Abstract
Klebsiella pneumoniae carbapenemase (KPC) production in Gram-negative bacilli is an increasing problem worldwide. Rectal swab surveillance is recommended as a component of infection prevention programs, yet few screening methods are published. We compared detection of KPC-producing Klebsiella pneumoniae and Escherichia coli in surveillance specimens by 2 methods: (i) inoculation of swabs in tryptic soy broth containing 2 μg/ml imipenem followed by plating to MacConkey agar (MAC) (method 1) and (ii) streaking swabs on MAC onto which a 10-μg ertapenem disk was then placed (method 2). Simulated rectal swab specimens of challenge isolates from a collection of well-characterized K. pneumoniae and E. coli strains and salvage rectal swab specimens collected from patients at 4 different health care facilities over a 7-month period were tested. The gold-standard comparator was blaKPC PCR testing of isolates. Method 1 detected 4/9 (44%) KPC-positive challenge isolates. By method 2, 9/9 KPC-positive challenge isolates exhibited zones of inhibition of ≤27 mm; all KPC-negative isolates exhibited zones of inhibition greater than 27 mm. The sensitivity and specificity of method 1 for detection of KPC-positive K. pneumoniae and E. coli in 149 rectal swab specimens were 65.6% (95% confidence interval [CI], 46.8% to 80.8%) and 49.6% (95% CI, 40.3% to 58.9%), respectively. With method 2, a zone diameter of ≤27 mm had a sensitivity of 97.0% (95% CI, 82.5% to 99.8%) and specificity of 90.5% (95% CI, 83.3% to 94.9%) for detection of KPC in rectal swab specimens. Direct ertapenem disk testing is simpler, more sensitive, and more specific than selective broth enrichment with imipenem for detection of KPC-producing K. pneumoniae and E. coli in surveillance specimens.
Klebsiella pneumoniae carbapenemase (KPC) is a class A beta-lactamase that confers resistance to all beta-lactam antibiotics, including carbapenems. KPC was first detected in 1996 in a K. pneumoniae isolate from North Carolina (28), and it remains most common in Enterobacteriaceae group bacteria, especially K. pneumoniae. Initially, KPC spread slowly; by 2005, the problem was limited almost exclusively to the northeastern United States (1, 9, 20, 30). Since then, the spread of KPC has accelerated worldwide, largely due to the emergence and dissemination of a hyperepidemic clone of K. pneumoniae designated multilocus sequence type 258 (ST258) (6, 13). Because KPC-producing bacteria, including ST258, are often resistant to all commonly used antibiotics, limiting their spread is critical.
The gastrointestinal tract serves as a reservoir for KPC-producing bacteria, and stool or rectal swab surveillance has been recommended as a component of infection prevention efforts (3). Although validated methods exist for the detection of carbapenemases, such as KPC, in bacterial isolates from clinical cultures (5), there are few published reports of surveillance methods. Real-time, rapid-cycle PCR testing is a fast, accurate means of identifying blaKPC in swab specimens (8, 19). Similarly, CHROMagar KPC has shown good sensitivity and specificity for screening (18). However, PCR requires equipment and expertise in molecular diagnostics that is not widely available, while CHROMagar KPC cannot be purchased in many locales, including in the United States.
Broth enrichment followed by selective culture on MacConkey agar is a method that has been suggested by the Centers for Disease Control and Prevention (CDC) as a screening test (2). Advantages of the 2-step culture method are that it uses materials and techniques available in most clinical microbiology laboratories. Limitations of this approach include the growth of a relatively large number of carbapenemase-negative bacteria that require confirmatory testing and a long turnaround time (29). In this study, we compare the performance of the 2-step selective broth enrichment method to a direct ertapenem disk test for identification of KPC-producing, lactose-fermenting, Gram-negative bacilli in surveillance swab specimens.
MATERIALS AND METHODS
Challenge isolate set testing.
In initial comparisons of the 2 methods, we utilized a challenge set of 9 KPC-positive bacterial isolates and 10 extended-spectrum beta-lactamase (ESBL)-positive, KPC-negative isolates. All strains were K. pneumoniae or Escherichia coli strains that were isolated from clinical cultures from various geographic locations (Table 1).
