Abstract
Reliability of microbial (starter) strains in terms of quality, functional properties, growth performance, and robustness is essential for industrial applications. In an industrial fermentation process, the bacterium should be able to successfully withstand various adverse conditions during processing, such as acid, osmotic, temperature, and oxidative stresses. Besides the evolved defense mechanisms, stress-induced mutations participate in adaptive evolution for survival under stress conditions. However, this may lead to accumulation of mutant strains, which may be accompanied by loss of desired functional properties. Defining the effects of specific fermentation or processing conditions on the mutation frequency is an important step toward preventing loss of genome integrity and maintaining the productivity of industrial strains. Therefore, a set of Lactobacillus plantarum mutator reporter strains suitable for qualitative and quantitative analysis of low-frequency mutation events was developed. The mutation reporter system constructed was validated by using chemical mutagenesis (N-methyl-N′-nitro-N-nitrosoguanidine) and by controlled expression of endogenous candidate mutator genes (e.g., a truncated derivative of the L. plantarum hexA gene). Growth at different temperatures, under low-pH conditions, at high salt concentrations, or under starvation conditions did not have a significant effect on the mutation frequency. However, incubation with sublethal levels of hydrogen peroxide resulted in a 100-fold increase in the mutation frequency compared to the background mutation frequency. Importantly, when cells of L. plantarum were adapted to 42°C prior to treatment with sublethal levels of hydrogen peroxide, there was a 10-fold increase in survival after peroxide treatment, and there was a concomitant 50-fold decrease in the mutation frequency. These results show that specific environmental conditions encountered by bacteria may significantly influence the genetic stability of strains, while protection against mutagenic conditions may be obtained by pretreatment of cultures with other, nonmutagenic stress conditions.
Lactobacillus plantarum is a common inhabitant of the human gastrointestinal tract (3), but it is also encountered in a variety of environmental niches, such as dairy, meat, and vegetable fermentations (25, 39). L. plantarum is widely used in industrial and traditional production of fermented plant, food, and feed products, such as sauerkraut, sausage, cabbage, olives, and silage (14, 31, 41). In addition, L. plantarum is employed as a starter culture, and it contributes to the conservation, flavor, and texture of fermented foods. The growth performance and robustness of this bacterium are key factors that determine the characteristics of the final products. Moreover, properties that improve consumer health have been attributed to various L. plantarum strains, and some strains are marketed as probiotics (10, 14, 35).
Reliability of (starter) strains in terms of quality, functional properties, growth performance, and robustness is very important for industrial applications. In an industrial fermentation process, the bacterium should be able to withstand various adverse conditions during processing, such as acid, osmotic, temperature, and oxidative stresses. In their natural environments bacteria are also constantly exposed to fluctuating environmental conditions, and many of these conditions are potentially detrimental and negatively affect the physiological state and growth rate. Many bacteria possess multiple regulatory networks of stress response systems that allow them to withstand harsh conditions and sudden changes in environmental conditions (49). Moreover, mutations may favor generation of strain variants that are better adapted to survive under these stress conditions (9, 13, 21). However, this process may not be desirable for maintaining the reliability of strains used in industrial processes. Especially when high productivity of strains results in decreased fitness, mutants with decreased productivity and loss of desired functional properties may be readily obtained (46, 53). In addition, DNA instability can even result in a so-called mutator phenotype, where sharply elevated spontaneous mutation rates (transiently) enhance the ability of strains to adapt to radical changes in the environment (19, 20, 51). The lactic acid bacterium Oenococcus oeni is an example of an organism that positively adapted to a harsh environment due to an elevated mutation rate. O. oeni lacks the mutS and mutL mismatch repair genes, which has been suggested to contribute to its hypermutable status and its accelerated evolution. It is likely that this factor contributed to the unique adaptation of O. oeni to acidic and alcoholic environments that made it an ideal organism for the malolactic fermentation during the production of wine (7, 29). However, elevated mutation rates may also result in deterioration of industrial fermentation properties (19, 20, 51). Therefore, defining the effects of conditions encountered during industrial fermentation processes on the mutation frequency is an important step toward preventing loss of genome integrity and maintaining the productivity of industrial strains. It has been shown that adaptation to mild stress conditions (e.g., mild heat shock) can induce cross-protection against more lethal stress conditions, resulting in an increased survival rate. Consequently, establishing the effect of cross-protection on the mutation frequency could also be an important step in maintaining the productivity of industrial strains (11, 49).
In this study we focused on selected environmental challenges that are relevant for fermentation and processing conditions and their effects on the mutation frequency of L. plantarum. To this end, a mutation reporter strain set was constructed, which was validated using chemical mutagenesis and controlled expression of candidate genes that may lead to mutator phenotypes upon overexpression. The frequencies of mutation under various environmental conditions were subsequently studied, which revealed that especially oxidative stress challenge (challenge with hydrogen peroxide, which can be produced as a by-product by L. plantarum or other lactobacilli during fermentation [4, 30, 36, 40, 49, 52]) increased the mutation frequency. Interestingly, pretreatment of cells under nonmutagenic conditions (42°C) appeared to protect them against subsequent increases in the mutation frequency due to oxidative stress, showing that sequential exposure to stresses may be used to protect industrial strains against elevated mutation rates and in this way may enhance the stability of these strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study and their relevant features are shown in Table 1 (see Table S1 in the supplemental material for a list of the primers used in this study).
TABLE 1.
