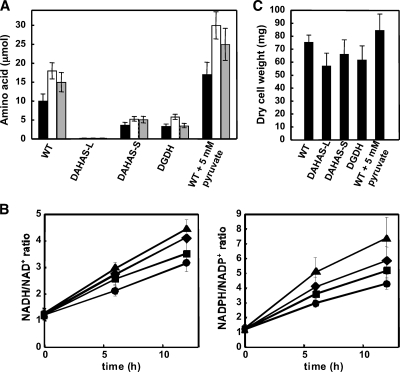
Extracellular metabolites produced by the strains DAHAS-L and DAHAS-S were measured. When the two strains and the WT were cultured in MMEN medium under normoxic conditions for 12 h, the amounts of metabolites differed little between them; that is, they produced malonate, maleate, and succinate but few amino acids (data not shown). Under hypoxic conditions DAHAS-L and DAHAS-S accumulated the same levels of lactate and malonate as the WT. Like the WT, the strains produced little maleate, succinate, citrate, and malate (Table 1; see Table S2 in the supplemental material), indicating that the lack of AHAS did not affect production of these organic acids. Levels of the hypoxically produced amino acids were altered in DAHAS-L and DAHAS-S. Both strains accumulated <50% of the amino acids compared with the WT in culture medium (Fig. 4A and Table S2). Among the amino acids, Val, Leu, and Ile production levels were the most obviously affected. DAHAS-S accumulated only 30% to 35% of these amino acids compared with the WT, and DAHAS-L accumulation was under the detection limit (Fig. 4A), indicating that both AHAS-L and AHAS-S are involved in branched-chain amino acid production under hypoxic conditions. The involvement of AHAS-S was in sharp contrast to the normoxically grown fungus that produced enough branched-chain amino acids for growth (Fig. 3). These results also indicated that the branched-chain amino acids produced under the hypoxic conditions were synthesized de novo. Adding pyruvate to hypoxic WT cultures resulted in the accumulation of 1.7-fold more Val, Leu, and Ile in the medium (Fig. 4A). These findings support the notion that branched-chain amino acids are synthesized via pyruvate.
FIG. 4.
Metabolic profiles and cell growth of A. nidulans strains. A. nidulans strains were cultured as described in the legend to Fig. 2 except that the DAHAS-L strain was precultured in MMDN medium supplemented with valine, leucine, and isoleucine (10 mM each). (A) Production of Val (black bars), Leu (white bars), and Ile (gray bars) in culture medium after a 12-h incubation. (B) Ratio of intracellular NADH and NAD+ (left) and NADPH and NADP+ (right) in WT (circles), DAHAS-L (triangles), DAHAS-S (squares), and DGDH (diamonds). (C) Dry cell weights of mycelia after a 12-h incubation. Data are means of results of three experiments. Error bars represent standard errors.
Cellular ratio of NAD(P)H and NAD(P)+.
The intracellular ratio of NADPH and NADP+ in A. nidulans cells cultured for 16 h under normoxic conditions was 1.2 (0-h culture in Fig. 4B). This ratio did not change after incubation for a further 6 or 12 h (data not shown), indicating that the cells balanced NADH/NAD+ during normoxic growth. When the normoxic culture (16 h) was shifted to hypoxic conditions for a further 12 h, the ratio of NADPH to NADP+ gradually increased to 4.3 (Fig. 4B). This indicated that electron acceptors are limited to reoxidizing NADH generated by oxidation of the carbon source under hypoxic conditions, which afforded the cells a more detrimental circumstance. The ratio of NADH to NAD+ was also increased under hypoxic conditions and reached 3.2 after 12 h (Fig. 4B).
