Abstract
Soil is exposed to hydrogen when symbiotic rhizobia in legume root nodules cannot recycle the hydrogen that is generated during nitrogen fixation. The hydrogen emitted is most likely taken up by free-living soil bacteria that use hydrogen as an energy source, though the bacteria that do this in situ remain unclear. In this study, we investigated the effect of hydrogen exposure on the bacteria of two different soils in a microcosm setup designed to simulate hydrogen-emitting root nodules. Although the size and overall composition of the soil bacterial community did not significantly alter after hydrogen exposure, one ribotype increased in relative abundance within each soil. This single-ribotype shift was identified by generating multiple terminal restriction fragment length polymorphism (T-RFLP) profiles of 16S rRNA genes from each soil sample, with gene sequence confirmation to identify terminal restriction fragments. The increased abundance of a single ribotype after hydrogen exposure, within an otherwise similar community, was found in replicate samples taken from each microcosm and was reproducible across replicate experiments. Similarly, only one member of the soil bacterial community increased in abundance in response to hydrogen exposure in soil surrounding the root nodules of field-grown soybean (Glycine max). The ribotypes that increased after hydrogen exposure in each soil system tested were all from known hydrogen-oxidizing lineages within the order Actinomycetales. We suggest that soil actinomycetes are important utilizers of hydrogen at relevant concentrations in soil and could be key contributors to soil's function as a sink in the global hydrogen cycle.
Soil is the major sink in the global hydrogen cycle and accounts for approximately 75 to 80% of uptake from the atmosphere (7, 11). Soil is such a strong sink that the atmospheric mixing ratio of molecular hydrogen, H2, is hemispherically asymmetric because of the greater landmass in the Northern Hemisphere (11). Many nitrogen-fixing bacteria that form symbiotic relationships with legume plants cannot recycle the H2 that is generated during N2 fixation (2, 13). Most of the H2 emitted from legume root nodules is taken up by the surrounding soil, within a few centimeters of the nodule surface, and is not released to the atmosphere (20). Although the H2 emitted by the rhizobial symbionts costs the legume approximately 5% of its daily photosynthate and “represents a significant investment by the plant” (9), there is growing evidence to suggest that soil exposed to H2 is beneficial to plant growth, separate from the benefits derived from N2 fixation (8, 10, 28). Previously, La Favre and Focht have hypothesized that “the hydrogen which is evolved during N2 fixation represents an additional energy input into the plant-soil ecosystem… since metabolism of H2 by chemolithotrophic bacteria results in an input of fixed carbon to the system” (20). A number of studies have found that when H2 is taken up by soil, net CO2 fixation occurs at the rate of 7 to 8 nmol CO2 per g of soil per h (22, 34). For a legume fixing 200 kg of atmospheric N2 per hectare, over 200,000 liters of H2 could be released into the legume's rhizosphere over the duration of the growing season and CO2 fixation could result in an extra 25 kg of soil carbon fixed per hectare (9, 10, 28).
Many bacteria isolated from soil are able to utilize H2 as an energy source (2, 5-7, 21), and free-living bacteria are most likely responsible for the H2 uptake observed by soil surrounding legume roots (22). Adding a bacterial energy source, such as H2, could affect the microbial population size, as has been observed previously (34), but more specific shifts within the bacterial community may occur if just the microorganisms able to utilize the energy source multiply. Their activity could also have downstream consequences specifically on other members of the community. Most H2-oxidizing cultures have required enrichment with concentrations of H2 that are not environmentally relevant and therefore cannot be assumed to be carrying out H2 oxidation at much lower, naturally occurring concentrations (5-7). Recent surveys of microbes present in soil samples, via their nucleic acids, have revealed many novel bacterial inhabitants that have been little studied and thus may also be contributing to the repertoire of bacterial soil processes, such as H2 uptake (16). A recent study into the effect of H2 on soil bacteria focused on a few groups of H2-oxidizing, autotrophic bacteria and thus ignored many other H2 utilizers potentially present in soil (34).
There are now many ways of characterizing the entire microbial community in environmental samples, either via their entire genomic content, though metagenomic analysis of soil is difficult at present, or via analysis of the lineages present according to 16S rRNA gene sequences, or ribotypes (36). A recent study comparing high-throughput pyrosequencing of 16S rRNA genes and an easily accessible profiling method, known as terminal restriction fragment length polymorphism (T-RFLP), found the simpler profiles were appropriate for comparing the dominant ribotypes in multiple samples (24). Although T-RFLP profiles only provide a simplified snapshot of the dominant members in microbial communities, compared to the deeper analyses provided by microarrays or high-throughput sequencing technologies, T-RFLP profiling is a cost-effective, reproducible, and robust method of “fingerprinting” many soil samples rapidly and efficiently (14, 24, 25, 32).
In this study, we chose to assess the dominant members of the soil bacterial community via T-RFLP profiles of ribotypes present in H2-treated and control soils to avoid a narrow focus on well-studied H2 oxidizers. We investigated the bacterial community structure in two different soils, utilizing a microcosm setup with concentrations of H2 calculated to occur in the rhizosphere of N2-fixing legumes, to determine whether common responses to H2 exposure could be predicted from soils that differ by climate, edaphic characteristics, and starting communities. Soil in microcosms has previously been shown to have similar H2 uptake properties to soil close to H2-emitting legume nodules (9), but we also complemented our plant-free microcosm work with an examination of soil collected from the root systems of field-grown soybean (Glycine max (L.) Merr.).
MATERIALS AND METHODS
Soils for the microcosm experiments.
Bulk soil (0 to 10 cm deep) for the microcosm experiments was collected from two field sites in Australia and stored in airtight containers. Soil was collected from CSIRO Plant Industry's Ginninderra Experimental Station (GES), in the Australian Capital Territory. The GES site is located 12 km northwest of Canberra (149°05′E, 35°12′S) in the temperate farming zone of the Southern Tablelands with a mean annual precipitation of 665 mm. Soil was also collected 1,700 km further north from the Queensland Department of Primary Industries' Research Station (QRS). The QRS site is located at Ayr (147°23′E, 19°37′S), 90 km north of Townsville in the Burdekin Delta in tropical north Queensland with a mean annual precipitation of 950 mm. Descriptions of the soil physical and chemical characteristics at both locations can be found in previous publications (28, 37).
Hydrogen treatment in the soil microcosms.