Strains and plasmids used in this study, their relevant characteristics, and references
| Strain or plasmid | Relevant featuresa | Reference |
|---|---|---|
| E. coli strains | ||
| Top10 | Cloning host | |
| XL1-blue | Cloning host | |
| L. lactis MG1363 | Cloning host | 18 |
| L. plantarum strains | ||
| NZ7100 | L. plantarum WCFS1 with chromosomal integration of plasmid pEMnisRK | 44 |
| NZ7140 | L. plantarum NZ7100 with ΔmelA obtained by double-crossover integration of pNZ5340 | This study |
| NZ1800 | Integration of L. lactis usp45 promoter in front of L. plantarum NZ7100 melA | This study |
| NZ1801 | NZ1800 with substitution mutation in the catalytic site of melA, constructed using the pNZ1801 mutagenesis vector | This study |
| NZ1802 | NZ1800 with substitution mutation in the catalytic site of melA, constructed using the pNZ1802 mutagenesis vector | This study |
| NZ1803 | NZ1800 with 1-bp deletion in the catalytic site of melA, constructed using the pNZ1803 mutagenesis vector | This study |
| NZ1804 | NZ1800 with 2-bp deletion in the catalytic site of melA, constructed using the pNZ1804 mutagenesis vector | This study |
| NZ1805 | NZ1800 with 1-bp insertion in the catalytic site of melA, constructed using the pNZ1805 mutagenesis vector | This study |
| NZ1806 | NZ1800 with 2-bp insertion in the catalytic site of melA, constructed using the pNZ1806 mutagenesis vector | This study |
| NZ1807 | NZ1800 with G stretch and 1-bp insertion in melA, constructed using the pNZ1807 mutagenesis vector | This study |
| Plasmids | ||
| pNZ5340 | Cmr Emr; integration vector with homologous regions up- and downstream of L. plantarum WCFS1 melA | 27 |
| pNZ5516 | Kmr; pCRblunt containing Pusp45-melA | 5 |
| pNZ1825 | Cmr; pNZ5516 derivative containing Pusp45-melA-cat-lox71 downstream region of melA | This study |
| pNZ1826 | Cmr; pNZ1825 derivative containing upstream region of melA | This study |
| pNZ1800 | Cmr; pNZ1826 derivative, insertion of a lox66 region; integration vector for inserting Pusp45 in front of melA based on cre-lox system | This study |
| pNZ5348 | Emr; pGID023 derivative containing cre under control of the lp_1144 promoter | 27 |
| pNZ1801 | Cmr; pNZ1800 derivative; substitution mutation in the catalytic site of melA | This study |
| pNZ1802 | Cmr; pNZ1800 derivative; substitution mutation in the catalytic site of melA | This study |
| pNZ1803 | Cmr; pNZ1800 derivative; 1-bp deletion in the catalytic site of melA | This study |
| pNZ1804 | Cmr; pNZ1800 derivative; 2-bp deletion in the catalytic site of melA | This study |
| pNZ1805 | Cmr; pNZ1800 derivative; 1-bp insertion in the catalytic site of melA | This study |
| pNZ1806 | Cmr; pNZ1800 derivative; 2-bp insertion in the catalytic site of melA | This study |
| pNZ1817 | Cmr; pNZ1800 derivative; G stretch and 1-bp insertion in melA | This study |
| pIL252 | Emr; low-copy-number vector | 45 |
| pNZ8048 | Cmr; high-copy-number vector with inducible PnisA from L. lactis | 12 |
| pNZ8154 | Emr; pIL252 derivative; mutator vector with PnisA-TpepN fragment of pNZ8048 | This study |
| pCRblunt | Kmr; positive-selection cloning vector | 6 |
| pNZ1813 | Emr; pNZ8154 derivative; insertion of L. plantarum WCFS1 hexA | This study |
| pNZ1814 | Emr; pNZ8154 derivative; insertion of L. plantarum WCFS1 truncated hexA | This study |
| pNZ1820 | Emr; pNZ8154 derivative; insertion of L. plantarum WCFS1 dinP | This study |
Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Kmr, kanamycin resistant.
L. plantarum was grown at 37°C in MRS medium (Difco, West Molesey, United Kingdom) without aeration. L. plantarum was also grown at 30°C in CDM (47) supplemented with 1% maltose, plated on CDM with 1% melibiose and 0.004% bromocresol purple (BCP) (Merck, Darmstadt, Germany) for determination of mutation frequencies, and plated on CDM with 1% maltose for determination of viable counts. The plates were incubated anaerobically at 30°C. Escherichia coli strain Top10 (Invitrogen, Breda, the Netherlands) was used as a cloning host for construction of pNZ1800. E. coli strain XL1-blue (Stratagene, La Jolla, CA) was used as a cloning host for construction of pNZ1801, pNZ1802, pNZ1803, pNZ1804, pNZ1805, pNZ1806, and pNZ1817. Both E. coli strains were grown aerobically at 37°C in TY broth (42). Lactococcus lactis MG1363 was used as an intermediate cloning host for plasmids pNZ8154, pNZ1813, and pNZ1814 and was grown without aeration at 30°C in M17 medium (Merck, Darmstadt, Germany) supplemented with 0.5% glucose. When appropriate, antibiotics were added to the media at the following concentrations: 10 μg/ml chloramphenicol and 10 μg/ml erythromycin for L. plantarum, 10 μg/ml chloramphenicol and 50 μg/ml kanamycin for E. coli, and 10 μg/ml chloramphenicol and 5 μg/ml erythromycin for L. lactis.
DNA techniques.
Plasmid DNA was isolated from E. coli on a small scale using the alkaline lysis method (8). Large-scale plasmid DNA isolation was performed using Jetstar columns according to the manufacturer's recommendations (Genomed GmbH, Bad Oberhausen, Germany).
Isolation of L. lactis plasmid DNA was performed using Jetstar columns according to the manufacturer's recommendations, with modifications described previously (5). Transformation of L. lactis was performed as described by Holo and Nes (23). For DNA techniques with E. coli and L. lactis, standard procedures were performed as described by Sambrook et al. (42).
L. plantarum total DNA was isolated and strains were transformed as described previously (27). The anticipated genetic organization of the transformants was verified by PCR analysis.
Restriction endonucleases, DNA polymerases, T4 DNA ligase, and the Klenow enzyme were purchased from Invitrogen (Breda, the Netherlands), New England Biolabs (Leusden, the Netherlands), and Stratagene (La Jolla, CA) and used according to the manufacturer's recommendations. Oligonucleotides were purchased from Invitrogen (Breda, the Netherlands). DNA sequencing analysis was performed by BaseClear B.V. (Leiden, the Netherlands).
Construction of L. plantarum NZ1800.
The regulated promoter of the melA α-galactosidase gene of L. plantarum NZ7100 was replaced by the strong and constitutive usp45 promoter from L. lactis MG1363 (48). To replace the promoter, a plasmid harboring Pusp45, melA, lox66, cat, and lox71 between the upstream and downstream regions of melA was constructed. The ori-Pusp45-melA fragment (5) was isolated from plasmid pNZ5516 by DraI and XbaI digestion and treated with the Klenow enzyme. This fragment was ligated to a Klenow enzyme-treated NruI-SalI fragment from pNZ5340 (27) with the cat-lox71 downstream region of melA, resulting in plasmid pNZ1825. The upstream region of melA was PCR amplified with Pwo polymerase using primers LacS2forPciI and LacS2revSacI and L. plantarum WCFS1 genomic DNA as the template. The PCR fragment was digested with PciI and SacI and ligated in similarly digested pNZ1825. The resulting plasmid (pNZ1826) was digested with DraI and NotI to introduce a lox66 region (composed of oligonucleotides N-lox66-D and D-lox66-N) by oligonucleotide linker ligation. The resulting plasmid, pNZ1800, was used for double-crossover integration in L. plantarum NZ7140. L. plantarum NZ7140 is a ΔmelA derivative of L. plantarum NZ7100 that was constructed by double-crossover integration of plasmid pNZ5340 using the method described previously for the L. plantarum WCFS1 ΔmelA derivative NZ5335 (27). Pusp45-melA-positive colonies were selected by growth on agar with the substrate 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal). The colonies of single-crossover mutants were light blue, and the colonies of double-crossover mutants were dark blue, which was confirmed by PCR analysis (with primers 93 and LK17 and primers LK1 and LK4). To remove the cat gene after integration, the Cre expression plasmid pNZ5348 (27) was transformed into the NZ7140 double-crossover mutant. The resulting Cre expression led to recombination of the lox sites and excision of the intermediate DNA sequence (1, 22, 27). Since plasmid pNZ5348 is very unstable in L. plantarum, it could readily be cured from L. plantarum transformants by growth in the absence of erythromycin selection pressure for 10 generations (27). The anticipated genetic structure of the Pusp45-melA locus in a single colony obtained after excision (designated L. plantarum NZ1800) was confirmed by PCR analysis and Southern blotting.