The ratios of NADPH and NADP+ were compared among WT, DAHAS-L, and DAHAS-S strains. Cellular nucleotide levels did not significantly differ under normoxic conditions, indicating that AHAS is not essential to maintain the cellular NADPH/NADP+ balance in the presence of O2 (0-h culture in Fig. 4B). As in the WT, exposure to hypoxia for 12 h increased the NADPH/NADP+ ratio in DAHS-L. The ratio reached 7.3, which was 2.2-fold higher than that in the WT strain (Fig. 4B). The NADPH/NADP+ ratio of hypoxic DAHS-S cells was 4.4, which was slightly higher than that of the WT. The extent of the increase in the ratio of NADPH to NADP+ corresponded to intracellular levels of AHAS activity (Table 3). These results indicated that AHAS contributed to the reoxidization of intracellular NADPH to NADP+ under hypoxic conditions. Although no NADPH participated in the AHAS reaction, the absence of AHAS activity decreased the cellular concentrations of 2-acetolactate, which is a substrate of AHAIR that follows AHAS in the branched-chain amino acid synthetic pathway (Fig. 1). Since the AHAIR reaction oxidized NADPH, this causes a low AHAIR turnover and a high NADPH/NADP+ ratio. The cellular NADH/NAD+ ratio also corresponded to the levels of AHAS activity (Fig. 4B). Since AHAIR utilizes NADPH (but not NADH) as an electron donor, NAD(P)+ transhydrogenase (AN8130.3) is likely to catalyze NADPH-dependent NAD+ reduction and increase the NADH/NAD+ ratio when the NADPH level is high. Transcription of the gene was upregulated under the hypoxic conditions (Table 2). NAD(P)+ transhydrogenase activity was 1.3-fold higher under hypoxic (17 ± 5 nmol min−1 mg−1) than under normoxic (13 ± 5 nmol min−1 mg−1) conditions. These data agree with the notion that NAD(P)+ transhydrogenase regulates the NADH/NAD+ ratio under hypoxia.
The dry cell weight of the DAHAS-L and DAHAS-S strains was 78% and 87% of that of the WT strain (Fig. 4C). Adding pyruvate increased the dry cell weight of WT after the hypoxic culture (Fig. 4C). These results indicated that levels of the branched-chain amino acid production closely correlated with the cell mass under hypoxic conditions. Exogenous Val, Leu, and Ile did not restore the lower cell mass in AHAS-deficient strains (data not shown), indicating that the growth defect was not due to levels of available branched-chain amino acids; rather, their biosynthetic pathway contributed to the increase in the cell mass under hypoxic conditions. These results are consistent with the notion that the mechanism of hypoxic branched-chain amino acid synthesis reoxidizes NAD(P)H and functions as an electron sink through which the cells prevent an unbalanced NAD(P)H/NAD(P)+ ratio that in turn impairs cellular metabolism and hence cell growth.
Glutamate dehydrogenase and glutamate metabolism.
A. nidulans produces NADPH-glutamate dehydrogenase (GDH) that generates Glu from 2-oxoglutarate and ammonium (16). Expression of the gene encoding GDH (gdhA) was upregulated (Table 2), and intracellular GDH activity was 2.5-fold higher under hypoxic than under normoxic conditions (Table 3). The proteomic differential display showed that the spot for the gene product of gdhA was 2.1-fold more dense in the hypoxic than in the normoxic cells (see Fig. S1 in the supplemental material). The gene disruptant of gdhA (strain DGDH) accumulated 22 μmol of Glu in the culture medium after 12 h of hypoxic incubation (see Table S2 in the supplemental material), which was only 65% of that accumulated by the WT (Table 1). These results indicated that the gene product of gdhA is involved in the hypoxic production of Glu. The remainder of the observed Glu is likely to have been generated by Glu synthase (24). Comparable amounts of Ala, Thr, Pro, Asp, and Phe were produced by WT and DGDH (see Table S2 in the supplemental material), while 32% to 33% of the branched-chain amino acids produced by DGDH were detected in the culture medium of hypoxic cells (Fig. 4A). This suggested that GDH supplied Glu for branched-chain amino acid production under hypoxic conditions (Fig. 1). The NAD(P)H/NAD(P)+ ratio of the DGDH strain was higher under hypoxic conditions than that of the WT (Fig. 4B). This indicated that GDH oxidizes NADPH and contributes to maintaining the NAD(P)H/NAD(P)+ ratio under hypoxic conditions as discussed below.