The soil microcosms consisted of two 1-liter polypropylene jars (10-cm diameter; Nalgene, Rochester, NY), each connected to a gas cylinder (Fig. 1). One of the gas cylinders contained 500 ppm H2 in an artificial air mixture of 360 ppm CO2 and 21% O2, balanced with N2 (BOC Gases, Tullamarine, Victoria, Australia). The other gas cylinder contained just the artificial air mixture of 360 ppm CO2 and 21% O2, balanced with N2 (BOC Gases). Gas flowed via a humidifier into the bottom of the microcosms through Tygon tubing (6-mm outer diameter, 3-mm inner diameter). Three pieces of fine-mesh screen (made from thin, flexible black plastic, with 35 regular rectangular holes cm−2) created a layer 2 cm above the gas inlet in the microcosms. At the beginning of each microcosm experiment, soil was sieved through a clean brass sieve with 2-mm aperture size (Endecotts, London, United Kingdom) and placed into the two microcosms, on top of the fine-mesh layer with equivalent depth and compaction, approximately half-filling the jars. Before it was added to the two microcosms, a sample of the sieved soil, termed the “original” soil for that microcosm experiment (or time = 0 days), was frozen at −20°C until its community DNA was extracted. In every microcosm experiment, one control soil microcosm was treated with the artificial air-gas mixture (labeled the “air-treated” or “control” soil microcosm), and a second microcosm was treated with the artificial air mixture enriched with 500 ppm H2 (labeled the “H2-treated” soil microcosm). The two gas lines were opened, and the flow of gas through both soil microcosms was adjusted to approximately 45 ml min−1. The screw-top lids for the microcosms were tightly sealed, so the only gas outlet was a 5-mm hole in the top of the lid. The H2 concentrations in the microcosms were sampled daily from the outlet at the top of the jars using an H2 sensor (model S121; Qubit Systems Inc., Kingston, Ontario, Canada) interfaced with a LabPro data logger (Vernier Software, Portland, OR).
FIG. 1.
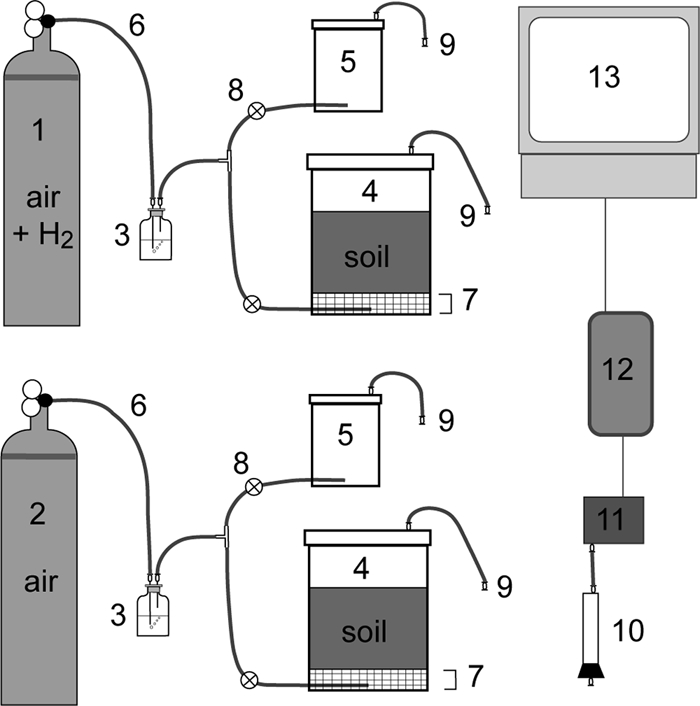
Schematic representation of the soil microcosm setup (not to scale). The experimental set up consisted of two gas cylinders (no. 1 and 2), each connected to a humidifier (no. 3), the 1-liter soil microcosm jar (no. 4), and an empty 250-ml jar (no. 5). Gas from the cylinders could flow through the humidifier and into the jars via Tygon tubing (no. 6). A plastic fine-mesh screen was positioned 2 cm above the gas inlet in the 1-liter soil microcosm (no. 7), to allow the gas from the inlet to expand evenly across the bottom of the jar before flowing through the sieved soil. The 250-ml jars (no. 5) remained empty of objects and soil during the experiments, and gas was prevented from going from the T-intersection to the smaller jar during most of the experiments by a clamp (no. 8). The gas line was directed through the empty jar once a day to measure the H2 concentration in the inlet gas line. The outlet from the lids, consisting of Tygon tubing and luer fittings (no. 9), could be connected via a column filled with magnesium perchlorate desiccant (no. 10) to an H2 sensor (no. 11). H2 readings were collected via a data logger (no. 12) and saved onto a computer (no. 13).
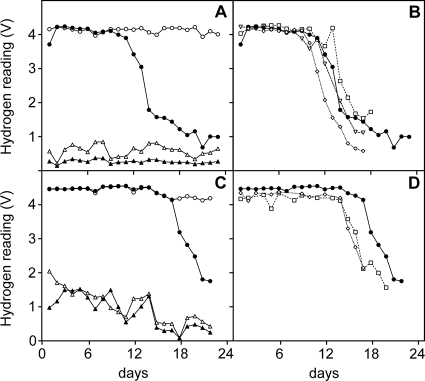
A total of four separate microcosm experiments were conducted with the GES soil, and three separate experiments were conducted with the QRS soil. The first experiment with the GES soil was a pilot experiment, and only one soil core was collected from both microcosms at the end of this experiment. At the completion of the other six microcosm experiments, triplicate soil cores from both the H2-treated and control microcosms were collected. All microcosm experiments were run for 17 to 23 days, when the majority of the H2 entering the system was taken up by the soil, which is the last point on each graph in Fig. 2. On the final day of the experiment, the three soil cores were obtained by driving 50-ml Falcon tubes into the soil from the top down to the mesh. The soil within 2 cm of the mesh layer, nearest the inlet gas layer and at the mouth of the Falcon tube after sampling, was used for all analyses. The three soil cores collected from each microcosm were analyzed separately, to allow detection of spatial heterogeneity within a microcosm. Soil samples for DNA extraction were frozen at −20°C.
FIG. 2.
Kinetics of H2 utilization in the soil microcosm experiments. All readings taken from the second experiment with the GES soil (A) and the second experiment with the QRS soil (C) are shown. The four readings shown in panels A and C are as follows: ○, artificial air enriched with 500 ppm H2 at the inlet to the microcosm; •, the H2-enriched gas mixture sampled from the outlet of the soil microcosm; ▵, the artificial air mixture at the inlet to the microcosm; and ▴, the artificial air mixture sampled from the outlet of the soil microcosm. Also shown are the H2 measurements from the outlet of the soil microcosms treated with the H2-enriched gas mixture from the four experiments using the GES soil (B) and the three experiments using QRS soil (D). The experiment numbers shown in panels B and D are as follows: ⋄, 1; •, 2 (the same as displayed in panels A and C); □, 3; and ▿, 4.
To enumerate the microbial population size, soil samples were dual stained with acridine orange and 4′,6-diamidino-2-phenylindole (DAPI) for microscopic cell counts as described previously (30), plus a real-time PCR assay of 16S rRNA genes (23) and heterotrophic plate counts on DNB media (17) were used. Soil pH was determined using 1:5 soil-water (reported) and 1:5 soil-0.01 M CaCl2 suspensions (29), and gravimetric water content was determined using standard protocols (17, 30).