Construction of the mutator reporter strains.
Mutations in melA in plasmid pNZ1800 were constructed using a QuikChange II mutagenesis kit obtained from Stratagene (La Jolla, CA). Six primer sets were designed to make substitution, deletion, and insertion mutations in the catalytic site of melA (Table 2) . One primer set was designed to make the mutations necessary to change two amino acid residues to glycine residues and to insert one extra G residue, resulting in a frameshift mutant and a stretch of G residues (Table 2). Sequence analysis (with primers LK34, LK35, and LK36) of the resulting plasmids (pNZ1801, pNZ1802, pNZ1803, pNZ1804, pNZ1805, pNZ1806, and pNZ1817 [Tables 1 and 2]) confirmed the presence of the correct mutations. The plasmids were transformed into L. plantarum NZ7100, and double-crossover mutants were selected to obtain a melA mutant phenotype. Removal of the cat gene and verification of the correct genetic organization of the Pusp45-melA locus in these strains (strains NZ1801 to NZ1807 [Table 1]) were performed as described above for strain NZ1800. In addition, the sequence of the mutated region of the melA gene was confirmed by PCR amplicon (primers LK4 and LK36) sequencing.
TABLE 2.
Mutations in the reporter strains
| Strain | Sequencea | Mutation | Primers |
|---|---|---|---|
| NZ7100 | ATTGATTATATC AAG TGG GAT ATGAAC | Wild typeb | |
| NZ1801 | ATTGATTATATC TAG TGG GAT ATGAAC | Substitution stop codon | 1L, 1R |
| NZ1802 | ATTGATTATATC AAG TGG TAT ATGAAC | Substitution Asp → Tyr | 2L, 2R |
| NZ1803 | ATTGATTATATC AAG GG GAT ATGAAC | 1-bp deletion frameshift | 3L, 3R |
| NZ1804 | ATTGATTATATC AAG G GAT ATGAAC | 2-bp deletion frameshift | 4L, 4R |
| NZ1805 | ATTGATTATATC GAAG TGG GAT ATGAAC | 1-bp insertion frameshift | 5L, 5R |
| NZ1806 | ATTGATTATATC AAG TGG CTG ATATGAAC | 2-bp insertion frameshift | 6L, 6R |
| NZ7100 | AAGTTACCA GGA GGC TTAGCGGATATTAG | Wild typec | |
| NZ1807 | AAGTTACCA GGG GGGG TTAGCGGATATTAG | 1-bp insertion frameshift | 7L, 7R |
Underlining indicates the catalytic site of the melA gene; bold type indicates the mutation site.
Positions 1438 to 1464 of the melA gene.
Positions 1189 to 1216 of the melA gene.
Construction of the mutator vector pNZ8154.
For controlled expression of potential mutator genes (e.g., hexA, dinP, and tr-hexA, an N-terminally truncated derivative of hexA [see below]), a low-copy-number vector with an inducible promoter was constructed. The inducible nisA promoter from plasmid pNZ8048 (26) was cloned into the low-copy-number plasmid pIL252 (45). Plasmid pNZ8048 was digested with BglII and XhoI and treated with the Klenow enzyme. The 576-bp PnisA-MCS-TpepN (where MCS stands for the multiple cloning site and T stands for the terminator sequence derived from the downstream region) fragment was ligated into SmaI-digested pIL252. PCR analysis (with primers Nis1 and RB148) showed that the PnisA-TpepN fragment was oriented in the same direction as the replication genes in all of the transformants analyzed. The resulting vector, pNZ8154, was introduced into NZ1800, NZ7140, and the mutator reporter strains. For all experiments performed with one of these strains, the strain harboring plasmid pNZ8154 was used as a control.
Cloning of hexA, tr-hexA, and dinP in pNZ8154.
The L. plantarum WCFS1 hexA gene (lp_2298) (25) was amplified with Pwo polymerase using chromosomal DNA as the template and primers LK20 and LK30. An N-terminally truncated derivative of hexA, designated tr-hexA, which lacks the sequence encoding amino acids 2 to 94 (279 bp) compared to the full-length hexA, was amplified with primers LK44 and LK30. The L. plantarum WCFS1 dinP gene (lp_2280) (25) was amplified with primers LK48 and LK51. The amplicons obtained were cloned directly in pCRblunt (Invitrogen, Breda, the Netherlands), and insert sequences were verified by sequence analyses. The hexA and dinP fragments were recovered from these plasmids as a NcoI-SpeI fragment and ligated into similarly digested pNZ8154. The resulting plasmids, pNZ1813 (hexA), pNZ1814 (tr-hexA), and pNZ1820 (dinP), were transformed into all seven mutator reporter strains.
Screening of MelA activity.
α-Galactosidase can convert the substrate 5- bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal) (MP Biomedicals, Amsterdam, the Netherlands), forming a blue color. The E. coli transformants and L. plantarum mutated strains were screened on agar plates with 40 μg/ml X-α-Gal for white (MelA−) or blue (MelA+) colonies. Alternatively, cells were plated on CDM agar plates with 1% melibiose and 0.004% BCP, and growth was scored by counting melibiose-fermenting clones that appeared as yellow colonies. The latter method was used for quantification of the mutation frequency.
Determination of the mutation frequency by using the mutator reporter system.
The frequency of spontaneous revertants of the mutator reporter strains was determined by plating preparations on plates used for selection of MelA activity. The strains were inoculated into CDM with 1% maltose, and after 6 h of growth at 30°C 1% of each culture was inoculated into fresh medium. The resulting cultures were grown for 16 h at 30°C until the optical density at 600 nm (OD600) was approximately 1.0. Based on the OD600, 1010, 109, and 108 CFU were washed three times with CDM without a carbon source and plated on selective CDM agar plates containing 1% melibiose and 0.004% BCP, as described above for screening of MelA activity. The total viable counts of each culture were also determined by nonselective plating on CDM agar with 1% maltose. The plates were incubated anaerobically at 30°C for 5 days, after which the colonies were counted.