Glucose represses hypoxic production of branched-chain amino acids.
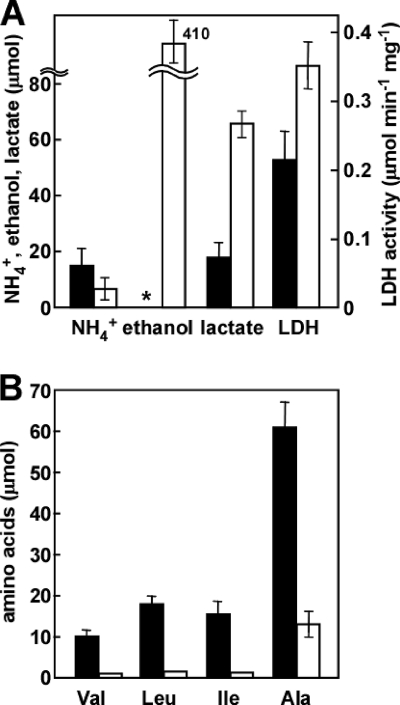
Hypoxic A. nidulans WT cells produced 3.4-fold more lactate (65 μmol) after culture for 12 h with glucose as the carbon source compared with ethanol (Fig. 5A; see Table S3 in the supplemental material). The culture grown in glucose also produced 410 μmol of ethanol in the medium. The intracellular activity of lactate dehydrogenase (LDH) was upregulated in the culture grown in glucose (Fig. 5A), indicating that the hypoxic fungus reoxidized NADH by reducing pyruvate to lactate and ethanol; that is, it fermented lactate and ethanol. The WT strain produced 1.3, 2.0, and 1.4 μmol of Val, Leu, and Ile, respectively, after culture for 12 h in medium using glucose as a carbon source (Fig. 5B). These amounts were 14%, 11%, and 9%, respectively, of those produced when ethanol was the carbon source (Table 1). The hypoxic induction of AHAS-L, AHAS-S, AHAIR, DHAD, and transaminase B gene expression was decreased or absent in cultures grown in glucose compared with those grown in ethanol (Table 2). This indicated that the hypoxic mechanism for branched-chain amino acid synthesis is an alternative mechanism to lactate and ethanol fermentation when glucose is unavailable under hypoxic conditions. Cultures grown in glucose generated less ammonium, which is a dissimilation product of nitrate (Fig. 5A) (45). These results imply that A. nidulans regulates the mechanisms for regenerating NAD(P)+ depending on the availability of carbon sources.
FIG. 5.
Effect of glucose on branched-chain amino acid production. A. nidulans WT (1 g wet weight) precultured in MMDN medium was transferred to 500-ml Erlenmeyer flasks containing 100 ml of MMEN medium (closed bars) or 100 ml of MMDN medium (open bars) and incubated at 30°C for 12 h under hypoxic conditions. (A) Ammonium, ethanol, and lactate production in culture medium and cell-free lactate dehydrogenase (LDH) activity. *, unable to determine due to vigorous consumption of ethanol added as a carbon source. (B) Branched-chain amino acid production in culture medium. Experiments were repeated three times. Mean values with standard deviations are shown.