Soybean field trial.
Soybeans (Glycine max (L.) Merr., cv. Leichhardt) were sown on 15 June 2005 at the Ayr Research Station, Queensland, Australia (described above). The soybean seeds were uninoculated or inoculated with either Bradyrhizobium japonicum strain CB1809, which possesses a hydrogenase uptake enzyme (designated Hup+) and recycles much of the H2 generated during N2 fixation, or strain USDA442, which lacks the hydrogenase uptake system (Hup−) and emits H2 from the root nodules (13). Full details of this field trial have been described previously (28).
Soybean root systems and the surrounding soil were excavated as an intact block from the central two rows from two replicate plots of each of the uninoculated, Hup+ and Hup− treatments on 16 August 2005 (62 days after sowing), 1 week after commencement of flowering, and again on 19 September 2005 (96 days after sowing), around the time of peak plant biomass. The blocks of soil were immediately placed in a portable refrigerator and frozen within a few hours of collection. Three samples of soil from each root system were collected and subsequently analyzed: distant soil (greater than 5 cm from the roots and nodules), proximal soil (within 5 mm of the nodules), and nodule surface (washings of the soil attached to five nodules per inoculated root system, in 1 ml 120 mM Na-PO4 buffer [pH 8.0]).
Generation and analysis of T-RFLP profiles.
DNA was extracted from frozen soil samples, PCR targeting bacterial 16S rRNA genes was performed using the primers FAM27f and 519r, and six T-RFLP profiles were generated from each soil sample as described previously (25). The restriction endonucleases (REs) used to generate six T-RFLP profiles for each soil sample were BstUI, HaeIII, HhaI, HinfI, MspI, and Sau96I (New England Biolabs, Inc., Beverly, MA). Matrices of Sorensen-Dice pairwise similarity coefficients of the T-RFLP profiles were calculated, after application of the optimal variable percentage threshold, and represented graphically as a consensus dendrogram, using the Fitch-Margoliash least-squares algorithm, as described previously (25).
The contribution of all peaks, or terminal restriction fragments (T-RFs), to the total area under the T-RFLP profiles was calculated as a percentage of the profile, on the raw profile RE data sets with no threshold applied. For each soil system and RE, the mean contribution of all T-RFs was compared in the H2-exposed soil samples and the control soil samples, to determine if any T-RFs were specific to the H2-exposed soil profiles. Two-tailed t tests were used to test the significance of T-RFs with an increased proportion of the H2-exposed soil T-RFLP profiles, compared to the control treatment for each soil system, in all six RE data sets. All Student's t tests were performed with Excel 10.1.4 software (Microsoft, Redmond, WA), employing a two-tailed test, assuming unequal variance. The P values and number of soil samples compared (n) are reported for each comparison.
Clone library construction.
Clone libraries were created by substituting the FAM27f primer with the unlabeled 27f primer in the PCR used to generate T-RFLP profiles and cloned, screened, and sequenced as described previously (26). Sequences were edited and in silico restriction digests were carried out using Sequencher version 4.1. Clone library sequences of interest were compared to GenBank database sequences using BLASTn (1).
Nucleotide sequence accession numbers.
Partial 16S rRNA sequences of the clones identified in this study have been submitted to the GenBank database under accession no. FJ878891 to FJ878936.
RESULTS
Hydrogen uptake in soil microcosms.
Soil microcosms were treated with a constant concentration and flow of H2 (500 ppm in artificial air, added at 45 ml min−1), to expose the soil nearest to the gas source to approximately 250 nmol H2 cm−3 h−1. This approximates H2 emission from legume nodules (9). Two soils collected from different climatic regions were tested for the ability to take up H2 in these soil microcosms. At least three separate microcosm experiments were conducted with each soil, and an air-treated control microcosm was run in parallel in each experiment. Exposure to H2 in the microcosms did not significantly alter the pH or moisture content of either soil, though both soils were significantly different from each other in these edaphic characteristics (GES, pH 5.73 and 20.3% moisture; QRS, pH 6.95 and 4.7% moisture).
Uptake of H2 by the soils was observed as a steady decline in the output reading of the H2 sensor located in the outlet gas stream of the soil microcosms, compared to the relatively constant reading of the H2-enriched inlet gas line (Fig. 2A and C). In all four experiments conducted with the GES soil, measurable H2 uptake began 10 to 14 days after starting the experiments (Fig. 2B). In the three experiments conducted with the QRS soil, uptake began 14 to 18 days after H2 exposure began (Fig. 2D). The microcosms were sacrificed and triplicate soil samples collected when the majority of the H2 entering the system was taken up by the soil. These times represent the last point on each graph in Fig. 2.
Comparison of the bacterial community size and composition in the soil microcosms.
The size of the microbial population was not significantly different according to microscopic cells counts of the 2 cm of soil closest to the gas source in a GES soil microcosm experiment (P = 0.23), though the counts were 12% higher in the microcosm soil treated with H2-enriched air (1.36 × 109) than those in the parallel air-treated soil (1.22 × 109) and the original soil (1.25 × 109). Heterotrophic plate counts and real-time PCR quantification of bacterial 16S rRNA genes in microcosm samples from both soils also failed to confirm any statistically significant difference between the population sizes in the H2-treated and control soils (data not shown).
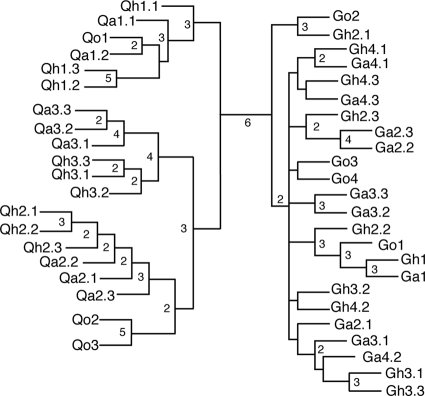
The bacterial communities in the original soils, in the 2 cm of soil closest to the H2 source in the H2-treated microcosms, and in the equivalent 2-cm zone in parallel air-treated microcosms from the seven experiments were compared using T-RFLP profiles. Profiles of PCR-amplified 16S rRNA genes were generated with six different REs for each soil sample, from the four separate experiments using the GES soil (4 original soil samples, 10 air-treated soil cores, and 10 H2-treated soil cores, for a total of 24 soil samples and 144 T-RFLP profiles), and the three separate experiments using the QRS soil (3 original soil samples, 8 air-treated soil cores, and 9 H2-treated soil cores, for a total of 20 soil samples and 120 T-RFLP profiles). The use of six different REs to generate multiple profiles from each sample was shown previously to be very important for generating levels of confidence when determining the similarity of samples (25). These confidence levels are easily represented by consensus values at branch points of a consensus dendrogram. The consensus dendrogram representing the similarity of the T-RFLP profiles in this study indicates that the bacterial communities in the two soils are very different, because they are separated with high confidence, i.e., by all six REs (25), and neither the H2 treatment nor the air treatment had a discernible effect on the dominant bacterial community composition (Fig. 3).