To determine the effect of (over)expression of a gene cloned in pNZ8154 under control of the nisin promoter, a strain containing a specific plasmid (pNZ1813 [hexA], pNZ1814 [tr-hexA], or pNZ1820 [dinP]) was inoculated into CDM with 1% maltose and grown overnight. The culture was diluted in fresh medium to obtain an OD600 of 0.1, and 40 ng/ml nisin (Sigma, Zwijndrecht, the Netherlands) was added to induce candidate mutator gene expression. The culture was incubated at 30°C until the OD600 was approximately 1.0 and then was inoculated (0.1%) into fresh medium containing 40 ng/ml nisin. After 16 h of growth the OD600 was measured, and then 109 CFU, as calculated based on the OD600 value, was washed three times with CDM without a carbon source and appropriate dilutions were plated on selective CDM agar plates with 1% melibiose and 0.004% BCP. Miller et al. (34) determined the mutation frequency of E. coli and observed that plating efficiency was strongly influenced by the number of cells that were actually plated; therefore, a scavenger strain had to be added when fewer than 109 cells from the actual culture were plated to prevent measurement artifacts due to so-called crowding effects (34). Our results confirmed the observations made by Miller et al. (34), and 108 CFU of the L. plantarum melA mutant strain NZ7140 was added routinely to each plate, which allowed accurate and reproducible quantification of MelA-positive revertants without interference by crowding effects. The total viable count of each culture was also determined by nonselective plating on CDM agar with 1% maltose. The plates were incubated anaerobically at 30°C for 5 days, after which the colonies were counted.
Determination of the mutational spectrum of tr-hexA.
Strains NZ1807(pNZ1814) (tr-hexA) and NZ1807(pNZ8154) (empty vector; spontaneous mutant) were grown overnight in CDM with 1% maltose. The cultures were diluted until the OD600 was 0.1, and 40 ng/ml nisin was added. The cultures were incubated at 30°C until the OD600 was approximately 1.0 and then were diluted (0.1%) in fresh medium with 40 ng/ml nisin. After 16 h of growth, the OD600 was measured, and, based on the OD600, 109 CFU was washed three times with CDM without a carbon source and appropriate dilutions were plated on CDM plates with 1% maltose and 50 ng/ml rifampin (Duchefa Biochemie, Haarlem, the Netherlands). The total viable count for each culture was also determined by nonselective plating on CDM agar with 1% maltose. The plates were incubated anaerobically at 30°C for 5 days, after which the colonies were counted. Chromosomal DNA was isolated from individual rifampin-resistant colonies obtained either after induced expression of tr-hexA [NZ1807(pNZ1814); 35 colonies] or spontaneously [NZ1807(pNZ8154); 31 colonies].
Garibyan et al. (17) determined the region in the E. coli rpoB gene where the main mutations that confer resistance to the antibiotic rifampin occur. The location of this region in the L. plantarum rpoB gene was determined by alignment of the sequences of the E. coli rpoB gene (b3987; accession no. NC_000913; positions 4179268 to 4183296) and the L. plantarum WCFS1 rpoB gene (lp_1021; accession no. NC_004567; positions 939961 to 943575). The sequences were aligned using the CLUSTAL program.
The analogous region of the rpoB gene (lp_1021; bp 1200 to 1800 of the coding sequence) was amplified with Pwo polymerase using the chromosomal DNA of the selected colonies as a template and primers LK54 and LK59 and was subjected to sequence analysis using primer LK54 as a sequencing primer.
Chemical mutagenesis with NTG.
To compare the mutation frequency of a mutator gene with the mutagenic capacity of N-methyl-N′-nitro-N-nitrosoguanidine (NTG), a pregrown culture of NZ1807(pNZ8154) with an OD600 of 1.0 was centrifuged, and the cells were resuspended in an equal volume of CDM without a carbon source. Then 300 μg/ml NTG (Sigma, Zwijndrecht, the Netherlands) was added, and the culture was incubated for 1 h at 30°C. Subsequently, the cells were washed three times with CDM without a carbon source, and appropriate dilutions were plated on selective plates. The total viable count for each culture was also determined by nonselective plating on CDM agar with 1% maltose. The plates were incubated anaerobically at 30°C for 5 days, after which the colonies were counted.
Effects of environmental conditions on the mutation frequency.
To determine whether environmental conditions modulate the mutation frequency of L. plantarum NZ1807(pNZ8154), the number of MelA-positive revertant colonies was determined as described above following exposure of the strain to several stress conditions. For all conditions the cultures were washed three times with CDM without a carbon source, and appropriate dilutions were plated after addition of 108 CFU of the melA mutant strain NZ7140 on selective CDM agar plates with 1% melibiose and 0.004% BCP. The total viable count for each culture was also determined by nonselective plating on CDM agar with 1% maltose. The plates were incubated anaerobically at 30°C for 5 days, after which the colonies were counted.
(i) Temperature.
A pregrown culture (OD600, 1.0) was inoculated (1%) into CDM with 1% maltose and incubated at 25°C, 30°C, 37°C and 42°C. After 16 h of growth the cultures were washed and plated.
(ii) High salt concentrations.
A pregrown culture was inoculated (1%) into CDM with and without 0.8 M NaCl and incubated at 30°C. After 24 h of growth the cultures were washed and plated.
(iii) Starvation.
A pregrown culture was inoculated (1%) into CDM with 0.2% maltose. After 4 days of incubation at 30°C, the culture was washed and plated.
(iv) Stationary phase.
A pregrown culture was inoculated (1%) into CDM with 1% maltose. After 4 days of incubation at 30°C, the culture was washed and plated.
(v) Low pH.
A pregrown culture was inoculated (1%) into CDM (pH 3.9) and into CDM (pH 5.6) and incubated at 30°C. After 24 h of growth the cultures were washed and plated.
(vi) Hydrogen peroxide.
A pregrown culture in CDM with 1% maltose (OD600, 1.0) was incubated with hydrogen peroxide (H2O2) at concentrations ranging from 0 to 6 mM for 30 min at 30°C. Subsequently, the cells were washed and plated.
(vii) Cross-protection.
A pregrown culture was inoculated (2%) into CDM with 1% maltose and incubated at 42°C. After 16 h of growth at 42°C, the OD600 of the culture was approximately 1.0, and culture was then incubated with H2O2 at concentrations ranging from 0 to 6 mM for 30 min at 30°C. Subsequently, the cells were washed and plated.
RESULTS
Mutator reporter system.
A set of L. plantarum mutator reporter strains suitable for qualitative and quantitative analysis of low-frequency mutation events was developed. In these strains the unique chromosomal α-galactosidase-encoding gene, melA, was replaced by a mutated gene under the control of the strong constitutive L. lactis usp45 promoter. A versatile set of reporter constructs was obtained in which the melA gene is not functional due to a disruptive mutation (see Materials and Methods and Tables 1 and 2) and can be reactivated by a single mutation event, either a base substitution or single-base frameshift. The first six mutants were created by mutation of the region of the melA gene that encodes the catalytic site of the cognate enzyme MelA. The location of the mutation in the seventh mutant was chosen because of the possibility of creating a stretch of G residues at this locus without changing the amino acid sequence of the encoded (wild-type) protein (Table 2). In each of the mutants a specific 1-bp mutation can result in a melA gene that encodes a functional MelA enzyme, and thus this versatile system should allow growth-based selection of such revertants on the basis of their capacity to grow on media that contain melibiose as a sole carbon source, while nonselective analysis of revertants is possible by identification of blue colonies on nonselective media containing the chromogenic MelA substrate X-α-Gal.