TABLE 1.
Challenge isolate set
| Strain designation | Species | Country of origin | Description | MIC (μg/ml) |
Source or reference | ||
|---|---|---|---|---|---|---|---|
| Ertapenem | Imipenem | Meropenem | |||||
| 428024011 | E. coli | Colombia | KPC (undefined), pulsotype EC3 | 2 | 4 | 2 | Gift of John Quinn |
| KPN2303 | K. pneumoniae | Colombia | KPC-2 pulsotype, KP2 | >256 | >256 | >256 | 24 |
| H680 | K. pneumoniae | Colombia | KPC (undefined), pulsotype KP3 | 8 | 8 | 4 | Gift of John Quinn |
| KPN2020 | K. pneumoniae | Colombia | KPC (undefined), pulsotype KP4 | 64 | 16 | 8 | Gift of John Quinn |
| CH9B | K. pneumoniae | USA | KPC-3, pulsotype KP1 | 32 | 8 | 16 | This publication |
| EC18 | E. coli | USA | KPC (undefined), pulsotype EC1 | 32 | 8 | 8 | This publication |
| EC11 | E. coli | USA | KPC (undefined), pulsotype EC2 | 4 | 4 | 4 | Gift of Michael Costello |
| KP11 | K. pneumoniae | USA | KPC (undefined), pulsotype KP1 | 32 | 8 | 16 | This study |
| KP19 | K. pneumoniae | USA | KPC (undefined), pulsotype KP1 | 64 | 8 | 8 | This study |
| 39197 | K. pneumoniae | Colombia | TEM, CTX-M, SHV | 0.03 | 0.5 | Not tested | 23 |
| CH4 | E. coli | China | TEM-1, SHV-43 | 0.03 | 0.5 | Not tested | 26 |
| ATCC 700603 | K. pneumoniae | USA | SHV-18 | 0.03 | Not tested | Not tested | American Type Culture Collection |
| MG32 | E. coli | USA | TEM-12, porin alteration | 0.03 | 0.12 | Not tested | 27 |
| 1338 | K. pneumoniae | Colombia | CTX-M-12 | 0.06 | 0.12 | Not tested | 23 |
| KPLA-1 | K. pneumoniae | Switzerland | SHV-12 | 0.06 | 0.25 | Not tested | 14 |
| RHH-1/K-3 | K. pneumoniae | United Kingdom | TEM-9 | 0.125 | 0.12 | Not tested | 21 |
| KP-3 | K. pneumoniae | USA | TEM-10, SHV | 0.03 | 0.12 | Not tested | 15 |
| K1(KC-1) | K. pneumoniae | USA | TEM-10 | 0.03 | 0.12 | Not tested | 17 |
| CH6 | K. pneumoniae | China | SHV-43 | 0.06 | 0.12 | Not tested | 26 |
Serial 10-fold dilutions of each challenge isolate were made in 0.9% normal saline (final target concentrations, 5 × 103 CFU/ml to 5 × 105 CFU/ml). To each dilution of a challenge isolate, an inoculum of the following microbial strains was added to a final concentration of 1 × 104 CFU/ml: (i) a clinical vanB vancomycin-resistant Enterococcus faecium strain (7), (ii) C. albicans ATCC 14053, and (iii) a strain of Pseudomonas aeruginosa that was resistant to imipenem due to OmpD deficiency (16). These strains were chosen to represent the types of competing microbial flora that may be found in the stools of patients who are colonized with KPC-positive bacteria (10).
For testing by the 2-step selective broth enrichment method (method 1), 100 μl of each mixture was placed in 5 ml tryptic soy broth (Remel, Lenexa, KS) that contained a 10-μg imipenem disk (Becton-Dickinson, Sparks, MD). Assuming complete elution of imipenem from the disk, the concentration of imipenem in broth was 2 μg/ml. Following overnight incubation, a 25-μl aliquot of each broth culture mixture was streaked onto MacConkey agar (Remel) and then incubated overnight. The following day, the presence or absence of lactose-fermenting colonies at each dilution was recorded.