DISCUSSION
This study demonstrated that upregulated branched-chain amino acid synthesis functions as an electron sink and accompanies the reoxidation of NAD(P)H in hypoxic A. nidulans cells and that the de novo biosynthetic mechanism of these amino acids contributes to this phenomenon. Hypoxia presents a survival challenge for A. nidulans because of a limited electron acceptor (O2) supply. Under hypoxic conditions, insufficient mitochondrial respiration results in the accumulation of intracellular NAD(P)H or an excess of reducing equivalents, both of which cause metabolic disorders. Cells therefore produce mechanisms for reoxidizing NAD(P)H. The production of branched-chain amino acids is considered to be one such mechanism through which fungi respond and adapt to lower levels of environmental O2. Providing pyruvate, which is generated from catabolic reactions of carbon sources, and ammonium is required to exert the mechanism (Fig. 1). Exogenous pyruvate can also serve as a substrate for branched-chain amino acid production (Fig. 4A). Ammonium was simultaneously produced by the reduction of nitrate included in the culture medium as described previously (37, 45). Nitrate reduction to ammonium and subsequent excretion into the medium is specific under hypoxic conditions, and it is also a dissimilation mechanism that oxidizes NAD(P)H (37, 45). This implies that the fungus regenerates NAD(P)+ by using both pyruvate and nitrate as electron sinks to produce branched-chain amino acids and ammonium. A. nidulans respires with O2 under normoxic conditions. When exposed to hypoxia with available glucose, it uses pyruvate as an electron sink and ferments ethanol and lactate. These findings imply that the fungus produces more types of hypoxic mechanisms to balance the cellular redox state than has hitherto been imagined. Notably, they also imply that the fungus regulates the production of such hypoxic mechanisms by responding not only to environmental O2 but also to carbon sources.
This study demonstrated that A. nidulans produces AHAS in response to a hypoxic environment. Although AHAS-S is not required for branched-chain amino acid production, a regulatory role of AHAS-S in yeast that increases AHAS-L activity in vitro has been proposed (30, 31). The present study is the first to provide genetic evidence that AHAS-S increases cellular AHAS activity (Table 3), and this is consistent with the in vitro function of the yeast AHAS-S. Besides this evidence, the AHAS-S of A. nidulans is dispensable for branched-chain amino acid production that supports normoxic growth (Fig. 3), indicating that the fungus produces sufficient levels of these amino acids without activating AHAS activity via AHAS-S. The results in S. cerevisiae are similar (7, 19). Nevertheless, AHAS-S largely affected branched-chain amino acid production under hypoxic conditions (Fig. 3), whereas the fungus produced branched-chain amino acids for reoxidizing NAD(P)H. Thus, fungal AHAS-S regulates the cellular redox state in response to decreasing levels of available O2. Upregulation of the gene expression of AHAS-S (Table 2) agrees with this notion. Notably, AHAS-S is conserved in microbes that inhabit normoxic, hypoxic, and anoxic milieus, and its function as a hypoxic regulator is probably also conserved among these microbes, although future investigation is required for confirmation.
In the presence of ammonium, the rate of GDH transcription is high (22), and from this transcription, glutamate is synthesized through the reductive amination of 2-oxoglutarate (16). This process is physiologically as significant as ammonia assimilation in fungi. We also indicated that GDH oxidizes NADPH under hypoxic conditions and maintains the cellular NAD(P)H/NAD(P)+ ratio. Thus, the GDH reaction is considered to play a dissimilatory role under hypoxic conditions. Furthermore, GDH indirectly participates in NAD(P)H reoxidation by hypoxic cells, since coordinated activity of transaminase and GDH oxidizes NADPH. For example, coupled transaminase B and GDH reactions oxidize 1 mol of NADPH to generate 1 mol of Val (Fig. 1) and A. nidulans also excretes Ala under hypoxic conditions (Fig. 2). NAD(P)H should be reoxidized by the reaction, since Ala transaminase also utilizes Glu as a substrate (Fig. 1). Maintenance of the redox balance by the coordinated activity of Ala transaminase and GDH in fermenting Pyrococcus furiosus has been proposed (43). That the Ala transaminase of higher plants participates in NAD+ regeneration under hypoxia has also been reported, although it depends on Glu synthase instead of GDH (26). Whether or not the coordination between transaminase B (for branched-chain amino acid synthesis) and GDH contributes to NAD+ regeneration in plants remains unknown.