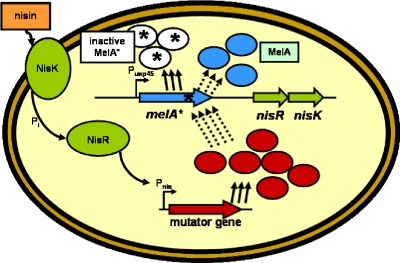
Moreover, a low-copy-number vector carrying the pAMβ1 replicon (45) was constructed, in which the nisA promoter was introduced, allowing controlled overexpression of target genes (12). This vector was used for analysis of mutation frequencies as a result of overexpression of potential mutator genes (Fig. 1).
FIG. 1.
Mutator reporter system. Mutated melA, nisR, and nisK genes are present in the genome, and the possible mutator gene (or chromosomal fragment) is present in the plasmid. Controlled expression of the mutator gene may enhance recovery of the MelA function by increasing the frequency of random mutations.
Development of a conditional mutator strain.
Yang et al. (51) identified 15 genes in E. coli which result in an elevated mutation frequency when they are overexpressed (mutator phenotype). Among these genes were a partially deleted mutS gene that generates an N-terminal truncated form of the methyl-directed mismatch repair protein MutS (approximately 940-fold increase in Lac+ revertant frequency and 5-fold increase in rifampin resistance) and the dinB gene encoding error-prone DNA polymerase IV (DinB; approximately 35-fold increase in Lac+ revertant frequency and 6-fold increase in rifampin resistance) (51). Homologs of the E. coli mutS and dinB genes were identified in the L. plantarum WCFS1 genome. The dinP gene (lp_2280) of L. plantarum WCFS1 was identified as a homolog of the E. coli dinB gene, and the lp_2298 gene of L. plantarum WCFS1 (referred to here as hexA) was identified as a homolog of the E. coli mutS gene.
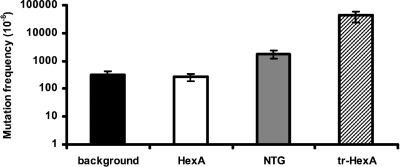
Based on an alignment of the E. coli mutS gene and the L. plantarum hexA gene, a truncated form of hexA (tr-hexA) was constructed. The N-terminal part of hexA encoding the sequence from amino acid 2 to amino acid 94 was deleted. Next, the dinP, hexA, and tr-hexA genes were overexpressed in the seven mutation reporter strains by means of the NICE system (33), and mutation frequencies were determined. Incubation with the known chemical mutagen NTG was included for comparison (Fig. 2). Before the effects of mutator genes and NTG on the mutation frequency were determined, the background revertant levels were measured. The background revertant level of L. plantarum reporter strain NZ1807 (G-stretch reporter strain) was 1 revertant per 106 cells. The other L. plantarum reporter strains could not be used to determine the mutator frequency efficiently, because the background revertant levels were below the detection limit, which was 1 revertant per 1011 cells. Therefore, the G-stretch reporter strain was used in all subsequent experiments.
FIG. 2.
Mutation frequency. L. plantarum strain NZ1807 was used as a G-stretch reporter strain. The mutation frequency is the number of revertants per 108 CFU. The background mutation frequency is the mutation frequency at 30°C with the empty vector pNZ8154. The effect of incubation with 300 μg/ml NTG for 1 h at 30°C on the mutation frequency was also determined with the empty vector pNZ8154. Plasmids pNZ1813 and pNZ1814 were used to determine the effects of HexA and tr-HexA, respectively, on the mutation frequency.
DinP and HexA did not have an effect on the mutation frequency compared to the background mutation frequency. Incubation with NTG resulted in an approximately 5-fold increase in the mutation frequency, and overexpression of the truncated hexA gene resulted in a >100-fold increase (Fig. 2) (data for DinP are not shown).
Experiments with overexpression of a hexA gene in which a larger fragment (encoding 282 amino acids) was deleted, also generating an N-terminally truncated form of HexA, did not result in an increased mutation frequency like that observed with tr-HexA (data not shown). Although overexpression by means of the NICE system is known to provide reliable and stable levels of expression in several Gram-positive bacteria, the possibility that differences in the expression levels between the genes examined could have occurred cannot be ruled out completely (32, 33).
Mutation spectrum of tr-HexA.
The truncated form of HexA resulted in a >100-fold increase in the mutation frequency, as determined with the G-stretch reporter strain. The mutated melA gene in this mutation reporter strain can be reactivated only by a single-base frameshift mutation. To determine if tr-HexA is also capable of inducing base substitution mutations, rifampin resistance experiments were conducted (16, 34).
Garibyan et al. (17) determined the region in the E. coli rpoB gene where the main set of mutations that result in resistance to the antibiotic rifampin occur. In their E. coli study all sequenced mutations were base pair substitutions. The location of this region in the L. plantarum rpoB gene was determined by alignment of the sequences of the E. coli rpoB gene and the L. plantarum WCFS1 rpoB gene. This region of the rpoB gene (bp 1200 to bp 1800 of the coding region) was sequenced for 35 individual rifampin-resistant colonies, which were obtained after overexpression of the tr-HexA strain NZ1807(pNZ1814), and 31 spontaneous rifampin-resistant colonies. Subsequently, the sequences were analyzed to ascertain the mutations responsible for the resistance to rifampin. Table 3 shows the mutations, their positions, the amino acid changes, and the distribution of the mutations.
TABLE 3.
Distribution of mutations in L. plantarum rpoB gene
| Site (bp) | Amino acid change | Base pair change | No. of colonies with mutation |
|
|---|---|---|---|---|
| Wild type (spontaneous) | tr-HexA | |||
| 1414-1422 | 9-bp deletion | 3 | 1 | |
| 1417-1425 | 9-bp deletion | 8 | 0 | |
| 1430 | Q477R | A·T → G·C | 0 | 13 |
| 1438 | D480Y | G·C → T·A | 12 | 0 |
| 1468 | H490Y | G·C → A·T | 3 | 6 |
| 1469 | H490R | A·T → G·C | 0 | 1 |
| 1478 | R493H | G·C → A·T | 3 | 5 |
| 1484 | S495Y | G·C → A·T | 2 | 9 |
| Total | 31 | 35 | ||
Overexpression of tr-HexA resulted in 10-fold more rifampin-resistant mutants. Deletions and base substitutions were observed for both the spontaneous and tr-HexA expression-induced mutants, although the distributions of the mutations were different. Most strikingly, the most abundant mutation observed in the revertants obtained after tr-hexA induction was not observed at all in the spontaneous mutant group and vice versa (Table 3). The rifampin resistance experiments suggest that tr-HexA has a clear preference for G·C → A·T and A·T → G·C transitions, while it does not stimulate G·C → T·A transitions, which appeared to occur only in the spontaneous mutant collection.