For direct ertapenem disk testing (method 2), 100 μl of each culture mixture was streaked for confluent growth onto MacConkey agar (Remel) and allowed to dry. A 10-μg ertapenem disk (Becton-Dickenson) was then applied to the plate. Following overnight incubation, the presence and number of lactose-fermenting colonies were recorded, and the diameter of any zone around the ertapenem disk that was clear of lactose-fermenting colonies was measured. For dilutions of challenge isolates that did not yield a measurable zone of inhibition, the distance of the closest lactose-fermenting colony to the ertapenem disk was measured and then doubled to estimate a zone diameter.
The same culture mixtures were tested on the same day by both methods. Testing was done in duplicate. All incubations were at 35°C in ambient air.
Rectal surveillance swab testing.
Salvage rectal swab specimens were obtained over a 7-month period (June 2008 through January 2009) from 3 sources: (i) specimens collected routinely at the time of admission to a long-term acute-care hospital to screen for multidrug-resistant organisms; (ii) specimens collected during 4-point prevalence surveys conducted as a component of an outbreak control program at the same long-term acute-care hospital (12); and (iii) swabs collected as part of routine point prevalence surveys to screen for KPCs at two different nursing homes. The long-term acute-care hospital and one nursing home were part of a regional outbreak of KPC-producing bacteria in northwest Indiana and south suburban Chicago (12, 28). The second nursing home was experiencing an unrelated outbreak of KPC-positive bacteria.
Each swab was tested for KPC by method 1 and method 2 as described above, with the following modifications. For both methods, rectal swab specimens were substituted for the 100-μl culture mixtures. For method 2, swab specimens were inoculated onto MacConkey agar and streaked to 4 quadrants for colony isolation. One 10-μg ertapenem disk (Becton-Dickenson) was placed at the junction of quadrants 1 and 2; a second 10 μg ertapenem disk was placed at the junction of quadrants 2 and 3. After overnight incubation, the diameter of any zone around either disk that was clear of lactose-fermenting colonies was measured. Two ertapenem disks were used in an effort to maximize the likelihood of obtaining a measurable zone. If the number of lactose-fermenting colonies that grew on a plate was so small that a zone of inhibition was not apparent, the colonies were picked and retested in the same manner as the original swab specimens. For both method 1 and method 2, all unique colony morphotypes of Gram-negative bacilli—both lactose fermenters and nonlactose fermenters—identified on the plates were isolated for subsequent testing for blaKPC.
Because method 1 included an initial broth enrichment step, it was not possible to randomly assign the order in which swabs were tested by the 2 methods. For each comparison, swabs were first streaked to MacConkey agar plates (method 2) followed immediately by inoculation into enrichment broth (method 1).
Additional microbiologic testing.
All lactose-fermenting, Gram-negative bacilli identified in rectal swab specimens underwent species determination by the MicroScan Walkaway System using 50 MS GNC panels (Siemens, New York, NY). Manual broth microdilution testing of ertapenem, imipenem, and meropenem and modified Hodge testing were performed according to Clinical Laboratory Standards Institute (CLSI) guidelines (4, 5).
PCR testing for blaKPC was done on each individual colony morphotype of a lactose-fermenting, Gram-negative bacillus identified by method 1 or method 2 using primers 5 and 10 as described by Yigit et al. (31). In addition, because KPC has been described for nonlactose fermenting bacteria (25), we also tested each morphotype of nonlactose fermenting colonies that was detected by either method for blaKPC.
Pulsed-field gel electrophoresis (PFGE) typing was performed by digesting chromosomal DNA with XbaI as previously described (11). XbaI-digested DNA was analyzed on a Chef Mapper system (Bio-Rad, Fremont, CA), with pulse times ramped linearly from 5 to 35 s for 23 h. Strain relatedness was determined according to the criteria of Tenover et al. (22).
Statistical analysis.
Receiver operator characteristic (ROC) curves were created using SAS version 9.1.3 (SAS Institute, Inc., Cary, NC).
RESULTS
Challenge isolate set testing.