As shown in the scheme (Fig. 1), production of 1 mol of the branched-chain amino acids generated 2 mol of NADP+ by using pyruvate and Glu. GDH produces Glu by using ammonium supplied by nitrate reduction, producing 4 mol of NAD(P)+ (Fig. 1). This indicates that production of 1 mol of the branched-chain amino acids from pyruvate and nitrate generates 6 mol of NAD(P)+. Similarly, 5 mol of NAD(P)+ is regenerated by producing 1 mol of Ala or Glu (Fig. 1). This enables us to estimate the contribution of each reaction to NAD(P)+ regeneration. For example, after a hypoxic incubation for 12 h in the presence of 10 mM nitrate, the culture produced Glu (41 μmol), Ala (72 μmol), Val (12 μmol), Leu (21 μmol), Ile (17 μmol), ammonium (15 μmol), and lactate (19 μmol) in the culture medium (Fig. 2 and Table 1). This estimates regeneration of 205, 360, 72, 132, 102, 60, and 21 μmol NAD(P)+ by each pathway. Total branched-chain amino acid production generates 304 μmol of NAD(P)+. The ratio of the regenerated NAD(P)+ by Glu, Ala, and the branched-chain amino acid productions is 2:36:30. These results indicate that branched-chain amino acid production is as relevant to regenerating NAD(P)+ and to an electron sink as the Glu and Ala production. The sum of the regenerated NAD(P)+ by the indicated pathways was 950 μmol, which accounted for reoxidation of 80% of the NADH (1,180 μmol) produced by oxidation of 590 μmol of ethanol. The remaining 230 μmol NADH is likely to be used for assimilation.
This study maintained hypoxic conditions in flasks sealed with butyl rubber stoppers. The initial oxygen concentration was set below 2 μM (less than 1% air) and we confirmed that after 6 h of incubation, the oxygen concentration decreased to less than 0.02 μM. This oxygen level is the lower limit our anaerobic system can handle without using an anaerobic chamber; that is, we cannot exclude possible contamination of such levels of air at the seals of the flasks and during the gas phase for measuring oxygen by gas chromatography. Under our conditions, A. nidulans is able to grow for 24 h with a lower (below 10%) growth rate than that under normoxic conditions (36), indicating that O2 limits the growth rate (hypoxic conditions). After the 24-h incubation, cell mass no longer increased, suggesting that A. nidulans almost completely consumed O2 and is not able to grow under anoxic conditions. To overcome the limitation of controlling aeration in our system, experiments using jar fermentors equipped with a continuous gas flow system are now under way.
The mechanism for branched-chain amino acid synthesis is evolutionarily conserved beyond biological domains, and it is recognized as an anabolic mechanism. Our findings are of significance in that they shed light on the catabolic (dissimilatory) role of the fungal mechanism of branched-chain amino acid synthesis. The fungal nitrate-reducing system of ammonia fermentation consists of NAD(P)H-nitrate and -nitrite reductases encoded by niaD and niiA, which were originally identified as anabolic enzymes for assimilating nitrate (37, 46). The involvement of reductive anabolic enzymes in the hypoxic catabolic mechanism is a common feature between hypoxic branched-chain amino acid production and ammonia fermentation. Under typical culture conditions with nitrate as a nitrogen source and no exogenous amino acids in the media, these enzymes are “housekeeping” mechanisms and are produced at considerable levels. Cells use these mechanisms to rapidly respond to sudden hypoxia without synthesizing other proteins that regenerate NAD+. These mechanisms are considered to constitute a distinct system for fungal survival under hypoxia.
Supplementary Material
Acknowledgments
We thank Norma Foster for critical reading of the manuscript.