FIG. 3.
Consensus dendrogram representing how many of the six REs grouped microcosm samples by Sorensen-Dice pairwise similarity coefficients, as indicated by the number at the nodes defining clusters. Nodes without a number were recovered only once. The microcosm samples are indicated by their labels at the branch termini: G, GES soil; Q, QRS soil; o, original soil; a, air-treated microcosm soil; h, H2-treated microcosm soil. The first number indicates the experiment number, and the number after the decimal point indicates the sample number (out of three), if replicate cores were taken from the microcosm.
Detection of a single-member shift.
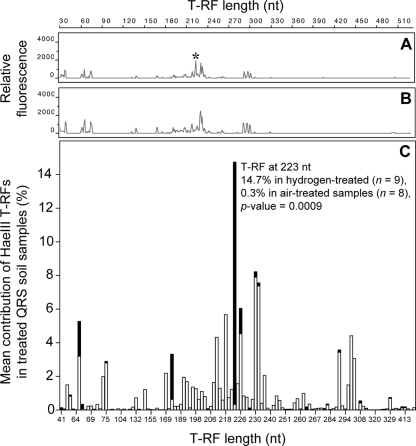
Visual inspection of the T-RFLP profiles confirmed that profiles generated from H2-treated microcosm soil samples were almost identical to profiles generated from the parallel control soils. (Fig. 4A and B show the T-RFLP profiles generated with the RE HaeIII from parallel air-treated and H2-treated QRS soil microcosm samples.) To investigate if there were subtle differences between treatments, the mean contribution of all T-RFs to the profiles was calculated from the replicate soil cores and compared for each treatment and RE. (The mean contribution of T-RFs from H2-treated and air-treated QRS soil T-RFLP profiles from the HaeIII data set are represented graphically in Fig. 4C.) The only statistically significant difference in the bacterial communities after H2 exposure was the increase of a single T-RF within each profile from each soil (Table 1). Each of the six REs used to generate the T-RFLP profiles resulted in the identification of a different T-RF because the different recognition cut sites of the six REs would produce fragments of different lengths from a single ribotype, which suggests that a single bacterial ribotype increased in abundance. No other changes in abundance were reproducible across replicate experiments with either soil, though there was one other significant difference found in the three cores from one of the four replicate GES soil microcosm experiments (see below). No T-RFs could be identified as constituting a reproducibly smaller proportion of the H2-treated profiles than the air-treated and original control soils.
FIG. 4.
(A and B) T-RFLP profiles generated with the RE HaeIII, from a QRS soil microcosm experiment, showing the similarity of an H2-treated soil T-RFLP profile (A) to an air-treated soil T-RFLP profile (B). The scale bar at the top indicates the length of the T-RFs in nucleotides (nt), and the markings on the y axes of the profiles indicate relative fluorescence units. The T-RF marked with an asterisk in panel A is 223 nt long. (C) Mean contribution of each T-RF to all of the air-treated QRS soil T-RFLP profiles (n = 8) and all of the H2-treated QRS soil T-RFLP profiles (n = 9), generated with the RE HaeIII. The mean contributions of each T-RF generated from air-treated samples (white bars) are superimposed on the mean contributions of each T-RF generated from the H2-treated samples (black bars). The T-RF at 223 nt, which was a significantly greater proportion of the HaeIII T-RFLP profiles generated from the H2-treated QRS soil (Table 1), is indicated.
TABLE 1.
Mean contribution of T-RFs that constituted a significantly greater proportion of the T-RFLP profiles from the H2-treated microcosms
| RE | Result fora: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GES soil |
QRS soil |
|||||||||
| H2-specific T-RF length (nt) | % Go (n = 4) | % Ga (n = 10) | % Gh (n = 10) | P value (Ga vs Gh) | H2-specific T-RF length (nt) | % Qo (n = 3) | % Qa (n = 8) | % Qh (n = 9) | P value (Qa vs Qh) | |
| BstUI | 223 | 3.7 | 4.1 | 12.0 | 0.0011 | 220 | 2.5 | 3.5 | 16.7 | 0.0014 |
| HaeIII | 65 | 3.6 | 4.1 | 8.5 | 0.0091 | 223b | 0.6 | 0.3 | 14.7 | 0.0009 |
| HhaI | 358 | 2.6 | 6.6 | 14.6 | 0.0112 | 177 | 0.03 | 0.02 | 9.2 | 0.0002 |
| HinfI | 323 | 17.8 | 16.6 | 26.8 | 0.0169 | 166 | 0.1 | 0.08 | 5.5 | 0.0010 |
| MspI | 140 | 2.1 | 3.0 | 6.0 | 0.0538 | 158 | 2.3 | 2.0 | 10.7 | 0.0044 |
| Sau96I | 64 | 2.1 | 3.0 | 6.1 | 0.0428 | 314 | 0 | 0 | 13.1 | 0.0017 |
The contributions are shown by soil type (where G represents GES soil microcosms and Q represents QRS soil microcosms) and treatment (where o represents original soil, a represents air-treated soil, and h represents H2-treated soil). The results of two-tailed t tests between the T-RFs' contributions to the treated samples from each soil are presented. n indicates the number of T-RFLP profiles analyzed for each soil and treatment for all six REs. In each case, the percentage shown represents the mean contribution of the T-RF to the total area of the T-RFLP profiles.
Comparisons of the HaeIII T-RFLP profiles generated from QRS soil are shown in Fig. 4.
Identification of the ribotype that increased with hydrogen exposure in each soil.
To identify the ribotype represented by the six H2-specific T-RFs in the two soils, clone libraries of 16S rRNA genes were generated from the H2-treated soil samples for each soil type. According to the in silico restriction analysis, 13 (19%) of the 68 cloned 16S rRNA genes from the H2-treated GES soil sample would yield T-RF sizes within 1 nucleotide (nt) of all six H2-specific T-RFs identified in the profiles generated from the H2-treated GES soil (Table 1). Therefore, the single T-RF that increased in each of the six profiles generated using different REs was from one or a very few similar 16S rRNA gene sequences, or ribotypes. According to BLASTn, these sequences had 98% identity to the 16S rRNA gene of Pseudonocardia petroleophila (GenBank accession no. AJ252828). Because of its dominance in the clone library and the correlation with each of the six H2-specific T-RFs generated using the six different REs, these 13 sequences most likely represent a Pseudonocardia sp. that increased in abundance after H2 exposure in the microcosm experiments conducted using GES soil. Five other inserts sequenced in this library were representative of Pseudonocardia spp. according to BLASTn, but the in silico restriction digests only matched between two and four of the six H2-specific T-RFs identified.