Effect of environmental conditions on the mutation frequency.
Next, the effect of environmental stress conditions on mutation frequencies was assessed. For this analysis the G-stretch reporter strain was exposed to several stress conditions that were chosen on the basis of their relevance for fermentation processes. Subsequently, the MelA-positive revertant frequency was determined.
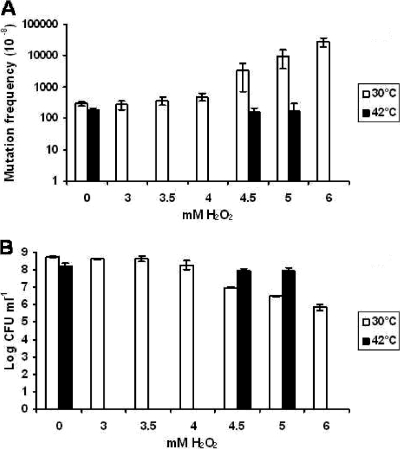
Growth at temperatures ranging from 25°C to 42°C (the optimum growth temperature for L. plantarum is 30°C, and no growth was observed at 45°C), under low-pH conditions, with high salt concentrations (NaCl), or under starvation conditions did not significantly alter the mutation frequency. Nor did stationary-phase incubation (72 h) have an effect on the mutation frequency. However, incubation with high levels (>4 mM) of hydrogen peroxide did result in a significant increase in the mutation frequency (Fig. 3A). Incubation for 30 min in a buffer containing 6 mM H2O2 resulted in a 100-fold increase in the mutation frequency, which was comparable to the effect of overexpression of tr-hexA. However, this increase coincided with a nearly 1,000-fold decrease in viability (Fig. 3B). These results indicate that the number of revertants per survivor and therefore the actual mutation frequency are increased dramatically by peroxide treatment, whereas the total number of revertants that could be recovered decreased substantially.
FIG. 3.
Effect of hydrogen peroxide and cross-protection. L. plantarum NZ1807(pNZ8154) grown at either 30°C or 42°C was incubated with hydrogen peroxide (0 to 6 mM, 30 min at 30°C), after which the mutation frequency (A) and viability (B) were determined. The positive effects of adaptation to 42°C (cross-protection) on the mutation frequency (A) and viability (B) when cells were exposed to high levels of hydrogen peroxide are shown.
Cross-protection.
De Angelis et al. (11) have shown that the viability of heat-adapted L. plantarum cells improves when they are exposed to stress conditions (e.g., heat shock). To determine if heat-adapted cells exhibit a similar viability response when they are exposed to hydrogen peroxide and to determine the effect of hydrogen peroxide on the mutation frequency, the L. plantarum G-stretch reporter strain was grown at 42°C and subsequently exposed to high levels of hydrogen peroxide.
When L. plantarum was adapted to 42°C, there was an approximately 100-fold-smaller decrease in viability upon exposure to hydrogen peroxide at a concentration of 5 mM (Fig. 3B). The effect of adaptation to 42°C on the mutation frequency was also dramatic. Incubation of the heat-adapted cells with 5 mM H2O2 resulted in a mutation frequency similar to the original background mutation frequency (at 30°C with no H2O2). This corresponds to a 50-fold decrease in the mutation frequency compared to the mutation frequency of non-heat-adapted cells exposed to 5 mM H2O2 (Fig. 3A). These results indicate that so-called cross-protection (11) not only is reflected by survival rates but also extends to the capacity of cells to prevent or counteract DNA damage, including increases in the mutation frequency due to environmental conditions.
DISCUSSION
A mutator reporter system for L. plantarum that can be used for accurate quantitative analysis of the mutation frequency was developed and used to assess the frequencies of spontaneous mutations, as well as the frequencies of mutations that are increased by genetic or environmental intervention. Unfortunately, only the G-stretch reporter strain could be used in this study. The other L. plantarum reporter strains, which would have enabled qualitative analysis of the mutations, could not be used to determine the mutator frequency efficiently, because the background revertant levels were below the detection limit, which was 1 revertant per 1011 cells. Nevertheless, these reporter strains could potentially be used in work on alternative mutator genes.
The L. plantarum hexA gene displays sequence homology with the mutS gene of E. coli, a truncated form of which was shown to increase the mutation frequency in this host (51). Conditional expression of the analogous truncated form of the hexA gene product in L. plantarum supported the hypothesis that this protein has a similar mutator role in this Gram-positive host. The increase in the mutation frequency due to tr-HexA was 20-fold greater than the increase resulting from incubation with the known chemical mutagen NTG. NTG induces a relatively wide spectrum of mutations by alkylating purines and pyrimidines. Most of the mutations induced by NTG are G·C → A·T transitions, but deletions and frameshifts occur to a lesser extent (24, 38). The mutation frequencies were determined with the G-stretch reporter strain, and the mutated melA gene in this reporter strain can be reactivated only by a single-base frameshift mutation. Therefore, the preference of NTG for G·C → A·T transitions could explain the lower mutation frequency compared to the frequency obtained with tr-HexA expression. However, the rifampin resistance experiments indicated that tr-HexA also has a preference for G·C → A·T and A·T → G·C transitions, which differs from the findings for its homolog in E. coli. Yang et al. determined that the truncated MutS protein of E. coli was a strong mutator with respect to frameshifts and a moderate mutator for base substitutions. It was also determined that the truncated MutS of E. coli increased the mutation frequency approximately 940-fold (Lac+ revertant reporter system), which is almost 10 times greater than the effect of tr-HexA. However, tr-HexA increased the number of rifampin-resistant mutants 10-fold and increased the number of truncated MutS mutants of E. coli only 5-fold (51).
The methyl-directed mismatch repair protein MutS is involved in recognition and repair of mismatched bases and small insertion or deletions and in this way limits spontaneous mutation. In E. coli, repair is initiated when the dimeric MutS protein recognizes and binds to mismatched base pairs or an insertion or deletion consisting of up to four nucleotides. Binding of MutS to a mismatch is followed by ATP-dependent binding of a homodimer of the ATPase MutL. Binding of the endonuclease MutH to the MutS-MutL complex triggers ATP binding by MutL, which enhances the endonuclease activity of MutH. The strand specificity of mismatch repair resides in MutH, which recognizes a hemimethylated GATC sequence and cleaves only the unmethylated daughter strand. The GATC site can be more than1,000 bp from and on either side of the mismatch. Both MutS and MutL are also essential in the subsequent excision and resynthesis of the daughter strand (2, 28, 37, 50). The N-terminal truncated form of the mismatch repair protein MutS lacks the intact mismatch recognition domain (28, 50). Therefore, overexpression of the truncated form of MutS could exert negative dominance over MutS, perhaps by competing with wild-type MutS for association with MutL (51). In the N-terminally truncated form of the L. plantarum hexA gene, the homolog of the E. coli mutS gene, the mismatch recognition domain was also removed, suggesting that this gene functions as a mutator gene via a mechanism similar to that of MutS.