This study was partly supported by the Bio-Oriented Technology Research Advancement Institution and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print on 15 January 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Blombach, B., M. E. Schreiner, J. Holatko, T. Bartek, M. Oldiges, and B. J. Eikmanns. 2007. l-Valine production with pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum. Appl. Environ. Biotechnol. 79:471-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruckmann, A., P. J. Hensbergen, C. I. Balog, A. M. Deelder, R. Brandt, I. S. Snoek, H. Y. Steensma, and G. P. van Heusden. 2009. Proteome analysis of aerobically and anaerobically grown Saccharomyces cerevisiae cells. J. Proteomics 30:662-669. [DOI] [PubMed] [Google Scholar]
- 3.Brunner, A., A. Devillers-Mire, and H. Robichon-Szulmajster. 1969. Regulation of isoleucine-valine biosynthesis in Saccharomyces cerevisiae. Altered threonine deaminase in an is-1 mutant responding to threonine. Eur. J. Biochem. 10:172-183. [PubMed] [Google Scholar]
- 4.Cano, N. J., D. Fouque, and X. M. Leverve. 2006. Application of branched-chain amino acids in human pathological states: renal failure. J. Nutr. 136:299S-307S. [DOI] [PubMed] [Google Scholar]
- 5.Caroline, D. F., R. W. Harding, H. Kuwana, T. Satyanarayana, and R. P. Wagner. 1969. The ilv-3 mutants of Neurospora crassa. II. Activity of acetohydroxy acid synthase. Genetics 62:487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chantrel, Y., M. Gaisne, C. Lions, and J. Verdiere. 1998. The transcriptional regulator Hap1p (Cyp1p) is essential for anaerobic or heme-deficient growth of Saccharomyces cerevisiae: genetic and molecular characterization of an extragenic suppressor that encodes a WD repeat protein. Genetics 148:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullin, C., A. Baudin-Baillieu, E. Guillemet, and O. Ozier-Kalogeropoulos. 1996. Functional analysis of YCL09C: evidence for a role as the regulatory subunit of acetolactate synthase. Yeast 12:1511-1518. [DOI] [PubMed] [Google Scholar]
- 8.Deckert, J., R. Perinei, B. Balasubramanian, and R. S. Zitomer. 1995. Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics 139:1149-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deckert, J., A. M. Rodriguez-Torres, J. T. Simon, and R. S. Zitomer. 1995. Mutational analysis of Rox1, a DNA-bending repressor of hypoxic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6109-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Groot, M. J., P. Daran-Lapujade, B. van Breukelen, T. J. Knijnenburg, E. A. de Hulster, M. J. Reinders, J. T. Pronk, A. J. Heck, and M. Slijper. 2007. Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveal post-transcriptional regulation of key cellular processes. Microbiology 153:3864-3878. [DOI] [PubMed] [Google Scholar]
- 11.Eden, A., G. Simchen, and N. Benvenisty. 1996. Two yeast homologs of ECA39, a target for c-Myc regulation, code for cytosolic and mitochondrial branched-chain amino acid aminotransferases. J. Biol. Chem. 271:20242-20245. [DOI] [PubMed] [Google Scholar]
- 12.Ernst, J. F., and D. Tielker. 2009. Responses to hypoxia in fungal pathogens. Cell Microbiol. 11:183-190. [DOI] [PubMed] [Google Scholar]
- 13.Falco, S. C., K. S. Dumas, and K. J. Livak. 1985. Nucleotide sequence of the yeast ILV2 gene which encodes acetolactate synthase. Nucleic Acids Res. 13:4011-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujii, T., and N. Takaya. 2008. Denitrification by the fungus Fusarium oxysporum involves NADH-nitrate reductase. Biosci. Biotechnol. Biochem. 72:412-420. [DOI] [PubMed] [Google Scholar]