Three of the 30 sequences (10%) generated from the H2-treated QRS soil sample were most similar to 16S rRNA genes of Streptomyces spp. Two of these sequences gave predicted T-RFs within 1 nt of all six H2-specific T-RFs identified in the profiles from microcosms using the QRS soil (Table 1), according to in silico restriction analysis. A third sequence had identical predicted cut sites to these two sequences, except that an additional BstUI cut site was introduced closer to the 5′ terminus at 177 nt, because of a single base substitution (A to C). The close correlation with all six H2-specific T-RFs indicates that a ribotype from the genus Streptomyces increased in response to the H2 input in three separate microcosm experiments with the QRS soil. Another three clones from this library had inserts that represented Streptomyces spp., but none of the predicted T-RFs for these clones were within 1 nt of the six H2-specific T-RFs. No other clone library sequences gave more than two predicted T-RFs within 1 nt of the H2-specific T-RFs.
A second ribotype increased in one microcosm experiment.
In one of the four GES soil microcosm experiments, both the reproducible Pseudonocardia-specific T-RF and an additional T-RF made up significantly greater proportions of the profiles generated from the three soil cores from the H2-treated microcosm compared to the three cores from the parallel control microcosm. However, the second significant T-RF was identified in only four of the six REs' data sets, with no additional T-RFs detected in the profiles generated using the REs HhaI and MspI (Table 2). A group of clone library sequences had predicted T-RFs within 1 nt of the secondary peaks for BstUI, HaeIII, HinfI, and Sau96I. None of these sequences had HhaI or MspI recognition cut sites in the region of the 16S rRNA gene sequence amplified for T-RFLP analysis, which explains why T-RFs corresponding to this ribotype did not increase in profiles generated with these REs. The sequences had 97% identity to a soil clone named TRE14 (GenBank accession no. AJ232811), which belongs to subdivision 3 of the candidate division TM7 (15). The four T-RFs representing this TM7 ribotype were either not detected or constituted only a very small proportion of the T-RFLP profiles generated from the other six microcosm experiments (Table 2).
TABLE 2.
Mean contribution of the T-RFs identified as increasing in abundance after H2 treatment in the second GES soil microcosm experiment
| RE | Result for T-RFa: |
P value (Ga2 vs Gh2) | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
Pseudonocardia specific |
Gh2 specific |
||||||||
| T-RF length (nt) | % Ga2 (n = 3) | % Gh2 (n = 3) | T-RF length (nt) | % Other Gh (n = 7) | % Go2 (n = 1) | % Ga2 (n = 3) | % Gh2 (n = 3) | ||
| BstUI | 223 | 3.1 | 14.1 | 111 | 0.2 | 0.1 | 0 | 9.0 | 0.0227 |
| HaeIII | 65 | 3.6 | 8.3 | 210 | 0.6 | 0 | 0 | 13.2 | 0.0001 |
| HhaI | 358 | 3.8 | 17.3 | ||||||
| HinfI | 323 | 9.8 | 23.1 | 155 | 0.2 | 0 | 0 | 5.8 | 0.0221 |
| MspI | 140 | 0.7 | 6.2 | ||||||
| Sau96I | 64 | 2.1 | 4.0 | 209 | 0.2 | 0 | 0 | 8.5 | 0.0098 |
The results of two-tailed t tests are reported for the contribution of the secondary T-RFs, indicative of a ribotype from the candidate phylum TM7, to the air-treated (Ga2) and H2-treated (Gh2) soil cores from the second GES soil experiment. The abbreviations used are the same as those in Table 1 and Fig. 3.
Bacterial community on soybean root nodules from a field trial.
To determine if the detection of a single-member shift in the microcosm bacterial community was a realistic simulation of natural H2 exposure, we analyzed soil collected from root systems of field-grown soybean inoculated with either H2-recycling (Hup+) or H2-emitting (Hup−) strains of rhizobia. Soil surrounding the roots of uninoculated soybean was also examined. Field measurements of H2 emission from excavated nodulated root systems undertaken during early reproduction confirmed the relative Hup status of the inoculated B. japonicum strains (7.7 and 119.5 liters H2 per hectare per hour for plants with nodules formed by CB1809 and USDA442, respectively [28]). The bacterial communities from the nodule surface, proximal soil (within 5 mm), and distant soil (greater than 5 cm from the nodules and roots) from the root systems, analyzed 62 and 96 days after sowing, did not group by either inoculant type or sampling time in a consensus dendrogram generated from the T-RFLP profiles (data not shown).
Cloned 16S rRNA gene sequences representing ribotypes from the genera Mesorhizobium, Sphingopyxis, and Rhizobium and the Bradyrhizobium inoculants all had six predicted RE cut sites, within 1 nt, that correlated with dominant T-RFs in the T-RFLP profiles generated from the nodule surface soil samples (Table 3). According to the presence of their indicative T-RFs, these ribotypes from the class Alphaproteobacteria were abundant on the surface of the field-grown soybean root nodules, regardless of inoculant or sampling time.
TABLE 3.
Mean contribution of the major T-RFs found in the soybean nodule surface samples, in each of the six data sets, for both inoculated treatments sampled at both time points
| RE | Major T-RF length (nt) | % Hup+ (n = 3) | % Hup− (n = 3) | Fold difference between Hup− and Hup+ | Phylogenetic affiliation (putative genus)a |
|---|---|---|---|---|---|
| BstUI | 93 | 7.7 | 14.1 | 1.8 | Mesorhizobium/Rhizobium |
| 95 | 9.4 | 15.1 | 1.6 | Sphingopyxis | |
| 97 | 6.2 | 11.8 | 1.9 | Bradyrhizobium | |
| 220 | 2.4 | 10.9 | 4.5 | Mycobacterium | |
| 223 | 6.6 | 6.8 | 1.0 | Kribbella | |
| HaeIII | 65 | 3.6 | 10.5 | 2.9 | Mycobacterium |
| 189 | 5.8 | 9.4 | 1.6 | Rhizobium | |
| 194 | 16.2 | 17.9 | 1.1 | Bradyrhizobium | |
| 224 | 13.9 | 11.6 | 0.8 | Mesorhizobium | |
| 226 | 23.0 | 10.5 | 0.5 | Paenibacillus | |
| 292 | 12.2 | 17.3 | 1.4 | Sphingopyxis | |
| HhaI | 59 | 2.9 | 5.4 | 1.9 | Mesorhizobium |
| 80 | 5.8 | 7.0 | 1.2 | Sphingopyxis | |
| 206 | 12.4 | 17.1 | 1.4 | ND | |
| 338 | 3.4 | 6.3 | 1.9 | Rhizobium | |
| 365 | 2.8 | 20.8 | 7.4 | Mycobacterium | |
| 472 | 14.9 | 22.1 | 1.5 | Bradyrhizobium | |
| HinfI | 99 | 2.6 | 3.3 | 1.3 | Mesorhizobium |
| 101 | 4.2 | 6.5 | 1.5 | Sphingopyxis | |
| 166 | 1.0 | 7.0 | 7.0 | Mycobacterium | |
| 321 | 7.3 | 6.8 | 0.9 | ND | |
| 472 | 13.4 | 10.9 | 0.8 | Bradyrhizobium | |
| MspI | 126 | 1.4 | 4.8 | 3.4 | Mesorhizobium/Rhizobium |
| 151 | 4.1 | 11.3 | 2.8 | Bradyrhizobium | |
| 160 | 2.9 | 3.5 | 1.2 | ND | |
| 170 | 0.3 | 9.5 | 31.7 | Mycobacterium | |
| 399 | 2.9 | 11.7 | 4.0 | ND | |
| Sau96I | 64 | 2.0 | 9.0 | 4.5 | Mycobacterium |
| 164 | 4.2 | 8.3 | 2.0 | Sphingopyxis | |
| 189 | 27.7 | 14.8 | 0.5 | Rhizobium | |
| 192 | 9.8 | 13.2 | 1.3 | Bradyrhizobium | |
| 289 | 8.2 | 9.1 | 1.1 | Mesorhizobium | |
| 291 | 3.4 | 5.6 | 1.6 | ND | |
| 317 | 5.6 | 2.7 | 0.5 | ND |
Phylogenetic affiliation by genus is given if it was able to be determined from the sequences in the parallel clone library. “ND” indicates that a phylogenetic affiliation was not able to be determined from the predicted T-RFs within 1 nt of the clone library sequences obtained.