Overexpression of the tr-hexA gene in the reporter strains is under control of the nisin-controlled gene expression system. Hence, the expression of the tr-hexA mutator gene and, as a result, also the mutator frequency can be regulated by addition of nisin (Fig. 1) (33). Consequently, these strains exhibit a conditional mutator phenotype and could be used as conditional mutators in future studies, e.g., for rapid strain improvement based on in vivo mutagenesis combined with high-throughput function-screening assays (43).
Stress-induced mutations can participate in adaptive evolution for survival under stress conditions encountered in industrial processes, and accumulation of such mutations can contribute to instability of desired functional properties of the microbial strains used. Therefore, the effect of conditions encountered during industrial fermentation processes on the frequencies of mutation was examined using the G-stretch mutation reporter strain. While growth at different temperatures, at low pH, with high salt concentrations, or under starvation conditions did not affect the mutation frequency significantly, incubation of cells with high levels of hydrogen peroxide led to a 100-fold increase in the mutation frequency. This increase in the mutation frequency can be attributed to the direct mutagenic effect of hydrogen peroxide, which belongs to a group of chemicals known as reactive oxygen species. Hydrogen peroxide can react with metal ions like Fe2+, yielding hydroxyl radicals that can damage DNA, proteins, and membranes (15). L. plantarum has been shown to produce hydrogen peroxide as a by-product during fermentation, and the concentration depends on the level of aeration during fermentation but can be up to 9 mM (4, 36, 40, 52). Many other lactobacilli that could be present with L. plantarum in an industrial process, such as Lactobacillus bulgaricus subsp. delbrueckii, are also known to produce hydrogen peroxide (30, 49, 52). Moreover, oxidative stress could also be encountered during processing due to the processing conditions used. Notably, the hydrogen peroxide effect on the mutation frequency in L. plantarum could be counteracted almost completely by adaptation of the cells to 42°C prior to exposure to hydrogen peroxide. Previously, heat adaptation has been shown to provide cross-protection in L. plantarum cells against subsequent stress challenges, and the effects include increased viability upon exposure to extreme heat shock (72°C for 90 s) and improved growth at pH 5 and in the presence of high salt concentrations (up to 6% NaCl) (11). Our findings expand the cross-protective effects of heat adaptation to protection against the specific mutagenic challenge resulting from exposure to hydrogen peroxide.
It is well established that bacterial stress responses rely on coordinated expression of genes that alter different cellular processes, like cell division, DNA metabolism, housekeeping, membrane composition, and transport (46). Integration of these stress responses is accomplished by networks of regulators which allow the cell to react to various and complex environmental shifts (46). Since stress defenses are coordinated by such integrated regulation systems, the general stress response to the mild heat shock when organisms are grown at 42°C results in cells that are protected better against high levels of hydrogen peroxide.
These results indicate that only a few stress conditions that may be encountered by L. plantarum during industrial fermentation processes appear to have a strong mutation frequency-enhancing effect (only one of the stress conditions tested had an effect). Nevertheless, specific conditions (e.g., exposure to oxidative stress conditions) may still negatively influence strain stability by increasing the random mutation frequency. However, cultures may (potentially) be protected against such detrimental consequences by specific pretreatments that harness the protective capacity of the general stress response networks to prevent or counteract the consequences of environmentally induced increased mutation frequencies. Thus, such pretreatments can be employed in industrial production to enhance the robustness of a strain, both through increasing its survival capacity under harmful conditions (classical cross-protection) and by increasing its genetic stability.
Supplementary Material
Acknowledgments
We thank Herwig Bachmann and Jolanda Lambert for providing plasmids pNZ5516 and pNZ5340, respectively.
Footnotes
Published ahead of print on 28 December 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abremski, K., R. Hoess, and N. Sternberg. 1983. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell 32:1301-1311. [DOI] [PubMed] [Google Scholar]
- 2.Acharya, S., P. L. Foster, P. Brooks, and R. Fishel. 2003. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell 12:233-246. [DOI] [PubMed] [Google Scholar]
- 3.Ahrné, S., S. Nobaek, B. Jeppsson, I. Adlerberth, A. E. Wold, and G. Molin. 1998. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Appl. Microbiol. 85:88-94. [DOI] [PubMed] [Google Scholar]
- 4.Archibald, F. S., and I. Fridovich. 1981. Manganese and defenses against oxygen toxicity in Lactobacillus plantarum. J. Bacteriol. 145:442-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachmann, H., M. Kleerebezem, and J. E. T. van Hylckama Vlieg. 2008. High-throughput identification and validation of in situ-expressed genes of Lactococcus lactis. Appl. Environ. Microbiol. 74:4727-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard, P., P. Gabant, E. M. Bahassi, and M. Couturier. 1994. Positive-selection vectors using the F plasmid ccdB killer gene. Gene 148:71-74. [DOI] [PubMed] [Google Scholar]
- 7.Bilhère, E., P. M. Lucas, O. Claisse, and A. Lonvaud-Funel. 2009. Multilocus sequence typing of Oenococcus oeni: detection of two subpopulations shaped by intergenic recombination. Appl. Environ. Microbiol. 75:1291-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjedov, I., O. Tenaillon, B. Gerard, V. Souza, E. Denamur, M. Radman, F. Taddei, and I. Matic. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404-1409. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham-Rundles, S., S. Ahrné, S. Bengmark, R. Johann-Liang, F. Marshall, L. Metakis, C. Califano, A.-M. Dunn, C. Grassey, G. Hinds, and J. Cervia. 2000. Probiotics and immune response. Am. J. Gastroenterol. 95:S22-S25. [DOI] [PubMed] [Google Scholar]
- 11.De Angelis, M., R. Di Cagno, C. Huet, C. Crecchio, P. F. Fox, and M. Gobbetti. 2004. Heat shock response in Lactobacillus plantarum. Appl. Environ. Microbiol. 70:1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Visser, J. A., A. D. Akkermans, R. F. Hoekstra, and W. M. de Vos. 2004. Insertion-sequence-mediated mutations isolated during adaptation to growth and starvation in Lactococcus lactis. Genetics 168:1145-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vries, M. C., E. E. Vaughan, M. Kleerebezem, and W. M. de Vos. 2006. Lactobacillus plantarum—survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 16:1018-1028. [Google Scholar]
- 15.Fridovich, I. 1978. The biology of oxygen radicals. Science 201:875-880. [DOI] [PubMed] [Google Scholar]