When tested by the 2-step selective broth enrichment method (method 1), 4 of 9 (44%) KPC-positive challenge strains were detected at the 2 higher bacterial concentrations; only 3 KPC-positive challenge strains were detected at the lowest bacterial concentration. None of 10 ESBL-positive, KPC-negative challenge strains grew when tested by method 1, regardless of bacterial concentration, i.e., there were no false-positive results.
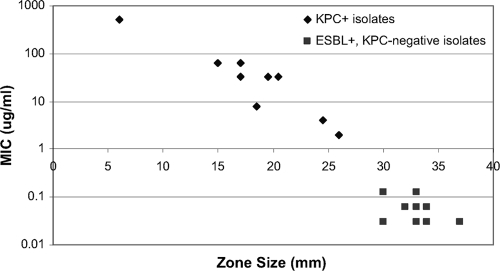
Results of testing the challenge isolate set by the direct ertapenem disk method (method 2) are shown in Fig. 1. By testing a range of serial dilutions, zone sizes were observed to increase as the bacterial concentration decreased. To analyze the results using the most stringent criteria, the data presented represent the final target concentration of 5 × 105 CFU/ml. At this concentration, there was a clear distinction between KPC-positive and KPC-negative isolates (Fig. 1). The average zone diameters of KPC-positive isolates ranged from 6 mm to 26 mm; average zone diameters of ESBL-positive, KPC-negative isolates ranged from 30 mm to 37 mm.
FIG. 1.
Relationship between MIC of ertapenem and zone of inhibition around a 10-μg ertapenem disk for 19 challenge strains of Klebsiella pneumoniae and Escherichia coli tested. Results shown are means of duplicate determinations.
Surveillance rectal swab testing.
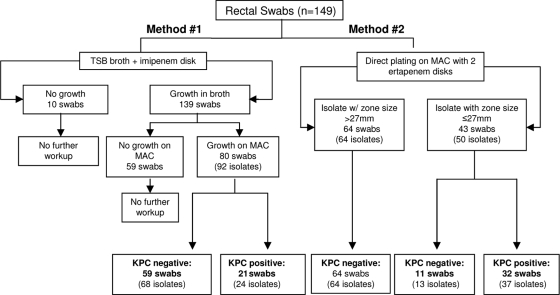
One hundred forty-nine salvage surveillance rectal swab specimens were tested, 17 immediately after collection and the remainder after 2 or 3 weeks of storage at 4°C (Fig. 2). By method 1, 80 specimens yielded 92 lactose-fermenting isolates that were considered to be presumptively positive for KPC; of these, 24 isolates from 21 specimens were confirmed KPC positive by PCR.
FIG. 2.
Testing scheme for surveillance rectal swab specimens. The presence or absence of KPC-producing bacteria was determined by PCR for blaKPC.
With method 2, 98 swabs yielded 105 unique lactose-fermenting colony morphotypes with measurable zones of inhibition. Nine additional swab specimens (6.0%) grew only a few isolated lactose-fermenting colonies. When these isolated colonies were retested, measurable zone diameters were apparent for each of them. Thus, a total of 107 swab specimens tested by method 2 grew 114 different lactose-fermenting colony morphotypes. Of these, 37 isolates from 32 swabs were confirmed KPC positive by PCR.
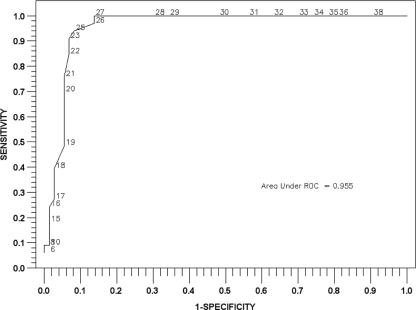
We analyzed ertapenem disk zone diameters and blaKPC PCR results for the 114 lactose-fermenting isolates identified by method 2 in a ROC curve. With this analysis, a zone diameter of 27 mm appeared to have the best sensitivity and specificity for detection of KPC-positive bacteria (Fig. 3). The area under the ROC curve was 0.955, indicating excellent ability of the test to discriminate between positive and negative results. When the 27-mm breakpoint was applied to method 2 results, 43/149 specimens were screen positive; 32 of these were confirmed positive by PCR.