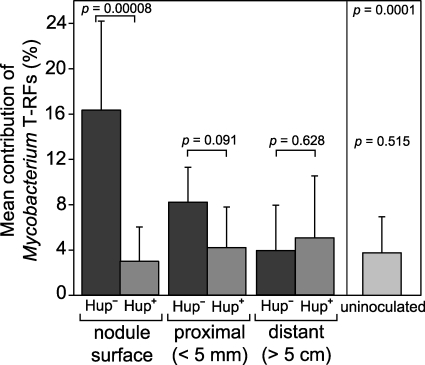
One T-RF in each of the six data sets (i.e., one per RE) made up a greater proportion (3- to 32-fold increase) of the T-RFLP profiles generated from the H2-emitting Hup− nodule surface compared to profiles generated from the control Hup+ nodules (Table 3). Those peaks that made up a greater proportion of the Hup− nodule T-RFLP profiles correlated with the six predicted T-RFs of a group of clone library sequences with 99% identity to the 16S rRNA gene of Mycobacterium cosmeticum (GenBank accession no. AY449729). No other ribotypes in the nodule surface samples, as represented by their T-RFs, constituted a consistently greater proportion of the H2-emitting Hup− nodule surface populations, at either time point across all six data sets. Significant increases in the contribution of Mycobacterium-specific T-RFs to the T-RFLP profiles were only observed in the soil collected at peak plant biomass, toward the end of growth (mean of 16.3% [n = 12], cf. mean of 1.2% [n = 6] in soil collected during the early reproductive stage). The mean proportion of T-RFs representing the Mycobacterium sp. in the bacterial community in these later soil samples was negatively correlated with distance from the Hup− nodule (Fig. 5).
FIG. 5.
Mean contribution of the Mycobacterium-specific T-RFs, calculated from all six REs, for all soybean field trial samples collected at peak plant biomass. The error bars represent 1 standard deviation, and the results from two-tailed t tests between comparable Hup− and Hup+ nodulated soybean root samples are indicated. The bar labeled “uninoculated” represents the mean contribution of the Mycobacterium-specific T-RFs from both proximal and distant soil samples of the uninoculated soybean root systems. The P values above the uninoculated mean are the results of two-tailed t tests comparing the uninoculated samples with the Hup− nodule surface samples (above) and comparison with the distant soil from both inoculated treatments (below).
DISCUSSION
Soil hydrogen uptake in the microcosm experiments.
The soil microcosm experiments were designed to simulate the flow of H2 from legume root nodules. The time required to initiate H2 uptake by both the Australian soils tested in this study was very similar to observations for Canadian and German soils exposed to similar levels of H2 in microcosms (9, 10, 34). The lag before detectable H2 uptake is consistent with a pure culture substrate utilization curve and may represent the time required to generate enough enzyme or biomass by the H2-utilizing microorganisms for uptake to be detected. Soil water content is known to influence soil H2 uptake (7, 11), which provides an explanation for the slightly longer lag before detectable uptake by the QRS soil, which had a lower water-holding capacity.
Identification of a single-member shift in diverse soil bacterial communities.
This study demonstrates the application of multiple T-RFLP profiles to confidently differentiate two very different soil communities and identify a subtle shift in complex soil bacterial communities in response to a factor, H2, that did not change the overall community structure. If there had been a general multiplication of many members of the bacterial community in response to the H2 input, then we would expect the analyses of the overall T-RFLP patterns or population size to show greater differences. Because all of the soil samples in this study were subjected to the same potential extraction and amplification biases during the generation of the T-RFLP profiles, we conclude that the differences detected are real. A high confidence value separated the two different soil types in the consensus dendrogram of the profile patterns, indicating that the soils from the two sites had different bacterial communities. Treatment with H2 did not change the overall community composition compared to that of the controls. There was, however, a statistically significant increase of a single ribotype, as represented by its T-RFs, specifically after H2 exposure in all replicate experiments. Our results strongly suggest that a single member increased in relative abundance within the bacterial community, but the other community members remained unchanged. “Sequence-aided” T-RFLP (35) is generally not encouraged on complex soil bacterial communities because a T-RF may be made up of many ribotypes. The generation of six T-RFLP profiles per soil sample allowed the increase in abundance of one bacterial ribotype to be identified by the unique “signature” of T-RFs from the various REs' recognition cut sites. The bacterial ribtoype was confirmed with sequence information obtained from the H2-treated soil. The generation of six profiles per sample also minimizes the identification of differences that are not real (a type I error).
Actinomycetes respond to hydrogen exposure.
Both ribotypes that increased significantly after H2 exposure in the replicate microcosm experiments were members of the order Actinomycetales and are closely related to known H2-oxidizing isolates. Members of the family Pseudonocardiaceae that can oxidize H2 have previously been isolated from the root nodule surface of N2-fixing alder trees (Alnus spp. [12]). A Streptomyces sp. was recently isolated that has a high-affinity uptake hydrogenase and is the first soil isolate able to utilize H2 at the very low concentrations found in the troposphere (530 ppb [6]). Although it is not conclusive from this study that the ribotypes that increased in abundance in the H2-treated samples are responsible for the H2 uptake in each of the soils, the only difference from the control microcosms was the H2 addition. The H2 uptake in the H2-treated soil microcosms (Fig. 2) is correlated with the increased abundance of the H2-specific ribotype, via the indicative T-RFs in all replicate cores from all replicate microcosm experiments (Fig. 4 and Table 1). Given that relatives of these actinomycete ribotypes are known H2-utilizing bacteria, this finding strongly suggests a growth response of these ribotypes to the H2 addition. This result could be confirmed by isolating bacteria corresponding to these ribotypes from these soils and characterizing their ability to use H2 at the concentration typically experienced in the rhizosphere of N2-fixing legumes, because the known H2-oxidizing strains from these lineages have not been tested for H2 utilization at these relatively low concentrations.
