Abstract
Reptile Campylobacter fetus isolates and closely related strains causing human disease were characterized by multilocus sequence typing. They shared ∼90% nucleotide sequence identity with classical mammalian C. fetus, and there was evidence of recombination among members of these two groups. The reptile group represents a possible separate genomospecies capable of infecting humans.
Campylobacter fetus is a human and animal pathogen which can be divided into two subspecies: subsp. fetus and subsp. venerealis (16). C. fetus subsp. fetus has a wide host range and causes abortions in sheep and cattle; C. fetus subsp. venerealis is host restricted, being isolated specifically from the bovine genital tract, and it causes fertility problems in cattle (5). C. fetus is an opportunistic pathogen in humans, particularly affecting severely immunocompromised patients. Initially, the bacterium can cause gastroenteritis; then, bacteremia can lead to septicemia and disseminated infections (1, 8). These two subspecies of mammalian C. fetus are referred to subsequently as “classical C. fetus.”
A multilocus sequence typing (MLST) scheme has been developed for classical C. fetus (http://pubmlst.org/cfetus/) and used to genotype 140 isolates from humans and animals (14). The data showed that classical C. fetus is genetically homogeneous and clonal. C. fetus has also been isolated from reptiles (7), and DNA hybridization and nucleotide sequence data indicate that these reptile C. fetus isolates are genetically distinct from classical C. fetus (12). Reptile-like C. fetus strains have also been isolated from cases of human disease (11). In the present study, both the reptile and the human reptile-like strains are referred to collectively as “reptile C. fetus strains.”
The MLST scheme for classical C. fetus was modified in this study to allow typing of reptile C. fetus (Table 1) and comparisons within and among Campylobacter species. The MLST method for classical C. fetus (15) was modified as follows. First, the annealing temperature of the PCR amplification was reduced to 47°C. Second, one of the oligonucleotide primers used to amplify the glyA locus (glyA2) was replaced with glyS4, 5′-AGGTGATTATCCGTTCCATCGC-3′, derived from the C. jejuni sequence. New allele and ST numbers were assigned, and the data were deposited at http://pubmlst.org/cfetus/. Data analysis was performed using the programs MEGA (http://www.megasoftware.net/) (9) and ClonalFrame (3). ClonalFrame is a model-based method for using multilocus sequence data to infer the clonal relationships of bacteria and the chromosomal position of homologous recombination events that disrupt a clonal pattern of inheritance.
TABLE 1.
MLST data for C. fetus reptile isolates recovered from humans and reptiles
| Strain | Source | Location | Yr isolated | ST | Allele no.: |
Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | ||||||
| 03-427 | Humana | NY, USA | 2003 | 15 | 5 | 6 | 6 | 4 | 6 | 6 | 6 | 11 |
| 03-445 | Humana | NY, USA | 2003 | 15 | 5 | 6 | 6 | 4 | 6 | 6 | 6 | 11 |
| 05-018 | Humanb | NY, USA | 2005 | 15 | 5 | 6 | 6 | 4 | 6 | 6 | 6 | Unpublished |
| D6659 | Humanb | MA, USA | 2005 | 15 | 5 | 6 | 6 | 4 | 6 | 6 | 6 | Unpublished |
| D6683 | Humanb | MA, USA | 2005 | 15 | 5 | 6 | 6 | 4 | 6 | 6 | 6 | Unpublished |
| 91-2 | Humanb | Denver, CO, USA | 1991 | 30 | 5 | 6 | 6 | 10 | 6 | 6 | 5 | Unpublished |
| 85-387 | Turtle | CA, USA | 1984 | 16 | 3 | 3 | 6 | 6 | 4 | 5 | 5 | 7 |
| 85-388 | Turtle | CA, USA | 1984 | 17 | 4 | 4 | 6 | 5 | 5 | 5 | 5 | 7 |
| 85-389 | Turtle | CA, USA | 1984 | 18 | 6 | 3 | 6 | 5 | 5 | 5 | 5 | 7 |
| CF78 | Skinkc | London Zoo, UK | 2003 | 26 | 10 | 3 | 6 | 8 | 7 | 9 | 5 | Unpublished |
| SP3 | Snakec | UK | 2006 | 27 | 11 | 8 | 6 | 9 | 7 | 10 | 5 | Unpublished |
Five reptile-derived and six human-derived (two from the same patient) reptile C. fetus strains were typed (Table 1). Allele sequences and therefore all sequence types (STs) differed from those described previously for classical C. fetus. A total of seven new STs were identified among the 11 reptile strains (Table 1). Compared to the classical strains, the reptile group was more variable, also confirmed by the presence of three STs in one turtle. The data were used to investigate the relationships among all known C. fetus STs (n = 30) including classical mammalian strains and reptile strains of both reptile and human origin. The classical STs differed by only 27 of 3,312 nucleotides (0.82%) and 7 of 1,104 amino acids (0.63%). Greater nucleotide sequence variation was detected within the reptile C. fetus STs, with 87 of 3,312 (2.62%) variable nucleotide sites and nine (0.82%) amino acid substitutions. When the classical and reptile groups were compared, there were 281 of 3,312 (8.48%) variable nucleotide sites and 15 of 1,104 (1.35%) amino acid substitutions, demonstrating that the two groups were distinct, each showing a high level of clonality. Strains with ST-16 and ST-26 were not included in this analysis, as they contained “imported alleles” and represented possible recombinants, as described below.