FIG. 3.
ROC curve for zones of inhibition around a 10-μg/ml ertapenem disk for 114 lactose-fermenting isolates identified from surveillance rectal swab specimens by method 2.
In total, 33 of 149 specimens (22.1%) grew 38 lactose-fermenting isolates that were confirmed KPC positive (Table 2). Twenty-three KPC-positive isolates were identified as K. pneumoniae, and 15 were identified as E. coli. Method 2 detected 11 more KPC-positive specimens than did method 1. When PCR testing of isolated colonies was considered to be the criterion standard, the sensitivity of method 1 was 65.6% (95% CI, 46.8% to 80.8%) and the specificity was 49.6% (95% CI, 40.3% to 58.9%) for detection of KPC-producing, lactose-fermenting, Gram-negative bacilli. For method 2, using the same criterion standard and a 27-mm breakpoint, the sensitivity and specificity were 97.0% (95% CI, 82.5% to 99.8%) and 90.5% (95% CI, 83.3% to 94.9%), respectively, for KPC detection.
TABLE 2.
Results of testing for blaKPC in lactose-fermenting isolates recovered from rectal swab specimens by method 1 and method 2a
| No. of swabs | Method 1 | Method 2 |
|---|---|---|
| 116 | Negative | Negative |
| 20 | Positive | Positive |
| 12 | Negative | Positive |
| 1 | Positive | Negative |
The presence of blaKPC was determined by PCR testing of isolated colonies (31).
The MIC50s and MIC90s for carbapenems were higher for K. pneumoniae isolates than for E. coli isolates (Table 3). By CLSI performance standard breakpoints, all isolates of K. pneumoniae tested were resistant to ertapenem and 23 (95.6%) were resistant to imipenem or meropenem (5). In contrast, only 5 of 15 (33.3%) E. coli isolates were resistant to ertapenem and none was resistant to imipenem or meropenem.
TABLE 3.
Comparison of MIC50s and MIC90s for KPC-positive isolates of Klebsiella pneumoniae and Escherichia coli identified in surveillance rectal swab specimens
| Organism | Parameter | MIC (μg/ml) |
||
|---|---|---|---|---|
| Ertapenem | Imipenem | Meropenem | ||
| Klebsiella pneumoniae (n = 23) | MIC50 | 32 | 16 | 16 |
| MIC90 | 256 | 256 | 128 | |
| Range | 8->256 | 4->256 | 4->256 | |
| Escherichia coli (n = 15) | MIC50 | 4 | 4 | 2 |
| MIC90 | 8 | 4 | 4 | |
| Range | 1-16 | 1-4 | 0.25-4 | |
PFGE analysis of KPC-producing E. coli isolates from 11 of 13 surveillance rectal specimens yielded 5 unique pulsotypes. The 19 K. pneumoniae isolates tested clustered into 4 related groups whose members were identical but that differed from one another by 1 to 8 bands.
Of the 59 isolates that gave presumptive false-positive results by method 1, none displayed an elevated carbapenem MIC (Table 4). In contrast, 5 of the 11 presumptive false-positive isolates identified by method 2 exhibited intermediate or resistant ertapenem MICs (Table 4). Mechanisms of carbapenem nonsusceptibility in these 5 isolates were not determined.
TABLE 4.
Susceptibility testing results of false-positive lactose-fermenting isolates identified by method 1 and method 2 in surveillance swab specimensc
| Method | Ertapenem (n [%]) |
Imipenem (n [%]) |
Meropenem (n [%]) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC range | S | I | R | MIC range | S | I | R | MIC range | S | I | R | |
| 1 (n = 60)a | <0.015-0.5 | 60 (100) | 0 (0) | 0 (0) | 0.06-1 | 60 (100) | 0 (0) | 0 (0) | <0.015-0.125 | 60 (100) | 0 (0) | 0 (0) |
| 2 (n = 11)b | 0.125-16 | 6 (55) | 1 (9) | 4 (36) | 0.125-4 | 11 (100) | 0 (0) | 0 (0) | 0.06-4 | 11 (100) | 0 (0) | 0 (0) |
Sixty of 68 lactose-fermenting isolates were available for testing. These included 42 Escherichia coli, 16 Klebsiella pneumoniae, 1 Citrobacter freundii complex, and 1 Enterobacter cloacae isolate.