It is unclear why the TM7 ribotype increased in abundance in only one microcosm experiment. It was not a localized increase in an isolated part of the H2-treated soil microcosm, because this ribotype made up a greater proportion of the community in all three cores taken from the microcosm. TM7 is a recently recognized phylum-level lineage that is found in many environments and has thus far eluded cultivation attempts, so little is known about the metabolic capabilities of members of this group (15). The increase of a TM7 ribotype in this one H2-treated GES soil microcosm could be a consequence of the growth of the Pseudonocardia sp. or a response to its metabolites or the slightly longer incubation for that experiment which may have allowed it to grow. It is also possible that this TM7 ribotype is able to utilize H2. If only one of the REs HhaI or MspI had been used to generate T-RFLP profiles in this study, the increase of this interesting group in one microcosm experiment would have gone undetected, which again highlights the value of generating multiple T-RFLP profiles with various REs.
Similar results were obtained on field-grown soybean nodules to those obtained with the microcosms. In this case, a Mycobacterium sp. increased in abundance. The genus Mycobacterium also belongs to the order Actinomycetales and includes species (M. gordonae and M. smegmatis) that have previously demonstrated H2 oxidation in pure culture (2, 19, 27). All of the H2 emitted from Hup− nodules is likely to have been consumed within 3 to 4.5 cm of the nodule surface (20), so the proportion of T-RFs representing the Mycobacterium sp. in the soil at different distances from the Hup− nodule most likely reflects the level of H2 in the soil as it diffuses away from the Hup− nodules. This, combined with the specific enrichment of the Mycobacterium sp. in only the Hup− nodule system, suggests it has increased in direct response to H2 exposure. Measurements of N2 fixation undertaken as part of the soybean field trial used in the present study (28), and elsewhere in Australia (3), indicate that rates of N2 fixation by soybean, and consequent rates of H2 emission, generally increase toward the end of the growing season. This may explain why no H2-specific differences were detected until the plants reached peak biomass, because sufficient amounts of H2 may not have been emitted into the soil during the first 2 months of growth to elicit a detectable response. The alphaproteobacterial groups detected on all of the root systems of the field-grown soybean have previously been detected on the roots of oilseed rape (Brassica napus [18]), Lodgepole pine (Pinus contorta [4]), and maize (Zea mays [31]).
Although a previous study postulated that actinomycetes were responsible for H2 uptake in a Canadian soil based on which antibacterial treatments knocked out the H2 uptake (22), the present study represents the first identification of actinomycete ribotypes increasing in abundance with H2 addition in situ using modern microbial ecology methods. A previous investigation based on fluorescence in situ hybridization (FISH) overlooked the actinomycetes when assessing the effects of H2 on soil bacteria (34), but our results indicate that the order Actinomycetales should not be ignored when assessing soils for H2 uptake. This finding highlights the value of first conducting a broad survey of the entire community, before more specific and targeted techniques, such as FISH, are employed to confirm and quantify the presence of particular bacterial groups.
Even though actinomycetes are generally considered heterotrophs, strains of Mycobacterium and Streptomyces have previously been found to fix CO2 or contain genes coding for RuBisCo (ribulose-1,5-bisphosphate carboxylase oxygenase) (27, 33), though CO2 or soil carbon measurements were not undertaken in the present study. Determination of whether the H2 oxidizers near legume root nodules are growing autotrophically in situ could provide novel insights into the possible promotion of soil bacterial carbon sequestration by legume crops in the emerging carbon economy.
Hydrogen exposure results in a common response.
This is the first study on the effect of H2 exposure on soil that has attempted to identify a common response in the bacterial communities across various soil systems. Analysis of two very different soils in replicate microcosms produced a singular and reproducible response to H2 addition in all microcosm experiments, which was also observed in the natural system of soil surrounding legume root nodules. We focused our experimental effort on the analysis of a range of soil samples, rather than conducting an in-depth analysis of a smaller number of samples with less statistical power. The results generated from three different soil systems indicate that a single actinomycete member of the bacterial community increased in abundance after H2 exposure, which was reproducible in replicate experiments and in all cores of each H2-treated soil microcosm, which are spatially-separated from the microbial point of view. Whether the increased abundance of different microorganisms in the field trial root systems and the microcosm experiments using soil from the same field site was due to biological effects, such as rhizosphere influences, or physical factors, such as aeration and moisture content, or simply reflected the influence of soil disturbance on microbial microsites, was not able to be elucidated in this study. Determining how the edaphic and climatic conditions influence which community member in the complex soil community proliferates with H2 exposure should allow greater understanding of the constraints on this key soil process across different soil types. Further work is also required to determine how far our conclusions can be applied to other soils around the globe.
The results from the present study, plus the recent isolation of an actinomycete that is the first bacterium able to utilize H2 at tropospheric levels (6), hint at the possibility that actinomycetes are major agents of H2 oxidation at the natural levels found in soil, be it the low levels emitted from N2-fixing legume root nodules or the even lower levels from the atmosphere. Therefore, soil actinomycetes might be key contributors to soil's function as the major sink in the global H2 cycle.
Acknowledgments
This work was supported by grants from the Grains Research and Development Corporation (GRDC) and the Australian Research Council (ARC).
We thank Paul McLennan, John Brockwell, and Barry Smith (CSIRO Plant Industry) for assistance with collection of the soil and root systems used in the field; Kathryn Davis (University of Melbourne) for technical assistance in the laboratory; Angelo Zaia (Microbiological Diagnostic Unit, University of Melbourne) for assistance with the real-time PCR assay; and Melinda Ziino at the Australian Genome Research Facility for conducting the fragment analysis. We also thank the many people who read the manuscript and provided useful feedback.