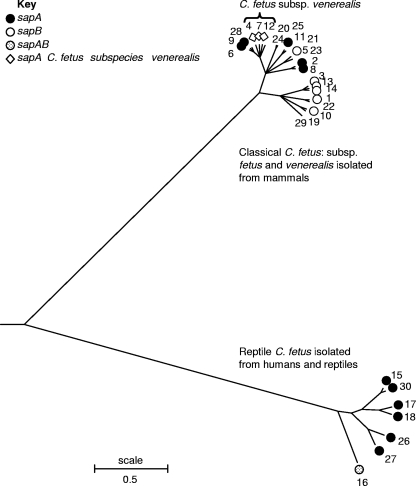
The genetic relationships among the classical and reptile C. fetus strains were investigated further. The nucleotide sequences of the alleles comprising the 30 STs were concatenated, and a consensus tree was constructed using ClonalFrame (3) and viewed using MEGA (9) (Fig. 1). The tree revealed two distinct clusters comprising (i) the classical C. fetus strains and (ii) the reptile C. fetus strains. The C. fetus subsp. venerealis strains formed a subgroup within the classical C. fetus group, as shown previously using a neighbor-joining tree (Fig. 1) (15).
FIG. 1.
Consensus tree (Newick tree) constructed using ClonalFrame (2) and viewed using MEGA (9) to show the two distinct groups formed by classical mammalian and reptile C. fetus. sap types associated with the STs are indicated. Input sequences comprised the concatenated sequences of the seven MLST loci.
The divergence in nucleotide sequence between reptile C. fetus and classical C. fetus (8.64%) is comparable to the divergence between C. jejuni and C. coli within these housekeeping gene loci. For example, the central genotypes of the most common C. jejuni and C. coli clonal complexes, ST-21 and ST-828 (4), are 13.5% divergent. In contrast, there were only 15 of 1,104 (1.35%) amino acid changes between classical and reptile C. fetus, compared to 5.16% for C. jejuni and C. coli (ST-21 and ST-828). This may indicate that the two C. fetus groups share a more recent common ancestor than C. jejuni and C. coli.
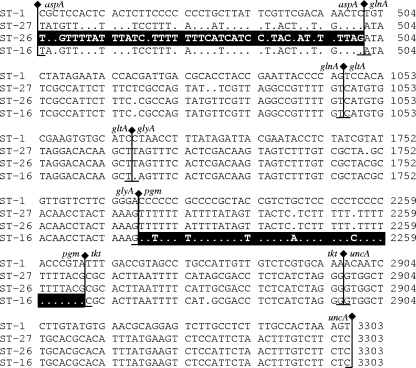
Sequence alignments of the variable sites in the 30 concatenated ST nucleotide sequences (four representatives shown in Fig. 2) indicated that ST-16 and ST-26 were potential recombinants, each containing an apparently imported “foreign” sequence at one of seven loci: aspA for ST-26 and pgm for ST-16. The program ClonalFrame confirmed that ST-16 was a recombinant between reptile C. fetus and a strain very closely related to classical C. fetus. This observation suggests that reptile and mammal C. fetus strains may have mixed at some point in an individual host. The closest relative of the “imported allele” in ST-26 in GenBank was classical C. fetus, with which the imported allele shares 92% identity, indicating that its precise species of origin has yet to be identified.
FIG. 2.
Nucleotide sequence alignment of concatenated STs showing the variable sites only. Dots indicate identity to ST-1. This illustrates the relationship between classical mammalian C. fetus, represented by ST-1, and reptile C. fetus, represented by ST-27. The confirmed recombinant reptile C. fetus strain, ST-16, has a high level of sequence identity with classical mammalian C. fetus in the pgm locus, shown by black shading. The possible recombinant ST-26 reptile C. fetus strain has a sequence divergent from those of reptile C. fetus ST-27 and ST-16 in the aspA locus, indicated by black shading.
The correlation of sap type with the two C. fetus groups was investigated (Fig. 1). sap type is determined by surface layer proteins, an orderly paracrystalline array and major virulence factor for host colonization and prevention of complement-mediated immune responses (2, 13). The sap genes can be rearranged on the chromosome, contributing to antigenic diversity of the S layer and inhibiting immune detection (14). The sap type of the possible recombinant ST-16 was unique, being sapAB, an observation supporting the hypothesis that it may have undergone a major recombinational event.