Eleven of 13 lactose-fermenting isolates were available for testing. All were identified as Escherichia coli.
S, susceptible; I, intermediate; R, resistant.
Results of modified Hodge testing of screen-positive isolates were 100% concordant with KPC PCR results. None of 281 nonlactose-fermenting Gram-negative bacillus isolates identified by either method was positive for blaKPC by PCR.
DISCUSSION
The recent acceleration in the spread of KPC-producing bacteria internationally and the emphasis on control of these pathogens predict that clinical microbiology laboratories worldwide will soon be faced with the challenge of screening surveillance specimens for KPC. In this report, we describe an ertapenem disk method to screen rectal swab specimens directly for KPC-producing K. pneumoniae and E. coli. The screening assay we describe is more sensitive and more specific than the 2-step selective enrichment broth method currently suggested by the CDC (2). If the modified Hodge test were used to confirm results of the ertapenem disk screening assay, both screening and confirmation could be done in most clinical microbiology laboratories, with materials and equipment already on hand.
Results of the ertapenem disk method (method 2) were available for 94% of rectal surveillance swab specimens tested in 16 to 24 h. Given the prevalence of KPC in the sample—22.1% of swabs with ≥1 KPC-positive isolate—the positive predictive value of a positive screening result was 74.4%. Although this value is substantially higher than the positive predictive value of the 2-step selective broth enrichment assay (method 1), which was 26.2%, infection control practitioners might still choose to wait for test confirmation before acting on results. Results of the modified Hodge test would typically be available 24 to 48 h after screening results, depending on whether isolates identified by screening required subculture to obtain a pure isolate. Thus, a usual turnaround time for screening by the ertapenem disk method followed by confirmation by modified Hodge testing would be 2 or 3 days.
It should be noted that the 27-mm breakpoint used in the screening assay we describe is greater than the 19- to 21-mm ertapenem zone diameter range proposed by CLSI to screen pure cultures of clinical isolates for KPC-producing Klebsiella spp. and E. coli. (5). Applying the 27-mm breakpoint to results of the diverse, well-characterized challenge isolate set tested would have resulted in detection of all KPC-positive strains and exclusion of all ESBL-producing, KPC-negative strains. These observations lend validity to our conclusion that a 27-mm zone diameter, or a distance of 13.5 mm from the center of the disk to a lactose-fermenting colony, is an appropriate criterion for direct screening for KPC-producing K. pneumoniae or E. coli isolates in rectal swab specimens.
The interpretation of this study warrants several considerations. First, we did not perform PCR directly on swabs. We chose to omit this evaluation so as to limit the total number of tests done on salvage specimens. If the results of KPC PCR performed directly on specimens were used as the criterion standard and if nonviable, blaKPC-positive isolates were present in any specimen, the sensitivity of method 1 and method 2 would be lower than calculated. Second, we consistently streaked swabs for method 2 before inoculating the selective broth in method 1, which may have biased results for sensitivity against method 1. However, method 1 also demonstrated very low sensitivity in challenge isolate testing, which would not have been affected by this bias. Finally, although we tested all recovered Gram-negative bacilli by KPC PCR, we identified blaKPC only in isolates of K. pneumoniae and E. coli. Therefore, we cannot comment on the performance of method 1 or method 2 in the detection of KPC in other bacterial species.
In conclusion, direct ertapenem disk testing followed by modified Hodge test confirmation is a simple, sensitive, and specific scheme for screening rectal swab specimens for KPC-producing K. pneumoniae and E. coli that could be implemented in most clinical microbiology laboratories. The performance characteristics of the ertapenem disk method appear to be superior to those of the 2-step selective broth enrichment method. While validation of this approach in other laboratories is required, we believe that the direct ertapenem disk screen test may be useful as a tool for identifying KPC-colonized patients in health care settings.