Footnotes
Published ahead of print on 8 January 2010.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aragno, M., and H. G. Schlegel. 1981. The hydrogen-oxidizing bacteria, p. 865-893. In M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes, vol. 1. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 3.Bergersen, F. J., J. Brockwell, R. R. Gault, L. Morthorpe, M. B. Peoples, and G. L. Turner. 1989. Effects of available soil nitrogen and rates of inoculation on nitrogen fixation by irrigated soybeans and evaluation of δ15N methods for measurement. Aust. J. Agric. Res. 40:763-780. [Google Scholar]
- 4.Chow, M. L., C. C. Radomski, J. M. McDermott, J. Davies, and P. E. Axelrood. 2002. Molecular characterization of bacterial diversity in Lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol. Ecol. 42:347-357. [DOI] [PubMed] [Google Scholar]
- 5.Conrad, R., and W. Seiler. 1979. The role of hydrogen bacteria during the decomposition of hydrogen by soil. FEMS Microbiol. Lett. 6:143-145. [Google Scholar]
- 6.Constant, P., L. Poissant, and R. Villemur. 2008. Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H2. ISME J. 2:1066-1076. [DOI] [PubMed] [Google Scholar]
- 7.Constant, P., L. Poissant, and R. Villemur. 2009. Tropospheric H2 budget and the response of its soil uptake under the changing environment. Sci. Total Environ. 407:1809-1823. [DOI] [PubMed] [Google Scholar]
- 8.Dean, C. A., W. Sun, Z. Dong, and C. D. Caldwell. 2006. Soybean nodule hydrogen metabolism affects soil hydrogen uptake and the growth of rotation crops. Can. J. Plant Sci. 86:1355-1359. [Google Scholar]
- 9.Dong, Z., and D. B. Layzell. 2001. H2 oxidation, O2 uptake and CO2 fixation in hydrogen treated soils. Plant Soil 229:1-12. [Google Scholar]
- 10.Dong, Z., L. Wu, B. Kettlewell, C. D. Caldwell, and D. B. Layzell. 2003. Hydrogen fertilization of soils—is this a benefit of legumes in rotation? Plant Cell Environ. 26:1875-1879. [Google Scholar]
- 11.Ehhalt, D. H., and F. Rohrer. 2009. The tropospheric cycle of H2: a critical review. Tellus B 61:500-535. [Google Scholar]
- 12.Embley, T. M. 1992. The family Pseudonocardiaceae, p. 996-1027. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 1. Springer-Verlag, New York, NY. [Google Scholar]
- 13.Evans, H. J., S. A. Russell, F. J. Hanus, and T. Ruiz-Argüeso. 1988. The importance of hydrogen recycling in nitrogen fixation by legumes, p. 777-791. In R. J. Summerfield (ed.), World crops: cool season food legumes. Kluwer Academic Publishers, Boston, MA.
- 14.Fierer, N., and R. B. Jackson. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of the candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssen, P. H. 2006. Identifying dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser, O., A. Pühler, and W. Selbitschka. 2001. Phylogenetic analysis of microbial diversity in the rhizoplane of oilseed rape (Brassica napus cv. Westar) employing cultivation-dependent and cultivation-independent approaches. Microb. Ecol. 42:136-149. [DOI] [PubMed] [Google Scholar]
- 19.King, G. M. 2003. Uptake of carbon monoxide and hydrogen at environmentally relevant concentrations by mycobacteria. Appl. Environ. Microbiol. 69:7266-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Favre, J. S., and D. D. Focht. 1983. Conservation in soil of H2 liberated from N2 fixation by Hup− nodules. Appl. Environ. Microbiol. 46:304-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maimaiti, J., Y. Zhang, J. Yang, Y.-P. Cen, D. B. Layzell, M. Peoples, and Z. Dong. 2007. Isolation and characterization of hydrogen-oxidizing bacteria induced following exposure of soil to hydrogen gas and their impact on plant growth. Environ. Microbiol. 9:435-444. [DOI] [PubMed] [Google Scholar]
- 22.McLearn, N., and Z. Dong. 2002. Microbial nature of the hydrogen-oxidizing agent in hydrogen-treated soil. Biol. Fertil. Soils 35:465-469. [Google Scholar]
- 23.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 24.Orcutt, B., B. Bailey, H. Staudigel, B. M. Tebo, and K. J. Edwards. 2009. An interlaboratory comparison of 16S rRNA gene-based terminal restriction fragment length polymorphism and sequencing methods for assessing microbial diversity of seafloor basalts. Environ. Microbiol. 11:1728-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborne, C. A., G. N. Rees, Y. Bernstein, and P. H. Janssen. 2006. New threshold and confidence estimates for terminal restriction fragment length polymorphism analysis of complex communities. Appl. Environ. Microbiol. 72:1270-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborne, C. A., M. Galic, P. Sangwan, and P. H. Janssen. 2005. PCR-generated artefact from 16S rRNA gene-specific primers. FEMS Microbiol. Lett. 248:183-187. [DOI] [PubMed] [Google Scholar]
- 27.Park, S. S., and B. T. DeCicco. 1976. Hydrogenase and ribulose diphosphate carboxylase during autotrophic, heterotrophic, and mixotrophic growth of scotochromogenic mycobacteria. J. Bacteriol. 127:731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peoples, M. B., P. D. McLennan, and J. Brockwell. 2008. Hydrogen emission from nodulated soybeans [Glycine max (L.) Merr.] and consequences for the productivity of a subsequent maize (Zea mays L.) crop. Plant Soil 307:67-82. [Google Scholar]
- 29.Rayment, G. E., and F. R. Higginson. 1992. Australian laboratory handbook of soil and water chemical methods, p. 17-23. Inkata Press, Melbourne, Australia.
- 30.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 31.Sanguin, H., B. Remenant, A. Deschesne, J. Thioulouse, T. M. Vogel, X. Nesme, Y. Moënne-Loccoz, and G. L. Grundmann. 2006. Potential of a 16S rRNA-based taxonomic microarray for analyzing the rhizosphere effects of maize on Agrobacterium spp. and bacterial communities. Appl. Environ. Microbiol. 72:4302-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schütte, U. M. E., Z. Abdo, S. J. Bent, C. Shyu, C. J. Williams, J. D. Pierson, and L. J. Forney. 2008. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 80:365-380. [DOI] [PubMed] [Google Scholar]
- 33.Selesi, D., M. Schmid, and A. Hartmann. 2005. Diversity of green-like and red-like ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes (cbbL) in differently managed agricultural soils. Appl. Environ. Microbiol. 71:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein, S., D. Selesi, R. Schilling, I. Pattis, M. Schmid, and A. Hartmann. 2005. Microbial activity and bacterial composition of H2-treated soils with net CO2 fixation. Soil Biol. Biochem. 37:1938-1945. [Google Scholar]
- 35.Székely, A., R. Sipos, B. Berta, B. Vajna, C. Hajdú, and K. Márialigeti. 2009. DGGE and T-RFLP analysis of bacterial succession during mushroom compost production and sequence-aided T-RFLP profile of mature compost. Microb. Ecol. 57:522-533. [DOI] [PubMed] [Google Scholar]
- 36.Tringe, S. G., and P. Hugenholtz. 2008. A renaissance for the pioneering 16S rRNA gene. Curr. Opin. Microbiol. 11:442-446. [DOI] [PubMed] [Google Scholar]
- 37.Webster, R., and B. E. Butler. 1976. Soil classification and survey studies at Ginninderra. Aust. J. Soil Res. 14:1-24. [Google Scholar]