- 16.Galan, J. C. M. Tato, M. R. Baquero, C. Turrientes, F. Baquero, and J. L. Martinez. 2004. Fosfomycin and rifampin disk diffusion tests for detection of Escherichia coli mutator strains. J. Clin. Microbiol. 42:4310-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garibyan, L., T. Huang, M. Kim, E. Wolff, A. Nguyen, T. Nguyen, A. Diep, K. Hu, A. Iverson, H. Yang, and J. H. Miller. 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2:593-608. [DOI] [PubMed] [Google Scholar]
- 18.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giraud, A., I. Matic, O. Tenaillon, A. Clara, M. Radman, M. Fons, and F. Taddei. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606-2608. [DOI] [PubMed] [Google Scholar]
- 20.Giraud, A., M. Radman, I. Matic, and F. Taddei. 2001. The rise and fall of mutator bacteria. Curr. Opin. Microbiol. 4:582-585. [DOI] [PubMed] [Google Scholar]
- 21.Hastings, P. J., A. Slack, J. F. Petrosino, and S. M. Rosenberg. 2004. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2:e399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoess, R. H., and K. Abremski. 1985. Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J. Mol. Biol. 181:351-362. [DOI] [PubMed] [Google Scholar]
- 23.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 24.Kim, M., E. Wolff, T. Huang, L. Garibyan, A. M. Earl, J. R. Battista, and J. H. Miller. 2004. Developing a genetic system in Deinococcus radiodurans for analyzing mutations. Genetics 166:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U. S. A. 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 27.Lambert, J. M., R. S. Bongers, and M. Kleerebezem. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 73:1126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamers, M. H., A. Perrakis, J. H. Enzlin, H. H. Winterwerp, N. de Wind, and T. K. Sixma. 2000. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature 407:711-717. [DOI] [PubMed] [Google Scholar]
- 29.Marcobal, A. M., D. A. Sela, Y. I. Wolf, K. S. Makarova, and D. A. Mills. 2008. Role of hypermutability in the evolution of the genus Oenococcus. J. Bacteriol. 190:564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marty-Teysset, C., F. de la Torre, and J. Garel. 2000. Increased production of hydrogen peroxide by Lactobacillus delbrueckii subsp. bulgaricus upon aeration: involvement of an NADH oxidase in oxidative stress. Appl. Environ. Microbiol. 66:262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merry, R. J., M. S. Dhanoa, and M. K. Theodorou. 1995. Use of freshly cultured lactic acid bacteria as silage inoculants. Grass Forage Sci. 50:112-123. [Google Scholar]
- 32.Mierau, I., and M. Kleerebezem. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705-717. [DOI] [PubMed] [Google Scholar]
- 33.Mierau, I., K. Olieman, J. Mond, and E. J. Smid. 2005. Optimization of the Lactococcus lactis nisin-controlled gene expression system NICE for industrial applications. Microb. Cell Fact. 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. H., P. Funchain, W. Clendenin, T. Huang, A. Nguyen, E. Wolff, A. Yeung, J. H. Chiang, L. Garibyan, M. M. Slupska, and H. Yang. 2002. Escherichia coli strains (ndk) lacking nucleoside diphosphate kinase are powerful mutators for base substitutions and frameshifts in mismatch-repair-deficient strains. Genetics 162:5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molin, G. 2001. Probiotics in foods not containing milk or milk constituents, with special reference to Lactobacillus plantarum 299v. Am. J. Clin. Nutr. 73:380S-385. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, M. G., and S. Condon. 1984. Comparison of aerobic and anaerobic growth of Lactobacillus plantarum in a glucose medium. Arch. Microbiol. 138:49-53. [Google Scholar]
- 37.Obmolova, G., C. Ban, P. Hsieh, and W. Yang. 2000. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature 407:703-710. [DOI] [PubMed] [Google Scholar]
- 38.Ohnishi, J., H. Mizoguchi, S. Takeno, and M. Ikeda. 2008. Characterization of mutations induced by N-methyl-N′-nitro-N-nitrosoguanidine in an industrial Corynebacterium glutamicum strain. Mutat. Res. 649:239-244. [DOI] [PubMed] [Google Scholar]
- 39.Oneca, M., A. Irigoyen, M. Ortigosa, and P. Torre. 2003. PCR and RAPD identification of L. plantarum strains isolated from ovine milk and cheese. Geographical distribution of strains. FEMS Microbiol. Lett. 227:271-277. [DOI] [PubMed] [Google Scholar]
- 40.Quatravaux, S., F. Remize, E. Bryckaert, D. Colavizza, and J. Guzzo. 2006. Examination of Lactobacillus plantarum lactate metabolism side effects in relation to the modulation of aeration parameters. J. Appl. Microbiol. 101:903-912. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Barba, J. L., D. P. Cathcart, P. J. Warner, and R. Jimenez-Diaz. 1994. Use of Lactobacillus plantarum LPCO10, a bacteriocin producer, as a starter culture in Spanish-style green olive fermentations. Appl. Environ. Microbiol. 60:2059-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Selifonova, O., F. Valle, and V. Schellenberger. 2001. Rapid evolution of novel traits in microorganisms. Appl. Environ. Microbiol. 67:3645-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serrano, L. M., D. Molenaar, M. Wels, B. Teusink, P. A. Bron, W. M. de Vos, and E. J. Smid. 2007. Thioredoxin reductase is a key factor in the oxidative stress response of Lactobacillus plantarum WCFS1. Microb. Cell. Fact. 6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 46.Smith, J. E. 2004. Biotechnology, 4th ed. Cambridge University Press, Cambridge, United Kingdom.
- 47.Teusink, B., F. H. van Enckevort, C. Francke, A. Wiersma, A. Wegkamp, E. J. Smid, and R. J. Siezen. 2005. In silico reconstruction of the metabolic pathways of Lactobacillus plantarum: comparing predictions of nutrient requirements with those from growth experiments. Appl. Environ. Microbiol. 71:7253-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Asseldonk M., G. Rutten, M. Oteman, R. J. Siezen, W. M. de Vos, and G. Simons. 1990. Cloning, expression in Escherichia coli and characterization of usp45, a gene encoding a highly secreted protein from Lactococcus lactis MG1363. Gene 95:155-160. [DOI] [PubMed] [Google Scholar]
- 49.Van de Guchte, M., P. Serror, C. Chervaux, T. Smokvina, S. Ehrlich, and E. Maguin. 2002. Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]
- 50.Wu, T. H., and M. G. Marinus. 1999. Deletion mutation analysis of the mutS gene in Escherichia coli. J. Biol. Chem. 274:5948-5952. [DOI] [PubMed] [Google Scholar]
- 51.Yang, H., E. Wolff, M. Kim, A. Diep, and J. H. Miller. 2004. Identification of mutator genes and mutational pathways in Escherichia coli using a multicopy cloning approach. Mol. Microbiol. 53:283-295. [DOI] [PubMed] [Google Scholar]
- 52.Zalán, Z., E. Németh, A. Baráth, and A. Halász. 2005. Influence of growth medium on hydrogen peroxide and bacteriocin production of Lactobacillus strains. Food Technol. Biotechnol. 43:219-225. [Google Scholar]
- 53.Zelder, O., and B. Hauer. 2000. Environmentally directed mutations and their impact on industrial biotransformation and fermentation processes. Curr. Opin. Microbiol. 3:248-251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.