Footnotes
Published ahead of print on 13 January 2010.
REFERENCES
- 1.Bradford, P. A., S. Bratu, C. Urban, M. Visalli, N. Mariano, D. Landman, J. J. Rahal, S. Brooks, S. Cebular, and J. Quale. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin. Infect. Dis. 39:55-60. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2009. Laboratory protocol for detection of carbapenem-resistant or carbapenemase-producing Klebsiella spp. and E. coli from rectal swabs. Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/NCIDOD/DHQP/pdf/ar/Klebsiella_or_Ecoli.pdf.
- 3.Centers for Disease Control and Prevention. 2009. Guidance for control of infections with carbapenem-resistant or carbapenemase-producing enterobacteriaceae in acute care facilities. MMWR Morb. Mortal. Wkly. Rep. 58:256-260. [PubMed] [Google Scholar]
- 4.CLSI. 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, eighth edition. Clinical Laboratory Standards Institute, Wayne, PA.
- 5.CLSI. 2009. Performance standards for antimicrobial susceptibility testing; nineteenth informational supplement. M100-S19. Clinical Laboratory Standards Institute, Wayne, PA.
- 6.Giakoupi, P., H. Maltezou, M. Polemis, O. Pappa, G. Saroglou, and A. Vatopoulos. 2009. KPC-2-producing Klebsiella pneumoniae infections in Greek hospitals are mainly due to a hyperepidemic clone. Euro Surveill. 14:19218. [DOI] [PubMed] [Google Scholar]
- 7.Hayden, M. K., G. M. Trenholme, J. E. Schultz, and D. F. Sahm. 1993. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J. Infect. Dis. 167:1224-1227. [DOI] [PubMed] [Google Scholar]
- 8.Hindiyeh, M., G. Smollen, Z. Grossman, D. Ram, Y. Davidson, F. Mileguir, M. Vax, D. Ben David, I. Tal, G. Rahav, A. Shamiss, E. Mendelson, and N. Keller. 2008. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J. Clin. Microbiol. 46:2879-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hossain, A., M. J. Ferraro, R. M. Pino, R. B. Dew III, E. S. Moland, T. J. Lockhart, K. S. Thomson, R. V. Goering, and N. D. Hanson. 2004. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 in an Enterobacter sp. Antimicrob. Agents Chemother. 48:4438-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landman, D., J. K. Salvani, S. Bratu, and J. Quale. 2005. Evaluation of techniques for detection of carbapenem-resistant Klebsiella pneumoniae in stool surveillance cultures. J. Clin. Microbiol. 43:5639-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matushek, M. G., M. J. Bonten, and M. K. Hayden. 1996. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J. Clin. Microbiol. 34:2598-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munoz-Price, L. S., M. K. Hayden, K. Lolans, S. Won, K. Calvert, M. Y. Lin, A. Stemer, and R. A. Weinstein. Successful control of an outbreak of KPC-producing Klebsiella pneumoniae at a long-term acute care hospital (LTACH). Infect. Control Hosp. Epidemiol., in press. [DOI] [PubMed]
- 13.Navon-Venezia, S., A. Leavitt, M. J. Schwaber, J. K. Rasheed, A. Srinivasan, J. B. Patel, and Y. Carmeli. 2009. First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob. Agents Chemother. 53:818-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nuesch-Inderbinen, M. T., F. H. Kayser, and H. Hachler. 1997. Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob. Agents Chemother. 41:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pattharachayakul, S., M. M. Neuhauser, J. P. Quinn, and S. L. Pendland. 2003. Extended-spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae: activity of single versus combination agents. J. Antimicrob. Chemother. 51:737-739. [DOI] [PubMed] [Google Scholar]
- 16.Quinn, J. P., D. Miyashiro, A. Darzins, and R. Miller. 1991. Imipenem resistance in Pseudomonas aeruginosa. Antibiot. Chemother. 44:240-244. [DOI] [PubMed] [Google Scholar]