- 15.Gunter, B. K. 2003. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 67:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurr, S. J., A. R. Hawkins, C. Drainas, and J. R. Kinghorn. 1986. Isolation and identification of the Aspergillus nidulans gdhA gene encoding NADP-linked glutamate dehydrogenase. Curr. Genet. 203:761-766. [DOI] [PubMed] [Google Scholar]
- 17.Halatko, J., V. Elisakova, M. Prouza, M. Sobotka, J. Nesvera, and M. Patek. 2009. Metabolic engineering of the L-valine biosynthesis pathway in Corynebacterium glutamicum using promoter activity modulation. J. Biotechnol. 139:203-210. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, A. L., B. Todd, and P. J. Espenshade. 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120:831-841. [DOI] [PubMed] [Google Scholar]
- 19.Kakar, S. N., and R. P. Wagner. 1964. Genetic and biochemical analysis of isoleucine-valine mutants of yeast. Genetics 49:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan, N. O. 1967. Beef heart TPNH-DPN pyridine nucleotide transhydrogenases. Methods Enzymol. 10:317-322. [Google Scholar]
- 21.Keng, T. 1992. HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:616-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinghorn, J. R., and J. A. Pateman. 1973. NAD and NADP l-glutamate dehydrogenase activity and ammonium regulation in Aspergillus nidulans. J. Gen. Microbiol. 78:39-46. [DOI] [PubMed] [Google Scholar]
- 23.Kispal, G., H. Steiner, D. A. Court, B. Rolinski, and R. Lill. 1996. Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J. Biol. Chem. 271:24458-24464. [DOI] [PubMed] [Google Scholar]
- 24.Kusnan, M. B., M. G. Berger, and H. P. Fock. 1987. The involvement of glutamine synthetase/glutamate synthase in ammonia assimilation by Aspergillus nidulans. J. Gen. Microbiol. 133:1235-1242. [DOI] [PubMed] [Google Scholar]
- 25.Leyval, D., D. Uy, S. Delaunay, J. L. Goergen, and J. M. Engasser. 2003. Characterisation of the enzyme activities involved in the valine biosynthetic pathway in a valine-producing strain of Corynebacterium glutamicum. J. Biotechnol. 104:241-252. [DOI] [PubMed] [Google Scholar]
- 26.Limami, A. M., G. Glevarec, C. Ricoult, J. B. Cliquet, and E. Planchet. 2008. Concerted modulation of alanine and glutamate metabolism in young Medicago truncatula seedlings under hypoxic stress. J. Exp. Bot. 59:2325-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacAlpine, D. M., P. S. Perlman, and R. A. Butow. 2000. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J. 19:767-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyashita, Y., R. Dolferus, K. P. Ismond, and A. G. Good. 2007. Alanine aminotransferase catalyses the breakdown of alanine after hypoxia in Arabidopsis thaliana. Plant J. 49:1108-1121. [DOI] [PubMed] [Google Scholar]
- 29.Panagiotou, G., S. G. Villas-Boas, P. Christakopoulos, J. Nilsen, and L. Olsson. 2005. Intracellular metabolite profiling of Fusarium oxysporum converting glucose to ethanol. J. Biotechnol. 115:425-434. [DOI] [PubMed] [Google Scholar]
- 30.Pang, S. S., and R. G. Duggleby. 1999. Expression, purification, characterization, and reconstitution of the large and small subunits of yeast acetohydroxyacid synthase. Biochemistry 38:5222-5231. [DOI] [PubMed] [Google Scholar]
- 31.Pang, S. S., and R. G. Duggleby. 2001. Regulation of yeast acetohydroxyacid synthase by valine and ATP. Biochem. J. 357:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piper, M. D., P. Daran-Lapujade, C. Bro, B. Regenberg, S. Knudsen, J. Nielsen, and J. K. Pronk. 2002. Reproducibility of oligonucleotide microarray transcriptome analyses. An interlaboratory comparison using chemostat cultures of Saccharomyces cerevisiae. J. Biol. Chem. 277:37001-37008. [DOI] [PubMed] [Google Scholar]