The C. fetus genome sequence (subsp. fetus strain 82-40; human isolate TIGR project ID 16293) was examined within the region of the recombinant loci. Both pgm and aspA are located near genes encoding either flagellin or Sap proteins (http://msc.tigr.org/campy/campylobacter_fetus_subsp_fetus_82_40/index.shtml), major antigens in C. fetus subject to selective pressure (2, 17). These genes are known to be prone to chromosomal rearrangements in campylobacters (6, 14, 17). Also, pgm is located about 2 kb from a putative site-specific recombinase (SSR) from the phage integrase family. This provides further evidence of the potential for these genomic regions to be involved in recombination events.
In conclusion, reptile C. fetus strains of both human and reptile origin are genetically distinct from classical mammalian C. fetus. The confirmed recombinant ST-16, identified in a turtle isolate, contained both reptile C. fetus and classical C. fetus-like sequences. Although they clustered together (Fig. 1), none of the STs of the reptile C. fetus strains isolated from humans were identical to those that have been isolated thus far from reptiles. Since the number of strains studied was low, it is not yet clear whether transmission of these strains occurs among reptiles and humans or whether the two hosts represent separate reservoirs. This reptile C. fetus cluster may represent a separate genomospecies.
Acknowledgments
This study used the Internet-accessible databases located at http://pubmlst.org/campylobacter/, the maintenance, development, and stewardship of which is supported by funding from the Wellcome Trust and the Department for Environment, Food and Rural Affairs Research Contract numbers OZ0611 and OZ0615. Kate Dingle is supported by the NIHR Oxford Biomedical Research Centre, John Radcliffe Hospital, Oxford, United Kingdom. This study was supported in part by the Diane Belfer Program in Human Microbial Ecology.
Footnotes
Published ahead of print on 6 January 2010.
REFERENCES
- 1.Blaser, M. J. 1998. Campylobacter fetus: emerging infection and model system for bacterial pathogenesis at mucosal surfaces. Clin. Infect. Dis. 27:256-258. [DOI] [PubMed] [Google Scholar]
- 2.Blaser, M. J., P. F. Smith, J. E. Repine, and K. A. Joiner. 1988. Pathogenesis of Campylobacter fetus infections. Failure of encapsulated Campylobacter fetus to bind C3b explains serum and phagocytosis resistance. J. Clin. Invest. 81:1434-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Didelot, X., and D. Falush. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia, M. M., M. D. Eaglesome, and C. Rigby. 1983. Campylobacters important in veterinary medicine. Vet. Bull. 53:793-818. [Google Scholar]
- 6.Harrington, C., F. M. Thomson-Carter, and P. E. Carter. 1997. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J. Clin. Microbiol. 35:2386-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey, S., and J. R. Greenwood. 1985. Isolation of Campylobacter fetus from a pet turtle. J. Clin. Microbiol. 21:260-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monno, R., M. Rendina, G. Ceci, C. Rizzo, I. Luzzi, A. Francavilla, G. Rizzo, and E. Ierardi. 2004. Campylobacter fetus bacteremia in an immunocompromised patient: case report and review of the literature. New Microbiol. 27:281-285. [PubMed] [Google Scholar]
- 9.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 10.Tu, Z. C., J. Hui, and M. J. Blaser. 2004. Conservation and diversity of sap homologues and their organization among Campylobacter fetus isolates. Infect. Immun. 72:1715-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu, Z. C., G. Zeitlin, J. P. Gagner, T. Keo, B. A. Hanna, and M. J. Blaser. 2004. Campylobacter fetus of reptile origin as a human pathogen. J. Clin. Microbiol. 42:4405-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tu, Z. C., W. Eisner, B. N. Kreiswirth, and M. J. Blaser. 2005. Genetic divergence of Campylobacter fetus strains of mammal and reptile origins. J. Clin. Microbiol. 43:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu, Z. C., C. Gaudreau, and M. J. Blaser. 2005. Mechanisms underlying Campylobacter fetus pathogenesis in humans: surface-layer protein variation in relapsing infections. J. Infect. Dis. 191:2082-2089. [DOI] [PubMed] [Google Scholar]
- 14.Tummuru, M. K., and M. J. Blaser. 1993. Rearrangement of sapA homologs with conserved and variable regions in Campylobacter fetus. Proc. Natl. Acad. Sci. U. S. A. 90:7265-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Bergen, M. A. P., K. E. Dingle, M. C. J. Maiden, D. G. Newell, L. van der Graaf-Van Bloois, J. P. M. van Putten, and J. A. Wagenaar. 2005. Clonal nature of Campylobacter fetus as defined by multilocus sequence typing. J. Clin. Microbiol. 43:5888-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Véron, M., and R. Chatelain. 1973. Taxonomic study of the genus Campylobacter and designation of the neotype strain for the type species, Campylobacter fetus (Smith and Taylor) Sebald and Véron. Int. J. Syst. Bacteriol. 23:122-134. [Google Scholar]
- 17.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1995. Variation of the flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology 141(Part 1):95-101. [DOI] [PubMed] [Google Scholar]