- 17.Quinn, J. P., D. Miyashiro, D. Sahm, R. Flamm, and K. Bush. 1989. Novel plasmid-mediated beta-lactamase (TEM-10) conferring selective resistance to ceftazidime and aztreonam in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 33:1451-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samra, Z., J. Bahar, L. Madar-Shapiro, N. Aziz, S. Israel, and J. Bishara. 2008. Evaluation of CHROMagar KPC for rapid detection of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 46:3110-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schechner, V., K. Straus-Robinson, D. Schwartz, I. Pfeffer, J. Tarabeia, R. Moskovich, I. Chmelnitsky, M. J. Schwaber, Y. Carmeli, and S. Navon-Venezia. 2009. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J. Clin. Microbiol. 47:3261-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith Moland, E., N. D. Hanson, V. L. Herrera, J. A. Black, T. J. Lockhart, A. Hossain, J. A. Johnson, R. V. Goering, and K. S. Thomson. 2003. Plasmid-mediated, carbapenem-hydrolysing beta-lactamase, KPC-2, in Klebsiella pneumoniae isolates. J. Antimicrob. Chemother. 51:711-714. [DOI] [PubMed] [Google Scholar]
- 21.Spencer, R. C., P. F. Wheat, T. G. Winstanley, D. M. Cox, and S. J. Plested. 1987. Novel beta-lactamase in a clinical isolate of Klebsiella pneumoniae conferring unusual resistance to beta-lactam antibiotics. J. Antimicrob. Chemother. 20:919-921. [DOI] [PubMed] [Google Scholar]
- 22.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villegas, M. V., A. Correa, F. Perez, M. C. Miranda, T. Zuluaga, and J. P. Quinn. 2004. Prevalence and characterization of extended-spectrum beta-lactamases in Klebsiella pneumoniae and Escherichia coli isolates from Colombian hospitals. Diagn. Microbiol. Infect. Dis. 49:217-222. [DOI] [PubMed] [Google Scholar]
- 24.Villegas, M. V., K. Lolans, A. Correa, C. J. Suarez, J. A. Lopez, M. Vallejo, and J. P. Quinn. 2006. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob. Agents Chemother. 50:2880-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walther-Rasmussen, J., and N. Hoiby. 2007. Class A carbapenemases. J. Antimicrob. Chemother. 60:470-482. [DOI] [PubMed] [Google Scholar]
- 26.Wang, H., S. Kelkar, W. Wu, M. Chen, and J. P. Quinn. 2003. Clinical isolates of Enterobacteriaceae producing extended-spectrum beta-lactamases: prevalence of CTX-M-3 at a hospital in China. Antimicrob. Agents Chemother. 47:790-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber, D. A., C. C. Sanders, J. S. Bakken, and J. P. Quinn. 1990. A novel chromosomal TEM derivative and alterations in outer membrane proteins together mediate selective ceftazidime resistance in Escherichia coli. J. Infect. Dis. 162:460-465. [DOI] [PubMed] [Google Scholar]
- 28.Won, S., L. S. Munoz-Price, K. Lolans, K. Calvert, M. Y. Lin, R. A. Weinstein, and M. K. Hayden. 2009. Multi-facility emergence and rapid spread of KPC+ Enterobacteriaceae in Northwest Indiana and Chicago area, abstr. 67. Abstr. 19th Annu. Sci. Meet. Soc. Healthc. Epidemiol. Am., San Diego, CA.
- 29.Wong, B., K. Anderson, and J. B. Patel. 2009. Limitations of a two-step culture method for isolation of carbapenem-resistant Enterobacteriaceae (CRE) from rectal specimens, p. D-153. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA.
- 30.Woodford, N., P. M. Tierno, Jr., K. Young, L. Tysall, M. F. Palepou, E. Ward, R. E. Painter, D. F. Suber, D. Shungu, L. L. Silver, K. Inglima, J. Kornblum, and D. M. Livermore. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York medical center. Antimicrob. Agents Chemother. 48:4793-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yigit, H., A. M. Queenan, G. J. Anderson, A. Domenech-Sanchez, J. W. Biddle, C. D. Steward, S. Alberti, K. Bush, and F. C. Tenover. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]