- 33.Ramil, E., C. Agrimonti, E. Shechter, M. Gervais, and B. Guiard. 2000. Regulation of the CYB2 gene expression: transcriptional co-ordination by the Hap1p, Hap2/3/4/5p and Adr1p transcription factors. Mol. Microbiol. 37:1116-1132. [DOI] [PubMed] [Google Scholar]
- 34.Rocha, S. 2007. Gene regulation under low oxygen: holding your breath for transcription. Trends Biochem. Sci. 32:389-397. [DOI] [PubMed] [Google Scholar]
- 35.Sato, I., M. Shimizu, M. T. Hoshino, and N. Takaya. 2009. The glutathione system of Aspergillus nidulans involves a fungus-specific glutathione S-transferase. J. Biol. Chem. 284:8042-8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu, M., T. Fujii, S. Masuo, K. Fujita, and N. Takaya. 2009. Proteomic analysis of Aspergillus nidulans cultured under hypoxic conditions. Proteomics 9:7-19. [DOI] [PubMed] [Google Scholar]
- 37.Takasaki, K., H. Shoun, M. Yamaguchi, K. Takeo, A. Nakamura, T. Hoshino, and N. Takaya. 2004. Fungal ammonia fermentation—a novel metabolic mechanism that couples the dissimilatory and assimilatory pathways of both nitrate and ethanol. J. Biol. Chem. 279:12414-12420. [DOI] [PubMed] [Google Scholar]
- 38.Takasaki, K., H. Shoun, A. Nakamura, T. Hoshino, and N. Takaya. 2004. Unusual transcription regulation of the niaD gene under anaerobic conditions supporting fungal ammonia fermentation. Biosci. Biotechnol. Biochem. 68:978-980. [DOI] [PubMed] [Google Scholar]
- 39.Ter Linde, J. J., and H. Y. Steensma. 2002. A microarray-assisted screen for potential HapI and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19:825-840. [DOI] [PubMed] [Google Scholar]
- 40.Todd, B. L., E. V. Stewart, J. S. Burg, A. L. Hughes, and P. J. Espenshade. 2006. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol. Cell. Biol. 26:2817-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vallas-Boas, S. G., D. G. Delicado, M. Akesson, and J. Nielsen. 2003. Simultaneous analysis of amino and nonamino organic acids as methyl chloroformate derivatives using gas chromatography-mass spectrometry. Anal. Biochem. 322:134-138. [DOI] [PubMed] [Google Scholar]
- 42.Velasco, J. A., J. Consado, M. C. Pena, T. Kawakami, J. Laborda, and V. Notario. 1993. Cloning of the dihydroxyacid dehydratase-encoding gene (ILV3) from Saccharomyces cerevisiae. Gene 137:179-185. [DOI] [PubMed] [Google Scholar]
- 43.Ward, D. E., S. W. Kengen, J. van Der Oost, and W. M. de Vos. 2000. Purification and characterization of the alanine aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus and its role in alanine production. J. Bacteriol. 182:2559-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westerfeld, W. W. 1945. A colorimetric determination of blood acetoin. J. Biol. Chem. 161:495-502. [PubMed] [Google Scholar]
- 45.Zhou, Z., N. Takaya, M. A. Sakairi, and H. Shoun. 2001. Oxygen requirement for the denitrification by the filamentous fungus Fusarium oxysporum. Arch. Microbiol. 175:19-25. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, Z., N. Takaya, A. Nakamura, M. Yamaguchi, K. Takeo, and H. Shoun. 2002. Ammonia fermentation, a novel anoxic metabolism of nitrate by fungi. J. Biol. Chem. 277:1892-1896. [DOI] [PubMed] [Google Scholar]
- 47.Znaidi, S., S. Weber, O. Z. Al-Abdin, P. Bomme, S. Saidane, S. Drouin, S. Lemieux, X. De Deken, F. Robert, and M. Raymond. 2008. Genomewide location analysis of Candida albicans Upc2p, a regulator of sterol metabolism and azole drug resistance. Eukaryot. Cell 7:836-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.